Abstract
Smad4 plays a pivotal role in all transforming growth factor β (TGF-β) signaling pathways. Here we describe six widely expressed alternatively spliced variants of human Smad4 with deletions of different exons in the linker, the region of Smad4 that separates the two well-conserved MH1 and MH2 domains. All these Smad4 variants form complexes with activated Smad2 and Smad3 and are incorporated into DNA-binding complexes with the transcription factor Fast-1, regardless of the amount of linker they contain. However, sequences encoded by exons 5 to 7 in the linker are essential for transcriptional activation. Most importantly, our observation that different Smad4 isoforms have different subcellular localizations has led us to the identification of a functional CRM1-dependent nuclear export signal in the Smad4 linker and a constitutively active nuclear localization signal in the N-terminal MH1 domain. In the absence of TGF-β signaling, we conclude that Smad4 is rapidly and continuously shuttling between the nucleus and the cytoplasm, the distribution of Smad4 between the nucleus and the cytoplasm being dictated by the relative strengths of the nuclear import and export signals. We demonstrate that inhibition of CRM1-mediated nuclear export by treatment of cells with leptomycin B results in endogenous Smad4 accumulating very rapidly in the nucleus. Endogenous Smad2 and Smad3 are completely unaffected by leptomycin B treatment, indicating that the nucleocytoplasmic shuttling is specific for Smad4. We propose that, upon TGF-β signaling, complex formation between Smad4 and activated Smad2 or -3 leads to nuclear accumulation of Smad4 through inhibition of its nuclear export. We demonstrate that after prolonged TGF-β signaling Smad2 becomes dephosphorylated and Smad2 and Smad4 accumulate back in the cytoplasm.
Members of the transforming growth factor β (TGF-β) superfamily of growth and differentiation factors regulate many diverse biological processes including cell proliferation, differentiation, migration, adhesion, survival, and specification of developmental fate (29). The ligands act by binding and activating pairs of serine/threonine kinase receptors (type I and type II), and the signals are transduced to the nucleus by the Smads (29). The receptor-regulated Smads (R-Smads), for example, Smad2 and Smad3, are directly phosphorylated by the activated type I receptors and consequently form activated complexes with a co-Smad (a Smad4 family member [29]). These complexes translocate to the nucleus, where they are involved in the regulation of transcription of target genes. Some Smad complexes, such as the TGF-β-activated Smad3-Smad4 complexes (46), bind DNA directly, but others, such as ligand-induced Smad2-Smad4 complexes or Smad1-Smad4 complexes, require specific transcription factors to recruit them to DNA (2, 10, 15).
A fundamental step in TGF-β signal transduction is the translocation of the Smads from the cytoplasm to the nucleus, but little is understood about its molecular mechanism or regulation. A membrane-associated protein, SARA, binds Smad2 and Smad3 in the cytoplasm and presents them to the activated type I receptors for phosphorylation (41). It is not clear whether SARA acts as a cytoplasmic anchor for these R-Smads, or whether the Smads associate only transiently prior to recruitment to the receptor. No such specific cytoplasmic anchor has been found for the other R-Smads or co-Smads. Other work has demonstrated that Smad2, Smad3, and Smad4 are bound to microtubules in unstimulated cells and that TGF-β triggers dissociation, allowing translocation of the Smads to the nucleus (8). Whether association with microtubules is sufficient to retain the Smads in the cytoplasm is unclear. Upon signaling, active complexes of R-Smads and co-Smads rapidly move to the nucleus, but little is known about the mechanism involved.
In the nucleus, activated Smad complexes regulate transcription. However, this is transient, indicating that after a certain elapsed time the signal must be terminated and the Smads must be deactivated. Several mechanisms have been proposed for this. Firstly, TGF-β family members upregulate the synthesis of inhibitory Smads, Smad6 and Smad7, which can bind the type I receptors, blocking their ability to phosphorylate the R-Smads and preventing further R-Smad activation (29). Secondly, phosphorylated nuclear Smad2 is ubiquitinated and targeted for destruction by the proteasome (28). However, the fate of the associated Smad4 in these circumstances is not known (16, 28). There is as yet no direct evidence for the most obvious termination mechanism, dephosphorylation of the R-Smads.
