Abstract
Unexplained recurrent spontaneous abortion (URSA) is one of the most intractable clinical challenges in reproduction. As a specific type of endogenous non-coding RNA, circular RNAs (circRNAs) have great pre-clinical diagnostic and therapeutic values in diseases. Recently, thousands of circRNAs were detected in human pre-implantation embryos, indicating that circRNAs potentially have important regulatory functions. However, the roles of circRNAs in URSA remain largely unknown. In this study, we elucidated deregulated circRNA expression and distinct competing endogenous RNA (ceRNA) networks by comparing URSA placental villus with that of patients with normal pregnancy using microarrays. We characterized a distinct circRNA, circRNA-0050703, which is downregulated in URSA placental villus (thus we named it circRNA-DURSA). Silencing of circRNA-DURSA results in trophoblast cell apoptosis in vitro. Furthermore, mechanistic dissection revealed that circRNA-DURSA exerts its effects by competitively binding to miR-760, which post-transcriptionally targets HIST1H2BE. Additionally, after circRNA-DURSA silencing in vivo, the numbers of implanted embryos decreased significantly. These results reveal the regulatory roles of circRNA-DURSA in trophoblasts and identified a distinct circRNA-DURSA/miR-760/HIST1H2BE axis as potentially important diagnostic and therapeutic targets for URSA treatment.
Keywords: circRNA, trophoblast, apoptosis, unexplained recurrent spontaneous abortion, miRNA
Graphical abstract
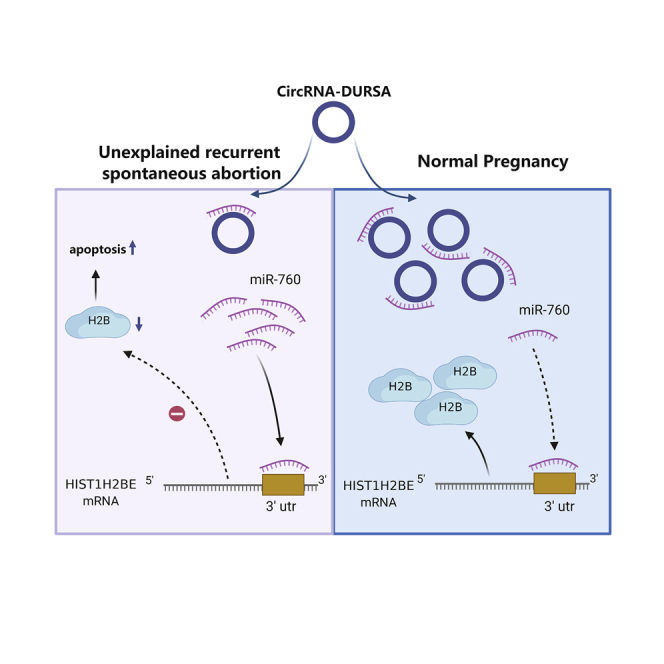
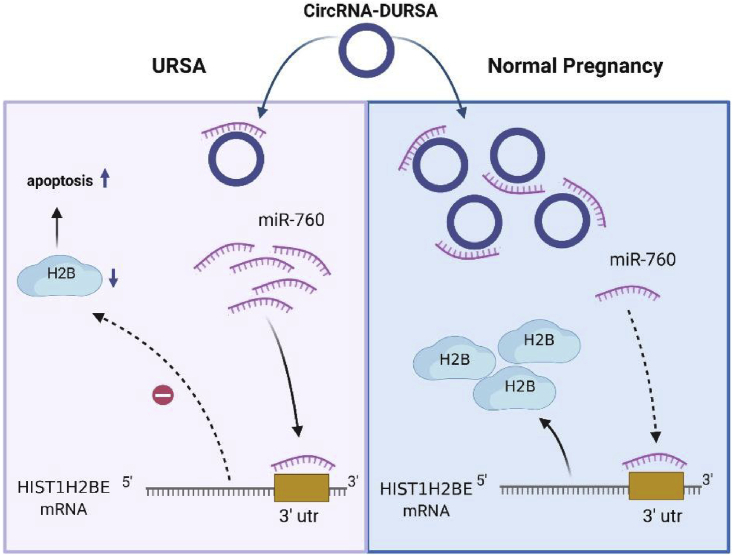
circRNAs have been found to have great pre-clinical diagnostic and therapeutic values in various diseases. In this study, we characterized a novel circRNA, circRNA-DURSA, which is downregulated in URSA placental villus and exerts regulatory roles through the circRNA-DURSA/miR-760/HIST1H2BE axis in trophoblasts. These findings may provide novel mechanistic and clinical insights for future management of URSA.
Introduction
Of all childbearing couples, 1%–5% suffer from great physical and psychological injury caused by recurrent spontaneous abortion (RSA).1 RSA usually refers to two or more failed clinical pregnancies according to RSA guidelines of the American Society for Reproductive Medicine (ASRM) and the European Society for Human Reproduction and Embryology (ESHRE).1,2 The causes of RSA involve genetic factors, maternal anatomy abnormalities, endocrine disorders, autoimmune diseases, environmental factors, and coagulopathy and psychological factors. However, in 30%–40% of RSA cases no cause can be identified.3,4 These cases are termed unexplained RSA (URSA) and lack effective clinical interventions and means of early diagnosis.5 As one of the most intractable clinical challenges in reproduction, URSA is a bewildering threat to childbearing couples. Therefore, determining the underlying pathogenesis of URSA is required for the development of effective treatments for patients with URSA.
There are many mechanisms involved in URSA, including embryo implantation and placentation, which are principal but complicated processes of pregnancy establishment. Trophoblasts play an integral role during implantation and placental development, and increased trophoblast apoptosis causes placenta dysregulation, which leads to URSA.6,7 The regulation of trophoblast apoptosis is partially understood at the transcriptional level,8 but little is known about its regulation at the post-transcriptional level. Therefore, a comprehensive understanding of the post-transcriptional mechanisms underlying trophoblast apoptosis is essential for improving URSA diagnosis and treatment.
Non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), are important regulators of gene expression, including post-transcriptional regulation, in various diseases.9, 10, 11, 12 Overwhelming evidence shows that miRNA alterations are closely related to various disease processes by binding to the 3′ untranslated regions (UTRs) of target mRNAs.13 In the placenta, at least 500 different miRNAs have been identified.14 These miRNAs are involved in placental formation and pregnancy maintenance.15,16 As a recently highlighted type of endogenous ncRNA, circRNAs are stable because they form a continuous covalently closed loop without a 5′ cap structure or 3′ poly(A) tail. circRNAs are involved in various diseases,17 including liver cancer,18 breast cancer,19 and cardiovascular diseases.20 circRNAs have tissue-specific and developmental stage-specific expression patterns, and they are found to distinctly participate in multiple cellular biological and pathological processes, including apoptosis, proliferation, angiogenesis, differentiation, and invasion.21, 22, 23, 24 In particular, researchers have identified more than 10,000 circRNAs in human pre-implantation embryos, with the proportion gradually increasing to 25% from the eight-cell stage to the morula stage, suggesting that they perform specific roles in human pre-implantation development.25 Recently, circRNAs have been shown to be involved in pathological pregnancies, such as in pre-eclampsia26 and repeated implantation failure,27 through regulating trophoblasts or endometrial functions. circRNA alterations may be a part of the dynamic change that occurs during human embryonic and placental development, and disruption of this function may be extensively involved in URSA.
In this study, circRNAs expression patterns were identified and their molecular biological features were characterized for the first time in the human URSA placental villus. We demonstrate that circRNA-0050703 is aberrantly downregulated in URSA placental villus (so we named it circRNA-DURSA), which subsequently induces trophoblast cell apoptosis. Furthermore, we identified that the circRNA-DURSA/miR-760/HIST1H2BE axis plays an important role in regulating trophoblast cell apoptosis. These findings may provide novel mechanistic and clinical insights and provide promising therapeutic targets for future management of URSA.
