Abstract
Transcripts harboring premature signals for translation termination are recognized and rapidly degraded by eukaryotic cells through a pathway known as nonsense-mediated mRNA decay (NMD). In addition to protecting cells by preventing the translation of potentially deleterious truncated peptides, studies have suggested that NMD plays a broader role in the regulation of the steady-state levels of physiologic transcripts. In Saccharomyces cerevisiae, three trans-acting factors (Upf1p to Upf3p) are required for NMD. Orthologues of Upf1p have been identified in numerous species, showing that the NMD machinery, at least in part, is conserved through evolution. In this study, we demonstrate additional functional conservation of the NMD pathway through the identification of Upf2p homologues in Schizosaccharomyces pombe and humans (rent2). Disruption of S. pombe UPF2 established that this gene is required for NMD in fission yeast. rent2 was demonstrated to interact directly with rent1, a known trans-effector of NMD in mammalian cells. Additionally, fragments of rent2 were shown to possess nuclear targeting activity, although the native protein localizes to the cytoplasmic compartment. Finally, novel functional domains of Upf2p and rent2 with homology to eukaryotic initiation factor 4G (eIF4G) and other translational regulatory proteins were identified. Directed mutations within these so-called eIF4G homology (4GH) domains were sufficient to abolish the function of S. pombe Upf2p. Furthermore, using the two-hybrid system, we obtained evidence for direct interaction between rent2 and human eIF4AI and Sui1, both components of the translation initiation complex. Based on these findings, a novel model in which Upf2p and rent2 effects decreased translation and accelerated decay of nonsense transcripts through competitive interactions with eIF4G-binding partners is proposed.
Contrary to intuition, the predominant consequence of nonsense mutations in eukaryotes is not the synthesis of truncated proteins. Rather, many nonsense transcripts are recognized and selectively degraded by the cell via a pathway known as nonsense-mediated mRNA decay (NMD) (17, 20, 47). The basic process has been most comprehensively studied in the yeast Saccharomyces cerevisiae. NMD requires at least three trans-acting factors, termed Upf1p, Upf2p, and Upf3p (15, 27, 40, 42, 43), which localize predominantly to the cytoplasm and associate with polysomes (2, 3). An emerging model for the mechanism of NMD proposes that the first translating ribosomes displace bound proteins as they traverse a nascent cytoplasmic messenger ribonucleoprotein (mRNP) molecule (22). Ribosomal pausing at a termination codon signals recruitment of a surveillance complex consisting of, at least, eukaryotic release factors eRF1 and eRF3 and Upf1p (18). Transcript decay is induced if the surveillance complex senses premature translation termination as indicated by inappropriately bound proteins remaining 3′ of the termination signal. Such proteins would normally be displaced by complete translation of the open reading frame (ORF). One such factor in yeast may be Hrp1p, a protein essential for NMD, which binds to the downstream sequence element (22), a cis-acting sequence that must be present 3′ of a premature termination codon in order to trigger NMD (57, 78). Hrp1p also interacts directly with Upf1p. An intriguing possibility is that recently identified protein complexes which are deposited at spliced exon-exon junctions might serve an analogous function in mammalian cells (44).
The role of the other two Upf proteins may be to recruit transcripts to the decay pathway. Upf3p is an attractive candidate for this purpose since it contains multiple nuclear localization signals (NLS) and nuclear export signals (NES) that allow shuttling across the nuclear envelope (65). Upf2p interacts with both Upf3p and Upf1p and may function as a bridge, allowing Upf3p to deliver the nonsense transcript to the surveillance machinery (25, 26). Despite the appealing nature of this model, little is known about the precise and coordinated function of these factors. The fact that Upf proteins have pleiotropic functions adds to this complexity. The Upf proteins not only participate in the decay of nonsense transcripts but also enhance the efficiency of translational termination (48, 75). These effects are genetically separable. Forms of Upf1p with mutations in the N-terminal Cys- and His-rich domain support NMD but allow nonsense suppression (readthrough). In contrast, forms of Upf1p with mutations in the helicase domain can promote efficient translational termination but fail to effect NMD.
One theory holds that NMD may preclude or perturb events that actively determine transcript stability. Normal mRNA turnover in S. cerevisiae initiates with shortening of the poly(A) tail, followed by 5′ decapping by Dcp1p and subsequent 5′-to-3′ decay by the exonuclease Xrn1p (5, 39). In contrast, NMD is characterized by deadenylation-independent decapping and decay by Dcp1p and Xrn1p, respectively (53). The poly(A) tail may influence cap stability through the interaction of poly(A)-binding protein (PABP) with eukaryotic initiation factor 4G (eIF4G) (31, 70), which additionally binds the cap-binding complex (eIF4A and eIF4E) (29). The synergistic interactions of this protein complex are thought to dictate a closed-loop transcript conformation, which enhances both stability and translational efficiency (21, 60). Progressive poly(A) tail shortening accompanies transcript maturation (5). Attainment of a threshold length would preclude PABP binding and disrupt the protein complex bridging the ends of the transcript, exposing the 5′ cap to the activity of Dcp1p. One model posits that NMD bypasses the need for deadenylation through prevention or disruption of the closed-loop conformation (34), although any concept of mechanism is lacking.
Many similarities, and a few apparent differences, exist between the NMD pathways in yeast and higher eukaryotes. For example, it appears that nonsense surveillance relies on conventional translation machinery in all organisms, as evidenced by sensitivity to pharmacologic translational inhibitors, hairpin structures that impair translation initiation, or suppressor tRNAs (20, 23, 47). Although the nuclear-cytoplasmic shuttling of Upf3p implies a role for the nuclear compartment in yeast NMD, even greater evidence exists for a nuclear role in mammalian cells. Subcellular fractionation studies revealed that nonsense mRNAs are reduced to the same extent in the nucleus and cytoplasm (11, 38). Furthermore, full stability was seen for cytoplasmic nonsense-containing mRNAs despite association with polysomes (66). One feature that distinguishes mammalian NMD from that in yeast is the general requirement for at least one intron downstream of the nonsense codon (10). An attractive hypothesis is that this intron serves an analogous function to the yeast downstream sequence element in defining a context within which a termination codon is considered premature. Other evidence suggests that the coding potential of a pre-mRNA can influence splicing decisions and induce either exon skipping or intron retention (9, 19). While nonsense-mediated perturbation of splicing may result via a different pathway from NMD, it suggests the ability to recognize a termination codon within an incompletely processed mRNA that is associated with the nucleus.
Very little is known about the NMD machinery in organisms other than S. cerevisiae. Although Upf1p orthologues have been identified in numerous species including Schizosaccharomyces pombe (Upf1p) (our unpublished observations), Caenorhabditis elegans (smg-2) (56), Mus musculus (rent1), and humans (rent1) (1, 58), homologues of Upf2p and Upf3p have not been described. The extent of structural and functional conservation of the NMD machinery in higher eukaryotes has therefore been left to speculation. A refined knowledge of the mechanism and role of NMD in mammals is of more than just academic importance. The function has been shown to be a potent modifier of selected dominant negative or gain-of-function phenotypes (16), presumably through the prevention of expression of deleterious truncated peptides. Furthermore, an emerging view holds that the NMD pathway regulates a significant proportion of physiologic transcripts, as evidenced by dysregulation of about 8% of the yeast transcriptome in Upf-deleted strains (45).
To further elucidate the mechanism of NMD in higher eukaryotes, we have identified and characterized homologues of Upf2p in S. pombe (Upf2p) and humans (rent2). We demonstrate that S. pombe Upf2p, a protein as structurally divergent from S. cerevisiae Upf2p as is rent2, is essential for NMD in fission yeast. rent2 interacts directly with rent1, a known trans-effector of mammalian NMD (67), utilizing regions that correspond to structurally and positionally defined domains within Upf1p and Upf2p in S. cerevisiae. rent2 harbors motifs that are capable of directing fusion peptides into the nucleus, but the native protein exists predominantly, if not exclusively, in the cytoplasm. Finally, we have identified two novel domains of Upf2p and rent2 that bear significant homology to eukaryotic proteins with a known function in the regulation of translational initiation including eukaryotic initiation factor 4G (eIF4G), poly(A)-binding protein-interacting protein 1 (PAIP-1), and NAT1 (also called DAP5 and p97) (13, 29, 32, 46, 76). Sequence conservation of these so-called eIF4G homology (4GH) domains is poorest in S. cerevisiae Upf2p, precluding recognition of this homology prior to our cloning of the S. pombe and human homologues. Regions of these proteins that mediate interactions proposed to be critical to the formation of the closed-loop conformation also span 4GH domains (33), and site-directed mutations within the 4GH domains of S. pombe Upf2p can abolish NMD activity. Additionally, we provide evidence that rent2 interacts with select components of the human translation initiation complex, eIF4AI and Sui1. The implications of these findings for the function of Upf2p and rent2 and the mechanism of NMD are discussed.
MATERIALS AND METHODS
Cloning of the RENT2 cDNA.
Following identification of a human expressed sequence tag (EST) (accession no. AA812020) with similarity to S. cerevisiae UPF2, the Genetrapper system (GIBCO BRL, Gaithersburg, Md.) was used to screen a SuperScript human heart cDNA library (GIBCO BRL) for additional clones. cDNA capture was performed with oligonucleotide Ob39f03.s1-1 (GCAGAAGCTGTAGCTTCCATCGTGG), and cDNA repair was carried out with Ob39f03.s1-2 (CTCTGATGTGAACTGTGCTGTGC) as specified by the manufacturer. Using this method, a clone containing 2.1 kb of coding sequence and 81 bp of the 5′ untranslated region (UTR) was isolated (clone R2-GT3). The remainder of the 3′ sequence was determined by sequencing EST64960 (accession no. AA356414). For 5′ rapid amplification of cDNA ends (RACE), the Marathon system (Clontech, Palo Alto, Calif.) was used to amplify adult heart cDNA with primer R2-5′RACE-NEST (CGTTCCCAAGCTTCCTGATGAAGCTG). 5′ RACE products were cloned into pCRII-TOPO (Invitrogen, Carlsbad, Calif.) and sequenced. The 5′ endpoints of RACE products ranged from positions −110 to −124 relative to the first ATG. For 3′ RACE, an initial round of amplification with primer GTR2-2A (ACACCAGAAGAACATGGGCCTGGA) was followed by nested PCR with primer GTR2-3A (CTGCATGTCTGATGTAGCAGAGG). All 3′ RACE products ended at nucleotide 5093 relative to the first ATG.
