Abstract
Transcription factors Myf5 and MyoD play critical roles in controlling myoblast identity and differentiation. In the myogenic cell line C2, we have found that Myf5 expression, unlike that of MyoD, is restricted to cycling cells and regulated by proteolysis at mitosis. In the present study, we have examined Myf5 proteolysis through stable transfection of myogenically convertible U20S cells with Myf5 derivatives under the control of a tetracycline-sensitive promoter. A motif within the basic helix-loop-helix domain of Myf5 (R93 to Q101) resembles the “destruction box” characteristic of substrates of mitotic proteolysis and thought to be recognized by the anaphase-promoting complex or cyclosome (APC). Mutation of this motif in Myf5 stabilizes the protein at mitosis but does not affect its constitutive turnover. Conversely, mutation of a serine residue (S158) stabilizes Myf5 in nonsynchronized cultures but not at mitosis. Thus, at least two proteolytic pathways control Myf5 levels in cycling cells. The mitotic proteolysis of Myf5 is unlike that which has been described for other destruction box-dependent substrates: down-regulation of Myf5 at mitosis appears to precede that of known targets of the APC and is not affected by a dominant-negative version of the ubiquitin carrier protein UbcH10, implicated in the APC-mediated pathway. Finally, we find that induction of Myf5 perturbs the passage of cells through mitosis, suggesting that regulation of Myf5 levels at mitosis may influence cell cycle progression of Myf5-expressing muscle precursor cells.
The myogenic regulatory factors (MRFs) are muscle-specific basic helix-loop-helix (bHLH) proteins which play essential roles in determination and differentiation of skeletal muscle cells both in vivo and in model myogenic cell lines (reviewed in reference 41). Ectopic expression of any one of these factors is sufficient to induce myogenic differentiation in various nonmuscle cells (4, 9) via mechanisms leading to cell cycle withdrawal and transactivation of muscle-specific promoters (8, 38). In vivo, expression of either Myf5 or MyoD is required for skeletal muscle lineage determination (34) and both are expressed in proliferating myoblasts in culture (27).
The muscle-specific transactivation functions of MyoD are repressed in proliferating myoblasts by numerous negative influences (reviewed in reference 3). Direct effects of the cell cycle include repression of MyoD function by cyclin D1 (36) in G1 phase, mediated by the direct interaction of MyoD with cyclin-dependent kinase 4 (cdk4) (43). In addition, it has recently been shown that MyoD expression is cell cycle regulated in proliferating C2 muscle cells such that its levels fall during G1 and recover following passage into S phase (24). The regulation of Myf5 factor activity is distinct from that of MyoD, since it does not interact with cdk4 (43) and shows a different periodicity in its expression through the cell cycle (24). Moreover, we have previously shown that Myf5 is regulated during the cell cycle by proteolysis, undergoing accelerated degradation at mitosis by a pathway which may involve the 26S proteasome (27). MyoD has been shown to undergo rapid, constitutive degradation by the ubiquitin-26S proteasome pathway (1) but does not appear to be additionally destabilized by passage into mitosis (27).
Proteolysis mediated by the 26S proteasome is an important mechanism for generating rapid irreversible transitions in diverse cellular processes such as cell cycle progression, signal transduction, and differentiation (reviewed in reference 16). Substrates are marked for degradation by the 26S proteasome by the conjugation of multiple ubiquitin molecules in a pathway requiring three distinct enzyme activities that include the activity of a ubiquitin carrier protein (Ubc, E2) and that of the ubiquitin ligase (E3), which mediates target recognition. Studies of proteolysis in cell cycle progression have so far implicated two major E3 activities in selection of cell cycle-specific substrates. Skp1–Cullin–F-box complex is active throughout the cell cycle, and targeting of substrates is generally dependent on their regulated phosphorylation (30). The anaphase-promoting complex or cyclosome (APC) was identified as a cell cycle-regulated E3 which is specifically activated at mitosis for destruction of mitotic cyclins and other mitotic regulators (23). Destruction of mitotic substrates depends on a loosely conserved motif known as the destruction box (D-box) (13, 22), which is thought to be recognized by the APC, although direct binding of the APC to its substrates has never been demonstrated (reviewed in reference 42).
We were interested in the possibility that the periodic destruction of Myf5 in the cell cycle might constitute a mechanism for regulating its myogenic functions in proliferating cells. In order to pursue this issue, we undertook to identify sequences involved in controlling the stability of Myf5. Here we describe point mutations which identify two distinct pathways regulating the turnover of Myf5. One is involved in constitutive Myf5 degradation and appears homologous to that described for MyoD (37). The other is mitosis specific and disrupted by mutation of a D-box-like motif immediately adjacent to the DNA-binding domain. However, despite the dependence of the mitosis-specific proteolytic pathway on an apparent D-box motif, further experiments indicate that it is not dependent on previously characterized APC activity. Further suggesting the existence of novel D-box-dependent pathways of proteolysis, we show that the onset of Myf5 destruction at mitosis can occur before that of known substrates of D-box-dependent proteolysis and that its precise timing appears to vary from cell to cell.
MATERIALS AND METHODS
Plasmid constructs.
Full-length cDNAs corresponding to mouse Myf5 (gift of S. Tajbahksh) was cloned into the tetracycline-regulatable vector pUHD10-3 (gift of H. Bujard) for transfection into UTA6 cells (see below).
Site-directed mutagenesis of Myf5 was carried out on the Myf5 cDNA cloned into T7plink (pT7βMyf5) by a whole-plasmid PCR based on the ExSite method described by Stratagene Cloning Systems (La Jolla, Calif.). Oligonucleotide primers bearing the required sequence alterations were phosphorylated at their 5′ ends using polynucleotide kinase. One microgram of each primer was used to amplify 300 ng of the plasmid template in the presence of 200 μM each deoxynucleoside triphosphate and 5% dimethyl sulfoxide by 15 cycles of PCR with Advantage cDNA polymerase (CLONTECH Laboratories Inc., Palo Alto, Calif.). Products were then treated with DpnI and with cloned Pfu DNA polymerase (Stratagene Cloning Systems) before ligation with T4 DNA ligase. Mutagenized sequences were subcloned into pUHD10-3-Myf5 as MscI-SphI fragments and confirmed by sequencing.
pJHEAUC+2 (which bears the gene encoding UbcH10) and pJHEAUC-CS2 (UbcH10-C114S) were the gift of J. Ruderman.
Cell culture and transfection.
The U20S-derived UTA6 clone has been previously described (10) and was cultured in a 50:50 (vol/vol) mixture of MCDB 202 medium and DME medium (both from CryoBiosystème, Angoulême, France) containing 10% (vol/vol) fetal calf serum (FCS; Jacques Boy, Reims, France), 500 μg of G418 (GIBCO-BRL) per ml, and 1 μg of tetracycline (Calbiochem, La Jolla, Calif.) per ml. Transfection of UTA6 cells with purified plasmids was carried out by a standard protocol for coprecipitation with calcium phosphate. pUHD10-3Myf5-derived constructs were cotransfected with a plasmid bearing the gene for hygromycin resistance (pCMVhygro, a gift of Frédéric Auradé). Transfected UTA6 cells were cultured for 10 to 12 days in the presence of 170 U of hygromycin B (Calbiochem) per ml, and selected clones were picked and subcultured for analysis of Myf5 induction by immunofluorescence analysis after 3 days with and without tetracycline.