Results
Differential expression profiles of circRNA and ceRNA networks in URSA and normal pregnancy
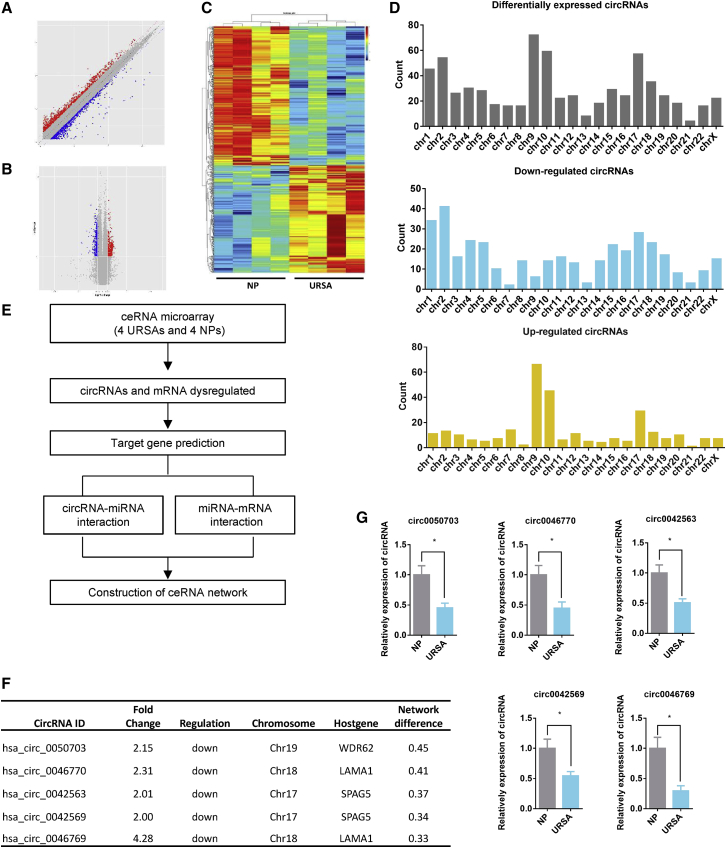
With the advances in microarray and sequencing technologies, circRNAs have been identified in many tissues across different species, prompting research into new circRNA-based diagnostic and therapeutic tools.11,24 To assess circRNA expression and distinct competing endogenous (ceRNA) networks in URSA, we performed a ceRNA microarray. The results revealed 290 upregulated and 374 downregulated circRNAs (fold change > 2.0 and p < 0.05) in URSA placental tissues (n = 4) compared with normal controls (n = 4) (Figures 1A–1C). A summary of the upregulated and downregulated circRNAs from different chromosomes is shown in Figure 1D. In recent studies, circRNAs have been found most commonly interacting with miRNAs through ceRNA mechanisms,28 so we predicted circRNA-miRNA-mRNA networks based on microarray results in these two groups (Figure 1E; Figure S1). We found that key circRNAs and their network targets always had the largest degrees. Therefore, we focused on the top five most significant circRNAs in URSA compared with normal controls (Figure 1F). Consistent with the microarray assay results, qRT-PCR analysis further confirmed the levels of these five top circRNAs (Figure 1G). To conclude, our analysis identified circRNA expression and distinct ceRNA networks in URSA.
Figure 1.
Differential expression profiles of circRNA and ceRNA networks in URSA and normal pregnancy
(A) Scatterplot of differential circRNA expression in placental villus of patients with normal pregnancy and URSA in early pregnancy. (B) Volcano plot shows the different circRNA expression in these two groups. (C) Hierarchical cluster analysis of upregulated and downregulated circRNAs in four pairs of normal pregnancy and URSA placental villus; red is higher expression and blue is lower expression. (D) Number of differentially expressed circRNAs in different chromosomes. (E) Flowchart of the steps used to build the ceRNA network in URSA villus. (F) Characterization of five top circRNAs in the ceRNA network. (G) Validation of microarray results by qRT-PCR analysis of top five circRNAs in the ceRNA network (circRNA-0050703, circRNA-0046770, circRNA-0042563, circRNA-0042569, circRNA-0046769) in four pair cases. Three different independent experiments with three technical repetitions were performed. Data are expressed as the mean ± SEM. ∗p < 0.05. NS, not significant.
The characteristics and clinical features of circRNA-DURSA
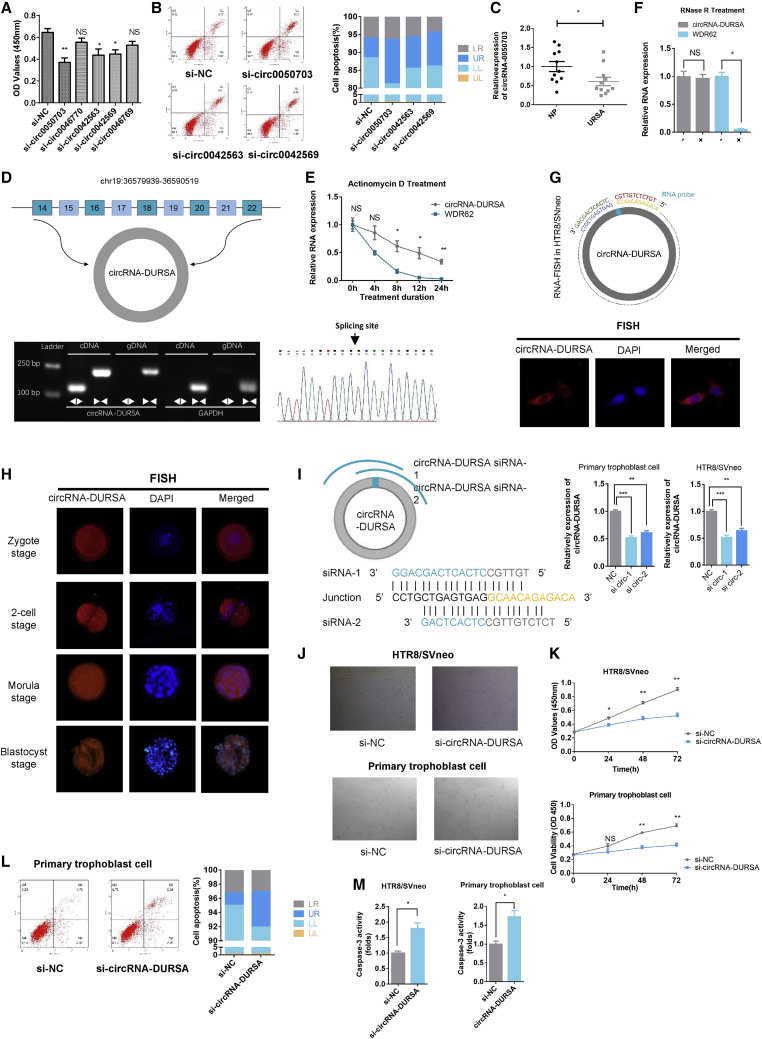
Functional screening of these circRNAs showed that silencing of circRNA-0050703, circRNA-0042563, or circRNA-0042569 significantly decreased cell viability in HTR8/SVneo cells (Figure 2A), and that circRNA-0050703 had the most impact. Propidium iodide (PI)/annexin V flow cytometry assays further showed that circRNA-0050703 silencing most significantly induced apoptosis in HTR8/SVneo cells (Figure 2B). Importantly, qRT-PCR of URSA (11 cases) and normal pregnant (11 cases) samples in early pregnancy showed that circRNA-DURSA levels were significantly decreased in URSA (Figure 2C). Hence, we renamed this molecule circRNA-DURSA (downregulated in URSA placental villus) and focused on its role in apoptosis in URSA. The circBase database revealed that circRNA-DURSA is located on chromosome 19, is 971 bp in length, and consists of nine WDR62 gene exons (exons 14–22) (Figure 2D). To avoid trans-splicing or genomic rearrangements, we designed convergent and divergent primers to amplify circRNA-DURSA using complementary DNA (cDNA) and genomic DNA (gDNA) from HTR8/SVneo cells. circRNA-DURSA was amplified from cDNA only by divergent primers, and no amplification product was observed from gDNA. Next, we confirmed head-to-tail splicing in the circRNA-DURSA RT-PCR product of the expected size using Sanger sequencing (Figure 2D). Then, we designed primers for WDR62 mRNA and circRNA-DURSA to evaluate their resistance to actinomycin D (an inhibitor of transcription) treatment. As shown in Figure 2E, circRNA-DURSA was more stable and resistant to actinomycin D treatment than was WDR62 mRNA. Moreover, circRNA-DURSA was resistant to RNase R exonuclease digestion (Figure 2F), while WDR62 mRNA was not. Taken together, these data confirm that circRNA-DURSA is a circular RNA.