The Stanford G3 radiation hybrid-mapping panel was obtained from Research Genetics (Huntsville, Ala.). Chromosomal localization of RENT2 was performed as specified by the manufacturer using primers T7-1/64960 (CATGTTGACGGGCATGTTT) and T7-1R/64960 (TCCAGACAAGGCGGAAAAG).
Plasmid construction.
For construction of the recombinant plasmids used in this study, all PCR amplifications were carried out with Pfu Turbo polymerase (Stratagene, La Jolla, Calif.) as specified by the manufacturer. All constructs were verified by direct sequencing.
The S. pombe UPF2 targeting vector (pUPF2Δ::URA4) was assembled by first amplifying 549 bp of genomic sequence 5′ of the UPF2 locus (nucleotides −15 to −564 relative to the first ATG) using primers pUPF2.5′flank.S (CCGGATCCCTGAGTATTGCTTAATTACCC) and pUPF2.5′flank.AS (CCGATATCGCATTGAAAAGCACTTCAATTC). The PCR product was cloned into pCR2.1 (Invitrogen), excised with BamHI and EcoRV, and ligated into the BamHI and EcoRV sites of pBluescript II SK(+) (Stratagene) to create pBS-5′UPF2. Then 377 bp of sequence 3′ of the UPF2 locus (nucleotides 3140 to 3517 relative to the first ATG) was amplified using primers pUPF2.3′flank.S (CGGATATCCTTCGAATAAGAGAAGCTCTTG) and pUPF2.3′flank.AS (CAACGTCGACGTTATGGTTTTCACCTTTGAC) and ligated into pCR2.1. The EcoRV-SalI fragment was excised and inserted into the EcoRV and SalI sites of pBS-5′UPF2 to create pBS-5′/3′UPF2. The S. pombe URA4 expression cassette was removed from pCG1 (a kind gift from J. Boeke, Johns Hopkins University, Baltimore, Md.) by digestion with HindIII, ends were filled in with the Klenow fragment of DNA polymerase I (New England Biolabs, Beverly, Mass.), and the fragment was inserted into the EcoRV site of pBS-5′/3′UPF2.
To construct a cDNA clone (pCMVSPORT-RENT2) containing the complete RENT2 ORF as well as 81 nucleotides of the 5′ UTR and the complete 3′UTR, the 1.4-kb XbaI fragment of R2-GT3 was replaced with the 2.4-kb XbaI fragment from EST64960. The newly created 474-bp ApaI fragment was then replaced with the 1.7-kb ApaI fragment from EST64960.
The RENT1 expression plasmid (pCMVSPORT-RENT1) was described previously as a full-length clone isolated from a human heart cDNA library (58).
Green fluorescent protein (GFP) fusions were constructed by cloning RENT2 PCR products into vector pcDNA3.1/CT-GFP-TOPO (Invitrogen). For the full-length RENT2-GFP fusion, primers GTR2-1A (GCTAATGTTGACAACAGGCTCGAG) and R2-3′GFP-AS (GACGTCTCCTCCCACCAGTC) were used to amplify the complete coding sequence. For amino acids 1 to 120 fused to GFP, primers GTR2-1A and R2-GFP1-AS (GCTGAGCAGCTGCTTCTTCTTC) were used to amplify the 5′ end of the gene. All PCRs were performed with pCMVSPORT-RENT2 as a template.
For two-hybrid analysis, rent1-GAL4AD fusions were constructed by ligating RENT1 cDNA fragments into pACT2 (Clontech). First, the EcoRI-BamHI fragment containing the complete coding sequence was excised from pCMVSPORT-RENT1 and ligated into the EcoRI and BamHI sites of pAS2-1 (Clontech) to create pAS21-RENT1. The NcoI-BamHI fragment from this plasmid was then inserted into the pACT2 NcoI and BamHI sites to fashion the full-length fusion [AD(R1:1–1118)]. For the fusion encompassing residues 1 to 415 [AD(R1:1–415)], the NcoI-SalI fragment from pAS21-RENT1 was cloned into the NcoI and XhoI sites of pACT2. For the fusion containing amino acids 120 to 890 [AD(R1:120–890)], primers Nco-S1 (ACCATGGCCGAAGGCATCCTGCAGAAC) and For-2 (ATCAGCAGGTGGTTCCAG) were used to amplify a fragment of RENT1 which was subsequently ligated into the TA cloning vector pCR2.1. The insert was excised with EcoRI and NcoI and cloned into the EcoRI and NcoI sites in pACT2.
rent2-GAL4BD fusion constructs were assembled by cloning RENT2 cDNA fragments into pGBKT7 (Clontech). The following primer pairs were used to amplify fragments from pCMVSPORT-RENT2, which were then directly cloned into the SmaI site of pGBKT7: for the full-length fusion [BD(R2:1–1272)], primers R2-1-EcoRI-S (GCCGGAATTCATGCCAGCTGAGCGTAAAAAGC) and R2-3′BstXI(PstI)-AS (CTGCAGAACCACTGCAGTGGACGTCTCCTCCCACCAGTC); for residues 1 to 656 [BD(R2:1–656)], primers R2-1-EcoRI-S and R2-656-BstXI(PstI)-AS (CTGCAGAACCACTGCAGTGGGTCCTTTTTCCGTACATGAAATC); for residues 1 to 757 [BD(R2:1–757)], primers R2-1-EcoRI-S and R2-757-BstXI(PstI)-AS (CTGCAGAACCACTGCAGTGGGTTGCAGTAGTAATATGCATTCTC); for residues 642 to 1095 [BD(R2:642–1095)], primers R2-642-BLUNT-S (GATGCTGAGGGGGGATTTCAG) and R2-1095-BstXI(PstI)-AS (CTGCAGAACCACTGCAGTGGTACCTCAGTATTCTCTTCATCG); for residues 757 to 1272 [BD(R2:757–1272)], primers R2-757-BLUNT-S (GCCACCTCCAGCTGAAAAAACC) and R2-3′BstXI(PstI)-AS; for residues 1084 to 1272 [BD(R2:1084–1272)], primers R2-1084-EcoRI-S (GCCGGAATTCAAGGAAAATGAAACCGATGAAG) and R2-3′BstXI(PstI)-AS.
Translation initiation factor-GAL4AD fusions were constructed by amplifying cDNA fragments from a human heart cDNA library and cloning them into pGADT7 (Clontech). For the full-length eIF4AI fusion [eIF4AI(1–407)], primers eIF4AI-BamHI-S (CGCGGATCCTTATGTCTGCGAGCCAGGATTCCCG) and eIF4AI-BamHI-AS (CGCGGATCCCAGAGATTGAGCCCTGGCTGGGG) were used and the resulting PCR product was digested and ligated into the BamHI site. For residues 1 to 325 of eIF4AI [eIF4AI(1–325)], primers eIF4AI-BLUNT-S (GATGTCTGCGAGCCAGGATTCCCG) and eIF4AI-BLUNT-AS (CAGAGATTGAGCCCTGGCTGGGG) were used to amplify the complete eIF4AI ORF, which was subsequently cloned into the SmaI site. Sequencing of this clone revealed that amplification had introduced a 5-nucleotide deletion at codon 325, leading to a premature stop 3 codons downstream. Human Sui1 (huISOSUI1) was amplified with primers hSui1-BLUNT-S (GATGTCCGCTATCCAGAACCTC) and hSui1-BLUNT-AS (TTAAAACCCATGAACCTTCAGC) and cloned into the SmaI site. hPrt1 was amplified using the Clontech Advantage-GC cDNA PCR kit with primers hPrt1-EcoRI-S (CCGGAATTCATGCAGGACGCGGAGAACG) and hPrt1-EcoRI-AS (CCGGAATTCTTAAATCCCCCACTGCAGACAC). Following EcoRI digestion, the cDNA fragment was ligated into the EcoRI site.
Hemagglutinin (HA)-tagged forms of rent2 were created by amplifying RENT2 cDNA fragments from pCMVSPORT-RENT2 with primers which incorporated an HA peptide. For the full-length N-terminal HA-tagged construct (HA-N-rent2), primers R2-5′HA-S (CCACCATGGCCTACCCCTACGACGTGCCCGACTACGCCGAAGAAAAAGACTCTTTACCAAAC) and R2-3′BLUNT-AS (TCAACGTCTCCTCCCACCAGTC) were used. For the C-terminal tagged constructs, the following primer pairs were used: residues 1 to 1095 [(R2:1–1095)HA], GTR2-1A and R2-1095HA-AS (TCAGGCGTAGTCGGGCACGTCGTAGGGGTATACCTCAGTATTCTCTTCATCG); for residues 757 to 1272 [(R2:757–1272)HA], R2-757(start)-S (GCCACCATGGCTCCACCTCCAGCTGAAAAAACC) and R2-3′HA-AS (TCAGGCGTAGTCGGGCACGTCGTAGGGGTAACGTCTCCTCCCACCAGTC). An N-terminally HA-tagged form of luciferase was constructed (HA-N-Lucif) by amplifying pGL2 (Promega, Madison, Wis.) with primers Luc-5′HA-S (CCACCATGGCCTACCCCTACGACGTGCCCGACTACGCCGAAGACGCCAAAAACATAAAGAAAG) and 3′Luc-HinDIII-AS (AAGCTTAAGAATTTCGTCATCGCTGAATACAG). All PCR products were cloned into pcDNA3.1/V5/HIS-TOPO (Invitrogen) as described by the manufacturer.
The S. pombe UPF2 expression plasmid was constructed by cloning the complete UPF2 ORF into pREP3 (obtained from J. Boeke). Genomic DNA was amplified with primers Upf2-MscI-S (TGGCCAGAATTGAAGTGCTTTTCAATGCA) and Upf2-MscI-AS (TGGCCACAAGAGCTTCTCTTATTCGAAG), and the resulting PCR product was ligated directly into the MscI site. For expressing c-myc-tagged forms of Upf2p, primers UPF2-N-myc-SaII-S (ACGCGT CGACAGAACAAAAATTGATTTCTGAAGAAGATTTGTCAAGAGAAG AACAAATAAAAAAAC) and UPF2-SaII-AS (ACGCGTCGACCAAGAGCTTCTCTTATTCGAAG) were used and the resulting PCR product was cloned into the SalI site of pREP3. Mutagenesis was performed with the QuickChange site-directed mutagenesis kit (Stratagene).
Disruption of S. pombe Upf2p.
