Abstract
Precise control over the assembly of biocompatible three-dimensional (3D) nanostructures would allow for programmed interactions within the cellular environment. Nucleic acids can be used as programmable crosslinkers to direct the assembly of quantum dots (QDs) and tuned to demonstrate different interparticle binding strategies. Morphologies of self-assembled QDs are evaluated via gel electrophoresis, transmission electron microscopy, small-angle X-ray scattering, and dissipative particle dynamics simulations, with all results being in good agreement. The controlled assembly of 3D QD organizations is demonstrated in cells via the colocalized emission of multiple assembled QDs, and their immunorecognition is assessed via enzyme-linked immunosorbent assays. RNA interference inducers are also embedded into the interparticle binding strategy to be released in human cells only upon QD assembly, which is demonstrated by specific gene silencing. The programmability and intracellular activity of QD assemblies offer a strategy for nucleic acids to imbue the structure and therapeutic function into the formation of complex networks of nanostructures, while the photoluminescent properties of the material allow for optical tracking in cells in vitro.
Keywords: quantum dots, therapeutic nucleic acids, 3D assemblies, programmable assembly, materials organization, bioimaging, therapeutic delivery
Graphical Abstract
1. INTRODUCTION
The organization of inorganic materials in diverse 3D nanostructures with hierarchical complexity mimics biological approaches by combining nanoscale components of distinct physicochemical properties with promising applications in imaging,1 sensing,2 drug delivery,3 and tissue engineering4 with a variety of assembly strategies. Generally, there are two approaches for the fabrication of materials with defined features and functionality at the nanometer scale: top-down and bottom-up. In several top-down approaches, inorganic structures of cellular origin can serve as templates for the organization of nanomaterials. Attachment to these template surfaces can be nonspecific, or microorganisms can be genetically engineered to express functional groups for selective interactions with nanoparticles or their adsorption with increased affinity.5–7 The top-down methods for nanoparticle organization on the cellular scale are intrinsically less controllable and highly depend on the structure/shape of the template. Therefore, the bottom-up approach for de novo organization of inorganic particles by natural polymers such as polypeptides and nucleic acids offers advantages for regulating the assembly behavior and morphology.8
Nucleic acids are materials with programmable, dynamic, and environmentally responsive functional components for hybrid nanoparticle systems. Due to their simple primary structure and known rules that guide the formation of their secondary and tertiary conformations, nucleic acids are robust materials for scaffolding in comparison to proteins or other biopolymers. The use of the Watson–Crick base pairing as a means of prescribing bottom-up assembly strategies has been shown to control and rationally program the 3D self-assembly of functional particles into well-ordered organizations from the nano- to microscale.9–12 Several experiments have demonstrated the versatility of nucleic acid scaffolds for displaying functional DNA/RNA motifs with intrinsic functionalities and promising applications in biotechnology or biomedicine.13–19 The ability to dynamically respond to the environment makes nucleic acids an attractive biomaterial for tailor-made structures with desired responsiveness.20,21 In recent decades, a wide array of artificially designed dynamic nucleic acid assemblies have been shown to react on the broad spectrum of physicochemical or biological stimuli (e.g., pH, light, ion concentration, small metabolites, enzymes, or nucleic acid strands).22–31
DNA oligonucleotides have been conjugated to a wide range of inorganic particles with distinct physicochemical properties.32,33 In particular, semiconductor nanocrystals or quantum dots (QDs) are attractive for the development of nano-theranostic concepts for simultaneous diagnostics and therapy.34 In comparison to organic fluorophores, QDs are strongly luminescent, have increased stability, and have higher brightness and resistance to photobleaching, as well as narrower and symmetric fluorescence spectra with tunable colors controlled by their size. Using DNA for linking, QDs can be utilized to create assemblies with controlled bonding, valency, and photoluminescence.35,36 Over the last 2 decades, numerous studies have developed approaches for modifying the surface of QDs with biomolecules for the attachment of functionalized moieties such as DNA/RNA oligonucleotides, antibodies, and peptides.2 Almost exclusively as optical labels, functionalized QDs have found many applications in biosensing and bioimaging.37–39 Instead of fluorescent dyes, QDs can be conjugated to aptamers for the visualization of aptamer binding and subsequent intracellular trafficking.40 Aptamer–QD complexes have been examined to detect a wide range of targets, from simple metal ions, drugs, or toxins to proteins.41
Although most biosensing and bioimaging applications of QDs rely on the measurement of changes in fluorescence (color or intensity), QDs offer additional properties for detection. The fluorescence intermittency or blinking is an inherent random fluctuation between ON (bright) and OFF (dark) states of individual QDs.42 The phenomenon is observable only in a single QD, while in aggregated QDs, the signal is semisteady. Therefore, differences in the signal between the single QDs versus an accumulated group of QDs can be distinguished and used for the detection of target molecules. The principle of this strategy is strand displacement triggered by a target sequence, leading to the reassociation of two split biotinylated oligonucleotides that subsequently promote the arrangement and assembly of streptavidin-decorated QDs.43
In this work, we set out to design a biocompatible nucleic acid-based scaffold for the assembly of QDs and delivery of functional therapeutics. The developed system uses both RNA and biotinylated DNA as a means to drive the 3D organization of streptavidin-decorated QDs. We first interrogated several approaches for the formation of bioresponsive QD 3D assemblies using sets of QDs functionalized with complementary single-stranded (ss)DNAs, combined with double-biotinylated DNA duplexes, or decorated with DNA/RNA hybrids that reassociate to release Dicer substrate (DS) RNAs via an isothermal strand displacement reaction.19,44–48 The results from each method of assembly were extensively characterized via electrophoretic mobility shift assays (EMSAs), transmission electron microscopy (TEM), and in situ small-angle X-ray scattering (SAXS). Next, we studied relative cellular uptake efficiencies, immunostimulatory properties, and intracellular colocalization of the 3D assemblies and their individual components. We have shown that intracellular formation of QD assemblies in human breast cancer cells releases DS RNAs and, upon dicing, triggers targeted gene silencing.
2. RESULTS
The assembly strategy, as dictated by the way nucleic acids are introduced, offers versatility in the resulting 3D assemblies with varying kinetics and functionalities. Three methods of QD assembly were evaluated. First, through the introduction of 39 bp double-stranded (ds)DNA oligonucleotides with a single biotin present on both ends of each duplex, binding with streptavidin-coated QDs which are 15–16 nm in diameter drives the rapid (~30 s) formation of assembled structures (Figure 1A). As seen in the agarose gel, the QDs alone begin to migrate upon the addition of an ssDNA. However, full assembly at a 1 QD/10 DNA molar ratio into larger-scale organization over time results in morphologies that are too large to enter the gel and can thus be observed in the loading wells only (Figure S1).43 In comparison, if individual fully complementary DNA strands are added separately to QDs and then combined, it takes closer to 30 min for the aggregate to fully assemble (Figure 1B). As the third approach, QDs were separately conjugated to complementary dsDNA/RNA hybrid duplexes via the biotinylated DNA. Once added together, complementary 12-nt ssDNA toeholds initiate the isothermal strand displacement reaction that promotes the formation of double-biotinylated DNA duplexes while releasing the RNA sequences to form functional DS RNAs (Figure 1C). The complete assembly of QDs and subsequent release of DS RNAs were achieved after ~10 min. In all three strategies of QD assembly, the addition of DNase to assembled 3D structures completely reversed the formation of assembled structures through the digestion of any DNA cross-linkers, resulting in the increased mobility of QDs. These results additionally confirm the DNA-driven organization of QDs.
Figure 1.
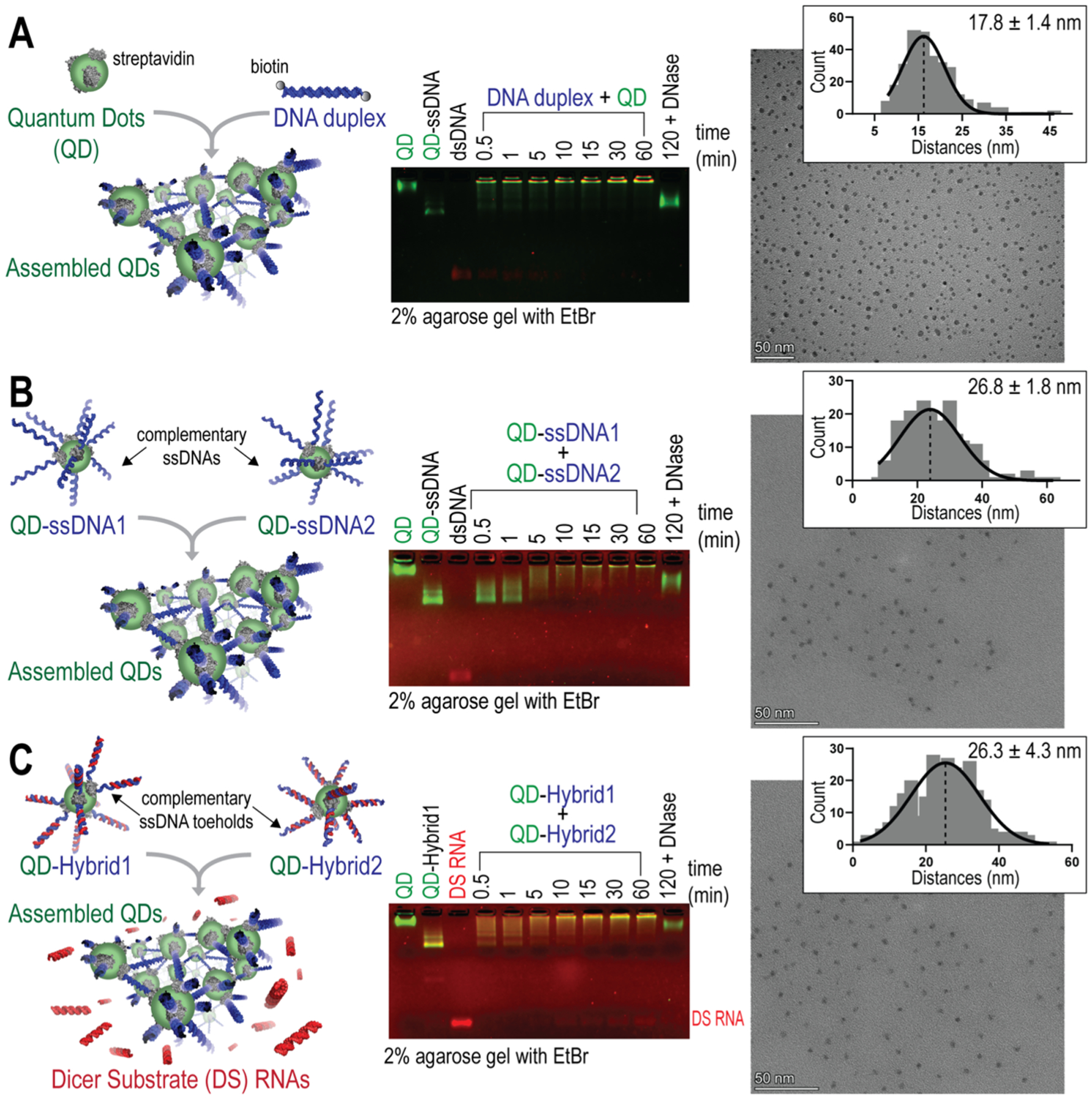
Three protocols of QD assembly with kinetics characterized by agarose gels. TEM images showing the distribution of QDs within their assemblies and measurements of the center-to-center distances presented in the histogram with their Gaussian fit for n = 100 QDs. Scale bar = 50 nm. Mean center-to-center distances calculated from n = 3 TEM images are shown with ± SEM. (A) QDs mixed with double-biotinylated DNA duplexes. (B) QDs decorated with complementary ssDNAs. (C) QDs decorated with RNA/DNA hybrids that reassociate via the complementary ssDNA toehold interactions and release DS RNAs.
