Abstract
The H/ACA small nucleolar RNAs (snoRNAs) are involved in pseudouridylation of pre-rRNAs. In the yeast Saccharomyces cerevisiae, four common proteins are associated with H/ACA snoRNAs: Gar1p, Cbf5p, Nhp2p, and Nop10p. In vitro reconstitution studies showed that four proteins also specifically interact with H/ACA snoRNAs in mammalian cell extracts. Two mammalian proteins, NAP57/dyskerin (the ortholog of Cbf5p) and hGAR1, have been characterized. In this work we describe properties of hNOP10 and hNHP2, human orthologs of yeast Nop10p and Nhp2p, respectively, and further characterize hGAR1. hNOP10 and hNHP2 complement yeast cells depleted of Nhp2p and Nop10p, respectively. Immunoprecipitation experiments with extracts from transfected HeLa cells indicated that epitope-tagged hNOP10 and hNHP2 specifically associate with hGAR1 and H/ACA RNAs; they also interact with the RNA subunit of telomerase, which contains an H/ACA-like domain in its 3′ moiety. Immunofluorescence microscopy experiments showed that hGAR1, hNOP10, and hNHP2 are localized in the dense fibrillar component of the nucleolus and in Cajal (coiled) bodies. Deletion analysis of hGAR1 indicated that its evolutionarily conserved core domain contains all the signals required for localization, but progressive deletions from either the N or the C terminus of the core domain abolish localization in the nucleolus and/or the Cajal bodies.
Synthesis, maturation, and also packaging of rRNAs into ribosomal particles in eukaryotes take place in the nucleolus. The 18S, 5.8S, and 25/28S rRNAs are generated from a single large precursor RNA by a complex series of endonucleolytic and exonucleolytic processing steps. In addition, rRNAs undergo extensive modifications: 2′-hydroxyl groups of many riboses are methylated, and specific uridines are converted into pseudouridines (reviewed in references 1, 51, 69, and 74). Cleavage and modification events are assisted by a large number of small nucleolar RNAs (snoRNAs), which function in the cell in the form of small nucleolar ribonucleoprotein particles (snoRNPs) (reviewed in references 1, 54, 72, 78, and 81). Two major classes of snoRNAs, box C/D and box H/ACA, have been identified based on conserved structural elements (3, 19, 73). Most C/D snoRNAs act as guides for the 2′-O-methylation of pre-rRNAs, whereas most H/ACA snoRNAs direct the site-specific pseudouridylation of pre-rRNAs, and each class is associated with specific proteins (10, 18, 39, 61; reviewed in references 1, 43, 72, 74, and 78). RNase MRP, an RNP involved in the endonucleolytic cleavage of pre-rRNA in yeast (references 13 and 50 and references therein), is structurally similar to RNase P (11, 16) and does not belong to either of the families.
Four common proteins associated with H/ACA snoRNAs have been identified in Saccharomyces cerevisiae: Gar1p (25 kDa), Cbf5p (65 kDa), Nhp2p (22 to 25 kDa), and Nop10p (10 kDa). All four are essential for growth; depletion of any of them causes inhibition of 18S rRNA production and impairs pseudouridylation of rRNA (6, 23, 29, 31, 44, 77). Gar1p and Nhp2p orthologs in the fission yeast Schizosaccharomyces pombe have also been described (24, 52). In vitro reconstitution studies with HeLa cell extracts showed that four proteins specifically interact with mammalian H/ACA snoRNAs (15). Two mammalian proteins have been characterized: NAP57/dyskerin, which is the ortholog of Cbf5p (27, 56, 58, 80), and hGAR1 (15).
Yeast Gar1p is characterized by the presence of glycine- and arginine-rich (GAR) domains flanking a highly conserved central core domain, which is necessary and sufficient for localization and function of the protein in vivo (23, 24, 25, 77). Although Gar1p has been shown to interact with snR10 and snR30 RNAs in vitro (2), it does not appear to be a primary binding protein. In yeast, depletion of Gar1p does not affect accumulation of H/ACA snoRNAs (6, 23). Moreover, in the mammalian in vitro reconstitution system, the presence of hGAR1 does not appear to be a prerequisite for the assembly of other H/ACA proteins (15). In contrast to depletion of Gar1p, depletion of Cbf5p affects the accumulation of H/ACA snoRNAs and also Gar1p (44). Cbf5p has sequence similarity with pseudouridine synthases involved in tRNA and bacterial rRNA modifications, which strongly suggests that it may represent the pseudouridine synthase of H/ACA snoRNPs (42, 44, 82). Orthologs of Cbf5p have been identified in rats (NAP57), humans (dyskerin), and flies (MFL or Nop60B) (22, 27, 56, 64). Mutations in the associated gene result in growth and developmental defects in Drosophila melanogaster (22), whereas in humans they cause dyskeratosis congenita, an X-linked recessive disease associated with bone marrow failure, skin defects, chromosome instability, and a predisposition to certain types of malignancy (27, 40). NAP57 was shown to interact with the phosphoprotein Nopp140, which is present in both the nucleolus and the Cajal (coiled) bodies (CBs) and which shuttles between the nucleolus and the cytoplasm on intranuclear tracks (30, 55, 80).
Nhp2p and Nop10p are two other core components of the yeast H/ACA snoRNPs. As for Cbf5p, their genetic depletion interferes with accumulation of H/ACA snoRNAs and Gar1p. Both proteins are highly conserved nucleolar proteins (29, 77). Interestingly, Nhp2p contains a putative RNA-binding domain shared by some spliceosomal and ribosomal proteins (29, 41, 52, 62, 70, 77).
The observation that mammalian telomerase RNA contains a 3′-terminal domain that structurally resembles H/ACA snoRNAs (57) has greatly stimulated interest in H/ACA snoRNPs and their components. Telomerase is a ribonucleoprotein enzyme responsible for maintaining telomeric ends of the chromosomes. The enzyme uses a short internal region of its RNA subunit as a template to synthesize sequence repeats that form telomeres (reviewed in references 4, 5, and 14). Mitchell et al. (57) have shown that the H/ACA domain is required for the accumulation and function of human telomerase RNA (hTR) in vivo. Moreover, both dyskerin and hGAR1 are associated with hTR in vivo (15, 58), and dyskerin was found to be a component of active telomerase (58). hGAR1 can also associate with hTR in vitro (15). It has been suggested that decreased telomerase activity, and not a failure in rRNA processing and/or pseudouridylation, is the main factor contributing to the development of dyskeratosis congenita (58).
In this work we describe the characterization of hNHP2 and hNOP10, the human orthologs of yeast Nhp2 and Nop10 proteins, and further analysis of hGAR1. Both hNHP2 and hNOP10 are functionally conserved proteins that can complement yeast cells depleted of Nhp2p or Nop10p. Immunoprecipitation experiments indicate that hNHP2 and hNOP10 specifically associate with hGAR1 and H/ACA snoRNAs. They also associate with hTR, as previously shown for dyskerin and hGAR1. Tagged hNHP2 and hNOP10 expressed in HeLa cells colocalize with hGAR1 in the dense fibrillar components (DFC) of nucleoli and in CBs. Finally, we demonstrate that short conserved sequences at the N and C termini of the hGAR1 core domain are essential for localization of the protein in HeLa cells.
MATERIALS AND METHODS
Cloning of hNHP2 and hNOP10 cDNAs.
Expressed sequence tag (EST) sequences encoding putative human orthologs of Nhp2p were identified in BLAST searches using the yeast Nhp2p sequence as a query. 5′ and 3′ rapid amplifications of cDNA ends (RACE) were performed on total RNA isolated from HeLa cells to determine the complete sequence of hNHP2 cDNA. Three different hNHP2-specific oligonucleotides, GGGCAGTGTGTCTCCTGCCAAAACC, CTGAACCTCTTTCACCCCGCGCCG, and CCGAATCTGCTTCTGCTTCACGC, were used for 5′ RACE, and primer GTCTATATCCCCTCTAAGACGGACCTGGG was used for the 3′ RACE, in accordance with the manufacturer's instructions (Boehringer Mannheim). Primers specific for the 5′ (CCGGAATTCATGACCAAAATAAAGGCAGATCCCGAC) and 3′ (GCGGAATTCTCATAGGGGTAGCAGGGAC) ends of the hNHP2 coding region (EcoRI sites are underlined) were used to amplify by PCR the hNHP2 coding region from a plasmid cDNA library derived from a human Namalwa (Burkitt lymphoma) cell line (71). The amplified fragment was cleaved with EcoRI and cloned into the EcoRI site of pcDNA3.1/HisC (Invitrogen) to generate plasmid pcDNA-hNHP2.
