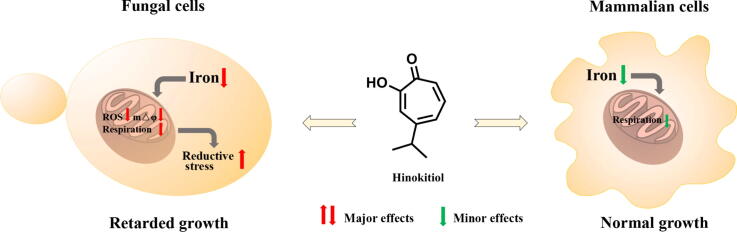
Graphical abstract
Keywords: Hinokitiol, Candida, Iron chelator, Mitochondria, Drug resistance
Abstract
Introduction
The increasing morbidity of fungal infections and the prevalence of drug resistance highlighted the discovery of novel antifungal agents and investigation of their modes of action. Iron chelators have been used to treat superficial fungal infections or potentiate the efficacy of certain antifungal drugs. Hinokitiol exhibits potent antifungal activity and iron-chelating ability. However, their relationships have not been established.
Objectives
This study aims to explore the selectivity of hinokitiol against fungal cells and mammalian cells and determine the role of iron-chelating for the antifungal activity of hinokitiol.
Methods
Iron probe FeRhonox-1 was used to determine intracellular Fe2+ content. 5-Cyano-2,3-ditolyl tetrazolium chloride probe and Cell Counting Kit-8 were used to detect the mitochondrial respiratory activities. Quantitative real-time PCR and rescue experiments were performed to determine the effect of iron on the antifungal activity of hinokitiol. The effects of hinokitiol on fungal mitochondria were further evaluated using reactive oxygen species probes and several commercial Assay Kits. The ability of hinokitiol to induce resistance in Candida species was carried out using a serial passage method. The in vivo therapeutic effect of hinokitiol was evaluated using Galleria mellonella as an infectious model.
Results
Hinokitiol was effective against a panel of Candida strains with multiple azole-resistant mechanisms and persistently inhibited Candida albicans growth. Mechanism investigations revealed that hinokitiol chelated fungal intracellular iron and inhibited the respiration of fungal cells but had minor effects on mammalian cells. Hinokitiol further inhibited the activities of mitochondrial respiratory chain complexes I and II and reduced mitochondrial membrane potential, thereby decreasing intracellular ATP synthesis and increasing detrimental intracellular reductive stress. Moreover, hinokitiol exhibited low potential for inducing resistance in several Candida species and greatly improved the survival of Candida-infected Galleria mellonella.
Conclusions
These findings suggested the potential application of hinokitiol as an iron chelator to treat fungal infections.
Introduction
Candida species cause various superficial and systemic fungal infections and are an important focus of antifungal research [1], [2]. Invasive candidiasis was globally estimated to occur in >250,000 patients each year with a mortality rate exceeding 40% [3]. Candida albicans is the leading cause of candidemia. Candida species have developed multiple resistant mechanisms to counteract antifungal agents, particularly the azole drugs [3]. The rapid evolution of resistance and the slow development of new therapies result in tough challenges to global health and are particularly grim in the developing world [4]. High mortality and the emergence of drug resistance highlight the need for novel antifungals. While synthetic efforts remain the mainstream in the antifungal drug discovery, natural products provide drugs including amphotericin B and caspofungin, which have novel mechanisms [5].
Hinokitiol, a tropolone monoterpene isolated from cupressaceous plants, exhibits diverse pharmacological effects, such as anti-inflammatory, antioxidant, antitumor and antimicrobial activities [6], [7], [8], [9]. Hinokitiol has been used in food preservation and has excellent biological safety, with low cytotoxicity towards many normal human cell lines, such as human pulp cells, oral epithelial cells and vascular smooth muscle cells, and low chronic toxicity in vivo [9], [10], [11], [12], [13]. Hinokitiol has been suggested to inhibit the growth and virulence factors of Candida species by regulating the Ras1-cAMP-PKA pathway and transcription factors associated with hyphal formation [14], but the exact molecular mechanisms of the antifungal action of hinokitiol remain elusive. In this study, we examined the role of iron chelation in the antifungal activity of hinokitiol.
Hinokitiol can bind iron via its hydroxyketone moiety [15] and has been reported to inhibit the progression of Parkinson's disease by chelating iron in susceptible cerebral regions [16]. Intracellular iron is essential for many vital physiological processes, such as electron transport chains and oxygen transport [17]. Some iron chelators have exhibited potential for the treatment of fungal infections either directly or by enhancing the sensitivity of cells to antifungal drugs such as fluconazole (FLC) and nystatin [18], [19], [20], [21].
Here, we observed that hinokitiol significantly inhibited the growth of Candida species, including a panel of drug-resistant isolates. Depletion of fungal intracellular iron by hinokitiol inhibited mitochondrial respiration and decreased mitochondrial membrane potential (mtΔψ), which in turn led to a decline in intracellular ATP generation and an increase in detrimental reductive stress. Hinokitiol exhibited very low potential in inducing drug resistance, probably due to its inability to induce cellular stress responses and provide enough energy support. Furthermore, hinokitiol greatly improved the survival of Candida-infected Galleria mellonella. These findings support further applications of hinokitiol in fighting against fungal infections.
Materials and methods
Strains, culture conditions and chemicals
All Candida strains used in this study are presented in Table 1 and Table 2. For all cultures, strains were propagated from frozen stocks at −80 °C onto yeast peptone dextrose (YPD) agar plates (1% yeast extract, 2% peptone, 2% glucose and 2% agar) using sterile inoculating loops, followed by incubation overnight at 30 °C. For further experiments, a selected colony was inoculated in 1 mL of YPD liquid medium overnight at 30 °C with agitation (200 rpm). Morpholinopropanesulphonic acid (MOPS)-buffered RPMI1640 (pH = 7.4) and synthetic dextrose (SD) medium (0.17% yeast nitrogen base without amino acids, 2% glucose, 0.5% ammonium sulfate and the appropriate amino acids) were used in some assays.
Table 1.
Antifungal activity of three hydroxyketones and FLC against Candida species.
| Strainsa | MICs (μg/mL) |
||||
|---|---|---|---|---|---|
| Hinokitiol | Tropolone | Maltol | FLC | ||
| C. albicans | SC5314 | 2 | 16 | >128 | 2 |
| ATCC10231 | 2 | 16 | >128 | 1 | |
| C. tropicalis | CT-LY1 | 2 | 32 | >128 | 1 |
| CT2 | 1 | 32 | >128 | 1 | |
| C. parapsilosis | CP-LY8 | 2 | 32 | >128 | 1 |
| CP001 | 1 | 8 | >128 | 0.5 | |
| C. glabrata | CG-LY12 | 0.5 | 8 | >128 | 1 |
| CG1 | 0.5 | 8 | >128 | 0.5 | |
| C. krusei | CK-LY16 | 1 | 16 | >128 | 2 |
| CK1 | 2 | 64 | >128 | 2 | |
ATCC10231 is a standard strain of C. albicans obtained from ATCC (American Type culture collection). SC5314 is a wild-type C. albicans strain. Other Candida strains were clinically derived.
Table 2.
Antifungal activity of hinokitiol against FLC-resistant or FLC-sensitive C. albicans strains.
| Strainsa | Characteristics | MICs (µg/mL) |
|
|---|---|---|---|
| Hinokitiol | FLC | ||
| DAY364 | FLC-sensitive strain with homozygous cnb1/cnb1 | 2 | 0.5 |
| DSY465 | FLC-sensitive strain with homozygous mdr1/mdr1 | 2 | 0.5 |
| YEM13 | FLC-resistant strain with hyperexpressed MDR1 | 8 | 64 |
| YEM15 | FLC-resistant strain with hyperexpressed CDR1 and CDR2 | 2 | 64 |
| SCMRR1R34MPG2A | FLC-resistant strain with P683S mutation in MRR1 | 8 | 128 |
| SCUPC2R14MPG2A | FLC-resistant strain with G648D mutation in UPC2 | 2 | >128 |
| G5 | FLC-resistant clinical isolate with G997V mutation in MRR1 | 8 | >128 |
| GU5 | FLC-resistant clinical isolate with G980E mutation in TAC1 | 1 | >128 |
| DSY296 | FLC-resistant clinical isolate with N977D mutation in TAC1 | 1 | 64 |
| DSY1751 | FLC-resistant strain with homozygous erg3/erg3 and heterozygous erg11/ERG11 | 2 | >128 |
| 28A | FLC-resistant clinical isolate | 2 | >128 |
| 28I | FLC-resistant clinical isolate | 2 | >128 |
| 28D | FLC-resistant clinical isolate | 2 | >128 |
All the strains in this Table were C. albicans. DAY364 is a constructed C. albicans strain with deletion of CNB1; DSY654 is a constructed C. albicans strain with deletion of MDR1; YEM13 is a constructed C. albicans strain with hyperexpression of MDR1; YEM15 is a constructed C. albicans strain with hyperexpression of both CDR1 and CDR2; SCMRR1R34MPG2A is a constructed C. albicans strain with a gain-of-function mutation in MRR1; SCUPC2R14MPG2A is a constructed C. albicans strain with a gain-of-function mutation in UPC2; G5 is a FLC-resistant clinical isolate from AIDS patient G with a gain-of-function mutation in MRR1; Gu5 is a FLC-resistant clinical isolate from AIDS patient Gu with a gain-of-function mutation in TAC1. DSY296 is a FLC-resistant clinical isolate with a gain-of-function mutation in of TAC1. DSY1751 is a constructed C. albicans strain with deletion of ERG3 and heterozygous erg11/ERG11. 28A, 28I and 28D are clinical pan-azole-resistant strains isolated from a cancer patient.
