Abstract
Porcine epidemic diarrhea virus (PEDV) is a reemerging Alphacoronavirus that causes lethal diarrhea in piglets. Coronavirus nonstructural protein 13 (nsp13) encodes helicase, which plays pivotal roles during viral replication by unwinding viral RNA. However, the biochemical characterization of PEDV nsp13 remains largely unknown. In this study, PEDV nsp13 was expressed in Escherichia coli and purified. The recombinant nsp13 possessed ATPase and helicase activities for binding and unwinding dsDNA/RNA substrates with 5′-overhangs, and Mg2+ and Mn2+ were critical for its ATPase and helicase activities. PEDV nsp13 also unwound dsDNA into ssDNA in the pH from 6.0–9.0, and used energy from all nucleoside triphosphates and deoxynucleoside triphosphates. Site-directed mutagenesis demonstrated that Lys289 (K289) of PEDV nsp13 was essential for its ATPase and helicase activities. These results provide new insights into the biochemical properties of PEDV nsp13, which is a potential target for developing antiviral drugs.
Keywords: Porcine epidemic diarrhea virus, nsp13, ATPase activity, Helicase activity
1. Introduction
Porcine epidemic diarrhea (PED) is caused by the PED virus (PEDV) and is characterized by vomiting, watery diarrhea, dehydration, and high morbidity and mortality in neonatal piglets (Pensaert and de Bouck, 1978; Coussement et al., 1982). The disease associated with PEDV was first observed in England in the late 1970s and then spread rapidly to other countries in Europe and Asia (Martelli et al., 2008). After the application of the CV777-derived strain vaccine, PED outbreaks became endemic in Asian countries. However, since the emergence of a highly virulent variant at the end of 2010, PEDV re-emerged in Thailand and Vietnam, and particularly in China. Later, a similar variant of PEDV was reported in the United States and quickly spread nationally within one year, resulting in the deaths of over 7 million piglets and huge economic losses (Cima, 2013; Schulz and Tonsor, 2015).
PEDV is an enveloped single-stranded positive-sense RNA virus, belonging to the family Coronaviridae, genus Alphacoronavirus (Cavanagh, 1997). The PEDV genome is approximately 28 kb and encodes two replicase polyproteins, pp1a and pp1ab, which are proteolytically cleaved into 16 mature nonstructural proteins (nsps) (Kocherhans et al., 2001). Among them, nsp3 (papain-like proteinase) and nsp5 (3C-like proteinase) mediate proteolysis, and nsp13 (helicase) and nsp12 (RNA-dependent RNA polymerase, RdRp) exert the conserved key replicative functions (van Hemert et al., 2008).
Helicases are motor proteins that use energy derived from nucleoside triphosphate (NTP) hydrolysis to unwind double-stranded nucleic acids into two single-stranded nucleic acids (Lohman, 1992; Singleton et al., 2007). Sequence conservation analysis indicates that coronavirus nsp13 belongs to helicase superfamily 1 (SF1) of the six helicase superfamilies, which includes many positive-sense RNA viral helicases. Coronavirus helicase is one of the most conserved proteins, making it a potential target for antiviral drug design. The betacoronavirus nsp13 has been biochemically characterized. The helicases from severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) were shown to unwind double-stranded (ds)RNA and dsDNA in a 5′-to-3′ direction, hydrolyze deoxynucleoside triphosphates (dNTPs) and NTPs, and have RNA 5′-triphosphatase activity (Tanner et al., 2003; Ivanov et al., 2004; Adedeji and Lazarus, 2016a). The NTPase and RNA helicase activities of SARS-CoV-2 nsp13 have also been determined recently (Shu et al., 2020). However, the biochemical characterization of nsp13 from the Alphacoronavirus, PEDV, remains largely unknown.
In this study, we demonstrated that PEDV nsp13 possessed strong ATPase activity that could hydrolyze adenosine triphosphate (ATP) in the presence of Mg2+ and Mn2+. The PEDV nsp13 could bind both dsRNA and dsDNA and unwind the substrates in a 5′-to-3′ direction using energy from the hydrolysis of all NTPs. We also demonstrated that lysine 289 (K289) of PEDV nsp13 was critical for its ATPase and helicase activities.
2. Materials and methods
2.1. Virus and cells
PEDV strain AJ1102 (GenBank accession no. JX188454.1), isolated from a suckling piglet with acute diarrhea in China in 2011 (Bi et al., 2012), was used in this study. Vero cells were cultured and maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10 % heat-inactivated fetal bovine serum at 37 °C in a humidified 5 % CO2 incubator.
2.2. Plasmid construction
Viral RNA was extracted from PEDV-infected Vero cells using TRIzol reagent (Omega Bio-Tek), then reverse-transcribed to cDNA. The full-length cDNA of PEDV nsp13 was amplified via PCR using the primers, 5′-CGGCCATGGGCCATCATCATCATCATCACTCTGCAGGGCTTTGTGTT-3′ and 5′-CGGCTCGAGTTACTGCAAATCAGACAATT-3′, then cloned into the prokaryotic expression vector, pET-28a, to generate the prokaryotic expression plasmid, pET-28a-nsp13. The nsp13-mutant K289A was generated via PCR mutagenesis and used to construct the prokaryotic expression plasmid, pET-28a-nsp13-K289A. All constructions were verified by DNA sequencing.