The amino acid sequence of the Nop10p-like protein (hNOP10), deduced from human ESTs has been reported previously (29). Primers GCGGAATTCATGTTTCTCCAGTATTACCTCAACGAGC and GCGGAATTCTCAGAGGACAGGGCGCGGTTGCTG, bearing EcoRI restriction sites (underlined), were used to amplify by PCR the open reading frame (ORF) encoding hNOP10 from a plasmid cDNA library (see above). The amplified fragment was cloned into the EcoRI site of pcDNA3.1/HisC to generate plasmid pcDNA-hNOP10.
The ORF of dyskerin (27) (accession no. AJ224481) was amplified by PCR from a plasmid cDNA library using primers CCGGAATTCGGTAACATGGCGGATGCGGAAG and CCGGAATTCTCACTACTCAGAAACCAATTC. The amplified fragment bearing terminal EcoRI sites was cloned into the EcoRI site of pcDNA3.1/HisC to generate plasmid pcDNA-DKC1. Recombinant proteins expressed from the cytomegalovirus promoter of pcDNA3.1/HisC contain an N-terminal six-His tag followed by the Xpress epitope. The inserts of all plasmids were sequenced on an ABI automated sequencer.
GFP fusion expression constructs.
The ORF of hGAR1 (15) (accession no. AJ276003) was amplified by PCR using primers CCCGATATCATGTCTTTTCGAGGCGGAGG and GGGGATATCTGGAGGTCCTTTCTCACCTG (EcoRV sites are underlined) and cloned in the EcoRV site of pβact-ECGFP (49) to generate pβhGAR1-EGFP, which encodes full-length hGAR1 fused to the N terminus of enhanced green fluorescent protein (EGFP). For simplicity, this and other fusions to EGFP are referred to as green fluorescent protein (GFP) fusions. The portion encoding the core domain of hGAR1 (amino acids 62 to 168) was cloned similarly, using appropriate primers, to generate pβcore-EGFP. Coding sequences for shorter forms of the core domain, truncated at either the N or C terminus, were likewise amplified by PCR and cloned in the EcoRV site of pβact-ECGFP. Plasmids pβcoreΔ3N-EGFP, pβcoreΔ6N-EGFP, pβcoreΔ8N-EGFP, and pβcoreΔ14N-EGFP encode the core domain N-terminal deletion mutant derivatives lacking 3, 6, 8, and 14 amino acids, respectively. Plasmids pβcoreΔ9C-EGFP, pβcoreΔ13C-EGFP, and pβcoreΔ20C-EGFP encode the core domain C-terminal deletion mutant derivatives lacking 9, 13, and 20 amino acids, respectively. The ORFs encoding hNHP2 and hNOP10 were amplified by PCR and cloned similarly to generate pβhNHP2-EGFP and pβhNOP10-EGFP, respectively. The primers used were CCCGATATCGCTGCGATGACCAAAATAAAGGC and GGGGATATCTAGGGGTAGGGGCAGGGACTG (hNHP2) and CCCGATATCATGTTTCTCCAGTATTACCTCAACGAG and GGGGATATCGAGGACAGGGCGCGGTTGCTG (hNOP10) (EcoRV sites are underlined). The ORF encoding dyskerin was amplified by PCR as described previously (see above) and cloned into the EcoRV site of pβact-mGFP6 (a kind gift from A. Matus, Friedrich Miescher Institute) to generate pβmGFP6-DKC1, which encodes dyskerin with GFP at its N terminus. The identities of all constructs were checked by sequencing. DNA manipulations were carried out as described by Sambrook et al. (67).
Subcellular localization.
HeLa cells were grown on glass coverslips in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (Gibco-BRL) in a 5% CO2 humidified atmosphere at 37°C. For transient transfection assays with GFP- or Xpress-tagged constructs, HeLa cells were transfected with plasmid DNA using FuGENE as recommended by the manufacturer (Boehringer Mannheim) and further incubated for 24 to 48 h. Indirect immunofluorescence microscopy was performed essentially as described by Genschik et al. (21), using purified rabbit anti-hGAR1 antibodies (Abs) (15) at 1/1,000 dilution, mouse antifibrillarin monoclonal antibody (MAb) 72B9 (a kind gift from J. A. Steitz and K. T. Tycowski, Yale University School of Medicine, and K. M. Pollard, The Scripps Research Institute) at 1/400 dilution, mouse anti-p80-coilin MAb (5P10-π; kindly provided by M. Carmo-Fonseca, University of Lisbon) at 1/100 dilution, rabbit polyclonal anti-p80-coilin (sera 204/5; a kind gift from A. Lamond, University of Dundee) at 1/2,000 dilution, mouse anti-p120 MAb (Ab-1; Calbiochem) at 1/200 dilution, mouse anti-U2B" MAb (4G3; Cappel) at 1/200 dilution, mouse anti-GFP MAbs (Boehringer Mannheim) at 1/400 dilution, mouse antiubiquitin MAb (Ubi-1; ZYMED Laboratories), and mouse anti-Xpress MAb (Invitrogen) at a dilution of 1/500. Appropriate secondary antibodies (Jackson ImmunoResearch Laboratories), specified in the figure legends, were used at 1/400 dilution. Samples were examined with a laser scanning confocal microscope from Leica. Direct GFP fluorescence microscopy on living cells was performed as described previously (33).
Immunoprecipitations.
Protein G-Sepharose (PGS) beads (Pierce) or protein A-Sepharose (PAS) beads (Amersham Pharmacia Biotech) were incubated with appropriate antibodies (mouse anti-GFP MAbs for the analysis of the coimmunoprecipitated RNAs with PGS and rabbit anti-hGAR1 Abs and mouse antifibrillarin MAb 72B9 for the analysis of the coimmunoprecipitated proteins with PAS) in NET-2 buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Nonidet P-40) with gentle agitation overnight at 4°C. HeLa cells were transfected using FuGENE as described above and grown for additional 24 to 48 h (90% confluency). Cells were scraped from the petri dishes, washed three times with phosphate-buffered saline and resuspended in NET-2 buffer supplemented with the protease inhibitor Complete cocktail (Boehringer Mannheim). Whole-cell extracts, prepared by sonication (three times for 30 s at 50 W; Braun sonicator), were spun at 15,000 × g for 10 min. For each immunoprecipitation, the lysate of ∼5 × 106 cells was used. Lysates (500 μl) were mixed with the antibody-coated beads and incubated with a gentle agitation for 1 h at 4°C. Beads were then washed five times with NET-2 buffer containing 400 mM NaCl. Immunoprecipitated RNAs and total RNA from the whole-cell sonicates (HeLa cells used for preparation of total RNA were grown in suspension cultures) were extracted with phenol-chloroform containing 0.5% sodium dodecyl sulfate (SDS) and precipitated with ethanol in the presence of 40 μg of glycogen. To analyze the immunoprecipitated proteins, beads were washed as described above and resuspended in SDS-gel loading buffer. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 12% gels and analyzed by Western blotting using anti-GFP MAbs.
RNase A/T1 protection assays.
Individual RNA species were identified by an RNase A/T1 protection assay as described previously (26) using antisense RNA probes complementary to U17 (37), U3 (35), U19 (38), U13 (36), and hTR (15). Probes were labeled with [α-32P]UTP (800 Ci/mmol; NEN). Immunoprecipitated RNA from 5 × 106 cells was used for three protection reactions. Protected RNA fragments were fractionated on 6% sequencing gels.
Heterocomplementation.
The region coding for hNHP2 was excised from pcDNA-hNHP2 by digestion with EcoRI and cloned into the EcoRI site of yeast 2 μm expression vector pYX242 (a kind gift from B. Séraphin, EMBL, Heidelberg, Germany), resulting in plasmid pYX-hNHP2. The sequence coding for hNOP10 was amplified by PCR from pcDNA-hNOP10 using primers GCGGAATTCATGTTTCTCCAGTATTACCTCAACGAGC and CCGCTCGAGTCAGAGGACAGGGCGCGGTTG (EcoRI and XhoI sites are underlined) and cloned in the EcoRI-XhoI sites of pYX242, resulting in pYX-hNOP10. Expression of both proteins is driven from the triose phosphate isomerase promoter.
The pGAL-NHP2 and pGAL-NOP10 yeast strains (29), conditionally expressing Nhp2p and Nop10p, were transformed with either pYX-hNHP2, pYX-hNOP10, or the empty vector pYX242. Transformants were selected on glucose- or galactose-containing (SD-L or SGal-L, respectively) plates. Drop tests were performed from independent colonies at either permissive conditions (YPGal or SGal plates) or nonpermissive conditions (YPD or SD plates). Heterocomplementation was monitored by analyzing the growth of transformants under nonpermissive conditions. Genetic manipulations and preparation of standard yeast media followed established procedures (7).
Nucleotide sequence accession number.
The sequence of the hNHP2 cDNA has been deposited in the EMBL database under accession no. HAJ293309.
RESULTS
cDNAs encoding hNHP2 and hNOP10.
