Abstract
Microbial communities are essential to fundamental processes on Earth. Underlying the compositions and functions of these communities are nutritional interdependencies among individual species. One class of nutrients, cobamides (the family of enzyme cofactors that includes vitamin B12), is widely used for a variety of microbial metabolic functions, but these structurally diverse cofactors are synthesized by only a subset of bacteria and archaea. Advances at different scales of study—from individual isolates, to synthetic consortia, to complex communities—have led to an improved understanding of cobamide sharing. Here, we discuss how cobamides affect microbes at each of these three scales and how integrating different approaches leads to a more complete understanding of microbial interactions.
Graphical Abstract
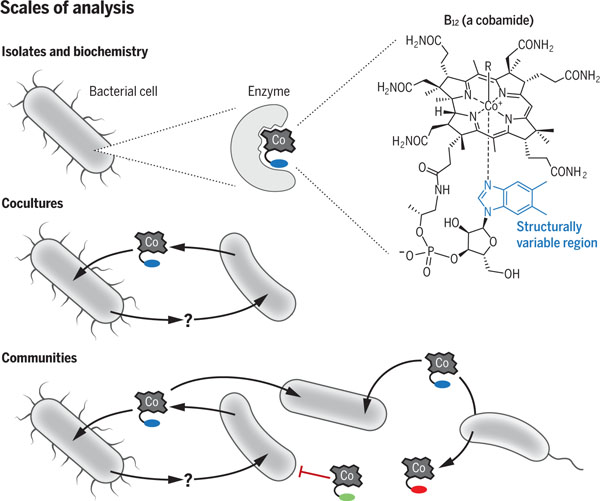
Cobamides as models for studying microbial interactions. Cobamides are a class of enzyme cofactors that are used for a wide variety of metabolic functions. They contain a catalytic upper ligand (R) and a structurally variable region (shown in blue, red, or green) that influences organisms’ metabolism and growth. Studies of cobamide biology on multiple scales—from enzymes to microbial communities—have revealed that cobamides constitute an effective model system for studying the complexity of microbial interactions.
Review Summary
BACKGROUND:
Nearly every plant, animal, and environment on earth is host to a diverse community of microorganisms that influence each other and their environment. Microorganisms within communities interact on a molecular level by competing for resources or sharing valuable nutrients (such as cobamides, which we highlight in this Review). Such molecular interactions influence the physiology of individual microorganisms as well as the overall function of communities. Therefore, studying how microbes interact with each other is essential for understanding, and potentially interfering with, microbial processes that influence human and environmental health.
Cobamides are structurally diverse, cobalt-containing cofactors, the most familiar of which is vitamin B12 (also known as cobalamin). Since the initial discovery of vitamin B12 as the treatment for the disease pernicious anemia in 1948, microbiologists have identified more than a dozen cobamides-B12 and analogs-that are produced exclusively by bacteria and archaea. Although vitamin B12 is most widely appreciated for its role in human health, B12 and other cobamides also play important roles in the context of microbial communities. Microbes use cobamides as catalysts for chemical reactions involved in amino acid synthesis, carbon metabolism, and many other functions. Importantly, microorganisms in all domains of life need cobamides, but most depend on surrounding species to produce this nutrient, which results in a network of cobamide-dependent interactions. A nuance of these interactions, derived from the structural diversity of cobamides, is that organisms are selective toward particular cobamides, and different species have distinct cobamide preferences. As a result, cobamides mediate specific associations among microorganisms and can have substantially different effects on the growth and metabolism of different species. Therefore, cobamide sharing can serve as a model for the complexity of microbial interactions and provide a useful system to study the mechanisms that influence community composition and function.
ADVANCES:
Our current understanding of the roles of cobamides in microbial communities is the result of multilayered approaches to studying cobamide biology. Historically, the differential effects of cobamides have been investigated using laboratory cultures of single species and the biochemical characterization of cobamide-dependent enzymes. However, it is only with comparative genomic analyses of thousands of microbial species that researchers have begun to fully recognize the prevalence of cobamide sharing among microorganisms. Several newly described cocultures of two to three microbial species bridge molecular analysis and community-wide studies, and these cocultures provide experimental systems for probing the mechanisms and dynamics of cobamide sharing. Integrating discoveries across these different scales of analysis is a valuable strategy for understanding the functions of important molecules in microbial communities.
OUTLOOK:
The structural diversity, functional specificity, and widespread use of cobamides by microorganisms have led researchers to speculate that cobamides could be used as tools to manipulate microbial community composition and function to improve environmental or human health. Performing cobamide-based manipulations in a controlled manner requires a greater understanding of how specific cobamides affect particular members of a community or might disrupt existing microbial interactions. Further integrating molecular approaches with community-wide studies will pave the way for understanding complex microbial communities in increasing mechanistic detail and may enable potential applications of cobamides in human health, agriculture, and industrial production.
Microbial communities inhabit all environments on Earth. They affect bio-geochemical cycles, modulate the health of macroscopic hosts, and are harnessed to produce compounds of human interest. To understand and potentially manipulate microbial communities in beneficial ways, scientists are actively researching the mechanisms that shape community assembly, composition, and resilience. Microbial communities are composed of up to thousands of member species that interact with each other on a molecular level. One type of interaction is nutrient sharing, wherein one species produces a metabolite that another requires. (Note, we use the term sharing here to describe the release of a metabolite that is taken up and used by another organism; it is not meant to imply a cooperative interaction.) Auxotrophs—those that cannot produce required metabolites—in microbial communities rely on other community members for essential nutrients, such as amino acids, nucleobases, or vitamins. Because of the prevalence of auxotrophy (1, 2), nutrient sharing between microorganisms is thought to be an important driver of microbial community structure.
Among the many shared nutrients are cobamides, a structurally diverse family of cofactors of which cobalamin (vitamin B12) is the most widely recognized (Fig. 1). Cobalamin is best known for its role as an essential micronutrient for humans. Less appreciated is the fact that many microorganisms use cobalamin and other cobamides for a variety of metabolic functions (Table 1) (3–6). Cobamides contribute to environmentally impactful processes including methanogenesis, the production of toxic methylmercury, and the decontamination of polluted lake waters (7–9). More commonly, microorganisms use cobamides for primary and secondary metabolism, such as in the catabolism of various carbon sources, nucleotide biosynthesis, and natural product biosynthesis (3–6). As such, cobamides are important in many microbial environments, including the human gut and marine habitats (10, 11). Moreover, most organisms that use cobamides rely on other species to acquire these cofactors because cobamides are exclusively produced by a subset of bacteria and archaea (5, 12, 13). This results in microbial interactions based on cobamide sharing. In this Review, we describe approaches used to study the biological roles of cobamides as shared nutrients, highlight notable recent discoveries about cobamide-dependent interactions, and discuss challenges and opportunities in the field of microbiology using cobamides as a model system.
Fig. 1. Structural diversity of cobamides.
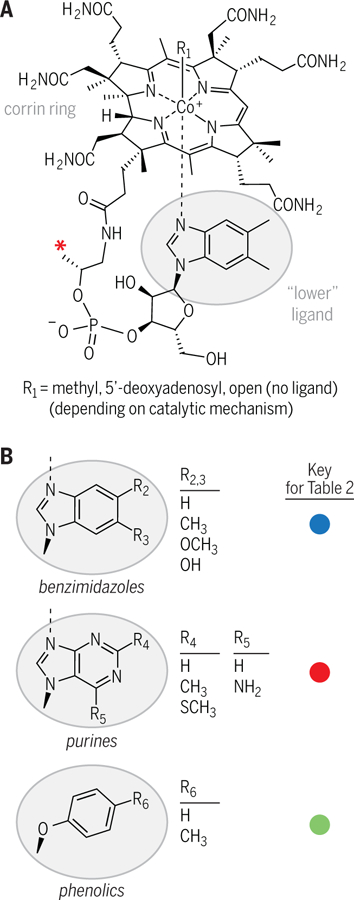
(A) The chemical structure of cobalamin (B12), the most well-known cobamide. Cobamides are characterized by a corrin ring (a contracted porphyrin, similar to the macrocycles of heme and chlorophyll) which houses a cobalt ion. Exchangeable upper ligands (R1) give cobamides versatile chemical reactivity (6). Lower ligands differ between cobalamin and other cobamides. The red asterisk indicates a methyl group that is absent in norcobamides (35, 36). (B) Three lower ligand structural classes are shown, which have multiple variable regions (R2 to R6).
Table 1. Metabolic functions and enzymes that use cobamide cofactors.