2.3. Expression and purification of recombinant proteins
pET-28a-nsp13 and pET-28a-nsp13-K289A were transformed into Transetta (DE3) cells (TransGen Biotech, China), grown at 37℃, and induced with IPTG (0.2 mM) when the optical density reached ∼0.8. Thereafter, the induced cells were grown at 18℃ for 16 h. Cells were harvested at 15,000×g by centrifugation at 4℃. After ultrahigh-pressure disruption and centrifugation at 20,000×g, the supernatants were filtered through a 0.45-μM filter (Millipore, MA, USA), then run through a Ni-affinity column. The proteins were then eluted with a linear-gradient concentration of imidazole from 20 to 250 mM. The eluates were concentrated, and the buffers were replaced with Buffer B2 (0.2 M Tris−HCl, 200 mM NaCl) using Amicon Ultra-15 filters (Millipore). The concentration of purified protein was determined using an enhanced BCA protein assay kit (Beyotime, China).
2.4. SDS-PAGE and Western blot analysis
Protein samples were separated on a 10 % SDS-PAGE gel, then stained with Coomassie brilliant blue or transferred to a polyvinylidene fluoride membrane (Millipore). The membranes were blocked with 5 % skim milk and immunoblotted with an anti-His monoclonal antibody (Cell Signaling Technology, #2366).
2.5. ATPase activity assay
The Kinase-Glo Plus Luminescent Kinase Assay kit (Promega, USA) was used to detect the ATPase activity of PEDV nsp13 or its mutant nsp13-K289A. Briefly, the purified recombinant proteins in reaction buffer (40 mM Tris−HCl at pH 7.5, 50 mM NaCl, 2 mM Mg2+, 200 μM ATP) were added to a 96-well black plate (Jet Bio-Filtration, Guangzhou, China) with deionized water to a total volume of 50 μL. The plate was then incubated in a 37 °C incubator for the indicated times. At the end of the reaction, 50 μL of Kinase-Glo reagent was added to the reaction mixture. After mixing and incubation at room temperature for 2 min, the luminescence of each well was measured via 1450 MicroBeta Trilux (Perkin Elmer, Waltham, MA, USA).
2.6. Nucleic acid substrates
Oligonucleotides with Cy5-labeled 5′-DNA and 5′-RNA were synthesized by TSINGKE (China). Table 1 lists the oligonucleotide sequences. The dsDNA duplexes with 5′- or 3′-overhangs were generated by annealing DNA* with 5′-DNA or 3′-DNA, respectively, at a ratio of 1:1.2 in the reaction mixture (50 mM Tris, pH 8.0, 50 mM NaCl). The mixture was heated at 95 °C for 5 min, then slowly cooled to RT. Similarly, 5′- and 3′-overhang dsRNA duplexes were produced using RNA* with 5′-RNA and 3′-RNA, respectively.
Table 1.
Oligonucleotide sequences used in this study.
| 5′-3′ | Label | |
|---|---|---|
| DNA* | GCTAGTCACTGTTCGAGCACCA | 5′-Cy5 |
| 3′-DNA | TAGATAGCCATAGATAGCATTGGTGCTCGAACAGTGACTAGC | |
| 5′-DNA | TGGTGCTCGAACAGTGACTAGCTAGATAGCCATAGATAGCAC | |
| RNA* | GCUAGUCACUGUUCGAGCACCA | 5′-Cy5 |
| 3′-RNA | UAGAUAGCCAUAGAUAGCAUUGGUGCUCGAACAGUGACUAGC | |
| 5′-RNA | UGGUGCUCGAACAGUGACUAGCUAGAUAGCCAUAGAUAGCAC |
2.7. Helicase activity assay
An electrophoretic mobility shift assay (EMSA) was conducted to evaluate the capacities of nsp13 and nsp13-K289A to bind and unwind the nucleic acid duplexes. To determine the binding activity, recombinant proteins were incubated with nucleic acid duplex substrates in a reaction buffer containing 20 mM HEPES at pH 7.5, 50 mM NaCl, and 5 mM Mg2+. After incubating for 30 min, 5× loading buffer (25 % glycerol and 100 mM HEPES) was added to terminate the reaction, and the mixture was prepared for electrophoretic mobility. Helicase activity was measured by incubating 20 nM of recombinant proteins with 0.3 μM of dsRNA/DNA substrates in a reaction buffer containing 20 mM HEPES at pH 7.5, 20 mM NaCl, 1 mM dithiothreitol, 0.1 mg/ml bovine serum albumin (BSA), 2 mM Mg2+, and 2 mM ATP at 30 °C for the indicated time. Reactions were quenched by adding an equal volume of loading buffer (0.2 % SDS and 20 % glycerol). EMSA was performed by electrophoresing the mixtures on 6 % or 8 % nondenaturing PAGE, then running them in buffer containing 89 mM Tris-borate at pH 8.2 at 4 °C with 110 V. The controls for measuring the maximum unwinding activity were dsRNA/DNA duplexes denatured by heat for 5 min at 95 °C. Images were obtained by scanning gels with the FLA-5000 imaging system (Fujifilm).