The Smads contain two well-conserved domains, the N-terminal MH1 domain and the C-terminal MH2 domain, which are separated by a proline-rich linker that differs substantially between the different Smad classes but has been well conserved through evolution. Whereas the MH1 and MH2 domains are functionally well characterized, much less is known about the role of the linker (29). In Smad4, the region of the linker adjacent to the MH2 domain, known as the Smad activation domain (SAD), is involved in transcriptional activation, mediated by the coactivator histone acetyltransferase p300/CBP (4, 5). In addition, other evidence points to a function for the linker in subcellular localization. Phosphorylation of the R-Smads in the linker by extracellular signal-regulated kinases (Erks) leads to sequestration of at least a proportion of the R-Smads in the cytoplasm, even in the presence of a TGF-β or bone morphogenetic protein (BMP) signal (25, 26). Moreover, two Smad4s that differ predominantly in the linker region have recently been found in Xenopus (18, 31). They both form complexes with activated R-Smads which have similar transcriptional activities. However, whereas XSmad4α, the Xenopus orthologue of human Smad4, moves from the cytoplasm to the nucleus upon ligand stimulation, XSmad4β, which has a divergent linker, is constitutively localized in the nucleus (18, 31).
Here we identify six widely expressed alternatively spliced variants of human Smad4 with deletions in different regions of the linker. Analysis of these Smad4 variants has allowed us to investigate the role of the Smad4 linker and to uncover a novel mode of regulation of Smad4. We demonstrate that all these Smad4 isoforms form complexes with activated Smad2 and Smad3 and are incorporated into DNA-binding complexes with the transcription factor Fast-1, regardless of the amount of linker they contain. However, the sequences encoded by exons 5 to 7 are required for full transcriptional activity. Our demonstration that different Smad4 isoforms have different subcellular localizations has led us to the identification of a functional CRM1-dependent nuclear export signal (NES) in the linker and a constitutively active nuclear localization signal (NLS) in the MH1 domain. We therefore propose that, in the absence of TGF-β, Smad4 shuttles continuously between the cytoplasm and the nucleus. We demonstrate that this nucleocytoplasmic shuttling is a property of endogenous Smad4 but not of endogenous Smad2 or Smad3. Upon TGF-β signaling, formation of complexes with activated Smad2 or -3 leads to nuclear accumulation of Smad4, possibly through inhibition of NES function. Our data also shed light on the mechanism of termination of TGF-β signaling. We demonstrate that following prolonged TGF-β signaling Smad2 is dephosphorylated and this coincides with the disappearance of Smad2 and Smad4 from the nucleus and their accumulation back in the cytoplasm.
MATERIALS AND METHODS
Plasmids.
Mixer, Fast-1, XSmad2, and hSmad4 in EF expression vectors (17) have been described previously (10). Human and mouse alternatively spliced Smad4 variants, obtained by reverse transcription-PCR (RT-PCR), were subcloned into pGEM-T (Promega) and subsequently into EF expression vectors. NES, NLS, and NES-NLS mutants of Smad4 (see Fig. 5A) were created using PCR. EF-[ALK5 (TD)] was generated by subcloning the coding sequence of the constitutively active ALK5 (32) into the EF expression vector. The activin-responsive element (ARE)- and distal element (DE)-driven luciferase reporter plasmids were generated by moving the AREs or DEs and minimal γ-actin promoter from the chloramphenicol acetyltransferase versions (10) into pGL3 (Promega).
FIG. 5.