Figure 2.
Characterization of circRNA-DURSA
(A) CCK-8 assays of HTR8/SVneo cells transfected with circRNA-0050703, circRNA-0046770, circRNA-0042563, circRNA-0042569, or circRNA-0046769 siRNA. (B) Flow cytometric analysis of cell apoptosis in HTR8/SVneo cells transfected with circRNA-0050703, circRNA-0042563, and circRNA-0042569 siRNA. (C) Relative expression of circRNA-DURSA in villus tissues of normal pregnancy and URSA (n = 22). (D) (Top) Schematic diagram of genomic location and splicing of circRNA-DURSA. (Bottom left) RT-PCR products using divergent and convergent primers showing circularization of circRNA-DURSA. GAPDH was used as a negative control. (Bottom right) Arrow represents the “head-to-tail” splicing sites of circRNA-DURSA according to Sanger sequencing. (E) circRNA-DURSA resistance to actinomycin D (ActD) was detected by qRT-PCR. (F) circRNA-DURSA and linear WDR62 mRNA expression in HTR8/SVneo cells treated with RNase R. (G) RNA FISH showed the expression and localization of circRNA-DURSA in HTR8/SVneo cells. DAPI solution was used to stain the nuclei. (H) RNA FISH showed the localization of circRNA-DURSA in human pre-implantation embryos. (I) Two different siRNAs were designed for targeting the back-splicing junction of circRNA-DURSA. HTR8/SVneo cells and human primary trophoblast cells were transfected with si-circRNA-DURSA or si-circRNA-NC, and the expression of circRNA-DURSA was detected by qRT-PCR to select the most effective siRNA. (J) The experiments of (J)–(M) were carried out in the groups transfected with si-circRNA-DURSA or si-NC. Cell morphology was determined by microscopy. (K) CCK-8 assays were used to determine cell viability. OD values were obtained at 24, 48, and 72 h after transfection. (L) Flow cytometric analysis of cell apoptosis in treated primary trophoblast cells. (M) Caspase-3 activity was measured to determine the activation level of the caspase-3 protein. Three different independent experiments with three technical repetitions were performed. Data are expressed as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. NS, not significant.
Fluorescence in situ hybridization (FISH) assays were used to assess circRNA-DURSA subcellular location. We found that circRNA-DURSA was predominantly and abundantly expressed in the cytoplasm (Figure 2G). Furthermore, we assessed the expression and localization of circRNA-DURSA in human pre-implantation embryos. At the zygote, two-cell, morula, and blastocyst stages, circRNA-DURSA was mainly expressed in the cytoplasm (Figure 2H).
circRNA-DURSA regulates apoptosis in trophoblast cells
We performed loss-of-function experiments in the HTR8/SVneo cell line and in human primary trophoblast cells. We designed two small interfering RNAs (siRNAs) targeting the back-splicing junction of circRNA-DURSA, and siRNA-1 showed better silencing efficiency (Figure 2I). Results showed that circRNA-DURSA silencing in HTR8/SVneo trophoblast cells and in human primary trophoblasts significantly promotes trophoblast apoptosis (Figures 2J–2L). At the molecular level, cell death is primarily executed via caspase-3 activation. Caspase-3 activity was measured and was drastically increased with circRNA-DURSA silencing (Figure 2M). In order to identify the collateral effect of siRNA-mediated circRNA-DURSA silencing, WDR62 mRNA and protein levels were detected while silencing circRNA-DURSA. As shown in Figure S2, circRNA-DURSA silencing did not affect WDR62 expression at mRNA and protein levels. Taken together, these results demonstrate that circRNA-DURSA is downregulated in URSA placental villus and can prevent trophoblast cell apoptosis.
Informatics reveals the circRNA-DURSA/miR-760-HIST1H2BE axis
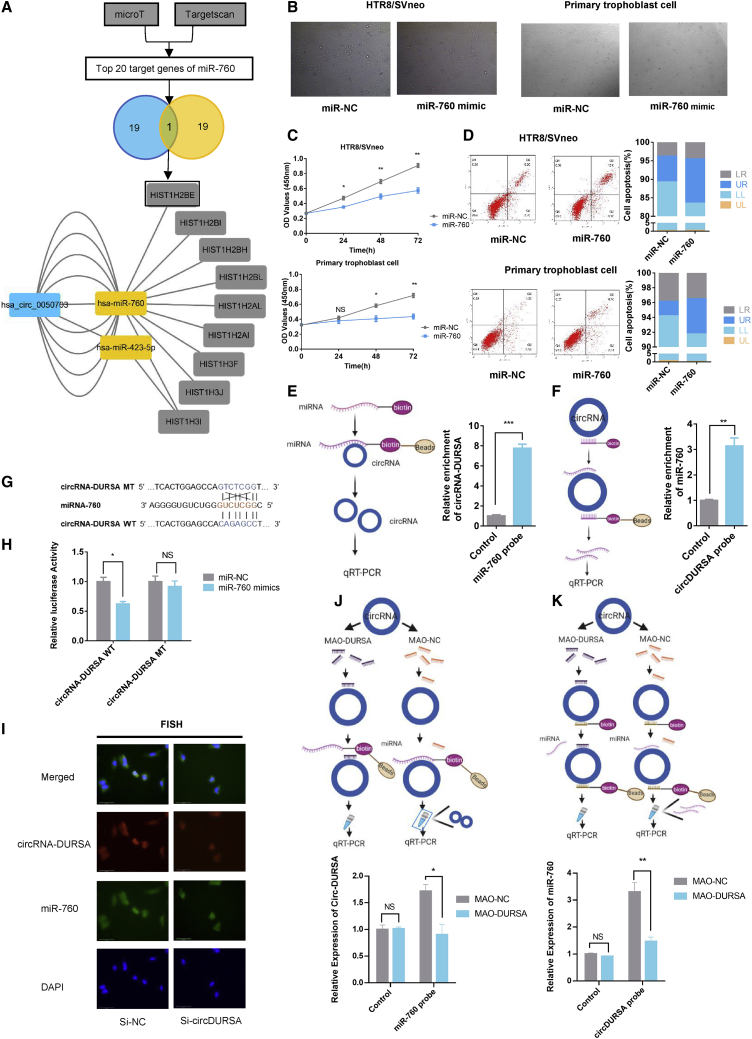
circRNAs were recently reported to function through ceRNA mechanisms in various diseases.29 Our predicted ceRNA network revealed that circRNA-DURSA was mainly able to bind to miR-760, which could target nine mRNAs. Among these nine mRNAs, HIST1H2BE was the only one that was also included in the top 20 target genes predicted using TargetScan and microT-CDS databases (Figure 3A). The binding sites of circRNA-DURSA and miR-760 were also confirmed by miRanda and RNAhybrid databases. Therefore, we hypothesized that circRNA-DURSA may retain miR-760 in the cytoplasm and indirectly promote HIST1H2BE expression. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed apoptosis among the top five significantly enriched pathways downstream of miR-760 (Figure S3A). Gene Ontology (GO) enrichment analyses further indicated that miR-760 might be involved in DNA binding and chromatin organization (Figure S3B).