3. Results
3.1. Expression and purification of PEDV nsp13
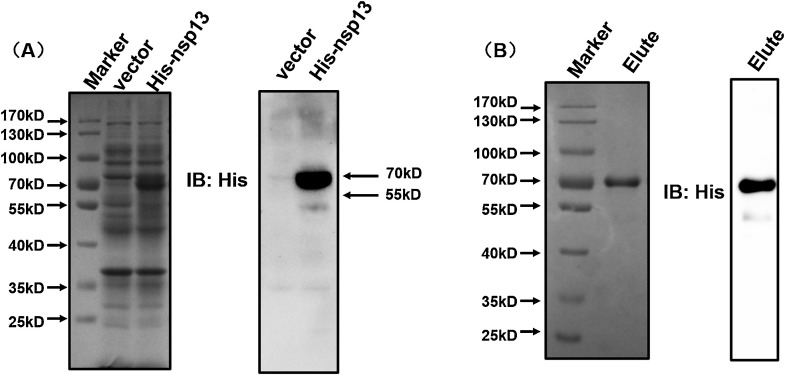
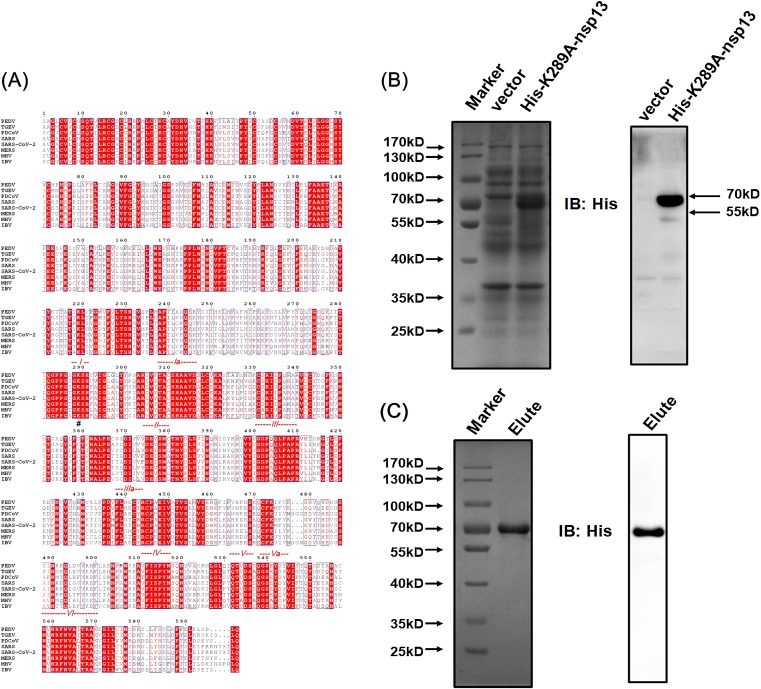
To investigate the biochemical activities of PEDV nsp13, recombinant His-nsp13 fusion protein was expressed using an Escherichia coli prokaryotic expression system and analyzed by SDS-PAGE and western blot. His-nsp13 fusion protein was expressed at the expected mass of 70 kD (Fig. 1 A). The expressed His-nsp13 in the supernatants was efficiently purified through Ni-affinity column chromatography. The purified His-nsp13 was also confirmed by SDS-PAGE and western blot (Fig. 1B). It should be noted that there exists an additional band lower than 55 kD in the western blot images (Fig. 1A and B). Because this additional band could be observed in both the expressed samples and the purified samples, we speculated that this additional band may be a degradation product of nsp13 protein.
Fig. 1.
Expression and purification of recombinant PEDV nsp13. The expressed (A) or purified (B) recombinant PEDV nsp13 was analyzed by electrophoresis on a 10 % SDS-PAGE gel and stained with Coomassie brilliant blue (left) or subjected to western blot with anti-His antibody (right). Elute indicates the affinity chromatography elusions of His-tagged nsp13.