UTA6-Myf5/ clones were maintained as the parental UTA6 cells, with the addition of 170 U of hygromycin B per ml.
For most of the experiments described (see Fig. 1, 2, 3A, and 4), UTA6 and derived clones were plated at 2 × 103 to 3 × 103 cells/cm2 in medium containing 10% FCS and 100 ng of tetracycline per ml. This reduced dose of tetracycline was sufficient to repress activity of the tetracycline-sensitive transactivator tTA and allowed more rapid induction of target genes than was observed in cells grown in 1 μg of tetracycline per ml. After 48 h with 100 ng of tetracycline per ml, cells were rinsed twice in tetracycline-free medium and the culture medium was replaced with medium containing 10% FCS only. For the experiments described in the legends of Fig. 1C and 5, cells were plated directly in the presence or absence of the doses of tetracycline indicated.
FIG. 1.
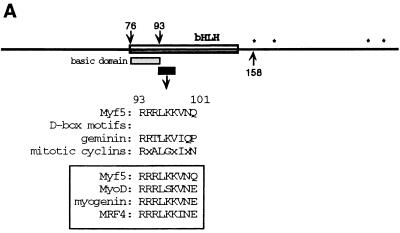
Stabilization of Myf5 at mitosis. (A) Schematic representation of Myf5 showing location of the D-box-like motif. The RXXL motif is present in all members of the MRF family (boxed) and resembles more closely the functional D-box in geminin than those in the mitotic cyclins. ∗, proline-directed serine/threonine residues that are possible phosphorylation sites at mitosis. (B) UTA6-derived cell lines were induced for expression of different versions of Myf5 by withdrawal of tetracycline from the culture medium for 16 h in the presence of nocodazole (to enrich the population of cells in mitosis). Parallel cultures were treated for 2 h with the proteasome inhibitor ALLN prior to preparation of extracts. Immunoblots of protein extracts prepared from the mitotic cells harvested by mitotic shake-off (M) and extracts prepared from the adherent, interphase (I) cells are compared for expression of Myf5. Levels of mitotic cyclins, and levels of transcription factor Sp1 as a loading control, are shown for a representative set of extracts (top). We note that steady-state levels of Myf5 proteins varied between clones, and we selected clones between which levels could most easily be compared. (C) UTA6-derived cell lines were induced for expression of different versions of Myf5 for 5 days. Cells were fixed and stained for expression of a skeletal muscle-specific marker, Troponin T, as described in Materials and Methods. All antibodies are described in Materials and Methods. tet, tetracycline.
FIG. 2.
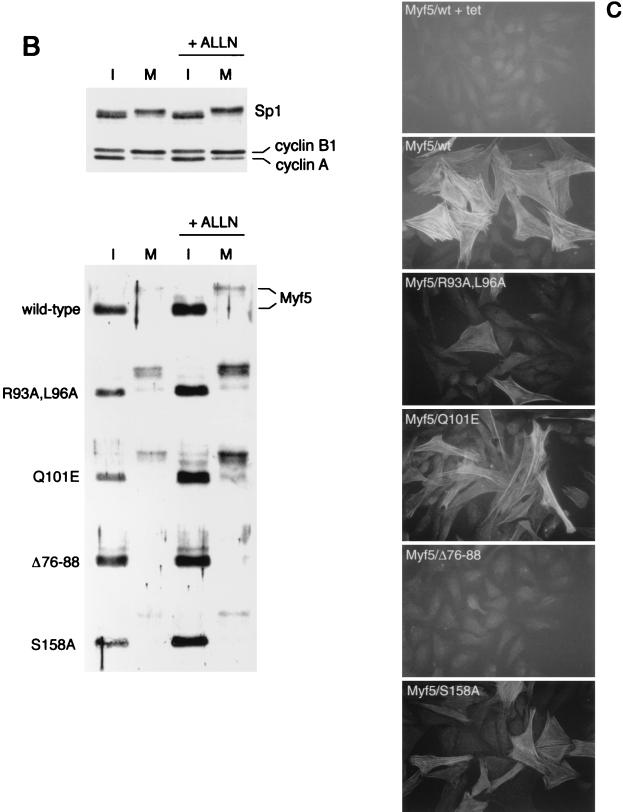
Stabilization of Myf5 in nonsynchronized cells. Cells were induced for 24 h and then CHX was added to cultures to block further protein synthesis. Protein extracts were prepared at the times indicated (in minutes following addition of CHX) and examined for Myf5 levels by immunoblot analysis.
FIG. 3.
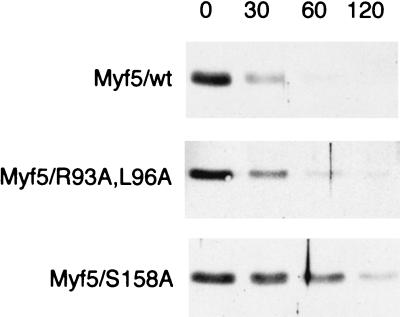
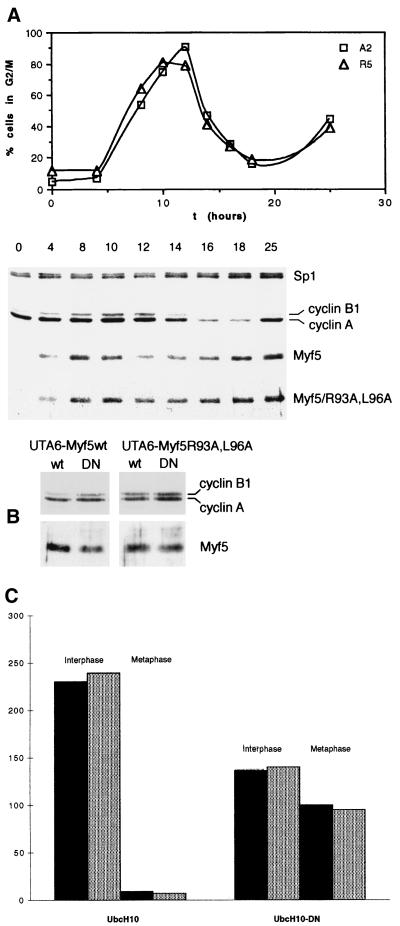
Myf5 proteolysis at mitosis is distinct from that of mitotic cyclins. (A) UTA6-Myf5/ cells were synchronized at the start of S phase by a double thymidine block protocol (Materials and Methods). Cells were simultaneously released from the second thymidine block and induced for expression of Myf5 or Myf5/R93A,L96A. Samples were prepared for flow cytometric analysis of DNA content (top) and immunoblot analyses (bottom) at the times indicated. (B) UTA6-Myf5/wt and UTA6-Myf5/R93A,L96A cells were transfected with expression vectors for wild-type UbcH10 (lanes wt) or a dominant-negative version, UbcH10-DN (lanes DN). Total cell extracts were prepared after 48 h and examined by immunoblot analysis for levels of Myf5 proteins and mitotic cyclins. Transfection efficiencies were assessed by cotransfection with a β-galactosidase-expressing plasmid and were the same for each transfection (approximately 20%), as assessed by in situ staining for β-galactosidase activity in parallel cultures. (C) The transfected cell populations described in the Fig. 3B legend and identified by in situ staining for β-galactosidase activity were examined for the number of mitotic versus interphase figures. At least 250 transfected cells in 10 different fields were scored for each transfection. Black histograms, UTA6-Myf5/wt cells. Gray histograms, UTA6-Myf5/R93A,L96A cells.
