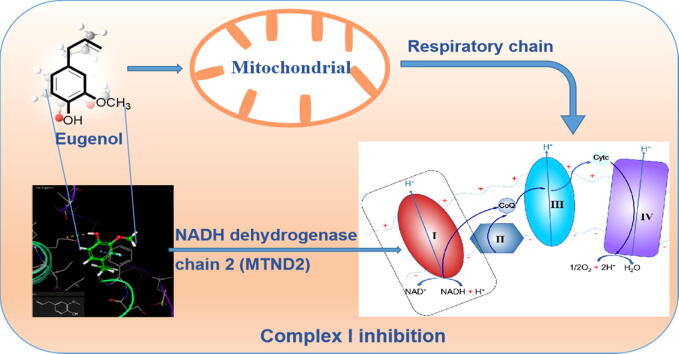
Graphical abstract
Keywords: Eugenol, Acaricidal activity, Complex I, Mitochondrial respiratory chain, Safety
Abstract
Introduction
Eugenol is a major component of essential oils of several plants, it exhibits significant antiparasitic and acaricidal activities, yet its molecular targets remain unknown.
Objectives
We aimed to systematically investigate the mechanism of action and the potential targets of eugenol against P. cuniculi, and evaluate the safety for laying the theoretical foundation for clinical application as an acaricide.
Methods
Using RNA-Seq analysis, surface plasmon resonance analysis and RNA interference assay, the mode of action of eugenol against Psoroptes cuniculi was investigated. The effect on the mitochondrial membrane potential and complex I of PC12 cells and C6/36 cells was assayed to investigate the species specificity of eugenol in insects and mammals. Finally, a safety evaluation of eugenol in vivo was performed.
Results
Eugenol inhibited complex I activity of the mitochondrial respiratory chain in the oxidative phosphorylation pathway by binding to NADH dehydrogenase chain 2 and resulted in the death of mites. The inhibition rates were 37.89% for 50 μg/mL and 60.26% for 100 μg/mL, respectively. Further experiments indicated that the difference in the complex I sequence between insects and mammals led to the different affinity of eugenol to specific peptide, resulting in species specificity. Eugenol exhibited significant inhibitory effects against the mitochondrial membrane potential and complex I in Aedes albopictus C6/36 cells but was not active in rat PC12 cells. Insect cells were particularly sensitive to eugenol. In contrast to the known inhibitor rotenone, eugenol had better safety and did not result in Parkinson’s disease or other diseases in rats.
Conclusion
This is the first report on acaricidal eugenol targeting complex I of the mitochondrial respiratory chain. This work lays the foundation for the development of eugenol as an environmentally alternative acaricidal agent.
Introduction
In recent decades, the development of drug resistance, environmental pollution and other disorders of commercial antiparasitic agents have hindered their applications [1], [2]. The development of ecofriendly alternative agents from sustainable natural products has become increasingly attractive and is much needed in both human and agricultural applications [2].
Plant essential oils have favorable ecotoxicological properties that make them suitable for managing parasites or insects [3], [4]. Eugenol (4-allyl-2-methoxyphenol), a major component of essential oils of several plants in the family Myrtaceae [5], has attracted the attention of researchers with its remarkably broad spectrum of activities [6], [7], [8], [9], [10]. Recent studies have shown that eugenol and its analogs exhibit significant antiparasitic activity against various parasites in both humans and animals (includes cattle and rabbits), such as Leishmania infantum chagasi, Psoroptes ovis and Psoroptes cuniculi [11], [12], [13], [14]. Knio et al. reported that eugenol exhibited larvicidal activity against Ochlerotatus caspius [15] by reducing the mitochondrial membrane potential as shown in isolated rat liver mitochondria [16]. Further studies showed that it could also be used in contact insecticides and fumigants against household pests to protect foods from microorganism contamination during storage and in herbicides against certain grassy and broad-leaved weeds [17]. Toxicity to plant pathogenic fungi and plant parasitic nematodes has also been found [18], [19]. In our previous studies, we found that eugenol and its analogs exhibited acaricidal activity [14], [20], [21]. Ma et al. studied the potential mechanism of eugenol on P. cuniculi using transcriptome analysis, and result showed that eugenol may affect the PPAR, NF-κB, TNF and Ras signaling pathways of mites [13]. However, the verification tests for these pathways were lacked, and the useful information for uncovering the mechanisms of eugenol were limited. The potential targets are still unknown. In this study, we aimed to systematically investigate the mechanism of action and the potential targets of eugenol against P. cuniculi, and evaluate the safety for establishing the theoretical foundation for clinical application as an acaricide.
Although mechanistic studies of drug action have a long history, the discovery of potential targets of compounds still relies on affinity chromatography and omics technologies [22], [23], [24], [25]. In the current study, surface plasmon resonance (SPR) and RNA-Seq analysis were combined to investigate the potential acaricidal targets of eugenol, and P. cuniculi was used as an example parasite. Subsequently, the enzyme-inhibitory activities of eugenol were studied to confirm the potential targets identified as well as RNA interference assay. Then, the effect on the mitochondrial membrane potential (MMP) and complex I of PC12 cells and C6/36 cells was assayed to investigate the species specificity of eugenol in insects and mammals. Finally, a safety evaluation of eugenol in rats was carried out.
Material and methods
Ethics statement
All experiments involving animals were reviewed and approved by the Institute Animal Care and Use Committee of Lanzhou Institute of Husbandry and Pharmaceutical Sciences of Chinese Academy of Agricultural Sciences (SYXK-2014–0002).
Chemicals
Eugenol (98%), acetylthiocholine iodine, 5,5′-dithiobis(2-nitrobenzoic acid), L-reduced glutathione and 1-chloro-2,4-dinitrobenzene were purchased from Sigma-Aldrich Co. Ltd. (St. Louis, USA); and ivermectin (95%) were purchased from Shanghai Yuanye Biotechnology Co. Ltd. (Shanghai, China).
Target discovery
RNA-Seq analysis
A total of 2000 adult Psoroptes cuniculi were isolated from infested rabbits according to methods previously described [20]. Briefly, eugenol was diluted with 10% DMSO to 100 μg/mL, and then 350 μL of the solution (100 μg/mL) was added to Petri dishes (Φ 60 mm) for the treatment group, while 350 μL of 10% DMSO was added for an untreated control group. The experiment was repeated three times. Subsequently, the mites were placed in the solution. All plates were incubated at 25 °C and 75% relative humidity. At 8 h and 16 h, 100 μL of eugenol (100 μg/mL) or 10% DMSO was added again to each well. After treatment for 24 h, treated and untreated mites (control group) were washed with cold phosphate-buffered saline (PBS) to remove any contaminants on the surface and immediately stored at −80 °C until use in RNA isolation [26].
Total RNA was extracted from the mites using TRIzol reagent (Invitrogen, USA) and the concentration and quality of RNA were determined using an Agilent Bioanalyzer 2100 system (Agilent Technologies, CA). Sequencing libraries were generated using an NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA). First strand cDNA was synthesized using random hexamer primer and M−MuLV Reverse Transcriptase (RNase H-), and second strand cDNA was synthesized using DNA Polymerase I and RNase H. The construction of a cDNA library and sequencing were performed using oligo (dT) magnetic beads, and mRNA was interrupted to form short fragments after the addition of fragmentation buffer, followed by cDNA synthesis. Double-stranded cDNA was purified using the QiaQuick PCR Purification Kit (Qiagen, Germany) and subjected to library preparation for sequencing analysis with an Illumina HiSeqTM 2000 platform.