In yeast, four protein components of H/ACA snoRNP have been identified: Gar1p, Cbf5p, Nhp2p, and Nop10p. Two H/ACA snoRNP proteins have also been characterized in mammals. NAP57/dyskerin, which is the ortholog of the putative pseudouridine synthase Cbf5 (44, 82), has been studied in mouse and human cells (27, 56, 80). hGAR1, the human ortholog of yeast Gar1p, has recently been described (15).
The in vitro reconstitution studies carried out with HeLa cell extracts suggested that four proteins interact specifically with human H/ACA snoRNAs (15). Indeed, database searches have previously identified human ESTs coding for proteins related to yeast Nop10p and Nhp2p (29, 52, 77). To functionally characterize human Nop10p- and Nhp2p-like proteins (hNOP10 and hNHP2, respectively), full-length cDNAs encoding the likely human orthologs (see Materials and Methods) were cloned into different expression vectors and their activity in S. cerevisiae and human HeLa cells was investigated. The selected hNOP10 sequence corresponds to the protein included in the alignment of Henras et al. (29). The hNHP2 protein is an extended and corrected version of sequences presented by other groups (29, 52, 77). hNHP2 and hNOP10 consist of 153 and 64 amino acids, respectively.
hNHP2 and hNOP10 complement yeast deletion mutants.
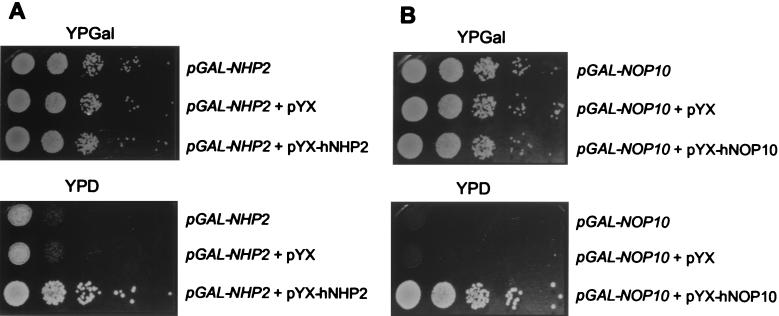
hNHP2 is 54.7% identical and 68.4% similar to yeast Nhp2p, while hNOP10 and yeast Nop10p are 60.3% identical and 67.2% similar. In order to find out whether the human proteins can complement yeast cells depleted of Nhp2p and Nop10p, the ORFs encoding hNHP2 and hNOP10 were cloned into the expression vector pYX242 to produce pYX-hNHP2 and pYX-hNOP10, respectively. Yeast strains pGAL-NHP2 and pGAL-NOP10 (29) were transformed with pYX-hNHP2, pYX-hNOP10, or the empty vector pYX242 and grown on either glucose- (YPD) or galactose-containing (YPGal) plates. As shown in Fig. 1, expression of human proteins complemented growth of pGAL-NHP2 and pGAL-NOP10 strains on YPD plates under conditions where the yeast proteins are not produced.
FIG. 1.
Complementation of the yeast pGAL-NHP2 and pGAL-NOP10 strains by the human hNHP2 and hNOP10 proteins. The pGAL-NHP2 (A) and pGAL-NOP10 (B) strains were transformed with either pYX-hNHP2 (A), pYXNOP10 (B), or the empty vector pYX, as indicated. Upper row (A and B), untransformed cells. Complementation was assayed by monitoring the growth of serially diluted yeast on YPD plates (YPD) and comparing it to the growth under permissive conditions (YPGal).
hNHP2, hNOP10, and dyskerin interact with hGAR1.
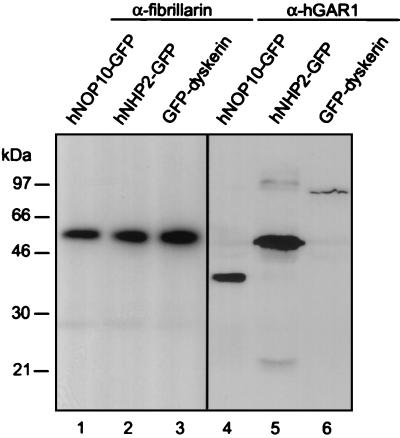
We investigated whether hNHP2, hNOP10, and dyskerin can be coimmunoprecipitated with hGAR1, the previously characterized protein component of human H/ACA snoRNPs. GFP fusions with hNHP2, hNOP10, and dyskerin were transiently expressed in HeLa cells, and whole-cell extracts were incubated with anti-hGAR1 Abs attached to PAS beads (for simplicity, we refer to these as anti-hGAR1 beads and we use this appellation for other antibody-bead complexes). As a control, the same extracts were incubated with antifibrillarin beads, since fibrillarin is a protein component of box C/D snoRNPs. The immunoprecipitated proteins were fractionated by SDS-PAGE. Western blotting with anti-GFP MAbs revealed that GFP fusions with hNHP2, hNOP10, and dyskerin were present in the anti-hGAR1 immunoprecipitates but not in the antifibrillarin immunoprecipitates (Fig. 2). Similar results were obtained when the association of hGAR1 with hNHP2, hNOP10, and dyskerin, expressed as fusions with the Xpress-tag (see Materials and Methods), was studied in HeLa cells (data not shown). These results indicate that hNHP2, hNOP10, dyskerin, and hGAR1 associate together in structures very likely corresponding to H/ACA snoRNPs.
FIG. 2.
GFP fusions with hNOP10, hNHP2, and dyskerin specifically associate with hGAR1. HeLa cells were transfected with plasmids expressing either hNOP10-GFP, hNHP2-GFP, or GFP-dyskerin, and protein extracts were incubated with antifibrillarin beads (lanes 1 to 3) or anti-hGAR1 beads (lanes 4 to 6). Immunoprecipitated proteins were separated by SDS-PAGE and analyzed by Western blotting using anti-GFP MAbs. The strong bands in lanes 1 to 3 correspond to the heavy chain of the antifibrillarin MAb recognized by the anti-mouse immunoglobulin G secondary Ab. The masses of proteins markers are indicated on the left.
hNHP2 and hNOP10 are specifically associated with H/ACA snoRNAs.
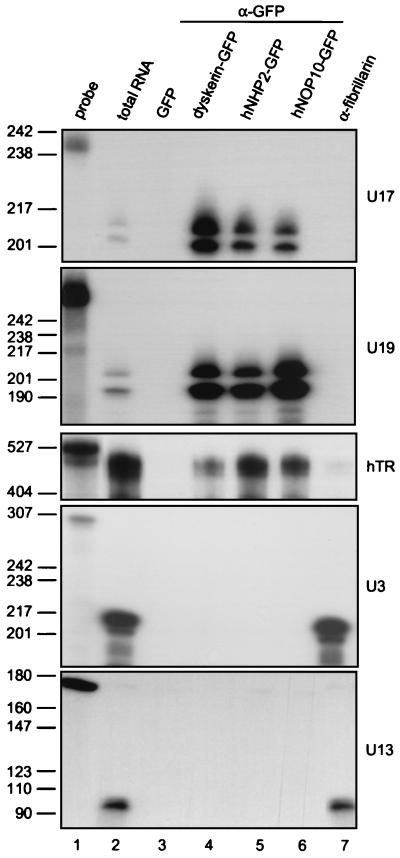
To determine if hNHP2 and hNOP10 are indeed components of the H/ACA snoRNPs, we investigated whether they are associated with H/ACA snoRNAs in human cells. GFP-tagged hNHP2, hNOP10, and dyskerin were transiently expressed in HeLa cells, and whole-cell extracts were incubated with anti-GFP beads. Immunoprecipitated RNAs were extracted, and individual RNA species were identified by RNase A/T1 mapping. H/ACA snoRNAs U17 and U19 were immunoprecipitated from cells expressing hNHP2-, hNOP10-, and dyskerin-GFP fusions but not from control cells expressing GFP alone (Fig. 3, lanes 4 to 6). Box C/D snoRNAs U3 and U13 were not detected in anti-GFP immunoprecipitates but were present in antifibrillarin immunoprecipitates isolated from control HeLa cells (Fig. 3, lane 7). hTR possesses an H/ACA-like domain and associates with dyskerin and hGAR1 (15, 57, 58). Mappings of RNAs isolated from different immunoprecipitates indicated that hNHP2 and hNOP10 also specifically associate with hTR (Fig. 3). Specific coprecipitation of H/ACA snoRNAs and hTR with hNHP2, hNOP10, and dyskerin expressed as fusions with the Xpress tag was also observed (data not shown). Taken together, our data suggest that hNHP2 and hNOP10 are components of H/ACA snoRNPs and telomerase in human cells.
FIG. 3.