FIG. 4.
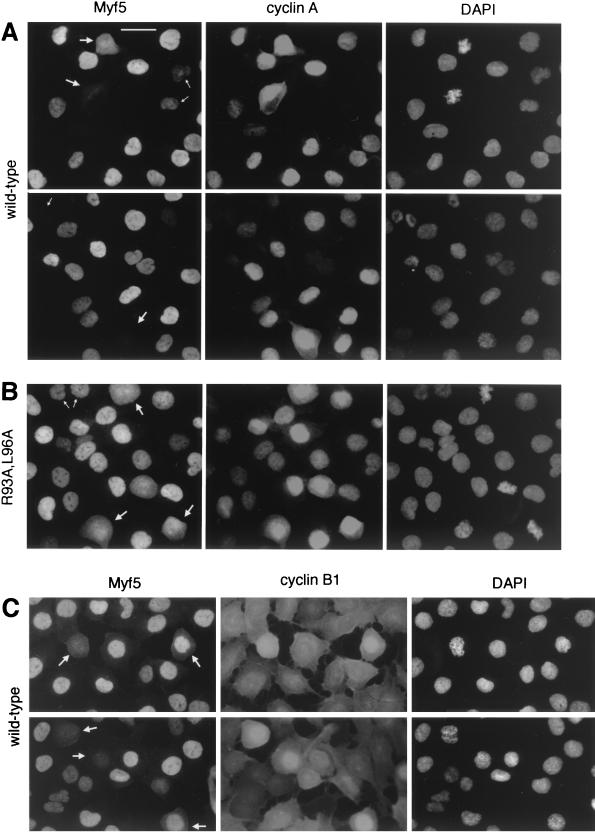
Cell-by-cell analyses of Myf5 levels at mitosis. UTA6-Myf5/ cells were prepared as described for Fig. 3A and, 12 h after release from double thymidine block, were fixed and prepared for immunofluorescence analyses of Myf5 and cyclin A (A and B) or Myf5 and cyclin B1 (C). Bar, 25 μm. (A) UTA6-Myf5/wt cells costained for Myf5, cyclin A, and DAPI. Two fields are shown. Most cells show strong nuclear staining for cyclin A, confirming that the majority of cells are in late S phase, G2, or prophase. The redistribution of cyclin A throughout the cell is observed following nuclear-envelope breakdown at the end of prophase, and its levels decrease gradually during subsequent prometaphase and metaphase (31). Cells with decondensed chromatin and which show no cyclin A staining are presumed to be in early G1 (having completed mitosis), and indeed, these cells systematically occur in pairs (top, small arrows). As expected, there is a clear correlation between the intensities of Myf5 and cyclin A stainings in cells outside of mitosis. That is, Myf5 staining is strongest in cells showing strong nuclear cyclin A staining (but before the onset of nuclear condensation). Cyclin A-negative cells in late mitosis or G1 (bottom and top, respectively; small arrows) typically show faint or no staining for Myf5. By contrast, in prophase and prometaphase cells (large arrows), there is no clear correlation between Myf5 and cyclin A stainings. (B) UTA6-Myf5/R93A,L96A cells costained for Myf5, cyclin A, and DAPI. Small arrows, G1 cells; large arrows, prometaphase cells. (C) UTA6-Myf5/wt cells costained for Myf5, cyclin B1, and DAPI. Cyclin B1 translocates to the nucleus from the cytoplasm during prophase (31). Cells clearly in late prophase or prometaphase (showing nuclear cyclin B1, or in which nuclear-envelope breakdown has occurred) are indicated with arrows.
FIG. 5.
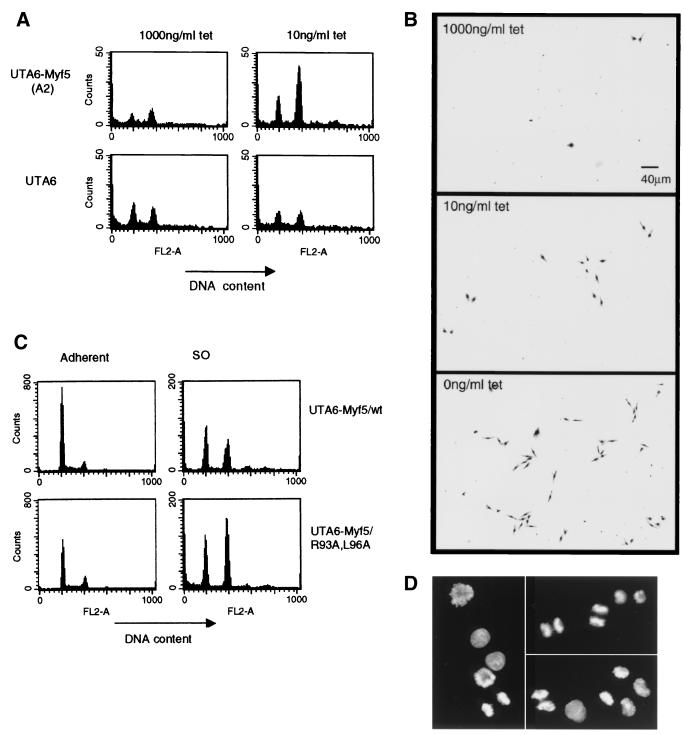
Flow cytometric analyses of Myf5-expressing cells. (A) Parental UTA6 cells or UTA6-Myf5/wt cells were cultured for 3 days at 1,000 or 10 ng of tetracycline (tet) per ml. The total cell population did not vary at these different doses of tetracycline. The culture medium was recovered after gentle shaking of the dish. Detached cells were harvested by centrifugation and prepared for flow cytometric analysis of DNA content. Samples were analyzed with a fixed time parameter to enable comparison of cell numbers in each sample. FL2-A (intensity of propidium iodide fluorescence) indicates relative DNA contents of cells, with a 4N cell population identified by an FL2-A of 400. Since the population of cells harvested resembles mitotic cells by phase-contrast microscopy, we suggest that the 2N fraction seen by flow cytometric analysis consists of cells which complete mitosis during the time of preparation of samples. (B) Detached cell populations harvested from UTA6-Myf5/ cells cultured at the doses of tetracycline indicated for 3 days were replated by transfer of the culture medium to fresh dishes. These cells were cultured for a further 24 h without change of medium and then fixed with 70% ethanol and stained with Giemsa (GIBCO-BRL). (C) Detached cell populations (SO cells) were prepared from UTA6-Myf/wt and UTA6/R93A,L96A cells grown for 3 days with 0 ng of tetracycline per ml. The adherent population of cells from each dish was recovered by trypsinization and prepared for flow cytometric analysis to allow quantification of the total cell population. Histograms from adherent and SO cells prepared with 0 ng of tetracycline per ml are shown (N.B., the adherent samples are diluted 50× compared to the SO samples). (D) Cells from the UTA6-Myf5/R93A,L96A SO population analyzed for panel C, fixed and stained with DAPI. Representative fields show cells in metaphase (left) and anaphase/telophase (top right) and undergoing cytokinesis (lower right).