Figure 3.
circRNA-DURSA regulates trophoblast apoptosis via sponging miR-760
(A) Flowchart showing the steps used to screen the target genes of miR-760. The interaction of the circRNA-DURSA (blue), two targeted miRNAs (yellow), and nine targeted mRNAs (gray) were exhibited. (B) Cell morphology by microscopy. HTR8/SVneo cells and human primary trophoblast cells were transfected with miR-760 mimic or miR-NC mimic. The experiments of (B)–(D) were carried out in the groups above. (C) CCK-8 assays were used to determine cell viability. OD values are obtained at 24, 48, and 72 h after transfection. (D) Flow cytometric analysis of cell apoptosis in treated HTR8/SVneo cells and primary trophoblast cells. (E) RNA pull-down assays for circRNA-DURSA using a miR-760 or control probe. (F) RNA pull-down assays for miR-760 using a circRNA-DURSA or control-probe. (G and H) HTR8/SVneo cells were co-transfected with psiCHECK-2-circDURSA-wild-type (WT) or psiCHECK-2-circDURSA mutant and miR-760, or miR-NC. Luciferase activity was detected with luciferase reporter assays, normalized with Renilla activity. (I) The co-localization of circRNA-RNA (red) and miR-760 (green) was observed by fluorescence in situ hybridization (FISH) in HTR8/SVneo cells transfected with si-NC or si-circRNA-DURSA. Cell nuclei were counterstained with DAPI (blue). (J) Sequence-specific morpholino antisense oligonucleotides (MAOs) targeting the miR-760 binding site on circRNA-SORE were designed. Pull-down assays treated with MAO-DURSA (MAO targeting circRNA-DURSA) or MAO-NC using a miR-760 or control probe. (K) Pull-down assays treated with MAO-DURSA or MAO-NC using a circRNA-DURSA or control probe. Three different independent experiments with three technical repetitions were performed. Data are expressed as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. NS, not significant.
circRNA-DURSA acts as an miR-760 sponge
To explore the function of miR-760 in trophoblast cells, an miR-760 mimic was used to overexpress miR-760. Results indicated that miR-760 overexpression in HTR8/SVneo trophoblast cells and in human primary trophoblasts significantly induced trophoblast apoptosis (Figures 3B–3D). RNA pull-down assays showed abundant enrichment of circRNA-DURSA pulled down by biotin-miR-760 in HTR8/SVneo cells (Figure 3E). Consistently, the biotinylated circRNA-DURSA probe captured more miR-760 than did the control probe, suggesting that circRNA-DURSA could bind to miR-760 (Figure 3F). To reveal the functional integration between circRNA-DURSA and miR-760, psiCHECK-2-circRNA-DURSA and a psiCHECK-2-circRNA-DURSA mutant were constructed; the mutation was based on the predicted miR-760 binding sites in circRNA-DURSA (Figure 3G). Dual-luciferase reporter assays revealed that miR-760 mimics could significantly attenuate luciferase activity in the wild-type group but not in the mutant group (Figure 3H). To explore the subcellular location of miR-760, FISH assays were performed. The results show that miR-760 is predominantly and abundantly expressed in the cytoplasm, and that circRNA-DURSA downregulation does not change miR-760 cellular localization (Figure 3I). To further verify the binding of circRNA-DURSA and miR-760, we designed sequence-specific morpholino antisense oligonucleotides (MAOs) targeting the miR-760 binding site on circRNA-SORE (5′-AGGCTCTGTGGCTCCAGTGA-3′). The biotinylated circRNA-DURSA probe captured less miR-760 in HTR8/SVneo cells treated with MAO targeting circRNA-DURSA (MAO-DURSA) than in those treated with control MAO (MAO-NC) (Figure 3J). Consistently, the biotinylated miR-760 probe captured less circRNA-DURSA in HTR8/SVneo cells treated with MAO-DURSA than in those treated with MAO-NC (Figure 3K). Taken together, these results show that circRNA-DURSA can regulate trophoblast apoptosis via miR-760 sponging to regulate trophoblast apoptosis.
circRNA-DURSA regulates trophoblast apoptosis via the miR-760-HIST1H2BE axis
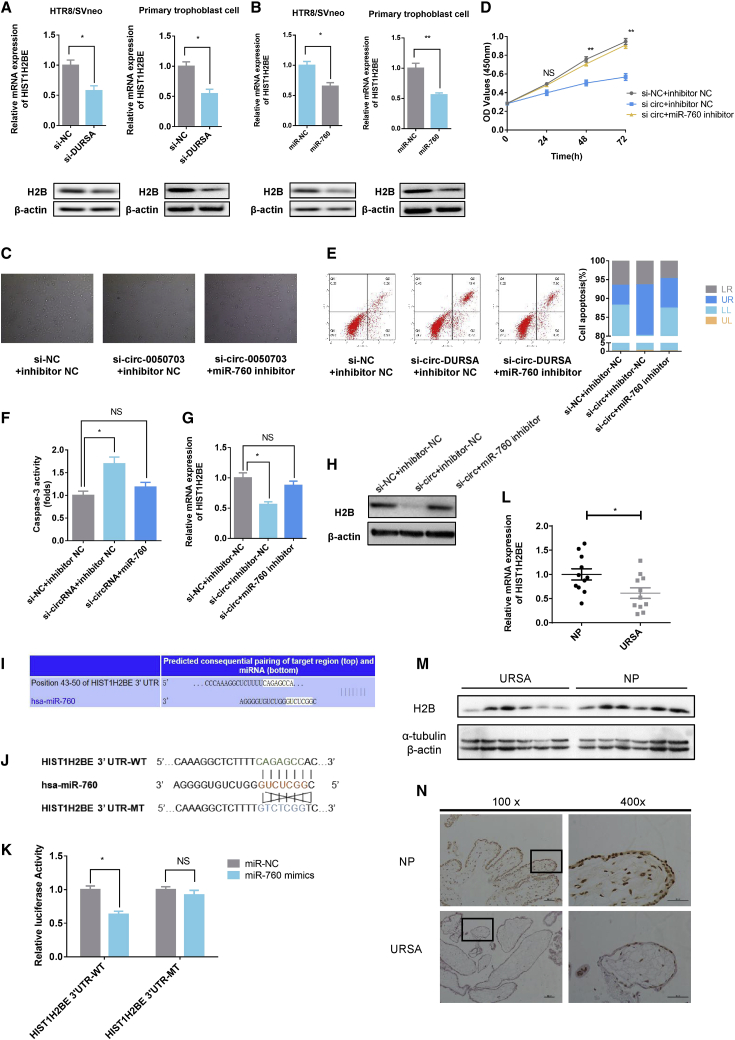
Next, we sought to identify the roles of miR-760 and HIST1H2BE in URSA. circRNA-DURSA silencing or miR-760 overexpression significantly decreased HIST1H2BE mRNA and protein levels in HTR8/SVneo cells and in human primary trophoblast cells (Figures 4A and 4B). Rescue assays showed that the miR-760 inhibitor could partially reverse the effect of siRNA (si-)circRNA-DURSA on cell viability, apoptosis, and caspase-3 activity in trophoblast cells (Figures 4C–4F). qRT-PCR and western blot analyses further indicated that miR-760 inhibitor could partially abolish si-circRNA-DURSA-mediated suppression of HIST1H2BE at both the mRNA and protein levels (Figures 4G and 4H).
Figure 4.
circRNA-DURSA regulates trophoblast apoptosis via the miR-760-HIST1H2BE axis
(A) HIST1H2BE mRNA and protein expression in HTR8/SVneo cells and primary human trophoblast cells transfected with si-circRNA-DURSA or si-circRNA-NC. (B) qRT-PCR and western blot analysis of HIST1H2BE mRNA expression and protein in HTR8/SVneo cells and primary human trophoblast cells transfected with miR-NC or miR-760 mimic. (C) Cell morphology by microscope. HTR8/SVneo cells were transfected with si-NC+inhibitor NC, si-circ-DURSA+inhibitor NC, or si-circ-DURSA+miR-760 inhibitor. The experiments of (C)–(H) were carried out in the three groups above. (D) Cell viability in treated HTR8/SVneo cells was assessed by CCK-8 assays. OD values were obtained at 24, 48, and 72 h after transfection. (E) Flow cytometric analysis of cell apoptosis in treated HTR8/SVneo cells and primary trophoblast cells of three groups. (F) Caspase-3 activity was measured to determine the activation level of the caspase-3 protein in the three groups. (G) HIST1H2BE mRNA expression levels were measured by qRT-PCR in treated HTR8/SVneo cells. (H) Western blot assays were performed to measure the H2B protein expression levels in treated HTR8/SVneo cells. (I) TargetScan database showing the putative binding sites of miR-760 related to HIST1H2BE. (J and K) HTR8/SVneo cells were co-transfected with psiCHECK-2-HIST1H2BE-3′ UTR WT or psiCHECK-2-HIST1H2BE-3′ UTR mutant and miR-760, or miR-NC. Luciferase activity was detected with luciferase reporter assays, normalized with Renilla activity. (L) HIST1H2BE mRNA expression in URSA and normal pregnancy placental villus (n = 22). (M and N) HIST1H2BE protein expression was detected by western blot and immunohistochemistry (IHC) in URSA and normal pregnancy placental villus (n = 22). Representative images of H2B expression detected by IHC assays are shown. Three different independent experiments with three technical repetitions were performed. Data are expressed as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01. NS, not significant.