Raw reads were transformed into clean reads and de novo RNA-Seq assembly was accomplished using Trinity software [26], [27], [28]. Databases were used to annotate all unigene sequences using BLASTx. After assembly, clean reads were mapped to unigenes using Bowtie2. Gene expression levels for each sample were calculated with RSEM. Differential expression analysis of two samples was performed using DEGseq. The P value was adjusted using the q value. Q < 0.005 and log2-fold change > 1 were set as the threshold criteria for significantly differential expression.
SPR-MS experiment
Small-molecule microarray (SMM) preparation and SPR experiment
Experiments were carried out using a PlexArray®HT system, which is based on the Three-dimensional (3D) SPR imaging technique. 3D Photo-cross-linker Sensor CHIP™ used in this part was provided by Betterways Inc. (Guangzhou, China). Firstly, the specificity of chips was validated, and DMSO and rapamycin (10 mM, FBBP 12 inhibitor) were fixed in chips as negative control and positive control, respectively. FKBP12 (100 nM), a FK506-binding protein of 12 kD, as running solution was injected into the SPR system and flowed through the rapamycin (FBBP 12 inhibitor) passage. The response signal of DMSO and rapamycin were detected. After the validation of 3D SPRi chips, eugenol solution (10 mM) were spotted in a multiplex separated block using a Genetix Q Array 2 spotter and left for complete evaporation of DMSO at room temperature in an N2 atmosphere for 2 h. The slides were exposed to UV irradiation 2.4 J/cm2 (365 nm) in a UV chamber (Amersham life science, USA). Next, the slides were washed with DMSO followed by DMF, ethanol and distilled water for 15 min each on a table concentrator to remove noncovalently bound compounds and dried with N2. Dried slides were assembled with a flow cell cover and then mounted on the SPR instrument for measurement, and the real-time binding signals were recorded and analyzed with a PlexArray SPRi system. Before the binding test, 100 µg/mL BSA in ddH2O was flowed through the surface at a rate of 1 µL/s for 15 min to block the remaining binding sites on the chip surface to decrease nonspecific absorption during the following procedures. The cell sample of mites was harvested by centrifugation at 1200 rpm, RT for 5 min. Before lysis, a final concentration of 1% (v:v) cocktail protease inhibition reagent (Thermo Fisher, United States) was added to the sample, fully mixed and re-suspended. Then BWLS-17 lysate buffer (Betterways Inc., China) was added for lysis at a volume ratio of 3:4. The mixture was incubated on ice bath for 30 min, then centrifuged at 12000 rpm, 4℃ for 15 min. The supernatant was transferred to a new EP tube and protein concentration was calibrated with BCA Protein Assay Kit (Thermo Fisher, United States). The concentration was adjusted using a lysate 1x stock solution to a final concentration of 200 μg /mL. Lysate cell solutions of mites with a concentration of 200 µg/mL in phosphate-buffered solution (PBST, pH 7.0) were used as analytes with a flow rate of 2 µL/s until the binding curve reached a plateau. The lysate solutions included 20 mM HEPES (pH 7.2), 150 mM NaCl, 0.05% Triton X-100, 1% sodium deoxycholate, 2 mM sodium pyrophosphate, 25 mM β-glycerol phosphate disodium salt, 1 mM EDTA and 1 mM sodium orthavanadium. Before using this solution, 50 µL of 0.5 µg/mL leupeptin was added. Lysate solutions of mites with a concentration of 200 µg/mL in phosphate-buffered solution (PBST, pH 7.0) were used as analytes with a flow rate of 2 µL/s until the binding curve reached a plateau. Then, the surface was washed with PBST with a flow rate of 0.5 µL/s for 120 s, and the capturing channels were sealed. After the capturing stage, the microfluidic valve on SPR detector was switched to quality control channel and 100 nM FKBP12 flowed through the rapamycin passage. The SPR detector automatically collects the binding signal of the FKBP12-rapamycin as a system positive control and PBST-rapamycin as a system negative control. All experiments were repeated at least three times to ensure data repeatability.
LC-MS experiment
Before identifying the possible mite proteins being targeted by eugenol using HPLC-MS/MS, the chips were digested in situ by trypsin to obtain the proteins or peptides captured by eugenol. In detail, after treatment of the chip surface with 30 µL of 10 mM dithiothreitol and 30 µL of 55 mM iodoacetamide and washing with 30 µL of 0.25 M triethylammonium bicarbonate, trypsin (Promega) was added for the digestion of captured proteins overnight at 37 °C according to the ratio of protein:trypsin = 20:1. Finally, the peptides were collected and dried under vacuum.
The peptides were desalted on Acclaim PepMap C18 Cartridges, then analyzed on Nano-C18 Cartridges (Thermo Fisher Scientific, United States). MS experiments were performed on a Q Exactive mass spectrometer coupled to a Nano Acquity UPLC system (Waters Corporation, United States) to the detection. Peptides were loaded onto a C18-reversed-phase column in solvent A (5% acetonitrile and 0.1% formic acid, pH 2.5) and separated with a linear gradient of solvent B (90% acetonitrile and 0.1% formic acid, pH 2.5) at a flow rate of 300 nL/min over 60 min. The gradient profile was from 2% B to 45% B at the given times. MS data were acquired using a data-dependent top ten method in which the most abundant precursor ions were dynamically chosen for HCD fragmentation. The instrument was run with peptide recognition mode enabled. MS data were collected by Xcalibur (Thermo Scientific, version 2.2.0), and MS experiments were performed triply for each sample. The MS data were analyzed using MaxQuant software (COX LAB, version 1.3.0.5). Peptides were identified by database searching and the MS2 results for selected proteins that changed quantity between sample types were annotated via BLASTP. The cut-off of the global false discovery rate (FDR) for peptide and protein identification was set to 0.01. The data was performed using 2.5 Da precursor and 0.7 Da fragment ion tolerance while the Q-TOF search used 10 ppm. The Andromeda search engine was used for the MS/MS spectra search against the adopted transcriptome, which was translated to protein code. Differential expression ratios for proteins were obtained using Mascot software (Matrix Science, version 2.4), which calculates protein ratios using only ratios from the spectra that are distinct for each protein and excluding the shared peptides of protein isoforms.
Because the P. cuniculi genome was not available, de novo transcriptome data were used for protein identification; that is, we translated the database into the amino acid code. The MS data were analyzed using MaxQuant software version 1.3.0.5. The cutoff of the global false discovery rate for peptide and protein identification was set to 0.01. The Andromeda search engine was used for the MS/MS spectra search against the adopted transcriptome. Differential expression ratios for proteins were obtained using Mascot software.
Enzyme activities
Isolation of the mitochondria
According to the manual of the Mitochondrial Isolation Kit (Solarbio, China), the mitochondria of mites were isolated for further tests. Briefly, 500 mites (P. cuniculi) were collected immediately from infested rabbits and then placed to glass homogenizer, and they were homogenized for 5 min with 600 μL of lysis buffer in ice water. The homogenates were transferred to a centrifuge tube and centrifuged at 1,000 × g at 4 °C for 10 min, and then the supernatant was collected and centrifuged again at 12,000 × g at 4 °C for 10 min. Subsequently, 0.5 mL of wash buffer was added to the sediments and centrifuged again at 1,000 × g at 4 °C for 5 min. Finally, the mitochondrial fractions of mites were collected and stored at −70 °C. The protein concentrations were assayed using the bicinchoninic acid method [29].