Transient transfection of UTA6-Myf5/ cells (see Fig. 3B) was carried out using Fugene 6 transfection reagent (Boehringer Mannheim) as recommended by the manufacturer. Expression of Myf5 was induced 1 h before transfection.
Synchronization of UTA6-Myf5/ cells (see Fig. 3A and 4) was achieved by a double thymidine block. Thymidine (2 mM; Sigma Chemical Co., St. Louis, Mo.) was added to cultures a few hours after cells were plated (in the presence of 100 ng of tetracycline per ml) for 24 h. This was followed by 14 h of incubation in thymidine-free culture medium (still in the presence of 100 ng of tetracycline per ml) and another 24-h incubation with 2 mM thymidine. Dishes were rinsed twice to remove thymidine and tetracycline at time zero.
Flow cytometric analysis.
Cells for analysis by flow cytometry were harvested by trypsinization, (except for the mitotic cells collected by “shake-off” shown in Fig. 5). Cells were washed once in ice-cold phosphate-buffered saline (PBS) and fixed for at least 24 h in 70% ethanol at 4°C. Cells were resuspended in PBS containing 50 μg of RNase A (Boehringer Mannheim) per ml and 10 μM propidium iodide (Sigma Chemical Co.), incubated for 1 h, and analyzed for propidium iodide fluorescence and/or cell number using a FACStar Plus cytometer (Becton Dickinson, Mountain View, Calif.).
Immunoblot analyses.
Extracts were prepared and analyzed as described previously (27). Myf5 polyclonal antiserum raised against the C-terminal peptide of Myf5 has been described previously (27) and was used at a dilution of 1:1,000. Cyclin A monoclonal antibody (clone CY-A1; Sigma Chemical Co.) was used at a 1:200 dilution. Cyclin B1 monoclonal antibody (clone V152; Neomarkers, Union City, Calif.) was used at 1:100. Sp1 polyclonal antibody (PEP2; Santa Cruz Biotechnology, Santa Cruz, Calif.) was used at 1:500.
Immunocytochemical analyses.
Cells were fixed with paraformaldehyde, permeabilized, and incubated with antibodies essentially as described previously (27). For Troponin T staining (see Fig. 1C), cells were incubated with mouse monoclonal antibody (clone JLT-12; Sigma Chemical Co.) at a 1:200 dilution followed by Alexa488-coupled goat anti-mouse antibody (Molecular Probes Inc., Eugene, Oreg.) at a 1:200 dilution. For Myf5-cyclin double labeling (see Fig. 4), cells were incubated with Myf5 antibody at a 1:1,000 dilution followed by Alexa488-coupled goat anti-rabbit antibody (Molecular Probes Inc.) at a 1:200 dilution. Following Myf5 staining, cells were refixed in 4% (wt/vol) paraformaldehyde for 5 min, washed in PBS, and then fixed in an ice-cold 1:1 (vol/vol) mixture of methanol and acetone. Cells then underwent sequential incubation with cyclin A or cyclin B1 antibody (1:200), biotin-coupled goat anti-mouse antibody (Sigma Chemical Co.), and Alexa594-coupled streptavidin (Molecular Probes, Inc.). Cells were rinsed briefly in PBS containing 4,6-diamidino-2-phenylindole (DAPI; Sigma Chemical Co.) and then mounted in Mowiol (Calbiochem) under glass coverslips, viewed, and photographed with a Zeiss Axiophot fluorescence microscope.
RESULTS
Our previous results demonstrated that Myf5 undergoes a mitosis-specific proteolytic degradation associated with phosphorylation of the protein (27). Preliminary study of the degradation of Myf5 in vitro using Xenopus ovocyte extracts (C. Lindon, unpublished data) indicated that removal of the bHLH domain protected Myf5 against mitotic degradation. Removal of the C-terminal domain, while abolishing the phosphorylation-associated mobility shift of Myf5 in mitotic extracts, had no such effect. Therefore, we investigated sequences within the bHLH domain which might regulate the mitotic stability of Myf5. Among these, we noticed a sequence adjacent to the basic domain of Myf5 which resembles the D-box motif (Fig. 1A). Since the D-box is required for substrate targeting in APC-mediated proteolysis (7, 11, 20, 22, 29, 35), we investigated whether this motif might play a role in regulating Myf5 stability.
Forced expression of Myf5 at readily detectable levels is frequently incompatible with proliferation (4; our unpublished observations). Therefore, we selected a system permitting conditional expression of stably transfected cDNA: the U20S osteosarcoma-derived clone UTA6 (10), which expresses the tetracycline-regulated transactivator tTA (14) and which we have previously found to express target genes in a highly inducible and dose-dependent fashion (2). U20S cells have previously been shown to undergo myogenic conversion in response to MyoD (15), and we find that the U20S-derived UTA6 cells undergo myogenic conversion when Myf5 expression is induced for more than 48 h (Fig. 1C and unpublished data). We transfected wild-type Myf5 (Myf5/wt) and other versions of Myf5 bearing point mutations in the bHLH region into UTA6 cells in the presence of tetracycline. After the cells were subjected to growth in selection medium, we identified clones in which the products of the transfected cDNAs were strongly expressed when tetracycline was withdrawn from the culture medium but which could not be detected by immunocytochemical or immunoblot analysis of cells grown in the presence of 1 μg of tetracycline per ml.
A D-box-like motif in the bHLH domain of Myf5 regulates Myf5 stability in mitotic cells.
