Abstract
Background:
Circulating tumor DNA (ctDNA) is a promising biomarker for non-invasive epidermal growth factor receptor mutations (EGFRm) detection in lung cancer patients, but existing methods have limitations in sensitivity and availability. In this study, we used the ΔCt value (mutant cycle threshold [Ct] value–internal control Ct value) generated during the polymerase chain reaction (PCR) assay to convert super-amplification-refractory mutation system (superARMS) from a qualitative method to a semi-quantitative method named reformed-superARMS (R-superARMS), and evaluated its performance in detecting EGFRm in plasma ctDNA in patients with advanced lung adenocarcinoma.
Methods:
A total of 41 pairs of tissues and plasma samples were obtained from lung adenocarcinoma patients who had known EGFRm in tumor tissue and were previously untreated. EGFRm in ctDNA was identified by using superARMS. Through making use of ΔCt value generated during the detection process of superARMS, we indirectly transform this qualitative detection method into a semi-quantitative PCR detection method, named R-superARMS. Both qualitative and quantitative analyses of the data were performed. Kaplan–Meier analysis was performed to estimate the progression-free survival (PFS) and overall survival (OS). Fisher exact test was used for categorical variables.
Results:
The concordance rate of EGFRm in tumor tissues and matched plasma samples was 68.3% (28/41). At baseline, EGFRm-positive patients were divided into two groups according to the cut-off ΔCt value of EGFRm set at 8.11. A significant difference in the median OS (mOS) between the two groups was observed (EGFRm ΔCt ≤8.11 vs. >8.11: not reached vs. 11.0 months; log-rank P = 0.024). Patients were divided into mutation clearance (MC) group and mutation incomplete clearance (MIC) group according to whether the ΔCt value of EGFRm test turned negative after 1 month of treatment. We found that there was also a significant difference in mOS (not reached vs. 10.4 months; log-rank P = 0.021) between MC group and MIC group. Although there was no significant difference in PFS between the two groups, the two curves were separated and the PFS of MC group tended to be higher than the MIC group (not reached vs. 27.5 months; log-rank P = 0.088). Furthermore, EGFRm-positive patients were divided into two groups according to the cut-off of the changes in ΔCt value of EGFRm after 1 month of treatment, which was set at 4.89. A significant difference in the mOS between the two groups was observed (change value of ΔCt >4.89 vs. ≤4.89: not reached vs. 11.0 months; log-rank P = 0.014).
Conclusions:
Detecting EGFRm in ctDNA using R-superARMS can identify patients who are more likely sensitive to targeted therapy, reflect the molecular load of patients, and predict the therapeutic efficacy and clinical outcomes of patients.
Keywords: Lung adenocarcinoma, Non-small cell lung adenocarcinoma, Liquid biopsy, Super-amplification-refractory mutation system, EGFR mutation
Introduction
The guidelines recommend non-small cell lung cancer (NSCLC) patients with epidermal growth factor receptor mutations (EGFRm) can be treated with tyrosine kinase inhibitors (TKIs), but in clinical practice, the effective rate of targeted therapy in patients with EGFR positive mutation was only about 70%, and most patients develop resistance within 9 to 12 months.[1–3] Therefore, detection of EGFRm and monitoring the efficacy of treatment are of great significance in such patients.[4] The majority of EGFR genotyping is assessed by conventional tumor biopsy, which however, has several shortcomings: the tissue specimen showed heterogeneity of time and space[5–7]; it is hard to obtain biopsy tissue after recurrence or metastasis; and intrathoracic biopsy is easy to cause complications.[8,9] In addition, the clinical evaluation of the therapeutic effect of solid tumor is currently based on alterations in imaging; however, patients with the same degree of imaging response have different treatment outcomes, supposing that there exist different degrees of tumor molecular response to guide treatment options and to predict prognosis. Recently, with the development of technology, liquid biopsy, especially circulating tumor DNA (ctDNA) detection, can bring us enough information, present information earlier than changes in imaging, and also overcome the limitations of tissue biopsy.[10–12]
At present, several molecular detection methods have been developed, such as amplification-refractory mutation system (ARMS), droplet digital polymerase chain reaction (ddPCR), and next generation sequencing (NGS).[13–15] The ARMS has been approved by the European Union and National Medical Products Administration (NMPA) for clinical testing of EGFRm using tissue or blood samples. However, it is only a qualitative detection technique and the sensitivity (48.2–67.4%) needs to be improved compared to the matched tumor tissue biopsy.[16,17] ddPCR and NGS have relatively high sensitivity and absolute quantification. But the development of ddPCR kit is not easy, and the construction of NGS database is complicated and the standardization of NGS detection platform needs to be improved, which makes these two methods more suitable for translational medical research[18,19] rather than clinical practice. Most recently, a newly developed method based on ARMS, super-amplification-refractory mutation system (superARMS), has been available for detecting EGFRm status in the plasma ctDNA with a qualitative polymerase chain reaction (PCR) method claimed to be highly selective, sensitive, and economic, and has been approved by (NMPA) for detecting the status of ctDNA EGFRm in patients with advanced NSCLC and conventionally applied to clinical practice.[20,21] However, it is still not a quantitative detection method, which limits its application in further clinical research.
