Abstract
The Schizosaccharomyces pombe DNA repair gene rhp51+ encodes a RecA-like protein with the DNA-dependent ATPase activity required for homologous recombination. The level of the rhp51+ transcript is increased by a variety of DNA-damaging agents. Its promoter has two cis-acting DNA damage-responsive elements (DREs) responsible for DNA damage inducibility. Here we report identification of Rdp1, which regulates rhp51+ expression through the DRE of rhp51+. The protein contains a zinc finger and a polyalanine tract similar to ones previously implicated in DNA binding and transactivation or repression, respectively. In vitro footprinting and competitive binding assays indicate that the core consensus sequences (NGG/TTG/A) of DRE are crucial for the binding of Rdp1. Mutations of both DRE1 and DRE2 affected the damage-induced expression of rhp51+, indicating that both DREs are required for transcriptional activation. In addition, mutations in the DREs significantly reduced survival rates after exposure to DNA-damaging agents, demonstrating that the damage response of rhp51+ enhances the cellular repair capacity. Surprisingly, haploid cells containing a complete rdp1 deletion could not be recovered, indicating that rdp1+ is essential for cell viability and implying the existence of other target genes. Furthermore, the DNA damage-dependent expression of rhp51+ was significantly reduced in checkpoint mutants, raising the possibility that Rdp1 may mediate damage checkpoint-dependent transcription of rhp51+.
All organisms have developed defense mechanisms to respond to genotoxic materials causing genetic injury. One response to DNA damage or DNA synthesis inhibition is to delay the cell cycle by blocking DNA replication and/or mitotic division. Another is the transcriptional induction of several genes whose products may contribute to DNA repair capacity (22).
During past decades, such damage-inducible genes have been identified and partially characterized for bacteria, yeasts, and higher eukaryotes (22). In particular, a large number of genes are induced in response to DNA damage and/or inhibition of DNA replication in Saccharomyces cerevisiae (22, 49). These include RNR2 (which encodes a small subunit of ribonucleotide reductase [19, 28]), RNR3 (which encodes a large subunit of ribonucleotide reductase [20]), CDC9 (which encodes DNA ligase [6]), and CDC17 (which encodes DNA polymerase I [21]), which are involved in DNA metabolism, and RAD2 (which encodes a DNA endonuclease required for nucleotide excision repair [NER] [48]), RAD7 (required for NER [48]), RAD18 (required for postreplication repair [34]), RAD23 (which encodes a uniquitin-like protein required for NER [41]), RAD51 (which encodes a RecA homolog required for double-strand break repair [7]), RAD54 (which encodes a putative DNA helicase required for double-strand break repair [13]), PHR1 (which encodes a photoreactivating enzyme [52]), and MAG (which encodes 3-methyladenine DNA glycosylase [12]), which are involved in DNA repair. However, the biological significance of the transcriptional induction of these genes has been uncovered only recently. A study has revealed that Dun1p serine/threonine protein kinase is involved in the transcriptional activation of RNR2 and has delineated a pathway by which the damage signal is transduced to a checkpoint and transcription response apparatus (65). The repressor protein Crt1p was found to bind to X boxes on the RNR2 and RNR3 promoters and to mediate repression of the genes by cooperating with the Tup1p-Ssn6p corepressor. DNA damage-induced hyperphosphorylation of Crt1p enables the protein to dissociate from X boxes, which leads to derepression of RNR2 transcription. This dissociation is also dependent on the MEC1-RAD53-DUN1 damage-signaling pathway (27). In another damage-inducible gene, PHR1, a 39-bp upstream repressing sequence (URS) is responsible for the damage induction (51). Rph1p and Gis1p have been identified as the regulators that bind PHR1's URS (29). Transcriptional regulation mediated by Rph1p, Gis1p, and Crt1p is similar in that derepression is responsible for their damage-inducible expression.
Despite a great effort to determine otherwise, it was revealed that the Rad53p-Dun1p-Crt1p cascade controls only a small set of genes, including RNR2 and RNR3 but not UBI4, a well-known damage-inducible gene encoding a single polypeptide consisting of multiple ubiquitin moieties. Presently, PHR1 is the only known target of Rph1p and Gis1p. The data strongly argue for the existence of multiple regulators involved in the DNA damage response. At present, the Spc1-Atf1 cascade involved in a general stress response is the best-characterized transcriptional response to damage in the fission yeast Schizosaccharomyces pombe (62). Atf1 factor is both a structural and a functional homolog of the mammalian bZip domain factor ATF-2 and is a key regulator of a number of target genes that are involved in stress responses (gpd1+, fbp1+, and catalase) and in the sexual differentiation pathway (ste11+) (53, 63). It appears that the S. pombe stress response closely resembles the mammalian stress response. It may therefore be a useful model system for studying stress-related events and the DNA damage response.
Thus, to identify novel regulators required for the activation of repair genes by DNA damage from fission yeast in addition to S. cerevisiae, we have been studying transcriptional regulation of rhp51+, a recA homolog from the fission yeast S. pombe. We have previously reported that rhp51+ expression is cell cycle regulated and induced by DNA damage but not by stress stimuli (30). The induction appears to require two decamer damage-responsive elements (DREs) commonly found in several DNA repair genes of both S. cerevisiae and S. pombe (31). To define the final effector involved in sensing and transducing the damage-inducible response, we attempted to identify a protein(s) that interacts with the DREs of rhp51+. We report here that a novel zinc finger protein, Rdp1, isolated by one-hybrid screening, specifically binds to the DRE in vitro and is essential for cell proliferation. Furthermore, we show that consensus sequences of the DRE are essential for Rdp1 binding and that the mutated DREs cause a significant reduction in the transcriptional induction of rhp51+, leading to a decrease in UV and methyl methanesulfonate (MMS) resistance. Our observations provide the first evidence that a novel transcriptional activator recognizing common consensus binding sequences regulates the damage-inducible response in S. pombe.
MATERIALS AND METHODS
Strains and cell culture.
S. cerevisiae strain RH1006 (MATa can1-100 his3-11 his3-15 leu2-3 leu2-112 trp1-1 ura3-52) was used as a host for one-hybrid screening (a gift from Mark Johnston). S. pombe JAC10 (h+ ade6-704 rhp51::ura4+ leu1-32::wtDRE-rhp51+-leu1+) and JAC20 (h+ ade6-704 rhp51::ura4+ leu1-32::mDRE-rhp51+-leu1+) were derived from transformation of JAC1/51Δ (h+ ade6-704 leu1-32 rhp51::ura4+) with pJKwt and pJKmD, respectively. The S. pombe diploid strain used for rpd1 disruption was obtained by a cross between ED665 (h− ade6-M210 leu1-32 ura4-D18) and ED668 (h+ ade6-M216 leu1-32 ura4-D18) from P. Fantes, Edinburgh, Scotland. S. pombe checkpoint mutant strains 1451 (ade6-704 ura4-D18 leu1-32 cds1::ura4+ chk1::ura4+), 1324 (ade6-704 ura4-D18 leu1-32 rad1::ura4+), 1378 (h− ade6-704 ura4-D18 leu1-32 rad3::ura4+), 1161 (ade6-M210/ade6-M216 on Ch16 ura4-D18 leu1-32 rad9::ura4+), 941 (h− ade6-704 ura4-D18 leu1-32 rad17::ura4+), 1123 (h− ade6-704 ura4-D18 leu1-32 rad26::ura4+), and Δcds1 (ura4-D18 leu1-32 cds1::ura4+) were generous gifts from Tony Carr (Sussex University, Falmer, United Kingdom). Strain TE484 (h− ura4-D18 leu1-32 hus1::LEU2) was obtained from T. Enoch (Harvard Medical School, Boston, Mass.). Strain NW158 (h+ ade6-M216 ura4-D18 leu1-32 chk1::ura4+) was kindly provided by N. Walworth (University of Medicine and Dentistry of New Jersey, Piscataway). Yeast cells were grown in YPD (1% yeast extract, 2% Bacto Peptone, 2% glucose) and YES (0.5% yeast extract plus 3% glucose, supplemented with appropriate amino acids) for S. cerevisiae and S. pombe, respectively. Selective media were prepared as described elsewhere (31).