To validate whether miR-760 directly targets the HIST1H2BE-3′ UTR, psiCHECK-2-HIST1H2BE-3′ UTR and the psiCHECK-2-HIST1H2BE-3′ UTR mutant were constructed based on TargetScan predicted binding sites (Figures 4I and 4J). Dual-luciferase reporter assays showed that miR-760 mimics significantly decreased the luciferase activity in the wild-type group, but not in the mutant group (Figure 4K). These results support the contention that HIST1H2BE-3′ UTR and miR-760 interact directly. Importantly, HIST1H2BE mRNA and protein levels were also significantly lower in URSA placenta villus than in placenta villus from patients with normal pregnancies (Figures 4L–4N).
Taken together, these experiments show that miR-760 can directly target the 3′ UTR of HIST1H2BE and that circRNA-DURSA could regulate trophoblast cell apoptosis via the miR-760-HIST1H2BE axis.
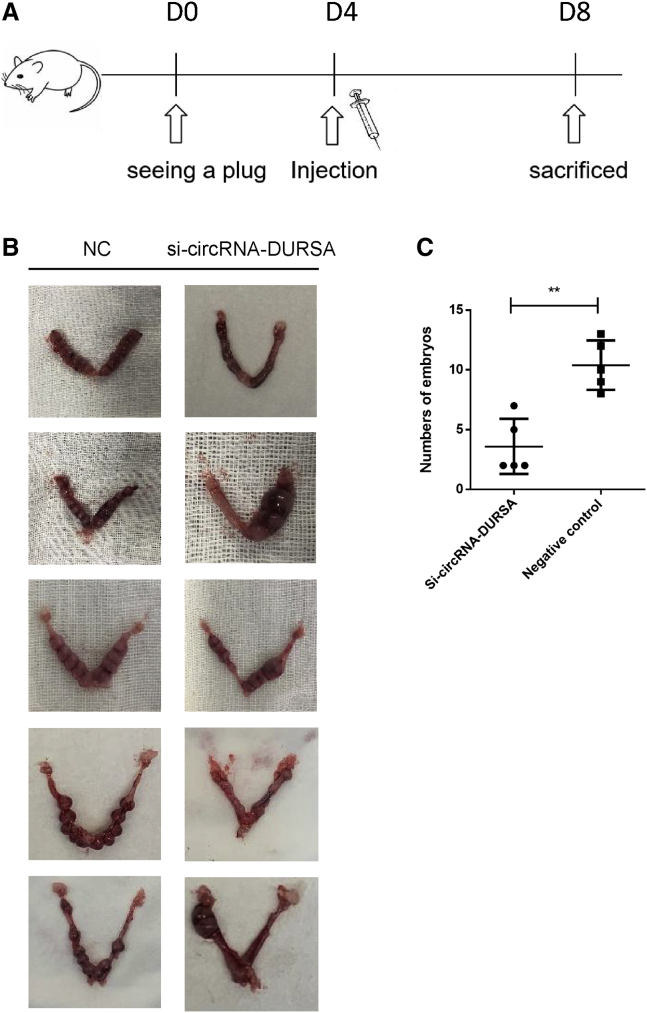
Silencing circRNA-DURSA inhibits embryo implantation and placentation in vivo
To further validate the findings in vitro, the in vivo pregnant ICR mice model was applied as described in Materials and methods and Figure 5A. Consistent with the in vitro results, the numbers of embryos implanted in si-circRNA-DURSA-treated mice were significantly fewer than those implanted in the si-control-treated mice (Figures 5B and 5C; p < 0.01), indicating that administration of 10 nmol of si-circRNA-DURSA significantly increases embryo resorption.
Figure 5.
circRNA-DURSA induces embryo resorption in vivo
(A) In vivo pregnant ICR mice model was applied as described in Materials and methods. Female mice were injected with 10 nmol of si-circRNA-DURSA and negative control agomir into the lumen of uterine horn adjacent to the oviduct 4 days after seeing a plug. (B and C) The numbers of embryos implanted in the two groups were calculated. All data are presented as the mean ± SEM. ∗∗p < 0.01.
Discussion
URSA is an intractable threat to women of reproductive age worldwide and lacks interventional targets for clinical management. Numerous studies have explored the molecular mechanisms of URSA, yet its pathophysiology remains largely unclear. To date, numerous theories on URSA pathogenesis have emerged, including poor decidualization, increased trophoblast cell apoptosis, impaired trophoblast invasion, and abnormal spiral artery remodeling.30,31 As the main constituent cells of the human placenta, the crucial role of trophoblasts has been known for decades. Deficiencies in trophoblast cell function can lead to pregnancy loss, pre-eclampsia, and intrauterine growth restriction.32,33 The balance between trophoblast proliferation and apoptosis contributes to the success of pregnancy, as trophoblast invasion appears to be controlled by apoptosis.8 Increasing numbers of studies have demonstrated that apoptosis is notably exaggerated in URSA.6,34 It is reported that Prdx2 plays a role in regulating trophoblast proliferation and apoptosis in the pathogenesis of RSA.31 Aberration of macrophage-induced FasL-mediated apoptosis may represent one cause of recurrent miscarriage.35 However, the regulatory mechanism of trophoblast cell apoptosis is very complex, and many aspects of this process remain unknown. Therefore, this study was designed to identify novel molecular diagnostic and therapeutic targets for URSA.
Recent studies indicate that ncRNAs are associated with pregnancy. circRNA-0004904 and circRNA-0001855 have been reported as potential biomarkers for the prediction of pre-eclampsia.26 Altered circRNA expression was also observed in the endometrium of patients with repeated implantation failure.27 circRNAs have also been found in the processes of early spontaneous abortion.16,36 Importantly, circRNAs are abundantly expressed in human pre-implantation embryos, prompting us to clarify the roles of circRNA in URSA. circRNAs are generally derived from one or more exons of known protein-coding genes through back-splicing. circRNAs often regulate target genes in human diseases through sponging miRNAs, which are suppressors of mRNA translation or stability.37 This understanding uncovers a new, complicated post-transcriptional regulatory network and mechanism. For example, circAGFG1 can promote triple-negative breast cancer progression by sponging miR-195-5p.19 circMTO1 acts as the sponge of miR-9 to suppress hepatocellular carcinoma progression.38 circMFACR mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression.39 It is well accepted that gene expression is altered in trophoblast cells of women with URSA.40,41 However, to date, the circRNAs and their related ceRNA networks in URSA are largely unknown. In this study, we demonstrate a global circRNA-miRNA-mRNA network for the first time, to the best of our knowledge. This network allows us to decipher the ceRNA mechanisms related to URSA. From these complex post-transcriptional regulatory networks, specific circRNAs are likely to be candidate biomarkers or molecular targets for URSA. Comparison of the circRNA interaction degrees in the ceRNA networks in placental villus of URSA and control allowed us to identify the top five circRNAs that significantly downregulated in the URSA group. Using this approach, we identified that circRNA-DURSA plays an anti-apoptotic role in the progression of URSA. FISH assay results indicated that circRNA-DURSA is located in cytoplasm in trophoblast cells and human pre-implantation embryos, suggesting that circRNA-DURSA function might occur through ceRNA mechanisms in URSA. Notably, circRNA-DURSA was also expressed in lower levels in URSA placental villus than in samples from normal pregnancies in the early pregnancy stage, suggesting that circRNA-DURSA has diagnostic and prognostic value. Importantly, silencing circRNA-DURSA significantly increased the embryo resorption rate in vivo in the pregnant ICR mice model, further supporting the potential therapeutic merits of circRNA-DURSA for patients with URSA.