SAM, S-adenosylmethionine.
| Functional category | Cobamide-dependent enzymes or enzyme families |
|---|---|
| (a) Amino acid metabolism | Methionine synthase*†, glutamate mutase, lysine/ornithine aminomutases |
| (b) DNA and RNA synthesis | Ribonucleotide reductase†, epoxyqueuosine reductase† |
| (c) Carbon and nitrogen metabolism | Isomerases† (e.g., methylmalonyl-CoA mutase*, methyleneglutarate mutase), ethanolamine ammonia lyase, glycerol/diol dehydratases, methyltransferases (e.g., O-demethylases, methylamine methyltransferases, Wood-Ljungdahl corrinoid protein) |
| (d) Secondary metabolite synthesis | Radical SAM-B12 enzymes (biosynthesis of gentamicin, oxetanocin A, fosfomycin, phosalacine, thiostrepton, thienamycin) |
| (e) Others | Reductive dehalogenases, bacteriochlorophyll cyclase, light-sensing transcription factor, methytransferases [(MT), e.g., mercury MT, estrogen MT, methanogenesis marker protein 10] |
Conserved in humans.
Among the four most common enzyme families in bacteria (5).
Cobamides as model nutrients for studying microbial interactions
The chemical diversity of cobamides, which sets them apart from other vitamins and most shared nutrients, makes cobamides particularly interesting in the context of microbial interactions. Cobamides are a family of more than a dozen enzyme cofactors that all contain a chemically reactive cobalt ion housed by a corrin ring, but these cofactors differ in the identity of a covalently tethered lower ligand (Fig. 1A). Cobamides are grouped into three chemical classes according to the structure of their lower ligand: benzimidazolyl, purinyl, and phenolyl cobamides (with benzimidazoles, purines, and phenolics, respectively, as lower ligands) (Fig. 1B). The structural diversity of lower ligands is important because, in most cases, cobamide-dependent metabolism varies on the basis of lower ligand structure, and the sets of cobamides that support growth or enzymatic activity in different organisms are distinct (Table 2) (8, 14–34). For example, Dehalococcoides mccartyi can respire chlorinated solvents with several benzimidazolyl cobamides but not with phenolyl cobamides, whereas Sporomusa ovata requires phenolyl cobamides for growth on multiple carbon sources, and its growth is inhibited by benzimidazolyl cobamides (Table 2) (21, 22). Additionally, one bacterial species is known to synthesize norcobamides, which lack a methyl group in the linker region (Fig. 1A, red asterisk); the impact of this modification on physiology has only been explored to a limited extent (35, 36). Though there are relatively few studies comparing the effects of different cobamides on microbial metabolism, it is clear that the cobamide selectivity of organisms is important in the context of microbial interactions, as we hope to illustrate in this Review.
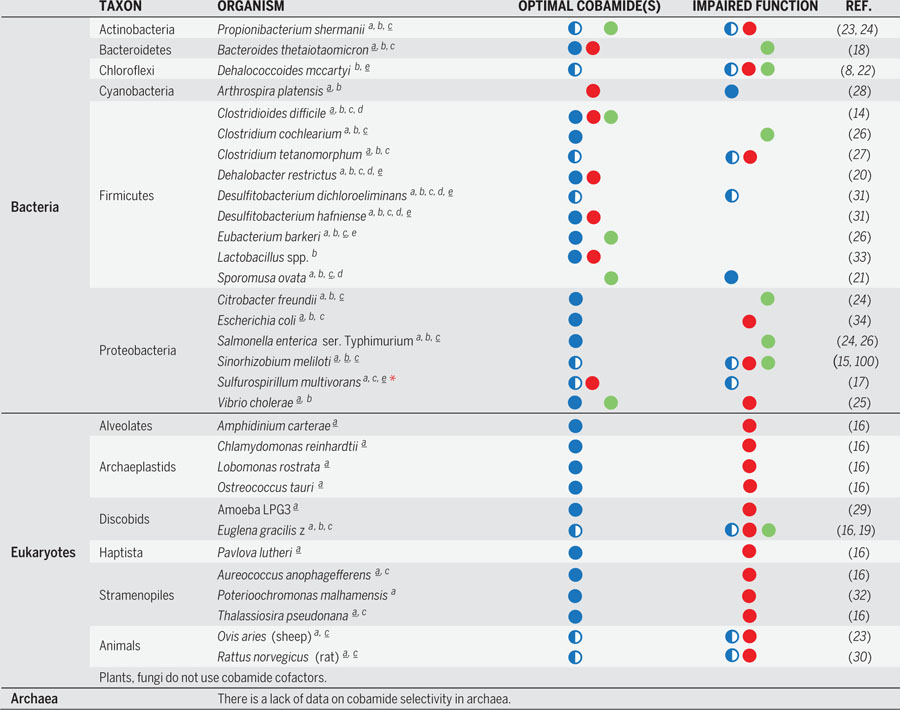
Table 2. Distinct cobamide use by different organisms.
Colored circles correspond to the lower ligand structural classes shown in Fig. 1B (blue, benzimidazolyl; red, purinyl; green, phenolyl cobamides); half-filled circles indicate a subset of the lower ligands within a class (in most cases only one member of each structural class has been tested for function, so the effect of diverse analogs is unknown). Superscripts after species names indicate the cobamide-dependent functions encoded in the genome of each organism, as defined in Table 1 (note, some species have not been sequenced, so they may be annotated incompletely in this table; radical SAM-B12 enzymes indicated by the superscript d are assumed to be involved in secondary metabolite biosynthesis, but not all have been experimentally characterized so some may have other functions). Underlined superscripts indicate the metabolisms that correspond to the selectivity data shown in this table, if known. S. multivorans uses norcobamides, indicated by the red asterisk. Data are from (8, 14–34, 100), as indicated in the rightmost column.
|
Like other metabolites studied in nutrient-sharing interactions (Table 3), cobamides are thought to be commonly shared (because many organisms that use cobamides do not produce them) and have well-characterized biosynthetic pathways that can be used to make predictions about cobamide-mediated interactions in natural systems. There are some technical benefits to using cobamides as models for nutrient sharing. For example, unlike amino acids, nucleotides, and most vitamins, cobamides have distinct spectral features that make them easily identifiable and that also provide information regarding chemical and conformational states that are related to function (37–39). Cobamides are notable from a nutrient-sharing perspective for several reasons. As catalytic cofactors, cobamides are effective at relatively low external concentrations [pico-molar to nanomolar (14, 18, 40, 41)] compared with nutrients that are incorporated into bio-mass. Thus, even small concentrations of cobamides can substantially affect microbial growth. Moreover, the functional diversity derived from the variation in lower ligand structure makes cobamides suitable models for various types of microbial interactions, in addition to nutrient sharing. For example, the inhibitory effects of phenolyl cobamides on the growth of D. mccartyi could constitute an antagonistic interaction that resembles competitive dynamics mediated by antimicrobial compounds. Cobamide-based partnerships are formed on the basis of the ability of microbes to produce and use a particular structural variant, as is the case for interactions based on the sharing of specific organic acids or carbohydrates (42). Generally, cobamides are a relevant model system for investigating how organisms evolve and adapt to structural variation in valuable metabolites, which is also pertinent to the study of structurally diverse secondary metabolites such as siderophores (43). By virtue of bridging the features of different classes of metabolites, cobamides are an effective and versatile model for studying microbial interactions.
Table 3. Comparison of nutrients that are shared in microbial communities, highlighting the qualities of cobamides as a model system.
Filled circles indicate that a class of metabolites has a particular feature; half-filled circles indicate that a subset of the metabolites have the corresponding feature.
| FEATURES | AMINO ACIDS | NUCLEOTIDES | ORGANIC COFACTORS | COBAMIDES |
|---|---|---|---|---|
| Evidence for widespread sharing | ● | ● | ● | ● |
| Well-characterized biosynthesis | ● | ● | ● | ● |
| Distinctive spectral features and colors | ◐ | ● | ||
| Effective in small quantities (catalytic) | ● | ● | ||
| Structurally diverse | ◐ | ● | ||
| Most are commercially available | ● | ● | ● |
Cobamide selectivity at the molecular and organismal levels
Cobamides modulate microbial growth and metabolism in structure-specific ways, which is the basis of cobamide selectivity. Nearly all of the diverse organisms tested for the ability to use different cobamides to date exhibit cobamide selectivity (Table 2), including both bacteria and eukaryotes. Understanding the mechanisms by which cobamides differentially affect metabolism is therefore critical to elucidating the roles of these cofactors in microbial interactions. Many molecular factors are thought to contribute to cobamide-dependent microbial growth, including selectivity in cobamide-dependent enzymes, differential cobamide import, and cobamide-specific gene regulatory responses. The extent to which each of these factors is a determinant of cobamide-dependent growth is still unclear.
