Abstract
The retinoblastoma protein (Rb) regulates both the cell cycle and tissue-specific transcription, by modulating the activity of factors that associate with its A-B and C pockets. In skeletal muscle, Rb has been reported to regulate irreversible cell cycle exit and muscle-specific transcription. To identify factors interacting with Rb in muscle cells, we utilized the yeast two-hybrid system, using the A-B and C pockets of Rb as bait. A novel protein we have designated E1A-like inhibitor of differentiation 1 (EID-1), was the predominant Rb-binding clone isolated. It is preferentially expressed in adult cardiac and skeletal muscle and encodes a 187-amino-acid protein, with a classic Rb-binding motif (LXCXE) in its C terminus. Overexpression of EID-1 in skeletal muscle inhibited tissue-specific transcription. Repression of skeletal muscle-restricted genes was mediated by a block to transactivation by MyoD independent of G1 exit and, surprisingly, was potentiated by a mutation that prevents EID-1 binding to Rb. Inhibition of MyoD may be explained by EID-1's ability to bind and inhibit p300's histone acetylase activity, an essential MyoD coactivator. Thus, EID-1 binds both Rb and p300 and is a novel repressor of MyoD function.
Terminal differentiation is a process whereby highly specialized cells undergo irreversible growth arrest and upregulate a panel of cell type-specific genes that are required for normal cellular function. The retinoblastoma gene product, Rb, has been implicated in mediating both the permanent cell cycle arrest and upregulation of tissue-specific genes associated with terminal differentiation in a wide variety of tissues (36, 38, 67, 74). Studies in skeletal muscle have determined that MyoD and other members of the basic helix-loop-helix family of myogenic transcription factors can induce both components of terminal differentiation, namely, myogenic gene transcription and cell cycle exit (44, 57). However, this ability appears to be dependent on the presence of Rb. Rb-deficient myotubes dedifferentiate and reenter the cell cycle in response to mitogens, despite the presence of the other pocket protein family members, p107 and p130 (57). Moreover, MyoD-transfected Rb−/− mouse embryonic fibroblasts fail to express late markers of myogenic differentiation and accumulate in the S and G2 phases of the cell cycle, implying that full MyoD transcriptional activity requires the coexpression of Rb (19, 44). The mechanism underlying this selective requirement for Rb is controversial; however, Rb dependence is also seen with C/EBP-mediated gene transcription in adipose differentiation (9). Interestingly, it has been proposed that Rb's ability to confer these two functions, namely, cell cycle arrest and differentiation, are mediated by separate domains within the B pocket, suggesting that Rb must interact with multiple cellular partners for full activity (58). Developmental studies that have correlated upregulation of Rb in myocardium in the neonatal period with growth arrest in ventricular myocytes support the premise that Rb may be the key to terminal differentiation in myocardium as well (16, 67).
Adenovirus E1A protein has been used in both skeletal (69) and cardiac (4, 32) myocytes to block pocket protein function and determine their importance in irreversible cell cycle exit and the control of muscle-specific gene expression (39). Cell cycle reactivation and a block to tissue-specific transcription were produced in both forms of striated muscle by E1A mutants that selectively block pocket protein function without binding the transcriptional coactivator, p300, a second pathway for dedifferentiation that is also targeted by E1A (15, 32, 49). The mechanism for dedifferentiation induced by E1A binding to pocket proteins is unknown but theoretically could be secondary to disruption of pocket protein family members' interaction with muscle-specific factors. Therefore, a requirement for Rb seems to be a general property in the terminal differentiation of multiple tissues, particularly striated muscle. To begin to understand the mechanism of this dependence, we sought to identify Rb-binding proteins in muscle tissue, utilizing the yeast two-hybrid system. Here, we describe the characterization of one clone isolated from a human cardiac tissue cDNA library, which we have called designated E1A-like inhibitor of differentiation 1 (EID-1). Like the adenoviral protein E1A, EID-1 binds the A-B pocket of Rb, impairs transactivation by the muscle determination protein, MyoD, and binds to p300, inhibiting its histone acetyltransferase (HAT) activity.
MATERIALS AND METHODS
Plasmids.
The bait plasmid, pASΔRb, was generated by ligating an EcoRI-MunI fragment (amino acids [aa] 379 to 835) of Rb into the EcoRI site of pAS2 (Clontech). pASRb2, pAS(H209), GST-Rb constructs, GAL4-SRF (aa 266 to 502), GAL4-MEF2C (aa 175 to 465), EMSV-MyoD, 4RTk-Luc, SkA-Luc, CaA-Luc, and CMV-LacZ constructs have been described previously (12, 40, 77). MCK-Luc was created by subcloning the 1,800-bp BglII-HindIII fragment of the MCK gene into pGL3 Basic (65). The p300 expression vectors and GST fusions have been previously reported (14, 27). CMV–EID-1 and CMV–EID-1(C180G) were constructed by cloning an EcoRI-AvaII fragment corresponding to the full-length coding sequences into pCDNA3 (Invitrogen). FLAG fusion proteins were constructed by ligating EID-1 (aa 3 to 187) or mutations in frame into CMV-FLAG2 (IBI Kodak). The GAL4-binding domain (GAL4-BD)–EID-1 vectors were constructed by cloning the EcoRI-XbaI fragments from the corresponding FLAG fusion constructs into p424, the GAL4-BD mammalian expression vector (53).
The point and deletion mutations in EID-1 were performed using site-directed mutagenesis as described by Deng and Nickoloff (11). Briefly, the Transformer Site-Directed Elimination Mutagenesis kit (Clontech) was used according to the manufacturer's instructions, with a synthetic oligonucleotide corresponding to the indicated mutation. The oligonucleotides used were as follows: EID-1(C180G), 5′-GAAGAACTCGGCGGTGATGAGATTATTG-3′; EID-1dl53–61, 5′-GGGGCCCAACAGCTCGGCCCAGCCAATGGCG-3′; and EID-1dl62–91, 5′-ATGGAGGAGGAGGAGGACTTCGAGAGCGAG-3′. Mutation EID-1dl92–115, kindly provided by W. Kaelin, is described in the accompanying manuscript (42). Combinatorial mutations were constructed by fusing the indicated fragments using convenient restriction sites. Rb–GAL4-BD fusion mutants in pAS2 have been previously described (12). All mutations were confirmed by DNA sequencing.
Interaction cloning.
Saccharomyces cerevisiae strain Y190 was transformed to Trp prototrophy with ASΔRb using lithium acetate (17). A single colony was grown in synthetic complete-Trp medium and transformed with human heart libraries cloned into pGAD10 or pACT2 (Clontech). The transformation mixture was then plated on media lacking tryptophan, leucine, and histidine but including 25 mM 3-amino-1,2,4-triazole (Sigma) and incubated for 5 to 7 days at 30°C. HIS+ colonies were then screened for β-galactosidase (β-gal) activity using the filter lift assay. Total DNA from true positive clones was isolated according to the method of Hoffman and Winston (26) and used to transform KC8 cells (Clontech) via electroporation with a Bio-Rad GenePulser according to the manufacturer's specifications. Transformants were plated on minimal media lacking leucine and containing ampicillin. Clones were sequenced by automated fluorescent dideoxy sequencing.
Quantification of LacZ activity in yeast.
S. cerevisiae cultures of 2 ml each were grown in the appropriate selective media to an optical density at 600 nm of ∼1.0. Yeast lysates were prepared according to the protocol of Guarente (20). Quantification of β-gal activity was achieved either with chlorophenyl-red-β-d-galactopyranoside (CPRG) (Boehringer Mannheim) and determining the amount of liberated chlorophenyl red at an optical density of 574 nm or with the luminescent LacZ substrate from Clontech.
Transfection assays.
The U2OS, C2C12, and CV-1 cell lines were obtained from the American Tissue Culture Collection and propagated in Dulbecco's modified Eagle's medium plus 10% fetal calf serum (Hyclone Laboratories). L17 cells were a gift from J. Massague. Human primary skeletal myocytes were purchased from Clonetics and cultured according to the manufacturer's specifications. All transfections were carried out using Lipofectamine (Life Technologies, Inc.) according to the manufacturer's specifications. Forty-eight hours after transfections, cell lysates were collected and analyzed for luciferase activity normalized to that of an internal CMV-LacZ control as previously described (40). For the experiments involving p300, luciferase activity was corrected for total protein as previously described (47). All experiments were repeated at least three times with two independent batches of plasmids. Results (mean ± standard error) were compared by analysis of variance and Fisher's PLSD tests, using a significance at a P value of <0.05.
Construction of adenoviruses.