Mitochondrial complex I activity of mites treated with eugenol
Firstly, 200 μL of eugenol (50 or 100 μg/mL), which was prepared in 10% DMSO was added to 24-well for the treatment group, and 10% DMSO (200 μL) was added for an untreated control group. Then, a total of 500 adult P. cuniculi were isolated from infested rabbits and were placed in the solution. The well were sealed by parafilm. All plates were incubated at 25 °C and 75% relative humidity. At 8 h and 16 h, 100 μL of eugenol (100 μg/mL) or 10% DMSO was added again to each well. After 24 h of treatment with eugenol, adult P. cuniculi mites were collected and the mitochondria isolated; 10% DMSO alone was applied to an untreated control group. After the isolation of 50 μg of mitochondria according to the above method, the mitochondria (2 μg) were used to evaluate the complex I activities of mites in each group using a Mitochondrial Activity Assay Kit (Solarbio, China). The absorbance of samples were determined using a Multiskan Go Microplate Spectrophotometer (Thermo Scientific., U.S. A) at 340 nm at room temperature for 2 min. The ΔA value measured the decrease in absorbance from 1 min to 2 min, and complex I activity (nmol/min/mg prot) was calculated to evaluate the effect of eugenol on mitochondrial complex I in mites. All treatments were replicated three times [30].
Effect of eugenol on the complex I activity of mitochondria isolated from mites
The inhibition of complex I activity in the electron transport chain was measured using a Mitochondrial Complex I Activity Colorimetric Assay Kit (Biovision, U.S.A.) following the manufacturer's protocol. Eugenol at final concentrations of 2, 10 and 20 ng/mL was used as an inhibitor in this test, rotenone (2 ng/mL) was used as a positive control, and 10% DMSO was used as a negative control. The activity was determined by measuring the decrease in absorbance/min at room temperature at 600 nm with an enzyme-linked immunosorbent assay microplate reader (Multiskan MK3, Thermo Scientific, U.S.A.). Absorbance was recorded every 1 min for 5 min. Three replicates were performed for each group.
To study the effect of succinate on the inhibition of complex I by eugenol, a polarographic study was performed. Eugenol at a final concentration of 20 ng/mL was used as an inhibitor in this test, succinate (2–20 ng/mL) was added independently or concurrently with eugenol, and 10% DMSO was used as a negative control. The activity was determined by measuring the decrease in absorbance in OD/min at room temperature at 600 nm. Absorbance was recorded for 5 min, and inhibition rates of chemicals against complex I activity were calculated. Three replicates were performed for each group.
RNA interference
Design and synthesis of dsRNA
Double stranded RNA (dsRNA) were synthesized by Yangling Tianaorunke Bio. (China) according to the sequence of the target gene NADH dehydrogenase subunit 2 (Psoroptes cuniculi) (NCBI NC_024675.1) (MTDN2). The forward primer was 5′-TAATACGACTCACTATAGGGTCCTCCCTACTCTCCTTCATAATC-3′, and the reverse primer was 5′-TAATACGACTCACTATAGGGAGGGAAGGTACACCATAGGT AG-3′. dsRNA was quantified using Nano-drop spectrophotometer (Thermo Fischer Scientific, Waltham, USA). Finally, the dsRNA was diluted in normal saline (Solarbio, China) to a final concentration of 500 nM and stored at −20˚C.
MTDN2 gene knockdown assays
P. cuniculi was isolated from the ear cerumen of naturally infested rabbits. After washing mites with distilled water two times, they were placed in 24-cell culture plates and incubated with 500 nM MTDN2-dsRNA (500 μL) as well as advanced transfection reagent (ZETA, U.S.A) at 25 ± 1 °C under 75% relative humidity for 48 h, and 500 μL of distilled water was added to control group.
Gene expression quantification by RT-qPCR
After washing dsRNA treated- mites and control with PBS two times, and the total RNA was extracted using total RNA extraction kit (Qiagen, Germany). After assaying the concentrations and purity, RNA were reverse transcribed into cDNAs according to the protocol of RT-PCR kit (Qiagen, Germany). PCR primers of MTDN2 gene for the analysis were forward: 5‘-TCCTCCCTACTCTCCTTCATAATC-3‘, reverse: 5‘-AGGGAAGGTACACCATAGGTAG-3‘.
Acaricidal activity in vitro
Mites of dsRNA treated group and control group were collected and placed in Petri dishes, and then 100 μg/mL eugenol were added into 24-cell culture plates, and the liquid excess was absorbed with filter paper. As well as above described, 50 μL eugenol were added again at 8 h and 16 h, respectively. For the next 24 h, the viability of the mites was checked, and the mites were recorded as dead if the body and appendages did not move under a microscope. Five replicates were performed [20].
Cytotoxicity test against PC12 and C6/36 cells in vitro
PC12 cells purchased from Procell Life Sci & Tech. Co. Ltd. (Wuhan, China) were grown in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented at 37 °C in a humidified CO2 (5%) incubator. Aedes albopictus clone C6/36 cells purchased from the China Center for Type Culture Collection (Wuhan, China) were grown in MEM (minimal essential medium) supplemented at 28 °C in a humidified CO2 (5%) incubator. PC12 cells (5 × 104) and C6/36 cells (1 × 105) were seeded in 96-well plates and cultivated for 24 h, and then a CCK-8 test was carried out to evaluate the cytotoxicity [31].
Effects of eugenol on MMP of PC12 and C6/36 cells
Briefly, PC12 cells (5 × 104) and C6/36 cells (1 × 105) were seeded in 96-well plates and cultivated for 24 h, respectively. Then, 50 μL of eugenol (0–100 μg/mL) was added to the cells and incubated for 36 h, and the cells were harvested, washed and incubated with Rhodamine 123 (2 μM) (Beyotime, China) in PBS for 60 min at 37 °C in darkness and washed with PBS. 1% DMSO was applied to an untreated group as control. The fluorescence was measured with a fluorescence spectrophotometer using 507 nm excitation and 529 nm emission filter settings [32]. The inhibition rate of eugenol on MMP was calculated by using the formula (1):
| (1) |
where ODc represents the optical density of the control and ODe represents the optical density of eugenol.
Mitochondrial complex I activity of PC12 and C6/36 cells treated with eugenol
In this test, PC12 cells (5 × 104/mL) and C6/36 cells (1 × 105/mL) were seeded in 6-well plates for 2 mL each well and cultivated for 24 h, and then eugenol was added to the medium to the final concentrations of 25 and 50 μg/mL for the incubation. Rotenone (50 μg/mL) was used as control. Each group was set to 6 multiple well. After treatment for 48 h, PC12 cells and C6/36 cells were collected. Then, the cells were harvested and combined to isolate the mitochondria; 1% DMSO was applied to an untreated group. According to the above methods, mitochondria (protein concentration 0.5 mg/mL) were isolated from PC12 and C6/36 cells, and 1 μg of mitochondria was used to determine the respective complex I activity using a Mitochondrial Complex I Activity Colorimetric Assay Kit (Biovision, U.S.A.). Five replicates were performed.