The biochemical selectivity of cobamide-dependent enzymes toward particular cobamide cofactors is hypothesized to be a major determinant of cobamide-dependent growth. Lower ligand selectivity has been primarily studied in two enzymes: reductive dehalogenases, a class of bacterial cobamide-dependent enzymes that degrade toxic halogenated by-products of the dry cleaning industry, and methylmalonyl–coenzyme A (CoA) mutase, an enzyme involved in propionate metabolism that is widespread among bacteria (5) and conserved in humans. In multiple bacterial homologs of methylmalonyl-CoA mutase, lower ligand structure modulates the binding affinity of cobamide cofactors, and selectivity is different between orthologs from bacteria that produce different cobamides (15). Notably, the cobamide-binding selectivity of methylmalonyl-CoA mutase from Sinorhizobium meliloti is largely reflected in the differential growth of S. meliloti in cobamide-dependent conditions (15). Despite the fact that lower ligand structure also substantially affects reductive dehalogenase activity, crystal structures of reductive dehalogenases bound to different cobamides do not reveal atomic interactions between protein residues and the lower ligand that could mediate selectivity (17). It is likely that processes such as protein folding or cofactor loading are sensitive to lower ligand structure (17, 44). Consistent with this possibility, factors other than binding selectivity likely contribute to cobamide-dependent growth in methylmalonyl-CoA mutase metabolism as well (15).
The cobamide selectivity of an organism may also be influenced by the effects of diverse cobamides on gene expression. Although expression of metabolic genes is most commonly regulated by transcription factors, the primary mechanisms of the regulation of genes related to cobamide metabolism in bacteria are riboswitches—structured elements within the 5′ untranslated region of mRNAs that directly bind cobamides and affect the expression of downstream genes (12, 45, 46). The ability to distinguish between structurally similar ligands has been demonstrated in riboswitches that recognize variants of amino acids, nucleotides, flavins, and various bacterial signaling molecules (47, 48). It is therefore possible that cobamide-responsive riboswitches also recognize small structural differences between cobamides. In support of this hypothesis, RNA-cobamide interactions have been found to vary modestly in vitro depending on lower ligand structure (37). Finding selectivity-encoding sequences in riboswitches would enable researchers to integrate gene regulatory responses to diverse cobamides into functional predictions generated from bacterial genomes.
A third factor that may contribute to the observed cobamide selectivity of organisms is cobamide uptake. Although the presence of a lower ligand is not required for transport by the model cobalamin importer BtuBFCD in Escherichia coli and Vibrio cholerae (49, 50), genetic studies have shown that three different BtuB homologs in Bacteroides thetaiotaomicron confer distinct competitive advantages in the presence of different cobamides (18), which suggests that transporters may preferentially import particular cobamides. Moreover, several organisms have yet-unidentified cobamide importers or noncanonical proteins implicated in cobalamin uptake (51–54), so the overall contribution of selective cobamide import to the physiology of bacteria remains largely unexplored. Integration of biochemical and physiological studies of transport, regulation, and cobamide-dependent enzymes will improve our mechanistic understanding of cobamide sharing in microbial communities. Additionally, the ability to infer cobamide-sharing interactions from genomes will require predicting cobamide selectivity from genome sequence, which remains a major challenge.
Cobamide-mediated interactions in laboratory cocultures
Cultures of two or more organisms are commonly used to study mechanisms and dynamics of nutrient sharing in the laboratory (55). In addition to experimentally demonstrating that microorganisms share cobamides, cocultures provide tractable systems to study the mechanisms of cobamide-mediated microbial interactions, which are difficult to untangle in whole communities. We highlight coculture systems from three distinct habitats that exemplify the nuances of cobamide sharing: the different types of interactions that involve cobamides, the importance of cobamide selectivity in microbial interactions, and the metabolic outcome of cobamide availability.
Long-standing models of cobamide-sharing interactions are algae-bacteria mutualistic cocultures, in which cobalamin is produced by bacteria in exchange for fixed carbon from the algae (40). These systems have enabled studies of nutrient-sharing mechanisms and auxotrophy evolution (56, 57). For example, in a coculture containing the alga Lobomonas rostrata and the bacterium Mesorhizobium loti, cobamide sharing is essential for the growth of both organisms in the experimental conditions of the study (Fig. 2A). If either cobalamin or a fixed carbon source is added to the coculture, the stable ratio of the two species across serial transfers of the coculture is disrupted (56). In recent years, new synthetic consortia have expanded our understanding of the types of nutrient-sharing relationships that may occur in nature. For example, algae-bacteria mutualisms can also be based on bidirectional sharing of multiple B vitamins (58). Thus, laboratory cocultures that model various nutritional interactions can be used to probe fundamental principles of nutrient sharing, including how interactions differ depending on the nature of the compounds being shared.
Fig. 2. Cobamide sharing in model microbial consortia.
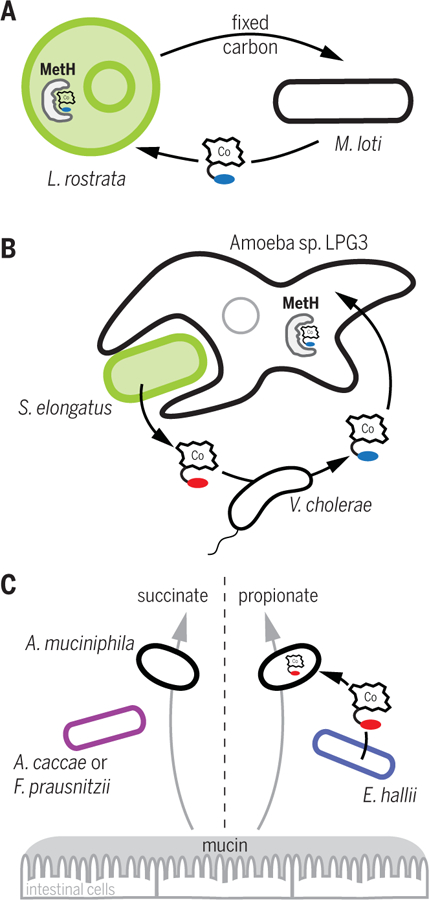
(A) Algae-bacteria mutualism. L. rostrata requires cobalamin produced by M. loti as a cofactor for its methionine synthase (MetH), and M. loti uses fixed carbon produced by L. rostrata (40, 56, 57). (B) Amoeba-prey consortium. V. cholerae remodels pseudocobalamin produced by Synechococcus elongatus to produce cobalamin, which can be used by the amoeba LPG3 for MetH activity (25, 29). (C) Cocultures of A. muciniphila with other human gut bacteria. Cobamide production by E. hallii causes a switch in the mucin degradation product generated by A. muciniphila, from succinate to propionate (62). Cobamide (Co-containing cartoon) with blue indicates cobalamin, and that with red indicates pseudocobalamin.
Cobamide-sharing microbial consortia have also provided valuable insights into the role of cobamide selectivity in cobamide sharing (29, 59, 60). For example, the predatory amoeba LPG3 notably requires both cyanobacterial prey and a bacterial co-isolate to meet its nutritional needs. Using a model triculture and molecular genetics, Ma et al. discovered that the second bacterial species is specifically essential for cobalamin production because the amoeba cannot use pseudocobalamin (a cobamide with adenine as the lower ligand) produced by the cyanobacterium (29). Furthermore, the cobalamin requirement of the amoeba could be satisfied by V. cholerae, a bacterium that cannot synthesize cobalamin de novo but was found to convert pseudocobalamin to cobalamin by cobamide remodeling—a process involving the removal and replacement of the lower ligand (22, 25, 61) (Fig. 2B). In addition to uncovering an unexpected three-member microbial interaction, Ma et al. discovered that an enzyme previously associated with a different branch of cobamide biosynthesis is involved in cobamide remodeling in V. cholerae (25). This example demonstrates how specificity for particular cobamides can drive a microbial association and illustrates how cocultures and tricultures provide useful systems for investigating the mechanisms of interactions.
The model systems described in the examples above focused on microbes that require cobamides for growth, but cobamides can also influence the metabolism of microbes for which cobamides are not essential. For example, although the human gut bacterium Akkermansia muciniphila produces succinate as a product of mucin degradation when cocultured with a butyrate-producing gut bacterium (Anaerostipes caccae or Faecalibacterium prausnitzii), A. muciniphila switches from succinate production to propionate production in the presence of a cobamide-producing bacterium, Eubacterium hallii. Analysis of metabolic pathways in A. muciniphila suggests a switch in fermentation strategy (predicted to involve methylmalonyl-CoA mutase) is elicited specifically by the cobamide production of E. hallii (62) (Fig. 2C). Thus, although A. muciniphila does not strictly require cobamides for growth, the presence of a cobamide-producing organism causes a physiological change that not only influences the metabolism of A. muciniphila itself but also might affect other microbes in the community. For example, increased succinate availability, which could result from fermentative metabolisms of human gut microbes, is linked to improved colonization by Clostridioides difficile in gnotobiotic mice (63). Studying the ways in which cobamide presence or absence modulates metabolism directly and indirectly may therefore reveal additional mechanisms by which cobamides affect microbial community structure.