Dimethylsulfoxide (DMSO) was purchased from Solarbio (Beijing, China). FLC, hinokitiol, tropolone, maltol, deferiprone (DFP), deferoxamine mesylate (DFO), ferrozine disodium salt hydrate, ferric chloride, ferrous chloride and adenosine 5′-triphosphate disodium salt hydrate (ATP-Na2) were purchased from Aladdin (Shanghai, China). Geldanamycin, FK506 and cyclosporine A were purchased from Sigma (St. Louis, MO, USA).
Determination of minimum inhibitory concentrations (MICs) against Candida species
The MICs of drugs against Candida species were determined according to Clinical and Laboratory Standards Institute document M27-A3 [22]. Briefly, overnight cultured fungal cells were diluted in MOPS-buffered RPMI1640 medium to yield approximately 1 × 103 cells/mL. A 100 μL aliquot of inoculum was inoculated into the wells of 96-well microtiter plates. The tested compounds were added into the medium at a range of concentrations from 0.25 μg/mL to 128 μg/mL and subsequently cultured at 35 °C for 24 h. The MIC endpoints were identified as the lowest concentration that led to no visible growth.
Observation of C. albicans cell growth in situ
An ImageXpress Micro 4 imaging system (Molecular Devices, Sunnyvale, CA, USA) was used to monitor C. albicans cell growth in situ in 96-well plates. Briefly, overnight cultured C. albicans SC5314 cells were diluted in RPMI 1640 medium to yield approximately 1 × 104 cells/mL, and hinokitiol or FLC was added into the cultures and incubated for 72 h at 30 °C. Images were acquired at three-hour intervals.
Propidium iodide (PI) staining and colony counting assays
In order to determine the survival rate of hinokitiol-treated C. albicans cells, C. albicans SC5314 strain with an initial concentration of 1 × 103 cells/mL was treated with 4 μg/mL of hinokitiol or equal volume of dimethyl sulfoxide (DMSO) in 10 mL RPMI 1640 medium at 30 °C. After 24 h, 48 h or 72 h of incubation, 100 μL aliquots were taken from the cell suspensions, diluted in phosphate buffer saline (PBS) buffer, and plated on YPD agar plates. These plates were incubated for 48 h at 30˚C and the survival colonies were counted. Another 500 μL aliquots were washed with PBS and stained by PI at a final concentration of 5 μM. After 20 min of staining, stained cells were imaged by an Olympus fluorescence microscope (Olympus IX71, Olympus, Tokyo, Japan). The dead cells emitted red fluorescence due to the intracellular accumulation of PI.
Quantitative real-time PCR
Overnight cultured C. albicans SC5314 cells were diluted in 20 mL RPMI 1640 medium to yield approximately 1 × 105 cells/mL and treated with 4 μg/mL or 8 μg/mL hinokitiol at 30 °C or 37 °C. The cells were harvested and washed twice with sterile PBS after incubation of 30 min, 1 h, 2 h or 10 h. Total RNA was obtained by using a hot phenol/chloroform method. After analysis of RNA quality by a biophotometer (Eppendorf, Hamburg, Germany), a cDNA Reverse Transcription kit (Takara Biotechnology, Beijing, China) was used for the synthesis of cDNA. Then the SYBR Green-based PCR assays were carried out in an Eppendorf Real-Time PCR System. The primer sequences used in this study were displayed in Table S1.
Measurement of intracellular Fe2+ and Ca2+
FeRhonox-1, a commercial fluorescent probe that specifically binds Fe2+, was used to determine intracellular Fe2+ content. The calcium ion probe Fluo-3AM was used to detect calcium in the cytoplasm. Overnight cultured C. albicans SC5314 cells were diluted in RPMI 1640 medium to yield approximately 1 × 106 cells/mL and challenged with various doses of hinokitiol at 30 °C or 37 °C for 30 min, 1 h, 2 h or 10 h. The cells were then immobilized with 4% paraformaldehyde for 1 h and washed thrice with PBS. After the addition of 5 μM FeRhonox-1, stained cells were placed in the dark for 20 min. The measurements of fluorescence intensities were performed in a flow cytometer (Becton Dickinson, San Jose, CA, USA), and images of cells were obtained using a fluorescence microscope. A similar protocol was used for the detection of Ca2+ using the probe Fluo-3AM. Hinokitiol-treated cells without immobilization with 4% paraformaldehyde were stained with 5 μM Fluo-3AM for 30 min. The measurements of fluorescence intensities were performed using flow cytometry.
Analysis of respiratory activity
5-Cyano-2,3-ditolyl tetrazolium chloride (CTC), a redox-sensitive dye, is mainly used to detect the metabolic activity of microorganisms [23]. CTC can be reduced by electron transport in respiration to CTC formazan, which produces red fluorescence. Overnight cultured C. albicans SC5314 cells were diluted in RPMI 1640 medium to yield approximately 1 × 106 cells/mL and challenged with different concentrations of hinokitiol at 30 °C or 37 °C for 30 min, 1 h, 2 h or 10 h. Cells were harvested and stained using 1 mM CTC in PBS. The measurements of fluorescence intensities were performed using flow cytometry, and images of cells were obtained by fluorescence microscopy.
Dehydrogenase activity in the mitochondria of mammalian or C. albicans cells was measured using Cell Counting Kit-8 (CCK-8). For mammalian cells, cell suspensions (1 × 104 cells/well) in DMEM medium plus 10% FBS were dispensed in a 96-well plate and placed in an incubator (37 °C, 5% CO2). After 24 h of incubation, cells were carefully washed thrice with PBS, then an aliquot of 100 μL DMEM medium plus 2% FBS was added into each well. After 12 h of incubation with various concentrations of hinokitiol, 10 μL of CCK-8 solution was added into each well. After another 2 h of incubation, the OD450 of each well was detected by microplate reader. For fungal cells, C. albicans SC5314 cells (5 × 106 cells/mL in RPMI 1640 medium) were treated with different concentrations of hinokitiol at 30 °C or 37 °C. After 12 h or 48 h of treatment, the cell suspension of each group was adjusted to 5 × 106 cells/mL with RPMI 1640 medium and dispensed into the wells of 96-well plates. Then CCK-8 assays were performed as described above.
Determination of the activities of mitochondrial respiratory chain complexes
The mitochondria of C. albicans SC5314 cells were isolated following previous method [24]. The mitochondria were diluted to an OD600 value of 1.0 by using the extraction buffer from the Electron Transport Chain Complex Activity Assay Kit for the mitochondrial respiratory chain complex of interest (Solarbio, Beijing, China). Hinokitiol was added to the suspensions to obtain various final concentrations. After 3 h of incubation at 30 °C, the enzyme activity assays were conducted according to the suggested instructions by the manufacturer. Protein quantity was determined using the BCA protein assay kit (Solarbio, Beijing, China). The activities of each respiratory chain complex were normalized by comparing the protein content in the samples.
Analysis of mtΔψ
Mitochondrial proton pump activity was measured by measuring mtΔψ using a rhodamine 123 (Rh123) probe. Overnight cultured C. albicans SC5314 cells were suspended in RPMI 1640 medium to yield approximately 1 × 106 cells/mL. Various concentrations of hinokitiol were added to the cultures. Cells were collected and resuspended in PBS with 5 μM of Rh123 after incubation at 30 °C for 10 h. The measurements of fluorescence intensities were performed in a flow cytometer.
Measurement of reactive oxygen species (ROS) and superoxide
The contents of intracellular ROS and superoxide in C. albicans SC5314 were assessed using 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA, Sigma-Aldrich, St. Louis, MO, USA) and MitoSOX Red (Thermo Fisher, Waltham, MA, USA). C. albicans SC5314 cells were diluted in RPMI 1640 medium to yield approximately 1 × 106 cells/mL and challenged with various doses of hinokitiol for 10 h at 30 °C or 37 °C for 1 h, 2 h or 10 h., followed by incubation with 40 μg/mL of DCFH-DA or 5 μM of MitoSOX Red in the dark for 30 min. The cells were washed twice with PBS, and the measurements of fluorescence intensities were performed by flow cytometry.
Evaluation of reductive stress in hinokitiol-treated C. albicans cells
For stress sensitivity analysis, overnight cultures of C. albicans SC5314 cells were harvested and adjusted to 5 × 104 cells/mL in SD medium containing 5 mM thiourea (Tu), 5 mM N-acetyl-L-cysteine (NAC) or 0.5 mM dithiothreitol (DTT). Various concentrations of hinokitiol were added to the cultures and incubated at 30 °C for 16 h. Cells that were not treated with hinokitiol served as a control. Growth curves were plotted based on the values recorded by a microplate reader at 600 nm.