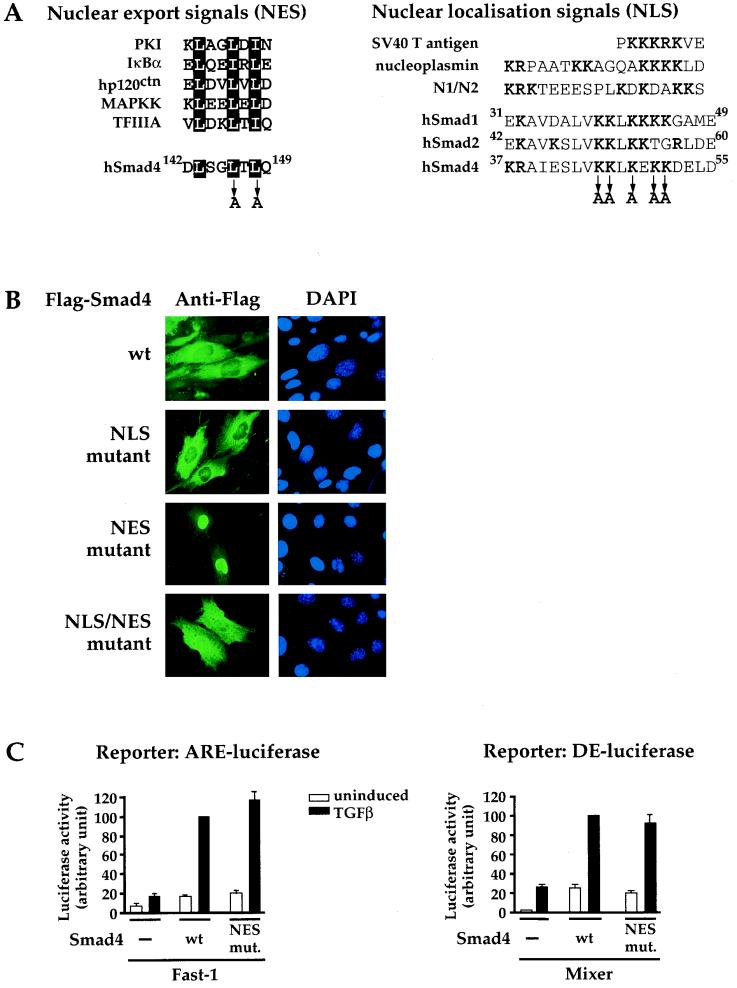
Smad4 contains an NES and an NLS. (A) The putative NES encoded by exon 3 of Smad4 is aligned with other previously characterized NESs (21, 43). Leucines 146 and 148 were mutated to alanine to generate the NES mutant as shown. The putative NLS of Smad4 is aligned with well-characterized NLSs (7) and with the putative NLSs of the R-Smads, Smad1 and Smad2. Lysines 45, 46, 48, 50, and 51 were mutated to alanine to generate the Smad4 NLS mutant. (B) The NES and NLS of Smad4 are functional. NIH 3T3 cells were transfected with Flag-tagged Smad4 derivatives, and their subcellular localization was determined as described for Fig. 4. (C) Smad4 forms functional complexes with activated Smad2 and Fast-1 or Mixer in the nucleus. MDA-MB468 cells were transfected with the appropriate luciferase reporters, plasmids expressing transcription factors (Fast-1 or Mixer), and wild-type or mutant Smad4 as shown. TGF-β inductions and luciferase assays were performed as described for Fig. 3. The data are averaged from at least three independent experiments, which were normalized by setting the TGF-β induction mediated by wild-type Smad4 to 100%. PKI, cyclic AMP-dependent protein kinase inhibitor; MAPKK, mitogen-activated protein kinase kinase; wt, wild type; mut., mutant; SV40, simian virus 40.
Oligonucleotides.
The oligonucleotides used were as follows: 1, ATGGACAATATGTCTATWACRAATAC (hSmad4 exon 1 forward); 2, ACCTGAACYTCCATTTCAAAGTAAGC (hSmad4 exon 8 reverse); 3, GTGTGAATCCATATCACTACG (hSmad4 exon 2); 4, CTCTCAGGATTAACACTGCAGAG (hSmad4 exon 3); 5, TATGTGCATGACTTTGAGGGAC (hSmad4 exon 4); 6, GGCAGCCATAGTGAAGGACTG (hSmad4 exon 5); 7, GGCGGGTGGTGCTGAAGATGG (hSmad4 exon 6); 8, CAGCCTCCCATTTCCAATCATCC (hSmad4 exon 7); 9, ATGTCGTCCATCTTGCCATTCAC (hSmad2 exon 2 forward); and 10, ATTGAACACCARAATGCAGGTTC (hSmad2 exon 8 reverse).
RNA isolation, RT-PCR, and RNase protections.
Isolation of total RNA from HaCaT cells and adult mouse tissues and RNase protection assays were performed as described previously (17, 18). The antisense probe for the loading control, γ-actin, was as described previously (9). The antisense probe that detects mouse Smad4 Δ5-6 protected 12 nucleotides of exon 3, exon 4, exon 7, and 48 nucleotides of exon 8; that designed to detect mouse Smad4 Δ4-7 protected 55 nucleotides of exon 1, exon 2, exon 3, and 48 nucleotides of exon 8. Note that each of these probes can also detect wild-type mouse Smad4. RT-PCR was performed as described previously (20). Oligonucleotides 1 and 2 were used as PCR primers to detect alternatively spliced hSmad4 variants, and oligonucleotides 9 and 10 were used for hSmad2.