In this study, all patients were routinely applied the superARMS method for plasma ctDNA detection. We proposed a reformed superARMS method named reformed-superARMS (R-superARMS). Through making use of ΔCt value generated during the detection process, we indirectly transform this qualitative detection method into a semi-quantitative PCR detection method. This research aimed to study the effects of different baseline ctDNA EGFRm levels and changes of ctDNA EGFRm with R-superARMS on the outcomes and prognosis of patients receiving targeted therapy.
Methods
Ethical approval
This study was approved by the Institutional Review Board of the First Hospital of Jilin University (No. NCT2017-444). Informed consent was obtained from all patients.
Patients and treatment
This study was conducted in the Cancer Center, the First Hospital of Jilin University in Changchun, China. A total of 41 Patients were enrolled from January 2017 to June 2018. Inclusion criteria are described as follows: (1) pathologically confirmed lung adenocarcinoma; (2) advanced clinical stage (stage IIIB or IV); (3) newly diagnosed or recurrence after surgery; (4) a subsequent treatment did not begin at the time of sample collection; and (5) tumor tissue and blood specimens were available and the interval between tissue collection and blood collection was less than 14 days.
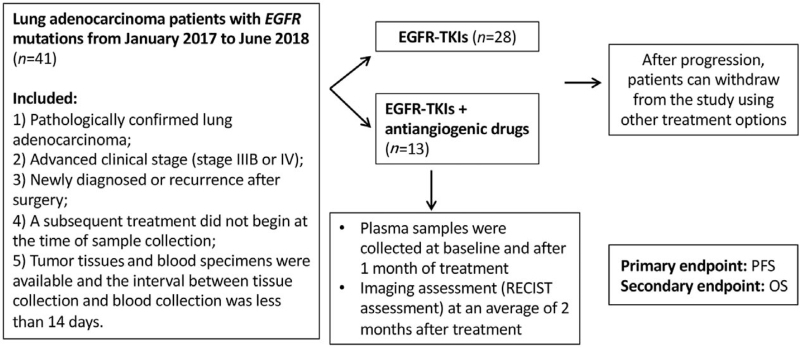
All enrolled patients received first-generation TKIs monotherapy (erlotinib, gefitinib, or ectinib) or combination with antiangiogenic drugs (bevacizumab or apatinib) as first-line treatment. Antitumor drugs were used according to the routine clinical practice (National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology: NSCLC). Patient data were collected from medical records and radiological images were used to assess baseline information, tumor response, and progression-free survival (PFS). After tumor progression, patients can withdraw from the study using other treatment options. Tumor assessment was performed using the response evaluation criteria in solid tumors (RECIST) Version 1.1, and EGFRm in plasma was assessed at baseline and 1 month after treatment. A schematic representation of the study is shown in Figure 1. The primary endpoint was PFS. Overall survival (OS) was evaluated as a secondary endpoint.
Figure 1.
Flow chart of the study. EGFR: Epidermal growth factor receptor; OS: Overall survival; PFS: Progression-free survival; RECIST: Response evaluation criteria in solid tumors; TKIs: Tyrosine kinase inhibitors.
DNA extraction
Plasma was separated from the whole blood within 2 h of collection. ctDNA was extracted from the plasma within 2 h by AmoyDx circulating DNA kit (Amoy Diagnostics, Xiamen, China) and stored at −80°C before use. The optical density of the extracted DNA samples was measured using a NanoDrop 2000 spectrophotometer (Thermofihier Scientific, Carlsbad, CA, USA). The A260/A280 value of all samples was 1.8 to 2.0. The extracted DNA was used for EGFR molecular testing.