GFP-tagged hNOP10, hNHP2, and dyskerin specifically associate with H/ACA snoRNAs and hTR. HeLa cells were transfected with plasmids expressing hNHP2-GFP, hNOP10-GFP, GFP-dyskerin fusion proteins or GFP alone (vector pβact-ECGFP), and protein extracts were incubated with anti-GFP beads. As an additional control, protein extracts from untransfected HeLa cells were incubated with antifibrillarin beads. Antisense RNA probes specific for U17, U19, U3, and U13 RNAs and hTR were used for RNase A/T1 mapping of RNA from anti-GFP (lanes 3 to 6) and antifibrillarin (lane 7) immunoprecipitates. Lane 1, aliquot of probe; lane 2, mapping of total RNA isolated from nontransfected HeLa cells; for mapping with the hTR probe, five times more total RNA was used. The hTR gel was exposed two times longer than other gels. The sizes of DNA markers in nucleotides are indicated on the left.
Comparison of hNOP10 and hNHP2 with likely orthologs from other organisms.
hNHP2 and hNOP10 have calculated molecular masses of 17.2 and 7.7 kDa, respectively, and their sizes are similar to those of yeast Nhp2p (calculated mass, 18.9 kDa) and Nop10p (6.6 kDa). However, as already observed for the yeast proteins (29, 77), the mobilities of hNHP2 and hNOP10 in SDS gels appear to be substantially slower then expected (data not shown). Hence, it is very likely that hNHP2 and hNOP10 correspond to p23 and p14, respectively, two of the four human proteins that can be specifically UV cross-linked to RNA in H/ACA snoRNPs reconstituted in vitro (15). We compared the sequences of these proteins with their likely orthologs from other organisms. Database searches indicated that sequences similar to hNOP10 are encoded not only in genomes of many other eukaryotes (29, 52, 77) but also in all archaeal genomes sequenced to date (Fig. 4); E values for the similarity of hNOP10 to different archaeal sequences, derived from BLAST searches, range from 0.011 to 1.2 (with the exception of a substantially higher value for the protein from Methanococcus jannaschii). Although these values are low, similar lengths and a nearly perfect colinearity of eukaryotic and archaeal NOP10-like proteins (four of the archaeal proteins differ from eukaryotic ones only by insertions of single Cys residues at two positions) add to the potential significance of this relationship (76) (see Discussion). No sequences with significant similarity to Nop10p or hNOP10 could be identified in Bacteria.
FIG. 4.
Comparison of amino acid sequences of Nop10p-like proteins from different organisms. Nop10p-like protein sequences from Homo sapiens (Hs) (29), Xenopus laevis (Xl; EST accession no. AW199609), Danio rerio (Dr; EST accession no. AI618239), D. melanogaster (Dm; AAF58753), Caenorhabditis elegans (Ce; AV201331), Arabidopsis thaliana (At; AAD25649), Zea mays (Zm; EST accession no. AI783103), S. cerevisiae (Sc) (29), S. pombe (Sp; CAB76034), Toxoplasma gondii (Tg; EST accession no. N82941), Brugia malayi (Bm; EST accession no. N41102), Archaeoglobus fulgidus (Af; O29724), Pyrococcus horikoshii (Ph; genomic DNA sequence accession no. AP000004), M. jannaschii (Mj; P81303), Methanobacterium thermoautotrophicum (Mt; O27362), and Aeropyrum pernix (Ap; AP000059) are shown. An additional archaeal sequence, corresponding to the Nop10p-like protein in Pyrococcus abysi, is identical to the P. horikoshii sequence. The alignment was generated with the PILEUP program (Genetics Computer Group, Madison, Wis.) and shaded using the Boxshade World Wide Web server (www.ch.embnet.org/software/BOX_form.html); amino acids identical or similar in more than 50% of analyzed sequences have black and gray backgrounds, respectively.
Yeast Nhp2p and human hNHP2 are 54.7% identical and 68.4% similar. As expected, the BLAST searches with the hNHP2 as a query identified orthologs in other metazoa, fungi, and plants, with E values ranging from 2.5 × 10−33 to 10−8 (data not shown). Since NHP2-like proteins are members of a large family of RNA-associated proteins, which include some ribosomal and spliceosomal proteins (references 29, 41, 52, 62, 70, and 77 and references therein), database searches with hNHP2 identified also other proteins in both eukaryotes and Archaea (data not shown). Archaeal sequences most probably represent ribosomal proteins (34, 52) (see Discussion).
Subcellular localization of human H/ACA snoRNP proteins.
Of the four identified protein components of the H/ACA snoRNPs in mammalian cells (15, 27, 58, 80; this work), only the cellular localization of NAP57/dyskerin was studied. This protein is localized in the DFC of the nucleolus and in CBs (28, 56). We investigated the intracellular distribution of hGAR1, hNHP2, and hNOP10 in HeLa cells.
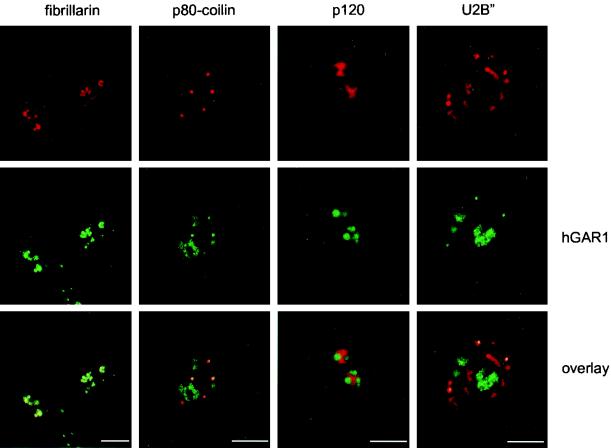
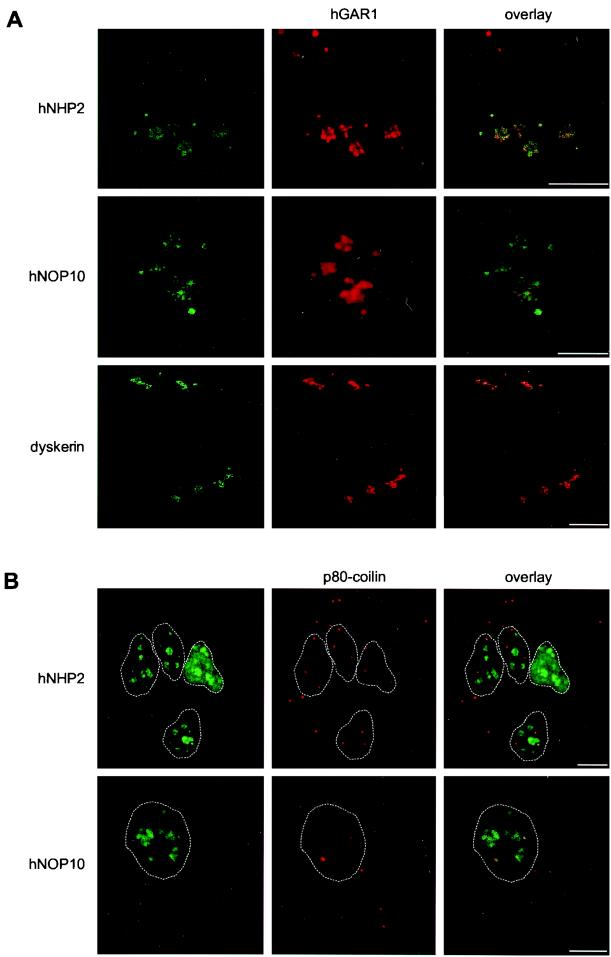
Localization of hGAR1 was studied by indirect immunofluorescence microscopy using affinity-purified anti-hGAR1 Abs (15). The following proteins were monitored in parallel as markers: (i) the box C/D snoRNP protein fibrillarin, which localizes to the DFC of nucleoli and in CBs (reviewed in references 45 and 68); (ii) p80-coilin, a marker for CBs (reviewed in references 17, 45, and 53); (iii) p120 nucleolar proliferation antigen localizing to the granular component of nucleoli (63); (iv) the splicesomal protein U2B" present in the nucleoplasmic speckles and in CBs (9). Double-staining experiments showed that hGAR1 colocalizes perfectly with fibrillarin in the DFC of nucleoli and in CBs (Fig. 5, left panels) and with p80-coilin in CBs (Fig. 5, second panels from left). Double labeling with p120 revealed that hGAR1 is excluded from nucleolar regions occupied by p120. Furthermore, hGAR1 did not colocalize with the U2-specific protein U2B" in the nucleoplasm; however, colocalization of the two proteins was observed in CBs.
FIG. 5.
Subcellular localization of endogenous hGAR1 studied by confocal microscopy. Double-staining experiments were performed on fixed HeLa cells using anti-hGAR1 Abs and fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G (IgG) (hGAR1; green channel) together with either antifibrillarin (fibrillarin), anti-p80-coilin (p80-coilin), anti-p120 (p120), or anti-U2B" (U2B") Abs, followed by Texas red-conjugated anti-mouse IgG (top row, red channel). Laser scanning confocal images were merged (overlay); regions of colocalization appear in yellow. Bar, 10 μm.