Measurement of ATP levels
The extraction of intracellular ATP was conducted using a previously described method [24]. Briefly, hinokitiol-treated C. albicans SC5314 cells were harvested and resuspended in cell lysate from an ATP Assay Kit (Beyotime, Shanghai, China). Glass beads vortexing was then used for disrupting fungal cells. The cell debris and glass beads were removed by centrifuging at 14,000 g. The ATP in the supernatant was quantified using the ATP Assay Kit with following the manufacturer’s instructions.
Rescue experiments
Overnight cultured C. albicans SC5314 cells were diluted in RPMI 1640 medium to yield approximately 1 × 103 cells/mL with the indicated concentrations of Fe2+, Fe3+, Cu2+ or ATP-Na2. Various concentrations of hinokitiol ranging from 1 μg/mL to 128 μg/mL were immediately added into the cultures. Cells incubated with the rescue agents but not hinokitiol were taken as the control. The MIC values were determined as described above.
Assessment of the potential for developing drug resistance
The drug resistance assessment was conducted by repeatedly exposing Candida strains to hinokitiol or FLC. Specifically, Candida cells challenged with a subinhibitory concentration (1/2 of MIC value) of hinokitiol or FLC acted as the inoculum for the MIC measurement of the next passage. The MICs were recorded throughout 30 passages.
The antifungal interaction of hinokitiol and Hsp90 or calcineurin inhibitors
The interaction of hinokitiol and Hsp90 or calcineurin inhibitors against C. albicans SC5314 isolate was assessed using the broth microdilution checkerboard assay. Briefly, the C. albicans culture was diluted to a final concentration of 0.5–2.5 × 103 cells/mL with RPMI 1640 medium. In a 96-well plate, 2-fold dilutions of hinokitiol, along the x-axis, and 2-fold dilutions of Hsp90 inhibitor geldanamycin, calcineurin inhibitors FK506 or cyclosporine A, along the y-axis, were mixed for a total volume of 100 μL per well. The concentrations of hinokitiol were set ranging from 0.25 μg/mL to 2 μg/mL. The concentrations of the Hsp90 inhibitor geldanamycin were set from 0.625 μg/mL to 5 μg/mL. The concentrations of the calcineurin inhibitors FK506 and cyclosporine A were set from 8 μg/mL to 64 μg/mL. The plates were incubated at 35 °C for 24 h, and the MIC was determined by using the broth microdilution method as described above.
In vivo evaluation using a G. mellonella infection model
G. mellonella larvae were used as an in vivo model to evaluate the effects of hinokitiol on the virulence of C. albicans [25]. Larvae with a body weight of about 0.25 g and without melanisation were selected for the subsequent experiment. Selected Larvae were then randomly divided into four groups: blank group, control group, FLC (2 μg/larva) group or hinokitiol (2 μg/larva) group. C. albicans SC5314 cells were diluted to 2 × 107 CFU/mL with PBS, and 20 larvae per group were injected with 10 μL of C. albicans suspension via the last right proleg of each larva. The blank group was injected with the same volume of PBS. Two hours later, the infected larvae were injected with 10 μL of the indicated drug solution for treatment. The blank group and the control group were injected with the same volume of PBS. The larvae were cultured at 30 °C and survival was recorded daily for 7 days.
Another four groups (each containing 9 larvae) treated as described above were used to assess the fungal burden of infected larvae. On the fourth day, the surface of each larva was disinfected with 75% ethanol. Then each larva was homogenized in 1 mL of sterile PBS by vortexing with glass beads. Candida cell counts were performed by smeared serial dilutions of the homogenate onto YPD plates. The surviving cells in each group were calculated by enumeration of colonies after 48 h of growth at 30 °C.
For histopathological analysis, 8 larvae were divided into 4 groups as described above. Two larvae were randomly picked from each group and fixed in 4% paraformaldehyde at 48 h post-infection. Samples were embedded in paraffin wax, sectioned and stained with periodic acid-Schiff (PAS). The tissue slides were examined under an Olympus microscope at 40 × magnification.
Statistical analysis
The statistical significance of differences between treated and control groups was evaluated by one-way ANOVA test or Student's t-test (two-tailed, unequal variance). For survival analysis, the Mantel-Cox test was used to compare differences between groups. Data were showed as mean ± standard deviation (SD). Statistical significance was determined according to the P value. *P < 0.05, **P < 0.01, ***P < 0.001.
Results
Hinokitiol is a persistent antifungal agent
Potent inhibitory activity of hinokitiol against Candida species, including several FLC-resistant strains, was reported previously [14]. Consistent with this report, hinokitiol exhibited MIC values of 0.5–2 μg/mL against our panel of clinically derived Candida strains (Table 1). We further utilized a group of C. albicans strains with definite resistance mechanisms to test the antifungal activity of hinokitiol. Gain-of-function mutations in Mrr1, Tac1 or Upc2 are considered to result in constitutive overexpressions of MDR1, CDR1/CDR2 or ERG11, respectively, and increase azole resistance [26], [27], [28]. We observed that the antifungal activities of hinokitiol against wild-type strains and azole-resistant C. albicans strains with gain-of-function mutations in Tac1 or Upc2, were similar (Table 1 and Table 2). However, two strains with an upregulated MDR1 showed an increase in the MIC against hinokitiol to 8 μg/mL (Table 2).
FLC is one of the most commonly administered antifungal drugs to treat candidiasis. However, trailing growth or tolerance by Candida strains can compromise the efficacy of FLC and cause persistent candidemia by allowing a subpopulation of cells to replicate at a slow rate in the presence of FLC, even at doses much higher than its MIC [29]. To determine if a similar phenomenon occurs in the use of hinokitiol as a fungistatic agent, we monitored C. albicans cell growth in situ by using an ImageXpress Micro 4 imaging system. Untreated C. albicans cells maintained a fast division rate and transformed to elongated hyphal cells in RPMI 1640 medium. When challenged with FLC, the cells still replicated, although the growth rate was slower than that of untreated cells. By contrast, hinokitiol-treated cells stopped dividing after one to two generations of cell division and did not form hyphae (Fig. S1). This inhibitory effect lasted >72 h, suggesting that hinokitiol treatment can persistently inhibit C. albicans growth. Moreover, the staining assay by using propidium iodide and colony counting assays revealed that hinokitiol-treated cells were still alive (Fig. S2), suggesting the fungistatic activity of hinokitiol.
The antifungal activity of hinokitiol is dependent on chelation of fungal intracellular iron
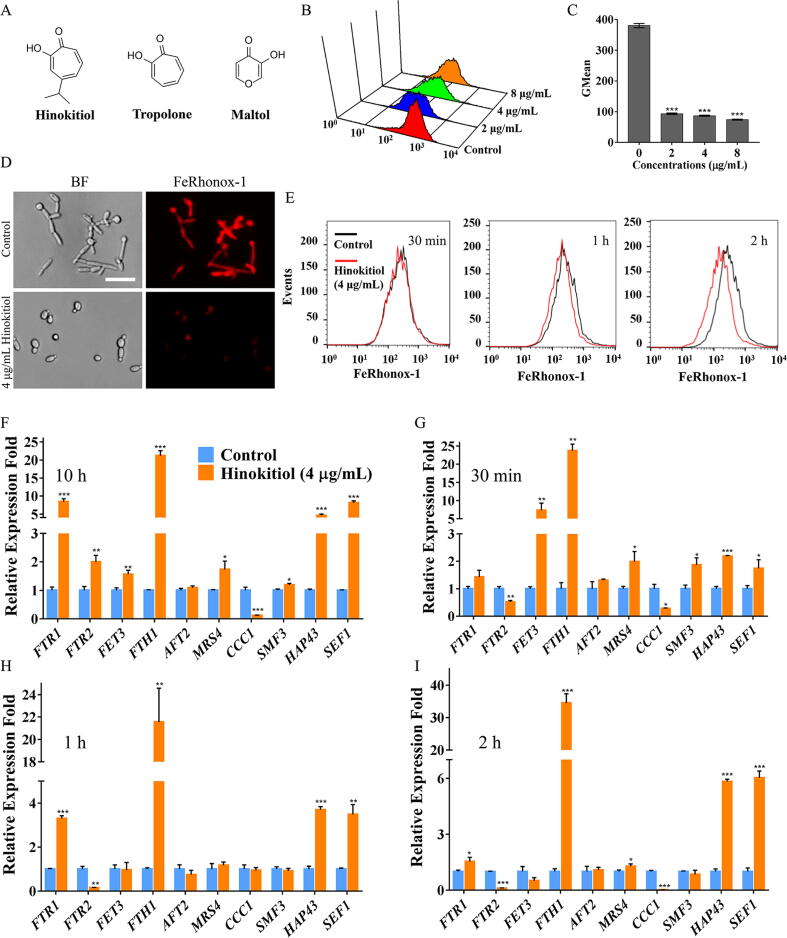
Hinokitiol contains a hydroxyketone moiety that chelates iron. Compared with tropolone, hinokitiol exhibited more potent iron-chelating ability in vitro (Fig. 1A) [15] and 8- to 32-fold greater antifungal activity (Table 1). Maltol exhibits much weaker iron binding than hinokitiol and tropolone [15], and consistent with this observation, maltol did not exhibit any antifungal activity even at concentrations up to 128 μg/mL (Fig. 1A and Table 1). Due to the lack of commercial probes to detect intracellular Fe3+, we utilized the probe FeRhonox-1, which specifically binds Fe2+, to measure intracellular iron in the presence of hinokitiol. Staining with FeRhonox-1 revealed that hinokitiol treatment decreased intracellular iron levels in C. albicans cells (Fig. 1B-D). Other common iron chelators with no apparent antifungal activities, such as DFO, DFP and ferrozine, did not elicit obvious drops in intracellular Fe2+ in C. albicans cells (Fig. S3). In order to exclude iron-chelation as a secondary effect, we measured the intracellular iron contents of hinokitiol-treated C. albicans cells within a short time. FeRhonox-1 staining assay revealed that the intracellular iron content had decreased after 1 h of hinokitiol treatment (Fig. 1E and Fig. S4). However, the decrease of intracellular iron content was more obvious at 37 °C compared with that at 30 °C. This is probably due to the differential intracellular hinokitiol uptake rates at different temperatures. When the treatment lasted for 2 h at 30 °C, greater decrease of intracellular iron was observed. DFP was reported to show antifungal activity at high dose with its MIC value of 160 μg/mL against C. albicans SC5314 [21]. Consistent with its MIC value, DFP did not exhibit potent ability of chelating fungal intracellular ferrous iron until the dose increased to 100 μg/mL. However, the intracellular ferrous iron chelating activity of DFP at 100 μg/mL or even at 200 μg/mL was still slightly lower than that of hinokitiol at the dose of 2 μg/mL (Fig. S5). These suggest that potent iron-chelation is an important determinant for the antifungal activity of hinokitiol.