Detection of ctDNA EGFRm
The EGFRm in plasma ctDNA was detected by superARMS EGFRm detection kit (Amoy Diagnostics, Xiamen, China), which can identify 41 types of the most common somatic EGFR mutations in exons 18 to 21. The assay was performed using a real-time PCR according to the manufacturer's instructions, while PCRs were performed using Stratagene Mx3000P quantitative PCR system (Agilent, Santa Clara, CA, USA). All reactions were carried out in a volume of 70 μL DNA sample (or negative/positive controls), plasma epidermal growth factor receptor (P-EGFR) enzyme mix, and P-EGFR reaction mix. PCR cycling conditions were performed with 10 min incubation at 95°C, followed by 15 cycles at 95°C for 40 s, 64°C for 40 s, and 72°C for 30 s, and then 28 cycles of 93°C for 40 s, 60°C for 45 s, and 72°C for 30 s, respectively. ΔCt value (mutant cycle threshold [Ct] value–internal control Ct value) was calculated to identify the presence of EGFRm, which was automatically calculated from PCR amplification plots of fluorescence and its corresponding number of cycles. The ΔCt cut-off value was defined as 11 (19del/L858R/20ins), 8 (T790M), or 12 (G719X/L861Q/S768I). When the ΔCt value was less than the cut-off value, the sample was considered as mutant. Mutation clearance (MC) group refers to that the ΔCt value of original EGFRm completely changed to the negative mutation value set above, and mutation incomplete clearance (MIC) group refers to that although the ΔCt value of EGFRm changed, it was still positive and did not completely turn negative.
Statistical analysis
The concordance rate was assessed by comparing the match between primary tissue and ctDNA sequencing results. The Kaplan–Meier analysis was performed to estimate survival probabilities. A receiver operating characteristic (ROC) curve was applied to calculate the cutoff values for the parameters (ΔCt value of EGFRm or the changes in ΔCt value of EGFRm) and area under the curve (AUC). The cutoff point was determined using Youden index. Fisher exact test was used for analysis of categorical variables. The hazard ratio for each cutoff point was calculated using the results from each of the corresponding survival analyses. The PFS was defined as the time from the start of the TKIs treatment to the first documentation of tumor progression or the last follow-up. OS was defined as the time from the start of follow-up to the death of patients or the end of follow-up. Throughout the analyses, all P values were based on a two-sided hypothesis, and those less than 0.05 were considered statistically significant. All statistical analyses were performed using GraphPad Prism version 6.0 for Mac and SPSS 22.0 (IBM Corp., Armonk, NY, USA).
Results
Characteristics of patients
A total of 41 lung adenocarcinoma patients met the criteria and were enrolled into the study. The patients’ clinical characteristics are listed in Table 1. The mean age was 63 years with a range of 39 to 75 years. Most patients were female and non-smokers. About 51.2% (21/41) had extrathoracic metastatic diseases (M1b; mainly for bone metastasis and brain metastasis) and 48.8% (20/41) only had intrathoracic metastatic diseases. About 68.3% (28/41) received first-generation epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) monotherapy, and others received first-generation EGFR-TKIs combined with antiangiogenic drugs.
Table 1.
Basic characteristics of all included patients with advanced lung adenocarcinoma (n = 41).
| Parameters | n (%) |
| Gender | |
| Male | 13 (31.7) |
| Female | 28 (68.3) |
| Age (years) | |
| <65 | 23 (56.1) |
| ≥65 | 18 (43.9) |
| Tumor stage | |
| M1a | 20 (48.8) |
| M1b | 21 (51.2) |
| ctDNA EGFRm status | |
| EGFRm-positive | 28 (68.3) |
| L858R | 12 (29.3) |
| 19del | 13 (31.7) |
| G719X | 2 (4.9) |
| T790M | 1 (2.4) |
| EGFRm-negative | 13 (31.7) |
| Treatment | |
| First-generation EGFR-TKIs | 28 (68.3) |
| First-generation EGFR-TKIs + anti-angiogenic drugs | 13 (31.7) |
ctDNA: Circulating tumor DNA; EGFR: Epidermal growth factor receptor; EGFRm: Epidermal growth factor receptor mutation; TKIs: Tyrosine kinase inhibitors.
Detection of ctDNA EGFRm using superARMS and R-superARMS
The EGFRm status in 41 pairs of tissue and matched plasma samples collected before the administration of TKIs treatment was tested by superARMS, and the concordance rate was 68.3% (28/41). Individual patient results are reported in Table 1. EGFR 19del (31.7%, 13/41) and L858R mutations (29.3%, 12/41) were the two most common type of mutations. Other rare mutation types included G719X and T790M. About 31.7% (13/41) were negative for EGFRm detected by superARMS in plasma samples but positive in the tissues.
Baseline levels of ctDNA EGFRm affect the outcomes of patients
The median PFS of all patients was 14.9 months. During a median follow-up period of 2.5 years, disease progression and death occurred in 36.6% (15/41) and 24.4% (10/41) of patients, respectively. All adverse events were tolerable and no treatment-related deaths occurred. Considering the variability in the effective rate of targeted therapy in patients with EGFR positive mutation, the semi-quantitative R-superARMS method was used in our study to detect the different values of EGFRm in EGFRm-positive patients. We set the cut-off ΔCt value of baseline EGFRm at 8.11 using ROC curve (AUC = 0.579; Youden index = 0.236).