Safety evaluation in vivo
Wistar rats (180–220 g) were randomly divided into two eugenol treatment groups (5 rats/group), a positive control group (rotenone, 5 rats), and a negative control group (3 rats). Eugenol was dissolved in distilled water and injected intraperitoneally (i.p.) or subcutaneously (i.h.) for 35 consecutive days at doses of 2.5 mg/kg, respectively, and the negative control group was orally administered distilled water. The positive control group was administered rotenone by intraperitoneal injection (2.5 mg/kg). Food and water were available ad libitum throughout the experiment. The behavioral changes, clinical signs of toxicity and mortality were observed and recorded, and the body weights of each animal were measured at 7, 14, 21, 28, and 35 days. At the end of the treatment, the animals were fasted overnight, and then they were anesthetized by intraperitoneal injection 10% chloral hydrate (3 mL/kg body weight), and blood samples were collected. After euthanasia with the over-dose 10% chloral hydrate, target organs were collected for subsequent analyses.
Hematological and biochemical parameters were assayed by an automatic hematology analyzer (BC-2800Vet, Mindray Co., China) and an automatic biochemistry analyzer (BS-420, Mindray Co., China). The brains and livers were collected, weighed, fixed in 10% buffered formalin and subjected to a routine histological process for paraffin embedding and light-microscopic examination. The brains were cut coronally using a frozen sliding microtome, and the sections were incubated with H2O2 (10%) for 30 min and then blocked with 10% normal goat serum for 1 h at room temperature. After that, sections were incubated in primary antibody for 24 h at 4 °C. Antibody detection was carried out with a Histostain-Plus Bulk Kit (Invitrogen) against rabbit IgG, and 3,3′diaminobenzidine was used to visualize the final product. All sections were photographed with a BA200 Digital camera (Motic Electric Group Co., Ltd) [33].
Results and discussion
RNA-Seq and SPR analysis of mites treated with eugenol
Importantly, mass spectrometry (MS) assays provide an unprecedented opportunity for integrating SPR and omics to reveal the potential targets and mechanisms of action [34]. To exploit the possible targets, RNA-Seq and SPR analysis were used. Expression profiles of genes with or without exposure to eugenol were mapped to provide valuable information on the functions of eugenol in parasites.
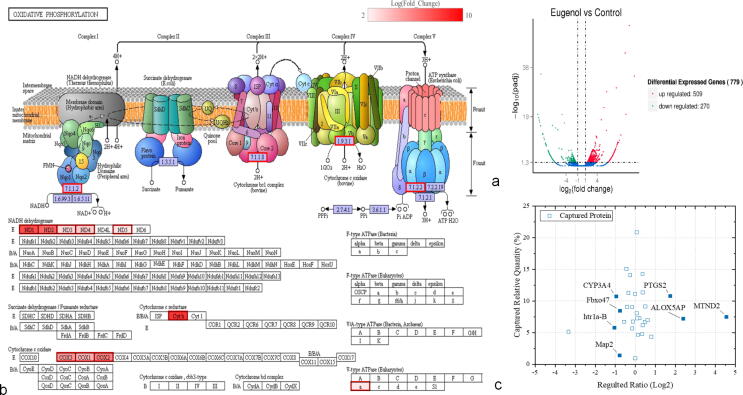
We used RNA-Seq analysis to identify 779 genes that were significantly regulated in response to eugenol exposure, including 270 downregulated genes and 509 upregulated genes (Fig. S1). The differentially expressed genes were enriched in 228 pathways, and 27 pathways were significantly enriched (P < 0.05), mainly associated with the ribosome (60 genes) and oxidative phosphorylation (28 genes) (Fig. S2). In the latter pathway, most of the significantly regulated genes were related to mitochondrial structure and function. Moreover, 12 genes encoding subunits of complex I (MTND1, MTND2, MTND3, MTND4, and MTND5), complex III (CYB), complex IV (MTCO1, MTCO2 and MTCO3), and complex V (ATP6V0C, ATP6V1F and TCIRG1) were upregulated by 2.07 ~ 8.42-fold by eugenol (Fig. 1a, b). This result is not consistent to the previous report [13]. Ma et al. found that to regulated pathways, the main up-regulated pathway was PPAR signaling pathway, and the down-regulated were associated with NF-κB and some inflammatory pathways [13]. However, authors did not list the more detailed information about the regulated genes, and if the differentially regulated genes were associated with oxidative phosphorylation? In this test, we found that after the treatment of eugenol, most of genes associated with oxidative phosphorylation were regulated, and the potential mechanism of action may be related to the mitochondria, not PPAR or NF-κB.
Fig. 1.
Integration of transcriptomics and surface plasmon resonance analysis for target discovery. (a: The number of regulated genes after treatment with eugenol. b. The oxidative phosphorylation pathway is enriched with upregulated genes after treatment with eugenol. c. SPR combined with RNA-Seq analysis).
An SPR assay was employed to investigate the affinity and interactive effect between eugenol and the body proteins of mites. After searching the proteins database and de novo transcriptome data, 239 proteins captured by eugenol were identified by LC-MS/MS (Table S1). Among these proteins, the binding socres of sar s11 allergen, paramyosin-like protein 2, actin, sar s11 allergen with eugenol were>1000, this result showed that eugenol has the higher affinity with these proteins than others. The affinity between proteins and compund is one of the factor for performing the activity, however, the relationship between affinity and activity is not direct and absolute. In this paper, to uncover the potential mechanism of action of eugenol against mites, more specific binding proteins (27) with binding scores over 200 were selected for the following experiments.
Integration of RNA-Seq analysis and SPR-MS
To validate and identify novel mechanisms of action of eugenol, the above RNA-Seq data and SPR-MS information (27 proteins) were compared and combined, and one promising protein was identified: NADH:ubiquinone oxidoreductase chain 2 (MTND2) (Table S2) (Fig. 1c). The SPR-MS results showed that this protein was bound to eugenol by affinity capture, and RNA-Seq sequencing showed that it was significantly upregulated at the mRNA level (log2 ratio > 4.56, E-value < 0.01) after treatment with eugenol. Hence, we hypothesized that as a part of NADH:ubiquinone oxidoreductase or mitochondrial respiratory complex I, MTND2 is the main potential target of eugenol. Usta et al. also reported that eugenol targeted respiratory chain complexes and interfered with mitochondrial function and subparticles in isolated rat liver and HepG2 cells [16], [34], and the result is consistent to our work. However, we, for the first time, thought that eugenol may be targeted to MTND2. The inhibition of complex I will destroy the gradient across the membrane and decrease the ATP level, resulting in impaired mitochondrial function and leading to cell death [35], [36].