Recombinant adenoviruses were constructed and propagated, and the titers of the viruses were determined as previously described by Graham and Prevec (18). Briefly, pJM17, constituting the adenovirus genome, was cotransfected with the pE1Asp1 shuttle vector containing the full-length EID-1 cDNA into 293 cells with Lipofectamine (GIBCO/BRL). Through homologous recombination, the EID-1 cDNA was integrated into the adenovirus genome. Viruses were propagated on 293 cells and purified using CsCl2 banding followed by dialysis against phosphate-buffered saline (PBS) and 10% glycerol. Titers were determined with 293 cells overlaid with Dulbecco's modified Eagle's medium plus 5% equine serum and 0.5% agarose. The AdLacZ virus was a gift from Frank Graham.
In vitro binding assays.
Expression of FLAG and glutathione S-transferase (GST) fusion proteins was induced in BL21 cells with 0.2 mM isopropylthio-β-galactoside (IPTG). After 4 h, the cells were pelleted and resuspended in PBS with 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 1 μg of aprotonin per ml, and 1 μg of antipain per ml. Cells were lysed by sonication and protein solubilized with 1% Triton X-100 for 30 min at 4°C. Cellular debris was removed by centrifugation. Proteins were purified from the supernatant with glutathione-coated beads or α-FLAG–agarose.
The in vitro binding assays were performed by adding 5 to 10 μl of [35S]methionine (Amersham) labeled, in vitro-transcribed and -translated EID-1 or mutations (Promega) to 1 μg of GST or GST fusion proteins in lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EGTA, 1 mM NaF, 0.25% Na deoxycholate, 0.1% NP-40). Mixtures were incubated for 1 h at room temperature and washed extensively with lysis buffer. Bound complexes were resolved on a sodium dodecyl sulfate–12% polyacrylamide gel (PAGE). The gels were dried, and autoradiography was performed overnight.
Antibodies and protein identification.
Immunoprecipitations were performed on total protein extracts prepared from L17 or U2OS cells in LS buffer (PBS plus 0.1% NP-40) plus protease inhibitors. The appropriate antibody was added, and the mixture was incubated for 11 h at 4°C. The bound proteins were collected for an additional 2 h with protein A/G-agarose and then washed four times with lysis buffer. The bound complexes were resolved by PAGE and immunoblotting was performed as previously described (7). Rb, recombinant proteins, and markers of muscle differentiation were detected with antibodies to Rb (Pharmingen), FLAG peptide (Kodak IBI), sarcomeric actin (Sigma), β-tubulin (Sigma), MyoD, Myf-5, myogenin, and p21 (Santa Cruz). Horseradish peroxidase-linked secondary antibodies were purchased from Amersham.
Rabbits were immunized subcutaneously with a synthetic peptide corresponding to the 15 C-terminal amino acids of EID-1, RLTEELGCDEIIDRE, since this was the most highly conserved domain across species, or ELYEESSDLQMDVMPGEG, resulting in antibodies GN735 and GN431, respectively. Immune serum was partially purified by affinity chromatography with the corresponding peptide and used for Western blot analysis. Western blotting was performed on total protein extracts from C2C12 cells differentiated for the indicated time points, according to established protocols (1).
Northern blot analysis.
A multiple-sample Northern blot prepared from human tissue (Clontech) was probed using a 32P-labeled 350-bp probe spanning the N-terminal coding sequences of human EID-1, by standard procedures (54). For Northern blots involving adenovirus, C2C12 cells were infected with 50 PFU of virus per cell and allowed to differentiate for 72 h. Total RNA was isolated and fractionated on a 1% denaturing agarose gel. The α-cardiac actin and α-skeletal actin probes used to monitor muscle-specific gene expression and the glyceraldehyde-3-phosphate dehydrogenase probe used as a constitutive control have been previously described (48).
Flow cytometry.
EID-1 (20 μg) was transfected into CV-1 cells along with CMV-CD20 (5 μg), a lymphocyte cell surface marker, at a ratio of 4:1 using Lipofectamine. Forty-eight hours after transfection, cells were stained with a fluorescein isothiocyanate (FITC)-labeled anti-CD20 antibody (Becton Dickinson) and analyzed by two-color flow cytometry. DNA content in the successfully transfected cells was quantified with propidium iodide as previously described (24). Parallel transfections were performed with CMV-E2F-1 and CMV-p21.
p300 HAT assays.
p300 assays were performed according to established protocols (23). Briefly, p300 complexes were immunoprecipitated from U2OS lysates harvested in lysate buffer plus 10 mM Na butyrate. p300 was incubated with recombinant protein in the presence of purified histones and [14C]acetyl coenzyme A at 30°C for 30 min. Acetylated proteins were resolved on an SDS–15% PAGE gel. Gels were fixed and incubated with Amplify (Amersham). Gels were dried, and autoradiography was performed. In vivo p300 acetylation assays were performed as described (23) using 3H-Na acetate on cardiac myocytes infected with 50 PFU of the indicated virus per cell.
RESULTS
Interaction cloning of a novel, muscle-restricted, Rb-binding protein.
pASΔRb, a yeast expression plasmid encoding aa 379 to 835 of Rb spanning the A-B and C pockets fused in frame to the GAL4 DNA-binding domain, was introduced into Y190. A single transformant was isolated and cotransformed with a library of cDNAs derived from human cardiac tissue and fused to the GAL4 activation domain (GAL4-AD). Approximately six million transformants were screened, and DNA from the initial HIS+ and LacZ-positive clones was purified for further analysis. Reintroducing the isolated GAL4-AD chimeric proteins into yeast along with pASΔRb or irrelevant GAL4-BD fusions revealed that 22 clones reproducibly and specifically interacted with Rb. These clones encoded nine unique proteins. Thirteen of the 22 clones encoded four overlapping cDNAs for the same protein, called EID-1, which was chosen for further study.
A consensus Kozak sequence was identified in frame with the GAL4-AD, revealing an open reading frame predicted to encode a novel 187-aa protein (Fig. 1A). Although the longest cDNA cloned, which was 1.3 kb, was expected to contain the complete open reading frame, analysis of mRNA suggested a transcript of 1.7 kb. Therefore, we screened conventional phage-based libraries and identified additional 3′ untranslated sequences but no further coding sequences, giving a complete cDNA of 1,638 nucleotides, which correlates well with the observed mRNA. A search of GenBank using BLAST did not reveal homology to any known proteins, although multiple homologous clones were identified from the GenBank library of expressed sequence tags and used to confirm the sequence. A search for potential functional motifs using the BLAST Enhanced Alignment Utility identified no known structural motifs. Analysis of the Human Gene Map maintained by the National Center for Biotechnology Information revealed the corresponding human expressed sequence tag has been mapped to chromosome 15q25.
FIG. 1.
Sequence analysis of EID-1. (A) The predicted amino acid sequence for EID-1. The LXCXE motif (shaded) and acidic domains (underlined) are indicated. (B) Comparison of the aligned amino acid sequence of the putative Rb-binding domain of EID-1 with the known Rb-binding sites of BRG1, Elf-1, SV40 large T Ag, adenovirus E1A, and HPV-16 E7 protein is shown. The conserved sequences in the consensus EnLXCXE motif are shaded.
Sequence analysis and homology searches did not reveal significant similarity to any known proteins, as mentioned. However, within the C terminus there was a consensus binding site for Rb (EXnLXCXE), which also is found in the Rb-binding domains of simian virus 40 (SV40) large T antigen (Ag) (34), E1A (13), human papillomavirus (28), and many cellular Rb-binding proteins, such as BRG1, hBRM, Elf-1, and the histone deacetylase, HDAC1 (68). Shown in Fig. 1B is the amino acid sequence of this motif compared with those of other Rb-binding proteins. The LXCXE motif of EID-1 and its surrounding sequences was 100% conserved across human, mouse, and rat species (data not shown), suggesting that this evolutionarily conserved domain is critical for normal function. The exact LGCDE motif seen in EID-1 was previously isolated from a library of random peptides as an Rb-binding peptide (75). This synthetic peptide sequence represented one of only seven peptides capable of interacting with Rb that were isolated from over three million transformants screened and 400 possible LXCXE combinations. This corroborates the specificity of the LXCXE-Rb interaction, in concurrence with the observation that only a small subset of all known proteins with LXCXE motifs has been demonstrated to interact with Rb (5).
EID-1 expression is enriched in muscle tissues.
To investigate the tissue distribution of EID-1, we probed a blot of poly(A)+ RNA prepared from multiple samples of adult human tissue. A single transcript was observed with an apparent size of 1.7 kb (Fig. 2A). As demonstrated, the highest levels of EID-1 were found in cardiac tissue. Significant expression was also seen in skeletal muscle and, to a lesser extent, in brain tissue. Low levels of expression were observed in most tissues; however, levels of expression in cardiac tissue are at least 10-fold higher than those seen in lung or liver. Preferential expression of EID-1 in adult striated muscle and brain likewise was seen in the mouse (not shown).