Through bioinformatics analyses, the specific circRNA-DURSA was predicted to function as a ceRNA by harboring miR-760, which eliminates miR-760-mediated HIST1H2BE suppression. As a H2B coding gene, HIST1H2BE is a potential target gene of miR-760 in the circRNA-DURSA ceRNA network. This was confirmed in two other databases. Histone proteins, including H3, H4, H2A, H2B, and H1, are the primary components of chromatin and are synthesized in large amounts to package newly replicated DNA. However, a histone shortage or excess blocks DNA replication and triggers genomic instability in mammals, which is closely associated with apoptosis. Indeed, DNA-templated processes, including DNA repair, are dependent on protein-DNA interactions that are restricted by histones in nucleosomes.42,43 HIST1H2BE levels are critical for cell growth and estrogen response in breast cancer.44 In this study, lower HIST1H2BE expression levels were detected in placental villus of URSA samples, suggesting that HIST1H2BE has unique roles in URSA. Of interest, another study found that histone mRNA was upregulated and miR-760 was downregulated, suggesting that the miR-760/histone mRNA axis has a crucial role in gastric cancer development.45 Recently, it was reported that miR-760 is involved in multiple cellular processes, including cell viability, proliferation, metastasis, and apoptosis.46,47 miR-760 has also been identified as a potential distinct biomarker for the early detection of colorectal cancer.48 However, the roles of miR-760 in URSA have remained elusive. Our pull-down assay and luciferase reporter gene assay results show that miR-760 directly binds circRNA-DURSA in vitro. Moreover, in silico prediction and KEGG pathway analyses revealed that apoptosis is the most significantly enriched miR-760 targeted pathway. We also demonstrated that miR-760 overexpression can promote trophoblast apoptosis in vitro, leading to URSA. Luciferase reporter gene assays confirmed that miR-760 can inhibit HIST1H2BE expression through binding to its 3′ UTR. Moreover, we identify the distinct circRNA-DURSA/miR-760/HIST1H2BE regulatory axis that is involved in trophoblast apoptosis.
There are several limitations to this study. circRNAs exert biological functions through protein binding,49 peptide coding,50 and transcription activation. Additionally, other modes of circRNA-DURSA function are still possible in URSA, rendering further investigation necessary in future studies. Finally, the pathogenesis of URSA is far more complicated than it appears to be, which needs further exploration.
In conclusion, we have provided the first evidence that the circRNA expression pattern is significantly altered in URSA placental villus. The circRNA-miRNA-mRNA network suggests that circRNA plays roles in the pathophysiological process of URSA. circRNA-DURSA, a distinct downregulated circRNA in URSA placental villus, functions as a ceRNA by harboring miR-760, eliminating its suppression of HIST1H2BE expression and protecting trophoblasts from apoptosis (Figure 6). Our results provide distinct insights into the roles and clinical values of circRNA-DURSA in URSA, and they suggest possible applications for URSA diagnosis and treatment.
Figure 6.
Schematic model of circRNA-DURSA/miR-760/HIST1H2BE functions in URSA
Materials and methods
Clinical samples
Placenta villus tissue samples from 15 URSA and 15 normal pregnant (NP) patients were collected from the Women’s Hospital, School of Medicine, Zhejiang University, China, between January 2017 and September 2019. All pregnant women had a gestational age of 6–12 weeks. Detailed patient information is summarized in the Table S1. The URSA group included patients with two or more losses of a clinically established intrauterine pregnancy, and excluded chromosomal abnormalities of embryo, abnormal maternal reproductive tract anatomy, symptoms of endocrine or metabolic diseases, history of autoimmune disease, and so forth. All tissues were placed immediately in liquid nitrogen after removal from patients and stored at −80°C or fixed in 4% formaldehyde and embedded in paraffin until use. Embryos with arrested development or poor preimplantation morphology that cannot to be used for transfer from in vitro fertilization embryo transfer patients were collected. This study was approved by the Ethical Review Committee of Women’s Hospital, School of Medicine, Zhejiang University, and all patients provided informed consent in accordance with the ethics guidelines.
Microarray analysis and construction of ceRNA network
Placenta villus tissue samples from four URSA and four NP patients were analyzed using the ceRNA microarray analysis. The microarray hybridization and collection of data were performed by Shanghai Biotechnology Cooperation, Shanghai, China. Figure 1E shows the methodology used to identify the ceRNA interacting genes. The miRanda method, which is based on dynamic programming (SW algorithm), and computing free energy was used to identify the target genes of miRNAs and ceRNAs.21, 22, 23, 24, 25 Based on this analysis, we built a circRNA-miRNA-mRNA interaction network. The relationship between the target and the miRNA was based on the adjacency matrix of miRNA and target A = [ai,j], where ai,j represents the weight of the relationship between the target (i) and its miRNA (j). In the circRNA-miRNA-mRNA network, the circle represents one edge, whereas the center of the network represents a degree. The degree denotes the contribution of a miRNA to the target gene around it or the contribution of a target to the miRNAs around it. The key miRNAs and their targets in the network always had the largest degrees.
Immunohistochemistry (IHC)
All tissues were fixed in a formalin solution, dehydrated in ethanol, embedded in paraffin, and sectioned. After blocking, the slides were incubated with anti-H2B (Abcam, ab52599) overnight at 4°C. After washing with PBS, the slides were incubated with goat anti-rabbit horseradish peroxidase (HRP) (Vector Laboratories, Burlingame, CA, USA) for 30 min at room temperature. A diaminobenzidine (DAB) kit (Sigma-Aldrich, St. Louis, MO, USA) was used to detect the immunohistochemical reactions. The slides were examined under a phase contrast light microscope (Nikon). Three areas were chosen randomly from each section for measurement.
Cell culture and treatment
The human trophoblast cell line HTR8/SVneo was cultured in DMEM (Life Technologies) with 10% FBS (Gibco), 100 U/mL penicillin, and 100 mg/mL streptomycin. Human primary trophoblast cells were isolated from chorionic villous explants as previously described.51 All cells were cultured at 37°C and 5% CO2 in a humidified atmosphere. For treatment with MAOs, each MAO (GeneTools, Philomath, OR, USA) was added to culture media at a final concentration of 10 μM along with Endo-Porter (GeneTools, Philomath, OR, USA) and incubated for 24 h. The sequence of MAOs targeting the predicted the binding site of miR-760 in circRNA-DURSA is 5′-AGGCTCTGTGGCTCCAGTGA-3′.
Western blotting
Total protein was extracted using radioimmunoprecipitation assay (RIPA) lysis buffer and determined using a bicinchoninic acid (BCA) protein assay kit (Beyotime). Equivalent amounts of protein (30 μg) were separated by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking membranes with 5% bovine serum albumin (Sigma, St. Louis, MO, USA), they were incubated with appropriate dilutions of specific primary antibodies against β-actin (FD0064, FUDE Biological Technology) and H2B (Abcam, ab52599, 1:10,000). The blots were incubated with HRP-conjugated secondary antibodies (1:2,000) and visualized using an electrochemiluminescence (ECL) system (Thermo Fisher Scientific, Rochester, NY, USA).
Cell Counting Kit-8 (CCK-8) assays
Treated HTR8/SVneo lines cells and human primary trophoblast cells were seeded into 96-well plates to ensure 2,000 cells/well and incubated in a 5% CO2 atmosphere at 37°C. CCK-8 solution (Yeasen Biotech, China) was added to each well at 0, 24, 48, and 72 h. The optical density (OD) values were measured with a microplate reader (BioTek, Winooski, VT, USA) at 450 nm.
RNA isolation and PCR
Total RNA was isolated from tissues and cell lines with TRIzol reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions. The quality and quantity of isolated RNA were detected. cDNA was synthesized from approximately 1 μg of RNA from each sample using Hifair II first-strand cDNA synthesis SuperMix for qPCR (gDNA digester plus) (Yeasen Biotech). Total DNAs from cells were extracted using an AxyPrep multisource gDNA miniprep kit (Axygen, USA). qRT-PCR was performed on a LightCycler 480 real-time PCR system (Roche Applied Science, Germany) using Hieff Unicon qPCR SYBR Green master mix (Yeasen Biotech). The relative mRNA expression or DNA content was calculated using the ΔΔCt method. The primers are listed in Table S2.