3.2. ATPase activity of PEDV nsp13
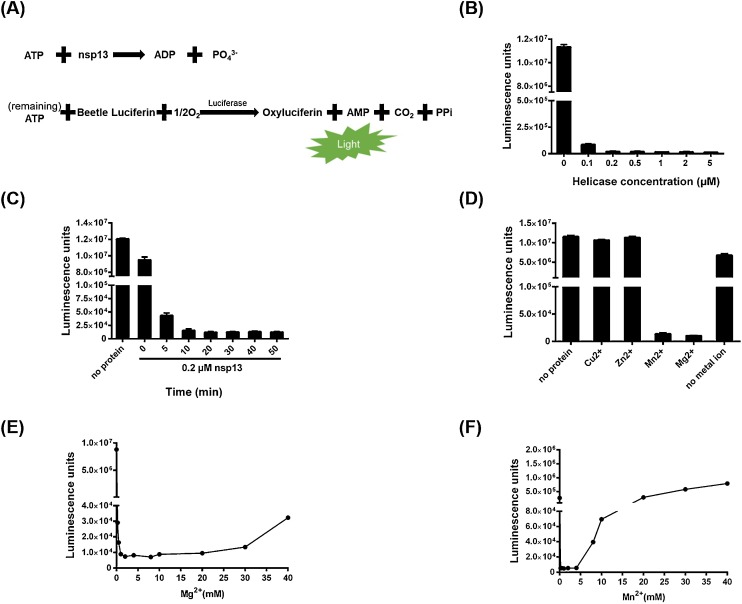
Because nucleic acid unwinding by helicase is energy-dependent, and ATP is the major energy resource in cells, we first detected the ATPase activity of PEDV nsp13 using a Kinase-Glo Plus Luminescent Kinase Assay kit, which measures the released luciferase-catalyzed photon-emitting luminescence to determine the remaining ATP in the reaction mixture (Fig. 2 A). The amount of ATP remaining in the reaction mixture sharply decreased as the His-nsp13 concentration increased (0, 0.1, 0.2, 0.5, 1, 2, and 5 μM; Fig. 2B). The nsp13 rapidly hydrolyzed the ATP. When nsp13 was added, the ATP immediately decreased and reached the bottom level after 10 min (Fig. 2C).
Fig. 2.
ATPase activity of PEDV nsp13. (A) Detection of the ATPase activity of PEDV nsp13 using the Kinase-Glo Plus Luminescent Kinase Assay kit. (B) Purified nsp13 recombinant proteins (0, 0.1, 0.2, 0.5, 1, 2, or 5 μM) were incubated with ATP in reaction buffer at 37℃ for 10 min, then Kinase-Glo reagent mix was added, and the ATPase activity was measured. (C) PEDV nsp13 (0.2 μM) was reacted with ATP at 37℃ for 5, 10, 20, 30, 40, or 50 min., then the ATPase activity was measured. (D) PEDV nsp13 (0.2 μM) was incubated in reaction buffer in the presence of MgCl2, MnCl2, CuCl2, or ZnCl2, then the ATPase activity was measured. (E, F) PEDV nsp13 (0.2 μM) was incubated in reaction buffer containing the indicated concentrations of MgCl2(E) or MnCl2(F), then the ATPase activity was measured.
To evaluate the metal requirements of nsp13 for ATPase activity, His-nsp13 was incubated with ATP (100 μM) in the presence of 2 mM of MgCl2, MnCl2, CuCl2, or ZnCl2. The luminescence measurements showed that nsp13 reached its optimal ATPase activity in the presence of MgCl2, followed by MnCl2, with no observable ATPase activity with CuCl2 or ZnCl2 (Fig. 2D). Furthermore, nsp13 could more efficiently hydrolyze ATP at 1–20 mM MgCl2 and 0.25−4 mM MnCl2; however, high (>20 mM) and low (<0.5 mM) concentrations of MgCl2, and high (>4 mM) concentrations of MnCl2 abolished the ATPase activity (Fig. 2E and F). These data indicate that nsp13 harbors ATPase activity and that Mg2+ and Mn2+ are critical for this activity.
3.3. Helicase activity of PEDV nsp13
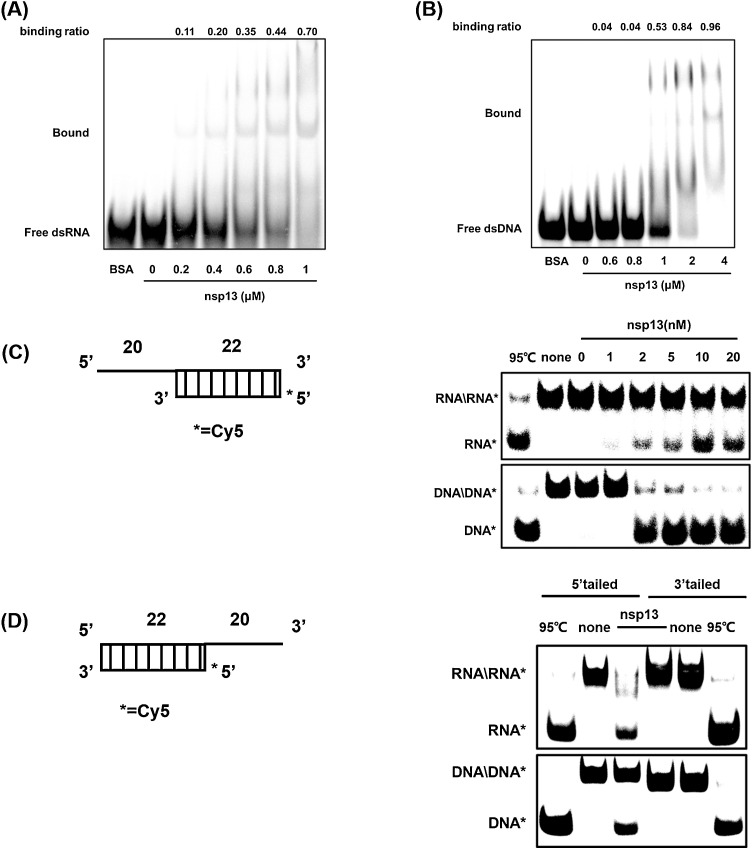
Previous studies reported that nidovirus helicases unwind in a 5′-to-3′ direction; thus, unless stated otherwise, 5′-overhang 20-nt dsRNA/dsDNA was used to analyze the helicase activity. To determine the helicase activity of His-nsp13, nucleic acid-binding and helix-unwinding assays were performed. The nucleic acid-binding assay was performed by incubating different concentrations of His-nsp13 with 0.3 μM of dsRNA/dsDNA for 30 min in binding-reaction buffer, and BSA was used as a negative control. His-nsp13 bound both dsRNA and dsDNA dose-dependently (Fig. 3 A and B). To monitor the helix-unwinding activity, 1, 2, 5, 10, and 20 nM of His-nsp13 were incubated with dsRNA/dsDNA substrates in a standard unwinding reaction mixture. The dsRNA/dsDNA reaction mixture at 95℃ was used as a positive control, and the reaction mixture without His-nsp13 was used as a negative control. Cy5-labeled single-strand RNA and DNA were efficiently released from the dsRNA/dsDNA substrate, respectively (Fig. 3C). The unwinding ability of the nsp13 3′-overhang 20-nt dsRNA/dsDNA was also tested. As expected, no Cy5-labeled single-strand RNA or DNA was observed (Fig. 3D). Thus, PEDV nsp13 possesses dsRNA/DNA unwinding activity with a 5′-to-3′ directionality, which is consistent with other coronavirus helicases.