FIG. 2.
Expression analysis of EID-1. (A) Blots containing poly(A)+ RNA from multiple samples of adult human tissue were hybridized with the radiolabeled 350-bp human EID-1 cDNA. A single 1.7-kb transcript was detected. High-level expression was detected in cardiac and skeletal muscle tissue. (B) Total RNA obtained from murine ventricles at the indicated developmental time points or adult murine atrial tissue was probed with a corresponding murine EID-1 probe. (C) Protein extracts from primary human skeletal myotubes, differentiated for the indicated times, were probed with polyclonal anti-EID-1 antibody (GN735) and antibodies to the indicated proteins.
Since expression of EID-1 was highest in cardiac tissue, we sought to determine the developmental expression and chamber specificity of EID-1 message. We prepared total RNA from murine ventricular tissue or from adult atrial tissue at the indicated time points (Fig. 2B). EID-1 mRNA abundance decreased with development in ventricular tissue while it remained highly expressed in adult atrial tissue, explaining the high levels of expression seen in the Northern blots of human tissue. To determine the expression of EID-1 during skeletal-muscle differentiation, protein extracts were prepared from primary cultures of human skeletal myocytes cultured under differentiating conditions (Fig. 2C). Using our affinity-purified polyclonal antibody generated against the highly conserved C terminus of EID-1, a specific protein product of 35 kDa was detected in differentiated human skeletal myocytes (Fig. 2C). This is consistent with the size of EID-1 determined by in vitro transcription and translation and overexpression of the cDNA in cultured cells (Fig. 3B; see below). Levels of EID-1 protein decreased during myogenic differentiation in contrast to that of muscle-specific α-sarcomeric actin. Equal levels of protein loading were confirmed by the uniform expression of α-tubulin. Therefore, EID-1 encodes a novel, developmentally regulated, muscle-enriched protein.
FIG. 3.
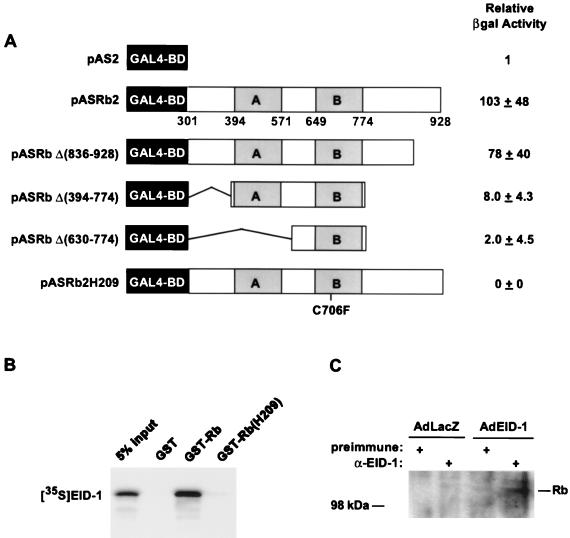
EID-1 interacts with Rb. (A) Yeast strain Y187 was transformed with the EID-1–AD fusion protein and either the GAL4-BD alone (pAS2) or indicated GAL4-Rb mutations. CPRG quantification of β-Gal activity was performed on six independent clones for each transformation. (B) In vitro-transcribed and -translated [35S]methionine-labeled EID-1 was incubated with 1 μg of either GST, GST-Rb, or GST-Rb(H209). Complexes were precipitated with glutathione-Sepharose beads and resolved by SDS-PAGE. (C) U2OS cells were infected with the indicated virus and immunoprecipitated with either preimmune or anti-EID-1 antibody (GN431). Immunoprecipitates were probed with anti-Rb antibody.
EID-1 interacts with Rb.
To confirm and further characterize the domains in Rb necessary for interaction with EID-1, more-extensive Rb deletional mutants were created. Deletion mutations were created either by using convenient restriction sites or by PCR-based mutagenesis. The respective constructs were subcloned into the yeast GAL4-BD expression vector, pAS2, for use in the modified yeast two-hybrid assay. Strength of interaction with EID-1 was quantified by determining the LacZ activity in liquid cultures of plasmid-transformed yeast (Fig. 3A). Results are expressed relative to activity in the GAL4-BD alone. Although EID-1 interacts very weakly with the minimal A-B pocket, consistent with the observation that this fragment of Rb mediates interactions with LXCXE-containing peptides such as E1A and SV40 large T Ag, high-affinity binding requires additional sequences within the C pocket. In contrast, EID-1 did not interact either with the B pocket alone or with full-length Rb containing a single amino acid substitution at codon 706 (Cys to Phe) (ASH209) within the B pocket, which is known to disrupt the binding of SV40 large T Ag (Fig. 3A). This corroborates the specificity of the EID-1-Rb interaction and is consistent with the observation that EID-1 demonstrated no interaction with any of the additional proteins tested (laminin C and p53). As an additional test of the interaction between EID-1 and Rb, in vitro binding assays were performed. In vitro-transcribed and -translated, 35S-labeled EID-1 was incubated with the indicated partially purified bacterially expressed GST fusion proteins. Consistent with the result of the two-hybrid assays, EID-1 bound to wild-type GST-Rb but was unable to bind a GST fusion protein containing the pocket protein mutant of Rb described above (C706F) (Fig. 3B). To confirm that EID-1 interacts with Rb in vivo, we infected U2OS osteosarcoma cells with adenovirus overexpressing EID-1 or an irrelevant protein, LacZ (Fig. 3C). Extracts were immunoprecipitated with either preimmune or anti-EID-1 antibody and blotted for Rb. As shown, only in cells infected with EID-1 was endogenous Rb coprecipitated.
EID-1 associates with Rb via its LXCXE motif, in vitro and in vivo.
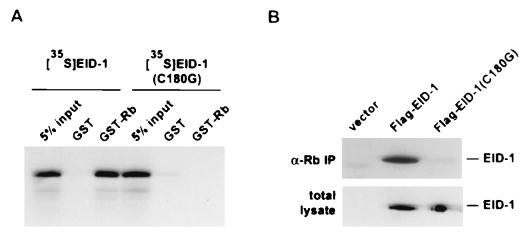
During the original library screening, one cDNA was isolated that corresponded to an N-terminal deletion of EID-1 encoding only the most C-terminal 80 aa. This suggested that the consensus LXCXE Rb-binding motif located in this C-terminal fragment was a likely candidate to mediate the Rb–EID-1 interaction. To confirm this, we created a single amino acid substitution by site-directed mutagenesis of EID-1, EID-1(C180G), which was predicted to disrupt Rb binding. This conversion of LXCXE to LXGXE has been shown to render E1A incapable of binding Rb (64).
In vitro binding assays were performed with in vitro-transcribed and -translated wild-type EID-1 or EID-1(C180G). [35S]methionine-labeled proteins were incubated with bacterially produced GST or GST-Rb, and complexes were resolved on polyacrylamide gels. As shown in Fig. 4A, a single amino acid substitution in the LXCXE motif abrogated the ability of EID-1 to interact with Rb. To confirm the specificity for interaction between EID-1 and Rb in vivo, we tested the binding of endogenous Rb to wild-type EID-1 versus the C180G mutation in transfected mammalian cells. Figure 4B shows the results of immunoprecipitation using anti-Rb antibody on total protein extracts from transiently transfected L17 cells, expressing FLAG fusion proteins of either EID-1 or EID-1(C180G). Immunoprecipitated complexes were resolved by PAGE, and immunoblots were probed with an anti-FLAG antibody. In a separate set of cultures, total extracts were probed with anti-FLAG antibody to confirm that the wild-type and mutant EID-1 proteins were expressed at comparable levels. Similar to the results seen in vitro, only wild-type EID-1 was able to associate with Rb. These data confirm the interaction of EID-1 and Rb by independent, complementary methods, implicating the LXCXE motif as the domain that mediates this association as anticipated.
FIG. 4.
EID-1 associates with Rb in mammalian cells. (A) In vitro-transcribed and -translated [35S]methionine-labeled EID-1 or EID-1(C180G) was incubated with 1 μg of either GST or GST-Rb. Complexes were precipitated with glutathione-Sepharose beads and resolved by SDS-PAGE. (B) L17 cells were transiently transfected with FLAG epitope-tagged EID-1 or EID-1(C180G). Cell lysates were immunoprecipitated with anti-Rb antibody, immunoprecipitates were resolved by SDS-PAGE and analyzed by immunoblotting with anti-FLAG antibody. Total cell lysates were immunoblotted with anti-FLAG antibody to ensure the equivalent expression of proteins.