We examined the stability of different versions of Myf5 in mitotic cells. Expression of Myf5 proteins was induced in UTA6-Myf5 clones in the presence of nocodazole. Nocodazole prevents formation of the mitotic spindle, causing cells to arrest in a metaphase-like state. Under these conditions, cyclin B1 is stable and cyclin A is degraded (Fig. 1B, top). Extracts were prepared both from the population of cells arrested in mitosis (M) and from the remaining nonmitotic cells (I). Parallel cultures were treated for 2 h with N-acetyl-Leu-Leu-norleucinal (ALLN), an inhibitor of protein degradation by the proteasome, before preparation of extracts. Immunoblot analysis of these extracts reveals that ectopically expressed Myf5/wt is absent from nocodazole-blocked U20S cells. Following treatment of cells with ALLN, Myf5 was detectable in mitotic extracts as a more slowly migrating form (Fig. 1B). This pattern is similar to that observed for endogenous Myf5 in C2-derived myoblasts, with which cells we demonstrated that the change in mobility of Myf5 from ALLN-treated mitotic cells is phosphorylation dependent (27).
Figure 1B shows that substitution of residues within the 9-amino-acid D-box-like motif has a significant effect on the mitotic stability of Myf5. Replacement of the minimum D-box consensus sequence RXXL by AXXA created a version of Myf5 (Myf5/R93A,L96A) readily detectable (in phosphorylated form) in extracts from mitotic cells without ALLN pretreatment (Fig. 1B). This result suggests that the R93-Q101 motif is a functional D-box. This putative D-box motif is highly conserved between the different members of the MRF family (Fig. 1A), yet it does not function as a D-box for MyoD, which is stable at mitosis (27), and is also unlikely to do so for the other members of the family, whose expression is associated with a differentiated state. The only position at which Myf5 differs from all other members of the family is at the final position of the putative 9-residue motif. We tested the possibility that this residue, Q101, contributes to the mitotic instability of Myf5 by substituting a glutamate (E) at this position, as is found in the other MRFs. We found that this version of Myf5 is also more stable than the wild-type protein in mitotic cells (Fig. 1B, Q101E), consistent with the ascription of Q101 to a putative D-box motif. Since the putative D-box lies within a region of the Myf5 protein implicated in DNA binding, we examined whether mutations in this region might affect Myf5 stability indirectly, through modification of its DNA-binding-associated functions. Figure 1C illustrates the myogenic conversion of U20S cells in response to expression of each of the mutant versions tested for their stability at mitosis (shown in Fig. 1B). Indeed, we have found that Myf5/R93A,L96A does show reduced activity in myogenic conversion assays (Fig. 1C), consistent with a reduced affinity for E-box sequences (unpublished data). However, Myf5/Q101E, which is also stabilized in mitotic cells, showed no loss in capacity to induce myogenic conversion (Fig. 1C). Thus, the mitotic stability of the different versions of Myf5 does not correlate with their myogenic activities. Moreover, we find that abrogation all myogenic activity does not increase the mitotic stability of Myf5 (Fig. 1B and C, Δ76-88).
Finally, we examined the appearance in mitotic extracts of a version of Myf5 analogous to MyoD/S200A, since it has been shown that S200 of MyoD is a site for phosphorylation by cdk's (25) and required for rapid turnover of the protein (25, 37). We find that the corresponding residue (S158) in Myf5, which does indeed regulate constitutive turnover (see below), is neither required for phosphorylation of Myf5 at mitosis nor involved in its mitotic destruction (Fig. 1B, S158A).
These data identify a D-box-like motif in Myf5 whose integrity is essential to the mitotic destruction of Myf5.
Myf5 stability is regulated by distinct pathways in mitotic and interphase cells.
We investigated whether the constitutive turnover of Myf5 was altered by mutation of the putative D-box motif. We assessed the turnover of different versions of Myf5, by addition of cycloheximide (CHX) to nonsynchronized cultures 24 h after induction of the proteins and immunoblot analysis of extracts prepared over a 2-h time course following CHX treatment (Fig. 2). Under these conditions, wild-type Myf5 is virtually undetectable within 1 h of CHX treatment (Fig. 2). The half-life of Myf5/R93A,L96A is similar to that of the wild-type protein. By contrast, Myf5/S158A appears considerably more stable, as has been described for the corresponding version of MyoD (25, 37). These results show that Myf5 is destabilized during interphase by phosphorylation on S158 but that the pathway of degradation disrupted by mutation in the R93-Q101 region is specific to mitotic cells. Thus, the proteolysis of Myf5 is regulated by distinct pathways in interphase and mitosis.
In addition, we note that Myf5/R93A,L96A in mitotic cells is further stabilized by the presence of ALLN and that Myf5/S158A is similarly stabilized in interphase cells (Fig. 1B). This finding indicates that these point mutations do not identify all of the determinants of Myf5 stability and that additional pathways target Myf5 for proteolysis.
The timing and mechanism of Myf5 destruction at mitosis appear distinct from those of known substrates of mitotic proteolysis.
The nocodazole block assay for mitotic instability does not indicate when degradation begins, and we had observed that the reduction in Myf5 levels in nocodazole-blocked cells was even more marked than the reduction in cyclin A levels (Fig. 1B), suggesting that the timings of their destructions might be different. In order to examine more precisely the onset of Myf5 instability at mitosis, we synchronized Myf5/wt and Myf5/R93A,L96A cells for progress through the cell cycle by release from a double thymidine block. Since Myf5 induces significant G1 arrest in U2OS cells (data not shown), Myf5 expression was induced at the time of release from the G1/S block to ensure that most, if not all, cells in the culture pass through mitosis.
We prepared cell extracts at various times after release of cultures from the early S-phase block and Myf5 induction and examined expression of Myf5 and mitotic cyclins by immunoblot analyses (Fig. 3A). Myf5 levels were found to fall before those of the endogenous mitotic cyclins. Destruction of cyclins A and B1 correlates with the exit of cells from mitosis 12 to 14 h following release from the double thymidine block. The fall in Myf5/wt levels 10 to 12 h after release from the block suggests that mitotic degradation of Myf5 begins before that of the mitotic cyclins and at a time which may correspond to the entry of cells into mitosis. However, we cannot exclude the possibility that generalized protein synthesis inhibition at mitosis (32), coupled to its short constitutive half-life, contributes to the early disappearance of Myf5 at mitosis. The level of Myf5/R93A,L96A protein is more stable than that of the wild-type protein, although (as also indicated by the nocodazole block assay [Fig. 1B]) there is still a decrease in the level of the protein between S phase and G2/M. Levels of Myf5 proteins recover rapidly after mitosis, unlike those of mitotic cyclins, whose degradation continues in G1 (6). Therefore, the timing of Myf5 degradation, although dependent on a D-box-like motif, appears distinct from that of known substrates of the APC, of which cyclin A is the only example shown to be degraded before the end of metaphase.