Direct validation and enzyme activity
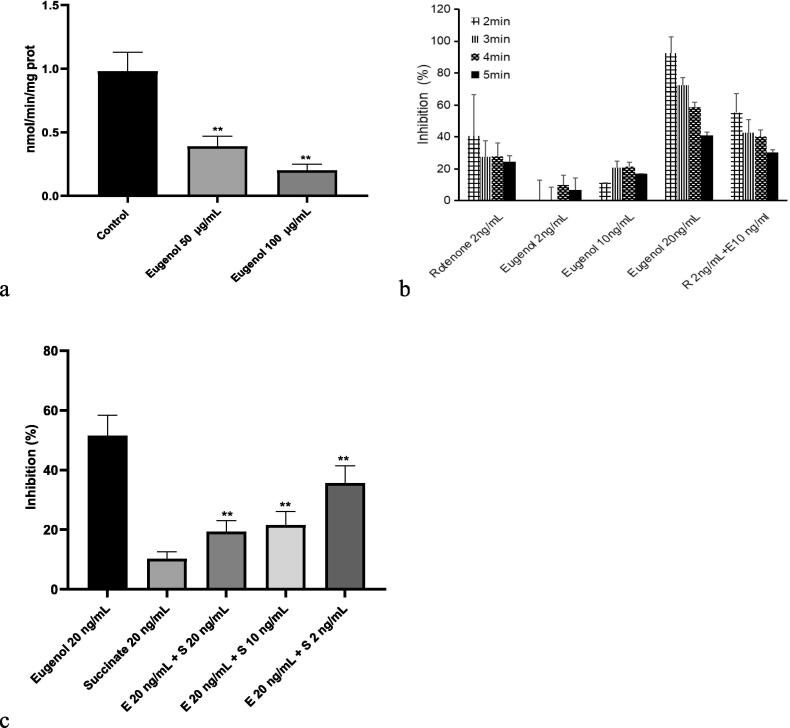
NADH dehydrogenase is the complex one of the respiratory chain (complex I), and is identified as an electron carrier in the electron transport chain in the mitochondrial membrane. This complex oxidizes NADH from the tricarboxylic acid cycle and by β-oxidation and transports protons across the inner membrane to generate proton-motive force [37], [38]. We therefore examined the inhibitory effect of eugenol on mitochondrial respiratory complex I activity. Because the mortality rates were 71.67% for 100 μg/mL and 26.67% for 50.00 μg/mL, we chose 50 and 100 μg/mL as the concentrations of eugenol in this assay. Eugenol addition resulted in a significant, concentration-dependent decrease in complex I activity in the mitochondrial respiratory chain in parasites (P < 0.05), and the inhibition rates were 37.89% for 50 μg/mL and 60.26% for 100 μg/mL (Fig. 2a). There is a positive relationship between the acaricidal activity of eugenol and its inhibitory effect on complex I activity. The inhibitory effect of eugenol on complex I in isolated rat liver mitochondria was also reported by Usta et al. [35].
Fig. 2.
Mitochondrial complex I activities of mites after treatment with eugenol at different concentrations. (a. The complex I activity of mitochondria isolated from treated mites as residual activity. b. The complex I activity of mitochondria isolated and then treated with eugenol. c. The complex I activity of mitochondria isolated and then treated with eugenol and/or succinate). Asterisks indicate significant differences from the control for a, n = 3, average ± SD, P < 0.05, ANOVA with Tukey’s HSD test; compared with the positive control at the same time point for b, n = 3, average ± SD, P < 0.05, independent samples t test; compared with eugenol for c, n = 3, average ± SD, P < 0.05, ANOVA with Tukey’s HSD test.
After a large number of pre-experiments, the concentrations of eugenol in the in vitro assay were set to 2, 10 and 20 ng/mL. Further experiments also demonstrated that eugenol (at 2, 10 and 20 ng/mL) inhibited complex I activity in isolated mitochondria in vitro and blocked the oxidation reaction from NADH to NAD+. At 2 min, eugenol (20 ng/mL) showed a significant inhibitory effect on complex I activity, with an inhibition rate of 92.59% (P < 0.05). Interestingly, the inhibitory effect was decreased to 40.88% at 5 min (Fig. 2b). Hence, we hypothesized that eugenol may bind the complex I with noncovalent link and then inhibited its activity. Due to the volatileness, with the extension of treatment time, the concentration of eugenol was decreased, and the complex I was released. This hypothesis also explained the reason why the different tests existed the significant different acaricidal activity [12], [13], [14]. As Chen et al. [12] claimed, the inconsistency of the results would be attributed to the different experimental design. In our assay, mites were placed into dishes to incubate with eugenol. When the test was started, dishes would be sealed by parafilm. In addition, eugenol would be added into dishes again at 8 h and 16 h, respectively. Above method used in the acaricidal test would prevent the volatilization of eugenol and keep the constant concentration. LC50 of eugenol and other volatile substances against mites become to low compared to other regular test [12], [13]. Meanwhile, the drug resistance of mites obtained from the infested rabbits also may contribute to the different acaricidal activity of eugenol against mites, the different standards for evaluating the death of mites under a microscope also should be noticed.
After adding both rotenone and eugenol, the inhibitory effect on complex I activity was significantly enhanced compared with that of rotenone or eugenol alone; however, there was no synergistic activity (Fig. 2b). Moreover, we found that after adding different concentrations of succinate (2–20 ng/mL), the inhibitory effect of eugenol (20 ng/mL) against complex I activity was significantly decreased in a concentration-dependent manner (P < 0.05) (Fig. 2c). For this reason, we hypothesized that complex I is a potential target of the acaricidal activity of eugenol in which the compound binds to MTND2 of complex I and results in death by inhibiting the activity of this enzyme.
RNA interference
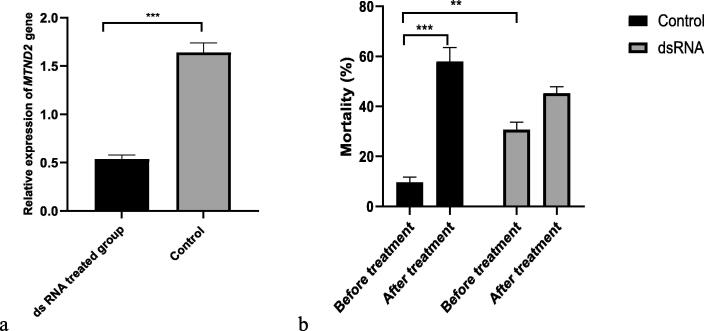
Compared to control group, the mRNA transcript levels of mites immersed in dsRNA which is targeted to MTND2, were significant reduced (P < 0.0001), and the mean reduction in transcript levels of gene relative to control was 67.21% (Fig. 3a). Meanwhile, we also noticed that after the treatment with dsRNA for 48 h, the mortality of mites was up to 30.67% (P < 0.01), and the control was 10.00%, dsRNA may affect the vitality of mites. However, when all mites were treated wth eugenol again for 24 h, the mortality of control group were remarkable increased from 10.00% to 58.00% (P < 0.001), and the mortality of dsRNA treated mites with silence gene have not significant raised (P > 0.05) (Fig. 3b). These results indicated that complex I is a potential target of the acaricidal activity of eugenol and results in death by inhibiting the activity of this enzyme.
Fig. 3.
RNA interference (a. the mRNA of MTND2 expression of ds RNA treated mites and control; b. the change of mite‘s mortality between before treatment and after treatment with eugenol at the concentration of 100 μg/mL) Asterisks indicate significant differences compared to control n = 3, average ± SD, P < 0.05, independent samples t test.