EID-1 inhibits muscle-specific gene expression.
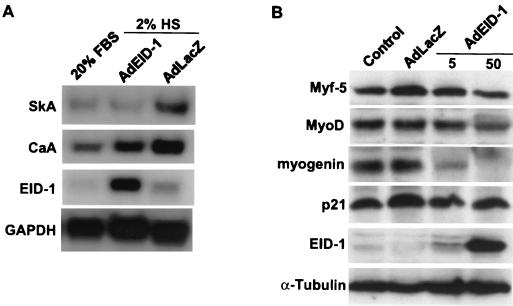
To determine the effects of EID-1 on endogenous skeletal muscle-specific gene expression, we used an adenovirus vector that overexpresses human EID-1 (AdEID-1) to uniformly infect C2C12 cells, a murine skeletal muscle cell line. Undifferentiated myoblasts were infected with AdEID-1 or an adenovirus vector expressing an irrelevant protein, AdLacZ, and allowed to differentiate in low-serum medium for 72 h. Northern blot analysis was performed on total RNA with the indicated probes, and RNA from undifferentiated C2C12 cells was included for comparison. Relative to levels seen in mitogenic serum (Fig. 4, lane 1), adenovirus delivery of exogenous human EID-1 blocked the increase of skeletal α-actin mRNA completely and the increase of cardiac α-actin mRNA by 51% (Fig. 5A). In this myogenic cell line, cardiac α-actin is an earlier marker of differentiation than skeletal α-actin (3), suggesting that EID-1 was disproportionately inhibiting later markers of differentiation. The human-specific EID-1 probe detected an appropriate increase in expression of human EID-1 in AdEID-1-infected cells. Levels of glyceraldehyde-3-phosphate dehydrogenase were similar in the three groups; thus, the effects of EID-1 were selective to the differentiation-specific genes tested.
FIG. 5.
EID-1 inhibits muscle-specific gene expression. (A) To determine the effects of EID-1 on endogenous skeletal muscle-specific gene expression, we used an adenovirus vector that overexpresses EID-1 (AdEID-1) to uniformly infect C2C12 cells, a murine skeletal muscle cell line. Cells were infected with 50 PFU of AdEID-1 or AdLacZ per cell and allowed to differentiate in low-serum medium (2% horse serum) for 72 h. Northern blot analysis was performed of total RNA with the indicated probes. RNA from undifferentiated C2C12 cells (in 20% fetal bovine serum) was included for comparison. (B) To determine the effects of EID-1 on MyoD family members, skeletal myoblasts were infected with 5 or 50 PFU of AdEID-1 or AdLacZ per cell and allowed to differentiate for 72 h. Protein extracts were probed with polyclonal anti-EID-1 antibody (GN735) and antibodies to the indicated proteins.
Skeletal-muscle differentiation is a highly controlled process involving the coordination of cell cycle exit and expression of tissue-specific genes that is regulated by the hierarchical expression of MyoD family members. To determine the effects of EID-1 on these muscle-determining factors, we infected skeletal myoblasts with differing amounts of EID-1 and characterized their expression (Fig. 5B). Levels of Myf-5, the functionally earliest member (52, 66), were unaffected by EID-1. MyoD expression was minimally affected and only with the highest doses of EID-1. In contrast, myogenin expression was inhibited by even the lowest doses of EID-1. This data suggest that although EID-1 inhibits skeletal muscle differentiation, the myogenic phenotype itself is not completely reversed, since both Myf-5 and MyoD remained expressed. Genetic studies with mice have confirmed the critical role of myogenin in skeletal muscle development, particularly late aspects of myogenesis (25, 71). Interestingly, levels of p21 were not affected by EID-1, which supports the lack of an effect of EID-1 on cell cycle progression (see Fig. 8).
FIG. 8.
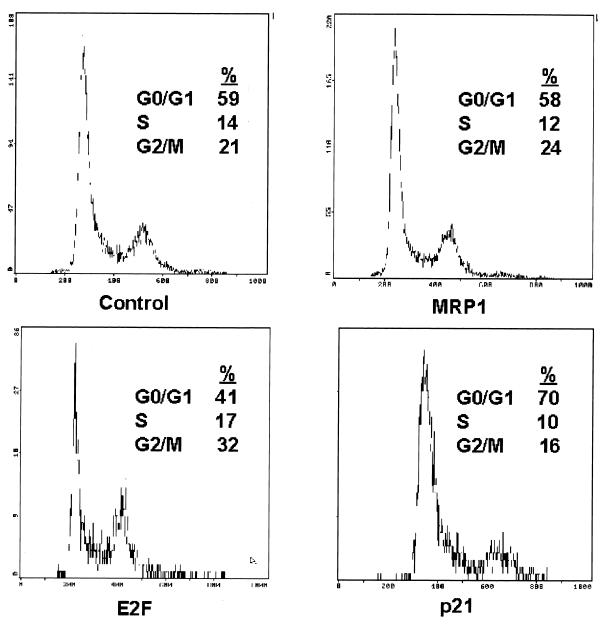
Overexpression of EID-1 does not cause cell cycle reentry. To determine the effects of EID-1 on cell cycle progression, vectors encoding EID-1, E2F-1, or p21 were transfected along with CMV-CD20, a lymphocyte cell surface marker, at a ratio of 4:1 into CV-1 cells, a highly transfectable clonal cell line. Transfected cells were identified with an FITC-labeled anti-CD20 antibody by flow cytometry, and DNA content was quantified with propidium iodide.
EID-1 inhibits MyoD-dependent transcription.
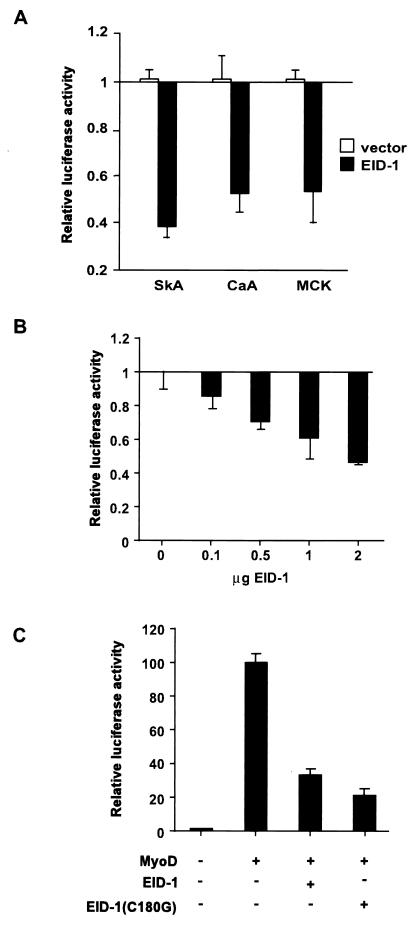
To confirm that EID-1 was directly inhibiting muscle-specific transcription, we tested its effect on a panel of skeletal muscle-specific promoters. As shown in Fig. 6A, with primary skeletal muscle cells transfected with EID-1 plus appropriate luciferase reporter genes, EID-1 inhibits the transcription of both the skeletal α-actin and the cardiac α-actin promoter. Similar inhibition was seen with an 1,800-bp fragment of the muscle creatine kinase (MCK) gene, including the MCK enhancer, which contains a canonical MyoD binding site (Fig. 6A).
FIG. 6.
EID-1 inhibits muscle-specific gene expression. (A) Primary cultures of skeletal myoblasts were transfected with SkA-Luc, CaA-Luc, or MCK-Luc along with 1 μg of vector or CMV–EID-1. The percent repression is compared to that of control cultures without CMV–EID-1. (B) U2OS cells were transfected with EMSV-MyoD, 4RTk-luc, CMV-LacZ, and increasing amounts of the CMV–EID-1 expression vector. Results are shown relative to vector-transfected cultures. (C) Cells were transfected as described along with 1 μg of vector, CMV–EID-1, or EID-1(C180G). The fold induction is compared to control cultures without EMSV-MyoD.