We investigated more directly whether the APC might be implicated in Myf5 proteolysis. We transiently transfected UTA6-Myf5 clones with wild-type and dominant-negative versions of the ubiquitin carrier protein UbcH10, originally identified as the human homologue of cyclin-selective E2 from clam ovocytes (39). The dominant-negative E2 (UbcH10-DN) blocks destruction of all APC substrates which have been tested (5) and arrests cells at the metaphase-anaphase transition (39). Total cell extracts prepared from UbcH10-DN-transfected cells contained somewhat elevated levels of cyclins A and B1 (Fig. 3B, top), as described by Townsley et al. (39), and we verified that these elevated levels correspond to an efficient metaphase block of transfected cells (Fig. 3C). In contrast, we found that Myf5 is not stabilized by the presence of UbcH10-DN (Fig. 3B, bottom). Moreover, we found a slight decrease in the level of Myf5/wt (but not in that of Myf5/R93A,L96A) in extracts from UbcH10-DN-transfected cells. This result is consistent with an increased rate of Myf5/wt degradation in metaphase-blocked cells. Thus, while Myf5 mitotic degradation depends upon the integrity of a D-box-like motif, the pathway of its proteolysis appears distinct with respect to both timing and mechanism from that of known APC targets.
Mitotic destruction of Myf5 is not synchronized.
We carried out a cell-by-cell analysis of Myf5 levels in order to further understand the timing of its destruction at mitosis. Populations of cells were prepared for immunocytochemical analysis 12 h after release from the double thymidine block (when the maximum number of cells have a 4N DNA content [Fig. 3A]) and stained simultaneously for Myf5 and cyclin A (Fig. 4A and B) or for Myf5 and cyclin B1 (Fig. 4C).
We found a clear correlation between the intensities of Myf5/wt and cyclin A stainings in cells which were in S, G2, or G1 phase (Fig. 4A). However, when we examined the different phases of mitosis, we found Myf5 staining to be highly variable. Approximately half of cells in prophase or prometaphase showed a complete absence of Myf5 staining. This heterogeneous pattern of Myf5 staining before metaphase does not result solely from a gradual loss of Myf5, since prophase cells (Fig. 4A, bottom) may stain more weakly than prometaphase cells (Fig. 4A, top). Cells showing strong staining for Myf5 could indeed be found at both prophase and prometaphase (Fig. 4A, top), and a fraction of metaphase cells were also positive for Myf5. This observation suggests that the onset of Myf5 destruction at mitosis is not a strictly synchronized event. As expected, Myf5/R93A,L96A was found to give a more homogeneous staining pattern: the protein was detected at similar levels in G2, prophase, and metaphase cells that stained for Myf5 (Fig. 4B), although cells showing no Myf5 staining could also be found in all phases. A statistical analysis of the distribution of Myf5-positive cells through mitosis in the populations illustrated is shown in Table 1.
TABLE 1.
| Phase | % Myf5-positive cells
|
|
|---|---|---|
| Wild type (A2) | R93A,L96A (R5) | |
| S/G2 | 86 | 88 |
| M (prophase) | 58 | 80 |
| M (prometaphase/metaphase/anaphase) | 36 | 72 |
| M (telophase)/G1 | 36 | 75 |
At least 200 UTA6-Myf5/wt cells (clone A2) or 120 UTA6-Myf5/R93A,L96A cells (clone R5) were counted for each phase, which was identified by the combination of cyclin A and DAPI staining patterns.
We also compared the pattern of Myf5 staining at mitosis to that of cyclin B1 (Fig. 4C). The cellular distribution of cyclin B1 is distinct from that of cyclin A, since it translocates from the cytoplasm to the nucleus following the onset of mitosis (31). Examination of Myf5 staining in cells showing nuclear cyclin B1 staining confirmed that prometaphase cells are more likely to be Myf5 negative than prophase cells (Fig. 4A, top) but, again, indicated that Myf5 staining is highly variable in early prophase (and does not correlate with the nuclear translocation of cyclin B1 [Fig. 4C, bottom]). Myf5/R93A,L96A-negative cells were present at all stages of mitosis, but in general, mitotic cells in this population showed stronger staining with the Myf5 antibody than those in the population expressing wild-type protein (data not shown).
These results do not allow us to distinguish between the possibilities that (i) Myf5 destruction occurs throughout prophase and prometaphase at a rate that varies from cell to cell and (ii) Myf5 destruction at mitosis is rapid but initiated at different moments throughout mitosis. However, from the images presented in Fig. 4, it is clear that—in cells where Myf5 undergoes mitotic degradation—the onset of this proteolysis can occur in prophase or earlier.
In conclusion, we find that the destruction of Myf5 accompanies entry into mitosis but is heterogeneous, suggesting that additional signals, and not only progression through the cell cycle, regulate Myf5 destruction at mitosis.
Myf5 perturbs the passage of cells through mitosis.
We noticed that induction of UTA6-Myf5 clones raised the apparent mitotic index of cultures, even at levels of Myf5 expression where there was no measurable effect on the overall rate of proliferation (10 ng of tetracycline per ml [data not shown]). Since many UTA6 cells detach from the substratum, or remain loosely attached, at mitosis, this population of cells can be harvested from the culture medium. We found that induction of Myf5/wt even at low levels of expression (equivalent to that of the endogenous protein in C2 myoblast cells [data not shown]) led to a threefold increase in the number of mitotic (4N) cells collected from the culture medium but that no such effect occurred in the parental UTA6 cell line (Fig. 5A). We have confirmed that this increased mitotic population represents viable cells, which reattach to the substratum and proliferate when replated (Fig. 5B). Thus, Myf5 perturbs the passage of cells through mitosis, even at low levels of expression. This effect is even more marked at higher levels of Myf5/wt expression (0 ng of tetracycline per ml) (Fig. 5B) but difficult to quantify due to the simultaneous G1 arrest induced by Myf5/wt at this level of expression (data not shown).
We compared the effects of Myf5/wt and Myf5/R93A,L96A on the yield of 4N cells collected. We found that Myf5/R93A,L96A had a stronger effect (Fig. 5C and Table 2). This effect is presumably direct (that is, does not rely on transactivation properties of Myf5), since Myf5/R93A,L96A shows a gain of function with respect to accumulation of 4N cells (Fig. 5C) but partial loss of function with respect to DNA binding and transactivation properties (unpublished data). Moreover, we have observed this effect in cells expressing Myf5/Δ76–88, which shows no DNA-binding activity at all (data not shown).
TABLE 2.
Relative cell numbers in shake-off and adherent populations of clones A2 and R5 analyzed by flow cytometry
| Tetracycline concn (ng/ml) | No. of cells counted/sa
|
|||
|---|---|---|---|---|
| Myf5/wt (A2)
|
Myf5/R93A,L96A (R5)
|
|||
| Adherent | SO | Adherent | SO | |
| 100 | 6.7 | 10.9 | 4.2 | 3.9 |
| 0 | 5.2 | 36.3 | 3.8 | 58.1 |
Adherent samples were diluted 1,000-fold compared to SO samples. SO, shake-off cells.
We examined the population of 4N cells stained with DAPI, in order to assess whether the expression of Myf5, or its stable derivative, might interfere with the degradation of substrates controlling mitosis progression; competition for intracellular ubiquitin and other components of APC-mediated proteolysis has been shown to limit entry into anaphase in the presence of excess APC substrate (17, 40). However, we found 4N cells in all phases of mitosis (representative fields of UTA6-Myf5/R93A,L96A cells are shown [Fig. 5D]), including many cells in anaphase and telophase. These observations suggest that the presence of Myf5 in cells does not perturb mitosis by titrating components required for APC-mediated proteolysis but does interfere with some other aspect of progression in mitosis.