Fig. 3.
Helicase activity of PEDV nsp13. (A, B) dsRNA (A) or dsDNA (B) substrates (0.3 μM) were incubated with PEDV nsp13 at the indicated concentrations for 30 min at room temperature, then loaded on 6 % nondenaturing PAGE at 4℃ for 60 min. Pictures were acquired via the FLA-5000 imaging system and the binding ratios were quantified by grayscale analysis with Image J. (C–D) Schematics of the nucleic acid substrates 5′-overhang (C) and 3′-overhang (D) dsRNA/DNA (20 ss, 22 ds) helix substrates. 5′-overhang dsRNA/DNA (C) or 3′-overhang dsRNA/DNA (D) substrates (0.3 μM) were incubated with the indicated concentrations of PEDV nsp13 in reaction buffer at 30℃ for 10 min, then loaded on 8 % nondenaturing PAGE and visualized using an FLA-5000 imaging system. RNA* = 5′-Cy5-ssRNA; DNA* = 5′-Cy5-ssDNA.
3.4. Metal, energy, and pH requirements for PEDV nsp13 helicase activity
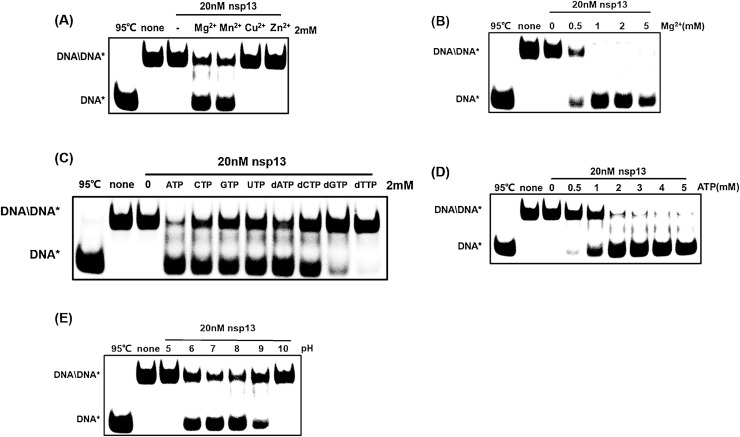
As shown in Fig. 3C and D, PEDV nsp13 has a stronger capacity to unwind dsDNA helices than dsRNA helices. Considering that the functions of coronavirus nsp13 to winding dsRNA had been reported in previous studies on other coronaviruses (Tanner et al., 2003; Ivanov et al., 2004; Adedeji and Lazarus, 2016; Shu et al., 2020), and that the advantages of dsDNA with simple manipulation and good dependability, we focused on the function of PEDV nsp13 to unwind 5′-overhang dsDNA substrates in the subsequent experiments. To evaluate the metal dependence of nsp13 helicase activity, His-nsp13 (20 nM) was reacted with 5′-overhang-dsDNA (0.3 μM) in the presence of 2 mM of MgCl2, MnCl2, CuCl2, or ZnCl2 for 10 min. Similar to the results of the ATPase activity analysis, MgCl2 or MnCl2 were required for nsp13 helicase activity (Fig. 4 A). The accumulation of unwound ssDNA was augmented as the MgCl2 concentration increased (≤2 mM) and decreased when the MgCl2 concentration reached 5 mM (Fig. 4B), indicating that His-nsp13 has optimal helicase activity with 2 mM of MgCl2. NTP hydrolysis provides energy for helicase translocation along ssRNA/DNA and duplex-RNA/DNA structures during unwinding. Therefore, we further investigated the activity of nsp13 to unwind dsDNA, where energy is derived from four types of NTPs. nsp13 could use all NTPs and dNTPs as energy sources but showed a preference for ATP (Fig. 4C). Furthermore, increasing the ATP dose-dependently promoted the efficiency of nsp13 to unwind the dsDNA helix (Fig. 4D). Additionally, the optimal pH for nsp13 unwinding activity was investigated in reactions similar to the aforementioned reactions but at pH 5, 6, 7, 8, 9, or 10. His-nsp13 efficiently unwound the DNA helix substrate at pHs 6–9, with higher efficiencies at pH 7 and 8 (Fig. 4E).