Inhibitory effects of eugenol against mitochondrial complex I in insects and mammals
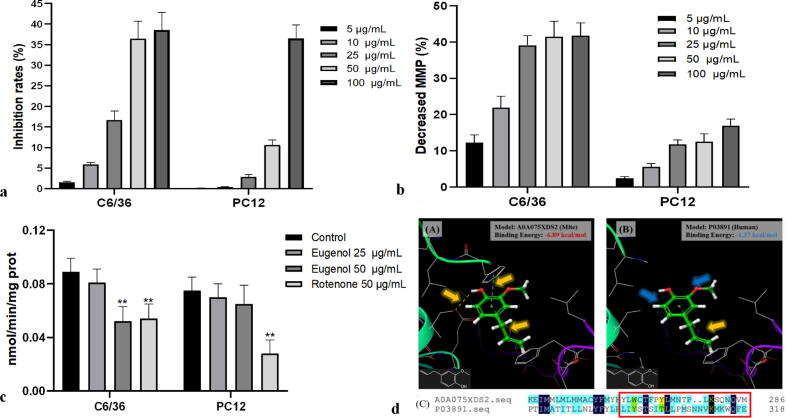
Regarding species specificity, insect and fish mitochondria are particularly sensitive to complex I inhibition [37]. To investigate the difference in eugenol sensitivity between insects and mammals and its possible mechanism of action, we compared eugenol’s cytotoxicity and the effects on MMP and complex I activity in Aedes albopictus clone C6/36 cells and rat PC12 cells. C6/36 cells, as an insect cell line, were used to study the mode of action for the insecticidal and antiparasitic activities of compounds. Due to the lack of mite cells in the test, this cell line was applied to study the inhibitory effects of eugenol against mitochondrial complex I of insects (mites), and a rat pheochromocytoma tumor cell line (PC12), as a mammalian animal cell line, was used to study the inhibitory effects against mitochondrial complex I of mammals and to investigate the potential neurotoxicity of eugenol for the subsequent safety evaluation. The results showed that eugenol was moderately toxic to C6/36 cells, with a dose-dependent effect (0–100 µg/mL) (Fig. 4a). Along with the increase in cytotoxicity of eugenol, MMP in C6/36 cells was significantly decreased (Fig. 4b), and there was a positive relationship between cytotoxicity and the decrease in MMP (Fig. 4a,b). A decrease in membrane potential would lead to cell death. However, as shown in Fig. 4a,b, although the toxicity to PC12 cells was markedly increased along with the increasing concentrations of eugenol, MMP in cultured PC12 cells was not significantly decreased, especially at the high concentration (100 µg/mL). Eugenol did not decrease the MMP of mammalian cells or lead to the death of PC12 cells. Then, we examined whether eugenol inhibited complex I activity in C6/36 and PC12 cells to further investigate the potential mechanism underlying the difference in the effects on MMP and thus the differences in sensitivity between insects and mammals. Compared with the control, treatment with eugenol (10–50 μg/mL) did not decrease the complex I activity in PC12 cells, but in C6/36 cells, the complex I activity was significantly inhibited by eugenol. The positive control agent rotenone presented an inhibitory effect on complex I in both PC12 and C6/36 cells, with inhibition rates of 43.75% and 64.91% at a concentration of 50 μg/mL (Fig. 4c). These results indicated that eugenol could damage the MMP by inhibiting complex I activity in insect cells, resulting in the death of insects.
Fig. 4.
The cytotoxicity and effects of eugenol on complex I and mitochondrial membrane potential in PC12 and C6/36 cells. (a. The cytotoxicity of eugenol to PC12 and C6/36 cells. b: The effect of eugenol on the MMP in PC12 and C6/36 cells. c. Inhibitory effect of eugenol on complex I in PC12 and C6/36 cells. d. Molecular docking assay for determining the affinity of eugenol to A0A075XDS2 (mite) and P03891 peptides (human)). Asterisks indicate significant differences in the inhibition rates of cytotoxicity for a and from PC12 cells for c; n = 3, average ± SD, P < 0.05, independent samples t test.
Subsequently, we investigated the reason the inhibitory effects on complex I differed between insects and mammals by comparing the different binding sequences of eugenol between insects and mammals and their affinities. Considering that the binding peptide in the parasite was NADH:ubiquinone oxidoreductase chain 2 of complex I, we searched UniProt (https://www.uniprot.org) for similar sequences in humans, and P03891 (human: NADH:ubiquinone oxidoreductase chain 2) was identified (Table S3). However, compared with 12 feasible binding sites on mite peptides, we could not find a binding site for eugenol on this peptide when we performed a molecular docking assay (Fig. 4d). These findings suggest that eugenol can bind to MTND2 of complex I in insects and parasites, thereby inhibiting complex I activity. However, in mammals, due to the different complex I sequences, eugenol could not bind a specific peptide to inhibit it. The different affinity of eugenol to complex I results in the difference in sensitivity to eugenol between insects and mammals.
Safety evaluation
Parkinson’s disease (PD) is the most common neurodegenerative movement disorder [38], [39], [40]. Although the cause of PD is unknown, some studies indicated pesticides and other environmental toxins would induce a systemic defect in mitochondrial complex I [39]. These toxins include rotenone, a high-affinity and specific inhibitor of complex I. However, rotenone was withdrawn from the market in many countries due to its toxicity [41]. Long-term exposure of rats to rotenone results in PD, and rats subjected to this effect have been developed as a model system. Therefore, the safety of acaricide or insecticide compounds is as important as their efficacy.
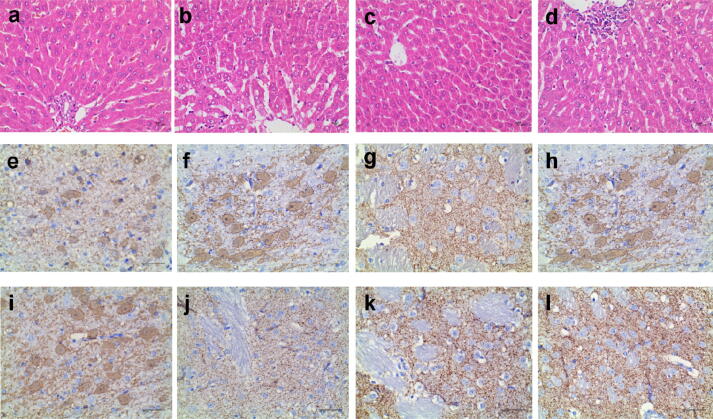
Considering that eugenol is a potential inhibitor of respiratory chain complex I, we therefore used a rat model of PD to determine whether complex I inhibition by eugenol causes mitochondrial dysfunction. We studied the neurological impact of eugenol with methods similar to those used for rotenone. The dose of 2.5 mg/kg was chosen to evaluate eugenol and the positive control rotenone because this dose is commonly used to generate the experimental PD model with rotenone [42], [43], [44], [45]. Eugenol was administered (i.p. and i.h., 2.5 mg/kg) to rats for 5 weeks to evaluate changes in nigral and striatal sections and blood biochemistry. The behaviors and body weight of rats treated with eugenol showed no significant change (Table S4). In addition, during the experimental period, no mortality or abnormal clinical signs were observed in rats subjected to eugenol treatment. Some hematological and serum biochemical parameters were slightly changed in the eugenol group vs. the control group, such as a reduction in white blood cells and increases in lymphocytes, red blood cells, platelets, alkaline phosphatase and lactate dehydrogenase. However, these differences did not reach statistical significance (Table S5, S6). Fig. 5a-d present microphotographs of livers in the eugenol-treated groups and control group, respectively. Minor histopathological changes in the liver were found in all three groups, such as liver cell degeneration and small focal necrosis, but these changes were caused by other faults (e.g. the fasted time is not enough), not long-term eugenol exposure and metabolism. Additionally, the complex I inhibitor rotenone (2.5 mg/kg, i.p.) caused loss of nigral and other neurological disabilities in rats, leading to the death of all rats in the test, but eugenol did not degenerate TH-immunoreactive neurons in the striatum of rats and therefore did not cause PD or related symptoms (Fig. 5 e-i) (Table S6). These exciting data suggest that eugenol has better safety than rotenone at the given concentrations and may serve as an acaricidal lead for future drug discovery efforts.