Plasmids.
The DNA structures of all plasmids were confirmed by restriction analysis and in some cases by sequencing.
The reporter plasmids for one-hybrid screening were constructed as follows. Three tandem copies of the annealed complementary oligonucleotide corresponding to the sequences from −236 to −199 bp containing the DREs of the rhp51+ promoter (DRErhp51+) (Fig. 1A) were inserted into the BamHI site of pRS315HIS (61) to generate the DRErhp51+-HIS3 plasmid pHis-F3. The 2.0-kb BamHI-SalI fragment of pHis-F3 was then ligated into the CEN-ARS-URA3 plasmid YCplac33 (23) to create pHis33-F3. The DRErhp51+-lacZ reporter plasmid pRW3-F3 was constructed by inserting a 150-bp EcoRI fragment of pHis33-F3 containing three copies of the DREs immediately upstream of lacZ in the CEN-ARS-TRP1 plasmid pRW95-3 (64). The S. pombe cDNA expression library based on a 2μm-LEU2 plasmid, pGAD GH, was purchased from Clontech (catalog no. XL4000AA).
FIG. 1.
Screening for DRErhp51+-binding protein. (A) Schematic representation of the rhp51+ promoter. Numbering is relative to the first base in the rhp51+ coding sequence. Filled rectangles indicate two decamer DRE sequences (−233 to −224 and −213 to −204), and hatched diamonds indicate two MCBs (−192 to −187 and −183 to −178). (B) Identification of a positive clone by an X-Gal plate assay. Levels of expression of the lacZ reporter gene between the empty vector and putative clones were compared by X-Gal assays. A positive clone, pGAD236, became dark blue, while the parental empty vector (pGAD424) and other putative clones did not show enhanced β-galactosidase activity.
The truncated cDNA sequence of rdp1+ was generated by PCR using oligonucleotides OR1F-Bam (5′-CACGGGATCCAACACTCCCACCGTAG-3′) and OR540R-EcoR (5′-GGGGTTGGAATCAGGCACTTGAC-3′) as primers and pGAD236 as the template. The 0.5-kb BamHI-EcoRI PCR product was subcloned into pGEX4T-1 and then used for expression of glutathione S-transferase (GST)-fused Rdp1.
To disrupt the rdp1+ gene, a 5.3-kb EcoRI-XhoI fragment containing the entire gene was derived from the cosmid SPAC1B1 (a gift from Rhian Gwilliam at The Sanger Centre) and subcloned into pBSIIKS(+) to create plasmid prdp1-830. The 1.1-kb BalI-BclI fragment from prdp1-830 was replaced with a 1.8-kb HincII-BamHI fragment of ura4+ to create pBS-rdp1::ura4+. The 2.3-kb SacI-HpaI fragment of the disruption cassette was used for transformation of yeast.
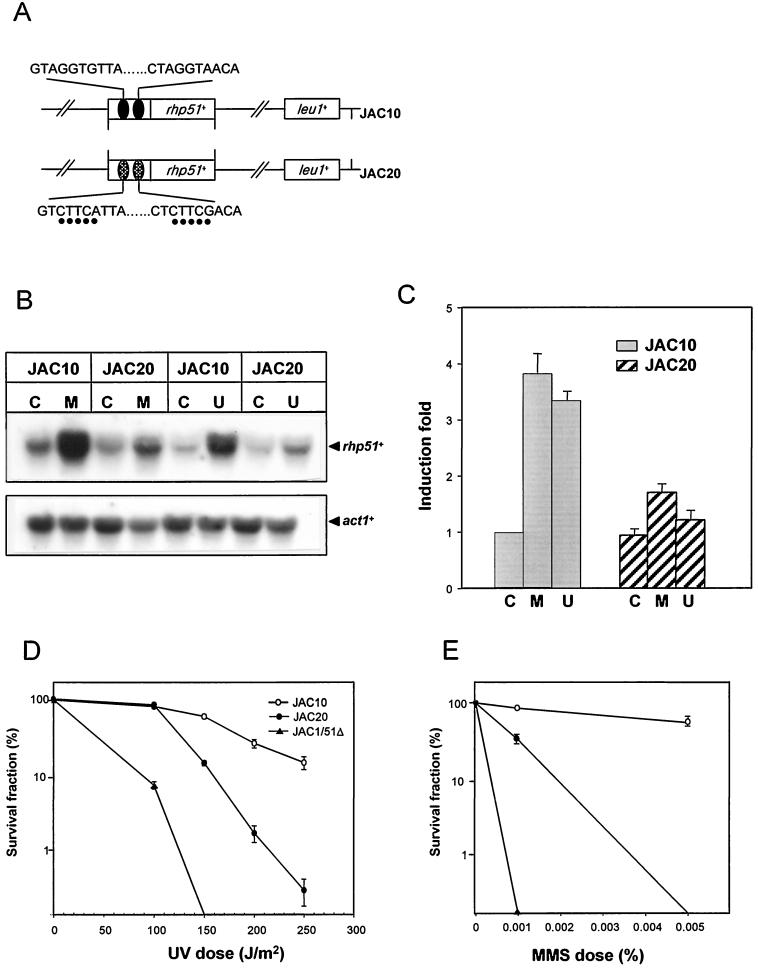
pJKwt and pJKmD are derivatives of an integration vector, pJK148, containing wild-type-DRE- and mutated-DRE-driven rhp51+ genes, respectively. Site-directed mutagenesis of the DRE sequence was carried out using the single-strand DNA of the pBluescript-rhp51+ phagemid and a primer containing DRE mutations (5′-TGTGTCTATTTAGTCTTCATTACCTTGCTAGTACTCTTCAACAATTGAAATCGCGTCGGACGCCTTTTAA-3′) that changed the DRE core sequence from AGGTG to CTTCA (italics) as described elsewhere (50). The resulting plasmid was confirmed by nucleotide sequencing. Wild-type rhp51+ and mutated DRE-regulated rhp51+ copies were subcloned into the pJK148 vector for chromosomal integration and named pJKwt and pJKmD, respectively. The pJK148 derivatives were used for integration at the leu1-32 locus to create a stable single-copy background. For integration, the plasmid pJKwt or pJKmD was linearized with NruI and used for transformation of the rhp51Δ null mutant strain JAC1/51Δ. To confirm stable and precise single-copy integration, robust Leu+ transformants were assessed by Southern blotting by using the 1-kb EcoRV fragment of the leu1+ gene as a probe as described elsewhere (36). For each construct, only the single-copy integrant was selected, and they were named JAC10 (wtDRE-rhp51+) and JAC20 (mDRE-rhp51+) as described in the legend to Fig. 5A.
FIG. 5.