Since expression of skeletal muscle-specific genes is dependent on the activity of MyoD or alternative MyoD family members, we determined the effects of EID-1 on MyoD-dependent transcription in a heterologous background (77). MyoD, along with a luciferase reporter construct under the control of multiple MyoD binding sites, was transfected into U2OS cells, and the effect of increasing amounts of EID-1 was determined. As shown, EID-1 caused a dose-dependent reduction in MyoD transcriptional activity (Fig. 6B). MyoD activity has been reported to be dependent on the coexpression of Rb (44). Therefore, if MyoD transcriptional activity requires a direct interaction with Rb, one explanation for the observed effects of EID-1 might be that overexpression of any Rb-binding protein could conceivably displace MyoD from the pocket, resulting in reduced activity. To examine if EID-1's inhibition of MyoD-dependent transcription required an intact Rb-binding domain, EID-1(C180G) was likewise tested for its effect on MyoD-dependent transcription (Fig. 6C). MyoD produced an 80-fold induction of the E-box reporter, which was inhibited 65% by EID-1 (P < 0.0001) and 79% by the Rb-binding mutant of EID-1 (P < 0.0001). Surprisingly, the mutation that abolished Rb binding was a more potent inhibitor of MyoD-dependent transcription than EID-1 at all doses (P < 0.02). This strongly argues against the possibility that EID-1 effects on skeletal muscle-specific transcription are directly related to its ability to compete for Rb pocket binding. Conversely, it suggests that pocket proteins normally function to attenuate EID-1's inhibitory effects. These results raise the possibility that EID-1 could exerts its effect on MyoD-dependent transcription indirectly, by a mechanism distinct from interference with Rb, perhaps by blocking the interaction of MyoD with other factors that are critical for the full transcriptional activity of MyoD.
EID-1 inhibits MyoD-dependent transcription through multiple domains.
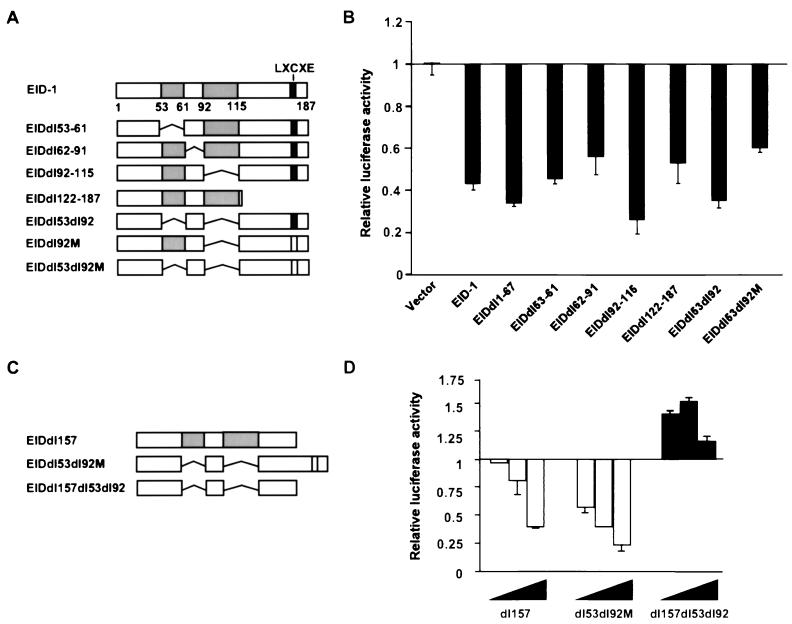
To determine the domain or domains within EID-1 responsible for its inhibitory effects on MyoD-dependent transcription, we created multiple-deletion mutations spanning the coding sequence of EID-1 (Fig. 7). As demonstrated, neither the C-terminal deletion (aa 122 to 187), the N-terminal deletion (aa 1 to 67), or the internal deletions singly or in combination (aa 53 to 61, 62 to 91, and 92 to 115) were able to completely block inhibition by EID-1 (Fig. 7A and B). However, deleting the acidic domains in concert with the C terminus of EID-1 alleviated inhibition of MyoD function (dl53dl92dl157; Fig. 7C and D) (42). In contrast, deleting the acidic domains in combination with the point mutation in the LXCXE motif was not sufficient to relieve this inhibitory effect. This implies that the C terminus interacts with factors or mediates effects distinct from its association with Rb. Equal expression of all constructs was confirmed by Western blotting (data not shown). These data suggest that multiple domains within EID-1 contribute to its inhibitory effects and that a block to MyoD function can be imposed by either of the two regions alternatively.
FIG. 7.
The C-terminal and acidic domains mediate inhibition of MyoD-dependent transcription by EID-1. To determine the domain or domains within EID-1 responsible for its inhibitory effects, we created multiple-deletion mutations spanning the coding sequences of EID-1. U2OS cells were transfected with 1 μg each of MyoD and 4Rtk-Luc and indicated mutations as described below. Luciferase activity, corrected for that of LacZ, is shown relative to expression in cells receiving MyoD in the absence of EID-1. Amount of EID-1 vectors, 1 μg (A and C) and 0.5, 1, or 2 μg (B and D).
Overexpression of EID-1 does not cause cell cycle reentry.
Many interventions that inhibit myogenic differentiation, such as mitogens, E1A, and SV40 large T Ag, concomitantly provoke cell cycle reentry. In skeletal muscle, substantial precedent has been established for the pivotal role played in these reciprocal events by developmentally regulated expression of cyclins, Cdks, and Cdk inhibitors, especially G1 cyclins (21, 51, 63), although differing domains within Rb are now thought to mediate growth control versus differentiation (58). Likewise, exogenous E2F-1 impairs both growth arrest and differentiation in skeletal and cardiac muscle cells (31, 72). Therefore, repression of MyoD activity could conceivably occur if overexpression of EID-1 resulted in cell cycle progression with a concomitant increase in G1 cyclin activity or with the release of E2F from the pocket. Therefore, to determine the effects of EID-1 on cell cycle progression, EID-1 was transfected along with CMV-CD20, a lymphocyte cell surface marker, at a ratio of 4:1 into CV-1 cells, a highly transfectable clonal cell line. Successfully transfected cells were identified with a FITC-labeled anti-CD20 antibody by flow cytometry, and DNA content was quantified using propidium iodide (24). Parallel transfections with E2F-1 and p21 were performed to demonstrate the fidelity of this technique, where E2F-1 and p21 increase or decrease the percentage of cells with DNA content greater than G0/G1, respectively. As illustrated by a representative experiment (Fig. 8), EID-1 had no effect on G1 exit whereas E2F-1 and p21 evoked the expected responses. Similar results were seen in independent studies, using adenovirus to express EID-1 in ventricular muscle cells (data not shown). Thus, forced expression of EID-1 had no discernable effects on cell cycle reentry in either cell background.
EID-1 preferentially inhibits p300-dependent transcription.
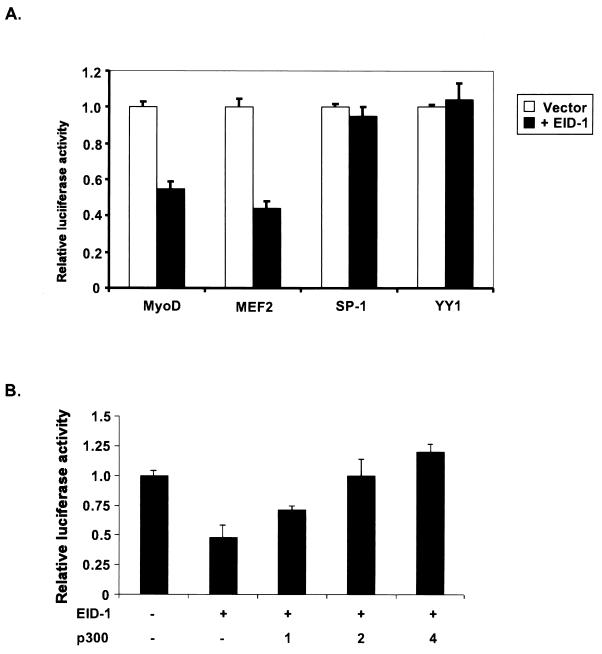
Muscle-specific transcription is dependent on the actions of both ubiquitous and cell type-specific transcription factors. Muscle-specific transcription factors such as MyoD (77) or MEF2 (43) require interaction with p300 for full transcriptional activity. Ubiquitously expressed factors, Sp-1 and YY1, have been implicated in regulating skeletal and cardiac muscle-specific transcription both positively (22) and negatively (37, 40), respectively. However, their transcriptional activity has been suggested to be p300 independent (2, 23). Therefore, selective inhibition of p300-dependent transcription would account for EID-1's preferential effect on tissue-restricted genes. To test EID-1's effect on both p300-dependent tissue-restricted factors such as MyoD or MEF2 or ubiquitously expressed p300-independent factors like Sp-1 or YY1, EID-1 was cotransfected along with GAL4-BD fusion proteins of these transcription factors into U2OS cells (Fig. 9A). EID-1 significantly inhibited MyoD or MEF2 transcriptional activity while activity of Sp-1 or YY1 was unaffected. To confirm that EID-1's inhibitory actions were p300 dependent, we cotransfected increasing amounts of p300 along with EID-1. Overexpression of p300 rescued EID-1's inhibitory effects (Fig. 9B). These data support the concept that EID-1's inhibitory effects are attributable to disrupting p300 function.