Fig. 1.
Effects of hinokitiol on iron homeostasis. (A) Chemical structures of hinokitiol and its two analogues. (B, C) C. albicans SC5314 cells were cultured in RPMI 1640 medium with hinokitiol for 10 h. The cells were then stained with FeRhonox-1, fluorescence intensity was detected by flow cytometry, and the Gmean value was calculated. (D) FeRhonox-1-stained C. albicans cells were further observed by fluorescence microscopy (scale bars: 50 μm). (E) C. albicans SC5314 cells were cultured in RPMI 1640 medium with hinokitiol for 30 min, 1 h or 2 h. The cells were then stained with FeRhonox-1 for measurement of intracellular iron through determining the fluorescence intensity by using flow cytometry. (F) C. albicans SC5314 cells were challenged with hinokitiol for 10 h at 30 °C. The transcription levels of genes encoding proteins in iron metabolism or regulators of iron homeostasis were determined by qPCR and are shown as the fold change relative to the control group. (G-I) C. albicans SC5314 cells were challenged with hinokitiol for 30 min, 1 h or 2 h at 30 °C. The transcription levels of genes associated with iron metabolism or regulators of iron homeostasis were determined by qPCR and were shown as the fold change relative to the control group. The bars represent means ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
We also assessed the transcriptional response to the iron deficiency caused by hinokitiol. Sef1 and Hap43 are two important transcription factors required for C. albicans cell growth in an iron-depleted environment [30]. Under the treatment of 4 μg/mL hinokitiol for 10 h, the transcriptional expression of SEF1 and HAP43 was upregulated by an average of 4.59-fold and 8.13-fold, respectively, reflecting regulatory feedback on iron depletion (Fig. 1F). To alleviate iron deficiency stress, C. albicans activates the expression of iron transporter genes including FTR1, FTR2, and FTH1 (Fig. 1F). Transcriptions of the high-affinity iron permease encoding gene FTR1 and high-affinity iron transporter encoding gene FTH1 were strongly up-regulated by an average of 8.44-fold and 21.21-fold, respectively, in the presence of 4 μg/mL hinokitiol. In addition, the transcription of the mitochondrial iron transporter gene MRS4 was upregulated by an average of 1.73-fold, and the transcription of the vacuolar iron importer gene CCC1 was downregulated by an average of 8.33-fold (Fig. 1F). These changes indicate a re-allocation of iron, probably due to a lack of iron in the mitochondria [31]. We further measured the iron depleted responses of Candida cells upon the treatment of hinokitiol within a short time scale at both 30 °C and 37 °C. The transcriptional expression of most iron transporters encoding genes was responsively upregulated and CCC1 transcriptional expression was downregulated after 30 min of hinokitiol treatment (Fig. 1G-I and Fig. S6), although the expression of some genes varied as the time elapsed (Fig. S6).
To further confirm that iron deprivation is a key factor in the antifungal activity of hinokitiol, we performed rescue experiments. Exogenous addition of non-cytotoxic concentrations of Fe2+ or Fe3+ deactivated hinokitiol (Table 3, Table S2 and S3). By contrast, exogenous addition of Cu2+ had no effect on the antifungal activity of hinokitiol although hinokitiol can chelate copper ions (Table 3). We further noticed that the iron in the culture medium also influenced the activity of hinokitiol. When C. albicans cells were cultured in SD medium, the activity of hinokitiol was reduced by 4-fold compared with cultured in RPMI1460 medium. When DFO at non-toxic dose of 100 μΜ was added into the SD medium to deplete the extracellular iron, the antifungal activity of hinokitiol restored to the same as in RPMI1640 medium (Table S4). Similar phenomenon was observed in Saccharomyces cerevisiae YPH499 strain (Table S4). Iron depletion in C. albicans decreases ergosterol content through the reciprocally regulated expression of ERG11 and ERG3, resulting in increased membrane fluidity and sensitivity to FLC [19]. Consistent with these previous observations, hinokitiol treatment downregulated ERG11 and upregulated ERG3 transcription in C. albicans and potentiated the antifungal activity of FLC (Fig. S7). These results reveal that the antifungal activity of hinokitiol involves chelation of fungal intracellular iron.
Table 3.
Effects of Fe3+, Fe2+ or Cu2+ on the antifungal activity of hinokitiol against C. albicans SC5314.
| Concentrations of metal ions (μM) | MICs (µg/mL) |
||
|---|---|---|---|
| Addition of Fe3+ | Addition of Fe2+ | Addition of Cu2+ | |
| 0 | 2 | 2 | 2 |
| 0.1 | 2 | 2 | 2 |
| 1 | 4 | 4 | 2 |
| 10 | 16 | 16 | 2 |
| 100 | 64 | 64 | 2 |
| 250 | >128 | >128 | 2 |
Hinokitiol inhibits fungal mitochondrial respiration
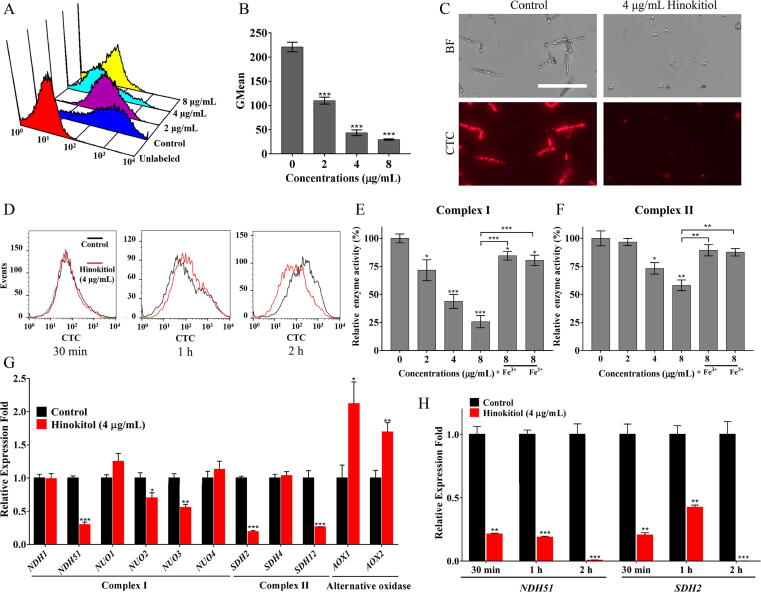
Many genes associated with the mitochondrial respiratory chain are regulated by the transcription factor Hap43 under iron-deprivation conditions [30]. The upregulation of HAP43 by hinokitiol treatment implied a potential influence of hinokitiol on mitochondrial respiration. We first studied the effect of hinokitiol on respiratory activity using a CTC staining assay. As shown in Fig. 2A-C, CTC was largely converted to fluorescent formazan in cells that were not treated with hinokitiol. Hinokitiol treatment dose-dependently reduced the fluorescence intensity in C. albicans cells, with very faint fluorescence when the dose was increased to 4 μg/mL or higher (Fig. 2A-C), suggesting that the respiratory activity of fungal cells was significantly retarded by hinokitiol. We also evaluated the effect of hinokitiol on the respiratory activity within a short time at both 30 °C and 37 °C. The retarded mitochondrial respiration at 37 °C was clearly observed after 2 h of treatment, slightly later than the incidence of iron chelation although these two events almost simultaneously occurred at 30 °C (Fig. 1E, Fig. 2D and Fig. S4), suggesting that inhibited respiration is a result of the iron-chelating effect caused by hinokitiol.
Fig. 2.