Effect of mutated DREs on rhp51+ expression and survival after treatment with UV and MMS. (A) Illustration of the rhp51+ gene structure in a host strain harboring wild-type DRE (JAC10) or mutated DRE (JAC20). Mutated bases are indicated by dots under the bases. (B) mRNA levels of rhp51+ following UV irradiation or MMS treatment. Exponentially growing cells were exposed to 0.1% MMS or 180 J of UV light per m2 and postincubated for 1 h. Total RNAs were extracted, and rhp51+ mRNA levels were assessed by Northern blotting. Symbols: C, mock treatment; M, 0.1% MMS treatment; U, 180 J of UV irradiation per m2. (C) Relative rhp51+ mRNA levels after DNA damage. The data were obtained from five independent experiments and normalized to data with act1+. The error bars indicate standard deviations. Symbols: C, mock treatment; M, 0.1% MMS treatment; U, 180 J of UV irradiation per m2. (D) Comparison of UV sensitivities. Cells were exposed to UV light at the indicated doses on YES plates, and the surviving colonies were counted after 4 to 5 days. The data points are averages from at least three independent experiments, and the error bars indicate standard deviations. (E) Comparison of MMS sensitivities. The MMS survival test was performed as described in Materials and Methods.
Yeast one-hybrid screening.
Two reporter plasmids, pHis33-F3 and pRW3-F3, were first introduced into RH1006. The Ura+ Trp+ transformants were then transformed with the cDNA expression library to which the activation domain of GAL4 was fused and screened for histidine prototrophy. The initial Ura+ Trp+ His+ colonies were rescreened for increased β-galactosidase activity by using an X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) assay (17). Plasmids from positive clones were recovered in DH5α cells and used to transform naive RH1006 cells harboring two reporter plasmids to confirm the Ura+ Leu+ His+ phenotype and increased β-galactosidase production. Finally, the insert DNA from the positive clone was sequenced at the 5′ and 3′ fusion sites using a Sequenase version 2.0 kit (Amersham).
Expression and purification of the GST fusion protein.
Escherichia coli BL21 was used for expression of GST fusion proteins. Expression and purification of the fusion protein were performed under conditions recommended by the manufacturer (Pharmacia). Briefly, following induction of mid-log-phase cultures of BL21 with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the cells were lysed with breaking solution (2% Triton X-100, 100 mM NaCl, 10 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]). Fusion proteins were incubated with glutathione-Sepharose 4B for 1 h, washed, and eluted with 10 mM reduced glutathione–50 mM Tris (pH 8.0).
Electrophoretic gel mobility shift assay (EMSA) and DNase I footprinting.
One hundred nanograms of GST-Rdp1 was incubated with a 5 nM concentration of the 32P-end-labeled double-stranded oligonucleotide in a volume of 20 μl. Unlabeled competitors were prepared by hybridization of oligonucleotide pairs (see Fig. 4A). The final binding solution contained 4 mM Tris-HCl (pH 8.0), 40 mM MgCl2, 40 mM NaCl, 1 μM ZnCl2, 10% glycerol, 100 μg of bovine serum albumin per ml, and 5 mM dithiothreitol. Competition experiments with unlabeled oligonucleotides typically employed a 5- to 100-fold molar excess of DNA relative to the concentrations of radiolabled probes. After 20 min of incubation on ice, DNA-protein complexes were separated on a 4% native polyacrylamide gel containing 0.25× Tris-borate-EDTA.
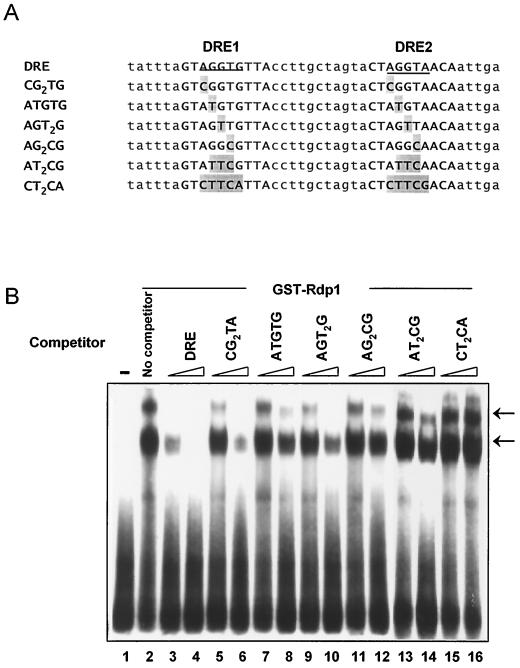
FIG. 4.
Determination of the binding consensus sequences by EMSA. (A) Nucleotide sequences of competitors used. Sites changed relative to the sequence of wild-type DRE are indicated by gray boxes. (B) Competition assay. The radiolabeled DRE was incubated with GST-Rdp1 with or without the indicated unlabeled competitor. Two concentrations are shown for each competitor, 100 nM (lanes 3, 5, 7, 9, 11, 13, and 15) and 500 nM (lanes 4, 6, 8, 10, 12, 14, and 16). Lane 1 contains the DNA substrate only, and lane 2 contains the substrate and GST-Rdp1 protein without competitor. Arrows indicate the bound DNA-protein complexes.
DNA footprinting was performed as previously described (31). 32P-labeled probes were prepared by treating the oligonucleotide 51SB (5′-AGTAGGGATGTGAGG-3′) with T4 polynucleotide kinase. PCR was performed using labeled 51SB and unlabeled UASIa (5′-AGCTTCGTTCCCTATCTCGTGA-3′) as primers and p51-420 (30) as the template. The 140-bp PCR product was cleaned and gel purified using a ProbeQuant G-50 micro column (Pharmacia), followed by electrophoresis in 6% polyacrylamide gel. Binding reactions were carried out in 20 μl with a 10 nM concentration of the 32P-labeled probe, 500 ng of poly(dA-dT), and 200 ng of GST-Rdp1. The binding buffer was as the same as that used in the EMSA. Following 20 min of incubation at room temperature, 1 U of DNase I (Promega) and 1 μl of 50 mM CaCl2 were added to the reaction. The reaction was allowed to proceed on ice for 30 s to 2 min and then stopped by the addition of 20 μl of stop solution containing 1% sodium dodecyl sulfate (SDS), 200 mM NaCl, 20 mM EDTA, and 40 μg of tRNA per ml. After extraction with phenol and precipitation with ethanol, the products were analyzed on an 8% polyacrylamide gel containing 7 M urea. The gel was dried and exposed to a phosphorimage analyzer (model BAS1500; Fuji).
Northern blot analysis and UV survival test.
Total RNA from S. pombe cells was isolated by extraction with phenol-chloroform-SDS (33). A 30-μg sample of the total RNA was separated on a 1.2% agarose gel containing 0.67 M formaldehyde and transferred onto Nytran membrane. After stringent washes, the blot was exposed to X-ray film or the phosphorimage analyzer. To detect the rhp51+ transcripts, a 0.4-kb EcoRI fragment corresponding to an internal region of the rhp51+ open reading frame (ORF) was labeled by the random primer method (31) and then used as a probe.
A survival test was performed as previously described (32). For UV survival, mid-log-phase cells were serially diluted to a final density of 4 × 103 cells/ml in distilled water. Four hundred cells were plated on YES and irradiated with various doses of UV using a Stratalinker 1800 (Stratagene). Plates were incubated at 30°C for 4 to 5 days, and colonies were counted. The relative survival of strains was calculated as the ratio of the number of colonies on UV-irradiated plates relative to the number of colonies on unirradiated plates. For MMS (Sigma-Aldrich, St. Louis, Mo.) survival, exponentially growing cells were directly plated onto rich medium in the presence of MMS at doses indicated in the figures. Colonies were counted after 4 to 5 days of growth at 30°C.
Selective spore germination analysis.