Before survival analysis, we analyzed the equilibrium of clinical characteristics (including gender, age, treatment, and tumor stage) between different groups with baseline ΔCt values of EGFRm ≤8.11 and >8.11, and found P > 0.05 for all comparisons, demonstrating that the balance was good [Table 2].
Table 2.
Comparison of clinical characteristics between groups with different ctDNA EGFRm ΔCt values.
| Baseline ctDNA EGFRm ΔCt values | Qualitative changes of ctDNA EGFRm 1 month after treatment | Quantitative changes in ctDNA EGFRm ΔCt values 1 month after treatment | |||||||
| Items | ≤8.11(n = 23) | >8.11(n = 5) | P | MC group (n = 19) | MIC group (n = 9) | P | ≤4.89 (n = 12) | >4.89 (n = 16) | P |
| Age (years) | |||||||||
| <65 | 12 (52.2) | 4 (80.0) | 0.36 | 12 (63.2) | 4 (44.4) | 0.43 | 8 (66.7) | 8 (50.0) | 0.46 |
| ≥65 | 11 (47.8) | 1 (20.0) | 7 (36.8) | 5 (55.6) | 4 (33.3) | 8 (50.0) | |||
| Gender | |||||||||
| Male | 8 (34.8) | 2 (40.0) | 1.00 | 6 (31.6) | 4 (44.4) | 0.68 | 5 (41.7) | 5 (31.2) | 0.70 |
| Female | 15 (65.2) | 3 (60.0) | 13 (68.4) | 5 (55.6) | 7 (58.3) | 11 (68.8) | |||
| Tumor stage | |||||||||
| M1a | 10 (43.5) | 2 (40.0) | 1.00 | 8 (42.1) | 4 (44.4) | 1.00 | 7 (58.3) | 5 (31.2) | 0.25 |
| M1b | 13 (56.5) | 3 (60.0) | 11 (57.9) | 5 (55.6) | 5 (41.7) | 11 (68.8) | |||
| Treatment | |||||||||
| First-generation EGFR-TKIs | 12 (52.2) | 4 (80.0) | 0.36 | 11 (57.9) | 5 (55.6) | 1.00 | 9 (75.0) | 7 (43.8) | 0.14 |
| First-generation EGFR-TKIs + anti-angiogenic drugs | 11 (47.8) | 1 (20.0) | 8 (42.1) | 4 (44.4) | 3 (25.0) | 9 (56.2) | |||
Data were presented as n (%). ctDNA: Circulating tumor DNA; EGFR: Epidermal growth factor receptor; EGFRm: Epidermal growth factor receptor mutation; MC: Mutation clearance; MIC: Mutation incomplete clearance; TKIs: Tyrosine kinase inhibitors.
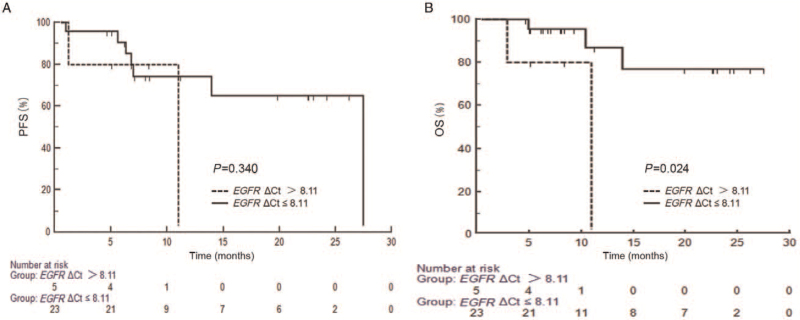
Firstly, all patients were divided into EGFRm-negative and EGFRm-positive groups by detecting baseline ctDNA EGFRm status in plasma samples using superARMS, and found that there was no significant difference in the median progression-free survival (mPFS) (22.2 vs. 27.5 months; log-rank P = 0.914) and median OS (mOS) (22.2 months vs. not reached; log-rank P = 0.683) between the two groups. Then the OS was compared between ΔCt value >8.11 group and ≤8.11 group. As shown in Figure 2, the patients with ΔCt ≤ 8.11 had better OS (not reached vs. 11.0 months; log-rank P = 0.024).
Figure 2.
Survival analysis (PFS [A] and OS [B]) of the groups with baseline ΔCt values of ctDNA EGFRm >8.11 and ≤8.11. ctDNA: Circulating tumor DNA; EGFR: Epidermal growth factor receptor; EGFRm: Epidermal growth factor receptor mutations; OS: Overall survival; PFS: Progression-free survival.