All three methods of QD assembly were conducted, and the products were imaged by TEM. Analysis of the center-to-center distances between a given QD and its three nearest neighbors was assessed to compare QD distributions (Figure S2). For the dsDNA duplex-driven QD assembly, the mean center-to-center distance was 17.8 ± 1.4 nm. For the ssDNA-driven QD assembly, the mean center-to-center distance was 26.8 ± 1.8 nm. Finally, for hybrid-driven QD assemblies, the mean center-to-center distance was 26.3 ± 4.3 nm. The distributions of distances for 100 QDs are shown for each TEM image in Figure 1A–C. The assembled distances show much less distribution than free QDs in solution imaged at the same concentrations, for which the mean center-to-center distance was 76.6 ± 17.2 nm (Figure S2).
While dynamic light scattering is often widely used to assess the morphologies of particles in preclinical studies, the high polydispersity and broad size distributions of the aggregated QD assemblies faced limitations by this analysis. Therefore, to investigate the ordered three-dimensional morphologies of the assemblies of the QDs in their native states for each of the assembly strategies described previously, synchrotron-based SAXS was utilized for probing the structures of each of these systems in situ. The scattering profiles for each of the designs showed a single broad peak as a function of the scattering vector, q, which corresponds to a disordered, aggregate system with a characteristic average center-to-center (DCC) distance of the assembled QDs. Figure 2 summarizes the structure factor plots, S(q), for each of the designs, and the corresponding real-space distances are indicated on the respective plots based on the center of the peak fit with a Lorentzian function. For the dsDNA-, ssDNA-, and hybrid-driven QD assemblies, these center-to-center distances were calculated to be 20.3, 20.5, and 20.3 nm, respectively. Compared to the values derived from TEM images, the SAXS-derived values for the dsDNA-driven QD assembly were the most similar, differing only by 2.5 nm between averages. Otherwise, there is good agreement between the center-to-center distances of the three assemblies between each method of analysis. The values are also feasible given the lengths of the materials. Additional SAXS plots are available in Figures S3 and S4.
Figure 2.
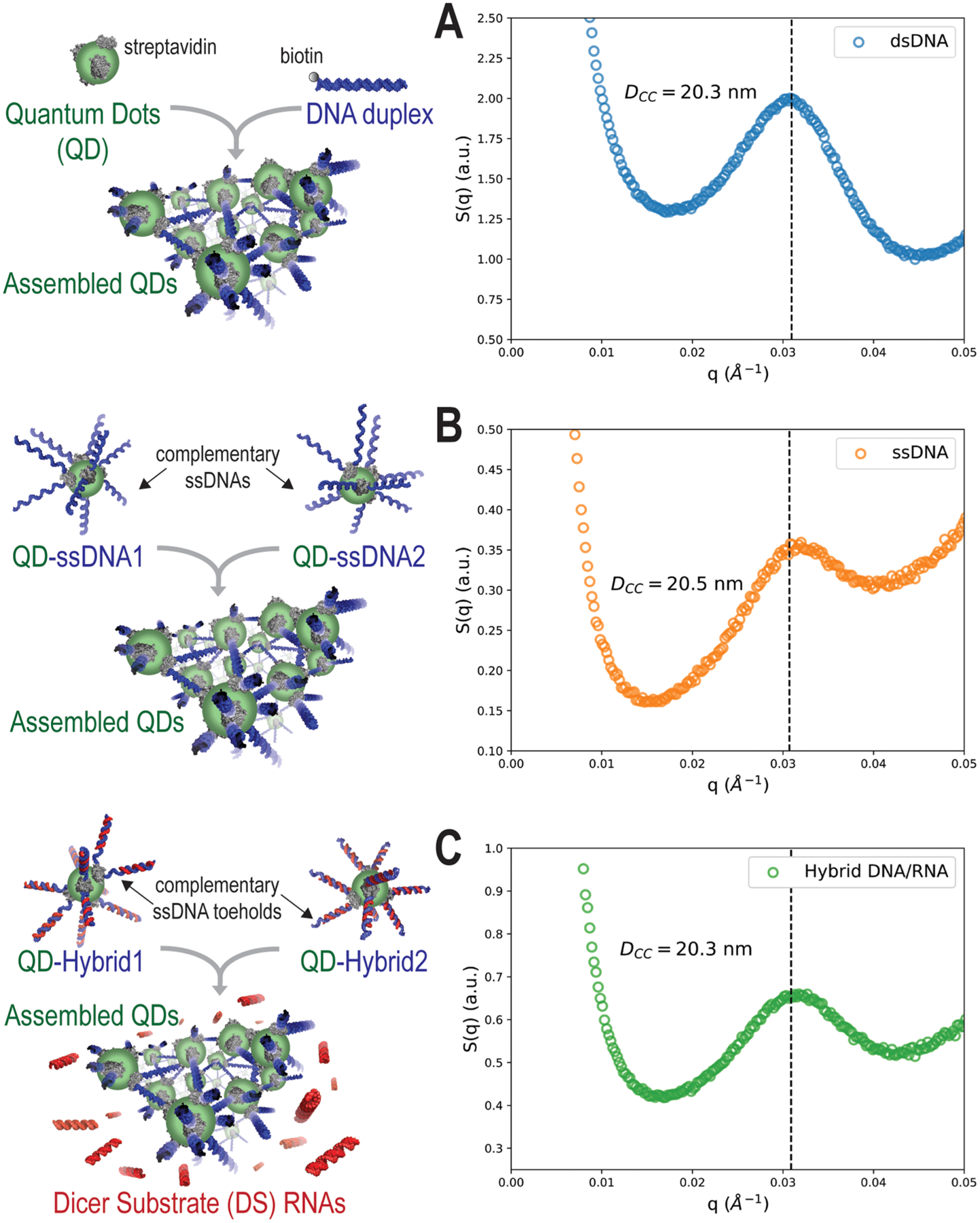
SAXS analysis of the assembly of (A) dsDNA-, (B) ssDNA-, and (C) hybrid DNA/RNA-driven QD formations shown by structure factor plots.
Computational modeling provides fundamental understanding of how experimental and environmental variables control the self-assembly process, resulting in changes in the organization of the QDs. Here, we use the mesoscale modeling technique dissipative particle dynamics (DPD) to understand these materials on the size and time scales relevant to the experiment.49,50 The technique has been successful in predicting and explaining the self-assembly of polymer-based materials, including block copolymers, polyelectrolytes, and DNA.51–58 Computational modeling of QD and DNA assemblies as a function of DNA length and salt concentration was performed using our implicit solvent ionic strength method (ISIS-DPD) model58 and provides key insights into the driving forces of the resulting morphologies. The initial system (Figure 3A) consists of 24 QDs decorated with 12 ssDNAs each based on the experimentally determined number of binding sites per QD.43 Figure 3B presents the snapshots of the final morphologies of the simulations conducted as a function of DNA length and salt concentration. As the length increases across any of the salt concentrations, the decorated QDs have an increased tendency to pack versus assembling in a more linear fashion for the shorter DNA. This is quantified in the integration of the radial distribution function (int-RDF) of the QD center of mass in Figure 3C, where each DNA length is averaged and then compared against the other lengths simulated. As the DNA length increases, the QDs are pushed further apart but not in proportion to the increase in the length of the DNA. This results in the more packed configurations that are seen in the phase diagram in Figure 3B. Lastly, it is worth noting that while salt was varied across a broad range of concentrations, the length of the DNA strand was the predominant variable that determined the morphology.
Figure 3.