For analysis of the rdp1Δ phenotype in liquid culture, a wild-type strain (ura4-D18/ura4-D18 ade6-M210/ade6-M216 leu1-32/leu1-32 h+/h+) and the strain with an rdp1 deletion (rdp1+/rdp1::ura4+ ura4-D18/ura4-D18 ade6-M210/ade6-M216 leu1-32/leu1-32 h+/h−) were grown in YES to mid-log phase and then sporulated in ammonium-free minimal media for 4 days. These cells were treated with 1% glusulase (Sigma) at 25°C overnight and washed two times with distilled water. Spores were collected by centrifugation at 1,500 × g for 20 min. For germination, the spores (2 × 107 to 2 × 108) were inoculated into minimal medium and incubated at 30°C and then harvested at various times. These cells were fixed in 70% ethanol and stained with 4′,6-diamidino-2-phenylindole (DAPI) (0.1 mg/ml). The nuclear morphology was examined using an inverted system microscope, model IX150, supplemented with fluorescence accessories (Olympus Optical Co. Ltd., Tokyo, Japan).
RESULTS
Identification of Rdp1 as a putative regulator acting through the DRErhp51+.
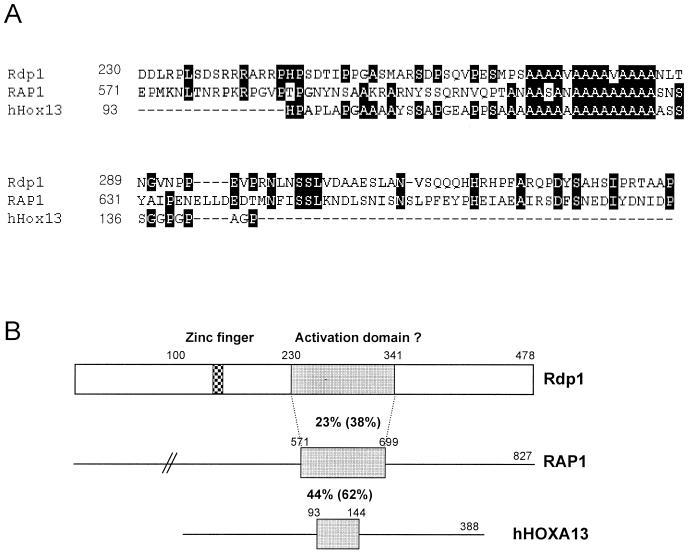
We have previously shown that the damage-inducible expression of rhp51+ requires the 69-bp region between positions −254 and −185 of its promoter. The 69-bp region contains two DRE elements (5′-GTAGGTGTTA-3′ and 5′-CTAGGTAACA-3′) (Fig. 1A) (33), to which DNA-binding proteins bind (31). To identify these DRErhp51+-binding proteins, we used the yeast one-hybrid system. Three copies of the DRE as bait were inserted into the minimal promoter regions of two reporter genes, HIS3 and lacZ, creating plasmids pHis33-F3 (URA3 marker) and pRW3-F3 (TRP1 marker), respectively. The reporter plasmids were used to transform RH1006 together with the S. pombe cDNA library (LEU2 marker). Approximately 3 million Ura+ Leu+ Trp+ transformants were tested for histidine prototrophy, and 25 His+ clones were obtained and examined for β-galactosidase activity. Only one of these clones, designated pGAD236, turned dark blue on X-Gal indicator plates (Fig. 1B). The plasmid with a 1.5-kb cDNA insert was recovered, and its nucleotide sequence was determined. Its ORF was identical to the sequence of the cosmid SPAC1B1.01 available from the Sanger Centre (http://www.sanger.ac.uk/pombe.html), which had not been characterized previously. We have named it rdp1+ (stands for rhp51+-DRE-binding protein). rdp1+ encodes a C2H2 zinc finger protein of 478 amino acid residues, with a calculated molecular mass of 53 kDa. The deduced amino acid sequence of the Rdp1 protein included several putative phosphorylation sites by protein kinase C and casein kinase II but did not show significant overall homology to any of the known proteins in the protein database. In Fig. 2A and B, the deduced ORF of 478 amino acids is aligned with S. cerevisiae RAP1, which is involved in transcription and telomeric silencing (54), and with the human homeodomain gene HOXA13 (35). Here, we have aligned the ORF only with these two genes because their deduced amino acid sequences showed the highest similarities among many proteins showing homology with Rdp1. Interestingly, the region having obvious sequence similarity among them was restricted to the ∼100-amino-acid stretch surrounding the polyalanine tract implicated in the control of transcription (24).
FIG. 2.
Protein domains of Rdp1. (A) Alignment of Rdp1 with other transcription factors. The Rdp1 shows a limited but significant homology with RAP1 (S. cerevisiae) and many homeodomain proteins from higher eukaryotes. The amino acid sequences from RAP1 and human HOXA13 (hHox13) were aligned with Rdp1 by the CLUSTAL W program, and output was generated by Genedoc. Black and gray shadows indicate identical and homologous amino acids, respectively. (B) Schematic diagram of the Rdp1 protein domain. The gray rectangle of Rdp1 indicates a region homologous with the activation domain of RAP1 and the polyalanine tract of HOXA13. Percentages indicate sequence identities (and similarities).
Rdp1 specifically binds to the DRErhp51+ in vitro.
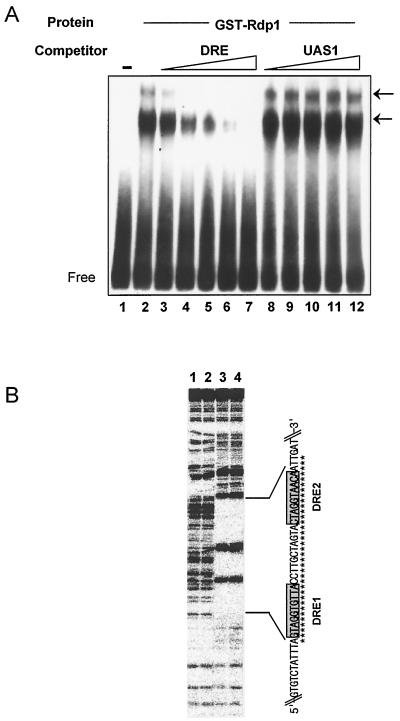
To verify that Rdp1 can bind to the DRE sequence in vitro, we examined its DNA-binding properties using EMSA. Recombinant GST-fused Rdp1 protein was expressed in E. coli, purified, and tested for specific high-affinity binding to the DRErhp51+. EMSA was performed with 32P-labeled DRErhp51+ and 100 ng of GST-Rdp1 protein. The results of EMSA demonstrate that Rdp1 indeed binds to the DRE with high affinity (Fig. 3A, lane 2). The sequence-specific binding was confirmed by competition assays in which a homologous oligonucleotide competed much more efficiently for the binding than the nonspecific competitor (Fig. 3A, compare results with DRE and UAS1). Multiple DNA-protein complexes were detected in EMSA, which might be due to Rdp1 binding at each DRE sequence or to oligomerization of Rdp1 at a single binding site (Fig. 3A).
FIG. 3.