For vegetative growth, S. pombe was grown in YEC medium (5 g of yeast extract and 2 g of Casamino Acids per liter). Selective growth was carried out in EMM supplemented with the appropriate amino acids (Bio 101, Vista, Calif.). All transformations were performed as previously described (55).
pupf2Δ::URA4 was linearized with BamHI and transformed into strains BP425 (h− leu1-32 ura4-D18), GP1540 (h− ade-M26 leu1-32 ura4-D18), GP1541 (h+ ade6-M375 leu1-32 ura4-D18), GP1594 (h+ ade6-469 leu1-32 ura4-D18), and GP937 (h− ade6-M216 leu1-32 ura4-D18). BP425 was obtained from J. Boeke, and all other strains were kind gifts from G. Smith (Fred Hutchinson Cancer Research Center, Seattle, Wash.). To confirm correct targeting of UPF2, genomic DNA was isolated as described previously (52), digested with BsaI, and analyzed by Southern blotting as described previously (61). Probes used were generated by PCR with the following primer pairs: 5′-flank probe, pombe-UPF2-5′probe-S (GATATAACTTCGATGCCACG) and pombe-UPF2-5′probe-AS (ATACGAGAAATGTTTCTCTCC); 3′-flank probe, pombe-UPF2-3′probe-S (TTGGATGGTCCCAAAGTGC) and pombe-UPF2-3′probe-AS (GAAGTTTCGACTGGTGAAG).
Analysis of S. pombe, mouse, and human RNA.
Total RNA was isolated from logarithmically growing S. pombe using the hot-phenol method (42). For measurement of transcript decay rates, S. pombe cultures were grown to mid-log phase before inhibiting transcription with 10 μg of thiolutin (Pfizer, Groton, Conn.) per ml. Aliquots of cells were then removed at appropriate time points and immediately centrifuged, and the pellets were frozen in a dry-ice–ethanol bath. Transcript half-lives were calculated as described previously (42). For analysis of Gus transcript abundance, total RNA was isolated from mouse tissues pooled from four littermates by using Trizol reagent (GIBCO BRL). Poly(A) extraction was then performed using the Oligotex Midi kit (Qiagen). For Northern blotting, RNA was electrophoresed in 1.2% agarose–formaldehyde gels, transferred to nylon filters (GeneScreen Plus; NEN, Boston, Mass.), and hybridized with randomly primed radiolabeled probes. All hybridizations were carried out in ExpressHyb (Clontech) as specified by the manufacturer. Radioactive signals were directly quantified using an Instant Imager (Packard, Downers Grove, Ill.).
The expression pattern of RENT2 was determined by probing the adult human 12-lane multi-tissue Northern blot (Clontech) with a PCR fragment generated from the pCMVSPORT-RENT2 template with primers SP6-5/64960 (TTACCGAATGGTGGAATCA) and T7-4/64960 (CCAAACTTTTCTTCCACCA). The mouse Rent2 cDNA fragment used to probe a mouse multitissue Northern blot (Clontech) was generated by amplifying mouse brain cDNA with primers mR2-probe-S (TAATTACAGAAATGGTCGAATCAGC) and mR2-probe-AS (CTCCAAACTTTTCTTCCACCAAAC). The β-actin cDNA probe was obtained from Clontech.
The 700-bp XhoI-HindIII fragment of the Gus cDNA (a generous gift from Mark Sands, Washington University) was used as a probe for Gus Northern blots. The glyceraldehyde-3-phosphate dehydrogenase (G3PDH) control probe was obtained from Clontech.
The ADE6 probe used to measure transcript levels in S. pombe was generated by PCR with primers Spade6-S (GTTGAACTGTCTAAGAAGTGC) and Spade6-AS (CATCAACGCATGAGTTGTG) using genomic DNA as a template. The YPT5 control probe was also amplified from genomic DNA, using primers pYPT5e7-S (GCATCTCTAGAAAAGGCC) and pYPT5e8-AS (CAAGAGCATGAACCGCTTG).
Tissue culture, transfections, and immunofluorescence.
HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and an antibiotic-antimycotic mixture (GIBCO BRL). Standard tissue culture practices were followed. Transfections were carried out with Lipofectin and PLUS reagent (GIBCO BRL) as described by the manufacturer. For transfection of HA-N-rent2 or the rent2-GFP fusions, 6 μg of plasmid was used per 10-cm-diameter dish. At 48 h following transfection, appropriate cells were treated for 3 h with tissue culture medium supplemented with 2 ng of leptomycin B (a kind gift from M. Yoshida, University of Tokyo, Tokyo, Japan) per ml. Living cells expressing the GFP fusions were imaged directly. For visualization of HA-rent2 or cyclin B1, cells were fixed for 20 min in 3.7% paraformaldehyde, permeabilized for 15 min in 0.1% Triton X-100, and blocked for 30 min in 1% bovine serum albumin in phosphate-buffered saline. Anti-HA staining was carried out with a 1:1,000 dilution of affinity-purified monoclonal anti-HA antibody HA.11 (Covance, Denver, Pa.) followed by a 1:200 dilution of fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G (Sigma, St. Louis, Mo.). A fluorescein isothiocyanate-conjugated monoclonal anti-cyclin B1 antibody (GNS1; Santa Cruz Biotechnology, Santa Cruz, Calif.) was used at a 1:50 dilution. All cells were imaged by confocal microscopy at the Johns Hopkins School of Medicine Microscopy Facility.
Two-hybrid analysis.
All plasmids and strains for two-hybrid analysis were obtained from the Matchmaker system 3 kit (Clontech) with the exception of pACT2 and pAS2-1, which are components of the Matchmaker system 2 kit. YPDA and the appropriate synthetic dropout (SD) media, also obtained from Clontech, were prepared as specified by the manufacturer. Yeast transformations were performed by the lithium acetate method (63). The plasmid encoding AD(R1:1–415) led to restricted growth and mating of cells when present. For this reason, cotransformation was performed to combine this plasmid with GAL4BD fusions. All other combinations of GAL4AD and GAL4BD fusions were generated by mating. For yeast matings, GAL4AD fusions were transformed into strain Y187 and GAL4BD fusions were transformed into AH109. Single colonies from each transformed strain were mixed in 0.5 ml of YPDA medium, incubated overnight at 30°C, and plated on SD −Trp, −Leu plates. For conditional growth assays, colonies containing combinations of GAL4AD and GAL4BD fusions were grown overnight in liquid SD −Trp, −Leu medium. Stationary-phase cultures (10-μl volumes) were spotted on the appropriate selective medium and incubated at 30°C for 3 to 5 days.
Protein preparation, immunoprecipitation, and Western analysis.
Total protein was isolated from logarithmically growing S. pombe strains harboring the c-myc-tagged wild-type or mutant Upf2p expression constructs as described previously (52). Protein was isolated from S. cerevisiae strains AH109 or Y187 harboring GAL4BD or GAL4AD fusions, respectively, as described previously (35). Equal amounts of protein, as confirmed by Coomassie staining, were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (8 to 10% polyacrylamide) and transferred to nitrocellulose for immunoblotting (see below).
For coimmunoprecipitation experiments, cells growing in 10-cm dishes were cotransfected with 3 μg of pCMVSPORT-RENT1 alone or in combination with 3 μg of (R2:1–1095)HA, (R2:757–1272)HA, or HA-N-Lucif. Cell extracts were prepared 48 h following transfection. All steps were performed at 4°C. Following two washes with phosphate-buffered saline, 1 ml of lysis buffer JM (50 mM HEPES [pH 7.6], 150 mM NaCl, 5 mM MgCl2, 0.1% Nonidet P-40, EDTA-free complete protease inhibitor [Roche]) was added per 10-cm dish. After a 30-min incubation, the cells were collected by scraping and the lysate was cleared by centrifugation. A 2-μg portion of anti-HA antibody HA.11 was added to 300 μl of lysate and agitated for 3 h. A 50-μl volume of a 50% slurry of protein G-Sepharose beads (Amersham, Piscataway, N.J.), washed twice in lysis buffer JM, was then added, and the samples were agitated for an additional 2 h. Following collection by centrifugation, beads were washed sequentially in 1 ml of wash buffers W1 (50 mM HEPES [pH 7.6], 300 mM NaCl, 0.5% Triton X-100), W2 (50 mM HEPES [pH 7.6], 500 mM LiCl, 0.5% Triton X-100), W3 (50 mM HEPES [pH 7.6], 40 mM NaCl, 500 mM LiCl), and W4 (50 mM HEPES [pH 7.6], 150 mM NaCl, 1 mM dithiothreitol). After being resuspended in 40 μl of 2× SDS sample buffer (61), samples were boiled and centrifuged briefly. A 20-μl volume of each sample was then subjected to SDS-PAGE (7.5% polyacrylamide) and transferred to nitrocellulose.
Immunoblotting with anti-HA (monoclonal antibody HA.11) or anti-c-myc (clone 9E10 [Roche]) was carried out as specified by the manufacturer. Affinity-purified rent1 antiserum was used as described previously (67). Immunoreactive bands were visualized using the Supersignal West Dura chemiluminescent substrate (Pierce, Rockford, Ill.).
Nucleotide sequence accession numbers.
The human Upf2p orthologue rent2 and the S. pombe Upf2p sequences have been deposited in GenBank under accession no. AF301013 and AF301014, respectively.
RESULTS
Identification of UPF2 homologues in S. pombe and humans.
The sequences for all known S. cerevisiae trans-effectors of NMD were submitted to the X-REF database genome cross-referencing effort (4). In this manner, we identified sequences from S. pombe and humans that exhibited homology to S. cerevisiae UPF2. An S. pombe chromosome I cosmid (accession no. Z98974) contained the complete genomic sequence encoding a putative Upf2p homologue (P = 1.4 × 10−34). The 3,150-bp ORF was not interrupted by introns. A human EST derived from tonsillar germinal B-cell cDNA (accession no. AA812010) encoded a short peptide with homology to Upf2p (P = 4.5 × 10−5). This sequence was used to design oligonucleotides for screening a human heart cDNA library using the Genetrapper system. This resulted in the isolation of a clone containing 2.1 kb of coding sequence. An overlapping EST derived from a Jurkat cDNA library (accession no. AA356414) contained 3.1 kb of additional 3′ sequence which included a termination codon and a polyadenylation signal. 5′ and 3′ RACE were then performed using human heart cDNA to complete and verify the terminal sequences of the cDNA. Radiation hybrid mapping using the Stanford G3 panel localized RENT2 to the human chromosomal subregion 10p13-10p15 with a logarithm of the odds (LOD) score of 13.64 at a distance of 5 centiRays from marker D10S2376 (data not shown).
Conceptual translation of the complete human cDNA predicts an ORF of 3,816 bp with a 124-bp 5′ UTR and a 1,276-bp 3′ UTR, excluding the poly(A) tail. Complete identification of the 5′ end of the coding sequence was confirmed by the presence of an in-frame termination codon encoded by nucleotides −123 to −121, relative to the most 5′ ATG. The first putative initiator and an ATG occurring 10 codons downstream both conform to the Kozak consensus (36).