Changes of ctDNA EGFRm during treatment affect the outcomes of patients
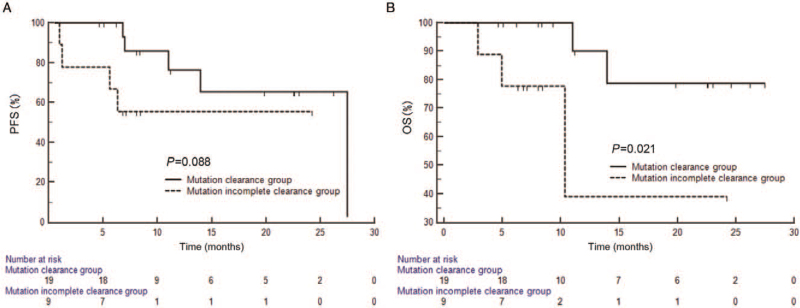
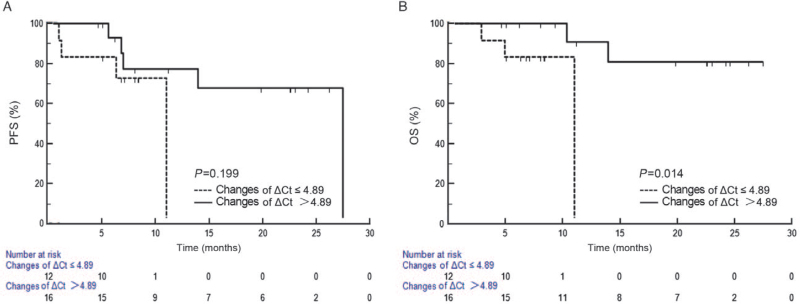
Next, we detected the changes of ctDNA EGFRm during treatment by qualitative and quantitative analysis with R-superARMS, and observed the effects of EGFRm changes on patients’ outcomes. According to the MC status after one month of treatment, 28 patients with positive EGFRm were divided into MC group (67.9%, 19/28) and MIC group (32.1%, 9/28). No significant difference was detected between the MC and MIC groups in clinical characteristics (all P>0.05) [Table 2]. The results showed that mOS between two groups (not reached vs. 10.4 months; log-rank P = 0.021) had significant difference. Although there was no statistical difference in PFS between the two groups, the two curves were separated and the PFS of MC group tended to be higher than the MIC group (not reached vs. 27.5 months; log-rank P = 0.088) [Figure 3]. Moreover, quantitative analysis of the data was performed to further clarify whether changes in the amount of mutation affect the outcomes of the patients. The cut-off value of the changes in ΔCt value of EGFRm after one month of treatment derived from the ROC curve was set at 4.89 (AUC = 0.737; Youden index = 0.600). There was no significant differences between the two groups in clinical characteristics (all P>0.05) [Table 2]. In the group with the change value of ΔCt >4.89, mOS was not reached, while in the group with the change value of ΔCt ≤4.89, mOS was 11.0 months, showing a significant difference (log-rank P = 0.014). However, there was no significant difference in mPFS (27.5 vs. 11.0 months; log-rank P = 0.199) [Figure 4].
Figure 3.
Survival analysis (PFS [A] and OS [B]) of mutation clearance group and mutation incomplete clearance group. OS: Overall survival; PFS: Progression-free survival.
Figure 4.
Survival analysis (PFS [A] and OS [B]) of the groups with changes in ΔCt values of ctDNA EGFRm >4.89 and ≤ 4.89. ctDNA: Circulating tumor DNA; EGFRm: Epidermal growth factor receptor mutations; OS: Overall survival; PFS: Progression-free survival.
In addition, we also analyzed the relationship between the change in the amount of mutations and the degree of response based on imaging in EGFRm-positive patients. It was found that patients in the MC group showed higher proportions of stable disease (SD) (4/12, 33.3%) and partial response (PR) (5/12, 41.7%) upon RECIST standard (about 2 months after treatment) than those of patients in the MIC group (SD: 1/5, 20.0%; PR: 1/5, 20.0%), with limited available data. This phenomenon was also seen when the group with changed value of ΔCt > 4.89 (SD: 3/12, 25.0%; PR: 6/12, 50.0%) was compared with the group with changed value of ΔCt ≤ 4.89 (SD: 1/5, 20.0%; PR: 1/5, 20.0%).