Effects of hinokitiol on respiratory activity. (A, B) C. albicans SC5314 cells were cultured in RPMI 1640 medium with hinokitiol for 10 h. The cells were then stained with CTC, and the fluorescence intensity was measured by flow cytometry. (C) C. albicans SC5314 cells stained with CTC were observed by fluorescence microscopy (scale bars: 100 μm). (D) C. albicans SC5314 cells were cultured in RPMI 1640 medium and treated by 4 μg/mL of hinokitiol for 30 min, 1 h or 2 h at 30 °C. The cells were then stained with CTC, and the fluorescence intensity was detected by flow cytometry. (E, F) Extracted mitochondria were challenged with 2, 4 or 8 μg/mL hinokitiol for 3 h. 100 μM of Fe2+ or Fe3+ was used in the rescue experiments. Enzyme activity assays of mitochondrial respiratory chains complexes I and II were then carried out. The results of enzyme activities of mitochondrial respiratory chains complexes III and IV were shown in Fig. S8. (G) C. albicans SC5314 cells were treated with 4 μg/mL hinokitiol for 10 h at 30 °C. The transcription levels of genes related to the mitochondrial respiratory chains complex I and II and alternative oxidases were determined by qPCR and are shown as the fold change relative to the control group. (H) C. albicans SC5314 cells were challenged with 4 μg/mL of hinokitiol for 30 min, 1 h or 2 h at 30 °C. The transcription levels of NDH51 and SDH2 were determined by qPCR and were shown as the fold change relative to the control group. The bars represent means ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Given its inhibitory effect on respiratory activity, we further investigated the effect of hinokitiol on the activities of mitochondrial respiratory chain complexes using isolated fungal mitochondria. As shown in Fig. 2E, treatment with 2 μg/mL, 4 μg/mL or 8 μg/mL hinokitiol reduced the activity of complex I in C. albicans to 71.65%, 43.81% and 25.77% compared with the control group, respectively. When exogenous Fe3+ or Fe2+ was added, the inhibitory effects of hinokitiol were reversed (Fig. 2E). Complex II enzyme activity was reduced by half at hinokitiol concentrations up to 8 μg/mL (Fig. 2F). Complex III and complex IV activities remained above 70% even at 8 μg/mL hinokitiol (Fig. S8). Consistent with these effects, hinokitiol treatment downregulated the transcription levels of genes encoding subunits of respiratory chain complexes I and II, namely, NDH51, NUO2, NUO3, SDH2 and SDH12 (Fig. 2G). Moreover, the transcription of alternative oxidase encoding genes AOX1 and AOX2 was upregulated by hinokitiol treatment, possibly as feedback from inhibition of the classical respiratory chain, as AOX-based respiration allows respiration when the classical chain is suppressed [32]. We further evaluated the transcription levels of selected two genes NDH51 and SDH2 of respiratory chain complexes I and II under the treatment of hinokitiol within a short time at both 30 °C and 37 °C. The transcriptional expression of both genes was downregulated after 0.5 h, 1 h, or 2 h of hinokitiol treatment (Fig. 2H and Fig. S6), consistent with the observation of 10 h treatment by hinokitiol.
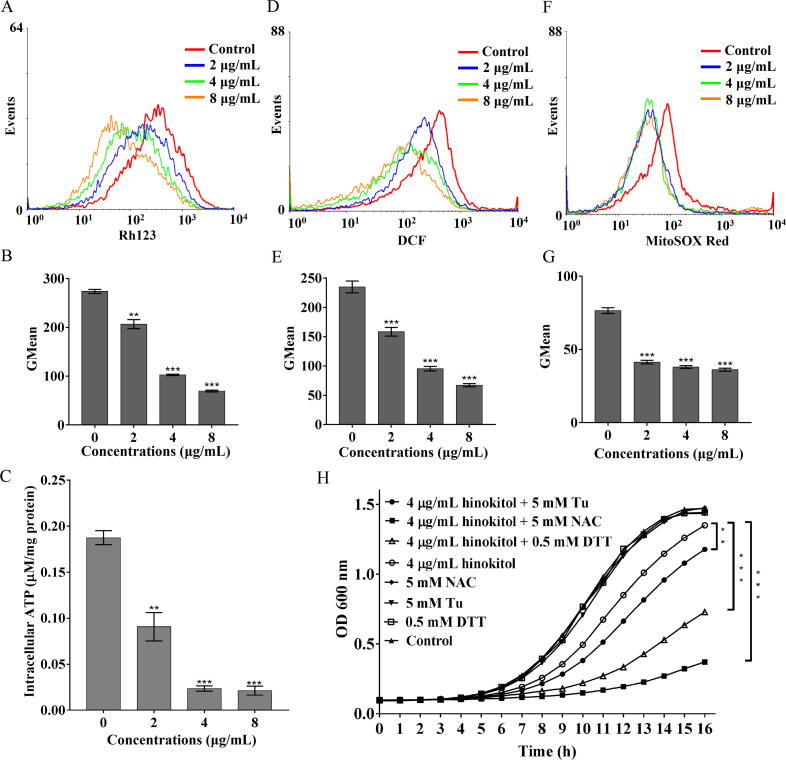
mtΔψ fluctuation and ATP generation are two important indicators of the functional status of the mitochondrion. As shown in Fig. 3A and 3B, hinokitiol significantly decreased the fraction of cells with high Rh123 fluorescence intensity, and the geometric mean (GMean) value decreased from 274.5 (control) to 206.74, 101.86 and 70.28 upon exposure to 2, 4 and 8 μg/mL hinokitiol, respectively, suggesting mtΔψ collapse. The effects of hinokitiol on intracellular ATP content were similar to the effects on mtΔψ (Fig. 3C). However, in the presence of 10 mM ATP-Na2, the MIC of hinokitiol against C. albicans SC5314 increased from 2 μg/mL to 8 μg/mL (Table S5).
Fig. 3.
Effects of hinokitiol on mitochondrial function. C. albicans SC5314 cells were cultured in RPMI 1640 medium with hinokitiol for 10 h. The cells were then stained with Rh123 (A, B) for measurement of mtΔψ. The cells were lysed for measurement of intracellular ATP content (C). The cells were further stained with DCFH-DA (D, E) or stained with MitoSOX Red (F, G) to determine the intracellular or mitochondrial ROS contents. The fluorescence intensity of the stained cells was detected by flow cytometry and the Gmean value was calculated. The bars represent means ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (H) C. albicans SC5314 cells were adjusted to 5 × 104 cells/mL in SC medium containing 5 mM Tu, 5 mM NAC or 0.5 mM DTT. 4 μg/mL of hinokitiol were added to the cultures and incubated at 30 °C for 16 h. Growth was measured by a microplate reader at 600 nm. Cells that were not treated with hinokitiol and reducing agents served as the control. Each point represents the mean of three replicated measurements. The OD600 values at the final detection point are used for significant difference analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
Consistent with the decrease in mtΔψ, we observed drops in ROS and superoxide in C. albicans challenged with hinokitiol at both short time (1 h or 2 h) and long time (10 h) treatment (Fig. 3D-G and Fig. S9). However, the decrease of both ROS and superoxide was more obvious at 37 °C compared with that at 30 °C within short time scales (Fig. S9). This is consistent with above findings of slight decrease of intracellular iron content upon hinokitiol treatment for 1 h at 30 °C (Fig. 1E). ROS levels below the physiological requirement induce “reductive stress”, which is as detrimental to cells as oxidative stress [33]. We utilized SD medium to evaluate the reductive stress in hinokitiol-treated cells because of the potent inhibitory activity of hinokitiol and low growth rates of fungal cells in RPMI 1640 medium. As expected, non-cytotoxic concentrations of the antioxidants Tu, NAC and DTT potentiated the inhibitory effect of hinokitiol on C. albicans growth (Fig. 3H).
Hinokitiol exhibits cell selectivity in chelating intracellular iron
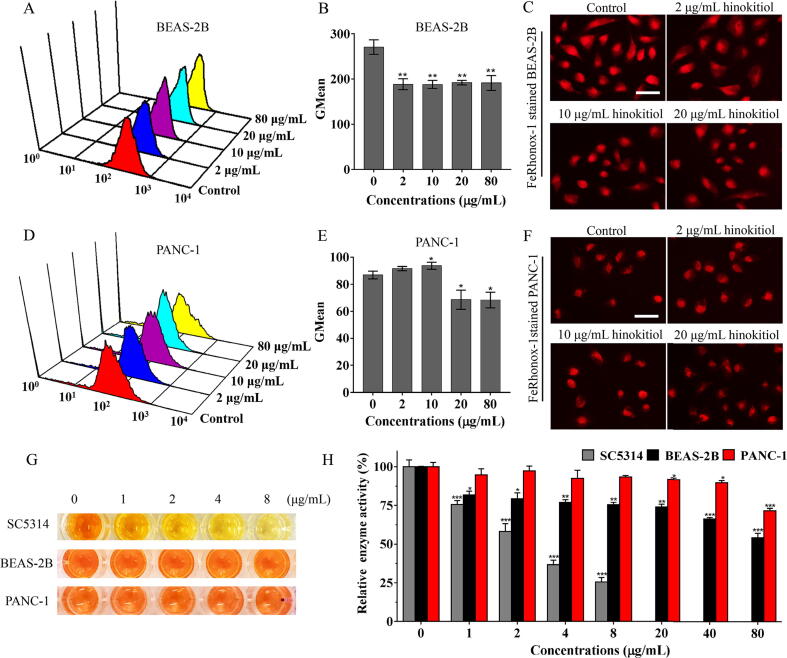
In contrast to the large decrease in intracellular Fe2+ in C. albicans cells, hinokitiol had minor effects on intracellular Fe2+ in mammalian cells, including BEAS-2B human bronchial epithelial cells, and PANC-1 human pancreatic cancer cells (Fig. 4A-F). These results suggest that hinokitiol exhibits selectivity in chelating intracellular iron. We further utilized CCK-8 assays to evaluate the influence of hinokitiol on mitochondrial dehydrogenase activity in fungal cells or mammalian cells. As shown in Fig. 4G and 4H, the reagent WST8 in CCK8 was converted to orange formazan by non-treated fungal cells or mammalian cells. However, in hinokitiol-treated C. albicans cells, the orange color was faint, and the absorbance at 450 nm decreased dose-dependently as the concentration of hinokitiol increased from 1 to 8 μg/mL. By contrast, hinokitiol even at 80 μg/mL had minor effects on the mitochondrial dehydrogenase activity of mammalian cells after 12 h of treatment (Fig. 4G and 4H). Due to the slower growth of mammalian cells compared with fungal cells, we further evaluated the cytotoxicities of hinokitiol after 48 h of culture of mammalian cells. Hinokitiol caused limited harm on the growth and mitochondrial function of mammalian PANC-1 cells even at high dose (Fig. S10). These results suggest biological safety of hinokitiol in antifungal applications due to its selectivity.