Southern blotting and silver staining.
RT-PCR products were separated on a nondenaturing 5% polyacrylamide gel, which was silver stained. For Southern blotting, the PCR products were transferred to a Hybond-N membrane and hybridized with specific oligonucleotide probes directed against particular exons, which were prepared by end labeling oligonucleotides 1 to 8 (above).
Cell culture and transfections.
HaCaT, NIH 3T3, and MDA-MB468 cells were all maintained in Dulbecco's modified Eagle medium containing 10% fetal calf serum (FCS). NIH 3T3 cells were transfected using Lipofectamine (Life Technologies) and MDA-MB468 cells were transfected using Superfect reagent (Qiagen) with the plasmids indicated in the figure legends.
Immunoprecipitation-Western blotting, band shift, and transcriptional assays.
Immunoprecipitation assays followed by Western blotting were performed essentially as described previously (10) except that the NaCl concentration in the lysis buffer was 400 mM, and the lysates were diluted to a final concentration of 160 mM NaCl before immunoprecipitation. Band shift assays using the ARE probe were as previously described (10). For transcriptional assays, cells were lysed in reporter lysis buffer (Promega), and luciferase assays were performed as described previously (22). β-Galactosidase assays were performed using chlorophenol red-β-d-galactopyranoside (Calbiochem) as a substrate and quantitated spectrophotometrically.
Nuclear and cytoplasmic extracts, TGF-β time courses, and cycloheximide treatment.
For the TGF-β time courses, cultures of HaCaT and NIH 3T3 cells growing in Dulbecco's modified Eagle medium–10% FCS were treated with 20 μg of cycloheximide per ml to prevent further protein synthesis and TGF-β was added at different times. All cells were harvested together at the final time point. Nuclear extracts were prepared as described previously (46). The cytoplasmic extracts corresponded to the initial low-salt fraction that was concentrated in a centrifugal filter (Millipore) to the same volume as the nuclear extracts. Equal volumes of these extracts were loaded on the sodium dodecyl sulfate (SDS)-polyacrylamide gel for Western blotting with monoclonal antibodies against Smad4 (B8; Santa Cruz), Smad2 (which also recognizes Smad3; Transduction Laboratories), PCNA (18), or GRB2 (Transduction Laboratories) or polyclonal antibodies against phosphorylated Smad2 (Upstate Biotechnology), the ERM proteins (Santa Cruz), or poly(ADP-ribose) polymerase (Roche). Visualization was performed by ECL (Amersham). To assess the efficiency with which cycloheximide inhibited protein synthesis, HaCaT and NIH 3T3 cells were incubated with 100 μCi of Pro-Mix (Amersham) for 9 h in the absence or presence of 20 μg of cycloheximide per ml. Cells were lysed in SDS-gel sample buffer and fractionated in an SDS-polyacrylamide gel which was stained with Coomassie blue, scanned, and quantitated. [35S]methionine and [35S]cysteine incorporation was quantitated using a PhosphorImager, and this was normalized to the Coomassie blue stain. From these data, the percent inhibition of translation was calculated.
Indirect immunofluorescence microscopy.
Immunofluorescence to detect transfected Flag-tagged Smad4 isoforms was performed as described previously (37) using the mouse anti-Flag monoclonal antibody (M2; Sigma) as the primary antibody and the rabbit anti-mouse immunoglobulin antibody coupled to fluorescein isothiocyanate (DAKO) as the secondary antibody. To detect the endogenous Smad2, Smad3, and Smad4 in HaCaT cells, the cells were treated as described in the figure legends, fixed in 4% formaldehyde in phosphate-buffered saline (PBS), permeabilized with 0.3% Triton X-100–PBS, and then blocked in 5% milk–10% FCS–0.3% bovine serum albumin–0.3% Triton X-100 in PBS for 30 min at room temperature. The primary antibodies against Smad2 and Smad3 or Smad4 were as described for Western blotting above, and the secondary antibody was as described above. Antibodies were used at a dilution of 1 in 250 (anti-Smad2 and -Smad3), 1 in 100 (anti-Smad4), or 1 in 200 (secondary antibody) in 10% FCS–0.3% bovine serum albumin–0.3% Triton X-100 in PBS. The washes were done with 0.1% Triton X-100–PBS. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted in Vectashield (Vector Laboratories, Inc.). Fluorescence was observed immediately either with a Zeiss Axioplan upright fluorescence microscope or with a Zeiss confocal LSM 510 microscope.
