Abstract
Background
In the context of the Covid-19 pandemic and following the increasing number of suspicious Covid-19 cases in Madagascar, Malagasy laboratories are overflowed mainly due to lack of human resource and available material restriction. The development and validation of rapid and easy-to-perform diagnostic methods are worth of interest and high priority. The aim of this prospective study was to evaluate the performances of a rapid immunochromatographic test for the detection of SARS-CoV-2 antigen, in comparison to Reverse transcription polymerase chain reaction (RT-PCR).
Methods
The fluorescence immunochromatographic SARS-CoV-2 antigen test StandardTM Q COVID-19 Ag Test (SD Biosensor Republic Korea) was evaluated in samples derived from patients who were examined for disease categories. Diagnostic accuracy was determined in comparison to SARS-CoV-2 RT-PCR considered as gold standard.
Results
A total of 200 samples were included; 94 were RT-PCR positive. Median patients’ age was 38.36 years, 63.5 % were male. Overall sensitivity and specificity of the Standard TM Q COVID-19 Ag (SD Biosensor® Republic Korea) were 62.66 % and 100 %, the sensitivity was significantly higher (100 %) in samples with high viral loads (Ct<29).
Conclusions
This antigen-based immunofluorescence RDT could be the potential to become an important tool for the early diagnosis of SARS-CoV-2 particularly in situations with limited access to molecular methods particularly in rural area of Madagascar.
Keywords: SARS-CoV-2, Covid-19, Diagnosis, Rapid diagnostic test, Antigen, Madagascar
1. Background
Coronavirus disease 2019 (COVID-19) is a newly emerging human infectious disease associated with severe respiratory distress caused by SARS-CoV-2. This new strain was first identified in the city of Wuhan in Hubei province in China at the end of 2019. The virus then spread rapidly through various countries including Madagascar. The virus appears to be transmitted mainly by droplets transmitted by the respiratory route and/or carried between 2 individuals. Most often at the origin of a mild infectious syndrome, associating to varying degrees mild symptoms (fever, cough, myalgia, headache and digestive problems), SARS-CoV-2 can be the cause of serious pulmonary pathologies and sometimes death (Patel and Jernigan, 2020).
For rapid treatment and to control the epidemic, the biology laboratories take on a real challenge since the confirmation of the infection is based on virological diagnosis by molecular biology (extraction of the viral genome followed by amplification and revelation by RT-PCR) (Corman et al., 2020).
Currently, facing the increase number of cases, the human and material capacities available at the laboratory level are exceeded. The use of rapid tests of viral antigens which potentially detect early cases is currently proposed in order to identify the outbreaks, expand screening and follow-up treatment. However, these kits should be evaluated before their routine use.
Among possible test formats, rapid diagnostic tests (RDTs) should be prioritized, since they are timely, easy to perform, and can serve as point-of-care testing (POCT). Here we present the evaluation of antigen-based RDT for the detection of SARS-CoV-2 in respiratory specimens from suspected Covid-19 cases.
2. Material and methods
2.1. Clinical specimens
We conducted a prospective study which aimed to assess the performance of the RDT SARS-CoV2- StandardTM Q COVID-19 Ag Test (SD Biosensor® Republic Korea) compared to RT-PCR which is the main biological testing used in Madagascar in order to confirm Covid-19.
Two hundred (200) nasopharyngeal samples derived from adult patients COVID-19 suspected were selected into following severity of illness categories at the Joseph Ravoahangy Andrianavalona University Hospital Center (JRA UHC) Antananarivo, Madagascar.
-
•
Asymptomatic or Presymptomatic Infection: Eighty-four (83) COVID-19 suspected without any apparent symptoms.
-
•
Mild Illness: Sixty-five (65) COVID-19 suspected who have any of the various signs and symptoms of COVID-19 (e.g., fever, cough, sore throat, malaise, headache, muscle pain) without shortness of breath, dyspnea, or abnormal chest imaging.
-
•
Moderate Illness: Twenty-nine (29) COVID-19 suspected who have evidence of lower respiratory disease by clinical assessment or imaging and a saturation of oxygen (SpO2) ≥94 % on room air at sea level.