Fig. 4.
The differential effects of hinokitiol on iron chelation and mitochondrial respiratory activity between fungal cells and mammalian cells. (A-F) Mammalian BEAS-2B (A-C) and PANC-1 (D-F) cells were cultured with hinokitiol for 10 h. The cells were then stained with FeRhonox-1, fluorescence intensity was detected by flow cytometry (A, D), and the Gmean value was calculated (B, E). FeRhonox-1-stained cells were further observed by fluorescence microscopy (scale bars: 100 μm) (C, F). (G, H) C. albicans SC5314, mammalian BEAS-2B or PANC-1 cells were treated with different concentrations of hinokitiol for 12 h at 37 °C. CCK-8 assays were performed to reveal the effects of hinokitiol on mitochondrial dehydrogenase activities in fungal cells or mammalian cells. The detailed procedures were seen in Materials and methods section. The wells with addition of CCK-8 solutions were imaged by a camera (G), and the absorbance at 450 nm was measured by a microplate reader (H). The bars represent means ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Hinokitiol has low capacity to develop resistance
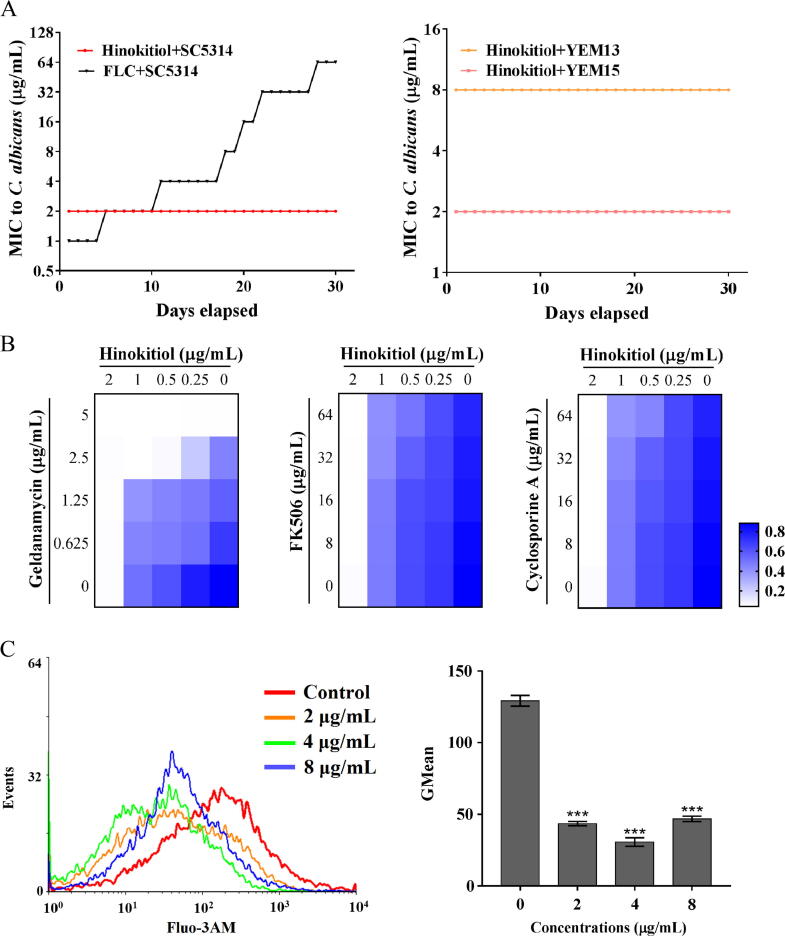
Candida species tend to develop resistance to azole drugs [34]. To evaluate the ability of hinokitiol to induce resistance in Candida species, C. albicans SC5314 cells were exposed to a sub-MIC level of hinokitiol or FLC for 30 days using a serial passage method in 96-well plates. After 30 passages of induced resistance, the MICs of hinokitiol against all Candida species remained constant. By comparison, the MIC values of FLC against SC5314 increased by 32 fold and up to 64 μg/mL after 30 passages (Fig. 5A), and similar results were obtained for other Candida species (Fig. S11). These results demonstrate that hinokitiol has a very low potential of inducing drug resistance in Candida species and suggest that iron restriction by chelators is a promising method to copy with the challenge of drug resistance in antifungal therapy.
Fig. 5.
The potential of hinokitiol to induce resistance. (A) The organisms of indicated C. albicans strains at a sub-lethal dose of hinokitiol or FLC were taken as the inoculum for the MIC measurement of the next passage. These passages lasted for 30 days. (B) A dose–response matrix was performed in RPMI 1640 medium with gradients of hinokitiol and Hsp90 inhibitor (geldanamycin) or calcineurin inhibitors (FK506 and cyclosporine A). Data were analyzed after incubation for 24 h at 35 °C and are displayed as a heat map. (C) C. albicans cells were cultured in RPMI 1640 medium with hinokitiol for 10 h. The cells were then stained with Fluo-3AM, and the fluorescence intensity was measured by flow cytometry. The bars represent means ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
C. albicans has developed multiple stress response pathways to adapt to host conditions and promote the evolution of drug resistance [35]. The molecular chaperone Hsp90 and its effector protein, the phosphatase calcineurin, are stress response regulators whose inhibition can enhance the antifungal activity of azole or echinocandin drugs against diverse fungal pathogens. Therefore, we evaluated the roles of Hsp90 and calcineurin in the response of C. albicans to hinokitiol treatment. Deletion of the calcineurin catalytic subunit Cnb1 or pharmaceutical inhibition of Hsp90 or calcineurin function had no obvious effect on the activity of hinokitiol (Fig. 5B and Table 2), implying no stress response via the Hsp90-calcineurin pathway. Cytoplasmic levels of calcium, which is a stimulator of the Hsp90-calcineurin pathway, were reduced by hinokitiol treatment (Fig. 5C). These results suggest that the lack of activation of Hsp90-calcineurin stress response signaling pathways at least reduces the potential for Candida species to develop resistance to hinokitiol.
Hinokitiol shows therapeutic effects in C. albicans-infected larvae
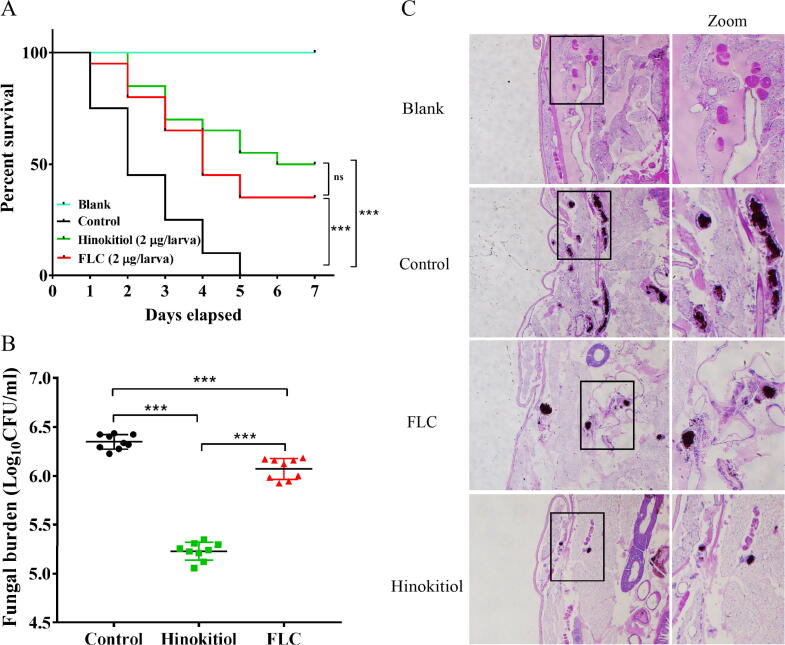
In view of the potent antifungal activity of hinokitiol in vitro, we further used G. mellonella as an infection model to assess the therapeutic potential of hinokitiol in vivo. Without drug treatment, infected larvae died within five days. By contrast, 50% of the larvae treated with hinokitiol (2 μg/larva) survived after 7 days (Fig. 6A), indicating therapeutic effectiveness of hinokitiol in treating C. albicans infection. Moreover, the fungal burden of hinokitiol-treated larvae was much lower than that of untreated larvae (Fig. 6B). Consistent with these observations, histological analysis using PAS staining revealed a higher number of melanized nodules and greater tissue infiltration in the untreated group compared with the hinokitiol-treated group (Fig. 6C). We further utilized uninfected larvae to test the toxicity of hinokitiol. Injection with 20 μg of hinokitiol into each larva did not influence the survival of larvae, suggesting the low toxicity of hinokitiol (Fig. S12). These in vivo results suggest that hinokitiol holds potential for treating fungal infections although its in vivo efficacy can also be diminished by excess iron (Fig. S12).