Discussion
In this study, we indirectly make use of ΔCt value generated during the detection process and transform superARMS from a qualitative into a semi-quantitative PCR detection method, and subsequently apply it to detect any change in ctDNA EGFRm. We have named it R-superARMS, and demonstrated its applicability for detecting EGFRm status in plasma as a simple, fast, and semi-quantitative PCR method that is claimed to be highly selective and sensitive. In our study, the concordance rate of EGFRm in tissue and matched plasma samples using superARMS was 68.3%, which was similar to previous research results.[19,22] However, given that there is a certain time interval between tissue collection and blood collection, our results may be biased. We also explored the effects of different baseline levels of EGFRm in plasma on the outcomes in patients receiving targeted drugs, and found that the baseline status of ctDNA EGFRm has an impact on the survival of all patients. Those with positive mutations detected in blood tests were more suitable for targeted therapy. Considering the effective rate of targeted therapy in patients with positive EGFRm is only 70%,[1–3] we supposed it is possibly related to the amount of initial EGFRm. We set the cut-off ΔCt value at 8.11 and found survival curves of the two groups were significantly different, which might partly explain why not all EGFRm-positive patients were responsive to TKIs. When the ΔCt value of baseline EGFRm is less than 8.11, the clinical outcome of patients receiving targeted therapy may be better, while for patients with positive EGFRm but ΔCt value greater than 8.11, we consider that the application of targeted therapy may be not effective. Therefore, quantitative detection methods for EGFRm may be a good strategy for screening and identifying patients who are more sensitive to targeted therapy in clinical practice. However, our study has a shortcoming that patients excluded from the study due to disease progression may receive other treatment regimens, which had an impact on the overall evaluation of OS. Therefore, the relationship between indicators, such as changes in ctDNA and OS should be interpreted with caution. More large-scale clinical trials are needed for further confirmation, while our study only provides an idea and direction for future prospective large-scale studies. Previously, Park et al[23] reported that the corrected delta cycle threshold value, which refers to EGFR quantification of small biopsy tissue or cytology specimens by peptide nucleic acid-mediated clamping PCR, can predict better objective response and clinical benefit rates of EGFR-TKIs therapy. This is also an important study of EGFRm quantitative detection method in identifying the efficacy of targeted therapy, but unlike our quantitative detection of liquid biopsy. This is because microdissecting the tiny tissue obtained by bronchoscopy or needle biopsy is not easy, while the study by Park et al[23] reflected the clinical limitations of tissue biopsy compared with liquid biopsy.
RECIST v1.1[24] is commonly used in clinical practice to evaluate the therapeutic efficacy in patients with solid tumor; however, it is well-known that imaging changes in tumor patients lag behind the occurrence of tumor molecular events.[13–15,18,25] The idea of using molecular response to evaluate the efficacy in patients is also reflected in the previously reported case of imatinib treatment in chronic myeloid leukemia,[26] which confirmed the rationale of our research. We detected ctDNA EGFRm by qualitative and quantitative methods using R-superARMS. In qualitative analysis, the TKI-treatment outcomes of patients in the MC group tended to be better than those in the MIC group, with OS between two groups being significantly different. Our study showed that ctDNA concentration of patients, who responded to treatment, decreased significantly since the initial treatment, with some decreasing to even undetectable levels. In the phase III AURA3 trial,[27] clearance of plasma EGFRm after 3/6 weeks of first-line treatment with TKIs was associated with a numerical improvement in PFS. Quantitative analysis of the data was performed using R-superARMS, and we found that when 4.89 was set as the cut-off of the changes in ΔCt value of EGFRm in EGFRm-positive patients. After 1-month treatment, PFS analysis showed that the results tended to be better in the group with changes in ΔCt value of EGFRm >4.89, suggesting that the changes of EGFRm may have a potential to predict disease progression. However, significant differences were not observed which may be due to the limited number of patients in this study. In the OS analysis, a statistically significant difference was found between the two groups, suggesting that quantitative detection of the changed value in ctDNA EGFRm before and after treatment may predict patient survival, although this may be affected by subsequent treatment. Using the above two analyses, we found the changes of mutations in plasma ctDNA, representing the degree of molecular responses, to be related to the degree of responses based on imaging. Patients in the MC group and in the group with changed value of ΔCt > 4.89 showed higher proportion of disease control (SD and PR) upon imaging assessment. Thus, compared to evaluation with imaging, the method with molecular responses has significant superiority in evaluating the efficacy and prognosis in patients. This approach may help identify the patients with better PFS or OS after treatment, providing insights for clinicians to tailor treatment strategies for specific patients.
Furthermore, conventional staging of malignant tumors follows the tumor-node-metastasis (TNM) notation system. Recently, routine analysis of ctDNA from liquid biopsy has been considered to possibly make cancer diagnosis, treatment, and prognosis more accurate for individual patients. Thus, researchers put forward the concept of TNMB (TNM staging by liquid biopsy [B]) tumor classification, opening a new horizon for precision medicine.[28] In our study, we demonstrated that the baseline status of ctDNA EGFRm has an impact on the survival of patients, confirming that it is valuable to propose the concept of TNMB staging for predicting clinical outcomes and prognosis. Additionally, we identified the clinical features of patients who could be more sensitive to drugs at an early stage, and based on the baseline ΔCt value, we could dynamically monitor the efficacy of TKIs. When changes in ΔCt value of ctDNA EGFRm did not reach cut-off level, we could recommend that clinicians change the medication or consider combination therapy. By using R-superARMS to detect the changes in EGFR mutations after 1 month treatment, we could predict the treatment response of patients. Then an NGS test can be further recommended for the patients with poor response, instead of performing an expensive and complex NGS testing at the time of initial diagnosis. At the same time, this method of measurement provides an opportunity to switch to NGS testing, rather than waiting for the disease progression. Based on the above considerations, ctDNA testing seems to provide us with a good and early detection method.