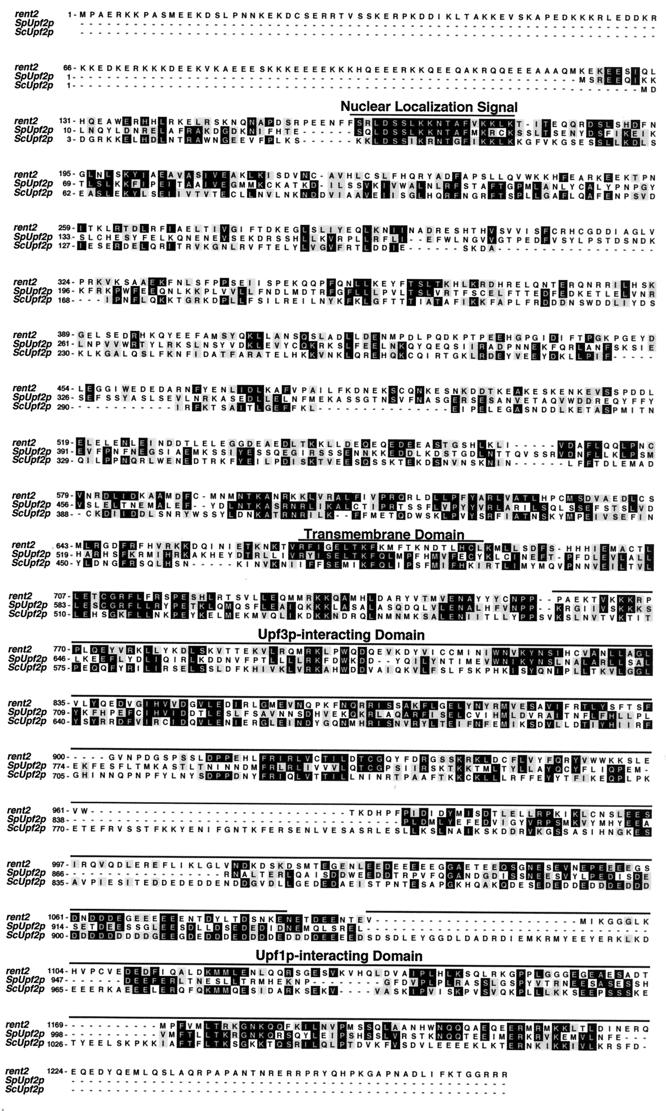
The previously identified mammalian orthologue of Upf1p is named rent1 (regulator of nonsense transcripts 1). Accordingly, we have assigned the name rent2 to the putative mammalian Upf2p orthologue described here. An alignment of the sequences of rent2 and S. pombe Upf2p with that of S. cerevisiae Upf2p is shown in Fig. 1. Compared to S. cerevisiae Upf2p, S. pombe Upf2p exhibits 22% amino acid identity and 43% amino acid similarity whereas rent2 exhibits 23% identity and 42% similarity. Thus, all three molecules are equally divergent. Regions of S. cerevisiae Upf2p with putative or known function show a measurable but small increase in the frequency of amino acids that are conserved through evolution. These include previously designated sequences without documented functionality, specifically a putative NLS (amino acids 26 to 46 of S. cerevisiae Upf2p) and a hydrophobic region termed the transmembrane domain (amino acids 470 to 490) (27), as well as the functionally defined Upf1p- and Upf3p-interacting domains (amino acids 933 to 1089 and 564 to 923, respectively) (25, 26). Additionally, rent2 includes a unique 120-amino-acid N-terminal extension which is absent is S. cerevisiae and S. pombe Upf2p. This highly charged domain shows no homology to any known protein and contains 12 putative NLSs, of both the simian virus 40 (SV40)-like and bipartite varieties, as defined by the PSORT algorithm (http://psort.nibb.ac.jp:8800/). Furthermore, rent2 contains a perfect match to the Rev-like leucine-rich NES consensus L(X)2–4L(X)2LXL at residues 1003 to 1013 and five matches to the looser consensus Φ(X)2–4Φ(X)2LXΦ, where Φ is a hydrophobic residue, X is any residue, and L is leucine (residues 189 to 198, 266 to 274, 518 to 527, 611 to 621, and 827 to 836) (28).
FIG. 1.
Sequence alignment of S. cerevisiae Upf2p (ScUpf2p), S. pombe Upf2p (SpUpf2p), and human rent2. A ClustalW alignment was performed using the MacVector 6.5.1 package of sequence analysis software. Identical residues shared between at least two of the three proteins are shaded in black; similar residues shared between at least two of the proteins are shaded in gray. The previously identified putative or documented functional domains of S. cerevisiae Upf2p are indicated by lines.
Upf2p is required for NMD in S. pombe.
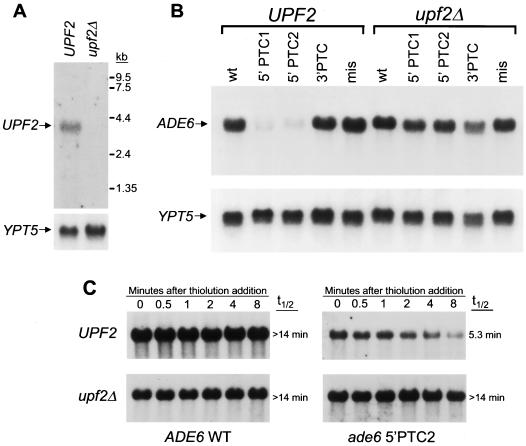
To determine the effect of UPF2 deletion on the efficiency of NMD in S. pombe, a targeting vector was constructed to completely replace the Upf2p coding sequence with a URA4 expression cassette (24). The gene was disrupted in S. pombe strains containing three different nonsense mutations in the nonessential ADE6 gene (ade6-M26, ade6-M375, and ade6-469) (68). Transcripts derived from the two mutant ade6 alleles harboring 5′ nonsense mutations (ade6-M26 and ade6-M375) were rapidly degraded, as inferred from low steady-state levels, while those harboring a 3′ nonsense mutation (ade6-469) were not substrates for NMD. To rule out the possibility of nonspecific effects on mRNA metabolism, UPF2 was also disrupted in S. pombe strains with wild-type ADE6 or ade6 with a missense mutation (ade6-M216) (68). Correct targeting was confirmed by Southern blot analysis (data not shown). Northern blotting established that UPF2 encodes a transcript of approximately 3.7 kb which is absent in upf2Δ strains (Fig. 2A). All strains were viable and showed no apparent growth abnormality. Quantitative Northern blot analysis was used to measure the steady-state levels of ADE6 transcripts in the UPF2 and upf2Δ strains (Fig. 2B). Deletion of UPF2 had no effect on the steady-state levels of wild-type ADE6 transcripts or ade6 transcripts containing a 3′ PTC or a missense mutation. In contrast, ade6 nonsense transcripts which exist at low steady-state levels in UPF2 strains accumulated to wild-type levels in upf2Δ strains, indicating loss of NMD function.
FIG. 2.
Upf2p is required for NMD in S. pombe. (A) S. pombe UPF2 encodes a single ∼3.7-kb transcript which is absent in upf2Δ strains. Total RNA was isolated from logarithmically growing S. pombe strains and analyzed for UPF2 expression by Northern blotting. The blot was stripped and rehybridized with a YPT5 probe to control for loading differences. (B) Deletion of UPF2 specifically stabilizes nonsense transcripts in S. pombe. The complete UPF2 ORF was disrupted in S. pombe strains which were wild type at the nonessential ADE6 locus (wt), harbored two different nonsense mutations near the 5′ end of the ADE6 gene which give rise to transcripts degraded by NMD (ade6-M26 and ade6-M375; 5′ PTC1 and 5′ PTC2, respectively), contained a nonsense mutation near the 3′ end of ADE6 which is not a substrate for NMD (ade6-469; 3′ PTC), or contained a missense mutation (ade6-M216; mis). To measure the steady-state levels of wild-type or mutant ADE6 transcripts, total RNA was analyzed by Northern blotting with an ADE6 probe. The blot was stripped and rehybridized with a YPT5 probe to control for loading differences. (C) The decreased steady-state level of ade6 nonsense transcripts is due to a UPF2-dependent accelerated mRNA decay rate. The half-life (t1/2) of wild-type (WT) ADE6 or mutant ade6 transcripts containing a nonsense mutation (5′PTC2; ade6-M375) was determined in UPF2 or upf2Δ strains. Transcription was inhibited with thiolutin, and transcript abundance was determined by Northern blot analysis at the indicated time points.
To confirm that these differences in steady-state abundance were due to changes in transcript decay rates, the half-lives of wild-type ADE6 or mutant ade6 transcripts containing a nonsense mutation (ade6-M375) were determined in a UPF2 or upf2Δ background (Fig. 2C). Transcription was inhibited with thiolutin, and transcript abundance was measured by Northern blot analysis. The wild-type ADE6 transcript had a half-life of longer than 14 min. The decay rate of this transcript was not significantly altered by UPF2 deletion. In contrast, the ade6 nonsense transcript decayed rapidly, with a half-life of 5.3 min, confirming that its low steady-state abundance is due to increased transcript lability. In the upf2Δ strain, this transcript was significantly stabilized, with a half-life of longer than 14 min. Thus, S. pombe Upf2p, a molecule as divergent from S. cerevisiae Upf2p as is rent2, is required for the accelerated degradation of nonsense transcripts in fission yeast.
Tissue-specific variation in RENT2 expression does not influence the efficiency of NMD.
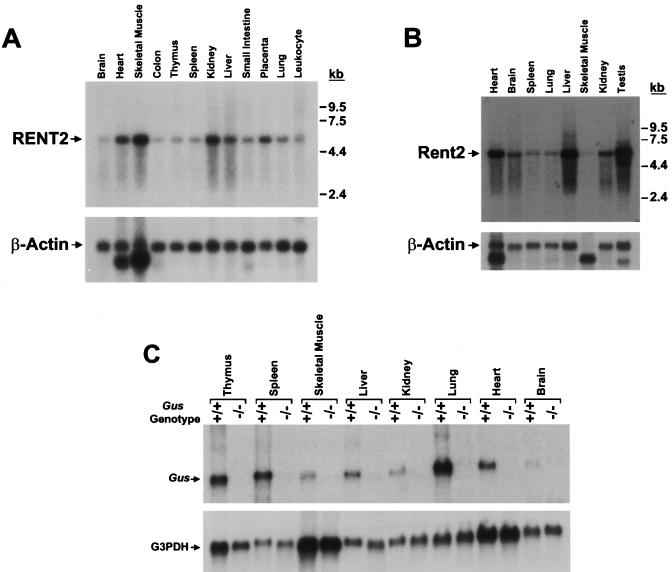
A Northern blot containing mRNA from multiple adult human tissues was hybridized with a RENT2 cDNA fragment corresponding to nucleotides 2638 to 2879 (Fig. 3A). All tissues tested exhibited a single signal of variable intensity corresponding to a transcript size of approximately 5.2 kb, as predicted from the cDNA sequence. Thus, both RENT1 (58) and RENT2 are ubiquitously expressed and both show tissue-specific variation in abundance. Both are highly expressed in the heart, skeletal muscles, kidneys, and placenta. The brain shows the lowest expression of RENT2 but relatively high expression of RENT1, whereas the inverse is true for the liver.