Rdp1 specifically binds to DRErhp51+ in vitro. (A) EMSA to test the binding specificity and affinity of Rdp1 to DRErhp51+. A 32P-labeled DRE oligonucleotide (5 nM), either without (lane 1) or with (lanes 2 to 12) GST-Rdp1, was incubated and electrophoresed as described in Materials and Methods. In lanes 3 to 12, the indicated unlabeled competitor was added. Competitor concentrations were as follows: in lanes 3 and 8, 25 nM; in lanes 4 and 9, 50 nM; in lanes 5 and 10, 100 nM; in lanes 6 and 11, 250 nM; and in lanes 7 and 12, 500 nM. Arrows indicate the DNA-protein complexes. (B). Footprinting of Rdp1 on the upstream regulatory region of rhp51+ containing the two DREs. End-labeled DNA fragments containing the two DRErhp51+s were incubated without (lanes 2 and 3) or with (lanes 4 and 5) GST-Rdp1 protein and subsequently subjected to DNase I digestion as described in Materials and Methods. The region protected from DNase I digestion is indicated by asterisks.
To determine the region within DRErhp51+ to which Rdp1 binds, DNase I footprinting was performed. As shown in Fig. 3B, GST-Rdp1 protected only the region between −234 and −201, which includes the two DREs. Together, these data strongly suggest that Rdp1 binds DRErhp51+ and regulates the damage response of rhp51+ by binding to the two DRE sequences.
Definition of Rdp1 binding specificity.
To further define the binding specificity of Rdp1, we compared the abilities of oligonucleotides containing mutations within the DRE to compete with the wild-type sequence in vitro. In our previous report, comparisons of sequences within the 5′ regulatory regions of damage-inducible genes, including rhp51+, RNR2, RAD51, PHR1, and MAG1, have revealed the presence of a consensus 10-bp sequence (5′-CGT/AGGT/ANGC/AC/A-3′) (31). Thus, we introduced several mutations into the highly conserved five bases within the DREs (AGGTG and AGGTA in DRE1 and in DRE2, respectively) (Fig. 4A). As shown in Fig. 4B, oligonucleotides containing a single mutation in each DRE (oligonucleotides CG2TG and AGT2G) were still able to compete significantly for Rdp1 binding, while oligonucleotides ATGTG, AG2CG, and AT2CG hardly competed. Also, those containing mutations in all five bases completely abolished the competition (oligonucleotide CT2CA [lanes 15 and 16 in Fig. 4B]), indicating that NGG/TTG/A is required for specific binding.
Mutations within the two DREs cause a significant decrease in both damage-dependent activation of rhp51+ and UV survival.
To determine if NGG/TTG/A within the DRE is functionally involved in the damage-inducible expression of rhp51+, we made plasmids harboring the rhp51+ gene with intact DRE (pJKwt) or with five-base-mutated DRE (pJKmD) and then integrated them at the leu1-32 locus in the rhp51-deficient strain JAC1/51Δ to generate the JAC10 and JAC20 strains, respectively (Fig. 5A). Precise single-copy integration was confirmed by Southern blot analysis (data not shown). The mRNA levels of rhp51+ in JAC10 and JAC20 with or without MMS treatment or UV irradiation were assessed by Northern blot analysis (Fig. 5B). The basal-level expression of rhp51+ was not influenced by the mutation in the DRE. In contrast, damage-inducible transcription was significantly reduced, by ∼50%, in JAC20, which contains the mutated DRE (Fig. 5C). These results indicated that the consensus binding sequences within the DREs mediate the transcriptional activation of rhp51+ in response to DNA damage.
To determine whether the down-regulation of rhp51+ with mutated DRE results in reduced repair capacity, we tested the survival rates of JAC10, JAC20, and JAC1/51Δ following UV irradiation or MMS treatment. As shown in Fig. 5D and E, DRE mutations decreased the UV and MMS survival rates and the relative reduction in survival was reflected in reduced rhp51+ expression. These results strongly argue for the notion that the damage response of rhp51+, controlled by the DREs, contributes to cell survival following DNA damage.
The rdp1+ is essential for cell growth and viability.
To examine the effect of loss of function on cell viability and growth, we constructed a strain by targeted disruption of rdp1+. The 1.1-kb BalI-BclI fragment containing almost the entire coding region of Rdp1 was replaced with the 1.8-kb ura4+ gene fragment (Fig. 6A). The disruption cassette for rdp1 was transformed into a diploid strain (ED665/ED668), and only stable Ura+ transformants were analyzed by Southern blotting for the heterozygous genotype rdp1+/rdp1::ura4+ (Fig. 6B). Unexpectedly, the tetrads of the rdp1+/rdp1::ura4+ heterozygotes revealed that only one or two of the four spores were viable (Fig. 6C, right plate) and they were all auxotrophs for uracil requirement (data not shown), indicating that the disruption of rdp1+ by the ura4+ fragment was lethal. To confirm the above data, we made two different additional sets of disruptions with the N-terminal third of the ORF or the entire ORF deleted and tested the effect of disruption on cell viability. The experiments also supported that rdp1::ura4+ disruption was lethal (data not shown). Thus, we concluded that the rdp1+ gene is essential for cell viability. The analysis by flow cytometry did not show a clear difference between the DNA profiles of wild-type and rdp1Δ spores (data not shown). Nevertheless, most of the nonviable germinating rdp1Δ spores arrested in an elongated and deformed shape with an abnormal nuclear structure, implying the possible involvement of Rdp1 in cell cycle progression (Fig. 6D, right plate).
FIG. 6.
Disruption of the rdp1+ gene and terminal phenotype of an rdp1Δ mutant. (A) Replacement of the rdp1+ gene by ura4+. The 1.1-kb BalI-BclI fragment was replaced with the 1.8-kb ura4+ gene as described in Materials and Methods. (B) Southern blot to confirm rdp1+/rdp1::ura4+ heterozygotes. The 3.3-, 2.17-, and 1.2-kb fragments were detected in the heterozygote when it was probed with the 3.3-kb EcoRV fragment of rdp1+. Lanes: 2 and 6, heterozygotes with rdp1+/rdp1::ura4+; 1, 3, 4, and 5, wild-type homozygotes. (C) Tetrad analysis of the rdp1+/rdp1::ura4+ heterozygote. The spores were microdissected onto YES plates and incubated for 4 days at 30°C. Heterozygotic tetrads produced only one or two viable spores with the Ura− phenotype, while most of tetrads from the wild-type diploid showed four viable spores with uracil auxotrophy. (D) Terminal morphology of wild-type and rdp1Δ spores after germination. The rdp1+/rdp1+ and rdp1+/rdp1::ura4+ diploid strains were sporulated, and the resulting spores were inoculated into minimal medium supplemented with uracil for the wild-type spores or uracil-free medium for the rdp1::ura4+ spores. Germinating cells were stained with DAPI and examined by fluorescence microscopy. Left plate, germinating wild-type spores (22 h); right plate, germinating rdp1Δ spores (22 and 24 h). Scale bar, 10 μm.
DNA damage checkpoints are involved in transcriptional induction of rhp51+ in response to DNA damage.