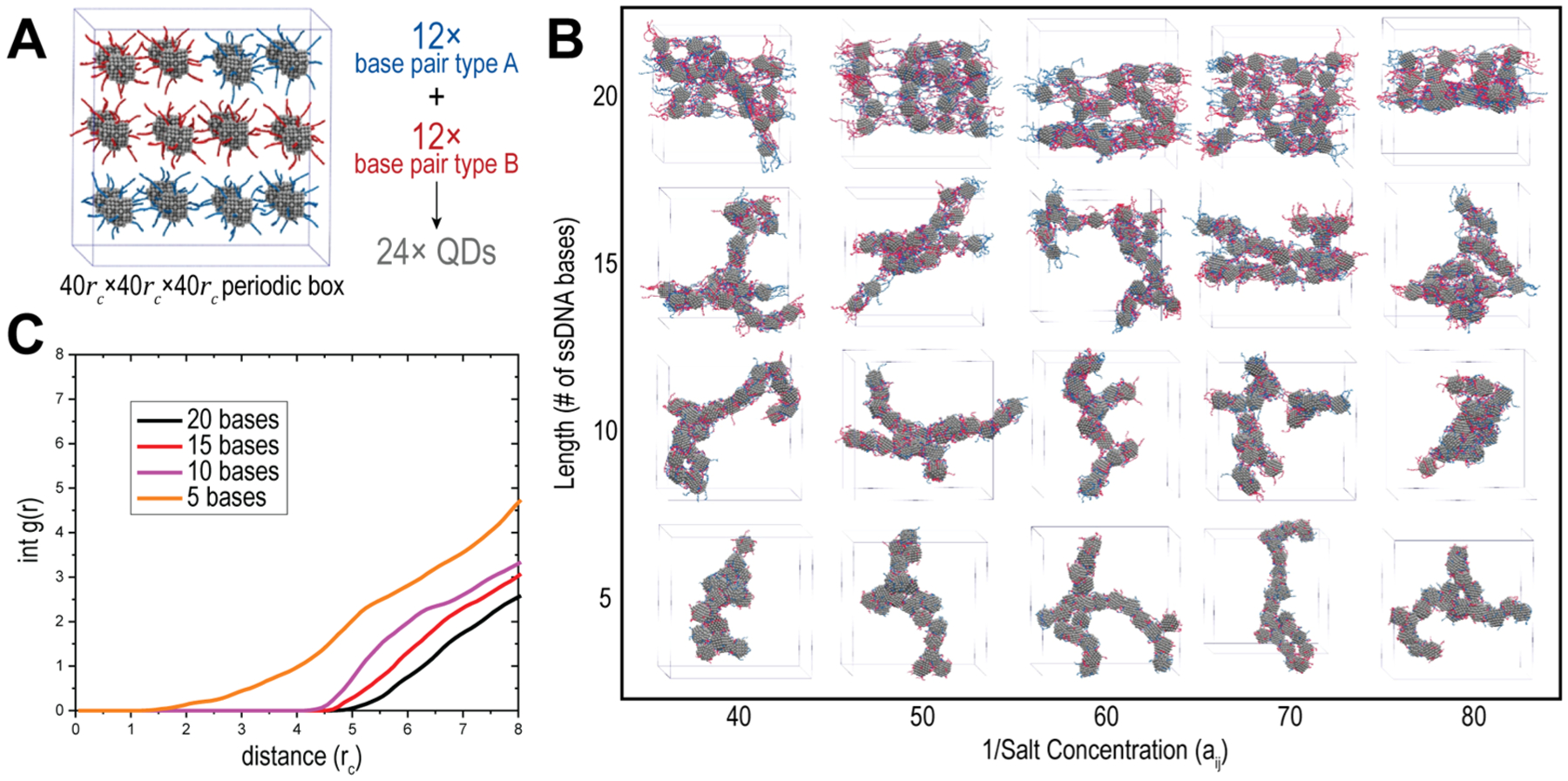
Computational modeling of QD–DNA formations. (A) Snapshot of the initial DPD configuration. (B) Phase diagram of the assembled QDs as a function of salt and DNA length. (C) int-RDF of averaged lengths.
To fully take advantage of the programmable assemblies of inorganic QDs, assemblies were introduced into cells using a lipid-based carrier for their intracellular delivery. Their relative uptake and intracellular assembly in MDA-MB-231 human breast cancer cells were assessed by separately introducing QDs carrying complementary DNA/RNA hybrids. Cells were then visualized via fluorescence microscopy and analyzed by flow cytometry (Figure 4A). Micrographs labeled a–d correspond with the geometric mean fluorescence intensity (gMFI) shown to the right, wherein the stepwise introduction of materials for their intracellular assembly resulted in higher gMFI than for either the QD component or assembly. To confirm that the cognate QDs can form intracellular assemblies and thus colocalize inside cells, QD545 and QD605 carrying complementary hybrid DNA/RNA duplexes were introduced and the cells were analyzed by confocal microscopy (Figure 4B). The colocalization of the emission of each QD shown on the superposition image (1 + 2 + 3) demonstrates the heterogeneous assembly of formations composed of both QDs and confirms the assembly of QDs in cells. Additional confocal microscopy images of the colocalization studies are shown in Figure S5.
Figure 4.
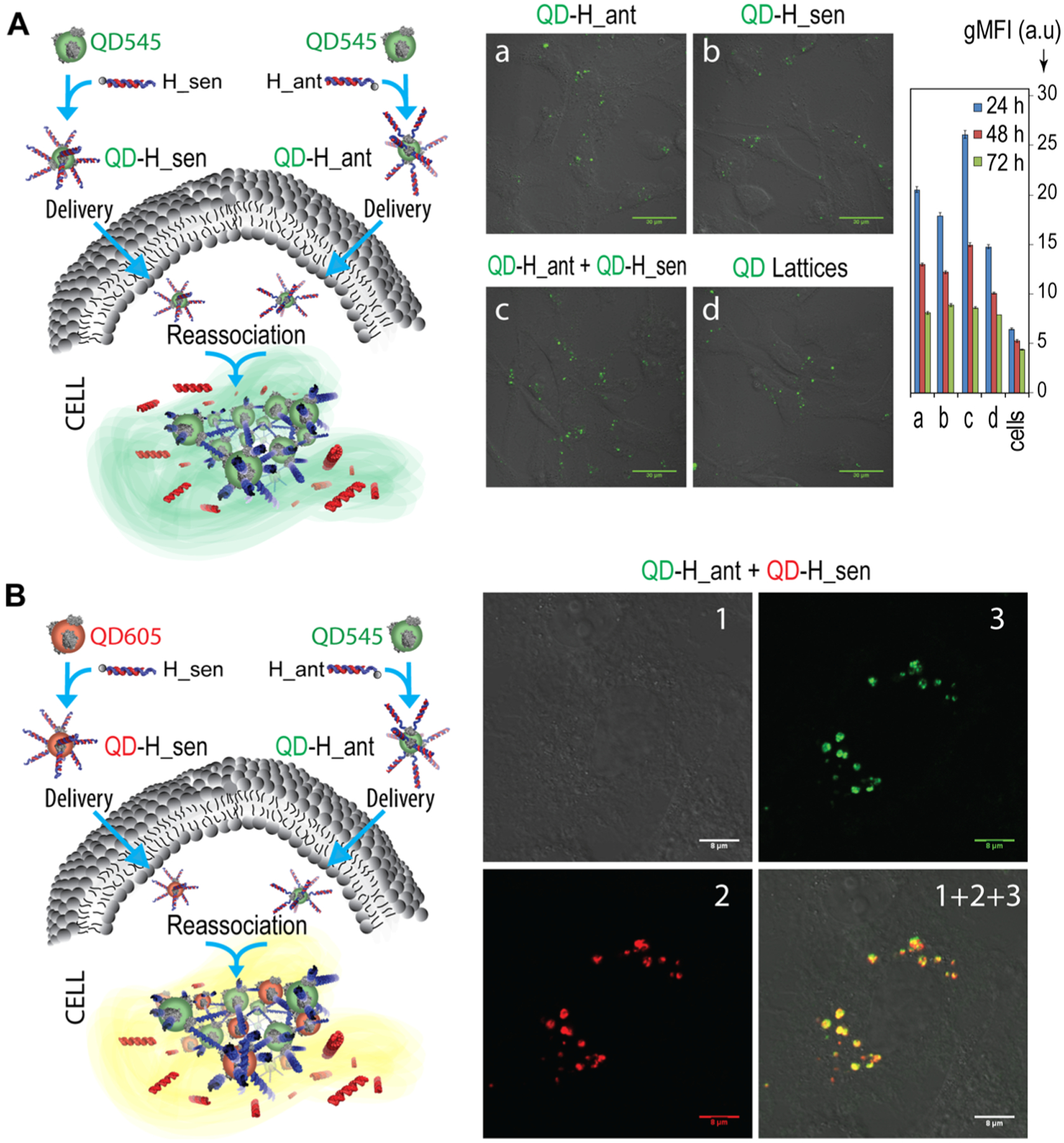
Relative uptake efficiencies and intracellular colocalization experiments. (A) The uptake of functionalized QD545 was analyzed by fluorescence microscopy and flow cytometry. Scale = 30 μm. Bars denote mean ± SEM of n = 20,000 individual events. (B) Colocalization of QD545 (green) and QD605 (red) entering the composition of QD assemblies as analyzed by confocal microscopy. Image numbers correspond to (1) differential interference contrast, (2) QD605 emission, and (3) QD545 emission. Image (1 + 2 + 3) is the superposition of three different images, scale = 8 μm.
With the uptake established, the potential for functional therapeutics imbued into the DNA/RNA assembly approach was further investigated. QDs carrying complementary hybrid duplexes were transfected stepwise into MDA-MB-231 cells engineered to express green fluorescent protein (GFP). The RNA sequences were designed to assemble into DS RNAs upon the reassociation of QDs inside the cell (Figure 5). With either the QDs or hybrid alone, the cells remained fluorescent with the expression of GFP. However, when both QDs were introduced to cells, the intracellular QD formation resulted in the silencing of GFP assessed after 72 h. Flow cytometry confirmed a statistically significant reduction in the gMFI as a result of the QD assembly. In time-course studies, silencing with 10 and 20 nM QD assemblies was observed even after up to 14 days (Figure S6).