Oligonucleotide transfection
The transient transfection was held out when cells reached 60%–70% confluence. si-circRNA, miR-760 mimic, and their related control oligonucleotides were designed and synthesized by RiboBio (Guangzhou, China). All transfections were performed by the final concentrations of 60 nM for miRNA mimics and 100 nM for miRNA inhibitor and si-circRNA. Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) was used as transfection medium according to the manufacturer’s instructions. The sequences used are listed in Table S2.
Flow cytometry
Treated HTR8/SVneo cells or human primary trophoblast cells (2 × 105 cells/well) were seeded into 24-well plates and incubated overnight at 37°C. Cells were harvested by trypsin digestion without EDTA. The apoptosis ratio was measured using an annexin V-fluorescein isothiocyanate (FITC)/PI apoptosis kit (Multi Science, China) according to the manufacturer’s protocol. Cells were incubated with 5 μL of annexin V-FITC and 10 μL of PI for 5 min in the dark condition. Apoptosis was analyzed by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
Luciferase reporter assay
Potential binding sites were predicted using the TargetScan database. The linear form of circRNA-DURSA or the 3′ UTR of HIST1H2BE containing the binding site of miR-760 or mutated was cloned downstream of the Renilla luciferase gene in the dual-luciferase plasmid psiCHECK-2 vector (Promega) to construct the psiCHECK-2-circRNA-DURSA vector or psiCHECK-2-HIST1H2BE-3′ UTR vector. HTR8/SVneo cells (1 × 105 cells/well) were added to a 24-well plate for transfection. Afterward, 0.8 μg of psiCHECK-2-circRNA-DURSA vector, psiCHECK-2-HIST1H2BE-3′ UTR vector, or psiCHECK-2 vector, with the addition of 20 pmol of miR-760 mimics or negative control mimics, was cotransfected into cells using Lipofectamine RNAiMAX (Invitrogen). After 48 h of transfection, the cells were harvested, and luciferase activities were measured with the Dual-Luciferase reporter assay system (Promega, Madison, WI, USA). The luciferase activity was normalized for transfection efficiency by comparing with Renilla luciferase activity.
Pull-down assay
The biotinylated circRNA-DURSA probe, biotinylated miRNA mimics, or their controls were synthesized by Tsingke Biological Technology (China) (Table S2). The biotinylated probe was incubated with magnetic beads (Life Technologies) at 25°C for 2 h to generate probe-coated beads. The cell lysates were incubated with probe at 4°C overnight. After washing with the wash buffer, the RNA complexes bound to the beads were eluted and extracted with TRIzol reagent (Invitrogen, Grand Island, NY, USA) for RT-PCR or real-time PCR.
Fluorescence in situ hybridization (FISH)
FISH was performed following the instructions of the probe manufacturer (RiboBio, Guangzhou, China). The cells and embryos were washed and fixed in 4% paraformaldehyde for 30 min. After 0.1% Triton X-100 treatment, the cells were incubated with 20 mg/mL circRNA or miRNA probe overnight at 37°C. Nuclei were dyed with DAPI. The intracellular localization of circRNA or miRNA was observed by using an FV1200 laser confocal microscope (Olympus). The sequence of the circRNA-DURSA probe for FISH is supplied in Figure 2G.
Caspase-3 activity assays
The activity of caspase-3 was determined by using a caspase-3 activity kit (Beyotime Institute of Biotechnology, China), which is based on the ability of caspase-3 to change acetyl-Asp-Glu-Val-Asp p-nitroanilide into the yellow formazan product, p-nitroaniline. The activity of caspase-3 was assessed by calculating the ratio of the OD405.
In vivo experiments
All mice used in our experiments were ICR (6–8 weeks of age). Female mice were mated with male mice. Female mice were injected with 10 nmol of si-circRNA-DURSA and negative control agomir (purchased from RiboBio Biotech) into the lumen of uterine horn adjacent to the oviduct 4 days after seeing a plug. Four days after the injection, the female mice were executed and the numbers of embryos were calculated. All animal experiments were performed humanely in compliance with guidelines reviewed by the Animal Ethics Committee of the Biological Resource Centre of the Agency for Science, Technology and Research at the Women’s Hospital, Zhejiang University School of Medicine.
Statistical analysis
Data analyses were performed with SPSS software version 23.0 (SPSS, Chicago, IL, USA) and GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, USA). A Student’s t test and variance analysis (ANOVA) were used to make the comparisons between two groups or among multiple groups, respectively. All experimental data are expressed as the mean ± SEM. p values <0.05 were considered statistically significant.
Acknowledgments
This research was supported by the Natural Science Foundation of China (no. 81873819), the National Key Research and Development Program of China (no. 2018YFC1003201), the Program for Key Subjects of Zhejiang Province in Medicine and Hygiene (to Y.Z.), the Natural Science Foundation of China (no. 81803245), and the Zhejiang Provincial Natural Science Foundation of China (LQ20H040010). We thank Rebecca Porter, PhD, from the Edanz Group for editing a draft of this manuscript.
Author contributions
M.T. conceived the project, and M.T., L.B., and Z.W. carried out most of the experiments. M.T. interpreted the results and wrote the manuscript. M.T., S.W., Y.X., Y.Q., and L.C. collected clinical samples and analyzed the data. X.H. and J.Y. contributed to the interpretation of the data and the critical review of the manuscript. Y.Z. and F.Q. directed the project. Y.Z., X.H., and J.Y. provided the funding for this study.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.06.012.
Contributor Information
Fan Qu, Email: syqufan@zju.edu.
Yimin Zhu, Email: zhuyim@zju.edu.cn.
Supplemental information
References
- 1.Practice Committee of the American Society for Reproductive Medicine Evaluation and treatment of recurrent pregnancy loss: a committee opinion. Fertil. Steril. 2012;98:1103–1111. doi: 10.1016/j.fertnstert.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 2.Bender Atik R., Christiansen O.B., Elson J., Kolte A.M., Lewis S., Middeldorp S., Nelen W., Peramo B., Quenby S., Vermeulen N. ESHRE guideline: Recurrent pregnancy loss. Hum. Reprod. 2018;Open 2:hoy004. doi: 10.1093/hropen/hoy004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephenson M.D., Awartani K.A., Robinson W.P. Cytogenetic analysis of miscarriages from couples with recurrent miscarriage: A case-control study. Hum. Reprod. 2002;17:446–451. doi: 10.1093/humrep/17.2.446. [DOI] [PubMed] [Google Scholar]
- 4.Alijotas-Reig J., Garrido-Gimenez C. Current concepts and new trends in the diagnosis and management of recurrent miscarriage. Obstet. Gynecol. Surv. 2013;68:445–466. doi: 10.1097/OGX.0b013e31828aca19. [DOI] [PubMed] [Google Scholar]
- 5.Kwak-Kim J., Park J.C., Ahn H.K., Kim J.W., Gilman-Sachs A. Immunological modes of pregnancy loss. Am. J. Reprod. Immunol. 2010;63:611–623. doi: 10.1111/j.1600-0897.2010.00847.x. [DOI] [PubMed] [Google Scholar]