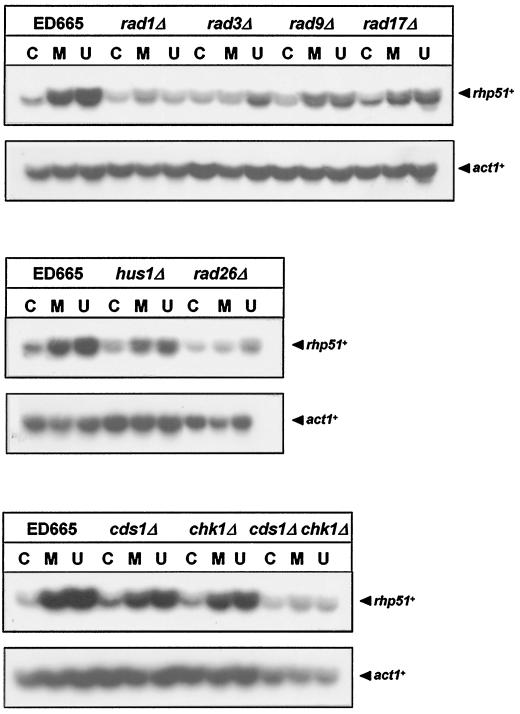
Recent reports demonstrated that DNA damage checkpoint genes control the transcriptional induction of a DNA damage regulon (DDR) in the budding yeast S. cerevisiae (1, 16). We questioned primarily if Rdp1 mediates rhp51+ expression controlled by the damage checkpoint. Before seeking an answer to this question, we aimed to understand whether DNA damage checkpoint genes are required for transcriptional induction of rhp51+ in response to DNA damage. Exponentially growing rad1Δ, rad3Δ, rad9Δ, rad17Δ, rad26Δ, hus1Δ, cds1Δ, chk1Δ, and cds1Δ-chk1Δ cells were treated with either UV irradiation or MMS. A significant decrease (∼30 to 50%) in the transcriptional induction of rhp51+ was observed in all checkpoint mutants except in cds1Δ and chk1Δ single mutants compared with the level of induction in the wild-type strain (Fig. 7). The data revealed that the DNA damage response of the DNA repair gene rhp51+ requires a damage checkpoint pathway, thus implying the existence of DDR control by checkpoints.
FIG. 7.
Defects in DNA damage checkpoints cause a significant decrease in transcriptional induction of rhp51+ in response to DNA damage. Total RNAs extracted from mid-log-phase cells of checkpoint mutant strains were electrophoresed on formaldehyde-agarose gels and transferred onto nitrocellulose membrane. The RNA blots were hybridized with 32P-labeled rhp51+ or act1+ DNA probes and autoradiographed. Symbols: C, mock treatment; M, 0.1% MMS treatment; U, 180 J of UV irradiation per m2.
DISCUSSION
In this paper, we have identified the protein encoded by rdp1+ as a DNA damage-responsive activator of rhp51+ expression. A C2H2 zinc finger protein, Rdp1, recognizes NGG/TTG/A sequences found in the previously defined DRErhp51+. Furthermore, we have shown that mutations of the Rdp1-binding sites in DRErhp51+ abolish Rdp1 binding in vitro and also reduce rhp51+ expression and cell survival in response to DNA damage. Surprisingly, despite the fact that rhp51+ is not an essential gene, loss of function of rdp1+ resulted in cell death, indicating that rdp1+ has an essential function in cell growth and viability in addition to regulation of rhp51+ expression.
The deduced amino acid sequence of Rdp1 shows one particularly interesting region of polyalanine tract near the center that has 30 to 40% identity with many homeodomain proteins, including human HOXA13 (24) and other transcription factors such as mouse Zic2 (3). Interestingly, we found that the transactivation domain of Rdp1, defined by the effect of the GAL4 binding domain-Rdp1 hybrids on the expression of lacZ fused to the upstream activating sequence of GAL, indeed contains a 12-residue polyalanine tract (data not shown). However, in another study, such tracts were proposed to repress transcription directly and the minimal repressor domains of Krüppel, Engrailed, and Evenskipped were also determined to contain alanine-rich sequences (24). In addition, it has been proposed that polyalanine tracts act as flexible spacer elements between functional domains (35). Taken together, the results of these studies provide interesting implications for the role of the polyalanine tract in the control of rhp51+ transcription.
Several S. pombe genes, including uvi15+, uvi31+, UVDE, and rhp16+, are DNA damage-inducible genes (5, 14). Of these, uvi31+, UVDE, and rhp16+ contain a putative sequence homologous to DRErhp51+ (5, 14, 37), suggesting the presence of a DDR controlled by DRErhp51+. Like the AG4 sequence, which has been defined as the URS of S. cerevisiae PHR1, the NGG/TTG/A sequence recognized by Rdp1 is found to be much too open in the fission yeast genome database to be meaningful. Nevertheless, it may be significant that one or more NGG/TTG/A sequences are found within 500 bp of the translational start sites of most of the DNA repair, checkpoint, and metabolism genes of fission yeast, even though they have not yet been tested for their damage inducibility (rad1+, rad8+, rad9+, rad13+, rad15+, rad17+, rad21+, rad22+, rad24+, rad26+, rad32+, rhp6+ rhp54+, rhp55+, rhp57+, cds1+, chk1+, and spdmc1+ [2, 8, 10, 11, 25, 39, 40, 43, 44, 46, 57, 58, 59]). As mentioned in a previous report, one or more NGG/TTG/A sequences are also found in the promoters of several of the known damage-inducible DNA repair and metabolism genes of S. cerevisiae (PHR1, RAD2, RAD16, RAD51, DDR48, RNR2, RNR3, and MAG3) (31). Thus, although a transcriptional regulator of the DNA damage response conserved between the two yeasts has not been found thus far and we failed to find an Rdp1 homolog in the S. cerevisiae genome, Rdp1 may be a candidate for this common type of regulator. Identification of another target(s) of Rdp1 would enable us to further understand the roles of this protein.
In S. cerevisiae, at least four different proteins, Rph1p, Gis1p, Crt1p, and Swi6p, are known as regulators of damage-inducible DNA repair genes (26, 27, 29, 55). The DNA damage response by S. pombe rdp1+ differs in one important aspect from that by the above-mentioned transcription factors. Rdp1 acts as a positive regulator of rhp51+ expression, while Rph1p, Gis1p, and Crt1p are damage-responsive repressors. However, one cannot exclude the possibility that Rdp1 may be switched to become an activator through modulation by other interacting proteins. The best example is the bZip domain factor Atf1, which is involved in the transcriptional regulation of stress-related genes (53, 62, 63). A recent study suggested that Atf1 is converted from a repressor to a transcriptional activator by Spc1 (mitogen-activated protein kinase) activity, at least in the response of the catalase gene to UV (15). Both Rdp1 and Atf1 seem to resemble each other in the fact that they are required for the increased levels of catalase or rhp51+ expression that is part of the UV response (15). Furthermore, Swi6p is a bifunctional regulator that acts depending on the promoter context of the target. The Hrr25p-Swi6p pathway controls the transcriptional activation of RNR2 and RNR3 (26), while Rad53-dependent phosphorylation of Swi6p is involved in down-regulation of CLN1 and CLN2 in response to DNA damage (55). Similar to the way Swi6p behaves in cyclin gene expression, the two MluI cell cycle box (MCB) elements adjacent to DRErhp51+ appear to act as upstream repressing sequences because mutations in the MCB caused derepression of rhp51+ and reduced damage inducibility (our unpublished observations). Furthermore, loss of function in the MCB-binding factors res1+, res2+, and rep2+ (4, 42, 45) results in the same phenotypes with respect to rhp51+ expression (unpublished data). Together, these data suggest that damage-dependent activation and repression of rhp51+ required Rdp1 and MCB-binding factors, respectively. Thus, it is possible not only that there are multiple DNA damage-responsive regulators but also that the signal transduction pathway involved in the regulation of the DDR differs depending on the promoter context of the target.
Despite a long-time interest and effort, the biological significance of the transcriptional induction of DNA repair genes is still unclear. In particular, failure to induce RAD54, a DNA repair gene in S. cerevisiae, appeared not to affect DNA repair or recombination phenotypes, raising significant questions about the physiology of damage-dependent induction (13). However, the present study indicates that the transcriptional activation of rhp51+ is required for cellular repair capacity, implying the presence of an SOS-like response in yeast.