Figure 5.
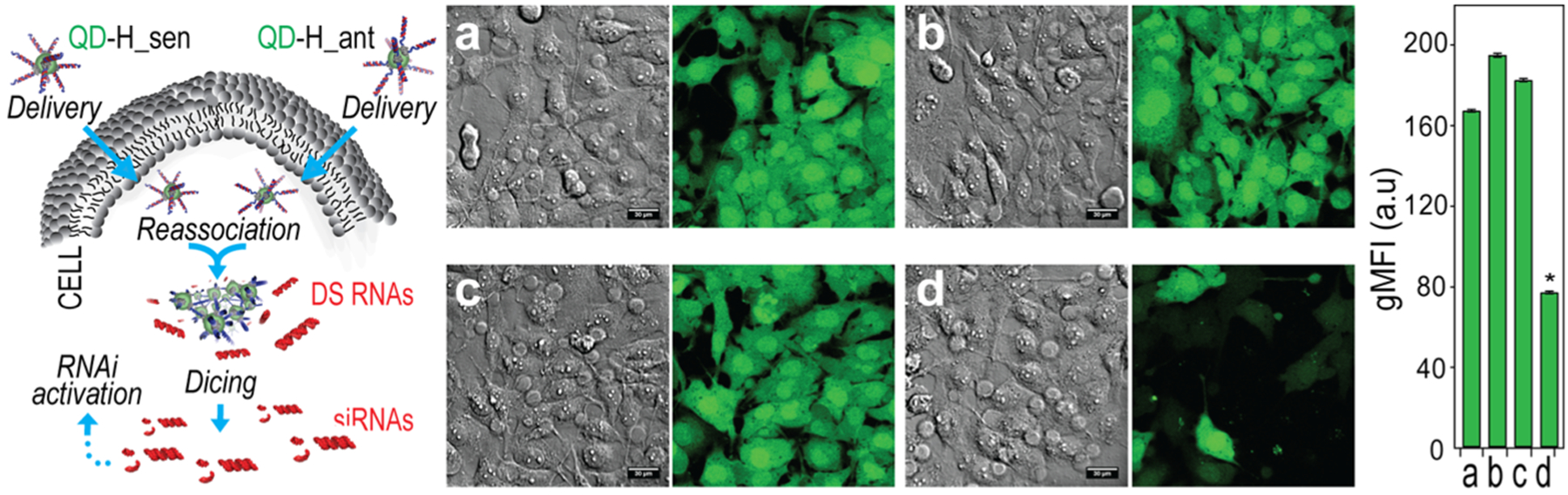
Activation of RNA interference in human breast cancer cells upon QD assembly formation. Intracellular QD assembly releases DS RNAs that trigger specific gene silencing upon dicing. Three days after the cotransfection of cells with QDs decorated with cognate hybrids, GFP silencing was confirmed by fluorescence microscopy and statistically analyzed with flow cytometry. Samples (a–d) have the same conditions for microscopy and flow cytometry experiments. (a) Untreated MDA-MB-231 eGFP cells show green fluorescence. Cells treated with either (b) QD-H_sen or (c) QD-H_ant show no fluorescence knockdown. Silencing is observed when (d) both QD-H_sen and QD-H_ant are transfected to reassociate in cells and drive the activation of RNAi. Final concentrations of nucleic acids and QDs are 10 nM and 1 nM, respectively. Bars denote mean ± SEM of n = 20,000 individual events. Statistically significant results are indicated with asterisks (* = P-value < 0.05).
As the last step to confirm the downstream biological applications of these materials, their immunostimulation in cell lines was assessed. Hybrid duplexes and their reassociation were compared with assembled QDs, free QDs, QDs with hybrid duplexes, and the reassociation of hybrid duplexes when one (QD-H_sen + H_ant) hybrid was bound to QDs. The relative production of cytokines hIL-1β, hIL-6, hIL-8, and hIFN-β in the human microglia-like cell line, hμglia, were assessed as normalized to cells treated only with a carrier, Lipofectamine 2000 (L2K) (Figure 6). The panel of these four cytokines was chosen due to their known roles as modulators of the inflammatory response: hIL-1β and hIL-6 as proinflammatory cytokines, hIL-8 as a chemokine, and hIFN-β as an interferon. Any recognition of exogenously introduced nucleic acids resulting in immunostimulation was expected to be observed from this representative panel as their release has been previously documented from microglia in response to pathogen-associated molecular patterns or pathogens.59,60 Overall, no components of the QD assembly were identified as potent immune activators. Only QD-H_sen showed statistical significance in regard to relative hIL-1β production. The same panel of cytokines was also investigated for a human astrocyte-like cell line, U87-MG (Figure S7), in which no conditions demonstrated statistically significant immune stimulation. Importantly, free QDs showed no statistically significant cytotoxicity in either hμglia or MDA-MB-231 cell lines (Figure S8).
Figure 6.
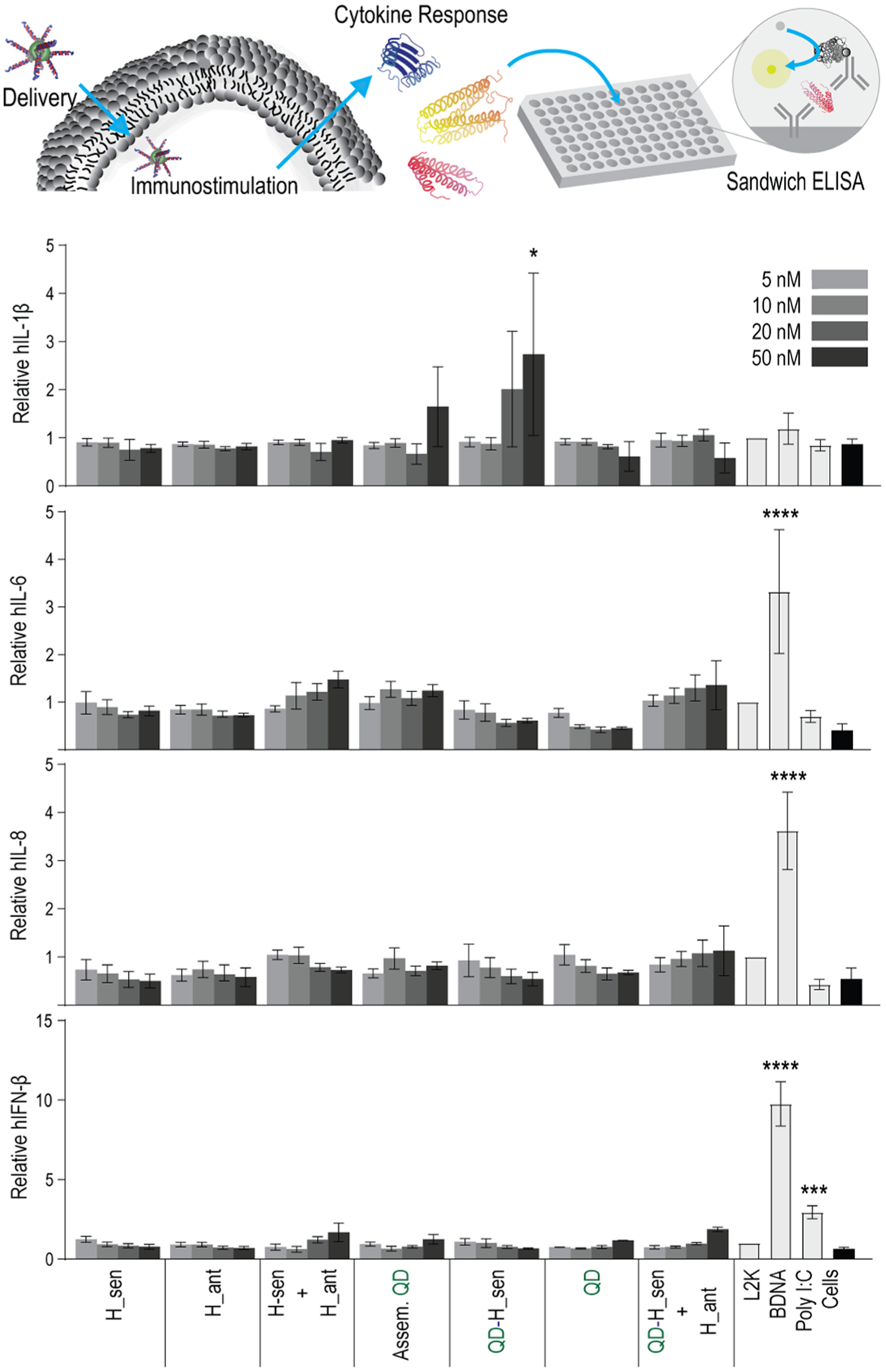
Immunostimulatory activity of QD assemblies. A human microglia-like cell line (hμglia) was transfected, and cell supernatants were collected 24 h later. Levels of hIL-1β, hIL-6, hIL-8, and hIFN-β were assessed by ELISA. Bars denote mean ± SEM of n = 3 independent repeats. Statistically significant results are indicated with asterisks (**** = P-value < 0.0001, *** = P-value < 0.001, and * = P-value < 0.05).
3. CONCLUSIONS
The colocalization of QDs 545 and 605 within human breast cancer cells demonstrates the assembly of QDs directly within the cellular environment, which is further validated by the significant fold knockdown of GFP in expressing cells via RNA interference upon QD formation. Importantly, despite the presence of a large number of dsDNAs in their structures, QD assemblies and their components do not invoke a significant difference in the production of cytokines, which makes this theranostic approach feasible in addition to biosensing. While QDs alone exhibit narrow emission, which is advantageous for tracking, assemblies of QDs offer more opportunities for sensing parameters.43 For example, one avenue is that the centrifugation of assembled QDs results in the formation of a precipitate, while individual monomers show no precipitation (Figure S9).