To determine the cellular localization of hNHP2 and hNOP10, Xpress epitope-tagged versions of these proteins were transiently expressed in HeLa cells. Double-immunostaining experiments were performed with an anti-Xpress MAb and anti-hGAR1 Abs. hNHP2 and hNOP10 fusion proteins (Fig. 6A, upper two rows) and also the Xpress-tagged dyskerin (bottom row) perfectly colocalized with hGAR1 in the DFC of nucleoli and in CBs. The same localization pattern was found when the distribution of C-terminal GFP fusions with hNHP2 and hNOP10 was investigated in costaining experiments with anti-p80-coilin and antifibrillarin Abs (Fig. 6B and data not shown).
FIG. 6.
Subcellular localization of tagged hNHP2, hNOP10, and dyskerin transiently expressed in HeLa cells. Double staining experiments were performed on fixed cells using anti-Xpress MAb and fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin G (IgG) (hNHP2, hNOP10, dyskerin; green; left column) together with anti-hGAR1 Abs and Texas red-conjugated anti-rabbit IgG (red; middle column) (A) or anti-GFP MAbs with FITC-conjugated anti-mouse IgG (hNHP2, hNOP10; green; left column) together with anti-p80-coilin Abs and Texas red-conjugated anti-rabbit IgG (p80-coilin; red; middle column) (B). Laser scanning confocal images were merged (overlay). Nucleoplasmic edges in panel B are highlighted with white dashed lanes. Bars, 10 μm.
In summary, these experiments indicate that all human H/ACA proteins localize to the DFC of the nucleolus and to CBs.
Localization of hGAR1 deletion mutants.
Previous work has indicated that GAR1 is not a primary snoRNA binding protein (see the introduction). It is possible that GAR1 is only required for the activity or localization of H/ACA snoRNPs. In order to get more insight into the function of hGAR1, we studied the intracellular localization of different mutant derivatives of the protein that were transiently expressed in HeLa cells.
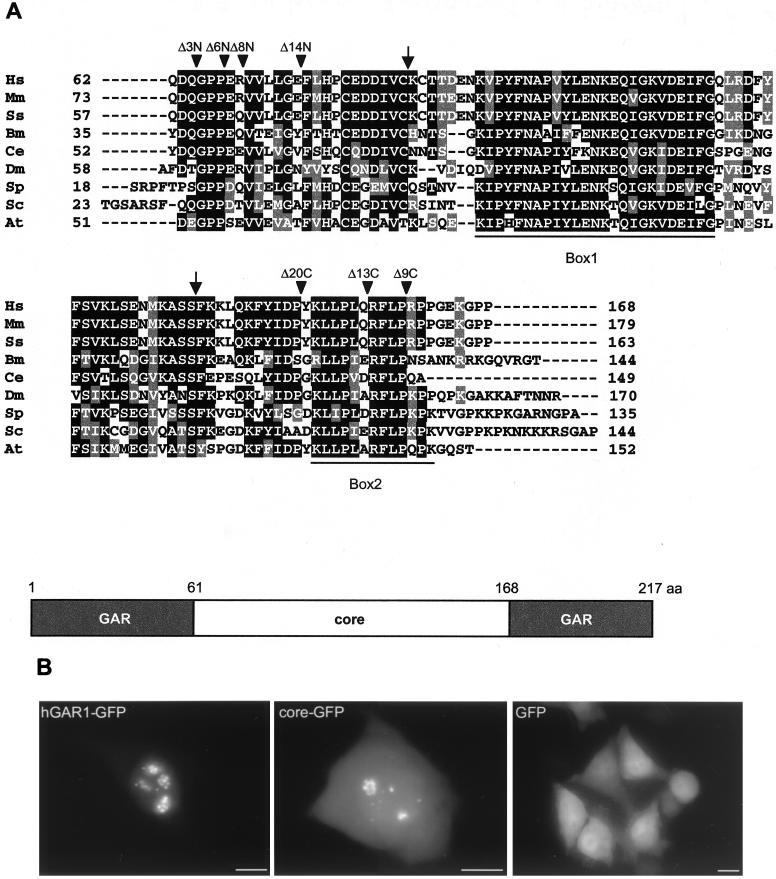
The overall structure of GAR1 is preserved among eukaryotes (15, 23, 24, 77). The GAR domains that flank the core domain vary in length and amino acid composition, while the core domain is very highly conserved at the primary sequence level (Fig. 7A). We have found that localization of the core domain fused to GFP (core-GFP) is similar to that of hGAR1-GFP, except that more staining is observed in the nucleoplasm and cytoplasm (Fig. 7B), indicating that GAR domains may help in targeting the protein to its functional sites. This result also suggests that the core-GFP hybrid protein is functional and that the core domain contains all the signals required for localization in the DFC of the nucleolus and in CBs. The data are consistent with the observation that the core domain of yeast Gar1p is sufficient for nucleolar targeting and function (25).
FIG. 7.
Sequences of GAR1 core domain and mutant derivatives. (A) Alignment of GAR1 core domain sequences from different organisms. Shown are sequences from Homo sapiens (Hs; CAB76563), Mus musculus (Mm; EST accession no. AL034727), Sus scrofa (Ss; EST accession no. Z84033), Brugia malayi (Bm; EST accession no. AI053139), Caenorhabditis elegans (Ce; AAD12858), D. melanogaster (Dm; S49193), S. pombe (Sp; CAA79628), S. cerevisiae (Sc; P28007), and Arabidopsis thaliana (At; AAF00626). The alignment and shading are as in Fig. 4. Arrowheads, sites of deletion in the core domain GFP fusions; arrows, sites of deletions performed on the core domain of S. cerevisiae (these deletions did not complement yeast growth) (25). The two highly conserved regions (Box1 and Box2) (2) are underlined. A schematic representation of hGAR1 is shown below the alignment. (B) Subcellular localization of the GFP-tagged hGAR1 and its core domain in living HeLa cells. hGAR1-GFP, core-GFP, and GFP alone were transiently expressed in HeLa cells and visualized in living cells by direct fluorescence microscopy. GFP is distributed throughout the cell, whereas both hybrid proteins accumulate in the nucleolus and CBs. Bars, 10 μm.
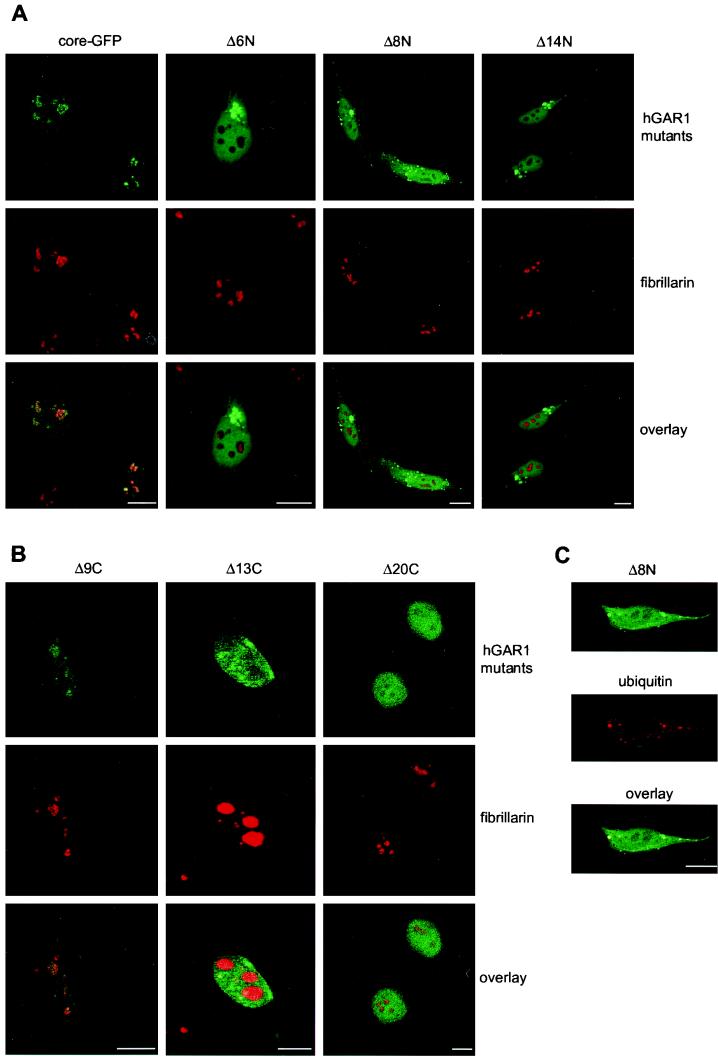
In an attempt to define the minimal core domain of hGAR1, the effect of short truncations at either end of the core domain fused to the N terminus of GFP was analyzed (Fig. 8). Deletion of the N-terminal tripeptide (QDQ) of the core domain (derivative Δ3N; for the structures of this and other mutant derivatives, see Fig. 7A) had no effect, but deleting three amino acids more (Δ6N), which eliminated the strictly conserved GPP motif, impaired localization of the hybrid protein. This mutant protein accumulated in cytoplasmic clumps or dot-like structures. A similar phenotype was observed with cells expressing the deletion mutant derivatives Δ8N and Δ14N, which are missing 8 and 14 amino acids at the N terminus of the core, respectively. In living cells, the cytoplasmic dots are mobile (data not shown). Moreover, the cytoplasmic clumps or dots were shown to be enriched in ubiquitin (Fig. 8C). These observations and the fact that the large clumps accumulated near the nuclear membrane (Fig. 8A) strongly suggest that the N-terminal deletions induce the formation of aggresomes in response to protein misfolding (20, 32, 79). Therefore, the GPP motif could be important for both protein folding and nucleolar localization, since the fraction of mutated proteins that could enter the nucleus remained excluded from nucleoli and CBs.