Fig. 5.
Microphotographs of liver and tyrosine hydroxylase immunoreactivity in the substantia nigra and striatum of eugenol-treated rats. (a. Microphotographs of liver at 400 × in the eugenol (2.5 mg/kg, i.p.)-treated group, b. control group; c. Substantia nigra (IHC × 400), eugenol (2.5 mg/kg, i.p.)-treated group, d. control group; e. Substantia nigra (IHC × 400), eugenol (2.5 mg/kg, i.h.)-treated group, f. control group; g: Microphotographs of liver at 400 × in the eugenol (2.5 mg/kg, i.h.)-treated group, h. control group; i. Striatum (IHC × 400), eugenol (2.5 mg/kg, i.p.)-treated group, j. control group; k. Striatum (IHC × 400), eugenol (2.5 mg/kg, i.h.)-treated group, l. control group).
Conclusion
This study shows that eugenol targets complex I of the mitochondrial respiratory chain. Future studies will focus on finding safer and more sustainable alternative agents by designing and synthesizing a series of eugenol derivatives and understanding the mechanisms of action of these compounds. In addition, certain chemical drawbacks of eugenol should also be improved, such as its volatile properties. This study lays an important foundation for the future development of eugenol as a relatively safe and sustainable alternative acaricidal agent.
Data deposition
The data reported in this paper have been deposited in the Sequence Read Archive database, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA531224).
CRediT authorship contribution statement
Xiao-Fei Shang: Conceptualization, Funding acquisition, Writing - original draft, Writing - review & editing. Li-Xia Dai: Data curation, Methodology. Chen-Jie Yang: Formal analysis, Methodology. Xiao Guo: Data curation, Methodology. Ying-Qian Liu: Conceptualization, Funding acquisition, Supervision, Writing - review & editing. Xiao-Lou Miao: Data curation, Investigation. Ji-Yu Zhang: Conceptualization, Funding acquisition, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported financially by the National Natural Science Foundation of China (31302136, 31772790, 31371975, 21672092), the National Key Research and Development Program of China (2017YFD0201404), and the Central Public-interest Scientific Institution Basal Research Fund (Y2019XK14).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.12.010.
Contributor Information
Ying-Qian Liu, Email: yqliu@lzu.edu.cn.
Ji-Yu Zhang, Email: shangxf928@126.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Anthony J.P., Fyfe L., Smith H. Plant active components - a resource for antiparasitic agents? Trends Parasitol. 2005;21:462–468. doi: 10.1016/j.pt.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Sun G.S., Xu X., Jin S.H., Lin L., Zhang J.J. Ovicidal and insecticidal activities of pyriproxyfen derivatives with an oxime ester group. Molecules. 2017;22 doi: 10.3390/molecules22060958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chermenskaya T.D., Stepanycheva E.A., Shchenikova A.V., Chakaeva A.S. Insecto acaricidal and deterrent activities of extracts of Kyrgyzstan plants against three agricultural pests. Ind. Crop. Prod. 2010;32:157–163. doi: 10.1016/j.indcrop.2010.04.009. [DOI] [Google Scholar]
- 4.Zanuncio J.C., Mourão S.A., Martínez L.C., Wilcken C.F., Ramalho F.S., Plata-Rueda A., et al. Toxic effects of the neem oil (Azadirachta indica) formulation on the stink bug predator, Podisus nigrispinus (Heteroptera: Pentatomidae) Sci. Rep. 2016;6:30261. doi: 10.1038/srep30261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prakash P., Gupta N. Therapeutic uses of Ocimum sanctum Linn (tulsi) with a note on eugenol and its pharmacological actions: a short review. Indian J. Phys. Pharmacol. 2005;49:125–131. [PubMed] [Google Scholar]
- 6.Nagababu E., Rifkind J.M., Boindala S., Nakka L. In: Free Radicals and Antioxidant Protocols. Methods in Molecular Biology (Methods and Protocols) Uppu R., Murthy S., Pryor W., Parinandi N., editors. Humana Press; U.S.A: 2010. Assessment of antioxidant activity of eugenol In vitro and In vivo; pp. 165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mastelić J., Jerković I., Blažević I., Poljak-Blaži M., Borović S., Ivančić-Baće I., et al. Comparative study on the antioxidant and biological activities of carvacrol, thymol, and eugenol derivatives. J. Agric. Food Chem. 2008;5:3989–3996. doi: 10.1021/jf073272v. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Wang Y., Zhu X., Cao P., Wei S., Lu Y. Antibacterial and antibiofilm activities of eugenol from essential oil of Syzygium aromaticum (L.) Merr. & L.M.Perry (clove) leaf against periodontal pathogen Porphyromonas gingivalis. Microb. Pathog. 2017;113:396–402. doi: 10.1016/j.micpath.2017.10.054. [DOI] [PubMed] [Google Scholar]
- 9.Benencia F., Courreges M.C. In vitro and in vivo activity of eugenol on human herpesvirus. Phytother. Res. 2000;14:495–500. doi: 10.1002/1099-1573(200011)14:7<495::AID-PTR650>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 10.Pisano M., Pagnan G., Loi M., Mura M.E., Tilocca M.G., Palmieri G., et al. Antiproliferative and pro-apoptotic activity of eugenol-related biphenyls on malignant melanoma cells. Mol. Cancer. 2007;6:8. doi: 10.1186/1476-4598-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Morais S.M., Vila-Nova N.S., Bevilaqua C.M., Rondon F.C., Lobo C.H., de Alencar Araripe Noronha Moura A., et al. Thymol and eugenol derivatives as potential antileishmanial agents. Bioorg. Med. Chem. 2014;22:6250–6255. doi: 10.1016/j.bmc.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z., van Mol W., Vanhecke M., Duchateau L., Claerebout E. Acaricidal activity of plant-derived essential oil components against Psoroptes ovis in vitro and in vivo. Parasit. Vect. 2019;12:425. doi: 10.1186/s13071-019-3654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma W., Fan Y., Liu Z., Hao Y., Mou Y., Liu Y., et al. The acaricidal activity and mechanism of eugenol on Psoroptes cuniculi. Vet. Parasitol. 2019;266:56–62. doi: 10.1016/j.vetpar.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Shang X.F., Dai L.X., Liu Y.Q., Zhao Z.M., Li J.C., Yang G.Z., et al. Acaricidal activity and enzyme inhibitory activity of active compounds of essential oils against Psoroptes cuniculi. Vet. Parasitol. 2019;267:54–59. doi: 10.1016/j.vetpar.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Knio K.M., Usta J., Dagher S., Zournajian H., Kreydiyyeh S. Larvicidal activity of essential oils extracted from commonly used herbs in Lebanon against the seaside mosquito. Ochlerotatus caspius, Bioresource Technol. 2008;99:763–768. doi: 10.1016/j.biortech.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 16.Usta J., Knio K., Dagher S., Barnabe P., Bou-Mouglabay Y., Kreydiyyeh S. Coriander sativum and its main oil ingredients: in vitro effect on hepG2 cells and mitochondrial respiratory chain complexes. Toxicol. Lett. 2005;15:118–119. [Google Scholar]