Fig. 4.
Influences of metal, energy and pH on PEDV nsp13 unwinding activity. 5′-overhang dsDNA substrates were incubated with nsp13 in reaction buffer containing 2 mM of MgCl2, MnCl2, CuCl2, or ZnCl2(A) or increasing concentrations (0, 0.5, 1, 2, or 5 mM) of MgCl2(B), or 2 mM of NTPs/dNTPs (C), or increasing concentrations (0, 0.5, 1, 2, 3, 4, or 5 mM) of ATP (D), or at the indicated pH (E) at 30℃ for 10 min, then loaded on 8 % nondenaturing PAGE and visualized using an FLA-5000 imaging system.
3.5. K289 of PEDV nsp13 is a key amino acid for its ATPase and helicase activities
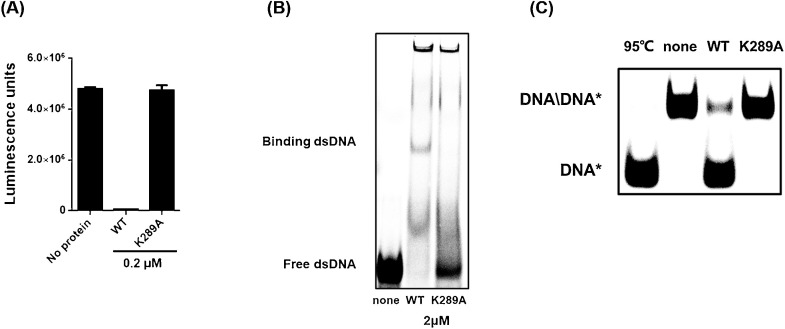
To identify the key amino acid that exerts ATPase and helicase functions, the PEDV nsp13 amino acid sequence was aligned with the nsp13s of some representative coronaviruses from different genera, including porcine transmissible gastroenteritis virus (TGEV) strain Purdue 46, porcine deltacoronavirus (PDCoV) isolate CHN-HN-2014, SARS-CoV isolate Frankfurt 1, MERS-CoV isolate Al-Hasa_7a_2013, murine hepatitis virus (MHV) strain JHM, avian infectious bronchitis virus (IBV) strain Beaudette, and SARS-CoV-2 isolate Wuhan-Hu-1. The nsp13 s from all tested coronaviruses shared conserved helicase motifs I–VI, including the K289 in motif I, which has been recognized as the key site for NTP binding and ATPase function in other coronaviruses such as SARS-CoV and MERS-CoV (Walker et al., 1982; Lehmann et al., 2015; Adedeji and Lazarus, 2016; Jia et al., 2019). To confirm the role of K289 in the ATPase and helicase function of PEDV nsp13, the nsp13-mutant K289A was constructed in which the K residue was replaced with alanine (A). The recombinant His-nsp13-K289A protein was expressed in the E. coli expression system as was His-nsp13. Recombinant His-nsp13-K289A expression and purification were confirmed by SDS-PAGE and western blot analyses (Fig. 5 B and 5C). Subsequently, ATPase, duplex DNA-binding, and DNA helix-unwinding assays were performed. Compared with the wild-type (WT) nsp13, K289A mutation almost completely abolished the ATP hydrolysis activity (Fig. 6 A), the duplex DNA-binding ability (Fig. 6B) and the unwinding ability (Fig. 6C), verifying that K289 is essential for PEDV nsp13 ATPase and helicase activities.
Fig. 5.
Expression and purification of the nsp13-K289A mutant. (A) Sequence alignments of nsp13 from different coronaviruses. Alignment was generated in ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi), and the nsp13 sequences of transmissible gastroenteritis virus (TGEV) strain Purdue 46 (accession number AJ271965), porcine deltacoronavirus (PDCoV) isolate CHN-HN-2014 (accession number KT336560), SARS-CoV isolate Frankfurt 1 (accession no. AY291315), MERS-CoV isolate Al-Hasa_7a_2013 (accession number KF600655), murine hepatitis virus (MHV) strain JHM (accession number AC_000192), avian infectious bronchitis virus (IBV) strain Beaudette (accession number NC_001451.1) and SARS-CoV-2 isolate Wuhan-Hu-1 (accession number NC_045512.2) were derived from GenBank. The red dotted lines indicate conserved helicase motifs I–VI. “#” indicates the conserved K289 residue. (B, C) The expressed (B) or purified (C) nsp13-K289A recombinant proteins were analyzed by electrophoresis on 10 % SDS-PAGE, then stained with Coomassie brilliant blue (left) or western blotted with anti-His antibody (right).