Cobamide biology on a community level
Genomic approaches have been powerful in illustrating the prevalence of cobamide sharing in microbial communities. Cobamide biosynthesis, requiring up to 30 genes, is well characterized and can be easily identified in genomes, and it is possible to predict the production of benzimidazolyl and phenolyl cobamides by gene content (5, 12, 64, 65). Discoveries of additional genes related to lower ligand biosynthesis, transport, and cobamide-dependent metabolism continue to refine our ability to make functional predictions about cobamide biology (4, 25, 36, 51, 53, 66–81). Genomic studies of all sequenced bacteria (5, 13) and of communities from specific environments (18, 82–84) estimate that 58 to 69% of bacteria are unable to produce cobamides de novo, yet up to 86% use cobamides (the use of cobamides is defined by the presence of at least one cobamide-dependent enzyme or importer) (Fig. 3); these data constitute genomic evidence of widespread cobamide sharing. In addition to bacteria, many eukaryotic microbes use cobamides; most eukaryotic algae have cobalamin-dependent methionine synthase, and for ~50% of algal species, cobalamin is essential for growth (40, 85–87). Notably, the genes required to produce cobamides and the families of enzymes that use them are unevenly distributed across bacteria and archaea (5, 12, 13, 88). For example, across diverse habitats, 96% of Bacteroidetes species are predicted to use but not produce cobamides (5). This suggests that certain taxa tend to provide cobamides to other taxa in their respective habitats.
Fig. 3. Distribution of cobamide production and use among bacteria.
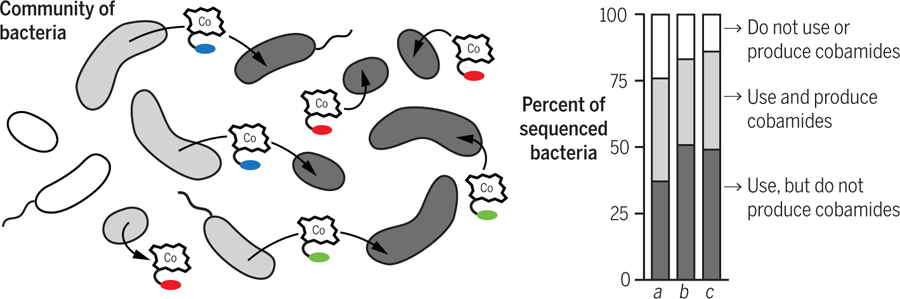
The percentages of species that use and produce cobamides are derived from published bioinformatic analyses of 540 bacterial genomes from diverse environments (column a) (13), 311 human gut bacterial genomes (column b) (18), and 11,463 bacterial genomes from diverse environments (column c) (5).
Genomic evidence for widespread cobamide sharing is corroborated by experimental analysis of cobamides from diverse microbial habitats. Bulk measurements of corrinoids—which include cobamides and corrin-containing cobamide precursors such as cobinamide—in the human gut, bovine rumen, mouse cecum, soil, and groundwater enrichment cultures show that each environment contains a diverse and distinct set of corrinoids, at concentrations ranging from picomolar to micromolar (83, 89–94). In such environments, microbes with a preference for particular cobamides must develop strategies to selectively acquire the cobamides that support their growth and avoid competitive inhibition by cobamides that do not. Moreover, the availability of different cobamides can vary spatially within a habitat. For example, in the North Pacific Ocean, cobalamin is detected throughout the surface waters, whereas pseudocobalamin is found primarily in the photic zone, correlating with the abundance of Cyano-bacteria, which are known to produce pseudocobalamin (91). Because abiotic factors can influence the spatial distribution of microbial species within communities in many environments, cobamide content, like other biological processes, is expected to vary across space.
The differential and distinct growth of bacteria in response to specific cobamides in pure culture, coupled with the prediction that most bacteria use cobamides, has inspired the idea that cobamides can be used to manipulate microbial community composition (10, 95). Several groups have tested the hypothesis that the addition of cobalamin to a community can alter its microbial composition, but the effect of cobamides with different lower ligands on communities has not been explored, likely because cobamides other than cobalamin are not commercially available. Studies in mice and in laboratory enrichment communities from the human gut, groundwater, and sea-water have shown that cobalamin addition can result in changes in community composition, corrinoid composition, and expression of cobamide salvaging and cobamide-dependent genes (11, 92, 93, 96–98). Limited sampling and the complexity of microbial communities have made it difficult to build mechanistic models of the growth and metabolic responses of members of a given community to specific perturbations. However, these results show promise for the potential of cobamides to modulate communities.
Open questions about cobamide sharing and challenges in the microbiome field
One major aim of the microbiome field has been to understand and predict microbial interactions using genomics. Because many organisms cannot be cultured and microbial communities can change dynamically over time and are often highly complex, genomics-based methods are useful for generating hypotheses about molecular interactions in a community. Owing to decades of work identifying the genes that encode cobamide-biosynthetic and cobamide-dependent enzymes, it is now possible to predict from genome sequences which organisms are capable of cobamide production and which carry cobamide-dependent functions. However, genomic predictions do not always match what occurs in situ. For example, whether cobamides are produced or released into the environment, and whether cobamide-dependent metabolic pathways are expressed in a community context, cannot necessarily be inferred from genomic potential. Additionally, cobamide-dependent metabolism occurs within a complex network of other metabolic pathways. Challenges remain in understanding how chemical environments influence cobamide-dependent metabolism, how direct effects of a nutrient indirectly affect metabolic networks, and how spatial and temporal dynamics influence nutrient sharing.
Recognizing environmental effects on cobamide biology
The chemical environment of an organism in its natural habitat can alter its physiology relative to what is observed in a laboratory setting (Fig. 4). Bacteria can synthesize different cobamides in different environments because lower ligand incorporation into cobamides depends not only on the selectivity of biosynthetic enzymes (20, 99–101) but also on the availability of exogenous lower ligand bases (102). Free lower ligand bases have been detected in several environments: dimethylbenzimidazole [(DMB), the lower ligand of cobalamin] has been detected in fresh water, soil, and rumen fluid (102), and alpha-ribazole, an activated form of DMB that can be taken up by some bacteria (103), is produced by marine bacteria (104, 105). Notably, organisms such as S. ovata and Sulfurospirillum multivorans, which produce phenolyl and purinyl cobamides, respectively, synthesize cobalamin when DMB is added to the culture medium, which leads to growth inhibition (21, 44). Other organisms are capable of remodeling suboptimal cobamides to cobalamin when DMB is available, including the bacteria D. mccartyi and V. cholerae (Fig. 2B) and the haptophyte Pavlova lutheri (16, 22, 25). These observations suggest that free lower ligand bases could potentially modulate microbial growth in natural settings. Growth conditions can alter cobamide production even in the absence of exogenous lower ligands; for example, Propionibacterium freudenreichii synthesizes pseudocobalamin under anoxic conditions and cobalamin under microoxic conditions (106). Thus, although we can often predict cobamide biosynthesis in pure culture, information about the chemical composition of natural environments is required to predict which cobamides are produced by organisms in situ and how that affects bacterial growth.
Fig. 4. Factors in complex chemical environments that can modulate cobamide physiology.
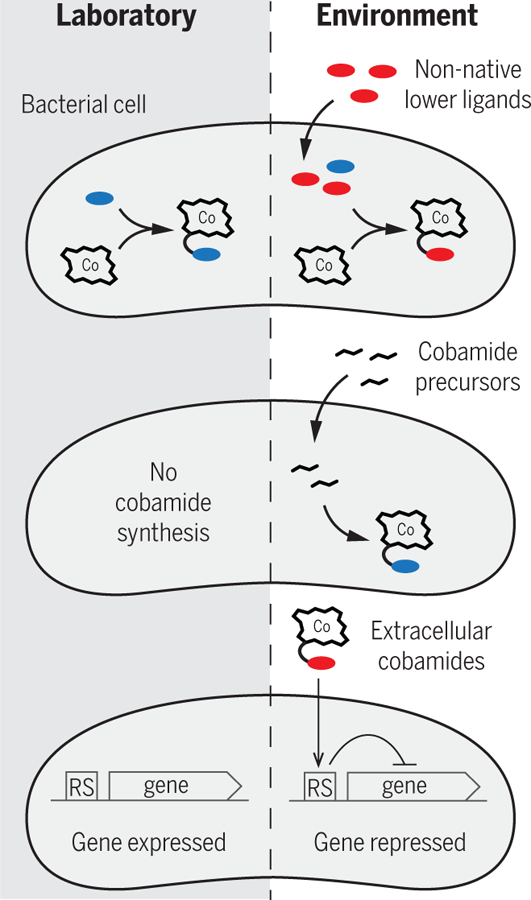
RS, riboswitch.