Several recent studies strongly argue that the DNA damage checkpoint is linked directly or indirectly to the DNA damage-dependent transcriptional response in addition to the delay of cell cycle progression (1, 16, 27, 29, 38). In particular, recent observations suggested that all damage-checkpoint genes, including RAD9, MEC1, and RAD53 of S. cerevisiae, control the induction of a large regulon of >15 genes whose roles are in DNA repair and metabolism, indicating that this DDR may be reminiscent of the SOS response of bacteria (1, 16, 60). For the fission yeast, a number of damage checkpoint genes involved in sensing abnormal DNA structures and transducing the damage signal to effector molecules have been identified and well characterized (9, 18, 47, 56). However, no one has ever tested whether the checkpoint pathway regulates the transcriptional induction of DNA damage-inducible genes in S. pombe. Interestingly, we also found that the transcriptional activation of rhp51+ in response to DNA damage was significantly reduced in all the checkpoint-defective strains of S. pombe tested, implying the existence of DDR control by the checkpoint pathway as in S. cerevisiae. Considering that Rdp1 was found to be a key regulator of DNA damage-dependent expression of rhp51+, Rdp1 may be one of the best candidates to act as a mediator that links the DNA damage-signaling cascade by means of checkpoints and damage-dependent induction of rhp51+ transcription. Our previous report and this study indicate that Rdp1 may cooperate with MCB-binding proteins for the maximal activation of damage-dependent transcription of rhp51+. To confirm this hypothesis, further experiments remain to be performed. Of particular interest is whether Rdp1 indeed mediates damage checkpoint-dependent induction of rhp51+ expression.
ACKNOWLEDGMENTS
We thank Mark Johnston, Tony Carr, Jonathan Millar, T. Enoch, Nancy Walworth, and P. Fantes for providing yeast strains and R. R. Reed, M. Schweizer, and R. Gwilliam for providing plasmids used for one-hybrid screening and those containing the rdp1 genomic clone. We also thank Gwen Sancar and Onyou Hwang for their invaluable comments on the manuscript and J. B. Seo for his technical assistance.
This research was supported in part by the grants from the Korea Science and Engineering Foundation through the Research Center for Cell Differentiation (grant 1999G0301-3). Y.S.S., Y.K.J., and S.D.P. are supported by a BK21 Research Fellowship from the Ministry of Education, Republic of Korea.
Y.S.S. and Y.K.J. contributed equally to this work.
REFERENCES
- 1.Aboussekhra A, Vialard J E, Morrison D E, de la Torre-Ruiz M A, Cernakova L, Fabre F, Lowndes N F. A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J. 1996;15:3912–3922. [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J, Lehmann A R, Carr A M. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aruga J, Nagai T, Tokuyama T, Hayashizaki Y, Okazaki Y, Chapman V M, Mikoshiba K. The mouse zic gene family. Homologues of the Drosophila pair-rule gene odd-paired. J Biol Chem. 1996;271:1043–1047. doi: 10.1074/jbc.271.2.1043. [DOI] [PubMed] [Google Scholar]
- 4.Ayte J, Leis J F, Decaprio J A. The fission yeast protein p73res2 is an essential component of the mitotic MBF complex and a master regulator of meiosis. Mol Cell Biol. 1997;17:6246–6254. doi: 10.1128/mcb.17.11.6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bang D D, Ketting R, Ruijter M, Brandsma J A, Verhage R A, Putte P, Brouwer J. Cloning of Schizosaccharomyces pombe rhp16+, a gene homologous to the Saccharomyces cerevisiae RAD16 gene. Mutat Res. 1996;364:57–71. doi: 10.1016/0921-8777(96)00010-9. [DOI] [PubMed] [Google Scholar]
- 6.Barker D G, White J H M, Johnston L H. The nucleotide sequence of the DNA ligase gene (CDC9) from Saccharomyces cerevisiae: a gene which is cell-cycle regulated and induced in response to DNA damage. Nucleic Acids Res. 1985;13:8323–8337. doi: 10.1093/nar/13.23.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basile G, Aker M, Mortimer R K. Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol Cell Biol. 1992;12:3235–3246. doi: 10.1128/mcb.12.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkenbihl R P, Subramani S. Cloning and characterization of rad21, an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr A M. DNA structure checkpoints in fission yeast. Semin Cell Biol. 1995;6:65–72. doi: 10.1016/1043-4682(95)90002-0. [DOI] [PubMed] [Google Scholar]
- 10.Carr A M, Schmidt H, Kirchoff S, Muriel W J, Sheldrick K S, Griffiths D J, Basmacioglu C, Subramani S, Clegg M, Nasim A, Lehmann A R. The rad16 gene of Schizosaccharomyces pombe: a homolog of the RAD1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:2029–2040. doi: 10.1128/mcb.14.3.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carr A M, Sheldrick K S, Murray J M, al-Harithy R, Watts F Z, Lehmann A R. Evolutionary conservation of excision repair in Schizosaccharomyces pombe: evidence for a family of sequences related to the Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:1345–1349. doi: 10.1093/nar/21.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Samson L. Induction of S. cerevisiae MAG 3-methyladenine DNA glycosylase transcript levels in response to DNA damage. Nucleic Acids Res. 1991;19:6427–6432. doi: 10.1093/nar/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole G M, Mortimer R K. Failure to induce a DNA repair gene, RAD54, in Saccharomyces cerevisiae does not affect DNA repair or recombination phenotypes. Mol Cell Biol. 1989;9:3314–3326. doi: 10.1128/mcb.9.8.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey S, Nass M L, Ferrer J V, Sidik K, Eisenberger A, Mitchell D L, Freyer G A. The fission yeast UVDR DNA repair pathway is inducible. Nucleic Acids Res. 1997;25:1002–1008. doi: 10.1093/nar/25.5.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Degols G, Russell P. Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:3356–3363. doi: 10.1128/mcb.17.6.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Torre-Ruiz M A, Green C M, Lowndes N F. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 1998;17:2687–2698. doi: 10.1093/emboj/17.9.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duttweiler H M. A highly sensitive and non-lethal beta-galactosidase plate assay for yeast. Trends Genet Sci. 1996;12:340–341. doi: 10.1016/s0168-9525(96)80008-4. [DOI] [PubMed] [Google Scholar]
- 18.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 19.Elledge S J, Davis R W. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol Cell Biol. 1987;7:2783–2793. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elledge S J, Davis R W. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990;4:740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- 21.Friedberg E C. Deoxyribonucleic acid repair in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:70–102. doi: 10.1128/mr.52.1.70-102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. pp. 595–631. [Google Scholar]
- 23.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 24.Goodman F R, Mundlos S, Muragaki Y, Donnai D, Giovannucci-Uzielli M L, Lapi E, Majewski F, McGaughran J, McKeown C, Reardon W, Upton J, Winter R M, Olsen B R, Scambler P J. Synpolydactyly phenotypes correlate with size of expansions in HOXD13 polyalanine tract. Proc Natl Acad Sci USA. 1997;94:7458–7463. doi: 10.1073/pnas.94.14.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths D J, Barbet N C, McCready S, Lehmann A R, Carr A M. Fission yeast rad17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho Y, Mason S, Kabayashi R, Hoekstra M, Andrews B. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:581–586. doi: 10.1073/pnas.94.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung M, Zhou Z, Elledge S J. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 28.Hurd H K, Roberts C W, Roberts J W. Identification of the gene for the yeast ribonucleotide reductase small subunit and its inducibility by methyl methanesulfonate. Mol Cell Biol. 1987;7:3673–3677. doi: 10.1128/mcb.7.10.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang Y K, Wang L, Sancar G B. RPH1 and GIS1 are damage-responsive repressors of PHR1. Mol Cell Biol. 1999;19:7630–7638. doi: 10.1128/mcb.19.11.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang Y K, Jin Y H, Myung K, Seong R H, Hong S H, Park S D. Differential expression of the rhp51+ gene, a recA and RAD51 homolog from the fission yeast Schizosaccharomyces pombe. Gene. 1996;169:125–130. doi: 10.1016/0378-1119(96)83099-x. [DOI] [PubMed] [Google Scholar]