FIG. 9.
EID-1 preferentially inhibits p300-dependent transcription. (A) U2OS cells were transiently transfected with 1 μg of GAL4-BD fusions of the indicated transcription factors with or without CMV–EID-1. After 48 h, cell lysates were harvested and luciferase activity was assayed and normalized to total protein. Results are shown, relative to the levels of GAL4-BD fusion alone. (B) To determine if overexpression of p300 could rescue EID-1's inhibitory effects, U2OS cells were transiently transfected with EMSV-MyoD, 4RTk-luc, CMV-LacZ, and CMV–EID-1, along with increasing doses of CMV-p300. After 48 h, cell lysates were harvested and luciferase activity was assayed and normalized to LacZ activity.
EID-1 interacts with p300.
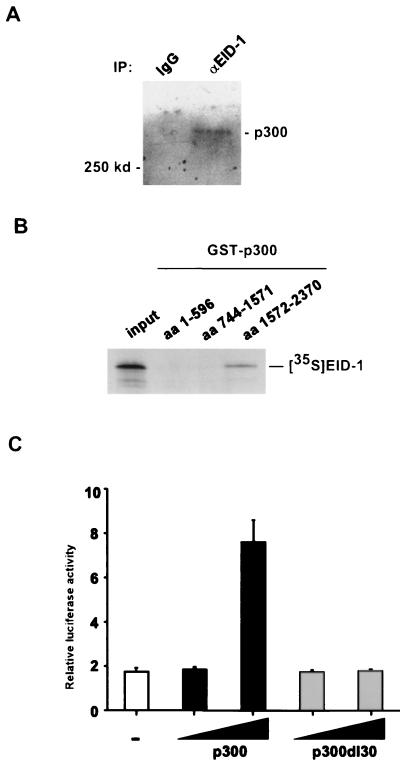
To test the ability of EID-1 to interact with p300 in vivo, we performed immunoprecipitations with lysates prepared from U2OS cells using an anti-EID-1 antibody (Fig. 10A). As shown, EID-1 immunoprecipitates specifically interacted with endogenous p300. To determine the domains in p300 that mediated this association, we performed in vitro binding assays utilizing [35S]methionine-labeled EID-1 and bacterially expressed GST fusion proteins spanning the entire p300 coding sequence. As shown in Fig. 10B, EID-1 interacts with the C-terminal fragment of p300 corresponding to aa 1572 to 2370. This fragment contains the C-H3 domain, which has previously been shown to mediate p300's interaction with E1A (14) and MyoD (15, 77). To confirm that EID-1 interacted with the C-H3 domain of p300 in vivo, U2OS cells were transfected with GAL4-BD–EID-1 along with wild-type p300 or an internal deletion mutant, p300dl30, removing residues 1738 to 1808, which are necessary for E1A and MyoD binding to the C-H3 domain (Fig. 10C). EID-1 was a weak transcriptional activator when fused to a heterologous DNA-binding domain. Wild-type p300 further potentiated EID-1-dependent transcription 7.6-fold. By contrast, the p300dl30 mutation, which retains full transcriptional activity (77), does not potentiate transcription via the GAL4-BD–EID-1 fusion protein, suggesting that the binding site for E1A, MyoD, and EID-1 all map to this C-terminal domain of p300. Adenovirus E1A, an inhibitor of muscle-specific gene expression, also transactivates transcription when fused to a heterologous DNA-binding domain, which is further potentiated by p300 (70). Superficially, this may seem paradoxical, since E1A is known to inhibit p300-dependent HAT activity (6). However, p300 is known to activate transcription by multiple mechanisms (33), which might account for the positive effect.
FIG. 10.
EID-1 interacts with p300. (A) Total protein lysates were prepared from U2OS cells, and EID-1-associated proteins were immunoprecipitated with α-EID-1 antibody. Complexes were resolved by SDS-PAGE and probed with α-p300 antibody. (B) In vitro-transcribed and -translated [35S]methionine-labeled EID-1 was incubated with 1 μg of either GST or the indicated GST-p300 fusion proteins. Complexes were precipitated with glutathione-Sepharose beads and resolved by SDS-PAGE. (C) U2OS cells were transiently transfected with either 0.5 μg of GAL4-BD or GAL4–EID-1, 2 or 4 μg of CMV-p300 or CMV-p300del30, and a luciferase reporter gene under the control of multiple GAL4-binding sites. After 48 h, cell lysates were harvested and luciferase activity was assayed and normalized to that of total protein. Results are shown relative to the amount of GAL4-BD alone.
EID-1 inhibits p300 HAT activity.
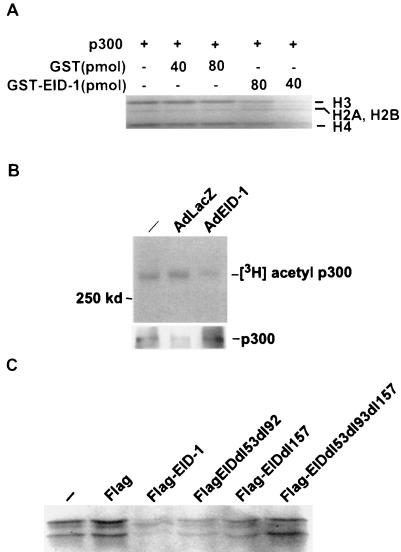
To address the theoretical possibility that EID-1, as a p300-binding protein, might merely interfere with MyoD by sequestering this essential cofactor under conditions of forced expression, we analyzed EID-1's effects on p300 function. Recently, several inhibitors of muscle-specific transcription including E1A that interact with p300 have been shown to inhibit p300-dependent HAT activity (6, 23). Therefore, to determine if EID-1 also represses p300 HAT activity we performed in vitro p300-dependent HAT assays in the presence of recombinant EID-1. As shown in Fig. 11A, GST–EID-1 was a potent inhibitor of p300-dependent HAT activity while GST alone had no effect. To confirm EID-1's ability to inhibit p300-dependent HAT activity, we ascertained its effect on p300's ability to autoacetylate itself in vivo with our recombinant adenovirus vectors. Cardiac myocytes were infected with AdEID-1 or AdLacZ, and the extent of p300-dependent autoacetylation was determined. As shown, EID-1 was a potent inhibitor of endogenous p300 HAT activity in vivo (Fig. 11B). To clarify the domains within EID-1 that mediated this effect, we utilized recombinant Flag-tagged EID-1 or indicated deletion mutants (Fig. 11C). Wild-type EID-1 or deletion of the acidic domains (dl53dl92) or the C-terminal deletion (dl157) retained the ability to inhibit p300 HAT activity. In contrast, the compound deletion mutation (dl53dl92dl157) that was incapable of inhibiting MyoD-dependent transcription had no effect on p300 HAT activity. These data suggest that EID-1's inhibitory properties are dependent on its ability to inhibit p300 HAT activity.
FIG. 11.
EID-1 inhibits p300 HAT activity. To determine if EID-1 inhibited HAT activity, immunoprecipitated p300 was incubated with purified histones and [14C]acetyl coenzyme A. Complexes were resolved by SDS-PAGE and subjected to autoradiography. (A) Purified GST or GST–EID-1 was added to the indicated reaction mixtures, and the effect on p300 HAT activity was determined. (B) Cardiac myocytes were infected with the indicated viruses. Thirty-six hours after infection, cells were exposed to 3H-Na acetate for 1 h. Total protein lysates were harvested in lysate buffer plus 10 mM Na butyrate, and p300 was immunoprecipitated. Shown are autoradiograms documenting p300 autoacetylation in vivo. One-tenth of the immunoprecipitated complexes was probed for total p300 expression to ensure that results were not related to EID-1 effects on total p300 levels. (C) Purified Flag peptide or Flag fusion proteins (80 pmol) were coincubated with the HAT assay mixture to determine the domains within EID-1 that mediate this inhibitory effect.