In addition to lower ligand bases, the presence of other cobamide precursors can affect cobamide biosynthesis. Some organisms have partial cobamide-biosynthetic pathways and can only produce cobamides when an appropriate precursor is present (Fig. 4) (5, 14, 107). For example, the human gut pathogen C. difficile requires either the early cobamide precursor 5-aminolevulinic acid (ALA) or the late precursor cobinamide to produce cobamides (14). Predicting whether C. difficile produces cobamides in the gut therefore requires knowing which precursors are present in its surroundings. Several different cobamide precursors are hypothesized to be shared in microbial communities on the basis of the presence of partial biosynthetic pathways in bacterial genomes (5, 14, 18, 95, 108), but few of these predictions have been tested, likely because of the limited commercial availability of precursor compounds other than ALA and cobinamide. The quantities of cobamide precursors in environments are also largely unknown, which makes it difficult to speculate about their availability to microbes in natural environments. Moreover, the mechanisms by which cobamides and their precursors are released by producers have yet to be identified (97).
Cobamides in the extracellular environment can also affect metabolism in situ by modulating the expression of cobamide-related pathways in an organism. In some cases, the gene regulatory responses of bacteria to the presence of cobamides can be predicted from sequence. Propionibacterium acnes down-regulates cobamide biosynthesis genes when cultured with cobalamin (109), which could be predicted on the basis of the presence of a cobalamin riboswitch in the biosynthesis operon (Fig. 4). Notably, this phenotype is reproduced in a natural habitat of P. acnes—on the skin of individuals taking cobalamin supplements— which supports the idea that functional predictions can be made for natural systems (109). Cobalamin riboswitches also regulate enzymes or pathways that are alternatives to cobamide-dependent processes. For example, Caulobacter crescentus has two methionine synthase enzymes (isozymes), one of which does not require a cobamide cofactor. The cobalamin-independent isozyme is regulated by a cobalamin riboswitch, and as predicted, a reporter system showed that its expression is down-regulated by cobalamin addition (110). This enzyme allows C. crescentus to tolerate fluctuations in environmental cobalamin levels, but there appears to be a fitness advantage to down-regulating its expression and using the cobalamin-dependent isozyme when cobalamin is available (110). The importance of the regulation of cobamide biosynthesis and isozymes for individuals in microbial communities has not been explored, and as mentioned previously, the effects of cobamides other than cobalamin on gene expression are mostly unknown.
Understanding direct and indirect effects of cobamide sharing
Cobamides may mediate microbial interactions not only through cobamide-dependent metabolism, but also through changes in a metabolic network elicited by cobamide-dependent reactions. The idea that products of cobamide-dependent reactions can have downstream effects on metabolism was highlighted earlier using the example of A. muciniphila (Fig. 2C). Additionally, metabolic pathways that do not directly require cobamides are affected by cobamide-dependent reactions. For example, cobalamin-dependent methionine synthesis is tightly linked to folate cycling in humans, such that cobalamin deficiency leads to an imbalance in folate pools, consequently affecting pathways such as purine synthesis that are not directly cobalamin dependent (111, 112). In humans, cobalamin deficiency therefore leads to complex and potentially severe pathologies including anemia and neurological disorders (113, 114). Similar connections between cobamide metabolism and other metabolic pathways are found in microbial metabolic networks but have been less studied in microbes than in humans. Thus, predicting cobamide-based interactions may require accounting for not only the sharing of cobamides themselves but also the changes to a metabolic map of an organism—or potentially a whole community—that result from cobamide-catalyzed reactions.
Investigating cobamide physiology in situ
Laboratory experiments are limited by their ability to accurately mimic environmental conditions and by the challenges associated with culturing microorganisms. In addition to meta-genomics, tools such as metatranscriptomics, metaproteomics, and metabolomics can help interrogate cobamide interactions in situ. Recently developed cobalamin-based chemical probes have been used to visualize cobalamin trafficking from E. coli to its nematode predator Caenorhabditis elegans and to capture cobalamin-binding proteins that were previously not known to interact with cobalamin in bacteria and eukaryotes (115–117). These probes may allow researchers to visualize how cobamides are used and shared in situ, which may help validate phenotypes that would be predicted from sequence. Furthermore, catalytically inactive chemical analogs of cobamides (antivitamins) (118–121) could potentially be used to perturb communities by inhibiting cobamide-dependent enzymes. Development and application of such tools for use in complex communities could be a way to study cobamide biology mechanistically on a community level.
Investigating cobamide interactions in situ could also address the importance of spatial structure in microbial interactions. Two organisms from the same environmental sample may or may not be able to interact, depending on their location in the sample. Thus, future predictions of microbial interactions should account for the spatial distribution of microbes. Unlike other shared nutrients, there has been little work on spatial constraints in natural or synthetic cobamide-sharing systems. Chemical probes, such as the fluorescent cobalamin analog described by Lawrence et al. (117), may be useful in this regard, as would be the application of recently developed techniques that have been used to study the spatial distributions of proteins, small molecules, and metals in other systems (122–127).
Looking forward
Focusing on a single class of shared nutrients—cobamides—across multiple scales has enabled researchers to begin to dissect the molecular interactions that shape microbial communities (128). One of the benefits of uncovering mechanistic details of microbial interactions is the potential opportunity to disrupt these interactions for the purpose of basic research or for environmental, health-related, or industrial applications. Because of their structural and functional diversity, cobamides make up a natural toolkit for manipulating microbial metabolism and growth, and the widespread use of cobamides in the bacterial domain makes them promising candidates for modulating community composition and function. Whether used as tools to perturb communities, or as a model system for asking basic questions about the nature of microbial interactions, cobamides exemplify the complexity and nuance of microbial life.
ACKNOWLEDGMENTS
We thank S. Gude, G. Pherribo, and K. Kennedy for critical reading of the manuscript; S. Gude, Z. Hallberg, K. Kennedy, A. Nicolas, B. Nguyen, D. Lauko, B. Bonet, and M. Orman for feedback on the accompanying Review summary; and our anonymous peer reviewers for their suggestions, particularly regarding Table 2 and Fig. 3.
Funding:
We acknowledge National Institutes of Health grant R01GM114535.
Footnotes
Competing interests: The authors declare no competing interests.
REFERENCES AND NOTES
- 1.D’Souza G et al. , Less is more: Selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution 68, 2559–2570 (2014). doi: 10.1111/evo.12468 [DOI] [PubMed] [Google Scholar]
- 2.Mee MT, Collins JJ, Church GM, Wang HH, Syntrophic exchange in synthetic microbial communities. Proc. Natl. Acad. Sci. U.S.A 111, E2149–E2156 (2014). doi: 10.1073/pnas.1405641111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roth JR, Lawrence JG, Bobik TA, Cobalamin (coenzyme B12): Synthesis and biological significance. Annu. Rev. Microbiol 50, 137–181 (1996). doi: 10.1146/annurev.micro.50.1.137 [DOI] [PubMed] [Google Scholar]
- 4.Bridwell-Rabb J, Drennan CL, Vitamin B12 in the spotlight again. Curr. Opin. Chem. Biol 37, 63–70 (2017). doi: 10.1016/j.cbpa.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shelton AN et al. , Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J 13, 789–804 (2019). doi: 10.1038/s41396-018-0304-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee R, Ragsdale SW, The many faces of vitamin B12: Catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem 72, 209–247 (2003). doi: 10.1146/annurev.biochem.72.121801.161828 [DOI] [PubMed] [Google Scholar]