Fig. 6.
ATPase and helicase activity of nsp13-K289A. (A) wild-type (WT) nsp13 or mutant nsp13-K289A proteins were incubated with ATP in reaction buffer at 37℃ for 10 min, then the ATPase activity was measured. (B) 0.3 μM of dsDNA substrates and 2 μM of WT nsp13 or nsp13-K289A proteins were mixed for 30 min at room temperature, then loaded into 6 % nondenaturing PAGE at 4℃ and run for 60 min. BSA was set as the negative control. (C) 0.3 μM of 5′-overhang dsDNA and 20 nM of WT nsp13 or nsp13-K289A proteins were mixed in reaction buffer at 30℃ for 10 min, then loaded on 8 % nondenaturing PAGE at 4℃ and run for 45 min.
4. Discussion
Viral helicases are involved in multiple parts of the infection process, including replication, transcription, translation, and encapsidation (Kadaré and Haenni, 1997; Bleichert and Baserga, 2007; Musier-Forsyth, 2010). During RNA replication among RNA viruses, helicase must efficiently unwind viral dsRNA replicative intermediates to release nascently synthesized progeny viral RNAs from template RNAs (Yang et al., 2014; Xia et al., 2015). PEDV nsp13 and other coronavirus nsp13 s belong to a small group of helicase families and share some conserved functions, including NTPase, dNTPase and 5′-to-3′ RNA and DNA duplex-unwinding activities, for which Mg2+ or Mn2+ are critical. In this study, PEDV nsp13 with a high concentration of Mg2+ and Mn2+ exhibited inhibitory ATPase activity for unknown reasons. Additionally, PEDV nsp13 had a stronger ability to unwind DNA than to unwind RNA in vitro. Thus, we hypothesized that, in addition to its role in viral RNA replication, PEDV nsp13 might act on host cell DNA. No preference for either RNA or DNA was observed in previous studies on other coronavirus nsp13 s. A previous study showed that SARS-CoV nsp12 (RNA polymerase) and nsp13 interact biophysically, enhancing the helicase activity of SARS-CoV nsp13 (van Hemert et al., 2008). More recently, the structure of the SARS-CoV-2 replication-transcription complex was determined, suggesting a possible role of nsp13 in generating backtracked replication-transcription complexes for proofreading or template switching during subgenomic RNA transcription or both. During viral RNA synthesis, nsp13 and nsp12 work together, and nsp12 must transcribe RNA in a 3′-to-5′ template direction opposite that of nsp13 (Chen et al., 2020). Whether PEDV nsp13 interacts with nsp12 and works synergistically during PEDV infection requires further study.
The nsp13 helicase is highly conserved among CoVs, and sequence alignment has shown that PEDV nsp13 has a similar core structure consisting of multiple functional domains, i.e., the N-terminal zinc-binding (1–113 aa), RecA1 (242–442 aa), and RecA2 (443–597 aa) domains. RecA1 contains motifs I, Ia, II, and III, and RecA2 includes motifs IV, V, and VI. Previous mutagenesis studies of nsp13 CoVs have identified several residues essential to the activity. K289 in the RecA1 domain of SARS-CoV nsp13 plays an essential role in ATPase and helicase activities (Ivanov et al., 2004; Jia et al., 2019). In the current study, K289 in PEDV nsp13 was also verified as being important for ATPase and helicase activities. We attempted to generate recombinant PEDV with a K289A mutation in nsp13 via the infectious clone of PEDV strain AJ1102 (Peng et al., 2020), the recombinant PEDV with a K289A mutation could not be successfully rescued, further demonstrating that K289 is essential for nsp13 helicase activity and is pivotal for viral survival.
In addition to the ATPase and helicase activities for viral replication, some viral helicases can regulate the host’s innate immune response. For example, the helicase domain of the West Nile virus nsp3 plays a role in inhibiting type I interferon signaling (Setoh et al., 2017), and SARS-CoV-2 helicase potently suppresses primary interferon production and interferon signaling (Yuen et al., 2020). Additionally, interaction between the host helicase/polymerase and viral helicase reportedly promotes viral replication. For example, the interaction between porcine reproductive and respiratory syndrome virus (PRRSV) helicase (nsp10) and host DEAD-box RNA helicase 18 can promote PRRSV replication (Jin et al., 2017) and the interaction of IBV helicase nsp13 and the host DNA polymerase subunit δ is conducive to viral replication (Xu et al., 2011). The role of PEDV nsp13 in regulating type I interferon production is being investigated in our laboratory.