-
•
Severe Illness: Twenty-two (22) COVID-19 suspected Individuals who have respiratory frequency>30 breaths per minute, SpO2 <94 % on room air at sea level, ratio of arterial partial pressure of oxygen to fraction of inspired oxygen (PaO2/FiO2) <300 mmHg, or lung infiltrates >50 %
2.2. Molecular analysis by RT-PCR
The Nasopharyngeal samples stored in an appropriate virus transport medium (e.g. UTM Viral Transport, Copan Diagnostics Inc., Brescia, Italy) were tested immediatly at the laboratory of molecular biology of JRA UHC Madagascar and the epidemiological surveillance lab of Madagascar (LA2M). The COVID-19 laboratory diagnosis relies on quantitative Real-Time RT-PCR assay. For extraction step, the Advansure™ nucleic Acid EX kit (LG Chem®, Ltd Republic of Korea) were used for RNA and DNA extractions, respectively, as instructed by the manufacturer. For all RNA extractions, RNA was extracted from 200 μL clinical samples using the AdvanSure E3 extraction system (LG Life Sciences®, Korea) and the final elution volume was 100 μl for each sample. For amplification step, we did reverse transcriptase quantitative PCR targeting the SARS-CoV-2 by using the Detection kit for 2019 Novel Coronavirus (2019-nCoV) Da An Gene Co® Ltd. of Sun Yat–Sen University China. The kit is based on one step RT-PCR technique. In practice, 2019 Novel Coronavirus (2019-nCoV) ORF1ab and N genes are selected as amplification target regions. A typical 20 μL monoplex RT-PCR assay contained 17 μL of NC (ORF1ab/N) PCR reaction solution A (specific primers, probes, trisaminomethane hydrochloric acid buffer) and 3 μL NC (ORF1ab/N) PCR reaction solution A (hot start Taq DNA polymerase, c-MMLV reverse transcriptase), and 5 μl of RNA sample. RT-PCR reactions were conducted by a thermal cycler (CFX 96 Biorad® real time PCR) with the following conditions: reverse transcription at 50 °C for 15 min, inactivation of reverse transcriptase at 95 °C for 15 min, 45 cycles of PCR amplification (Denaturing at 95 °C for 15 s; Annealing/Extending at 55 °C for 45 s). The time for each RT-PCR run lasted for approximately 1 h and 15 min.
The result was considered valid only when the cycle threshold (Ct) value of the reference gene was less than 40. The result was considered positive when the Ct values of both target genes were less than 40 and negative when they were both greater than 40. If only one of the target genes had a Ct value of 38 or less and the other was more than 38, it was interpreted as a single-gene positive.
2.3. Antigen-based rapid detection test
StandardTM Q COVID-19 Ag Test (SD Biosensor® Republic Korea), is a rapid chromatographic immunoassay ready to use test which allows rapid and qualitative detection of SARS-CoV-2 antigen in nasopharyngeal secretions. As instructed by the manufacturer, the specimen should be tested as soon as after collection or at room temperature for up to 1 h. All test procedures except the reading of the cassette were performed under a BSL2 cabinet. Results of the RDT were compared to those of RT-PCR as reference method. The demographic and clinical data were obtained from the mandatory national Covid-19 notification forms and were analyzed anonymously.
2.4. Statistics
Results of the Standard TM Q COVID-19 Ag Test (SD Biosensor® Republic Korea) were compared to those of RT-PCR as reference method; for samples with discordant result, tests were repeated. RT-PCR was considered as the gold standard for this evaluation, therefore positive and negative samples by molecular techniques were considered to be true positive and true negative samples, respectively. Statistical analysis considered the calculation of sensitivity, specificity, and Kappa coefficient using standard formulas. Test performance was analyzed as recommended by current CLSI guidelines. Sensitivity was further analyzed for certain subgroups such as severity of illness and RT-PCR Ct values.
Steps were taken to maintain strict confidentiality when preparing the files. The prospective study was performed with the agreement signed by the patient.