FIG. 3.
Variation in RENT2 expression does not influence the efficiency of NMD. (A) Expression of RENT2 was assessed in adult human tissues by probing a multi-tissue Northern blot (Clontech) with a human RENT2 cDNA fragment. The blot was stripped and rehybridized with a β-actin cDNA probe to control for loading differences. (B) Expression of mouse Rent2 in adult tissues was analyzed by hybridizing a mouse multi-tissue Northern blot (Clontech) with a mouse Rent2 cDNA probe. β-Actin was again used as a loading control. (C) Northern blot analysis of poly(A) RNA from wild-type or homozygous mutant gusmps mice. To measure steady-state Gus transcript levels, 2 μg of poly(A) RNA from the indicated adult tissues was probed with a Gus cDNA fragment. The blot was stripped and rehybridized with a G3PDH probe to control for loading differences.
The expression pattern of mouse Rent2 was determined by probing a Northern blot containing mRNA from multiple adult mouse tissues (Fig. 3B). A mouse EST (accession no. AUO23401) was identified which exhibited significant homology to nucleotides 2696 to 3174 of human RENT2 (95% nucleotide identity), and this sequence was used to generate a cDNA probe. The observed expression pattern differed somewhat from that determined for human RENT2. Most tissues express a single Rent2 isoform of approximately 5.2 kb. The testes, however, show an additional smaller isoform of approximately 4.8 kb. Mouse Rent2 shows the highest expression in the heart, liver, and testes. Surprisingly, expression is lowest in skeletal muscle, the tissue which exhibits the highest human RENT2 expression.
In light of the variable expression of RENT1 and RENT2, we sought to measure the efficiency of NMD in multiple mammalian tissues. To do this, we used the gusmps mouse, a model of mucopolysaccharidosis type VII, which harbors a 1-bp deletion in exon 10 of the ubiquitously expressed β-glucuronidase gene (62). The transcript derived from this mutant allele contains a PTC and is a substrate of the NMD pathway (6). Steady-state levels of the Gus transcript from adult wild-type and homozygous mutant littermates were measured in multiple tissues using quantitative Northern blot analysis (Fig. 3C). Despite the wide variation in expression of RENT1 and RENT2, all tissues examined showed efficient degradation of the mutant transcript.
RENT2 localizes to the cytoplasmic compartment.
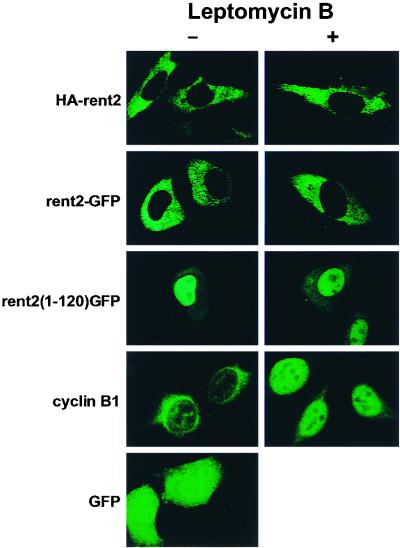
As an initial attempt to determine the subcellular localization of rent2, an N-terminal HA-tagged full-length rent2 expression construct was fashioned and transiently transfected into HeLa cells. Western blotting confirmed the expression of a recombinant protein of the appropriate size (data not shown). Indirect immunofluorescence performed with a monoclonal anti-HA antibody followed by confocal microscopy revealed an exclusively cytoplasmic distribution of the tagged protein (Fig. 4).
FIG. 4.
rent2 localizes to the cytoplasmic compartment of mammalian cells despite possessing functional NLSs. The subcellular localization of N-terminal HA-tagged full-length rent2 (HA-rent2), a full-length C-terminal rent2-GFP fusion (rent2-GFP), or the N-terminal 120 amino acids of rent2 fused to GFP [rent2(1–120)GFP] was determined in transiently transfected HeLa cells in the presence or absence of the inhibitor of nuclear export leptomycin B. As a positive control for leptomycin B treatment, the subcellular localization of cyclin B1 was determined in the presence or absence of the drug.
To examine the rent2 subcellular localization further, a chimeric protein consisting of GFP fused to the C terminus of rent2 was transiently expressed in HeLa cells. As seen with the N-terminal HA-tagged rent2 construct, confocal microscopy performed on cells expressing a full-length rent2-GFP fusion revealed an exclusively cytoplasmic distribution of the hybrid protein (Fig. 4). In contrast, a GFP fusion containing only the unique N-terminal 120 amino acids of rent2, which includes 12 putative NLSs, was significantly concentrated in the nucleus compared to the distribution of GFP alone (Fig. 4). These data demonstrate that the N terminus of rent2 is sufficient to direct nuclear targeting of a reporter protein. Two possibilities exist to explain the cytoplasmic distribution of full-length rent2: either the functional NLSs are masked or inhibited in the context of the complete protein or rent2 only transiently enters the nucleus before being rapidly exported. To test the latter hypothesis, we used the specific inhibitor of nuclear export, leptomycin B. This compound inactivates CRM1/exportin 1 (37), thus preventing the recognition and function of Rev-like leucine-rich NESs. HeLa cells transiently expressing the N-terminal HA-tagged rent2 construct or either the full-length or N-terminal 120 amino acids of rent2 fused to GFP were treated with leptomycin B and visualized by confocal microscopy (Fig. 4). As a positive control for leptomycin B treatment, the subcellular distribution of cyclin B1, a protein previously shown to shuttle between the nucleus and cytoplasm in a CRM1-dependent manner (72, 77), was determined by immunofluorescence. Whereas cyclin B1 was significantly concentrated in the nucleus following treatment with the drug, the distribution of the rent2 fusion proteins was not altered, indicating that rent2, if exported, does not utilize the CRM1/exportin 1 pathway.
Conserved rent1-rent2 interactions.
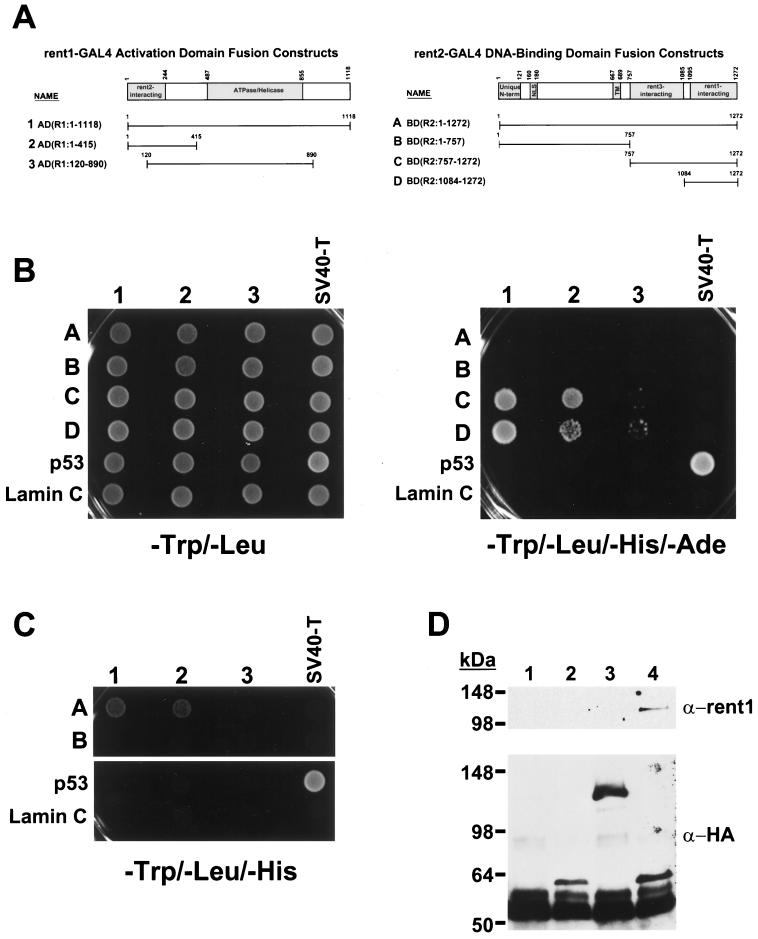
In S. cerevisiae, interaction between Upf1p and Upf2p is required for the formation of a functional nonsense surveillance complex (25, 26). If the mechanism of NMD is conserved between yeast and mammals, rent1, a known trans effector of mammalian NMD and orthologue of Upf1p (1, 58, 67), should interact with rent2 in an analogous fashion. To test this hypothesis, we used the yeast two-hybrid system to assess for interactions between full-length and truncated forms of rent1 and rent2 (Fig. 5). rent1–GAL4 activating-domain (GAL4AD) fusion constructs and rent2–GAL4 DNA-binding-domain (GAL4BD) fusion constructs (Fig. 5A) were prepared and cointroduced into yeast strain AH109, which contains the ADE2 and HIS3 reporter genes driven by a GAL4 upstream activating sequence. The known interaction between SV40 large T antigen and p53 (51) served as a positive control, and the absence of interaction with lamin C served as a negative control. All fusion proteins were expressed as myc- or HA-tagged forms, and all showed comparable levels of expression by Western blot analysis except for the full-length rent2-GAL4BD fusion, which was produced at a significantly reduced level (data not shown).
FIG. 5.