However, till date, we have rarely found a study like ours using quantitative methods to analyze EGFR changes. Although we obtained meaningful results, a limitation of this study was the small cohort of patients. Additionally, considering the effect of subsequent treatment after the progression of patients on the evaluation of OS, our study still has some shortcomings, and the relationship between changes in ctDNA and OS should be carefully interpreted. Nevertheless, our research as an initial exploration can provide some directions and ideas for larger, more rationally designed clinical studies in the future, including the effect of baseline levels and different changes of ctDNA EGFRm values on patient outcomes, the impact of different drug treatments on the degree of molecular response, and the function of different molecular response intensities in predicting patient outcomes. We have proposed a semi-quantitative EGFR blood testing with R-superARMS to identify the people who respond to treatment more accurately and sensitively. As a supplement to the RECIST standard, R-superARMS has offered a good option to address the yet unmet clinical needs, such as early molecular responses, early detection of micrometastasis, prediction of tumor treatment efficacy and prognosis, etc. We look forward to conducting a large randomized controlled trial in the future to confirm the findings of our present study.
In conclusion, R-superARMS is likely to be a simple, fast, economic, and semi-quantitative PCR method, that is, claimed to be highly selective and sensitive for detecting EGFRm levels in plasma. Detecting ctDNA EGFRm using this method can identify patients who are more likely sensitive to targeted therapy, reflect the molecular load of patients, and predict the therapeutic efficacy and clinical outcomes of patients.
Acknowledgements
The authors are grateful to Amoy Diagnostics Co., Ltd, Xiamen, China for providing technical support to the research.
Funding
This study was supported by the National Key Research & Development Program of China (No. 2016YFC1303800), the Innovation Project of Health and Technology in Jilin Province (No. 2017J064), the Special Project of Development and Reform Commission in Jilin Province (No. 2017C022), and the Key Laboratory Construction Project of Department of Science and Technology of Jilin Province (No. 20170622011JC).
Conflicts of interest
None.
Footnotes
How to cite this article: Liu XL, Bai RL, Chen X, Zhao YG, Wang X, Ma KW, Tian HM, Han FJ, Liu ZL, Yang L, Li W, Gai F, Cui JW. Correlation of circulating tumor DNA EGFR mutation levels with clinical outcomes in patients with advanced lung adenocarcinoma. Chin Med J 2021;134:2430–2437. doi: 10.1097/CM9.0000000000001760
Xiang-Liang Liu and Ri-Lan Bai contributed equally to the work.
References
- 1.Petrelli F, Borgonovo K, Cabiddu M, Barni S. Efficacy of EGFR tyrosine kinase inhibitors in patients with EGFR-mutated non-small-cell lung cancer: a meta-analysis of 13 randomized trials. Clin Lung Cancer 2012; 13:107–114. doi: 10.1016/j.cllc.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Minguet J, Smith KH, Bramlage P. Targeted therapies for treatment of non-small cell lung cancer -- recent advances and future perspectives. Int J Cancer 2016; 138:2549–2561. doi: 10.1002/ijc.29915. [DOI] [PubMed] [Google Scholar]
- 3.Ni J, Zhang L. Evaluation of three small molecular drugs for targeted therapy to treat nonsmall cell lung cancer. Chin Med J (Engl) 2016; 129:332–340. doi: 10.4103/0366-6999.174484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn 2018; 20:129–159. doi: 10.1016/j.jmoldx.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Turajlic S, Sottoriva A, Graham T, Swanton C. Resolving genetic heterogeneity in cancer. Nat Rev Genet 2019; 20:404–416. doi: 10.1038/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science 2013; 339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Errico A. Lung cancer: heterogeneity in space and time. Nat Rev Clin Oncol 2014; 11:684.doi: 10.1038/nrclinonc.2014.186. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Liu D, Li S, Zheng Y, Yang X, Li X, et al. Comparison of EGFR signaling pathway somatic DNA mutations derived from peripheral blood and corresponding tumor tissue of patients with advanced non-small-cell lung cancer using liquidchip technology. J Mol Diagn 2013; 15:819–826. doi: 10.1016/j.jmoldx.