- 7.Podar M et al. , Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci. Adv 1, e1500675 (2015). doi: 10.1126/sciadv.1500675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan J et al. , The corrinoid cofactor of reductive dehalogenases affects dechlorination rates and extents in organohalide-respiring Dehalococcoides mccartyi. ISME J 10, 1092–1101 (2016). doi: 10.1038/ismej.2015.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul L, Ferguson DJ Jr., Krzycki JA, The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J. Bacteriol 182, 2520–2529 (2000). doi: 10.1128/JB.182.9.2520-2529.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degnan PH, Taga ME, Goodman AL, Vitamin B12 as a modulator of gut microbial ecology. Cell Metab 20, 769–778 (2014). doi: 10.1016/j.cmet.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertrand EM et al. , Phytoplankton-bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc. Natl. Acad. Sci. U.S.A 112, 9938–9943 (2015). doi: 10.1073/pnas.1501615112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS, Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem 278, 41148–41159 (2003). doi: 10.1074/jbc.M305837200 [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Rodionov DA, Gelfand MS, Gladyshev VN, Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics 10, 78 (2009). doi: 10.1186/1471-2164-10-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shelton AN, Lyu X, Taga ME, Flexible cobamide metabolism in Clostridioides (Clostridium) difficile 630 Derm. J. Bacteriol 202, e00584–19 (2020). doi: 10.1128/JB.00584-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokolovskaya OM et al. , Cofactor selectivity in methylmalonyl coenzyme A mutase, a model cobamide-dependent enzyme. mBio 10, e01303–19 (2019). doi: 10.1128/mBio.01303-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helliwell KE et al. , Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12. Curr. Biol 26, 999–1008 (2016). doi: 10.1016/j.cub.2016.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller S et al. , Selective utilization of benzimidazolyl-norcobamides as cofactors by the tetrachloroethene reductive dehalogenase of Sulfurospirillum multivorans. J. Bacteriol 200, e00584–17 (2018). doi: 10.1128/JB.00584-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL, Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell Host Microbe 15, 47–57 (2014). doi: 10.1016/j.chom.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe F, Nakano Y, Stupperich E, Different corrinoid specificities for cell growth and cobalamin uptake in Euglena gracilis z. J. Gen. Microbiol 138, 1807–1813 (1992). doi: 10.1099/00221287-138-9-1807 [DOI] [Google Scholar]
- 20.Yan J et al. , Purinyl-cobamide is a native prosthetic group of reductive dehalogenases. Nat. Chem. Biol 14, 8–14 (2018). doi: 10.1038/nchembio.2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mok KC, Taga ME, Growth inhibition of Sporomusa ovata by incorporation of benzimidazole bases into cobamides. J. Bacteriol 195, 1902–1911 (2013). doi: 10.1128/JB.01282-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi S et al. , Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl. Environ. Microbiol 78, 7745–7752 (2012). doi: 10.1128/AEM.02150-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lengyel P, Mazumder R, Ochoa S, Mammalian methylmalonyl isomerase and vitamin B12 coenzymes. Proc. Natl. Acad. Sci. U.S.A 46, 1312–1318 (1960). doi: 10.1073/pnas.46.10.1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poppe L, Stupperich E, Hull WE, Buckel T, Rétey J, A base-off analogue of coenzyme-B12 with a modified nucleotide loop: 1H-NMR structure analysis and kinetic studies with (R)-methylmalonyl-CoA mutase, glycerol dehydratase, and diol dehydratase. Eur. J. Biochem 250, 303–307 (1997). doi: 10.1111/j.1432-1033.1997.0303a.x [DOI] [PubMed] [Google Scholar]
- 25.Ma AT, Tyrell B, Beld J, Specificity of cobamide remodeling, uptake and utilization in Vibrio cholerae. Mol. Microbiol 113, 89–102 (2020). doi: 10.1111/mmi.14402 [DOI] [PubMed] [Google Scholar]
- 26.Poppe L et al. , Elucidation of the coenzyme binding mode of further B12-dependent enzymes using a base-off analogue of coenzyme B12. J. Mol. Catal., B Enzym 10, 345–350 (2000). doi: 10.1016/S1381-1177(00)00136-3 [DOI] [Google Scholar]
- 27.Barker HA et al. , Isolation and properties of crystalline cobamide coenzymes containing benzimidazole or 5, 6-dimethylbenzimidazole. J. Biol. Chem 235, 480–488 (1960). [PubMed] [Google Scholar]
- 28.Tanioka Y et al. , Methyladeninylcobamide functions as the cofactor of methionine synthase in a Cyanobacterium, Spirulina platensis NIES-39. FEBS Lett 584, 3223–3226 (2010). doi: 10.1016/j.febslet.2010.06.013 [DOI] [PubMed] [Google Scholar]
- 29.Ma AT, Beld J, Brahamsha B, An amoebal grazer of cyanobacteria requires cobalamin produced by heterotrophic bacteria. Appl. Environ. Microbiol 83, e00035–17 (2017). doi: 10.1128/AEM.00035-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stabler SP, Brass EP, Marcell PD, Allen RH, Inhibition of cobalamin-dependent enzymes by cobalamin analogues in rats. J. Clin. Invest 87, 1422–1430 (1991). doi: 10.1172/JCI115148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schubert T et al. , Guided cobamide biosynthesis for heterologous production of reductive dehalogenases. Microb. Biotechnol 12, 346–359 (2019). doi: 10.1111/1751-7915.13339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ford JE, B12-vitamins and growth of the flagellate Ochromonas malhamensis. Microbiology 19, 161–172 (1958). doi: 10.1099/00221287-19-1-161 [DOI] [PubMed] [Google Scholar]
- 33.Berman D, Yacowitz H, Weiser HH, A differential microbiological assay for vitamin B12 and pseudovitamin B12. Appl. Microbiol 4, 49–52 (1956). doi: 10.1128/AEM.4.1.49-52.1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiao JS, Peterson WH, Microbiological assay of vitamin B12 with a mutant strain of Escherichia coli. Appl. Microbiol 1, 42–46 (1953). doi: 10.1128/AEM.1.1.42-46.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kräutler B et al. , The cofactor of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans is norpseudo‐B12, a new type of a natural corrinoid. Helv. Chim. Acta 86, 3698–3716 (2003). doi: 10.1002/hlca.200390313 [DOI] [Google Scholar]
- 36.Keller S, Treder A, von Reuss SH, Escalante-Semerena JC, Schubert T, The SMUL_1544 gene product governs norcobamide biosynthesis in the tetrachloroethene-respiring bacterium Sulfurospirillum multivorans. J. Bacteriol 198, 2236–2243 (2016). doi: 10.1128/JB.00289-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallo S, Oberhuber M, Sigel RK, Kräutler B, The corrin moiety of coenzyme B12 is the determinant for switching the btuB riboswitch of E. coli. ChemBioChem 9, 1408–1414 (2008). doi: 10.1002/cbic.200800099 [DOI] [PubMed] [Google Scholar]
- 38.Campanello GC et al. , Sacrificial cobalt-carbon bond homolysis in coenzyme B12 as a cofactor conservation strategy. J. Am. Chem. Soc 140, 13205–13208 (2018). doi: 10.1021/jacs.8b08659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deery E et al. , An enzyme-trap approach allows isolation of intermediates in cobalamin biosynthesis. Nat. Chem. Biol 8, 933–940 (2012). doi: 10.1038/nchembio.1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG, Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93 (2005). doi: 10.1038/nature04056 [DOI] [PubMed] [Google Scholar]
- 41.Bassford PJ Jr., Kadner RJ, Genetic analysis of components involved in vitamin B12 uptake in Escherichia coli. J. Bacteriol 132, 796–805 (1977). doi: 10.1128/JB.132.3.796-805.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Souza G et al. , Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep 35, 455–488 (2018). doi: 10.1039/C8NP00009C [DOI] [PubMed] [Google Scholar]
- 43.Miethke M, Marahiel MA, Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev 71, 413–451 (2007). doi: 10.1128/MMBR.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keller S et al. , Exogenous 5,6-dimethylbenzimidazole caused production of a non-functional tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Environ. Microbiol 16, 3361–3369 (2014). doi: 10.1111/1462-2920.12268 [DOI] [PubMed] [Google Scholar]
- 45.Mukherjee S et al. , RiboD: A comprehensive database for prokaryotic riboswitches. Bioinformatics 35, 3541–3543 (2019). doi: 10.1093/bioinformatics/btz093 [DOI] [PubMed] [Google Scholar]
- 46.Nahvi A et al. , Genetic control by a metabolite binding mRNA. Chem. Biol 9, 1043–1049 (2002). doi: 10.1016/S1074-5521(02)00224-7 [DOI] [PubMed] [Google Scholar]
- 47.Weinberg Z, Nelson JW, Lünse CE, Sherlock ME, Breaker RR, Bioinformatic analysis of riboswitch structures uncovers variant classes with altered ligand specificity. Proc. Natl. Acad. Sci. U.S.A 114, E2077–E2085 (2017). doi: 10.1073/pnas.1619581114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang JX, Lee ER, Morales DR, Lim J, Breaker RR, Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol. Cell 29, 691–702 (2008). doi: 10.1016/j.molcel.2008.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mireku SA et al. , Conformational change of a tryptophan residue in BtuF facilitates binding and transport of cobinamide by the vitamin B12 transporter BtuCD-F. Sci. Rep 7, 41575 (2017). doi: 10.1038/srep41575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agarwal S, Dey S, Ghosh B, Biswas M, Dasgupta J, Mechanistic basis of vitamin B12 and cobinamide salvaging by the Vibrio species. Biochim. Biophys. Acta Proteins Proteom 1867, 140–151 (2019). doi: 10.1016/j.bbapap.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 51.Wexler AG et al. , Human gut Bacteroides capture vitamin B12 via cell surface-exposed lipoproteins. eLife 7, e37138 (2018). doi: 10.7554/eLife.37138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gopinath K et al. , A vitamin B12 transporter in Mycobacterium tuberculosis. Open Biol 3, 120175 (2013). doi: 10.1098/rsob.120175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santos JA et al. , Functional and structural characterization of an ECF-type ABC transporter for vitamin B12. eLife 7, e35828 (2018). doi: 10.7554/eLife.35828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rempel S et al. , A mycobacterial ABC transporter mediates the uptake of hydrophilic compounds. Nature 580, 409–412 (2020). doi: 10.1038/s41586-020-2072-8 [DOI] [PubMed] [Google Scholar]