We have been unable to detect any sufficiently significant differences in the overall timings of mitosis between synchronized cultures of UTA6, UTA6-Myf5/wt, and UTA6-Myf5/R93A,L96A cells (Fig. 3A and data not shown) or in their rates of proliferation to explain the apparent threefold increase in mitotic cell number.
Thus, ectopic expression of Myf5 in U20S cells perturbs these cells in mitosis by a mechanism that remains to be defined. We do not know at present if the increased population of mitotic cells harvested from induced cultures corresponds to those which do not down-regulate Myf5 before or during prophase (Fig. 4). However, these observations suggest that the disappearance of Myf5 early in mitosis, a regulated event itself, controls some aspect of passage through mitosis.
DISCUSSION
In this paper we show that at least two distinct mechanisms regulate the turnover of the MRF Myf5 in cycling cells. One of these mechanisms operates specifically at mitosis, giving rise to the destruction of Myf5 which we have observed in myoblasts blocked at mitosis (27). Those previous results, showing that Myf5 is detected in phosphorylated form in extracts from mitotic cells treated with a proteasome inhibitor (27), suggested that Myf5 may be targeted for destruction by a phosphorylation state specific to mitosis. Here we show that, in addition to any requirement for phosphorylation of the protein, destruction of Myf5 at mitosis is sensitive to mutation of a motif in its “hinge” region (between the basic and bHLH domains) which resembles the D-box implicated in mitotic destruction of substrates of the APC (7, 11, 20, 22, 29, 35). The putative D-box motif (minimal consensus RXXLXXXXX) in Myf5 is identical in four of nine residues with the D-box of the S-phase inhibitor geminin (29). Substitution of the consensus residues impairs M-phase destruction of Myf5 in U2OS cells. Significantly, a version of this motif mutated by substitution of a single residue (Q101) to resemble more closely the homologous motif present in MyoD also has a stabilizing effect on Myf5 at mitosis. This finding suggests that sequence differences within the hinge region of the MRFs might determine their stability at mitosis. Since the hinge region is thought to be important for DNA-binding and transactivation properties of the MRFs (26), this increased stability may accompany modified interactions of Myf5 with DNA and/or protein targets, although we have not found a direct correlation between DNA-binding affinity and stability at mitosis of different versions of Myf5.
Although D-box-mediated proteolysis is thought to be the hallmark of APC activity, we find that the degradation of Myf5 in mitotic cells involves a mechanism distinct from that which regulates known substrates of the APC. Not only does the onset of Myf5 destruction precede that of known substrates, but Myf5 is also not stabilized in the presence of a dominant-negative version of an E2 activity (UbcH10) involved in APC-mediated destruction of several mitotic substrates, including cyclins A and B1 and geminin (5). Thus, the D-box-like motif we have described may participate in recognition of phosphorylated Myf5 by a different ubiquitin ligase activity. On the other hand, there is some evidence that the APC may be active in a broader context than is usually described: the D-box-dependent degradation of budding yeast Cdc25p has been shown to be cell cycle independent (21), and APC activity in postmitotic neurons has recently been reported (12). In conclusion, it appears that D-box motifs can be recognized at times outside of the metaphase-G1 window by proteolytic pathways which may or may not involve APC activity. It remains to be shown whether Myf5 destruction at mitosis requires some form of the APC; this question awaits detailed biochemical analysis of Myf5 degradation. The turnover of the MyoD protein is regulated by the ubiquitin-26S proteasome pathway (1) and is sensitive to phosphorylation on serine residue S200 (25, 37). Although there is reduced sequence conservation between MyoD and Myf5 in this region of the proteins, we altered a serine residue in Myf5 (S158) which appeared to lie in a homologous position when the downstream conserved region of the proteins was taken as a reference. We find that although this mutation does strongly diminish the turnover of Myf5 in nonsynchronized cultures, it does not allow accumulation of the protein in mitotic cells. Thus, at least two pathways exist by which Myf5 is degraded. A high level of turnover is superimposed by a distinct, D-box-dependent mechanism at G2/M.
Are there functional implications to the instability of Myf5? The idea that degradation of Myf5 is a highly regulated event is consistent with the idea that Myf5 destruction may be a mechanism for regulation of its functions in determination and differentiation. Indeed, a specific defect in differentiation of a clonal cell line isolated from C2 has been correlated with the increased rate of MRF turnover in these myoblasts (18). Several transcription factors have been reported to be phosphorylated at mitosis (reference 28 and references therein); this is shown to correlate with a loss of the DNA-binding activities of these factors in vitro (see, for example, reference 33) or their exclusion from condensed chromatin in mitotic cells (28) and may be a mechanism to enable the reprogramming of target promoters as cells enter G1 (discussed in references 19 and 28). Since Myf5 is expressed predominantly in proliferating cells where its presumed target genes are silent, perhaps the absence of Myf5 during M phase is critical to prevent reprogramming of muscle-specific E-box-dependent promoters in cells which have not yet received a signal to differentiate. Transcriptional targets of Myf5 prior to the onset of differentiation have not been identified, and its functions in proliferating myoblasts—if any—are unknown. Here we show that Myf5 perturbs cells in mitosis, leading to the accumulation of detached mitotic cells. This effect of Myf5 does not appear to result from any measurable accelerated entry into, delayed exit from, or decreased viability during mitosis. We are investigating the possibility that Myf5 might influence the attachment of mitotic cells. Together with our recent observation that Myf5 plays a positive role in the proliferation of primary myoblast cultures (D. Montarras, C. Lindon, P. Domeyne, and C. Pinset, unpublished data), these results indicate that novel functions of Myf5 in the cell cycle might control the balance between proliferation and differentiation in determined myoblasts.
ACKNOWLEDGMENTS
We thank Christoph Englert for UTA6 cells; Shahragim Tajbakhsh for Myf5 cDNA; Hermann Bujard, Frédéric Auradé, and Joan Ruderman for plasmids; Olivier Coux, Gustavo Gutierrez, Thierry Lorca, and Eric Karsenti for insightful comments during the course of this work; and Olivier Coux and Gustavo Gutierrez for critical reading of the manuscript.
This work was supported by the Association Française contre les Myopathies (AFM) and by grants to C.L. from the AFM and the Fondation pour la Recherche Medicale (FRM).