Conserved rent1 and rent2 interactions. (A) Schematic representation of rent1–GAL4 activation domain (GAL4AD) and rent2–GAL4 DNA-binding domain (GAL4BD) fusion constructs. Predicted functional domains of rent1 and rent2 based on homology to S. cerevisiae Upf1p and Upf2p, respectively, are shaded. Overlying numbers indicate amino acid position. (B) Stringent growth assay for interaction. The indicated GAL4AD and GAL4BD fusion constructs were co-introduced into strain AH109 and plated on minimal medium lacking Trp and Leu (left) to select for cotransformants or plated on medium lacking Trp, Leu, His, and Ade (right) to select for interacting proteins. As a positive control, SV40 large T antigen (SV40-T)–GAL4AD and p53-GAL4BD fusion constructs were used to test a known interaction between these proteins. The lack of interactions with a lamin C-GAL4BD fusion peptide served as a negative control. (C) Reduced-stringency growth assay for interaction. rent2-GAL4BD fusions which failed to interact with rent1-GAL4AD fusions were tested on minimal media lacking Trp, Leu, and His. (D) Coimmunoprecipitation of rent1 and rent2. Cell extracts from HeLa cells overexpressing rent1 alone (lane 1) or rent1 plus either HA-tagged luciferase (lane 2), residues 1 to 1095 of rent2 with a C-terminal HA tag (lane 3), or residues 757 to 1272 of rent2 with a C-terminal HA tag (lane 4) were precipitated with an anti-HA monoclonal antibody (Covance) and analyzed by Western blotting. Duplicate blots from the same experiment are shown, probed with anti-rent1 antiserum (upper panel) or an anti-HA monoclonal antibody (lower panel). The intense lower band in all lanes is the heavy chain of the anti-HA antibody used for immunoprecipitation.
Full-length rent2 showed no apparent interaction with any rent1 fragments when a high-stringency assay was applied (Fig. 5B, growth on −Trp/−Leu/−His/−Ade medium). A similar result was obtained with a fragment of rent2 spanning residues 1 to 757. In contrast, rent2 fragments containing amino acids 757 to 1272 or 1084 to 1272 supported growth when combined with full-length rent1 or fragments spanning the first 415 amino acids of rent1. Reduced but detectable growth was also observed when these rent2 fragments were combined with amino acids 120 to 890 of rent1 (Fig. 5B).
We next tested for evidence of interactions using less stringent requirements for conditional growth (Fig. 5C, −Trp/−Leu/−His). Under these conditions, full-length rent2 supported rapid growth when combined with full-length rent1 or amino acids 1 to 415 of rent1. Residues 1 to 757 of rent2 again did not show evidence of interaction with any tested fragments of rent1. These data indicate that full-length rent2 interacts with rent1, albeit less strongly than the C-terminal fragments of rent2. It is likely that this weaker interaction is due to the reduced expression and/or stability of the large rent2-GAL4BD fusion protein. Furthermore, these data document that residues 1084 to 1272 of rent2 encompass the domain required for rent1 interaction. In rent1, residues 1 to 415 are required for rent2 interaction, with amino acids 1 to 120 providing a positive but not essential influence on binding. These data correlate well with the predicted position of interacting domains based on homology to the Upf proteins. Residues 933 to 1089 of S. cerevisiae Upf2p mediate interaction with residues 1 to 181 of Upf1p (26). The corresponding residues are 1095 to 1272 and 1 to 244 in rent2 and rent1, respectively.
To confirm that these interactions occurred in mammalian cells, expression constructs encoding HA-tagged forms of rent2 with or without the rent1-interacting domain were constructed. A plasmid which overexpresses rent1 was transfected into HeLa cells alone or in combination with plasmids expressing HA-tagged luciferase, residues 1 to 1095 of rent2 with a C-terminal HA tag, or residues 757 to 1272 of rent2 with a C-terminal HA tag. Cell extracts were precipitated with an anti-HA monoclonal antibody and analyzed for rent1 coprecipitation by Western blot analysis (Fig. 5D). As predicted from the two-hybrid data, rent1 specifically copurified with the rent2 fragment encompassing residues 757 to 1272. These data provide compelling evidence that rent1, a known trans effector of the mammalian NMD pathway, and rent2 interact in mammalian cells using structurally conserved domains.
Novel functional domains of Upf2p/rent2 exhibit homology to eIF4G.
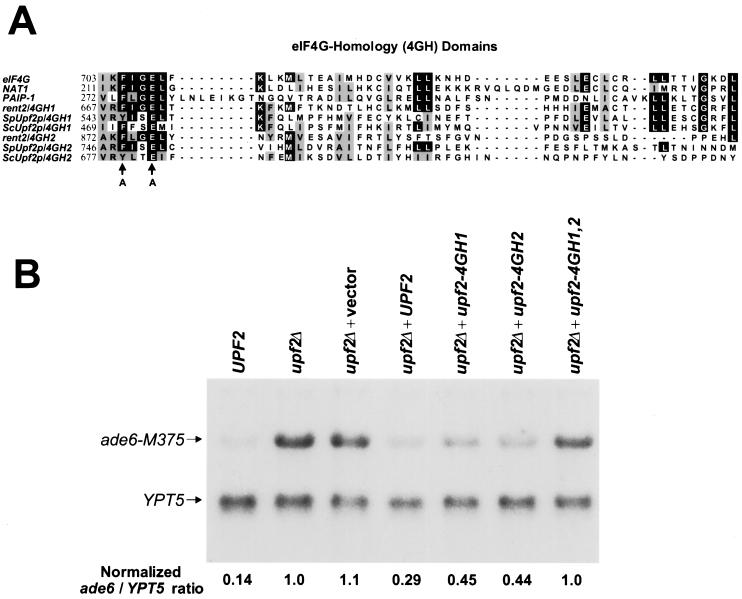
BLAST analysis of the rent2 amino acid sequence revealed a domain with homology to proteins with a known function in the regulation of translation initiation, including eIF4G, PAIP-1, and NAT1/DAP5/p97 (Fig. 6A) (13, 29, 32, 46, 76). This motif, which we have termed the 4GH domain, is repeated twice in S. cerevisiae Upf2p (residues 469 to 517 and 677 to 725), S. pombe Upf2p (residues 543 to 590 and 746 to 794), and rent2 (residues 667 to 714 and 872 to 916). Sequence conservation is poorest in S. cerevisiae Upf2p, precluding recognition of this homology prior to our cloning of the S. pombe and human homologues. Interestingly, the more N-terminal 4GH domain, which we refer to as 4GH1, falls within the previously designated transmembrane domain of Upf2p (27). Deletion of this region of S. cerevisiae Upf2p is sufficient to abolish the function of this protein (25). The more C-terminal 4GH domain, termed 4GH2, falls within the Upf3p-interacting domain. All 4GH domains, including those contained within Upf2p homologues as well as eukaryotic translational regulatory proteins, show strict preservation of an aromatic residue (F or Y) at position 3 and a glutamic acid (E) at position 6 (Fig. 6A).
FIG. 6.
The 4GH domains of S. pombe Upf2p are required for NMD. (A) Alignment of all known 4GH domains. With the exception of S. cerevisiae Upf2p (ScUpf2p) and S. pombe Upf2p (SpUpf2p), all proteins shown are human. Arrows indicate residues mutated in the upf2-4GH1 and upf2-4GH2 S. pombe Upf2p expression constructs. (B) Effect of 4GH mutations on the function of S. pombe Upf2p. A upf2Δ S. pombe strain harboring a nonsense mutation in ADE6 (ade6-M375) was transformed with an empty expression vector or vector containing wild-type S. pombe UPF2 (UPF2), UPF2 mutated at positions 3 and 6 of 4GH1 (upf2-4GH1), UPF2 mutated at positions 3 and 6 of 4GH2 (upf2-4GH2), or UPF2 mutated at positions 3 and 6 of both 4GH domains (upf2-4GH1,2). The steady-state levels of the ade6 nonsense transcript were determined by quantitative Northern blot analysis. Following hybridization with an ADE6 probe, the blot was reprobed for YPT5 to standardize for loading differences. The ade6/YPT5 ratio was calculated for each sample and normalized to the ratio measured in the upfΔ strain.
To directly examine the importance of 4GH domains to Upf2p function, we determined the consequence of directed mutations at position 3 (F-to-A) and position 6 (E-to-A) of the 4GH domains of S. pombe Upf2p. The complete coding region of S. pombe Upf2p was cloned into the fission yeast expression vector pREP3 (50). Site-directed mutagenesis was then performed to produce forms of Upf2p with alterations at positions 3 and 6 of 4GH1 (upf2-4GH1), 4GH2 (upf2-4GH2), or both 4GH domains (upf2-4GH1,2). Mutant forms were expressed in upf2Δ strains of S. pombe, and the efficiency of NMD was assessed by measuring the steady-state levels of ade6 nonsense transcripts (ade6-M375) (68) using quantitative Northern blot analysis (Fig. 6B). Abundant expression of all recombinant forms of Upf2p was confirmed by Western blot analysis of c-myc fusion proteins (data not shown). Transformation with empty vector had no effect on the steady-state level of the ade6 nonsense transcript. In contrast, expression of wild-type recombinant Upf2p led to significant, although not complete, restoration of NMD efficiency. Failure to fully restore NMD function may represent the consequence of overexpression of this molecule. Strains expressing forms of Upf2p with isolated mutations in 4GH1 or 4GH2 showed a further reduction in NMD efficiency, with an approximately 1.5-fold increase in the steady-state abundance of the mutant transcript, compared to that resulting from expression of recombinant wild-type Upf2p. Importantly, this result demonstrates that the 4GH2 mutation, which falls within the putative Upf3p-interacting domain, is not sufficient to fully abolish the function of the protein. This argues against the possibility that this mutation simply prevents Upf3p binding, which would preclude functional restoration of the NMD pathway. Finally, a strain expressing a double-mutant form of Upf2p showed a complete loss of NMD function. Thus, the two 4GH domains of S. pombe Upf2p appear to synergistically contribute to protein function.