- 6.Sharp A.N., Heazell A.E., Crocker I.P., Mor G. Placental apoptosis in health and disease. Am. J. Reprod. Immunol. 2010;64:159–169. doi: 10.1111/j.1600-0897.2010.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huppertz B., Kadyrov M., Kingdom J.C. Apoptosis and its role in the trophoblast. Am. J. Obstet. Gynecol. 2006;195:29–39. doi: 10.1016/j.ajog.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 8.Huppertz B., Hemmings D., Renaud S.J., Bulmer J.N., Dash P., Chamley L.W. Extravillous trophoblast apoptosis—A workshop report. Placenta. 2005;26(Suppl):S46–S48. doi: 10.1016/j.placenta.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Amaral P.P., Mattick J.S. Noncoding RNA in development. Mamm. Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 10.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Lin Z., Xia S., Liang Y., Ji L., Pan Y., Jiang S., Wan Z., Tao L., Chen J., Lin C., et al. LXR activation potentiates sorafenib sensitivity in HCC by activating microRNA-378a transcription. Theranostics. 2020;10:8834–8850. doi: 10.7150/thno.45158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel D.P. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouillet J.F., Ouyang Y., Coyne C.B., Sadovsky Y. MicroRNAs in placental health and disease. Am. J. Obstet. Gynecol. 2015;213(4, Suppl):S163–S172. doi: 10.1016/j.ajog.2015.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou Y., Jiang Z., Yu X., Zhang Y., Sun M., Wang W., Ge Z., De W., Sun L. miR-101 regulates apoptosis of trophoblast HTR-8/SVneo cells by targeting endoplasmic reticulum (ER) protein 44 during preeclampsia. J. Hum. Hypertens. 2014;28:610–616. doi: 10.1038/jhh.2014.35. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Zhou J., Li M.Q., Xu J., Zhang J.P. MicroRNA-184 promotes apoptosis of trophoblast cells via targeting WIG1 and induces early spontaneous abortion. Cell Death Dis. 2019;10:223. doi: 10.1038/s41419-019-1443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang Y. Circular RNAs as novel biomarkers with regulatory potency in human diseases. Future Sci. OA. 2018;4:FSO314. doi: 10.4155/fsoa-2018-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang J., Hong H., Xue X., Zhu X., Jiang L., Qin M., Liang H., Gao L. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222–232. doi: 10.1016/j.canlet.2018.10.040. [DOI] [PubMed] [Google Scholar]
- 19.Yang R., Xing L., Zheng X., Sun Y., Wang X., Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol. Cancer. 2019;18:4. doi: 10.1186/s12943-018-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Zeng Y., Du W.W., Wu Y., Yang Z., Awan F.M., Li X., Yang W., Zhang C., Yang Q., Yee A., et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7:3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukiw W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD) Front. Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., Wu M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dang Y., Yan L., Hu B., Fan X., Ren Y., Li R., Lian Y., Yan J., Li Q., Zhang Y., et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17:130. doi: 10.1186/s13059-016-0991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang M., Lash G.E., Zhao X., Long Y., Guo C., Yang H. circRNA-0004904, circRNA-0001855, and PAPP-A: Potential novel biomarkers for the prediction of preeclampsia. Cell. Physiol. Biochem. 2018;46:2576–2586. doi: 10.1159/000489685. [DOI] [PubMed] [Google Scholar]
- 27.Liu L., Li L., Ma X., Yue F., Wang Y., Wang L., Jin P., Zhang X. Altered circular RNA expression in patients with repeated implantation failure. Cell. Physiol. Biochem. 2017;44:303–313. doi: 10.1159/000484887. [DOI] [PubMed] [Google Scholar]
- 28.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labarrere C.A., DiCarlo H.L., Bammerlin E., Hardin J.W., Kim Y.M., Chaemsaithong P., Haas D.M., Kassab G.S., Romero R. Failure of physiologic transformation of spiral arteries, endothelial and trophoblast cell activation, and acute atherosis in the basal plate of the placenta. Am. J. Obstet. Gynecol. 2017;216:287.e1–287.e16. doi: 10.1016/j.ajog.2016.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu F., Tian F., Zeng W., Liu X., Fan J., Lin Y., Zhang Y. Role of peroxiredoxin2 downregulation in recurrent miscarriage through regulation of trophoblast proliferation and apoptosis. Cell Death Dis. 2017;8:e2908. doi: 10.1038/cddis.2017.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang M., You J., Wang W., Lu Y., Hu X., Wang C., Liu A., Zhu Y. Impact of galectin-1 on trophoblast stem cell differentiation and invasion in in vitro implantation model. Reprod. Sci. 2018;25:700–711. doi: 10.1177/1933719117725816. [DOI] [PubMed] [Google Scholar]
- 33.Niu Z.R., Han T., Sun X.L., Luan L.X., Gou W.L., Zhu X.M. MicroRNA-30a-3p is overexpressed in the placentas of patients with preeclampsia and affects trophoblast invasion and apoptosis by its effects on IGF-1. Am. J. Obstet. Gynecol. 2018;218:249.e1–249.e12. doi: 10.1016/j.ajog.2017.11.568. [DOI] [PubMed] [Google Scholar]
- 34.Nair R.R., Khanna A., Singh K. Association of FAS −1377 G>A and FAS −670 A>G functional polymorphisms of FAS gene of cell death pathway with recurrent early pregnancy loss risk. J. Reprod. Immunol. 2012;93:114–118. doi: 10.1016/j.jri.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Ding J., Yin T., Yan N., Cheng Y., Yang J. FasL on decidual macrophages mediates trophoblast apoptosis: A potential cause of recurrent miscarriage. Int. J. Mol. Med. 2019;43:2376–2386. doi: 10.3892/ijmm.2019.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong X., Yang L., Wang H. miR-520 promotes DNA-damage-induced trophoblast cell apoptosis by targeting PARP1 in recurrent spontaneous abortion (RSA) Gynecol. Endocrinol. 2017;33:274–278. doi: 10.1080/09513590.2016.1266476. [DOI] [PubMed] [Google Scholar]
- 37.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 38.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M., et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 39.Wang K., Gan T.Y., Li N., Liu C.Y., Zhou L.Y., Gao J.N., Chen C., Yan K.W., Ponnusamy M., Zhang Y.H., Li P.F. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo W., Zhu X., Yan L., Qiao J. The present and future of whole-exome sequencing in studying and treating human reproductive disorders. J. Genet. Genomics. 2018;45:517–525. doi: 10.1016/j.jgg.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 41.Jia N., Li J. Noncoding RNAs in unexplained recurrent spontaneous abortions and their diagnostic potential. Dis. Markers. 2019;2019:7090767. doi: 10.1155/2019/7090767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guintini L., Charton R., Peyresaubes F., Thoma F., Conconi A. Nucleosome positioning, nucleotide excision repair and photoreactivation in Saccharomyces cerevisiae. DNA Repair (Amst.) 2015;36:98–104. doi: 10.1016/j.dnarep.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez Y., Hinz J.M., Smerdon M.J. Accessing DNA damage in chromatin: Preparing the chromatin landscape for base excision repair. DNA Repair (Amst.) 2015;32:113–119. doi: 10.1016/j.dnarep.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nayak S.R., Harrington E., Boone D., Hartmaier R., Chen J., Pathiraja T.N., Cooper K.L., Fine J.L., Sanfilippo J., Davidson N.E., et al. A role for histone H2B variants in endocrine-resistant breast cancer. Horm. Cancer. 2015;6:214–224. doi: 10.1007/s12672-015-0230-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwaya T., Fukagawa T., Suzuki Y., Takahashi Y., Sawada G., Ishibashi M., Kurashige J., Sudo T., Tanaka F., Shibata K., Endo F., et al. Contrasting expression patterns of histone mRNA and microRNA 760 in patients with gastric cancer. Clin. Cancer Res. 2013;19:6438–6449. doi: 10.1158/1078-0432.CCR-12-3186. [DOI] [PubMed] [Google Scholar]
- 46.Yan C., Zhang W., Shi X., Zheng J., Jin X., Huo J. miR-760 suppresses non-small cell lung cancer proliferation and metastasis by targeting ROS1. Environ. Sci. Pollut. Res. Int. 2018;25:18385–18391. doi: 10.1007/s11356-017-1138-0. [DOI] [PubMed] [Google Scholar]
- 47.Cao L., Liu Y., Wang D., Huang L., Li F., Liu J., Zhang C., Shen Z., Gao Q., Yuan W., Zhang Y. miR-760 suppresses human colorectal cancer growth by targeting BATF3/AP-1/cyclinD1 signaling. J. Exp. Clin. Cancer Res. 2018;37:83. doi: 10.1186/s13046-018-0757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q., Huang Z., Ni S., Xiao X., Xu Q., Wang L., Huang D., Tan C., Sheng W., Du X. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS ONE. 2012;7:e44398. doi: 10.1371/journal.pone.0044398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Q., Du W.W., Wu N., Yang W., Awan F.M., Fang L., Ma J., Li X., Zeng Y., Yang Z., et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y., et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y., Klausen C., Cheng J.C., Zhu H., Leung P.C. Activin A, B, and AB increase human trophoblast cell invasion by up-regulating N-cadherin. J. Clin. Endocrinol. Metab. 2014;99:E2216–E2225. doi: 10.1210/jc.2014-2118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.