DISCUSSION
In the present study, we have attempted to identify the factors in striated muscle that interact with Rb. We have cloned a novel protein, EID-1, expressed in cardiac and skeletal muscle that specifically interacts with Rb through a LXCXE motif located in its C terminus. Developmental expression patterns and overexpression studies of EID-1 suggested that this molecule represented a novel inhibitor of differentiation. While in vitro binding studies and two-hybrid assays strongly support the notion that EID-1 interacts with Rb, we have been unable to directly show an interaction of the endogenous proteins. This may be simply related to technical limitations of our reagents or the low levels of expression of the two factors. Despite this, our data suggest, at least indirectly, that the interaction of EID-1 with Rb is functionally important. The point mutation in EID-1 that abolishes Rb binding was a more potent inhibitor of MyoD-dependent transcription. Therefore, our data support an alternative model for the interplay of Rb and MyoD during skeletal muscle cell differentiation that would be dependent on the expression of Rb but does not require a direct Rb-MyoD physical association. In support of this premise, data are presented in the accompanying manuscript by Miyake et al. that not only can Rb rescue EID-1's inhibitory effects but that overexpression of EID-1 can disrupt some aspects of Rb function (42). EID-1 joins a growing list of inhibitors of differentiation, including p202 and HBP1, that have been shown to interact with Rb (10, 35). The fact that several inhibitory factors have been identified which interact with Rb suggests that this protein may represent a differentiation checkpoint, linking factors regulating cell cycle exit and tissue-specific gene expression (60).
Interestingly, Rb has also recently been shown to be critical for MEF2-dependent transcriptional activity (45). This potentiation of MEF2 activity was in part independent of Rb's effects on cell cycle progression, but the basis of this cell cycle-independent effect was not determined. Since MEF2 transcriptional activity (55), like that of MyoD, has been reported to be p300 dependent, an indirect mechanism involving EID-1 might explain both the defect in skeletal muscle gene expression in the absence of pocket proteins (19, 44) and our and others' inability to confirm a physical interaction between Rb and MyoD in vivo (45). Additionally, it might explain the paradoxical observation that Rb can potentiate the transcriptional activity of certain factors (8, 30, 62) despite its inherent transcriptional repressor-like activity (59, 73). This link between Rb and p300 provides a cogent hypothesis to explain these discordant results.
The similarities between EID-1 and E1A are obvious; however, several differences are also apparent. The most obvious is EID-1's lack of effect on cell cycle progression. A priori, based on results with classic LXCXE proteins, E1A and SV40 large T Ag, one would have predicted cell cycle reentry as the default hypothesis; however, there are now multiple reports that endogenous cellular proteins with LXCXE motifs can have divergent effects on the cell cycle (29, 50). This may in part be explained by the ability of pocket proteins to bind multiple partners simultaneously (61). A model for this regulatory activity has been proposed whereby Rb effects on cell cycle are separable from its differentiation-promoting properties (58): Rb-dependent growth arrest requires an intact E2F binding site, while E2F binding was dispensable for Rb to promote differentiation, suggesting that Rb was binding and modulating a second, functionally distinct class of proteins.
HATs play a critical role in tissue-specific transcription by relieving repressive effects of chromatin condensation. p300 is a structural and functional homologue of CREB-binding protein, a transcriptional coactivator that not only has intrinsic HAT activity (46) but is capable of recruiting additional HAT factors to the transcriptional complex (76). p300, which was originally cloned as an E1A-binding protein, was subsequently shown to be critical for normal skeletal muscle differentiation, since disruption of its function by neutralizing antibodies or dominant-negative mutations blocks both differentiation and cell cycle arrest (49, 55). E1A mutations that selectively block p300 function are capable of inhibiting skeletal and cardiac muscle differentiation (32, 41). Until recently, it was presumed that competitive binding was the basis for the ability of E1A to inhibit tissue-restricted expression in skeletal muscle, since E1A interacts with the same region of p300 as MyoD (C-H3 domain). However, E1A has now been shown to directly inhibit the HAT domain of p300 or of the p300- and/or CBP-associated factor, PCAF (6, 23). We have provisionally suggested an analogous mechanism for EID-1, whose interaction with p300 in two-hybrid assays was specifically contingent on the C-H3 domain (Fig. 10C) and could inhibit p300 HAT activity (Fig. 11). Whether this inhibition of HAT activity is a general model for differentiation inhibitors will need to be determined; however, Twist, a skeletal muscle inhibitor, has already been shown to interact with p300 and inhibit its HAT activity (23).
In summary, our data suggest a novel mechanism for the interplay of Rb and MyoD and for the dependence of skeletal muscle cell differentiation on p300 and/or CBP. Further studies of EID-1 are in progress to delineate whether EID-1 also directly effects MyoD, possibly by inhibiting PCAF-dependent acetylation which has recently been shown to be important for MyoD DNA binding and transcriptional activity (56). Although EID-1 appears to be highly expressed in striated muscle and brain tissue, its expression in adult tissues is widespread, albeit at lower levels. Since Rb has been postulated to play a role in the differentiation of a wide variety of tissues, EID-1 and the model proposed may represent a more generalized mechanism of differentiation regulation. The extent of this role will await studies detailing its developmental expression and the creation of a mouse model deficient in this protein.
ACKNOWLEDGMENTS
We thank T. Durfee, Y. Shi, and W. Kaelin for the indicated plasmids, Frank Graham for the CMV.βgal virus, and J. Kim and the Baylor Flow Cytometry Lab for their technical assistance. We thank Satoshi Miyake and Bill Kaelin for their discussions and for sharing data prior to publication.
This work was supported by a gift from the Laubisch Fund and by NIH grant K08 HL03671 to W.R.M. and NIH grants R01 HL47567 and R01 HL61668 to M.D.S.
REFERENCES
- 1.Abdellatif M, Schneider M D. An effector-like function of Ras GTPase-activating protein predominates in cardiac muscle cells. J Biol Chem. 1997;272:525–533. doi: 10.1074/jbc.272.1.525. [DOI] [PubMed] [Google Scholar]
- 2.Austen M, Luscher B, Luscher-Firzlaff J M. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J Biol Chem. 1997;272:1709–1717. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- 3.Bains W, Ponte P, Blau H, Kedes L. Cardiac actin is the major actin gene product in skeletal muscle cell differentiation in vitro. Mol Cell Biol. 1984;4:1449–1453. doi: 10.1128/mcb.4.8.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishopric N H, Zeng G Q, Sato B, Webster K A. Adenovirus E1A inhibits cardiac myocyte-specific gene expression through its amino terminus. J Biol Chem. 1997;272:20584–20594. doi: 10.1074/jbc.272.33.20584. [DOI] [PubMed] [Google Scholar]
- 5.Buyse I M, Shao G, Huang S. The retinoblastoma protein binds to RIZ, a zinc-finger protein that shares an epitope with the adenovirus E1A protein. Proc Natl Acad Sci USA. 1995;92:4467–4471. doi: 10.1073/pnas.92.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 7.Charng M J, Zhang D, Kinnunen P, Schneider M D. A novel protein distinguishes between quiescent and activated forms of the type I transforming growth factor beta receptor. J Biol Chem. 1998;273:9365–9368. doi: 10.1074/jbc.273.16.9365. [DOI] [PubMed] [Google Scholar]
- 8.Chen P L, Riley D J, Chen-Kiang S, Lee W H. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P L, Riley D J, Chen Y, Lee W H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 10.Choubey D, Lengyel P. Binding of an interferon-inducible protein (p202) to the retinoblastoma protein. J Biol Chem. 1995;270:6134–6140. doi: 10.1074/jbc.270.11.6134. [DOI] [PubMed] [Google Scholar]
- 11.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 12.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 13.Dyson N, Guida P, Munger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 15.Eckner R, Yao T P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 16.Flink I L, Oana S, Maitra N, Bahl J J, Morkin E. Changes in E2F complexes containing retinoblastoma protein family members and increased cyclin-dependent kinase inhibitor activities during terminal differentiation of cardiomyocytes. J Mol Cell Cardiol. 1998;30:563–578. doi: 10.1006/jmcc.1997.0620. [DOI] [PubMed] [Google Scholar]
- 17.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham F L, Prevec L. Methods in molecular biology. Vol. 7. Clifton, N.J: The Humana Press, Inc.; 1991. [DOI] [PubMed] [Google Scholar]