RESULTS
The Smad4 mRNA is alternatively spliced.
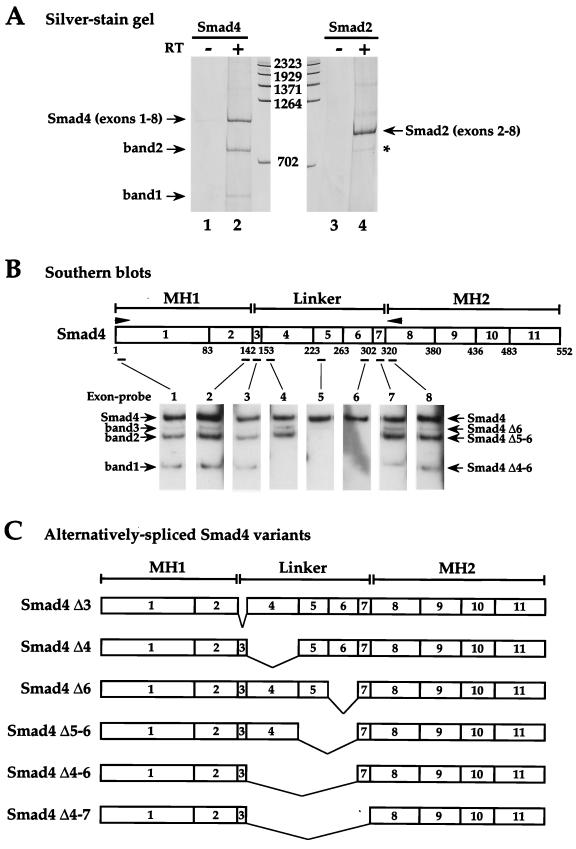
In the course of our studies of Smad4 expression, we performed RT-PCR with a forward primer centered on the initiation codon of Smad4 in exon 1 and a reverse primer at the beginning of the MH2 domain in exon 8, using total RNA from the TGF-β-responsive human keratinocyte cell line, HaCaT, as a template (Fig. 1A and B). In addition to the expected full-length product (1,004 bp; lane 2), two smaller fragments were also detected (lane 2, bands 1 and 2). These products corresponded to fragments of approximately 560 and 770 bp, respectively. Analysis of the human Smad4 genomic sequence reveals that any of the five exons between the end of the MH1 domain (exon 2) and the beginning of the MH2 domain (exon 8) can be deleted while maintaining the correct reading frame (13), suggesting that these smaller fragments may be derived from alternatively spliced variants of Smad4 containing different lengths of linker.
FIG. 1.
Human Smad4 mRNA is alternatively spliced. (A) Silver-stained polyacrylamide gel showing fragments obtained from RT-PCRs performed using total RNA from HaCaT cells as template and forward and reverse primers specific for exon 1 and exon 8, respectively, of human Smad4 (lanes 1 and 2) or exon 2 and exon 8, respectively, of human Smad2 (lanes 3 and 4). The reactions in lanes 1 and 3 were performed with no reverse transcriptase. DNA markers are measured in base pairs. The fragment derived from full-length Smad4 is designated Smad4 (exons 1-8), and that derived from full-length Smad2 is designated Smad2 (exons 2-8). Bands 1 and 2 are derived from alternatively spliced Smad4 mRNA. The asterisk denotes the Smad2 alternatively spliced variant (Smad2 Δexon 3) (49). (B) Identification of Smad4 isoforms by Southern blotting. A diagram of the Smad4 coding region is shown, denoting the MH1, linker, and MH2 domains. Arrowheads show PCR primers used in the RT-PCR. Exons are denoted as boxes with the exon numbers indicated. Numbers below are amino acids at the exon-intron boundaries (13). Positions of the oligonucleotide exon-specific probes are indicated. RT-PCR products similar to that in lane 2 above were Southern blotted with exon-specific probes as indicated. The bands 1 and 2 are as described above; band 3 is detected only by Southern blotting. The bands are identified as Smad4 Δ4-6, Δ5-6, and Δ6, respectively. (C) Schematics of the alternatively spliced Smad4 isoforms identified.