- 31.Jang Y K, Jin Y H, Shim Y S, Kim M J, Yoo E J, Choi L S, Lee J S, Seong R H, Hong S H, Park S D. Identification of the DNA damage-responsive elements of the rhp51+ gene, a recA and RAD51 homolog from the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1996;251:167–175. doi: 10.1007/BF02172915. [DOI] [PubMed] [Google Scholar]
- 32.Jang Y K, Jin Y H, Shim Y S, Kim M J, Yoo E J, Seong R H, Hong S H, Park S D. Evidences for possible involvement of Rhp51 protein in mitotic events including chromosome segregation. Biochem Mol Biol Int. 1995;37:329–337. [PubMed] [Google Scholar]
- 33.Jang Y K, Jin Y H, Kim M J, Seong R H, Hong S H, Park S D. A simple and efficient method for the isolation of total RNA from the fission yeast Schizosaccharomyces pombe. Biochem Mol Biol Int. 1995;37:339–344. [PubMed] [Google Scholar]
- 34.Jones J S, Prakash L. Transcript levels of the Saccharomyces cerevisiae DNA repair gene RAD18 increase in UV irradiated cells and during meiosis but not during the mitotic cell cycle. Nucleic Acids Res. 1991;19:893–898. doi: 10.1093/nar/19.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karlin S, Burge C. Trinucleotide repeats and long homopeptides in genes and proteins associated with nervous system disease and development. Proc Natl Acad Sci USA. 1996;93:1560–1565. doi: 10.1073/pnas.93.4.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keeney J B, Boeke J D. Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S H, Kim M, Lee J K, Kim M J, Jin Y H, Seong R H, Hong S H, Joe C O, Park S D. Identification and characterization of uvi31+, a UV-inducible gene from Schizosaccharomyces pombe. Environ Mol Mutagen. 1997;30:72–81. doi: 10.1002/(sici)1098-2280(1997)30:1<72::aid-em10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 38.Kiser G L, Weinert T A. Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol Biol Cell. 1996;7:703–718. doi: 10.1091/mbc.7.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman H B, Hopkins K M, Laverty M, Chu H M. Molecular cloning and analysis of Schizosaccharomyces pombe rad9, a gene involved in DNA repair and mutagenesis. Mol Gen Genet. 1992;232:367–376. doi: 10.1007/BF00266239. [DOI] [PubMed] [Google Scholar]
- 40.Lindsay H D, Griffiths D J, Edwards R J, Christensen P U, Murray J M, Osman F, Walworth N, Carr A M. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madura K, Prakash S. Transcript levels of the Saccharomyces cerevisiae DNA repair gene RAD23 increase in response to UV light and in meiosis but remain constant in the mitotic cell cycle. Nucleic Acids Res. 1990;18:4737–4742. doi: 10.1093/nar/18.16.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyamoto M, Tanaka K, Okayama H. res2+, a new member of the cdc10+/SWI4 family, controls the ‘start’ of mitotic and meiotic cycles in fission yeast. EMBO J. 1994;13:1873–1880. doi: 10.1002/j.1460-2075.1994.tb06456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muris D F, Vreeken K, Carr A M, Murray J M, Smit C, Lohman P H, Pastink A. Isolation of the Schizosaccharomyces pombe RAD54 homologue, rhp54+, a gene involved in the repair of radiation damage and replication fidelity. J Cell Sci. 1996;109:73–81. doi: 10.1242/jcs.109.1.73. [DOI] [PubMed] [Google Scholar]
- 44.Murray J M, Doe C L, Schenk P, Carr A M, Lehmann A R, Watts F Z. Cloning and characterization of the Schizosaccharomyces pombe rad15 gene, a homologue to the Saccharomyces cerevisiae RAD3 gene and human ERCC2 genes. Nucleic Acids Res. 1992;20:2673–2678. doi: 10.1093/nar/20.11.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakashima N, Tanaka K, Sturm S, Okayama H. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J. 1995;14:4794–4802. doi: 10.1002/j.1460-2075.1995.tb00161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds P, Koken M H, Hoeijimakers J H, Prakash S, Prakash L. The rhp6+ gene of Schizosaccharomyces pombe: a structural and functional homolog of the RAD6 gene from the distantly related yeast Saccharomyces cerevisiae. EMBO J. 1990;9:1423–1430. doi: 10.1002/j.1460-2075.1990.tb08258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson G W, Nicolet C M, Kalainov D, Friedberg E C. A yeast excision-repair gene is inducible by DNA damaging agents. Proc Natl Acad Sci USA. 1986;83:1842–1846. doi: 10.1073/pnas.83.6.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruby S W, Szostak J W. Specific Saccharomyces cerevisiae genes are expressed in response to DNA-damaging agents. Mol Cell Biol. 1985;5:75–84. doi: 10.1128/mcb.5.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Sancar G B, Ferris R, Smith F W, Vandeberg B. Promoter elements of the PHR1 gene of Saccharomyces cerevisiae and their roles in the response to DNA damage. Nucleic Acids Res. 1995;21:4320–4328. doi: 10.1093/nar/23.21.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sebastian J, Kraus B, Sancar G B. Expression of the yeast PHR1 gene is induced by DNA-damaging agents. Mol Cell Biol. 1990;10:4630–4637. doi: 10.1128/mcb.10.9.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- 54.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 55.Sidorova J M, Breeden L L. Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev. 1997;11:3032–3045. doi: 10.1101/gad.11.22.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stewart E, Enoch T. S-phase and DNA-damage checkpoints: a tale of two yeasts. Curr Opin Cell Biol. 1996;8:781–787. doi: 10.1016/s0955-0674(96)80078-0. [DOI] [PubMed] [Google Scholar]
- 57.Sunnerhagen P, Seaton B L, Nasim A, Subramani S. Cloning and analysis of a gene involved in DNA repair and recombination: the rad1 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1990;10:3750–3760. doi: 10.1128/mcb.10.7.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tavassoli M, Shayeghi M, Nasim A, Watts F Z. Cloning and characterization of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double-strand breaks and recombination. Nucleic Acids Res. 1995;23:383–388. doi: 10.1093/nar/23.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsutsui Y, Morishita T, Iwasaki H, Toh H, Shinagawa H. A recombination repair gene of Schizosaccharomyces pombe, rhp57+, is a functional homolog of the Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene. Genetics. 2000;154:1451–1461. doi: 10.1093/genetics/154.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walker G C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang M M, Reed R R. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature (London) 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 62.Wilkinson M G, Millar J B. SAPKs and transcription factors do the nucleocytoplasmic tango. Genes Dev. 1998;12:1391–1397. doi: 10.1101/gad.12.10.1391. [DOI] [PubMed] [Google Scholar]
- 63.Wilkinson M G, Samuels M, Takeda T, Toone W M, Shieh J C, Toda T, Millar J B, Jones N. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- 64.Wolf S S, Roder K, Schweizer M. Construction of a reporter plasmid that allows expression libraries to be exploited for the one-hybrid system. BioTechniques. 1996;20:567–574. doi: 10.2144/19962004568. [DOI] [PubMed] [Google Scholar]
- 65.Zhou Z, Elledge S J. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]