Fig. 6.
Hinokitiol has therapeutic effects on C. albicans-infected G. mellonella. The larvae in each group were infected with at 2 × 105C. albicans cells. After 2 h of infection, the larvae were treated with PBS (control), FLC (2 μg/larva) or hinokitiol (2 μg/larva). The larvae in blank group were not infected but injected with equal volume of PBS. (A) Survival curve of infected G. mellonella by C. albicans (n = 20 per group). The number of surviving larvae was recorded each day. The Mantel-Cox test was used to compare differences between the groups. *P < 0.05, **P < 0.01, ***P < 0.001. ns means non-significance. (B) Fungal burden of infected G. mellonella by C. albicans (n = 9 per group). (C) The larvae in different treated groups (n = 2 per group) were fixed by paraformaldehyde and processed for periodic acid-Schiff (PAS) staining. The histopathology of infected G. mellonella under different indicated treatments was examined by microscopic inspections. The rectangles highlighted the melanized nodules containing a mixture of yeast cells and filaments for closer observation (right lane).
Discussion
Perturbation of iron homeostasis is an established strategy for combatting infectious diseases. Iron chelators have been used for superficial fungal infections and improve the efficacy of certain drugs [19]. Hinokitiol has potent antifungal activity and can chelate iron. However, the relationship between the iron-chelating ability of hinokitiol and its antifungal activity has not been established.
In this study, we demonstrated that the strong antifungal activity of hinokitiol is mainly attributable to its iron-chelating ability. Compared with tropolone and maltol, hinokitiol binds iron more strongly [15], which explains the greater antifungal activity of hinokitiol. FeRhonox-1 staining revealed that some iron chelators, such as DFO, DFP and ferrozine at low dose (8 μg/mL), did not decrease intracellular Fe2+ content, probably due to the impermeability of the fungal cell membrane to these agents or their weaker ability to bind Fe2+. For DFP, it requires much higher dose to chelate fungal intracellular Fe2+ and achieve the antifungal activity. On the contrary, the intracellular iron chelating ability for hinokitiol at 2 μg/mL is comparable to that of DFP at 200 μg/mL. These findings together with intense iron-depletion responses in response to hinokitiol and its diminished activity by addition of iron suggest that intracellular iron chelation is an important determinant for the antifungal activity of hinokitiol.
Intracellular iron deficiency results in dysfunction of mitochondrial respiration [17], which is a fundamental function of mitochondria and stands as a potential therapeutic target [32], [36]. In the present study, hinokitiol treatment suppressed respiratory activity and significantly inhibited the enzyme activities of mitochondrial complexes I and II in C. albicans because of the important roles of iron-sulfur proteins in these complexes. We noticed that reduced intracellular iron contents and iron depletion responses of genes transcriptions occurred before the inhibition of mitochondrial respiration, suggesting that the repression of mitochondrial respiration is a downstream effect of fungal intracellular iron-chelation. The fungal mitochondrial respiration is closely linked with Ras1-cAMP-PKA pathway [37], [38], [39]. Elevated ATP levels correlate with Ras1-cAMP-PKA pathway activation and the downstream of Efg1-induced gene expressions. In hinokitiol treated C. albicans cells, the mitochondrial respiration was inhibited and ATP generation was reduced, which downregulated the Ras1-cAMP-PKA pathway. This result is consistent with previous findings that hinokitiol inhibits the Candida growth by regulating the Ras1-cAMP-PKA pathway [14].
On the other hand, hinokitiol-mediated suppression of mitochondrial respiration brings about reductive stress. Reductive stress is considered as an excess accumulation of reducing equivalents including NADH, NADPH, and GSH, which overwhelms the capacity of endogenous oxidoreductases [40]. Under the treatment of hinokitiol, NADH oxidation at complex I is inhibited, which leads to a buildup of NADH and consequent reductive stress. Moreover, fewer ROS generation is another source of reductive stress when the mitochondrial respiration is inhibited by hinokitiol. Reductive stress decreases cellular ROS levels and can also impair cellular functions [41]. For example, proper protein disulfide formation is perturbed under reductive stress and leads to endoplasmic reticulum (ER) stress. As we expected, hinokitiol-treated cells exhibited hypersusceptibility to reducing stresses induced by NAC, Tu, and ER stress inducer DTT.
By contrast, at the cellular level, we did not observe notable depletion of Fe2+ by hinokitiol in our tested mammalian cell lines. Accordingly, the respiratory activity of mammalian cells was not severely affected by hinokitiol. In previous study, hinokitiol is considered as an iron carrier to transport iron into or out of cells to maintain intracellular iron homeostasis and prevent ferritoxicity in animal cells [15]. However, the iron chelation for hinokitiol features in Candida cells. These distinct effects of hinokitiol in mammalian versus fungal cells may explain the good biological safety of hinokitiol and support its potential application as a selective inhibitor of respiratory activity in fungal cells.
Resistant strains of fungal pathogens can quickly be generated after the introduction of antifungal drugs, particularly fungistatic agents [4]. The fungistatic drug FLC imposes constant inhibitory pressure on fungal cells, eliciting stress responses such as activation of Hsp90-calcineurin signaling pathways for survival tolerance. Pharmacological inhibition of Hsp90-calcineurin signaling pathways can not only prevent the emergence of azole resistance but also render FLC fungicidal [26], [34]. Hinokitiol is also a fungistatic agent but shows low potential for the development of resistance, consistent with a previous finding that the iron chelator ciclopirox olamine has minimal ability to induce drug resistance [18]. One possible reason for the lack of development of resistance is that the low intracellular ATP levels induced by hinokitiol cannot sustain ATP-dependent efflux or mutations to complement the stress. Another potential reason is the non-activation of Hsp90-calcineurin stress response signaling pathways that reduces the possibility of drug resistance evolution. In addition, iron depletion had minimal effects on the expressions of genes encoding efflux pumps [19]. These findings suggest that iron chelators may be ideal agents for treating fungal infections.
Conclusion
We demonstrated that hinokitiol has promising antifungal activity by interfering with iron homeostasis. Respiratory chain dysfunction caused by the disruption of iron homeostasis by hinokitiol ultimately impeded the proliferation of fungal cells. Our findings reveal the underlying antifungal mechanism of hinokitiol and support its potential application to treat relevant fungal infections, particularly those resulting from resistant organisms.
Compliance with Ethics Requirements
No human or animal subjects are involved in this article.
CRediT authorship contribution statement
Xueyang Jin: Methodology, Software, Investigation, Writing - original draft. Ming Zhang: Visualization, Data curation. Jinghui Lu: Validation. Ximeng Duan: Investigation. Jinyao Chen: Investigation. Yue Liu: Investigation. Wenqiang Chang: Conceptualization, Resources, Writing - review & editing, Funding acquisition. Hongxiang Lou: Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We appreciate Professor Kim Lewis of Northeastern University, Professor Joachim Morschhäuser of the University of Würzburg, and Professor Qingguo Qi of Shandong University for kindly donating the C. albicans strains used in this study. This work was funded by National Natural Science Foundations [Nos. 81630093, 81773786]; Fund for Innovative Team of Shandong University to H.L.; Natural Science Fund for Excellent Young Scholars of Shandong Province of China [ZR2020YQ63]; and Young Scholars Program of Shandong University [2017WLJH41].
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.06.016.
Contributor Information
Wenqiang Chang, Email: changwenqiang@sdu.edu.cn.