3. Results
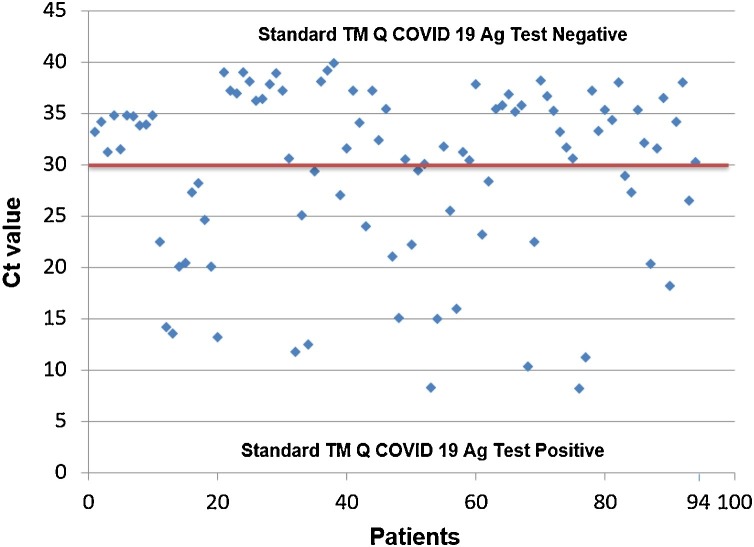
We collected 200 nasopharyngeal samples from 200 COVID-19 suspected patients. The median age of the studied population was 38.36 (range: 12–79) with a sex ratio of 1.73 (127 men and 73 women). According to RT-PCR results, 106 samples were negative and 94 were positive, with a median Cycle threshold (Ct) value of 30 (mean: 30.18; range:8–39) (Fig. 1 ).
Fig. 1.
Cycle threshold (Ct) values of 94 RT-PCT positive samples. Above the line red represent false negative and below true positive with Standard TM Q COVID-19 Ag Test (SD Biosensor® Republic Korea).
The overall sensitivity and specificity of the evaluated Standard TM Q COVID-19 Ag Test (SD Biosensor® Republic Korea) were 62.66 % and 100 %, respectively (Table 1 ). The Kappa coefficient is of 0.58, values between 0,40 and 0,75 can be considered as moderate and representative of a fair agreement beyond chance.Sensitivity was significantly reduced in the subgroup of samples with Ct values > 30.1, indicating lower viral loads. A subgroup analysis of Ct values revealed that the sensitivity increase with clinical gravity (Table 2 ). In the asymptomatic Ct mean 33.51 (range: 20.3–39.89) subgroup,the sensibility was 59.32 % in contrast with severe illness Ct mean 22.27 (range: 11.3–36.2) sensibility was 80.95 %. No significant difference within illness categories have been observed, the sensitivity was 100 % with Ct<30.
Table 1.
Diagnostic performance indicators of nasopharyngeal Antigen RDT Standard TM Q COVID-19 Ag Test (SD Biosensor® Republic Korea) compared to RT-PCR.
| RT-PCR positive | RT-PCR negative | ||
|---|---|---|---|
| Antigen RDT positive | 94 | 0 | Predicted positive: 100 % |
| Antigen RDT negative | 56 | 106 | Predicted negative: 65.43 % |
| Sensitivity: 62.66 % | Specificity: 100 % |
Table 2.
Sensitivity of Standard TM Q COVID-19 Ag (SD Biosensor® Republic Korea) in subgroups in different severity of illness categories.
| Ct Value | Mean Ct Value | Standard TM Q COVID-19 Ag positive | Standard TM Q COVID-19 Ag negative | Sensitivity % | |
|---|---|---|---|---|---|
| Asymptomatic | Ct [20.3–28.2] | 26.25 | 11 | 0 | 59.32 |
| Ct [34.7 – 39.] | 36.84 | 0 | 24 | ||
| Mildillness | Ct [8.24–30.21] | 21.96 | 4 | 0 | 54.54 |
| Ct [30.08–38.0] | 34.37 | 0 | 20 | ||
| Moderate illness | Ct [12.51–29.42] | 22.7 | 10 | 0 | 69.23 |
| Ct [34.08–38.2] | 35.43 | 0 | 8 | ||
| Severe illness | Ct [11.3–30.5] | 18.92 | 13 | 0 | 80.95 |
| Ct [30.5–36.2] | 33.17 | 0 | 4 |
4. Discussion
Facing the increased Covid-19 cases in Madagascar, the limited capacity of PCR laboratories drive to a longer delivery result time up to 10 days, delaying patient care and response measures. The search for a reliable rapid test that can be installed in remote areas even without electricity is worth of interest. One of the promising approaches is the rapid antigenic tests although they have not been recommended yet for the acute diagnosis of COVID-19 due to their low sensitivity.