- 55.Dolinšek J, Goldschmidt F, Johnson DR, Synthetic microbial ecology and the dynamic interplay between microbial genotypes. FEMS Microbiol. Rev 40, 961–979 (2016). doi: 10.1093/femsre/fuw024 [DOI] [PubMed] [Google Scholar]
- 56.Kazamia E et al. , Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ. Microbiol 14, 1466–1476 (2012). doi: 10.1111/j.1462-2920.2012.02733.x [DOI] [PubMed] [Google Scholar]
- 57.Helliwell KE et al. , Fundamental shift in vitamin B12 eco-physiology of a model alga demonstrated by experimental evolution. ISME J 9, 1446–1455 (2015). doi: 10.1038/ismej.2014.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cooper MB et al. , Cross-exchange of B-vitamins underpins a mutualistic interaction between Ostreococcus tauri and Dinoroseobacter shibae. ISME J 13, 334–345 (2019). doi: 10.1038/s41396-018-0274-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Men Y et al. , Sustainable growth of Dehalococcoides mccartyi 195 by corrinoid salvaging and remodeling in defined lactate-fermenting consortia. Appl. Environ. Microbiol 80, 2133–2141 (2014). doi: 10.1128/AEM.03477-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan J, Ritalahti KM, Wagner DD, Löffler FE, Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl. Environ. Microbiol 78, 6630–6636 (2012). doi: 10.1128/AEM.01535-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gray MJ, Escalante-Semerena JC, The cobinamide amidohydrolase (cobyric acid-forming) CbiZ enzyme: A critical activity of the cobamide remodelling system of Rhodobacter sphaeroides. Mol. Microbiol 74, 1198–1210 (2009). doi: 10.1111/j.1365-2958.2009.06928.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belzer C et al. , Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. mBio 8, e00770–17 (2017). doi: 10.1128/mBio.00770-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferreyra JA et al. , Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16, 770–777 (2014). doi: 10.1016/j.chom.2014.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC, The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep 19, 390–412 (2002). doi: 10.1039/b108967f [DOI] [PubMed] [Google Scholar]
- 65.Moore SJ, Warren MJ, The anaerobic biosynthesis of vitamin B12. Biochem. Soc. Trans 40, 581–586 (2012). doi: 10.1042/BST20120066 [DOI] [PubMed] [Google Scholar]
- 66.Hazra AB et al. , Anaerobic biosynthesis of the lower ligand of vitamin B12. Proc. Natl. Acad. Sci. U.S.A 112, 10792–10797 (2015). doi: 10.1073/pnas.1509132112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zallot R et al. , Identification of a novel epoxyqueuosine reductase family by comparative genomics. ACS Chem. Biol 12, 844–851 (2017). doi: 10.1021/acschembio.6b01100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeter VL, Mattes TA, Beattie NR, Escalante-Semerena JC, A new class of phosphoribosyltransferases involved in cobamide biosynthesis is found in methanogenic archaea and cyanobacteria. Biochemistry 58, 951–964 (2019). doi: 10.1021/acs.biochem.8b01253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang PH et al. , Retroconversion of estrogens into androgens by bacteria via a cobalamin-mediated methylation. Proc. Natl. Acad. Sci. U.S.A 117, 1395–1403 (2020). doi: 10.1073/pnas.1914380117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Radle MI, Miller DV, Laremore TN, Booker SJ, Methanogenesis marker protein 10 (Mmp10) from Methanosarcina acetivorans is a radical S-adenosylmethionine methylase that unexpectedly requires cobalamin. J. Biol. Chem 294, 11712–11725 (2019). doi: 10.1074/jbc.RA119.007609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Picking JW, Behrman EJ, Zhang L, Krzycki JA, MtpB, a member of the MttB superfamily from the human intestinal acetogen Eubacterium limosum, catalyzes proline betaine demethylation. J. Biol. Chem 294, 13697–13707 (2019). doi: 10.1074/jbc.RA119.009886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tavares NK, VanDrisse CM, Escalante-Semerena JC, Rhodobacterales use a unique L-threonine kinase for the assembly of the nucleotide loop of coenzyme B12. Mol. Microbiol 110, 239–261 (2018). doi: 10.1111/mmi.14100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mehta AP et al. , Anaerobic 5-hydroxybenzimidazole formation from aminoimidazole ribotide: An unanticipated intersection of thiamin and vitamin B12 biosynthesis. J. Am. Chem. Soc 137, 10444–10447 (2015). doi: 10.1021/jacs.5b03576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parks JM et al. , The genetic basis for bacterial mercury methylation. Science 339, 1332–1335 (2013). doi: 10.1126/science.1230667 [DOI] [PubMed] [Google Scholar]
- 75.Zarzycki J, Sutter M, Cortina NS, Erb TJ, Kerfeld CA, In vitro characterization and concerted function of three core enzymes of a glycyl radical enzyme - associated bacterial microcompartment. Sci. Rep 7, 42757 (2017). doi: 10.1038/srep42757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krysenko S et al. , Initial metabolic step of a novel ethanolamine utilization pathway and its regulation in Streptomyces coelicolor M145. mBio 10, e00326–19 (2019). doi: 10.1128/mBio.00326-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.LaMattina JW et al. , 1,2-Propanediol dehydration in Roseburia inulinivorans: Structural basis for substrate and enantiomer selectivity. J. Biol. Chem 291, 15515–15526 (2016). doi: 10.1074/jbc.M116.721142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vetting MW et al. , Experimental strategies for functional annotation and metabolism discovery: Targeted screening of solute binding proteins and unbiased panning of metabolomes. Biochemistry 54, 909–931 (2015). doi: 10.1021/bi501388y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tavares NK, Zayas CL, Escalante-Semerena JC, The Methanosarcina mazei MM2060 gene encodes a bifunctional kinase/decarboxylase enzyme involved in cobamide biosynthesis. Biochemistry 57, 4478–4495 (2018). doi: 10.1021/acs.biochem.8b00546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rempel S, Colucci E, de Gier JW, Guskov A, Slotboom DJ, Cysteine-mediated decyanation of vitamin B12 by the predicted membrane transporter BtuM. Nat. Commun 9, 3038 (2018). doi: 10.1038/s41467-018-05441-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deobald D, Hanna R, Shahryari S, Layer G, Adrian L, Identification and characterization of a bacterial core methionine synthase. Sci. Rep 10, 2100 (2020). doi: 10.1038/s41598-020-58873-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Romine MF, Rodionov DA, Maezato Y, Osterman AL, Nelson WC, Underlying mechanisms for syntrophic metabolism of essential enzyme cofactors in microbial communities. ISME J 11, 1434–1446 (2017). doi: 10.1038/ismej.2017.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu X, Heal KR, Ingalls AE, Doxey AC, Neufeld JD, Metagenomic and chemical characterization of soil cobalamin production. ISME J 14, 53–66 (2020). doi: 10.1038/s41396-019-0502-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Magnúsdóttir S, Ravcheev D, de Crécy-Lagard V, Thiele I, Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet 6, 148 (2015). doi: 10.3389/fgene.2015.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blaby-Haas CE, Merchant SS, Comparative and functional algal genomics. Annu. Rev. Plant Biol 70, 605–638 (2019). doi: 10.1146/annurev-arplant-050718-095841 [DOI] [PubMed] [Google Scholar]
- 86.Tang YZ, Koch F, Gobler CJ, Most harmful algal bloom species are vitamin B1 and B12 auxotrophs. Proc. Natl. Acad. Sci. U.S.A 107, 20756–20761 (2010). doi: 10.1073/pnas.1009566107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG, Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol. Biol. Evol 28, 2921–2933 (2011). doi: 10.1093/molbev/msr124 [DOI] [PubMed] [Google Scholar]
- 88.Doxey AC, Kurtz DA, Lynch MD, Sauder LA, Neufeld JD, Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J 9, 461–471 (2015). doi: 10.1038/ismej.2014.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Allen RH, Stabler SP, Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am. J. Clin. Nutr 87, 1324–1335 (2008). doi: 10.1093/ajcn/87.5.1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Girard CL, Santschi DE, Stabler SP, Allen RH, Apparent ruminal synthesis and intestinal disappearance of vitamin B12 and its analogs in dairy cows. J. Dairy Sci 92, 4524–4529 (2009). doi: 10.3168/jds.2009-2049 [DOI] [PubMed] [Google Scholar]