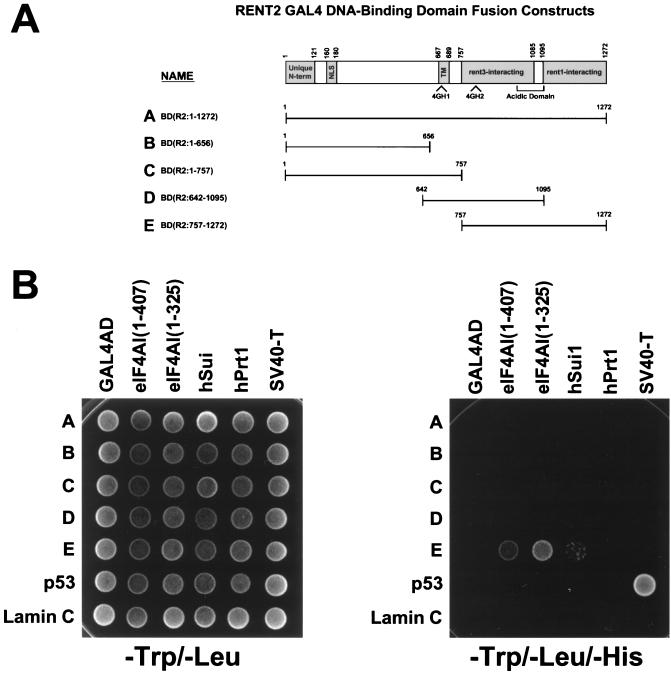
Regions of eIF4G, PAIP-1, and NAT1 that span 4GH domains participate in binding to eIF4A and eIF3. We thus hypothesized that rent2 may also interact with these proteins via its 4GH domains. To test this prediction, we again utilized the yeast two-hybrid system to assess for interactions between fragments of rent2 and full-length human eIF4AI [eIF4AI(1–407)], a truncated form of human eIF4AI [eIF4AI(1–325)], or two different components of human eIF3, Sui1 (huISOSUI1) and hPrt1 (Fig. 7). All fusion proteins showed high levels of expression by Western blot analysis, except for the full-length rent2-GAL4BD fusion (data not shown). By using low-stringency conditions, i.e., growth on minimal medium lacking Trp, Leu, and His, we obtained evidence for interaction between a fragment of rent2 containing 4GH2 [BD(R2:757–1272)] and both full-length and C-terminally truncated forms of eIFAI. Additionally, we observed evidence of interaction between the same rent2 fragment and Sui1. These data suggest that residues 757 to 1272 of rent2 encompass a domain capable of interacting with specific components of the translation initiation complex. No evidence for interaction was seen with any other tested rent2 fragment, perhaps signifying the presence of positive and/or negative regulatory sequences that modify binding-site recognition or affinity.
FIG. 7.
Two-hybrid analysis of rent2-translation initiation factor interactions. (A) Schematic representation of the rent2–GAL4 DNA-binding domain (GAL4BD) fusion constructs used in this study. (B) Growth assay for interaction. Full-length human eIF4AI [eIF4AI(1–407)], truncated human eIF4AI [eIF4AI(1–325)], human Sui1, and human Prt1 were fused to the GAL4AD and cointroduced into strain AH109 with the indicated GAL4BD fusions. Yeast strains were plated on minimal medium lacking Trp and Leu (left) to select for cotransformants or plated on medium lacking Trp, Leu, and His (right) to select for interacting proteins. SV40 large T antigen (SV40-T)–GAL4AD and p53-GAL4BD fusion constructs were used as a positive control, and a lamin C-GAL4BD fusion peptide served as a negative control.
DISCUSSION
The recognition and accelerated decay of transcripts harboring premature termination codons, a phenomenon known as NMD, is a ubiquitous feature of eukaryotic cells (17). Despite this complete conservation of function, very little is known about the mechanism of NMD in higher eukaryotes. In S. cerevisiae, at least three trans effectors, Upf1p to Upf3p, are required for NMD (15, 27, 40, 42, 43). In C. elegans, seven factors, termed smg-1 to smg-7, are necessary (7, 30, 59). smg-2, the C. elegans orthologue of the Upf1p protein, is the only smg gene with a known counterpart in yeast (56). The apparent minimal overlap between the proteins which mediate NMD in yeast and C. elegans has lead to the presumption that higher eukaryotes use an NMD apparatus based on a fundamentally different set of factors from those used by more primitive cells. Here, we provide evidence that the nonsense surveillance pathway in evolutionarily divergent organisms utilizes structurally and functionally related machinery.
Evolutionary conservation of Upf2p function.
In this report, we describe Upf2p homologues in S. pombe and humans (rent2). Although S. pombe Upf2p exhibits only 22% identity to S. cerevisiae Upf2p, disruption of the gene established that the orthologue is essential for NMD in fission yeast. rent2, a protein equally divergent from S. cerevisiae Upf2p as is S. pombe Upf2p, interacts with rent1, a known trans effector of NMD in mammalian cells (1, 58, 67). This interaction is mediated by residues in rent1 and rent2 which correspond well to the domains which mediate the association of S. cerevisiae Upf1p and Upf2p (26). These data strongly argue that the function of Upf2p in the decay of nonsense transcripts is conserved throughout evolution. Indeed, other species also appear to possess Upf2p homologues. In addition to the genes described in this report, GenBank contains sequences from zebra fish, Drosophila, C. elegans, and Arabidopsis that encode proteins with significant homology to Upf2p (our unpublished observations). These results imply significant mechanistic overlap of the NMD pathway in many, if not all, eukaryotes.
Subcellular localization of rent2.
It is widely accepted that cytoplasmic translation is required for nonsense surveillance in S. cerevisiae (49). Experimental perturbations or transcript conformations that are known to impair translation initiation or elongation also abrogate NMD in mammalian cells (20, 47). Nevertheless, careful subcellular fractionation of mammalian cells has established that for most nonsense mRNAs, any decrease in cytoplasmic steady-state abundance or stability can be fully attributed to a reduction observed in the nuclear fraction (11, 38). A role for the nucleus in yeast NMD is also suggested by the observation that Upf3p shuttles between nuclear and cytoplasmic compartments (65). The prevailing view contends that scanning and decay occur very early in the cytoplasmic life of a transcript, while the mRNA is still associated with, but not confined to, the nuclear compartment (73). Shuttling of Upf3p and perhaps additional NMD trans effectors would be an effective mechanism of positioning them at or near the nuclear pore, such that they could recruit the surveillance complex to the earliest-translating ribosomes.
In light of the controversial role of the nucleus in nonsense surveillance, the presence of a unique N-terminal extension in rent2 containing 12 putative NLSs is particularly interesting. Additionally, Upf2p includes a previously described NLS without documented functionality (27), which we have found to be conserved in all Upf2p homologues. We have demonstrated that the N terminus of rent2 is sufficient to target GFP to the nucleus but does not direct apparent nuclear localization in the context of the native protein. rent2 may contain sequences which mask or inhibit the function of the NLS-containing region. Alternatively, rent2 may be rapidly exported following transient nuclear entry. Several examples exist for which this type of nuclear-cytoplasmic shuttling established an apparently cytoplasmic distribution of protein (41, 72). Although rent2 contains multiple putative Rev-like NESs, its subcellular distribution is not altered on treatment with leptomycin B, a specific inhibitor of this nuclear export pathway (37). Nuclear export pathways which are insensitive to this drug exist and may account for our observations. Interestingly, the known proteins that utilize these alternate export mechanisms are involved in mRNA metabolism (28, 69). Further studies are required to identify the sequence elements in rent2 which give rise to a cytoplasmic distribution despite the presence of functional NLSs.
Mechanistic implications of novel functional domains of Upf2p/rent2.
The most stable mRNA conformation is believed to be a closed-loop structure which is mediated by poly(A) binding protein and eIFs (Fig. 8) (60). This arrangement is thought to promote translation and prevent removal of the 5′ cap and subsequent 5′-to-3′ degradation of transcripts (21, 34). Functional communication between the 5′ and 3′ termini of mRNAs is well established. For example, despite binding to the 3′ end of transcripts, poly(A)-binding protein (PABP in mammalian cells, Pab1p in yeast) appears to protect or stabilize the 5′ cap (5). Consistent with this function, generalized deadenylation-independent decapping occurs in pab1Δ strains of S. cerevisiae (8) and tethering of Pab1p to transcripts lacking poly(A) tails is sufficient to prevent premature decapping (12). The poly(A) tail is also known to have stimulatory effects on translation initiation (60). Additionally, several lines of evidence suggest a role for the translation initiation factors in the regulation of transcript stability. Temperature-sensitive mutations in eIF4G, eIF4A, eIF4E, and PRT1 (a subunit of eIF3) cause accelerated deadenylation and decay of wild-type transcripts at the nonpermissive temperature (64). Perhaps more relevant to the deadenylation-independent decay of nonsense transcripts, specific alleles of PRT1 (74) and SUI1 (another eIF3 subunit) (14) selectively impair the efficiency of NMD without affecting wild-type transcript stability.
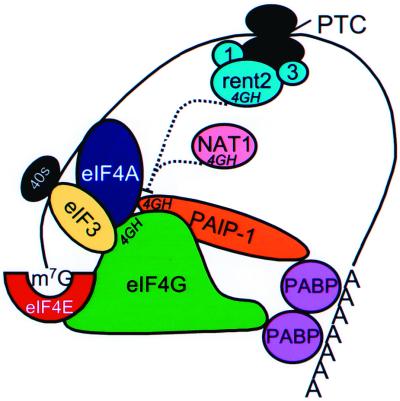
FIG. 8.
Proposed model of 4GH domain-mediated transcript destabilization. Stable, efficiently translated transcripts are believed to adopt a closed-loop conformation mediated by protein-protein interactions which bridge the 5′ cap (m7G) and the 3′ poly(A) tail. The model suggests that the 4GH domains of rent2 and NAT1 may compete with the corresponding domain in eIF4G (and perhaps PAIP-1) for interacting partners eIF3 and/or eFI4A. Such associations (dashed lines) may occur in cis when rent2 is a component of a mature surveillance complex (partnered with rent1 and rent3) but in trans upon induction of NAT1 expression.
Given that nonsense transcripts undergo deadenylation-independent decapping and are translated less efficiently than their wild-type counterparts (54), the factors which mediate communication between PABP or Pab1p and the cap-binding complex (eIF4E and eIF4A) are appealing putative targets for the effector arm of NMD. eIF4G is particularly attractive, since it binds PABP or Pab1p, eIF4E, and eIF4A simultaneously and synergistically, thus serving an important bridging function in the formation of the closed loop (29). Mammalian cells also express PAIP-1, which has homology to eIF4G and binds PABP and eIF4A (13). In this study, we have identified two novel functional domains of Upf2p and rent2, termed 4GH domains, which are similar to sequences found in eIF4G and PAIP-1. Both eIF4G and PAIP-1 bind to eIF4A via regions that encompass 4GH domains (13, 33). The 4GH domain region of eIF4G also participates in binding to eIF3. Point mutations in highly conserved residues of the 4GH domain of eIF4G are sufficient to abolish eIF4A and eIF3 binding (33). NAT1, the only other known protein to contain a 4GH domain, is induced during apoptosis and effects a generalized and severe decline in translational efficiency (32, 46, 76). Like eIF4G, NAT1 binds to eIF3 and eIF4A but fails to bind eIF4E (33). Current theory holds that NAT1 interferes with translation by competing for eIF4G (and perhaps PAIP-1) interactions.