Precise control over the assembly of complex networks of materials requires the coordination of all the individual components. Nucleic acids offer a straightforward route to scaffolding due to their programmable base pairing and also allow for biologically relevant sequences to be implemented for therapeutic applications, as demonstrated here with the incorporation of RNA interference inducers. While three methods of assembly were demonstrated, their characterization shows similar morphologies despite variations in their kinetics. Based on the predicted DPD models, variations in morphologies can also be achieved by changing the lengths of DNAs in the assembly composition, which could also allow for the integration of other functional nucleic acids. Further work to fine-tune the organization of QD assemblies may take advantage of nucleic acids’ programmability in order to control the size and shape of the network on a larger scale.
4. EXPERIMENTAL SECTION
4.1. Sequence Design and Preparation.
DS RNAs designed against GFP and their complementary DNA sequences with 12 nt toeholds were purchased from Integrated DNA Technologies (IDT), as shown in a previous work.45 All sequences are listed in the Supporting Information. Oligos were diluted in endotoxin-free HyClone HyPure cell culture-grade water (Cytiva) before use.
4.2. Assembly of QDs.
QD545s (Qdot 545 ITK streptavidin conjugate kit, catalog #Q10091MP) composed of a cadmium selenide core with a zinc sulfide shell and covalently attached streptavidin were purchased from Invitrogen. QD545s were reported by the manufacturer to be 15–16 nm in diameter with an emission maximum of 545 ± 4 nm and were used for all studies. QD545s were assembled with either dsDNA, ssDNA, or DNA/RNA hybrid duplexes in endotoxin-free water with all QDs at a 100 nM final concentration.
4.2.1. For (DNA Duplex + QD) Assembly.
DNA duplexes were made by mixing complementary DNA oligos in an equimolar ratio. The mixture was heated at 95 °C for 2 min before the assembly buffer [a final concentration of 89 mM tris-borate (pH 8.2), 50 mM KCl, and 2 mM MgCl2] was added, followed by incubation at room temperature for 20 min.61 Afterward, QDs were added with the DNA duplex at a 1:10 QD/duplex molar ratio and incubated at 37 °C for 30 min.
4.2.2. For (QD + ssDNA1) + (QD + ssDNA2) Assembly.
QDs were assembled in two separate tubes with each DNA oligo in a 1:10 QD/DNA molar ratio in the assembly buffer. Samples were incubated at 37 °C for 20 min. Afterward, the contents of the two tubes were mixed at a 1:1 volumetric ratio and incubated at 37 °C for 30 min.
4.2.3. For (QD + H_sen) + (QD + H_ant) Assembly.
Hybrid DNA/RNA duplexes were assembled in two separate tubes: H_sen (“DNA for Sense_12_Biotin” + “RNA Sense”) and H_ant (“DNA for Antisense_12_Biotin” + “RNA Antisense”). The hybrid duplexes were prepared by adding their constituent oligos in an equimolar ratio, heating at 95 °C for 2 min, and adding the assembly buffer, followed by incubation at room temperature for 20 min. QDs were added to each separate hybrid duplex tube in a 1:10 QD/duplex molar ratio and incubated at 37 °C for 30 min. Afterward, hybrid duplexes were mixed in a 1:1 volumetric ratio and incubated at 37 °C for 60 min. For cellular colocalization in Figure 4B, one hybrid duplex was added with QD545, while its conjugate was added with QD605 for the 30 min incubation at 37 °C. For this, the Qdot 605 streptavidin conjugate (catalog # Q10101MP) composed of a cadmium selenide core, a zinc sulfide shell, and polymer coating to allow for streptavidin conjugation was purchased from Invitrogen. QD605s were reported to be 15–20 nm in diameter with an emission maximum of 608 ± 4 nm.
4.3. Electrophoretic Mobility Shift Assays.
To confirm assembly, QD assemblies were analyzed on a 2% agarose gel stained with 0.5 μg/mL ethidium bromide. Gels were run in 89 mM tris-borate and 2 mM ethylenediaminetetraacetic acid (pH 8.2) for 20 min at 220 V and then visualized on a Bio-Rad ChemiDoc MP imaging system using the multichannel protocol for QD525 (used to view QD545) and QD605 (used to view ethidium bromide).
4.3.1. For EMSAs of the Kinetics of Assembly.
As shown in Figure 1, QDs were assembled in 40 μL volumes as described and incubated at 37 °C for 120 min as previously shown.43 At the 0.5, 1, 5, 10, 15, 30, and 60 min timepoints, 4 μL of assembling QDs were added to 4 μL of agarose loading buffer (30% glycerol, 0.25% bromophenol blue, and 0.25% xylene cyanol) in a tube, which was immediately placed on dry ice. At the 60 min timepoint, 1 μL of RQ1 RNase-Free DNase (Promega) was added and allowed to incubate for an additional 30 min. Samples were visualized via EMSA as mentioned above by loading 4.0 μL of each sample per well by descending timepoints, along with controls.
4.4. Precipitation of QD Assemblies.
50 μL samples of assembled QDs (with a final QD concentration of 100 nM) or controls were centrifuged at 4 °C for 5 min at 10, 5, 2.5, or 1 G on a Thermo Scientific Sorvall Legend Micro 21R centrifuge. All precipitates were immediately visualized on a Bio-Rad ChemiDoc MP imaging system.
4.5. Transmission Electron Microscopy.
The QD assemblies were diluted by a factor of 100 in deionized water to a final concentration of ~1 nM for preparing TEM samples. Immediately after dilution, a drop of the QD assembly in solution was allowed to dry on a TEM grid with an ultrathin amorphous carbon support film. Bright-field TEM images were acquired using a FEI Talos F200XG2 microscope with an accelerating voltage of 200 kV. Three representative images (Figure S2) chosen from each of the three methods of QD formation were assessed in ImageJ using the ND ImageJ plugin to calculate center-to-center distances.62,63 After excluding edges, the radius (r1) of each identified QD point was calculated as half the average of the width plus height. The distances from the edge of each QD to its three nearest neighbors (d) were averaged along with the average radii (r2) of the three nearest neighbors using the ND ImageJ plugin. To calculate each center-to-center distance, r1, r2, and d were added together and averaged for the first 100 events in each of the three TEM images. The three average center-to-center distances were then averaged, and the standard error of the mean (SEM) was calculated based on n = 3 images. For visualizing the distribution of center-to-center distances, the first 100 events in each of the images shown in Figure 1 were plotted in a histogram and fit with a Gaussian distribution using GraphPad Prism version 9.0.0 for Windows, GraphPad Software, San Diego, California, USA, www.graphpad.com. As a control, free QDs were analyzed at the same concentration.
4.6. Small-Angle X-ray Scattering.
40 μL of assembled QD samples was loaded into capillary tubes and then sealed with wax. Samples were then measured under vacuum conditions at the Complex Materials Scattering beamlines at the National Synchrotron Light Source II at Brookhaven National Laboratory (Upton, NY). The 2D scattering data were collected on area detectors downstream of the sample. The 2D data were then integrated into one-dimensional I(q) curves as a function of the scattering vector, q. The scattering vector is defined as , where λ and θ are the wavelength of the incident X-rays and the full scattering angle, respectively. The resultant 1D curves span from roughly 0.03 to 1 nm−1 with a resolution of 0.002 nm−1. The experimental S(q) was calculated by dividing the obtained I(q) 1D curves by the form factor or P(q) corresponding particles used in the sample preparation. Additional details of the experimental setup are provided in the Supporting Information. After the S(q) curves were obtained, peaks were fit with a Lorentzian distribution to obtain the center of the peak, which was used to calculate the center-to-center distances (DCC) of the assembled particles, where DCC = 2π/q.
4.7. Dissipative Particle Dynamics.
As already stated, DPD is a proven mesoscopic method for modeling materials such as DNA. For mesoscale simulations, DPD utilizes a coarse-grained approach where groups of atoms, among which their specific interactions are outside the scope of concern, are lumped together to form one bead. The movements of the DPD beads are dictated by Newton’s equations of motion and are subject to a soft potential comprising three non-bonded pairwise components along with a harmonic bonded spring force
| (1) |
where ri, vi, and mi are the position, velocity, and mass of bead i, respectively. The force between the two beads, fij, comprises three non-bonded components including a conservative force, , a dissipative force, , and a random force, . They are resolved by
| (2) |
| (3) |
| (4) |
where aij is the maximum repulsion between the two beads i and j; rij is the magnitude of the distance between i and j; vij = vi − vj, and is the unit vector along j to i. γ and σD are coefficients that determine the amount of the dissipative and random forces, respectively, where
| (5) |
Δt is the timestep, and θij (t) is a symmetric random number. Note that wD and wR are weight functions that must be related as
| (6) |
In addition to the three-component non-bonded force, there is an additional force for the connected beads that form the polymer, . This is a harmonic spring-type interaction that follows
| (7) |
where C is the spring constant.
To use DPD for this specific material, a combination of two approaches was utilized to predict the morphology of the material assembly. Svaneborg demonstrated that dynamic bonding and DPD could be used to model DNA, while Li et al. established that electrostatics could be calculated implicitly using the novel ISIS-DPD.53,54,58 The means of dynamic bonding used in this approach is based on a two-bead model resolution of DNA, where the phosphate and sugar ring comprises one bead and the other is the nucleobase. When two complementary bases are within the cutoff distance of each other, they will form a bond as described in eq 7, ultimately forming the assembled dsDNA. To complement the dynamic bonding, using the ISIS-DPD approach, the salt conditions of the solution can be scanned as a function of the DNA length to predict the resulting morphology. This unique combination of two DPD-centric methods allows for the assembly of these materials to be modeled and provides insights into the role of the solvent’s impact on the morphology.