- 91.Heal KR et al. , Two distinct pools of B12 analogs reveal community interdependencies in the ocean. Proc. Natl. Acad. Sci. U.S.A 114, 364–369 (2017). doi: 10.1073/pnas.1608462114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kelly CJ et al. , Oral vitamin B12 supplement is delivered to the distal gut, altering the corrinoid profile and selectively depleting Bacteroides in C57BL/6 mice. Gut Microbes 10, 654–662 (2019). doi: 10.1080/19490976.2019.1597667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Men Y et al. , Identification of specific corrinoids reveals corrinoid modification in dechlorinating microbial communities. Environ. Microbiol 17, 4873–4884 (2015). doi: 10.1111/1462-2920.12500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Suffridge CP et al. , B vitamins and their congeners as potential drivers of microbial community composition in an oligotrophic marine ecosystem. J. Geophys. Res. Biogeosci 123, 2890–2907 (2018). doi: 10.1029/2018JG004554 [DOI] [Google Scholar]
- 95.Seth EC, Taga ME, Nutrient cross-feeding in the microbial world. Front. Microbiol 5, 350 (2014). doi: 10.3389/fmicb.2014.00350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu Y et al. , Cobalamin (vitamin B12) induced a shift in microbial composition and metabolic activity in an in vitro colon simulation. Front. Microbiol 9, 2780 (2018). doi: 10.3389/fmicb.2018.02780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Men Y et al. , Metagenomic and metatranscriptomic analyses reveal the structure and dynamics of a dechlorinating community containing Dehalococcoides mccartyi and corrinoid-providing microorganisms under cobalamin-limited conditions. Appl. Environ. Microbiol 83, e03508–16 (2017). doi: 10.1128/AEM.03508-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhu X et al. , Impact of cyanocobalamin and methylcobalamin on inflammatory bowel disease and the intestinal microbiota composition. J. Agric. Food Chem 67, 916–926 (2019). doi: 10.1021/acs.jafc.8b05730 [DOI] [PubMed] [Google Scholar]
- 99.Hazra AB, Tran JLA, Crofts TS, Taga ME, Analysis of substrate specificity in CobT homologs reveals widespread preference for DMB, the lower axial ligand of vitamin B12. Chem. Biol 20, 1275–1285 (2013). doi: 10.1016/j.chembiol.2013.08.007 [DOI] [PubMed] [Google Scholar]
- 100.Crofts TS, Seth EC, Hazra AB, Taga ME, Cobamide structure depends on both lower ligand availability and CobT substrate specificity. Chem. Biol 20, 1265–1274 (2013). doi: 10.1016/j.chembiol.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 101.Chan CH, Escalante-Semerena JC, ArsAB, a novel enzyme from Sporomusa ovata activates phenolic bases for adenosylcobamide biosynthesis. Mol. Microbiol 81, 952–967 (2011). doi: 10.1111/j.1365-2958.2011.07741.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Crofts TS, Men Y, Alvarez-Cohen L, Taga ME, A bioassay for the detection of benzimidazoles reveals their presence in a range of environmental samples. Front. Microbiol 5, 592 (2014). doi: 10.3389/fmicb.2014.00592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gray MJ, Escalante-Semerena JC, A new pathway for the synthesis of a-ribazole-phosphate in Listeria innocua. Mol. Microbiol 77, 1429–1438 (2010). doi: 10.1111/j.1365-2958.2010.07294.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wienhausen G, Noriega-Ortega BE, Niggemann J, Dittmar T, Simon M, The exometabolome of two model strains of the Roseobacter group: A marketplace of microbial metabolites. Front. Microbiol 8, 1985 (2017). doi: 10.3389/fmicb.2017.01985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Johnson WM, Kido Soule MC, Kujawinski EB, Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J 10, 2304–2316 (2016). doi: 10.1038/ismej.2016.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deptula P et al. , BluB/CobT2 fusion enzyme activity reveals mechanisms responsible for production of active form of vitamin B12 by Propionibacterium freudenreichii. Microb. Cell Fact 14, 186 (2015). doi: 10.1186/s12934-015-0363-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lawrence JG, Roth JR, The cobalamin (coenzyme B12) biosynthetic genes of Escherichia coli. J. Bacteriol 177, 6371–6380 (1995). doi: 10.1128/JB.177.22.6371-6380.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roper JM et al. , The enigma of cobalamin (vitamin B12) biosynthesis in Porphyromonas gingivalis. Identification and characterization of a functional corrin pathway. J. Biol. Chem 275, 40316–40323 (2000). doi: 10.1074/jbc.M007146200 [DOI] [PubMed] [Google Scholar]
- 109.Kang D, Shi B, Erfe MC, Craft N, Li H, Vitamin B12 modulates the transcriptome of the skin microbiota in acne pathogenesis. Sci. Transl. Med 7, 293ra103 (2015). doi: 10.1126/scitranslmed.aab2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aretakis JR, Gega A, Schrader JM, Absolute measurements of mRNA translation in Caulobacter crescentus reveal important fitness costs of vitamin B12 scavenging. mSystems 4, e00170–19 (2019). doi: 10.1128/mSystems.00170-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Froese DS, Fowler B, Baumgartner MR, Vitamin B12, folate, and the methionine remethylation cycle—biochemistry, pathways, and regulation. J. Inherit. Metab. Dis 42, 673–685 (2019). doi: 10.1002/jimd.12009 [DOI] [PubMed] [Google Scholar]
- 112.Sauer H, Wilmanns W, Cobalamin dependent methionine synthesis and methyl-folate-trap in human vitamin B12 deficiency. Br. J. Haematol 36, 189–198 (1977). doi: 10.1111/j.1365-2141.1977.tb00639.x [DOI] [PubMed] [Google Scholar]
- 113.Huemer M, Baumgartner MR, The clinical presentation of cobalamin-related disorders: From acquired deficiencies to inborn errors of absorption and intracellular pathways. J. Inherit. Metab. Dis 42, 686–705 (2019). doi: 10.1002/jimd.12012 [DOI] [PubMed] [Google Scholar]
- 114.Shevell MI, Rosenblatt DS, The neurology of cobalamin. Can. J. Neurol. Sci 19, 472–486 (1992). doi: 10.1017/S0317167100041676 [DOI] [PubMed] [Google Scholar]
- 115.Romine MF et al. , Elucidation of roles for vitamin B12 in regulation of folate, ubiquinone, and methionine metabolism. Proc. Natl. Acad. Sci. U.S.A 114, E1205–E1214 (2017). doi: 10.1073/pnas.1612360114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rosnow JJ et al. , A cobalamin activity-based probe enables microbial cell growth and finds new cobalamin-protein interactions across domains. Appl. Environ. Microbiol 84, e00955–18 (2018). doi: 10.1128/AEM.00955-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lawrence AD et al. , Construction of fluorescent analogs to follow the uptake and distribution of cobalamin (vitamin B12) in bacteria, worms, and plants. Cell Chem. Biol 25, 941–951.e6 (2018). doi: 10.1016/j.chembiol.2018.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Widner FJ et al. , Total synthesis, structure, and biological activity of adenosylrhodibalamin, the non-natural rhodium homologue of coenzyme B12. Angew. Chem. Int. Ed 55, 11281–11286 (2016). doi: 10.1002/anie.201603738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Calafat AM et al. , Structural and electronic similarity but functional difference in methylmalonyl-CoA mutase between coenzyme B12 and the analog 2′,5′-dideoxyadenosylcobalamin. Biochemistry 34, 14125–14130 (1995). doi: 10.1021/bi00043a017 [DOI] [PubMed] [Google Scholar]
- 120.Fukuoka M, Nakanishi Y, Hannak RB, Kräutler B, Toraya T, Homoadenosylcobalamins as probes for exploring the active sites of coenzyme B12-dependent diol dehydratase and ethanolamine ammonia-lyase. FEBS J 272, 4787–4796 (2005). doi: 10.1111/j.1742-4658.2005.04892.x [DOI] [PubMed] [Google Scholar]
- 121.Zelder F, Sonnay M, Prieto L, Antivitamins for medicinal applications. ChemBioChem 16, 1264–1278 (2015). doi: 10.1002/cbic.201500072 [DOI] [PubMed] [Google Scholar]
- 122.DePas WH et al. , Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. mBio 7, e00796–16 (2016). doi: 10.1128/mBio.00796-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bellin DL et al. , Integrated circuit-based electrochemical sensor for spatially resolved detection of redox-active metabolites in biofilms. Nat. Commun 5, 3256 (2014). doi: 10.1038/ncomms4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schiessl KT et al. , Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nat. Commun 10, 762 (2019). doi: 10.1038/s41467-019-08733-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ryan DJ et al. , MicroLESA: Integrating autofluorescence microscopy, in situ micro-digestions, and liquid extraction surface analysis for high spatial resolution targeted proteomic studies. Anal. Chem 91, 7578–7585 (2019). doi: 10.1021/acs.analchem.8b05889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cassat JE et al. , Integrated molecular imaging reveals tissue heterogeneity driving host-pathogen interactions. Sci. Transl. Med 10, eaan6361 (2018). doi: 10.1126/scitranslmed.aan6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Herrmann AM et al. , Nano-scale secondary ion mass spectrometry — A new analytical tool in biogeochemistry and soil ecology: A review article. Soil Biol. Biochem 39, 1835–1850 (2007). doi: 10.1016/j.soilbio.2007.03.011 [DOI] [Google Scholar]
- 128.Abreu NA, Taga ME, Decoding molecular interactions in microbial communities. FEMS Microbiol. Rev 40, 648–663 (2016). doi: 10.1093/femsre/fuw019 [DOI] [PMC free article] [PubMed] [Google Scholar]