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Kawamura T, Kenmotsu H, Taira T, Omori S, Nakashima K, Wakuda K, et al. Rebiopsy for patients with non-small-cell lung cancer after epidermal growth factor receptor-tyrosine kinase inhibitor failure. Cancer Sci 2016; 107:1001–1005. doi: 10.1111/cas.12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nurwidya F, Zaini J, Putra AC, Andarini S, Hudoyo A, Syahruddin E, et al. Circulating tumor cell and cell-free circulating tumor DNA in lung cancer. Chonnam Med J 2016; 52:151–158. doi: 10.4068/cmj.2016.52.3.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim M, Kim CJ, Sunkara V, Kim MH, Cho YK. Liquid biopsy in lung cancer: clinical applications of circulating biomarkers (CTCs and ctDNA). Micromachines (Basel) 2018; 9:100.doi: 10.3390/mi9030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coco S, Alama A, Vanni I, Fontana V, Genova C, Dal Bello MG, et al. Circulating cell-free DNA and circulating tumor cells as prognostic and predictive biomarkers in advanced non-small cell lung cancer patients treated with first-line chemotherapy. Int J Mol Sci 2017; 18:1035.doi: 10.3390/ijms18051035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res 2015; 21:3196–3203. doi: 10.1158/1078-0432.CCR-14-2594. [DOI] [PubMed] [Google Scholar]
- 14.Sorber L, Zwaenepoel K, Deschoolmeester V, Roeyen G, Lardon F, Rolfo C, et al. A Comparison of cell-free DNA isolation kits: isolation and quantification of cell-free DNA in plasma. J Mol Diagn 2017; 19:162–168. doi: 10.1016/j.jmoldx.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 15.De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci 2019; 40:172–186. doi: 10.1016/j.tips.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Qian X, Liu J, Sun Y, Wang M, Lei H, Luo G, et al. Circulating cell-free DNA has a high degree of specificity to detect exon 19 deletions and the single-point substitution mutation L858R in non-small cell lung cancer. Oncotarget 2016; 7:29154–29165. doi: 10.18632/oncotarget.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma M, Shi C, Qian J, Teng J, Zhong H, Han B. Comparison of plasma and tissue samples in epidermal growth factor receptor mutation by ARMS in advanced non-small cell lung cancer. Gene 2016; 591:58–64. doi: 10.1016/j.gene.2016.06.053. [DOI] [PubMed] [Google Scholar]
- 18.Bennett CW, Berchem G, Kim YJ, El-Khoury V. Cell-free DNA and next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget 2016; 7:71013–71035. doi: 10.18632/oncotarget.11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y, et al. Highly sensitive droplet digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. J Mol Diagn 2015; 17:265–272. doi: 10.1016/j.jmoldx.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Xu H, Su S, Ye J, Chen J, Jin X, et al. Clinical validation of a highly sensitive assay to detect EGFR mutations in plasma cell-free DNA from patients with advanced lung adenocarcinoma. PLoS One 2017; 12:e0183331.doi: 10.1371/journal.pone.0183331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui S, Ye L, Wang H, Chu T, Zhao Y, Gu A, et al. Use of SuperARMS EGFR mutation detection kit to detect EGFR in plasma cell-free DNA of patients with lung adenocarcinoma. Clin Lung Cancer 2018; 19:e313–e322. doi: 10.1016/j.cllc.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Lu Y, Zhu G, Lei Y, Zheng L, Qin H, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol 2013; 66:1065–1069. doi: 10.1136/jclinpath-20∗13-201728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park HY, Oh HJ, Kim KH, Kim TO, Park CK, Shin HJ, et al. Quantification of epidermal growth factor receptor (EGFR) mutation may be a predictor of EGFR-tyrosine kinase inhibitor treatment response. Thorac Cancer 2016; 7:639–647. doi: 10.1111/1759-7714.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 25.Pérez-Barrios C, Nieto-Alcolado I, Torrente M, Jiménez-Sánchez C, Calvo V, Gutierrez-Sanz L, et al. Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: impact on biomarker testing. Transl Lung Cancer Res 2016; 5:665–672. doi: 10.21037/tlcr.2016.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breccia M, Molica M, Colafigli G, Massaro F, Alimena G. Early molecular response in chronic myeloid leukemia and halving time: latest evidences. Leuk Res 2016; 48:20–25. doi: 10.1016/j.leukres.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Shepherd FA, Papadimitrakopoulou V, Mok T, Wu YL, Han JY, Ahn MJ, et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib in the AURA3 trial. J Clin Oncol 2018; 36: (15_suppl): 9027.doi: 10.1200/JCO.2018.36.15_suppl.9027. [Google Scholar]
- 28.Yang M, Forbes ME, Bitting RL, O’Neill SS, Chou PC, Topaloglu U, et al. Incorporating blood-based liquid biopsy information into cancer staging: time for a TNMB system. Ann Oncol 2018; 29:311–323. doi: 10.1093/annonc/mdx766. [DOI] [PMC free article] [PubMed] [Google Scholar]