- 19.Gu W, Schneider J W, Conderstil G, Kaushai S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 20.Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 21.Guo K, Walsh K. Inhibition of myogenesis by multiple cyclin-Cdk complexes. Coordinate regulation of myogenesis and cell cycle activity at the level of E2F. J Biol Chem. 1997;272:791–797. doi: 10.1074/jbc.272.2.791. [DOI] [PubMed] [Google Scholar]
- 22.Gustafson T A, Kedes L. Identification of multiple proteins that interact with functional regions of the human cardiac alpha-actin promoter. Mol Cell Biol. 1989;9:3269–3283. doi: 10.1128/mcb.9.8.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H Y, Wang J Y, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 24.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 25.Hasty P, Bradley A, Morris J H, Edmondson D G, Venuti J M, Olson E N, Klein W H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 27.Ip H S, Wilson D B, Heikinheimo M, Tang Z, Ting C-N, Simon M C, Leiden J M, Parmacek M S. The GATA-4 transcription factor transactivates the cardiac muscle-specific troponin C promoter-enhancer in nonmuscle cells. Mol Cell Biol. 1994;14:7517–7526. doi: 10.1128/mcb.14.11.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones R E, Wegrzyn R J, Patrick D R, Balishin N L, Vuocolo G A, Riemen M W, Defeo-Jones D, Garsky V M, Heimbrook D C, Oliff A. Identification of HPV-16 E7 peptides that are potent antagonists of E7 binding to the retinoblastoma suppressor protein. J Biol Chem. 1990;265:12782–12785. [PubMed] [Google Scholar]
- 29.Kato J, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 30.Kim S J, Onwuta U S, Lee Y I, Li R, Botchan M R, Robbins P D. The retinoblastoma gene product regulates Sp1-mediated transcription. Mol Cell Biol. 1992;12:2455–2463. doi: 10.1128/mcb.12.6.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirshenbaum L A, Chakraborty S, Schneider M D. Human E2F-1 reactivates cell cycle progression in ventricular myocytes and represses cardiac gene transcription. Dev Biol. 1996;179:402–411. doi: 10.1006/dbio.1996.0270. [DOI] [PubMed] [Google Scholar]
- 32.Kirshenbaum L A, Schneider M D. Adenovirus E1A represses cardiac gene transcription and reactivates DNA synthesis in ventricular myocytes, via alternative pocket protein- and p300-binding domains. J Biol Chem. 1995;270:7791–7794. doi: 10.1074/jbc.270.14.7791. [DOI] [PubMed] [Google Scholar]
- 33.Kraus W L, Manning E T, Kadonaga J T. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larose A, Dyson N, Sullivan M, Harlow E, Bastin M. Polyomavirus large T mutants affected in retinoblastoma protein binding are defective in immortalization. J Virol. 1991;65:2308–2313. doi: 10.1128/jvi.65.5.2308-2313.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavender P, Vandel L, Bannister A J, Kouzarides T. The HMG-box transcription factor HBP1 is targeted by the pocket proteins and E1A. Oncogene. 1997;14:2721–2728. doi: 10.1038/sj.onc.1201243. [DOI] [PubMed] [Google Scholar]
- 36.Lee E Y, Hu N, Yuan S S, Cox L A, Bradley A, Lee W-H, Herrup K. Dual roles of the retinoblastoma protein in cell cycle regulation and neuron differentiation. Genes Dev. 1994;8:2008–2021. doi: 10.1101/gad.8.17.2008. [DOI] [PubMed] [Google Scholar]
- 37.Lee T C, Zhang Y, Schwartz R J. Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene. 1994;9:1047–1052. [PubMed] [Google Scholar]
- 38.Lipinski M M, Jacks T. The retinoblastoma gene family in differentiation and development. Oncogene. 1999;18:7873–7882. doi: 10.1038/sj.onc.1203244. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Kitsis R N. Induction of DNA synthesis and apoptosis in cardiac myocytes by E1A oncoprotein. J Cell Biol. 1996;133:325–334. doi: 10.1083/jcb.133.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacLellan W R, Lee T-C, Schwartz R J, Schneider M D. Transforming growth factor-β response elements of the skeletal α-actin gene. J Biol Chem. 1994;269:16754–16760. [PubMed] [Google Scholar]
- 41.Missero C, Calautti E, Eckner R, Chin J, Tsai L H, Livingston D M, Dotto G P. Involvement of the cell-cycle inhibitor Cip1/WAF1 and the E1A-associated p300 protein in terminal differentiation. Proc Natl Acad Sci USA. 1995;92:5451–5455. doi: 10.1073/pnas.92.12.5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyake S, Sellers W R, Safran M, Li X, Zhao W, Grossman S R, Gan J, DeCaprio J A, Adams P D, Kaelin W G., Jr Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol Cell Biol. 2000;20:8889–8902. doi: 10.1128/mcb.20.23.8889-8902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 44.Novitch B G, Mulligan G J, Jacks T, Lassar A B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novitch B G, Spicer D B, Kim P S, Cheung W L, Lassar A B. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr Biol. 1999;9:449–459. doi: 10.1016/s0960-9822(99)80210-3. [DOI] [PubMed] [Google Scholar]
- 46.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 47.Paradis P, MacLellan W R, Belaguli N S, Schwartz R J, Schneider M D. Serum response factor mediates AP-1-dependent induction of the skeletal α-actin promoter in ventricular myocytes. J Biol Chem. 1996;271:10827–10833. doi: 10.1074/jbc.271.18.10827. [DOI] [PubMed] [Google Scholar]
- 48.Parker T G, Packer S E, Schneider M D. Peptide growth factors can provoke “fetal” contractile protein gene expression in rat cardiac myocytes. J Clin Investig. 1990;85:507–514. doi: 10.1172/JCI114466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puri P L, Avantaggiati M L, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin X-Q, Livingston D M, Ewen M, Sellers W R, Arany Z, Kaelin W G., Jr The transcription factor E2F-1 is a downstream target of RB action. Mol Cell Biol. 1995;15:742–755. doi: 10.1128/mcb.15.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao S S, Chu C, Kohtz D S. Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic helix-loop-helix regulators. Mol Cell Biol. 1994;14:5259–5267. doi: 10.1128/mcb.14.8.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rawls A, Olson E N. MyoD meets its maker. Cell. 1997;89:5–8. doi: 10.1016/s0092-8674(00)80175-0. [DOI] [PubMed] [Google Scholar]
- 53.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sartorelli V, Puri P L, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang J Y, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 57.Schneider J W, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- 58.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G J. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sellers W R, Rodgers J W, Kaelin W G J. A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shih H H, Tevosian S G, Yee A S. Regulation of differentiation by HBP1, a target of the retinoblastoma protein. Mol Cell Biol. 1998;18:4732–4743. doi: 10.1128/mcb.18.8.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shirodkar S, Ewen M, DeCaprio J A, Morgan J, Livingston D M, Chittenden T. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992;68:157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 62.Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 63.Skapek S X, Rhee J, Kim P S, Novitch B G, Lassar A B. Cyclin-mediated inhibition of muscle gene expression via a mechanism that is independent of pRB hyperphosphorylation. Mol Cell Biol. 1996;16:7043–7053. doi: 10.1128/mcb.16.12.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stein R W, Corrigan M, Yaciuk P, Whelan J, Moran E. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sternberg E A, Spizz G, Perry W M, Vizard D, Weil T, Olson E N. Identification of upstream and intragenic regulatory elements that confer cell-type-restricted and differentiation-specific expression on the muscle creatine kinase gene. Mol Cell Biol. 1988;8:2896–2909. doi: 10.1128/mcb.8.7.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- 67.Tam S K C, Gu W, Mahdavi V, Nadal-Ginard B. Cardiac myocyte terminal differentiation. Potential for cardiac regeneration. Ann N Y Acad Sci. 1995;752:72–79. doi: 10.1111/j.1749-6632.1995.tb17407.x. [DOI] [PubMed] [Google Scholar]
- 68.Taya Y. RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 69.Tiainen M, Spitkovsky D, Jansen-Durr P, Sacchi A, Crescenzi M. Expression of E1A in terminally differentiated muscle cells reactivates the cell cycle and suppresses tissue-specific genes by separable mechanisms. Mol Cell Biol. 1996;16:5302–5312. doi: 10.1128/mcb.16.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trouche D, Kouzarides T. E2F1 and E1A(12S) have a homologous activation domain regulated by RB and CBP. Proc Natl Acad Sci USA. 1996;93:1439–1442. doi: 10.1073/pnas.93.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Venuti J M, Morris J H, Vivian J L, Olson E N, Klein W H. Myogenin is required for late but not early aspects of myogenesis during mouse development. J Cell Biol. 1995;128:563–576. doi: 10.1083/jcb.128.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Helin K, Jin P, Nadal-Ginard B. Inhibition of in vitro myogenic differentiation by cellular transcription factor E2F1. Cell Growth Differ. 1995;6:1299–1306. [PubMed] [Google Scholar]
- 73.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 74.Wiman K G. The retinoblastoma gene: role in cell cycle control and cell differentiation. FASEB J. 1993;7:841–845. doi: 10.1096/fasebj.7.10.8393817. [DOI] [PubMed] [Google Scholar]
- 75.Yang M, Wu Z, Fields S. Protein-peptide interactions analyzed with the yeast two-hybrid system. Nucleic Acids Res. 1995;23:1152–1156. doi: 10.1093/nar/23.7.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 77.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]