To investigate whether this was the case, the PCR fragments were blotted onto nitrocellulose and hybridized with oligonucleotide probes specific for each of the eight amplified exons. The product derived from full-length Smad4 hybridized with all eight probes as expected (Fig. 1B). Band 1 hybridized with all the probes except those directed against exons 4, 5, and 6, suggesting that it corresponds to an alternatively spliced Smad4 variant with deletions of exons 4, 5, and 6 (Smad4 Δ4-6). Similarly, band 2 was identified as Smad4 with deletions of exons 5 and 6 (Smad4 Δ5-6). An additional very weak band (Fig. 1B, band 3) was detected by Southern blotting and was not visualized by silver staining. It corresponded to Smad4 with a deletion of exon 6 (see below). These shorter PCR products were therefore derived from alternatively spliced variants of Smad4 lacking specific sequences in the linker.
Cloning and sequencing of these products (bands 1 to 3) confirmed their identity (data not shown). In addition, three more Smad4 isoforms (Smad4 Δ3, Smad4 Δ4, and Smad4 Δ4-7) were detected by sequencing the PCR products (data not shown). They were not detected by Southern blotting or with silver stain because their sizes are such that they are not resolved from other spliced variants of similar size by polyacrylamide gel electrophoresis.
Alternative splicing of the region encoding the linker is not a general property of all Smads. A similar RT-PCR analysis of the receptor-regulated Smad Smad2 (Fig. 1A, lane 4) generated only a major product (858 bp) corresponding to full-length Smad2 and a shorter fragment (768 bp, marked with an asterisk) that arises from an alternatively spliced variant lacking sequences in the MH1 domain (Smad2 Δexon 3) (49). No fragments corresponding to Smad2 derivatives lacking linker sequences were detected.
Taken together, these results indicate that the Smad4 mRNA is alternatively spliced in the linker region to give six different mRNA isoforms (Smad4 Δ3, Δ4, Δ5-6, Δ6, Δ4-6, and Δ4-7) (Fig. 1C).
The alternatively spliced variants of Smad4 are widely expressed.
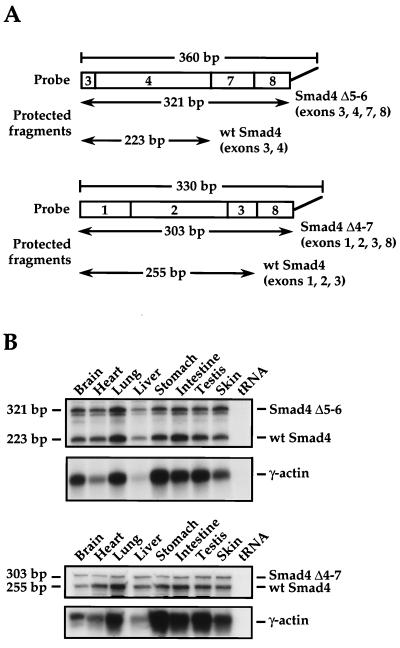
One of the Smad4 isoforms (Smad4 Δ5-6) was previously identified in a breast tumor cell line, MDA-MB231, thought to arise from a mutation leading to inappropriate splicing (6), and Smad4 Δ5-6 and Smad4 Δ4-6 have also been detected in neuroblastomas (24). To determine whether differential splicing of the Smad4 linker region was a widespread phenomenon, we generated mouse RNase protection probes to investigate the presence of alternatively spliced Smad4 variants in a panel of mouse tissues (Fig. 2A). Smad4 Δ5-6 and Smad4 Δ4-7 were used as examples of abundant and less abundant isoforms, respectively. These Smad4 isoforms are present in every mouse tissue examined (Fig. 2B). All six alternatively spliced Smad4 variants are also present in embryonic stem cells and mouse embryo fibroblast cells (data not shown). Thus, these alternatively spliced Smad4 variants are widely expressed in mammalian tissues and cell lines with full-length Smad4.
FIG. 2.
The alternatively spliced Smad4 variants are widely expressed in the adult mouse. (A) Diagram of the RNase protection probes designed to specifically protect Smad4 Δ5-6 mRNA and Smad4 Δ4-7 mRNA and the sizes of the resulting protected fragments. In both cases, the probes also protect full-length Smad4 mRNA, giving rise to a second, smaller fragment. (B) RNase protection assays detecting Smad4 Δ5-6 (upper panels) or Smad4 Δ4-7 (lower panels) in different adult mouse tissues. Protected fragments corresponding to the alternatively spliced mRNA variants and full-length Smad4 mRNA are indicated. γ-Actin was used as a loading control for all tissues. wt, wild type.