The presence of 4GH domains in Upf2p and rent2 suggests that this protein may participate in NMD through interaction with components of the translation initiation complex. We have demonstrated that the 4GH domains of S. pombe Upf2p are required for NMD function. Additionally, we have provided evidence that rent2 interacts with human eIF4AI and Sui1. The potential for interaction with Sui1 is particularly provocative since the yeast homologue of this molecule has previously been demonstrated to be a trans effector of NMD (14). These data suggest a model in which Upf2p or rent2 competes with eIF4G for 4GH domain-mediated interactions, thus disrupting or preventing the formation of the stable, closed-loop mRNP structure (Fig. 8). Unlike NAT1, which constitutively competes in trans on induction of expression, one could imagine that Upf2p and rent2 can compete only in cis upon the recognition of a PTC and recruitment and activation of a mature surveillance complex. These Upf2p- and rent2-mediated interactions would have to be tightly regulated, or a general inhibition of translation and/or decrease in mRNA stability would occur. Our two-hybrid analysis is consistent with this prediction. Only a specific fragment of rent2, spanning residues 757 to 1272 and containing 4GH2, is capable of interacting with the translation initiation factors. Perhaps only this fragment lacks regulatory sequences which normally serve to prevent inappropriate interactions. Additionally, compared to the full-length eIF4AI protein, a truncated form of eIF4AI lacking the C-terminal 82 amino acids appeared to interact more strongly with rent2. While this may simply reflect technical limitations of the two-hybrid system, it is also possible that the C terminus of eIF4AI serves to regulate this interaction. Additional biochemical analysis is required to further characterize the nature and precise molecular determinants of these interactions.
Remarkably, virtually nothing is known about the effects of NAT1 on mRNA stability. If competition with eIF4G interactions indeed disrupts the closed-loop conformation of transcripts, generalized mRNA lability is likely to result from expression of this protein. Moreover, if our model is correct, we would predict that the decay pathway induced by NAT1 would mimic NMD and occur independently of deadenylation.
This model of Upf2p and rent2 function reconciles a number of previous observations. Disruption of the closed-loop conformation in cis to a PTC would be predicted to uncouple decapping from deadenylation, decrease the efficiency of translation initiation in addition to accelerating transcript decay, and increase the accessibility of a transcript to the decapping enzyme Dcp1p. Each of these predictions is a fundamental characteristic of NMD (53, 54, 71). Upf2p and rent2 may provide a predicted link between the translation initiation apparatus and the nonsense surveillance complex.
In conclusion, we have demonstrated that Upf2p is an evolutionarily conserved component of the NMD machinery in S. pombe and humans. The study of conserved domains of this protein has provided novel insights into the basic mechanism of nonsense surveillance. Our improved understanding of this process will aid future efforts to delineate the physiologic role of NMD in mammalian cells and perhaps to manipulate the pathway for therapeutic purposes.
ACKNOWLEDGMENTS
We acknowledge J. Boeke and G. Smith for S. pombe strains and reagents. We thank M. Sands for gusmps mice, M. Yoshida for leptomycin B, M. Dellanoy for technical assistance with confocal microscopy, and D. Arking for assistance with manuscript preparation.
This work was supported by the NIH (grant GM55239), the Howard Hughes Medical Institute (to H.C.D.), and the Medical Scientist Training Program (to J.T.M.).
REFERENCES
- 1.Applequist S E, Selg M, Raman C, Jäck H M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:15–23. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkin A L, Altamura N, Leeds P, Culbertson M R. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkin A L, Schenkman L R, Eastham M, Dahlseid J N, Lelivelt M J, Culbertson M R. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 4.Bassett D E, Jr, Boguski M S, Spencer F, Reeves R, Goebl M, Hieter P. Comparative genomics, genome cross-referencing and XREFdb. Trends Genet. 1995;11:372–373. doi: 10.1016/s0168-9525(00)89109-x. [DOI] [PubMed] [Google Scholar]
- 5.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 6.Birkenmeier E H, Davisson M T, Beamer W G, Ganschow R E, Vogler C A, Gwynn B, Lyford K A, Maltais L M, Wawrzyniak C J. Murine mucopolysaccharidosis type VII. Characterization of a mouse with beta-glucuronidase deficiency. J Clin Investig. 1989;83:1258–1256. doi: 10.1172/JCI114010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cali B M, Kuchma S L, Latham J, Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics. 1999;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 9.Carter M S, Doskow J, Morris P, Li S, Nhim R P, Sandstedt S, Wilkinson M F. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 10.Cheng J, Belgrader P, Zhou X, Maquat L E. Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol Cell Biol. 1994;14:6317–6325. doi: 10.1128/mcb.14.9.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J, Maquat L E. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol. 1993;13:1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coller J M, Gray N K, Wickens M P. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig A W, Haghighat A, Yu A T, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 14.Cui Y, Gonzalez C I, Kinzy T G, Dinman J D, Peltz S W. Mutations in the MOF2/SUI1 gene affect both translation and nonsense-mediated mRNA decay. RNA. 1999;5:794–804. doi: 10.1017/s1355838299982055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 16.Culbertson M R. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 17.Czaplinski K, Ruiz-Echevarria M J, Gonzalez C I, Peltz S W. Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. Bioessays. 1999;21:685–696. doi: 10.1002/(SICI)1521-1878(199908)21:8<685::AID-BIES8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 18.Czaplinski K, Ruiz-Echevarria M J, Paushkin S V, Han X, Weng Y, Perlick H A, Dietz H C, Ter-Avanesyan M D, Peltz S W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietz H C, Valle D, Francomano C A, Kendzior R J, Jr, Pyeritz R E, Cutting G R. The skipping of constitutive exons in vivo induced by nonsense mutations. Science. 1993;259:680–683. doi: 10.1126/science.8430317. [DOI] [PubMed] [Google Scholar]
- 20.Frischmeyer P A, Dietz H C. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 21.Gallie D R. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez C I, Ruiz-Echevarria M J, Vasusevan S, Henry M F, Pletz S W. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol Cell. 2000;5:489–499. doi: 10.1016/s1097-2765(00)80443-8. [DOI] [PubMed] [Google Scholar]
- 23.Gozalbo D, Hohmann S. Nonsense suppressors partially revert the decrease of the mRNA level of a nonsense mutant allele in yeast. Curr Genet. 1990;17:77–79. doi: 10.1007/BF00313252. [DOI] [PubMed] [Google Scholar]
- 24.Grimm C, Kohli J, Murray J, Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- 25.He F, Brown A H, Jacobson A. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA. 1996;2:153–170. [PMC free article] [PubMed] [Google Scholar]
- 26.He F, Brown A H, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 28.Henderson B R, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res. 2000;256:213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- 29.Hentze M W. eIF4G: a multipurpose ribosome adapter? Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. . (Erratum, 275:1553.) [DOI] [PubMed] [Google Scholar]
- 30.Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imataka H, Olsen H S, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobson A. Poly(A) metabolism and translation: the closed-loop model. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 35.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 36.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner E P, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kugler W, Enssle J, Hentze M W, Kulozik A E. Nuclear degradation of nonsense mutated beta-globin mRNA: a post- transcriptional mechanism to protect heterozygotes from severe clinical manifestations of beta-thalassemia? Nucleic Acids Res. 1995;23:413–418. doi: 10.1093/nar/23.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaGrandeur T E, Parker R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee B S, Culbertson M R. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci USA. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S, Neumann M, Stearman R, Stauber R, Pause A, Pavlakis G N, Klausner R D. Transcription-dependent nuclear-cytoplasmic trafficking is required for the function of the von Hippel-Lindau tumor suppressor protein. Mol Cell Biol. 1999;19:1486–1497. doi: 10.1128/mcb.19.2.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 43.Leeds P, Wood J M, Lee B S, Culbertson M R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Hir H, Moore M J, Maquat L E. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 45.Lelivelt M J, Culbertson M R. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy-Strumpf N, Deiss L P, Berissi H, Kimchi A. DAP-5, a novel homolog of eukaryotic translation initiation factor 4G isolated as a putative modulator of gamma interferon-induced programmed cell death. Mol Cell Biol. 1997;17:1615–1625. doi: 10.1128/mcb.17.3.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci USA. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maderazo A B, He F, Mangus D A, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol. 2000;20:4591–4603. doi: 10.1128/mcb.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 50.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 51.Mietz J A, Unger T, Huibregtse J M, Howley P M. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 53.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 54.Muhlrad D, Parker R. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol Biol Cell. 1999;10:3971–3978. doi: 10.1091/mbc.10.11.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Page M F, Carr B, Anders K R, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peltz S W, Brown A H, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans- acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 58.Perlick H A, Medghalchi S M, Spencer F A, Kendzior R J, Jr, Dietz H C. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 60.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 62.Sands M S, Birkenmeier E H. A single-base-pair deletion in the beta-glucuronidase gene accounts for the phenotype of murine mucopolysaccharidosis type VII. Proc Natl Acad Sci USA. 1993;90:6567–6571. doi: 10.1073/pnas.90.14.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 64.Schwartz D C, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shirley R L, Lelivelt M J, Schenkman L R, Dahlseid J N, Culbertson M R. A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J Cell Sci. 1998;111:3129–3143. doi: 10.1242/jcs.111.21.3129. [DOI] [PubMed] [Google Scholar]
- 66.Stephenson L S, Maquat L E. Cytoplasmic mRNA for human triosephosphate isomerase is immune to nonsense-mediated decay despite forming polysomes. Biochimie. 1996;78:1043–1047. doi: 10.1016/s0300-9084(97)86728-4. [DOI] [PubMed] [Google Scholar]
- 67.Sun X, Perlick H A, Dietz H C, Maquat L E. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci USA. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szankasi P, Heyer W D, Schuchert P, Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination host spot allele ade6–M26. J Mol Biol. 1988;204:917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- 69.Tang H, McDonald D, Middlesworth T, Hope T J, Wong-Staal F. The carboxyl terminus of RNA helicase A contains a bidirectional nuclear transport domain. Mol Cell Biol. 1999;19:3540–3550. doi: 10.1128/mcb.19.5.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tarun S Z, Jr, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 71.Tharun S, Parker R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;151:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. Nuclear export of cyclin B1 and its possible role in the DNA damage- induced G2 checkpoint. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Urlaub G, Mitchell P J, Ciudad C J, Chasin L A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989;9:2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Welch E M, Jacobson A. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 1999;18:6134–6145. doi: 10.1093/emboj/18.21.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weng Y, Czaplinski K, Peltz S W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamanaka S, Poksay K S, Arnold K S, Innerarity T L. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997;11:321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]
- 77.Yang J, Bardes E S, Moore J D, Brennan J, Powers M A, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang S, Ruiz-Echevarria M J, Quan Y, Peltz S W. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol Cell Biol. 1995;15:2231–2244. doi: 10.1128/mcb.15.4.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]