Hongxiang Lou, Email: louhongxiang@sdu.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Kullberg B.J., Arendrup M.C. Invasive candidiasis. N Engl J Med. 2015;373(15):1445–1456. doi: 10.1056/NEJMra1315399. [DOI] [PubMed] [Google Scholar]
- 2.Millsop J.W., Fazel N. Oral candidiasis. Clin Dermatol. 2016;34(4):487–494. doi: 10.1016/j.clindermatol.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 3.Arendrup M.C., Patterson T.F. Multidrug-resistant Candida: epidemiology, molecular mechanisms, and treatment. J Infect Dis. 2017;216(suppl_3):S445–S4S51. doi: 10.1093/infdis/jix131. [DOI] [PubMed] [Google Scholar]
- 4.Fairlamb A.H., Gow N.A., Matthews K.R., Waters A.P. Drug resistance in eukaryotic microorganisms. Nat Microbiol. 2016;1(7):16092. doi: 10.1038/nmicrobiol.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vengurlekar S., Sharma R., Trivedi P. Efficacy of some natural compounds as antifungal agents. Pharmacogn Rev. 2012;6(12):91–99. doi: 10.4103/0973-7847.99942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domon H., Hiyoshi T., Maekawa T., Yonezawa D., Tamura H., Kawabata S., et al. Antibacterial activity of hinokitiol against both antibiotic-resistant and -susceptible pathogenic bacteria that predominate in the oral cavity and upper airways. Microbiol Immunol. 2019;63(6):213–222. doi: 10.1111/1348-0421.12688. [DOI] [PubMed] [Google Scholar]
- 7.Jayakumar T., Liu C.-H., Wu G.-Y., Lee T.-Y., Manubolu M., Hsieh C.-Y., et al. Hinokitiol inhibits migration of A549 lung cancer cells via suppression of MMPS and induction of antioxidant enzymes and apoptosis. Int J Mol Sci. 2018;19(4) doi: 10.3390/ijms19040939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyras D., Shih Y.-H., Lin D.-J., Chang K.-W., Hsia S.-M., Ko S.-Y., et al. Evaluation physical characteristics and comparison antimicrobial and anti-inflammation potentials of dental root canal sealers containing hinokitiol in vitro. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0094941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasumoto E., Nakano K., Nakayachi T., Morshed S.R.M., Hashimoto K., Kikuchi H., et al. Cytotoxic activity of deferiprone, maltol and related hydroxyketones against human tumor cell lines. Anticancer Res. 2004;24(2B):755–762. [PubMed] [Google Scholar]
- 10.Fallik E., Grinberg S. Hinokitiol: a natural substance that controls postharvest diseases in eggplant and pepper fruits. Postharvest Biol Tec. 1992;2(2):137–144. [Google Scholar]
- 11.Imai N., Doi Y., Nabae K., Tamano S., Hagiwara A., Kawabe M., et al. Lack of hinokitiol (beta-thujaplicin) carcinogenicity in F344/DuCrj rats. J Toxicol Sci. 2006;31(4):357–370. doi: 10.2131/jts.31.357. [DOI] [PubMed] [Google Scholar]
- 12.Li J.-Y., Liu C.-P., Shiao W.-C., Jayakumar T., Li Y.-S., Chang N.-C., et al. Inhibitory effect of PDGF-BB and serum-stimulated responses in vascular smooth muscle cell proliferation by hinokitiol via up-regulation of p21 and p53. Arch Med Sci. 2018;14(3):579–587. doi: 10.5114/aoms.2018.75085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura M., Fujibayashi T., Tominaga A., Satoh N., Kawarai T., Shinozuka O., et al. Hinokitiol Inhibits Candida albicans adherence to oral epithelial cells. J Biosci. 2010;52(1):42–50. [Google Scholar]
- 14.Kim D.J., Lee M.W., Choi J.S., Lee S.G., Park J.Y., Kim S.W. Inhibitory activity of hinokitiol against biofilm formation in fluconazole-resistant Candida species. PLoS ONE. 2017;12(2):e0171244. doi: 10.1371/journal.pone.0171244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grillo A.S., SantaMaria A.M., Kafina M.D., Cioffi A.G., Huston N.C., Han M., et al. Restored iron transport by a small molecule promotes absorption and hemoglobinization in animals. Science. 2017;356(6338):608–616. doi: 10.1126/science.aah3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjørklund G., Hofer T., Nurchi V.M., Aaseth J. Iron and other metals in the pathogenesis of Parkinson's disease: Toxic effects and possible detoxification. J Inorg Biochem. 2019;199 doi: 10.1016/j.jinorgbio.2019.110717. [DOI] [PubMed] [Google Scholar]
- 17.Li Y., Sun L., Lu C., Gong Y., Li M., Sun S. Promising antifungal targets against Candida albicans based on ion homeostasis. Front Cell Infect Microbiol. 2018;8:286. doi: 10.3389/fcimb.2018.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niewerth M., Kunze D., Seibold M., Schaller M., Korting H.C., Hube B. Ciclopirox olamine treatment affects the expression pattern of Candida albicans genes encoding virulence factors, iron metabolism proteins, and drug resistance factors. Antimicrob Agents Chemother. 2003;47(6):1805–1817. doi: 10.1128/AAC.47.6.1805-1817.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prasad T., Chandra A., Mukhopadhyay C.K., Prasad R. Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob Agents Chemother. 2006;50(11):3597–3606. doi: 10.1128/AAC.00653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puri S., Kumar R., Rojas I.G., Salvatori O., Edgerton M. Iron chelator deferasirox reduces Candida albicans invasion of oral epithelial cells and infection levels in murine oropharyngeal candidiasis. Antimicrob Agents Chemother. 2019;63(4):e02152–e2218. doi: 10.1128/AAC.02152-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savage K.A., Parquet Md.C., Allan D.S., Davidson R.J., Holbein B.E., Lilly E.A., et al. Iron restriction to clinical isolates of Candida albicans by the novel chelator DIBI inhibits growth and increases sensitivity to azoles in vivo in a murine model of experimental vaginitis. Antimicrob Agents Chemother. 2018;62(8):e02576–e2617. doi: 10.1128/AAC.02576-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute . CLSI; Wayne, PA, USA: 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts-Third Edition: Approved Standard M27–A3. [Google Scholar]
- 23.Sun L., Liao K., Wang D. Honokiol induces superoxide production by targeting mitochondrial respiratory chain complex I in Candida albicans. PLoS One. 2017;12(8):e0184003. doi: 10.1371/journal.pone.0184003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M., Chang W., Shi H., Li Y., Zheng S., Li W., et al. Floricolin C elicits intracellular reactive oxygen species accumulation and disrupts mitochondria to exert fungicidal action. FEMS Yeast Res. 2018;18(1):foy002. doi: 10.1093/femsyr/foy002. [DOI] [PubMed] [Google Scholar]
- 25.Champion O.L., Titball R.W., Bates S. Standardization of G. mellonella larvae to provide reliable and reproducible results in the study of fungal pathogens. J Fungi (Basel) 2018;4(3) doi: 10.3390/jof4030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coste A., Turner V., Ischer F., Morschhäuser J., Forche A., Selmecki A., et al. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics. 2006;172(4):2139–2156. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flowers S.A., Barker K.S., Berkow E.L., Toner G., Chadwick S.G., Gygax S.E., et al. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell. 2012;11(10):1289–1299. doi: 10.1128/EC.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morschhäuser J., Barker K.S., Liu T.T., Bla B.W.J., Homayouni R., Rogers P.D. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 2007;3(11):e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg A., Ene I.V., Bibi M., Zakin S., Segal E.S., Ziv N., et al. Antifungal tolerance is a subpopulation effect distinct from resistance and is associated with persistent candidemia. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-04926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh R.P., Prasad H.K., Sinha I., Agarwal N., Natarajan K. Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J Biol Chem. 2011;286(28):25154–25170. doi: 10.1074/jbc.M111.233569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen O.S., Kaplan J. CCC1 suppresses mitochondrial damage in the yeast model of Friedreich's ataxia by limiting mitochondrial iron accumulation. J Biol Chem. 2000;275(11):7626–7632. doi: 10.1074/jbc.275.11.7626. [DOI] [PubMed] [Google Scholar]
- 32.Duvenage L., Munro C.A., Gourlay C.W. The potential of respiration inhibition as a new approach to combat human fungal pathogens. Curr Genet. 2019;65(6):1347–1353. doi: 10.1007/s00294-019-01001-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zorova L.D., Popkov V.A., Plotnikov E.Y., Silachev D.N., Pevzner I.B., Jankauskas S.S., et al. Mitochondrial membrane potential. Anal Biochem. 2018;552:50–59. doi: 10.1016/j.ab.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morschhauser J. The development of fluconazole resistance in Candida albicans-an example of microevolution of a fungal pathogen. J Microbiol. 2016;54(3):192–201. doi: 10.1007/s12275-016-5628-4. [DOI] [PubMed] [Google Scholar]
- 35.Cowen L.E., Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309(5744):2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 36.Kulawiak B., Hopker J., Gebert M., Guiard B., Wiedemann N., Gebert N. The mitochondrial protein import machinery has multiple connections to the respiratory chain. BBA-Bioenergetics. 2013;1827(5):612–626. doi: 10.1016/j.bbabio.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Grahl N., Demers E.G., Lindsay A.K., Harty C.E., Willger S.D., Piispanen A.E., et al. Mitochondrial activity and cyr1 are key regulators of ras1 activation of C. albicans virulence pathways. PLoS Pathog. 2015;11(8):e1005133. doi: 10.1371/journal.ppat.1005133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silao F.G.S., Ward M., Ryman K., Wallström A., Brindefalk B., Udekwu K., et al. Mitochondrial proline catabolism activates Ras1/cAMP/PKA-induced filamentation in Candida albicans. PLoS Genet. 2019;15(2):e1007976. doi: 10.1371/journal.pgen.1007976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belenky P., Camacho D., Collins J.J. Fungicidal drugs induce a common oxidative-damage cellular death pathway. Cell Rep. 2013;3(2):350–358. doi: 10.1016/j.celrep.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao W., Loscalzo J. Metabolic responses to reductive stress. Antioxid Redox Sign. 2020;32(18):1330–1347. doi: 10.1089/ars.2019.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Handy D.E., Loscalzo J. Responses to reductive stress in the cardiovascular system. Free Radical Bio Med. 2017;109:114–124. doi: 10.1016/j.freeradbiomed.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.