We have also detected alternatively spliced Smad4 mRNAs in other vertebrates: in zebra fish, a highly abundant mRNA corresponding to Smad4 Δ5 was isolated, and in Xenopus laevis, an XSmad4α with a deletion of exon 6 was identified (data not shown and reference 18).
The Smad4 linker is not required for interaction with activated Smad2 or Smad3 or for complex formation with activated Smad2 and Fast-1.
These naturally occurring Smad4 isoforms containing different lengths of linker provide us with ideal molecular tools to investigate the function of the Smad4 linker in TGF-β signaling.
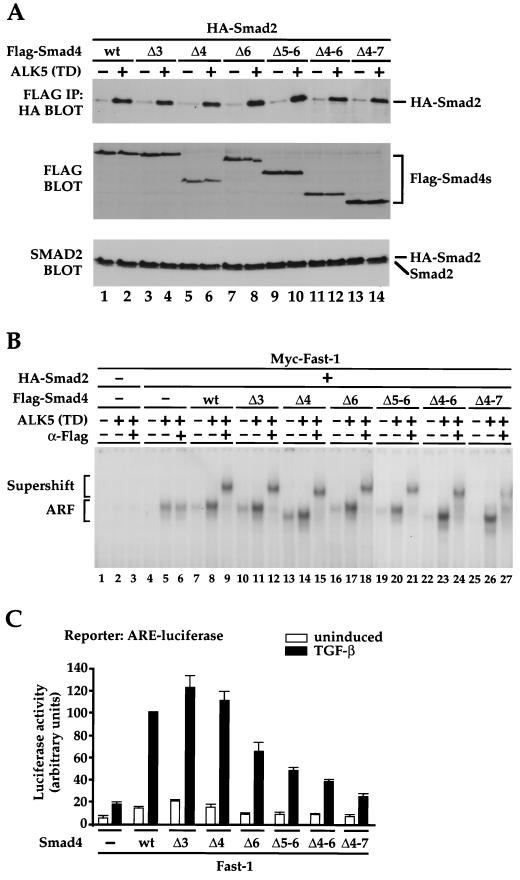
We first determined, using coimmunoprecipitation, whether the alternatively spliced Smad4 variants could associate with Smad2 in a signaling-dependent manner. This would indicate whether the linker was involved in this and whether the MH2 domain (which is known to be required for Smad2 interaction [14, 47]) was correctly folded in all the Smad4 isoforms. Flag-tagged Smad4 isoforms were coexpressed in NIH 3T3 cells with hemagglutinin (HA)-tagged Smad2, with or without the constitutively active TGF-β receptor [ALK5 (TD)] (45) to mimic TGF-β signaling. The Smad4 derivatives were immunoprecipitated with the anti-Flag antibody, and the immunoprecipitates were blotted with an anti-HA antibody to detect coprecipitating Smad2 (Fig. 3A, top panel). Equal loading of extracts and equal protein expression were confirmed by Western blotting of whole-cell extracts using anti-Flag or anti-Smad2 antibodies (Fig. 3A, middle and bottom panels). Smad2 clearly interacts with all the Smad4 isoforms in a signaling-dependent manner. Most strikingly, Smad4 Δ4-7, the isoform that has virtually no linker, interacts with activated Smad2 as efficiently as does full-length Smad4 (Fig. 3A, lanes 1, 2, 13, and 14). Thus, the Smad4 linker is not required for association with Smad2, and all six alternatively spliced Smad4 variants have correctly folded MH2 domains.
FIG. 3.