REFERENCES
- 1.Abu Hatoum O, Gross-Mesilaty S, Breitschopf K, Hoffman A, Gonen H, Ciechanover A, Bengal E. Degradation of myogenic transcription factor MyoD by the ubiquitin pathway in vivo and in vitro: regulation by specific DNA binding. Mol Cell Biol. 1998;18:5670–5677. doi: 10.1128/mcb.18.10.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albagli O, Lantoine D, Quief S, Quignon F, Kerckaert J, Montarras D, Pinset C, Lindon C. Overexpressed BCL6 (LAZ3) oncoprotein triggers apoptosis, delays S phase progression and associates with replication foci. Oncogene. 1999;18:5063–5075. doi: 10.1038/sj.onc.1202892. [DOI] [PubMed] [Google Scholar]
- 3.Arnold H-H, Winter B. Muscle differentiation: more complexity to the network of myogenic regulators. Curr Opin Genet Dev. 1998;8:539–544. doi: 10.1016/s0959-437x(98)80008-7. [DOI] [PubMed] [Google Scholar]
- 4.Auradé F, Pinset C, Chafey P, Gros F, Montarras D. Myf5, MyoD, myogenin and MRF4 myogenic derivatives of the embryonic mesenchymal cell line C3H10T1/2 exhibit the same adult muscle phenotype. Differentiation. 1994;55:185–192. doi: 10.1046/j.1432-0436.1994.5530185.x. [DOI] [PubMed] [Google Scholar]
- 5.Bastians H, Topper L M, Gorbsky G, Ruderman J V. Cell cycle-regulated proteolysis of mitotic target proteins. Mol Biol Cell. 1999;10:3927–3941. doi: 10.1091/mbc.10.11.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandeis M, Hunt T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 1996;15:5280–5289. [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen-Fix O, Peters J-M, Kirschner M, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 8.Crescenzi M, Fleming T, Lassar A, Weintraub H. MyoD induces growth arrest independent of differentiation in normal and transformed cells. Proc Natl Acad Sci USA. 1990;87:8442–8446. doi: 10.1073/pnas.87.21.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis R L, Weintraub H, Lassar A B. Expression of a single transfected cDNA converts fibroblast to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 10.Englert C, Hou X, Maheswaran S, Bennett P, Ngwu C, Re G G, Garvin A J, Rosner M R, Haber D A. WT1 suppresses synthesis of the epidermal factor receptor and induces apoptosis. EMBO J. 1995;14:4662–4675. doi: 10.1002/j.1460-2075.1995.tb00148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funabiki H, Yamano H, Nagao K, Tanaka H, Yasuda H, Hunt T, Yanagida M. Fission yeast Cut2 required for anaphase has two destruction boxes. EMBO J. 1997;16:5977–5987. doi: 10.1093/emboj/16.19.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gieffers C, Peters B H, Kramer E R, Dotti C G, Peters J-M. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc Natl Acad Sci USA. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glotzer M, Murray A M, Kirschner M W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 14.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu W, Schneider J, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 16.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 17.Holloway S L, Glotzer M, King R W, Murray A W. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;73:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- 18.Horwitz M. Hypermethylated myoblasts specifically deficient in MyoD autoactivation as a consequence of instability of MyoD. Exp Cell Res. 1996;226:170–182. doi: 10.1006/excr.1996.0216. [DOI] [PubMed] [Google Scholar]
- 19.John S, Workman J L. Bookmarking genes for activation in condensed mitotic chromosomes. Bioessays. 1998;20:275–279. doi: 10.1002/(SICI)1521-1878(199804)20:4<275::AID-BIES1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 20.Juang Y-L, Huang J, Peters J-M, McLaughlin M E, Tai C-Y, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- 21.Kaplon T, Jacquet M. The cellular content of Cdc25p, the Ras exchange factor in Saccharomyces cerevisiae, is regulated by destabilization through a cyclin destruction box. J Biol Chem. 1995;270:20742–20747. doi: 10.1074/jbc.270.35.20742. [DOI] [PubMed] [Google Scholar]
- 22.King R W, Glotzer M, Kirschner M W. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King R W, Peters J-M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 24.Kitzmann M, Carnac G, Vandromme M, Primig M, Lamb N J C, Fernandez A. The muscle regulatory factors MyoD and Myf-5 undergo distinct cell cycle-specific expression in muscle cells. J Cell Biol. 1998;142:1447–1459. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitzmann M, Vandromme M, Schaeffer V, Carnac G, Labbé J C, Lamb N, Fernandez A. Cdk1 and Cdk2 mediate phosphorylation of MyoD S200 in growing C2 myoblasts: role in modulating half-life and myogenic activity. Mol Cell Biol. 1999;19:3167–3176. doi: 10.1128/mcb.19.4.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kophengnavong T, Michnowicz J E, Blackwell T K. Establishment of distinct MyoD, E2A, and twist DNA binding specificities by different basic region-DNA conformations. Mol Cell Biol. 2000;20:261–272. doi: 10.1128/mcb.20.1.261-272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindon C, Montarras D, Pinset C. Cell cycle-regulated expression of the muscle determination factor Myf5 in proliferating myoblasts. J Cell Biol. 1998;140:111–118. doi: 10.1083/jcb.140.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Balbas M A, Dey A, Rabindran S K, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 29.McGarry T J, Kirschner M W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 30.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 31.Pines J, Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prescott D M, Bender M A. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- 33.Roberts S B, Segil N, Heintz N. Differential phosphorylation of the transcription factor Oct1 during the cell cycle. Science. 1991;253:1022–1026. doi: 10.1126/science.1887216. [DOI] [PubMed] [Google Scholar]
- 34.Rudnicki M A, Schnegelsberg P, Stead R, Braun T, Arnold H, Jaenisch R. MyoD or Myf5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 35.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skapek S X, Rhee J, Kim P S, Novitch B G, Lassar A B. Cyclin-mediated inhibition of muscle gene expression via a mechanism that is independent of pRB hyperphosphorylation. Mol Cell Biol. 1996;16:7043–7053. doi: 10.1128/mcb.16.12.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song A, Wang Q, Goebl M G, Harrington M A. Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol Cell Biol. 1998;18:4994–4999. doi: 10.1128/mcb.18.9.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorrentino V, Pepperkok R, Davis R, Ansorge W, Philipson L. Cell proliferation inhibited by MyoD1 independently of myogenic differentiation. Nature. 1990;345:813–815. doi: 10.1038/345813a0. [DOI] [PubMed] [Google Scholar]
- 39.Townsley F M, Aristarkhov A, Beck S, Hershko A, Ruderman J V. Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc Natl Acad Sci USA. 1997;94:2362–2367. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamano H, Tsurumi C, Gannon J, Hunt T. The role of the destruction box and its neighboring lysine residues in cyclin B for anaphase ubiquitin-dependent proteolysis in fission yeast: defining the D-box receptor. EMBO J. 1998;17:5670–5678. doi: 10.1093/emboj/17.19.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yun K, Wold B. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr Opin Cell Biol. 1996;8:877–889. doi: 10.1016/s0955-0674(96)80091-3. [DOI] [PubMed] [Google Scholar]
- 42.Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J-M, Wei Q, Zhao X, Paterson B M. Coupling of the cell cycle and myogenesis through the cyclin D1-dependent interaction of MyoD with cdk4. EMBO J. 1999;18:926–933. doi: 10.1093/emboj/18.4.926. [DOI] [PMC free article] [PubMed] [Google Scholar]