FIG. 8.
Subcellular localization of hGAR1 deletion mutants. HeLa cells were transfected with plasmids expressing hGAR1 core domain fused to GFP (A, core-GFP) or its derived N-terminal (A) and C-terminal (B) deletion mutant derivatives. Direct fluorescence from GFP-tagged constructs is shown in green (hGAR1 mutants). Fibrillarin was labeled with antifibrillarin MAb and Texas red-conjugated anti-mouse immunoglobulin G (IgG) (fibrillarin; red). Laser scanning confocal images were merged (overlay). The Δ13C panel was overexpressed in order to make CBs visible. (C) Colocalization of the N-terminal core deletion mutant Δ8N with ubiquitin. HeLa cells were transfected with pβcoreΔ8N-EGFP and visualized by laser scanning confocal microscopy. Direct fluorescence from the coreΔ8N-EGFP deletion mutant is shown in green (Δ8N). Ubiquitin was visualized with anti-ubiquitin MAb and Texas red-conjugated anti-mouse IgG (ubiquitin; red). Confocal images were merged (overlay). Identical results were obtained with HeLa cells transfected with either pβcoreΔ6N-EGFP or pβcoreΔ14N-EGFP (data not shown). Bars, 10 μm.
Cells expressing the C-terminal deletion mutant derivatives of the core domain had very different phenotypes (Fig. 8B). Truncation of nine amino acids (Δ9C) had no effect on the subcellular localization of the hybrid protein (Fig. 8B); however, deleting four amino acids more (Δ13C), which removed the RFLP tetrapeptide that is perfectly conserved in all GAR1 sequences (Fig. 7A), impaired accumulation of the fusion protein in the nucleolus but not in CBs. It is important to note that coreΔ13C-GFP was not excluded from the nucleolus, but it could not accumulate in this compartment. A longer C-terminal deletion (20 amino acids) that eliminated the KLLPL pentapeptide, another highly conserved portion of the core domain, completely abolished localization of the coreΔ20C-GFP hybrid protein in the nucleolus and CBs. Taken together these results indicate that highly conserved amino acids at the C-terminal border of the core domain are required for localization in the nucleolus and CBs.
Association of hGAR1 mutant derivatives with U17 RNP.
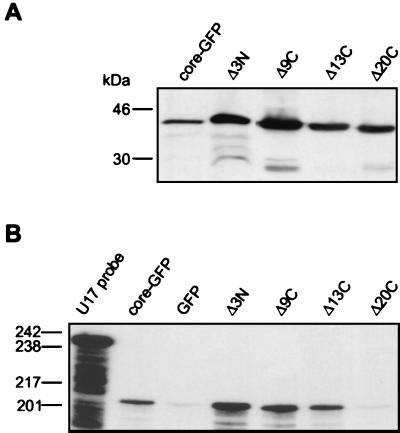
We investigated whether deletions in the core domain disrupted its potential to associate with U17 RNP. Western blotting analysis indicated that the deletion mutant derivatives Δ3N, Δ9C, Δ13C, and Δ20C accumulate to levels comparable to that of the core-GFP (Fig. 9A). Accumulation of the mutant derivatives Δ6N, Δ8N, and Δ14N was low (data not shown), supporting the suggestion that these mislocalizing hybrid proteins undergo ubiquitin-mediated proteolysis (Fig. 8), and their characterization was not pursued further. As shown in Fig. 9B, core-GFP and the mutant derivatives Δ3N and Δ9C, which accumulate in both the nucleolus and CBs, and the mutant derivative Δ13C, which accumulates in CBs but not the nucleolus, efficiently coprecipitated the U17 snoRNA, suggesting that they are competent in assembling into an RNP. The Δ20C mutant derivative, which could not accumulate in the nucleolus or CBs, was unable to associate with the U17 RNP.
FIG. 9.
Association of core-GFP mutant derivatives with U17 snoRNP. HeLa cells were transfected with plasmids expressing either core-GFP, or Δ3N, Δ9C, Δ13C, and Δ20C deletion mutants. Proteins extracts were separated by SDS-PAGE and analyzed by Western blotting using anti-GFP MAbs (A) or they were incubated with anti-GFP beads and immunoprecipitated RNAs were analyzed by an RNase A/T1 protection assay using a U17-specific RNA probe (B). Protein and DNA size markers are indicated on the left.
DISCUSSION
In the yeast S. cerevisiae, four protein components of the H/ACA snoRNPs have been identified: Gar1p, Cbf5p, Nhp2p, and Nop10p. Based on in vitro reconstitution studies, four proteins were also found to specifically interact with H/ACA snoRNAs in HeLa cell extracts. Two of the mammalian proteins have already been characterized, namely, hGAR1 and NAP57/dyskerin, the ortholog of Cbf5p. In this work we describe properties of hNOP10 and hNHP2, the human orthologs of yeast Nop10p and Nhp2p, and further characterize hGAR1. We find that hNOP10 and hNHP2 complement yeast cells depleted of endogenous Nop10p and Nhp2p. Moreover, transiently expressed epitope-tagged hNOP10 and hNHP2 specifically associate with hGAR1, H/ACA snoRNAs, and the telomerase RNA in HeLa cells. hNOP10, hNHP2, hGAR1, and dyskerin localize to the DFC of the nucleolus and in CBs. We demonstrate that the evolutionarily conserved core domain of hGAR1 contains all the signals required for localization but that progressive deletions from either the N or the C terminus of the core domain abolish localization in the nucleolus or both the nucleolus and CBs.
A comparison of hNOP10 and hNHP2 with the orthologs from S. cerevisiae and the recently characterized NHP2 of the fission yeast S. pombe and database searches indicate that both proteins are highly conserved among eukaryotes 29, 52, 77; this work; our unpublished results). NHP2 is likely to be a primary RNA-binding protein since it belongs to a family of proteins containing a novel type of RNA-binding domain (41); this family includes proteins highly similar to NHP2, such as some ribosomal proteins (rp) from both eukaryotes (reference 41 and references therein) and Archaea (34), and the spliceosomal 15.5-kDa protein associated with U4 snRNA (62) (for the most recent alignments and the phylogenetic analysis of NHP2 and related proteins, see references 52 and 62). Genetic depletion of Nhp2p, similar to depletion of Nop10p and Cbf5p, results in a reduction of the steady-state levels of the H/ACA snoRNAs in yeast, which is consistent with the notion that Nhp2p interacts directly with the RNA and that this interaction is required for stability. Importantly, the H/ACA-specific p23 protein, which most probably corresponds to hNHP2 (see below), was by far the most efficiently cross-linked to RNA (15). Moreover, NHP2 can bind directly to RNA in vitro (A. Henras, personal communication; N. Watkins, and R. Lührmann, personal communication; V. Pogačić, unpublished data). For two NHP2-related proteins, the yeast rpL30 (formerly rpL32) and the human 15.5-kDa protein associated with U4, the structure of the RNA-binding sites has been characterized (48, 62, 75). Henras et al. (29) suggested that there is some similarity between the rpL30 RNA-binding site and functionally important regions of H/ACA, but our attempts to model any conserved H/ACA regions as structures recognized by rpL30 or the 15.5-kDa protein have not been successful.
Interestingly, database searches indicated that sequences similar to that of the hNOP10 protein are encoded not only in eukaryotes but also in all archaeal genomes (Fig. 4). No sequences with similarity to that of NOP10 could be identified in Bacteria. While this study was being prepared for publication, Watanabe and Gray (76) also reported detection of NOP10-like sequences in Archaea. Moreover, these authors identified archaeal genes encoding proteins having weak but significant similarity to GAR1. Since the existence of Cbf5p- and dyskerin-like proteins in Archaea has also been proposed (reference 76 and references therein), it is possible that pseudouridylation of rRNA in these organisms involves particles related to eukaryotic H/ACA snoRNPs (76). To date not RNAs resembling H/ACA snoRNAs have been identified in Archaea. In addition, it is likely that archaeal NHP2-like sequences correspond to ribosomal proteins (34, 52) (see above); however, the possibility that these ribosomal proteins could also function as components of H/ACA-like RNPs cannot be excluded.