The computational calculations of DPD were conducted using the molecular dynamics simulator LAMMPS.55 The initial system consists of 24 QDs distributed within a 40rc × 40rc × 40rc periodic box. Each dot is decorated with 12 ssDNAs of varying lengths: 5, 10, 15, and 20 nucleobases. In other words, the system starts initially in a disassembled state and is iterated for 3 million timesteps, resulting in the decorated QDs assembled via complementary base pairing. Of the 24 QDs, 12 were decorated with base pair type A, and 12 were decorated with complementary base pair type B. An example of the initial system can be seen in Figure 3A. The interactions within the system consist of the non-bonded interactions described in eq 1, the bonded interactions within the DNA and complementary base pairs, and two angular harmonic potentials dictated by
| (8) |
where K is 2 times the harmonic prefactor, θ is the angle at the time of calculation, and θ0 is the equilibrium value of the angle. To reflect the physical orientation of DNA, the angles within the backbone beads consist of the phosphate and sugar ring, K = 20 and θ0 = 150°. The angles between the nucleobases and the backbone were K = 15 and θ0 = 100°. These angles were chosen based on mapping the centers of mass of each resolved bead on an atomic weight scale calculation. The bonded interactions established in eq 8 were harmonically set to C = 200 with an equilibrium distance of 0. The aij interactions established in eq 2 are referenced in Table 1. The bead composed of the phosphate and sugar ring is referred to as the backbone bead.
Table 1.
Non-Bonded Interactions
| interaction description | a ij |
|---|---|
| backbone to backbone | varied from 25 to 90 |
| backbone to nucleobases | 40 |
| backbone to QD | 27 |
| backbone to water | 25 |
| nucleobase type A to type A | 22 |
| nucleobase type A to type B | 5 |
| nucleobase to QD | 27 |
| nucleobase to water | 27 |
| water to water | 25 |
Lastly, the QD core was composed of 162 beads that were fix-grouped, meaning that for each QD, the forces exerted on each individual bead were distributed throughout the entire core. This results in a fixed dot that consists of beads moving in concert with each other.
Since computational modeling was conducted, calculations that quantify the morphological differences were conducted, including the int-RDF. The int-RDF is a convenient method to describe the spatial relative positioning between the QDs.
4.8. Cell Culture and Transfection.
The human breast cancer cell line MDA-MB-231(with or without GFP) was maintained in Dulbecco’s modified Eagle medium (DMEM), 10% heat-inactivated fetal bovine serum (FBS), and 1% PenStrep in incubators at 37 °C, 5% CO2. Cells were plated in 35 mm dishes to perform confocal visualization (Ibidi, Germany). Lipofectamine 2000 (L2K) was used for all experiments according to the manufacturer’s guidelines. Briefly, plated cells were transfected for 4 h in Opti-MEM using a 10 nM concentration of the nucleic acid transfected. Upon 4 h, the medium was replaced with DMEM, and the cells were further incubated for 3 days (silencing experiments) or 14 h (uptake experiments). The cells were then washed three times in phosphate-buffered saline and fixed with 4% paraformaldehyde before imaging.
4.9. Uptake and Colocalization.
Experiments were performed using a UV 510 confocal microscope (Carl Zeiss, Oberkochen) and a Plan-Apochromat 63×/1.4 oil lens. To image QD545, a 488 nm laser beam was used for excitation and a BP filter 505–550 nm for detection. To image QD605, a 561 nm laser beam was used for excitation and a BP filter 575–615 nm for detection. All images were taken with a pinhole adjusted to 1 Airy unit. Flow cytometry was performed on a BD Accuri C6 flow cytometer; CellQuest or the CFlow Sampler software was used to obtain the gMFI. Data are expressed as the mean ± SEM for n = 20,000 events per treatment.
4.10. Silencing Assays.
For silencing experiments, cells were visualized using a UV 510 confocal microscope (Carl Zeiss, Oberkochen) and a Plan-Neofluar 40×/1.3 oil lens. To image GFP fluorescence, a 488 nm laser beam was used for excitation, and detection was acquired using a BP filter 505–550 nm. Flow cytometry was performed on a BD Accuri C6 flow cytometer; CellQuest or the CFlow Sampler software was used to obtain the gMFI. To determine statistical significance, treatments were compared to cells-only using a one-way analysis of variance (ANOVA), followed by a Dunnett’s multiple comparisons test performed in GraphPad Prism version 9.0.0 for Windows, GraphPad Software, San Diego, California, USA, www.graphpad.com. Data are expressed as the mean ± SEM for n = 20,000 events per treatment. A P-value of <0.05 was considered statistically significant.
4.11. Immune Stimulation by ELISA.
Dr. Jonathan Karn (Case Western Reserve University) generously provided the human microglia cell line, hμglia. Cells were maintained in DMEM supplemented with 5% FBS and 100 U/mL penicillin-100 μg/mL streptomycin at 37 °C 5% CO2. The immortalized human astrocytic cell line, U87-MG, was obtained from the American Type Culture Collection (ATCC; HTB-14) and maintained in Eagle’s minimum essential media (EMEM) supplemented with 10% FBS and 100 U/mL penicillin-100 μg/mL streptomycin at 37 °C 5% CO2. Cells were transfected with final concentrations of 5, 10, 20, or 50 nM of each sample using lipofectamine 2000 (L2K, Invitrogen) according to the manufacturer’s guidelines. Ligands were incubated for 30 min at room temperature with lipofectamine 2000 before transfection of cells with the indicated samples. Cells were incubated at 37 °C, 5% CO2 for 4 h with the transfection reaction. Afterward, the medium was aspirated and replaced with 1 mL of fresh medium. Cell supernatants were collected at 24 h post transfection. IL-6, IFN-β, IL-1β, and IL-8 were quantified using specific-capture enzyme-linked immunosorbent assays (ELISAs). IL-6 was detected using a rat anti-human IL-6 capture antibody (BD PharMingen, cat # 554543; Clone Mq2-13A5) and a biotinylated rat anti-human IL-6 detection antibody (BD PharMingen, cat# 554546; Clone MQ2-39C3). IFN-β was detected using a polyclonal rabbit anti-human IFN-β capture antibody (Abcam, cat# ab186669) and a biotinylated polyclonal rabbit anti-human IFN-β detection antibody (Abcam, cat# ab84258). IL-8 (R&D systems, cat# DY208) and IL-1β (R&D systems, cat # DY201) were detected using DuoSet ELISA kits. For all specific-capture ELISAs, a bound detection antibody was detected using streptavidin–HRP (BD Biosciences), followed by incubation with a tetramethylbenzidine substrate. The cytokine concentration in cell supernatants was extrapolated from a standard curve generated using a dilution of recombinant cytokines. For each graph, the relative amount of cytokine production was normalized to the L2K-only treatment. To determine statistical significance, treatments were compared to the L2K-only treatment using a one-way ANOVA, followed by Dunnett’s multiple comparisons test performed in GraphPad Prism version 9.0.0 for Windows, GraphPad Software, San Diego, California, USA, www.graphpad.com. Data are expressed as the mean ± SEM for a minimum of three independent experimental replicates. A P-value of <0.05 was considered statistically significant.
4.12. Cell Viability Assay.
To assess the cytotoxicity of QDs, MDA-MB-231 cells and hμglia cells were seeded in 96-well plates in their respective media. Cells were transfected with final concentrations of 5, 10, 20, or 50 nM of each sample using L2K (Invitrogen) according to the manufacturer’s guidelines. QD545s were incubated for 30 min at room temperature with L2K before transfection of cells with the indicated samples. Cells were incubated at 37 °C, 5% CO2 for 4 h with the transfection reaction. Afterward, the medium was aspirated and replaced with fresh medium. Cells were assessed 24 h post-transfection using a CellTiter 96 AQueous one-solution cell proliferation assay (MTS, Promega) according to the manufacturer’s guidelines. For each graph, the relative percent cell viability was normalized to the cells-only treatment. To determine statistical significance, treatments were compared to the cells-only treatment using a one-way ANOVA, followed by a Dunnett’s multiple comparisons test performed in GraphPad Prism version 9.0.0 for Windows, GraphPad Software, San Diego, California, USA, www.graphpad.com. Data are expressed as the mean ± SEM for a minimum of n = 3 independent experimental replicates. A P-value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R01GM120487 and R35GM139587 (to K.A.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This research used resources of the Center for Functional Nanomaterials and the National Synchrotron Light Source II (Complex Material Scattering beamline 11-ID), which are the US DOE Office of Science Facilities at Brookhaven National Laboratory under contract no. DE-SC0012704. O.G. and B.M. were supported by the National Science Foundation under grant no. 1905920. Research reported in this publication was supported by the National Science Foundation award number CMMI-1763025 and a UNC System Inter-institutional Planning Grant. This work was performed in part at the Analytical Instrumentation Facility (AIF) at North Carolina State University, which is supported by the State of North Carolina and the National Science Foundation (award number ECCS-2025064). This work made use of the instrumentation at the AIF, acquired with the support from the National Science Foundation (DMR-1726294). The AIF is a member of the North Carolina Research Triangle Nanotechnology Network (RTNN), a site in the National Nanotechnology Coordinated Infrastructure (NNCI). This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government. This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), Frederick National Laboratory, Center for Cancer Research. M.P.’s involvement in this publication is the result of the project implementation: “Open scientific community for modern interdisciplinary research in medicine (OPENMED)” ITMS2014+: 313011V455 supported by the Operational Program Integrated Infrastructure, funded by the ERDF. The authors would like to thank Prof. Marcus Jones for helpful discussion at the early stages of this work. The authors would also like to thank Jose Castaneda (UNC Charlotte) and Brian Lynch (NCSU) for their contributions to the preliminary work on this project.