According to several studies, the analytical performance of these rapid antigenic tests depends on various factors including the viral load, the quality of the sample and the way it is processed. Scohy A et al. putted forward several advantages of rapid antigen detection such as (i) the ease and fast achievement of the test, (ii) the rapidity of response, (iii) the low cost and the non-requirement of special equipment or skills compared with molecular techniques. The rapid antigen detection test is able to detect SARS-CoV-2 with high sensitivity in nasopharyngeal samples with high viral load equivalent at least to 1.7 × 105 copies/mL (Ct < 25), but the sensitivity declines substantially when the viral load decreases with Ct values over 30, which is often the case in patients suffering of COVID-19 (Scohy et al., 2020). Their results were similar to our study, showing good sensitivity if the viral load was high (Ct <30) and a decrease in sensitivity after Ct values over 30. Considering our cohort of patients, the sensitivity of the test is increased in very symptomatic patients, even if the viral load that was detected in the asymptomatic patient was similar to that of symptomatic patients (Zou et al., 2020). This could be explained by the fact that symptomatic patients exhibit a longer high viral load (Wölfel et al., 2020).
Zou, L et al. pointed out that Ct is generally less than 30 in the first 10 days after the onset of symptoms and Just et al. highlighted that patients with anosmia and close contact with an infected person have shown more positive results (Zou et al., 2020; Just et al., 2020). Therefore the test remains fully appropriate during this particular period and for those symptomatic patients. Thus, it could potentially be used in priority as a triage test to quickly identify patients at high risk for COVID-19, reducing the need of expensive molecular confirmatory testing that is not readily available in some regions and basic health centers in Madagascar. Secondarily, patients with negative results and supposedly out of this period should be screened by PCR.
Considering that the viral loads in throat swabs and sputum have reached the maximum level after 5–6 days from the beginning of the infection, the SARS-CoV-2 Ag RDT has no indication in the follow-up of patients and the confirmation of viral clearance (Pan et al., 2020). Any Antigen negative RDT in a person suspected of COVID-19 must be confirmed by RT-PCR.
In low-income countries, the cost of the analysis certainly plays an important role in the number of screened people; interestingly the price of PCR turns around $ 50 while an Standard TM Q COVID-19 Ag (SD Biosensor® Republic Korea) is less than $ 5.However, as for PCR the Standard TM Q COVID-19 Ag (SD Biosensor® Republic Korea) is carried out from a nasopharyngeal sample; the handling of the collection sample must take into account the infectivity of the sample which may expose operator/environment and the management of the elimination of contaminated waste which oblige the handling in a biosafety cabinet following the WHO recommendation (Porte et al., 2020).
To date, there have been no reports of molecular tests or antigen-based immunofluorescence detection being affected by other mutations carried by the new variants. However, given the number of mutations in the S gene that each variant carries, laboratories that use tests with targeting the S gene are recommended to monitor for ‘dropout’ and consider implementing assays specific for other genomic targets.
In conclusion, strength of this antigen-based immunofluorescence detection can be resumed to the easy-perform technic and provided results in a timely manner and its high sensitivity and specificity in respiratory samples obtained from patients with high viral load usually during the first week of Covid-19. Thus, this Standard TM Q COVID-19 Ag (SD Biosensor® Republic Korea) could be an important tool for the early diagnosis of SARS-CoV-2 and consecutively helps reducing the delay in the establishment of adequate treatment particularly in rural area of Madagascar.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Toky Rakotomalala Randriamahazo: Conceptualization, Investigation, Formal analysis, Writing - original draft, Writing - review & editing. Andry Maharo Andrianarivelo: Methodology, Formal analysis. Andry Toky Rakotoarivo: Conceptualization, Methodology, Writing - review & editing. Todisoa Mahenina Raheritiana: Conceptualization, Methodology, Writing - review & editing. Luc Andriamiadana Rakotovao: Writing - review & editing. Zely Arivelo Randriamanantany: Writing - review & editing. Aimée Olivat Rakoto Alson: Conceptualization, Methodology, Writing - review & editing. Andry Rasamindrakotroka: Conceptualization, Methodology, Writing - review & editing.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
We thank the Ministry of Public Health of Madagascar for choosing our laboratory for the evaluation of Antigen-based rapid detection test.
References
- Corman V.M., Landt O., Kaiser M., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just Johannes, Puth Marie-Therese, Regenold Felix, et al. Distinguishing between COVID-19 and the common cold in a primary care setting-comparison of patients with positive and negative SARS-CoV-2 PCR results. medRxiv. 2020 doi: 10.1101/2020.04.27.20081877. [DOI] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Jernigan D.B. Initial public health response and interim clinical guidance for the 2019 novel coronavirus outbreak—United States, December 31, 2019–February 4, 2020. Morbid. Mortal. Weekly Rep. 2020;69(5):140. doi: 10.15585/mmwr.mm6905e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte L., Legarraga P., Vollrath V., Aguilera X., Munita J.M., Araos R., et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]