Immunoprecipitation experiments performed with epitope-tagged hNHP2 and hNOP10 expressed in HeLa cells revealed that these proteins associate with box H/ACA but not box C/D snoRNAs in vivo. Tagged hNHP2, hNOP10, and dyskerin also coprecipitated with hGAR1, providing additional evidence that all four proteins function together as components of human H/ACA snoRNPs. Four proteins, p60, p29, p23, and p14, were previously found by UV cross-linking to specifically interact with H/ACA snoRNAs in HeLa cell extracts, and immunodepletion experiments indicated that p29 corresponds to hGAR1 (15). Based on the mobility in SDS gels of proteins expressed in transfected cells or translated in vitro (data not shown), it is very likely that dyskerin, hNHP2, and hNOP10 correspond to p60, p23, and p14, respectively.
The telomerase RNA in vertebrates contains a 3′-terminal domain that structurally resembles H/ACA snoRNAs (12, 57). This domain is required for the accumulation and function of hTR in vivo. Two human H/ACA proteins, dyskerin (58) and hGAR1 (15), were previously shown to associate with hTR in vivo, and dyskerin was found to be a component of active telomerase. hGAR1 also associates with hTR in the in vitro reconstitution system (15). We showed that epitope-tagged hNHP2 and hNOP10 expressed in transfected HeLa cells specifically immunoprecipitate hTR (Fig. 3). We can thus conclude that all four core H/ACA proteins are components of telomerase in human cells. The function of the H/ACA domain in hTR is not well understood. Based on the recently derived structural model of vertebrate telomerase RNAs, the region resembling the pseudouridylation pocket of H/ACA snoRNAs is unlikely to act as a guide in RNA modification (12). More probably, association of H/ACA proteins with hTR is required for correct 3′-end processing and/or stabilization of hTR (57, 58). Indeed, mutations in dyskerin which lead to dyskeratosis congenita result in reduced hTR accumulation and shortening of telomeres (58). The H/ACA domain might also be required for bringing hTR to the nucleolus (57, 59), possibly for progressive assembly of the RNP, which would be analogous to the partial nucleolar assembly of the signal recognition particle (65). Finally, the observation that the replacement of the H/ACA-like domain of hTR with a conventional H/ACA snoRNA sequence does not produce the active telomerase in vivo (57) suggests that this domain contains some specific sequences likely to be recognized by telomerase proteins, a possibility supported by the results of previous in vitro assembly experiments (15).
In S. cerevisiae, Gar1p, Nhp2p, and Nop10p were shown to localize to the nucleolus, whereas for mammalian orthologs cellular localization was only studied for NAP57/dyskerin. This protein localizes to the DFC of the nucleolus and to CBs (28, 56). We have found that hGAR1, hNHP2, and hNOP10 also localize to the DFC of nucleoli and to CBs. Localization of all known H/ACA proteins to the DFC is consistent with the pseudouridylation of rRNA being an early maturation event in the nucleolus (reviewed in reference 51). Association of the H/ACA proteins with CBs supports a proposed role for these structures in biogenesis, trafficking, or recycling of small nucleoplasmic and nucleolar RNPs (reviewed in references 17, 45, 47, and 53). Importantly, the H/ACA snoRNA U17 localizes similarly to H/ACA proteins in HeLa cells; it is present in both the nucleolus and CBs (M. Hebert, G. Matera, P. Pelczar, and W. Filipowicz, unpublished results). However, it should be noted that localization of box C/D snoRNAs in CBs, but not box H/ACA snoRNAs, could be visualized in microinjected Xenopus oocytes (46, 59, 60).
We have shown that, as in yeast (25), the core domain of hGAR1 contains all the signals required for proper subcellular localization. In the absence of GAR domains, the core-GFP hybrid protein was found in the nucleolus and CBs, but residual staining in the nucleoplasm and cytoplasm indicates that GAR domains help in targeting the protein to, or retaining it in, its functional sites. It is noteworthy that GAR domains are found in many nucleolar proteins and might be required for optimal nucleolar targeting or retention. Additional deletions in the core domain revealed that the GPP motif at the N-terminal border of the core domain is required for proper folding and nucleolar localization. Removal of this motif induced the formation of aggresomes, a protein misfolding response that is activated when the proteasome is overwhelmed (20, 32, 79). An active transport mechanism is required for localization of GAR1 (25); since the GPP tripeptide is strictly conserved among Eucarya, this motif might be required at some stage during the cytosol-to-nucleolus itinerary.
Conserved motifs at the C-terminal border of the core domain are also required for proper targeting and accumulation in the nucleolus and CBs. Indeed, deletion of the RFLP motif, which is strictly conserved among Eucarya, impaired accumulation, but not targeting, of the Δ13C mutant derivative in the nucleolus. This protein could, however, accumulate in CBs. The RFLP motif might be required for function and/or interaction with other nucleolar components; in its absence, defective snoRNPs could be rejected and targeted to CBs for recycling (53). This view is in agreement with the observation that nucleolar targeting of dyskerin in microinjected mammalian cells precedes accumulation in CBs (28). Moreover, the nucleolar and CB protein Nopp140, which associates with H/ACA and C/D snoRNPs and which has been proposed to act as an snoRNP chaperone, also accumulates in the nucleolus prior to associating with CBs (30, 80). Immunoprecipitation experiments indicated that the coreΔ13C mutant derivative interacts with the U17 snoRNP. In contrast, the Δ20C mutant derivative, which is missing an additional motif (KLLPL), did not coprecipitate with U17 RNA and did not accumulate in the nucleolus or CBs. Thus, the conserved C-terminal motifs play an important role in assembly and localization of H/ACA snoRNPs. Our results further suggest that mutant proteins need to bind snoRNP before entering the nucleolus and CBs. Alternatively, it is possible that RNP binding occurs in the nucleolus and that proteins that fail to enter the nucleolus cannot form RNPs. However, we favor the former scenario since it is very likely that snoRNP assembly is concomitant with snoRNA transcription in the nucleoplasm (8, 66).
Bagni and Lapeyre (2) have shown that yeast Gar1p specifically binds H/ACA box snoRNAs snR10 and snR30 in vitro. The N-terminal half of the core domain is absolutely essential but not sufficient for this interaction to take place; it additionally requires the presence of either one of the GAR domains or the C-proximal half of the core domain. Based on the yeast Gar1p mutagenesis studies (2), the hGAR1 mutant derivatives Δ9C and Δ13C are unlikely to retain the potential to bind RNA. The ability of these mutant derivatives to associate with the U17 snoRNP may, therefore, indicate that protein-protein contacts rather than binding to RNA are responsible for the association of hGAR1 with the particle. Investigation of the RNA-binding potential of hGAR1 and other H/ACA proteins and of interactions between individual components of H/ACA snoRNPs will be important for a better understanding of the assembly and function of these particles.
ACKNOWLEDGMENTS
V. Pogačić and F. Dragon contributed equally to this work.
We are grateful to J. A. Steitz, K. T. Tycowski, K. M. Pollard, M. Carmo-Fonseca, and A. Lamond for generous gifts of antibodies and to A. K. Winteroe (The Royal Veterinary and Agricultural University, Denmark) and S. A. Williams (Clark Science Center, Smith College) for the Sus scrofa and Brugia malayi GAR1 cDNA clones, respectively. We thank our FMI colleagues Andrew Matus for plasmids, Patrick Matthias for the plasmid cDNA library, Peter Müller for oligonucleotide synthesis, Herbert Angliker for DNA sequencing, and Zdravko Lorković for help with art work.
The Friedrich Miescher Institute is part of the Novartis Research Foundation.