ABBREVIATIONS
- QD
quantum dot
- ss
single-stranded
- DS
Dicer substrate
- EMSA
electrophoretic mobility shift assay
- TEM
transmission electron microscopy
- SAXS
small-angle X-ray scattering
- ds
double-stranded
- DCC
center-to-center distance
- DPD
dissipative particle dynamics
- GFP
green fluorescent protein
- DIC
differential interference contrast
- gMFI
geometric mean fluorescence intensity
- SEM
standard error of the mean
- ANOVA
analysis of variance
- ATCC
American Type Culture Collection
- FBS
fetal bovine serum
- L2K
Lipofectamine 2000
- IFN
interferon
- ELISA
enzyme-linked immunosorbent assay
- MEM
minimal essential medium
- EMEM
Eagle’s minimum essential medium
- DMEM
Dulbecco’s modified Eagle medium
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.1c09230.
List of nucleic acid sequences, CMS beamline experimental setup, QD binding assay, TEM images used in analysis, structure factor plots, Lorentzian fits, intensity profiles, confocal microscopy of cell colocalization, flow cytometry of cell silencing up to 14 days, immunostimulation in the U87-MB cell line, cell viability assays, and precipitation of assembled QDs (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsami.1c09230
The authors declare no competing financial interest.
Contributor Information
Morgan Chandler, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, North Carolina 28223, United States;.
Brian Minevich, Department of Chemical Engineering, Columbia University, New York, New York 10027, United States.
Brandon Roark, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, North Carolina 28223, United States.
Mathias Viard, Laboratory of Integrative Cancer Immunology, Frederick National Laboratory for Cancer Research, Leidos Biomedical Research Inc., Frederick, Maryland 21702, United States.
M. Brittany Johnson, Department of Biological Sciences, University of North Carolina at Charlotte, Charlotte, North Carolina 28223, United States.
Mehedi H. Rizvi, Department of Materials Science and Engineering, North Carolina State University, Raleigh, North Carolina 27695, United States
Thomas A. Deaton, Department of Materials Science and Engineering, North Carolina State University, Raleigh, North Carolina 27695, United States
Seraphim Kozlov, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, North Carolina 28223, United States.
Martin Panigaj, Institute of Biology and Ecology, Faculty of Science, Pavol Jozef Safarik University in Kosice, Kosice 04154, Slovak Republic.
Joseph B. Tracy, Department of Materials Science and Engineering, North Carolina State University, Raleigh, North Carolina 27695, United States;.
Yaroslava G. Yingling, Department of Materials Science and Engineering, North Carolina State University, Raleigh, North Carolina 27695, United States;.
Oleg Gang, Department of Chemical Engineering and Department of Applied Physics and Applied Mathematics, Columbia University, New York, New York 10027, United States; Center for Functional Nanomaterials, Brookhaven National Laboratory, Upton, New York 11973, USA;.
Kirill A. Afonin, Nanoscale Science Program, Department of Chemistry, University of North Carolina at Charlotte, Charlotte, North Carolina 28223, United States;.
REFERENCES
- (1).Jooken S; de Coene Y; Deschaume O; Krylychkina O; Verbiest T; Clays K; Callewaert G; Bartic C Quantum Dot-Functionalized Extracellular Matrices for In Situ Monitoring of Cardiomyocyte Activity. ACS Appl. Nano Mater 2020, 3, 6118–6126. [Google Scholar]
- (2).Freeman R; Girsh J; Willner I Nucleic Acid/Quantum Dots (QDs) Hybrid Systems for Optical and Photoelectrochemical Sensing. ACS Appl. Mater. Interfaces 2013, 5, 2815–2834. [DOI] [PubMed] [Google Scholar]
- (3).Yuan Y; Zhang J; An L; Cao Q; Deng Y; Liang G Oligomeric Nanoparticles Functionalized with NIR-Emitting CdTe/CdS QDs and Folate for Tumor-Targeted Imaging. Biomaterials 2014, 35, 7881–7886. [DOI] [PubMed] [Google Scholar]
- (4).Jahed V; Vasheghani-Farahani E; Bagheri F; Zarrabi A; Jensen HH; Larsen KL Quantum Dots-βcyclodextrin-Histidine Labeled Human Adipose Stem Cells-Laden Chitosan Hydrogel for Bone Tissue Engineering. Nanomed. Nanotechnol. Biol. Med 2020, 27, 102217. [DOI] [PubMed] [Google Scholar]
- (5).Rosi NL; Thaxton CS; Mirkin CA Control of Nanoparticle Assembly by Using DNA-Modified Diatom Templates. Angew. Chem., Int. Ed 2004, 43, 5500–5503. [DOI] [PubMed] [Google Scholar]
- (6).Berry V; Rangaswamy S; Saraf RF Highly Selective, Electrically Conductive Monolayer of Nanoparticles on Live Bacteria. Nano Lett. 2004, 4, 939–942. [Google Scholar]
- (7).Dujardin E; Peet C; Stubbs G; Culver JN; Mann S Organization of Metallic Nanoparticles Using Tobacco Mosaic Virus Templates. Nano Lett. 2003, 3, 413–417. [Google Scholar]
- (8).Kahn JS; Minevich B; Gang O Three-Dimensional DNA-Programmable Nanoparticle Superlattices. Curr. Opin. Biotechnol 2020, 63, 142–150. [DOI] [PubMed] [Google Scholar]
- (9).Nykypanchuk D; Maye MM; van der Lelie D; Gang O DNA-Guided Crystallization of Colloidal Nanoparticles. Nature 2008, 451, 549–552. [DOI] [PubMed] [Google Scholar]
- (10).Park SY; Lytton-Jean AKR; Lee B; Weigand S; Schatz GC; Mirkin CA DNA-Programmable Nanoparticle Crystallization. Nature 2008, 451, 553–556. [DOI] [PubMed] [Google Scholar]
- (11).He M; Gales JP; Ducrot É; Gong Z; Yi G-R; Sacanna S; Pine DJ Colloidal diamond. Nature 2020, 585, 524–529. [DOI] [PubMed] [Google Scholar]
- (12).Laramy CR; O’Brien MN; Mirkin CA Crystal Engineering With DNA. Nat. Rev. Mater 2019, 4, 201–224. [Google Scholar]
- (13).Afonin KA; Viard M; Koyfman AY; Martins AN; Kasprzak WK; Panigaj M; Desai R; Santhanam A; Grabow WW; Jaeger L; Heldman E; Reiser J; Chiu W; Freed EO; Shapiro BA Multifunctional RNA Nanoparticles. Nano Lett. 2014, 14, 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Khaled A; Guo S; Li F; Guo P Controllable Self-Assembly of Nanoparticles for Specific Delivery of Multiple Therapeutic Molecules to Cancer Cells Using RNA Nanotechnology. Nano Lett. 2005, 5, 1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Goldsworthy V; LaForce G; Abels S; Khisamutdinov E Fluorogenic RNA Aptamers: A Nano-Platform for Fabrication of Simple and Combinatorial Logic Gates. Nanomaterials 2018, 8, 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Panigaj M; Johnson MB; Ke W; McMillan J; Goncharova EA; Chandler M; Afonin KA Aptamers as Modular Components of Therapeutic Nucleic Acid Nanotechnology. ACS Nano 2019, 13, 12301–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Johnson MB; Chandler M; Afonin KA Nucleic Acid Nanoparticles (NANPs) as Molecular Tools to Direct Desirable and Avoid Undesirable Immunological Effects. Adv. Drug Deliv. Rev 2021, 173, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ke W; Afonin KA Exosomes as Natural Delivery Carriers for Programmable Therapeutic Nucleic Acid Nanoparticles (NANPs). Adv. Drug Deliv. Rev 2021, 113835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Nordmeier S; Ke W; Afonin KA; Portnoy V Exosome Mediated Delivery of Functional Nucleic Acid Nanoparticles (NANPs). Nanomed. Nanotechnol. Biol. Med 2020, 30, 102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Bindewald E; Afonin KA; Viard M; Zakrevsky P; Kim T; Shapiro BA Multistrand Structure Prediction of Nucleic Acid Assemblies and Design of RNA Switches. Nano Lett. 2016, 16, 1726–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Zakrevsky P; Parlea L; Viard M; Bindewald E; Afonin KA; Shapiro BA Preparation of a Conditional RNA Switch. RNA Nanostructures; Springer, 2017; pp 303–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Achenbach JC; Nutiu R; Li Y Structure-Switching Allosteric Deoxyribozymes. Anal. Chim. Acta 2005, 534, 41–51. [Google Scholar]
- (23).Bath J; Green SJ; Turberfield AJ A Free-Running DNA Motor Powered by a Nicking Enzyme. Angew. Chem., Int. Ed. Engl 2005, 44, 4358–4361. [DOI] [PubMed] [Google Scholar]
- (24).Liu X; Lu C-H; Willner I Switchable Reconfiguration of Nucleic Acid Nanostructures by Stimuli-Responsive DNA Machines. Acc. Chem. Res 2014, 47, 1673–1680. [DOI] [PubMed] [Google Scholar]
- (25).Yurke B; Turberfield AJ; Mills AP; Simmel FC; Neumann JL A DNA-Fuelled Molecular Machine Made of DNA. Nature 2000, 406, 605–608. [DOI] [PubMed] [Google Scholar]