5. Conclusions
In summary, we expressed and purified PEDV nsp13 and revealed its biochemical characteristics: (1) PEDV nsp13 possesses ATPase and helicase activities for unwinding dsDNA or dsRNA with 5′-to-3′ directionality, and Mg2+ or Mn2+ are indispensable for both functions; (2) PEDV nsp13 exhibits better helicase activity with the NTPs than with the dNTPs, especially with ATP, at pHs 6–9; (3) K289 of PEDV nsp13 is essential for its ATPase and helicase activities. These findings extend our knowledge of the key viral replicative enzyme of PEDV and suggest that nsp13 is a valuable target for designing antiviral drugs against PEDV.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
This work is supported by the National Key R&D Plan of China (2016YFD0500103) and the National Natural Science Foundation of China (318607804, 31672569).
References
- Adedeji A.O., Lazarus H. Biochemical characterization of middle east respiratory syndrome coronavirus helicase. mSphere. 2016;1:e00235–16. doi: 10.1128/mSphere.00235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J., Zeng S., Xiao S., Chen H., Fang L. Complete genome sequence of porcine epidemic diarrhea virus strain AJ1102 isolated from a suckling piglet with acute diarrhea in China. J. Virol. 2012;86:10910–10911. doi: 10.1128/JVI.01919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F., Baserga S.J. The long unwinding road of RNA helicases. Mol. Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cima G. Viral disease affects U.S. pigs: porcine epidemic diarrhea found in at least 11 states. J. Am. Vet. Med. Assoc. 2013;243:30–31. [PubMed] [Google Scholar]
- Coussement W., Ducatelle R., Debouck P., Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets. I. Histological and histochemical study. Vet. Pathol. 1982;19:46–56. doi: 10.1177/030098588201900108. [DOI] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Zhou L., Ge X., Zhang H., Zhang R., Wang C., Wang L., Zhang Z., Yang H., Guo X. Cellular DEAD-box RNA helicase 18 (DDX18) promotes the PRRSV Replication via Interaction with Virus nsp2 and nsp10. Virus Res. 2017;238:204–212. doi: 10.1016/j.virusres.2017.05.028. [DOI] [PubMed] [Google Scholar]
- Kadaré G., Haenni A.L. Virus-encoded RNA helicases. J. Virol. 1997;71:2583–2590. doi: 10.1128/JVI.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23:137–144. doi: 10.1023/a:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K.C., Snijder E.J., Posthuma C.C., Gorbalenya A.E. What we know but do not understand about nidovirus helicases. Virus Res. 2015;202:12–32. doi: 10.1016/j.virusres.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman T.M. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol. Microbiol. 1992;6:5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- Martelli P., Lavazza A., Nigrelli A.D., Merialdi G., Alborali L.G., Pensaert M.B. Epidemic of diarrhoea caused by porcine epidemic diarrhoea virus in Italy. Vet. Rec. 2008;162:307–310. doi: 10.1136/vr.162.10.307. [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K. RNA remodeling by chaperones and helicases. RNA Biol. 2010;7:632–633. doi: 10.4161/rna.7.6.14467. [DOI] [PubMed] [Google Scholar]
- Peng Q., Fang L., Ding Z., Wang D., Peng G., Xiao S. Rapid manipulation of the porcine epidemic diarrhea virus genome by CRISPR/Cas9 technology. J. Virol. Methods. 2020;276 doi: 10.1016/j.jviromet.2019.113772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/bf01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz L.L., Tonsor G.T. Assessment of the economic impacts of porcine epidemic diarrhea virus in the United States. J. Anim. Sci. 2015;93:5111–5118. doi: 10.2527/jas.2015-9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoh Y.X., Periasamy P., Peng N.Y.G., Amarilla A.A., Slonchak A., Khromykh A.A. Helicase Domain of West Nile Virus NS3 Protein Plays a Role in Inhibition of Type I Interferon Signalling. Viruses. 2017;9:326. doi: 10.3390/v9110326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T., Huang M., Wu D., Ren Y., Zhang X., Han Y., Mu J., Wang R., Qiu Y., Zhang D.-Y., Zhou X. SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities That Can Be Inhibited by Bismuth Salts. Virol. Sin. 2020;35:321–329. doi: 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S.M., Poon L.L.M., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5’ to 3’ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert M.J., van den Worm S.H.E., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Wang P., Wang G.C., Yang J., Sun X., Wu W., Qiu Y., Shu T., Zhao X., Yin L., Qin C.F., Hu Y., Zhou X. Human enterovirus nonstructural protein 2CATPase functions as both an RNA helicase and ATP-Independent RNA chaperone. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.H., Huang M., Fang S.G., Liu D.X. Coronavirus infection induces DNA replication stress partly through interaction of its nonstructural protein 13 with the p125 subunit of DNA polymerase δ. J. Biol. Chem. 2011;286:39546–39559. doi: 10.1074/jbc.M111.242206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Cheng Z., Zhang S., Xiong W., Xia H., Qiu Y., Wang Z., Wu F., Qin C.-F., Yin L., Hu Y., Zhou X. A cypovirus VP5 displays the RNA chaperone-like activity that destabilizes RNA helices and accelerates strand annealing. Nucleic Acids Res. 2014;42:2538–2554. doi: 10.1093/nar/gkt1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H., Cai J.P., Jin D.Y., To K.K., Chan J.F., Yuen K.Y., Kok K.H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]