- 17.Ahuja N., Batish D.R., Singh H.P., Kohli R.K. Herbicidal activity of eugenol towards some grassy and broad-leaved weeds. J. Pest. Sci. 2013;88:209–218. doi: 10.1007/s10340-014-0570-x. [DOI] [Google Scholar]
- 18.Jing C., Gou J., Han X., Wu Q., Zhang C. In vitro and in vivo activities of eugenol against tobacco black shank caused by Phytophthora nicotianae. Pest. Bioch. Physiol. 2017;142:148–154. doi: 10.1016/j.pestbp.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Moreira L.C.B. Nematocidal effect of eugenol on tomato plants. Pesquisa Agropecuária Trop. 2013;43:286–291. [Google Scholar]
- 20.Shang X., Wang Y., Zhou X., Guo X., Dong S., Wang D., et al. Acaricidal activity of oregano oil and its major component, carvacrol, thymol and p-cymene against Psoroptes cuniculi in vitro and in vivo. Vet. Parasitol. 2016;226:93–96. doi: 10.1016/j.vetpar.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Guo X., Shang X., Li B., Zhou X.Z., Wen H., Zhang J. Acaricidal activities of the essential oil from Rhododendron nivale Hook. f. and its main compound, δ-cadinene against Psoroptes cuniculi. Vet. Parasitol. 2017;23:51–54. doi: 10.1016/j.vetpar.2017.01.028. [DOI] [PubMed] [Google Scholar]
- 22.Mao W., Schuler M.A., Berenbaum M.R. Disruption of quercetin metabolism by fungicide affects energy production in honey bees (Apis mellifera) Proc. Natl. Acad. Sci. U.S.A. 2017;114:2538–2543. doi: 10.1073/pnas.1614864114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piotrowski J.S., Okada H., Lu F., Li S.C., Hinchman L., Ranjan A., et al. Plant-derived antifungal agent poacic acid targets β-1,3-glucan. Proc. Natl. Acad. Sci. U.S.A. 2015;112:e1490–e1497. doi: 10.1073/pnas.1410400112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lomenick B., Olsen R.W., Huang J. Identification of direct protein targets of small molecules. ACS Chem. Biol. 2011;6:34–46. doi: 10.1021/cb100294v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maya A.F., Eric D.B. Strategies for target identification of antimicrobial natural products. Nat. Prod. Rep. 2016;33:668. doi: 10.1039/C5NP00127G. [DOI] [PubMed] [Google Scholar]
- 26.Song X., Chen Z., Jia R., Cao M., Zou Y., Li L., et al. Transcriptomics and proteomic studies reveal acaricidal mechanism of octadecanoic acid-3, 4-tetrahydrofuran diester against Sarcoptes scabiei var. cuniculi. Sci. Rep. 2017;7:45479. doi: 10.1038/srep45479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2011;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao S., Song Q., Liu J., Zhang X., Ji X., Wang P. E2F1 mediates the downregulation of POLD1 in replicative senescence. Cell. Mol. Life Sci. 2019;76:2833–2850. doi: 10.1007/s00018-019-03070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalghatgi S., Spina C.S., Costello J.C., Liesa M., Morones-Ramirez J.R., Slomovic S., et al. Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in mammalian cells. Sci. Transl. Med. 2013;5:192ra85. doi: 10.1126/scitranslmed.3006055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M., Chen J., Yu X., Xu S., Li D., Zheng Q., et al. Myricetin suppresses the propagation of hepatocellular carcinoma via down-regulating expression of YAP. Cells. 2019;8:358. doi: 10.3390/cells8040358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang M. Crocetin inhibits leukocyte adherence to vascular endothelial cells induced by AGEs. J. Ethnopharmacol. 2006;107:25–31. doi: 10.1016/j.jep.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 33.Erbaş O., Yılmaz M., Taşkıran D. Levetiracetam attenuates rotenone-induced toxicity: A rat model of Parkinson’s disease. Environ. Toxicol. Pharmacol. 2016;42:226–230. doi: 10.1016/j.etap.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Sun Q., Zhou G., Cai Y., Fan Y., Zhu X., Liu Y., et al. Transcriptome analysis of stem development in the tumourous stem mustard Brassica juncea var. tumida Tsen et Lee by RNA sequencing. BMC Plant Biol. 2012;12:53. doi: 10.1186/1471-2229-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Usta J., Kreydiyyeh S., Bajakian K., Nakkash-Chmaisse H. In vitro effect of eugenol and cinnamaldehyde on membrane potential and respiratory chain complexes in isolated rat liver mitochondria. Food Chem. Toxicol. 2002;40:935–940. doi: 10.1016/S0278-6915(02)00071-6. [DOI] [PubMed] [Google Scholar]
- 36.Shigenega M., Hagen T.M., Ames B. Oxidative damage and mitochondrial decay in aging. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esposti M.D. Inhibitors of NADH-ubiquinone reductase: an overview. BBA-Bioenergetics. 1998;1364:222–235. doi: 10.1016/S0005-2728(98)00029-2. [DOI] [PubMed] [Google Scholar]
- 38.Sarkar S., Malovic E., Harishchandra D.S., Ghaisas S., Panicker N., Charli A., et al. Mitochondrial impairment in microglia amplifies NLRP3 inflammasome proinflammatory signaling in cell culture and animal models of Parkinson’s disease, Npj Parkinson‘s. Dis. 2017;3:30. doi: 10.1038/s41531-017-0032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou J., Qu X.D., Li Z.Y., Liu Q., Ma Y.H., He J.J. Salvianolic acid B attenuates toxin-induced neuronal damage via Nrf2-dependent glial cells-mediated protective activity in Parkinson’s disease models. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0101668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pattanasiri T., Taparhudee W., Suppakul P. Acute toxicity and anesthetic effect of clove oil and eugenol on Siamese fighting fish, Betta splendens. Aquacul. Int. 2016;25:1–13. doi: 10.1007/s10499-016-0020-2. [DOI] [Google Scholar]
- 41.WHO Recommended Classification of Pesticides by Hazard. http://www.who.int/ipcs/ publications/pesticides_hazard/en/ (accessed Oct 2, 2013). ning these substances. L 335/ 91.13.12.2008 (2008c). 2013.
- 42.Betarbet R., Sherer T.B., MacKenzie G., Garcia-Osuna M., Panov A.V., Greenamyre J.T. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat. Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 43.Alam M., Schmidt W.J. Rotenone destroys dopaminergic neurons and induces parkinsonian symptoms in rats. Behavioural Brain Res. 2002;136:317–324. doi: 10.1016/S0166-4328(02)00180-8. [DOI] [PubMed] [Google Scholar]
- 44.Yan X., Yuan X., Zhao Q., Zhang Y., Zhang L., Zhang Z. Study on behavioral test and pathological changes in rotenone-induced Parkinson‘s disease rats. J. Taishan Med. Coll. 2019;40:7. [Google Scholar]
- 45.Wen X.X., Zhang N., Dou D.Q. Protective effect of arctigenin on organ damage of rotenone-induced Parkinson rats. J. Shengyang Pharmaceut. Univ. 2017;34:893. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.