- (26).Modi S; Swetha MG; Goswami D; Gupta GD; Mayor S; Krishnan Y A DNA Nanomachine That Maps Spatial and Temporal pH Changes Inside Living Cells. Nat. Nanotechnol 2009, 4, 325–330. [DOI] [PubMed] [Google Scholar]
- (27).Zhou M; Liang X; Mochizuki T; Asanuma H A Light-Driven DNA Nanomachine for the Efficient Photoswitching of RNA Digestion. Angew. Chem., Int. Ed. Engl 2010, 49, 2167–2170. [DOI] [PubMed] [Google Scholar]
- (28).Halman JR; Satterwhite E; Roark B; Chandler M; Viard M; Ivanina A; Bindewald E; Kasprzak WK; Panigaj M; Bui MN; Lu JS; Miller J; Khisamutdinov EF; Shapiro BA; Dobrovolskaia MA; Afonin KA Functionally-Interdependent Shape-Switching Nanoparticles with Controllable Properties. Nucleic Acids Res. 2017, 45, 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ke W; Hong E; Saito RF; Rangel MC; Wang J; Viard M; Richardson M; Khisamutdinov EF; Panigaj M; Dokholyan NV; Chammas R; Dobrovolskaia MA; Afonin KA RNA-DNA Fibers and Polygons with Controlled Immunorecognition Activate RNAi, FRET and Transcriptional Regulation of NF-κB in Human Cells. Nucleic Acids Res. 2019, 47, 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kahn JS; Hu Y; Willner I Stimuli-Responsive DNA-Based Hydrogels: From Basic Principles to Applications. Acc. Chem. Res 2017, 50, 680–690. [DOI] [PubMed] [Google Scholar]
- (31).De Fazio AF; El-Sagheer AH; Kahn JS; Nandhakumar I; Burton MR; Brown T; Muskens OL; Gang O; Kanaras AG Light-Induced Reversible DNA Ligation of Gold Nanoparticle Superlattices. ACS Nano 2019, 13, 5771–5777. [DOI] [PubMed] [Google Scholar]
- (32).Cutler JI; Auyeung E; Mirkin CA Spherical Nucleic Acids. J. Am. Chem. Soc 2012, 134, 1376–1391. [DOI] [PubMed] [Google Scholar]
- (33).Sun D; Gang O DNA-Functionalized Quantum Dots: Fabrication, Structural, and Physicochemical Properties. Langmuir 2013, 29, 7038–7046. [DOI] [PubMed] [Google Scholar]
- (34).Matea C; Mocan T; Tabaran F; Pop T; Mosteanu O; Puia C; Iancu C; Mocan L Quantum Dots in Imaging, Drug Delivery and Sensor Applications. Int. J. Nanomed 2017, 12, 5421–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tikhomirov G; Hoogland S; Lee PE; Fischer A; Sargent EH; Kelley SO DNA-Based Programming of Quantum Dot Valency, Self-Assembly and Luminescence. Nat. Nanotechnol 2011, 6, 485–490. [DOI] [PubMed] [Google Scholar]
- (36).Sun D; Gang O Binary Heterogeneous Superlattices Assembled from Quantum Dots and Gold Nanoparticles with DNA. J. Am. Chem. Soc 2011, 133, 5252–5254. [DOI] [PubMed] [Google Scholar]
- (37).Martynenko IV; Litvin AP; Purcell-Milton F; Baranov AV; Fedorov AV; Gun’ko YK Application of Semiconductor Quantum Dots in Bioimaging and Biosensing. J. Mater. Chem. B 2017, 5, 6701–6727. [DOI] [PubMed] [Google Scholar]
- (38).Gill R; Zayats M; Willner I Semiconductor Quantum Dots for Bioanalysis. Angew. Chem., Int. Ed. Engl 2008, 47, 7602–7625. [DOI] [PubMed] [Google Scholar]
- (39).Michalet X; Pinaud FF; Bentolila LA; Tsay JM; Doose S; Li JJ; Sundaresan G; Wu AM; Gambhir SS; Weiss S Quantum Dots for Live Cells, In Vivo Imaging, and Diagnostics. Science 2005, 307, 538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Opazo F; Eiden L; Hansen L; Rohrbach F; Wengel J; Kjems J; Mayer G Modular Assembly of Cell-Targeting Devices Based on an Uncommon G-Quadruplex Aptamer. Mol. Ther. Nucleic Acids 2015, 4, No. e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wen L; Qiu L; Wu Y; Hu X; Zhang X Aptamer-Modified Semiconductor Quantum Dots for Biosensing Applications. Sensors 2017, 17, 1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Efros AL; Nesbitt DJ Origin and Control of Blinking in Quantum Dots. Nat. Nanotechnol 2016, 11, 661–671. [DOI] [PubMed] [Google Scholar]
- (43).Roark B; Tan JA; Ivanina A; Chandler M; Castaneda J; Kim HS; Jawahar S; Viard M; Talic S; Wustholz KL; Yingling YG; Jones M; Afonin KA Fluorescence Blinking as an Output Signal for Biosensing. ACS Sens. 2016, 1, 1295–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhang DY; Seelig G Dynamic DNA Nanotechnology Using Strand-Displacement Reactions. Nat. Chem 2011, 3, 103–113. [DOI] [PubMed] [Google Scholar]
- (45).Afonin KA; Viard M; Martins AN; Lockett SJ; Maciag AE; Freed EO; Heldman E; Jaeger L; Blumenthal R; Shapiro BA Activation of Different Split Functionalities Upon Re-Association of RNA-DNA Hybrids. Nat. Nanotechnol 2013, 8, 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Qian L; Winfree E Scaling Up Digital Circuit Computation with DNA Strand Displacement Cascades. Science 2011, 332, 1196–1201. [DOI] [PubMed] [Google Scholar]
- (47).Afonin KA; Desai R; Viard M; Kireeva ML; Bindewald E; Case CL; Maciag AE; Kasprzak WK; Kim T; Sappe A; Stepler M; KewalRamani VN; Kashlev M; Blumenthal R; Shapiro BA Co-Transcriptional Production of RNA–DNA Hybrids for Simultaneous Release of Multiple Split Functionalities. Nucleic Acids Res. 2014, 42, 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Afonin KA; Viard M; Tedbury P; Bindewald E; Parlea L; Howington M; Valdman M; Johns-Boehme A; Brainerd C; Freed EO; Shapiro BA The Use of Minimal RNA Toeholds to Trigger the Activation of Multiple Functionalities. Nano Lett. 2016, 16, 1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Español P; Warren P Statistical Mechanics of Dissipative Particle Dynamics. Europhys. Lett 1995, 30, 191. [Google Scholar]
- (50).Warren PB Dissipative Particle Dynamics. Curr. Opin. Colloid Interface Sci 1998, 3, 620–624. [Google Scholar]
- (51).Deaton TA; Aydin F; Li NK; Chu X; Dutt M; Yingling YG Dissipative Particle Dynamics Appr0the Self-Assembly and Morphology of Neutral and Ionic Block Copolymers in Solution. In Foundations of Molecular Modeling and Simulation; Maginn EJ, Errington J, Eds.; Springer: Singapore, 2021; pp 75–100. [Google Scholar]
- (52).Li NK; Fuss WH; Tang L; Gu R; Chilkoti A; Zauscher S; Yingling YG Prediction of Solvent-Induced Morphological Changes of Polyelectrolyte Diblock Copolymer Micelles. Soft Matter 2015, 11, 8236–8245. [DOI] [PubMed] [Google Scholar]
- (53).Walker CL; Lukyanov KA; Yampolsky IV; Mishin AS; Bommarius AS; Duraj-Thatte AM; Azizi B; Tolbert LM; Solntsev KM Fluorescence Imaging Using Synthetic GFP Chromophores. Curr. Opin. Chem. Biol 2015, 27, 64–74. [DOI] [PubMed] [Google Scholar]
- (54).Svaneborg C LAMMPS Framework for Dynamic Bonding and an Application Modeling DNA. Comput. Phys. Commun 2012, 183, 1793–1802. [Google Scholar]
- (55).Plimpton S Fast Parallel Algorithms for Short-Range Molecular Dynamics. J. Comput. Phys 1995, 117, 1–19. [Google Scholar]
- (56).Li NK; Kuang H; Fuss WH; Zauscher S; Kokkoli E; Yingling YG Salt Responsive Morphologies of ssDNA-Based Triblock Polyelectrolytes in Semi-Dilute Regime: Effect of Volume Fractions and Polyelectrolyte Length. Macromol. Rapid Commun 2017, 38, 1700422. [DOI] [PubMed] [Google Scholar]
- (57).Ge W; Li NK; McCormick RD; Lichtenberg E; Yingling YG; Stiff-Roberts AD Emulsion-Based RIR-MAPLE Deposition of Conjugated Polymers: Primary Solvent Effect and Its Implications on Organic Solar Cell Performance. ACS Appl. Mater. Interfaces 2016, 8, 19494–19506. [DOI] [PubMed] [Google Scholar]
- (58).Li NK; Fuss WH; Yingling YG An Implicit Solvent Ionic Strength (ISIS) Method to Model Polyelectrolyte Systems with Dissipative Particle Dynamics. Macromol. Theory Simul 2015, 24, 7–12. [Google Scholar]
- (59).Burmeister AR; Marriott I The Interleukin-10 Family of Cytokines and Their Role in the CNS. Front. Cell. Neurosci 2018, 12, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Chauhan VS; Sterka DG; Gray DL; Bost KL; Marriott I Neurogenic Exacerbation of Microglial and Astrocyte Responses to Neisseria meningitidis and Borrelia burgdorferi. J. Immunol 2008, 180, 8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Afonin KA; Grabow WW; Walker FM; Bindewald E; Dobrovolskaia MA; Shapiro BA; Jaeger L Design and Self-Assembly of siRNA-Functionalized RNA Nanoparticles for Use in Automated Nanomedicine. Nat. Protoc 2011, 6, 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Schneider CA; Rasband WS; Eliceiri KW NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Haeri M; Haeri M ImageJ Plugin for Analysis of Porous Scaffolds Used in Tissue Engineering. J. Open Res. Software 2015, 3, No. e1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