REFERENCES
- 1.Bachellerie J-P, Cavaillé J, Qu L-H. Nucleotide modifications of eukaryotic rRNAs: the world of small nucleolar RNA guides revisited. In: Garrett R A, Douthwaite S R, Liljas A, Matheson A T, Moore P B, Noller H F, editors. The ribosome: structure, function, antibiotics, and cellular interactions. Washington, D.C.: ASM Press; 2000. pp. 191–203. [Google Scholar]
- 2.Bagni C, Lapeyre B. Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitrothrough a nontypical RNA binding element. J Biol Chem. 1998;273:10868–10873. doi: 10.1074/jbc.273.18.10868. [DOI] [PubMed] [Google Scholar]
- 3.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn E H, Greider C W. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 5.Blackburn E H. Telomerase. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 609–635. [Google Scholar]
- 6.Bousquet-Antonelli C, Henry Y, Gélugne J-P, Caizergues-Ferrer M, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 1997;16:4770–4776. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown A J P, Tuite M. Yeast gene analysis. London, United Kingdom: Academic Press; 1998. [Google Scholar]
- 8.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 9.Carmo-Fonseca M, Pepperkok R, Carvalho M T, Lamond A I. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavaillé J, Nicoloso M, Bachellerie J-P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 11.Chamberlain J R, Lee Y, Lane W S, Engelke D R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J-L, Blasco M A, Greider C W. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 13.Chu S, Archer R H, Zengel J M, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins K. Mammalian telomeres and telomerase. Curr Opin Cell Biol. 2000;12:378–383. doi: 10.1016/s0955-0674(00)00103-4. [DOI] [PubMed] [Google Scholar]
- 15.Dragon F, Pogačić V, Filipowicz W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol Cell Biol. 2000;20:3037–3048. doi: 10.1128/mcb.20.9.3037-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster A C, Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- 17.Gall J G, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 19.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 20.García-Mata R, Bebök Z, Sorscher E J, Sztul E J. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genschik P, Billy E, Swianiewicz M, Filipowicz W. The human RNA 3′-terminal phosphate cyclase is a member of a new family of proteins conserved in Eucarya, Bacteria and Archaea. EMBO J. 1997;16:2955–2967. doi: 10.1093/emboj/16.10.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giordano E, Peluso I, Senger S, Furia M. minifly, a Drosophilagene required for ribosome biogenesis. J Cell Biol. 1999;144:1123–1133. doi: 10.1083/jcb.144.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girard J-P, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girard J-P, Caizergues-Ferrer M, Lapeyre B. The SpGAR1 gene of Schizosaccharomyces pombe encodes the functional homologue of the snoRNP protein GAR1 of Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2149–2155. doi: 10.1093/nar/21.9.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girard J-P, Bagni C, Caizergues-Ferrer M, Amalric F, Lapeyre B. Identification of a segment of the small nucleolar ribonucleoprotein-associated protein GAR1 that is sufficient for nucleolar accumulation. J Biol Chem. 1994;269:18499–18506. [PubMed] [Google Scholar]
- 26.Goodall G J, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- 27.Heiss N S, Knight S W, Vulliamy T J, Klauck S M, Wiemann S, Mason P J, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 28.Heiss N S, Girod A, Salowsky R, Wiemann S, Pepperkok R, Poustka A. Dyskerin localizes to the nucleolus and its mislocalization is unlikely to play a role in the pathogenesis of dyskeratosis congenita. Hum Mol Genet. 1999;8:2515–2524. doi: 10.1093/hmg/8.13.2515. [DOI] [PubMed] [Google Scholar]
- 29.Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gélugne J-P, Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isaac C, Yang Y, Meier U T. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang W, Middleton K, Yoon H J, Fouquet C, Carbon J. An essential yeast protein, Cbf5p, binds in vitro to centromeres and microtubules. Mol Cell Biol. 1993;13:4884–4893. doi: 10.1128/mcb.13.8.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnston J A, Ward C L, Kopito R R. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaech S, Ludin B, Matus A. Cytoskeletal plasticity in cells expressing neuronal microtubule-associated proteins. Neuron. 1996;17:1189–1199. doi: 10.1016/s0896-6273(00)80249-4. [DOI] [PubMed] [Google Scholar]
- 34.Kimura J, Arndt E, Kimura M. Primary structures of three highly acidic ribosomal proteins S6, S12, and S15 from the archaebacterium Halobacterium marismortui. FEBS Lett. 1987;224:65–70. doi: 10.1016/0014-5793(87)80423-4. [DOI] [PubMed] [Google Scholar]
- 35.Kiss T, Filipowicz W. Evidence against a mitochondrial location of the 7-2/MRP RNA in mammalian cells. Cell. 1992;70:11–16. doi: 10.1016/0092-8674(92)90528-k. [DOI] [PubMed] [Google Scholar]
- 36.Kiss T, Filipowicz W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993;12:2913–2920. doi: 10.1002/j.1460-2075.1993.tb05953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 38.Kiss T, Bortolin M-L, Filipowicz W. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol Cell Biol. 1996;16:1391–1400. doi: 10.1128/mcb.16.4.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 40.Knight S W, Heiss N S, Vulliamy T J, Greschner S, Stavrides G, Pai G S, Lestringant G, Varma N, Mason P J, Dokal I, Poustka A. X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am J Hum Genet. 1999;65:50–58. doi: 10.1086/302446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koonin E V, Bork P, Sander C. A novel RNA-binding motif in omnipotent suppressor of translation termination, ribosomal proteins and a ribosomal modification enzyme? Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koonin E V. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lafontaine D L, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem Sci. 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- 44.Lafontaine D L J, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamond A I, Earnshaw W C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 46.Lange T S, Ezrokhi M, Amaldi F, Gerbi S A. Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA. Mol Biol Cell. 1999;10:3877–3890. doi: 10.1091/mbc.10.11.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewis J D, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- 48.Li H, White S A. RNA apatamers for yeast ribosomal protein L32 have a conserved purine-rich internal loop. RNA. 1997;3:245–254. [PMC free article] [PubMed] [Google Scholar]
- 49.Ludin B, Doll T, Meili R, Kaech S, Matus A. Application of novel vectors for GFP-tagging of proteins to study microtubule-associated proteins. Gene. 1996;173:107–111. doi: 10.1016/0378-1119(95)00899-3. [DOI] [PubMed] [Google Scholar]
- 50.Lygerou Z, Allmang C, Tollervey D, Séraphin B. Accurate processing of a eukaryotic pre-rRNA by RNase MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 51.Maden B E. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 52.Maiorano D, Brimage L J E, Leroy D, Kearsey S E. Functional conservation and cell cycle localization of the Nhp2 core component of H+ACA snoRNPs in fission and budding yeasts. Exp Cell Res. 1999;252:165–174. doi: 10.1006/excr.1999.4607. [DOI] [PubMed] [Google Scholar]
- 53.Matera A G. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- 54.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 55.Meier U T, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- 56.Meier U T, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell J R, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell J R, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 59.Narayanan A, Lukowiak A, Jády B E, Dragon F, Kiss T, Terns R M, Terns M P. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 1999;18:5120–5130. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narayanan A, Speckmann W, Terns R, Terns M P. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 62.Notrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Lührmann R. Functional interaction of a novel 15.5kDa [U4/U6 · U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 1999;18:6119–6133. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ochs R L, Reilly M T, Freeman J W, Busch H. Intranucleolar localization of human proliferating cell nucleolar antigen p120. Cancer Res. 1988;48:6523–6529. [PubMed] [Google Scholar]
- 64.Phillips B, Billin A N, Cadwell C, Buchholz R, Erickson C, Merriam J R, Carbon J, Poole S J. The Nop60B gene of Drosophilaencodes an essential nucleolar protein that functions in yeast. Mol Gen Genet. 1998;260:20–29. doi: 10.1007/s004380050866. [DOI] [PubMed] [Google Scholar]
- 65.Politz J C, Yarovoi S, Kilroy S M, Gowda K, Zwieb C, Pederson T. Signal recognition particle components in the nucleolus. Proc Natl Acad Sci USA. 2000;97:55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Samarsky D A, Fournier M J, Singer R H, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 68.Shaw P J, Jordan E G. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- 69.Sollner-Webb B, Tycowski K T, Steitz J A. Ribosomal RNA processing in eukaryotes. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA: structure, evolution, gene expression and function in protein synthesis. Boca Raton, Fla: CRC Press; 1996. pp. 469–490. [Google Scholar]
- 70.Stevens S W, Abelson J. Purification of the yeast U4/U6 · U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc Natl Acad Sci USA. 1999;96:7226–7231. doi: 10.1073/pnas.96.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strubin M, Newell J W, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 72.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 73.Tyc K, Steitz J A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 75.Vilardell J, Warner J R. Ribosomal protein L32 of Saccharomyces cerevisiaeinfluences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watanabe Y-I, Gray M W. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: Cbf5 in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res. 2000;28:2342–2352. doi: 10.1093/nar/28.12.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watkins N J, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Lührmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weinstein L B, Steitz J A. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 79.Wigley W C, Fabunmi R P, Lee M G, Marino C R, Muallem S, DeMartino G N, Thomas P J. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Y, Isaac C, Wang C, Dragon F, Pogacic V, Meier U T. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol Biol Cell. 2000;11:567–577. doi: 10.1091/mbc.11.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu Y-T, Scharl E C, Smith C M, Steitz J A. The growing world of small nuclear ribonucleoproteins. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 487–524. [Google Scholar]
- 82.Zebarjadian Y, King T, Fournier M J, Clarke L, Carbon J. Point mutations in yeast CBF5can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]