Sequences in the Smad4 linker are not required for formation of transcription factor complexes but are required for transcriptional activation. (A) The Smad4 linker is not required for interaction with activated Smad2. Extracts were prepared from NIH 3T3 cells transfected with the different Flag-tagged alternatively spliced Smad4 variants with HA-Smad2, with or without constitutively active ALK5 [ALK5 (TD)] as indicated. Extracts were assayed either by immunoprecipitation of complexes with anti-Flag antibody followed by Western blotting with anti-HA antibody (top panel) or by Western blotting the whole extract with anti-Flag antibody (middle panel) or with anti-Smad2 antibody (bottom panel). (B) The Smad4 linker is not required for formation of the Fast-1–Smad2–Smad4 complex ARF on the ARE. Extracts were prepared from NIH 3T3 cells transfected with Myc–Fast-1, HA-Smad2, and Flag-tagged alternatively spliced Smad4 variants and ALK5 (TD) as indicated. The extracts were analyzed by band shift using the ARE as probe in the presence or absence of anti-Flag antibody (α-Flag) as indicated to confirm the presence of the Flag-tagged Smad4 spliced variant in the complex. The Fast-1–Smad2–Smad4 complex ARF is indicated, as is the supershifted ARF complex. (C) Sequences encoded by exons 5, 6, and 7 are required for transcriptional activation mediated by Smad4. MDA-MB468 cells were transfected with the ARE-luciferase reporter, plasmids expressing Fast-1, and alternatively spliced Smad4 variants as indicated. Cells were cultured with or without TGF-β1 (2 ng/ml) for 6 h. Cells were harvested, and luciferase activity was measured relative to LacZ activity from the internal control. The data are averaged from at least four independent experiments and were normalized by setting the TGF-β induction mediated by wild-type Smad4 to 100%. IP, immunoprecipitation; wt, wild type.
Activated Smad2-Smad4 complexes are predominantly recruited to DNA through their interaction with other transcription factors (30, 40, 44). The best characterized is the Xenopus winged-helix/Forkhead transcription factor, Fast-1, which forms an activin-induced complex with Smad2 and Smad4 called ARF (for activin-responsive factor) at the ARE of the Xenopus Mix.2 promoter (2, 3, 19). We therefore investigated whether all the Smad4 isoforms could be incorporated into the ARE-bound ARF complex upon TGF-β signaling. Extracts were prepared from NIH 3T3 cells transfected with plasmids expressing Myc–Fast-1, HA-Smad2, and Flag-Smad4 isoforms, with or without activated ALK5. The ARF complex is barely detected upon TGF-β signaling when Fast-1 is expressed (Fig. 3B, lanes 1 to 3) but is enhanced when Smad2 is expressed and further induced by expression of wild-type Smad4 (lanes 5 and 8). Each of the Smad4 isoforms, regardless of the amount of linker sequences that they contain, is incorporated into the ARF complex, as seen by the high level of ARF complex formed, migrating according to the size of the Smad4 isoform that it contains and supershifting with the anti-Flag antibody (Fig. 3B, lanes 10 to 27; also compare mobilities of the complexes with the mobilities of the Smad4 isoforms on the SDS-polyacrylamide gel in Fig. 3A, middle panel). In addition, all the Smad4 isoforms formed nuclear complexes with Smad3 upon TGF-β signaling that bound to the Smad-binding element from the c-jun promoter (data not shown and reference 46). The linker therefore plays no role in Smad complex recruitment by transcription factors or in the formation of the DNA-binding Smad complexes.
Exons 5, 6, and 7 of Smad4 are required for its transcriptional activity.
Previous work has identified a region in the Smad4 linker adjacent to the MH2 domain (amino acids 275 to 322, encoded by exon 7 and part of exon 6) as a SAD, required for transcriptional activation (4, 5). To investigate whether other linker sequences might also be involved, we assayed the transcriptional activity of the alternatively spliced isoforms of Smad4 in the absence and presence of TGF-β, in the context of the ARF complex bound to the ARE. The Smad4-null cell line MDA-MB468 (36) was used, to avoid interference with endogenous Smad4.
In the absence of Smad4, Fast-1 elicited a weak TGF-β induction via the ARE (Fig. 3C). Overexpression of wild-type Smad4 increased the basal activity and substantially increased the TGF-β-induced level of transcription (Fig. 3C). The results indicated that neither exon 3 nor exon 4 contributed to TGF-β-induced transcriptional activity of Smad4 but that sequences encoded by exons 5, 6, and 7 are all required for full TGF-β-induced transcriptional activation by Smad4. Exactly analogous results were obtained when the Smad4 isoforms were assayed in the context of a different transcription factor-Smad complex, the Mixer-Smad2-Smad4 complex that binds the DE of the Xenopus goosecoid promoter (reference 10 and data not shown).
Smad4 is actively exported from the nucleus.
In the current model for TGF-β signaling, Smad4 is assumed to be retained in the cytoplasm in the absence of a signal (29), but nothing is known about the mechanism whereby this is achieved. To determine whether sequences within the linker are required, we investigated the subcellular localization of the alternatively spliced Smad4 variants.
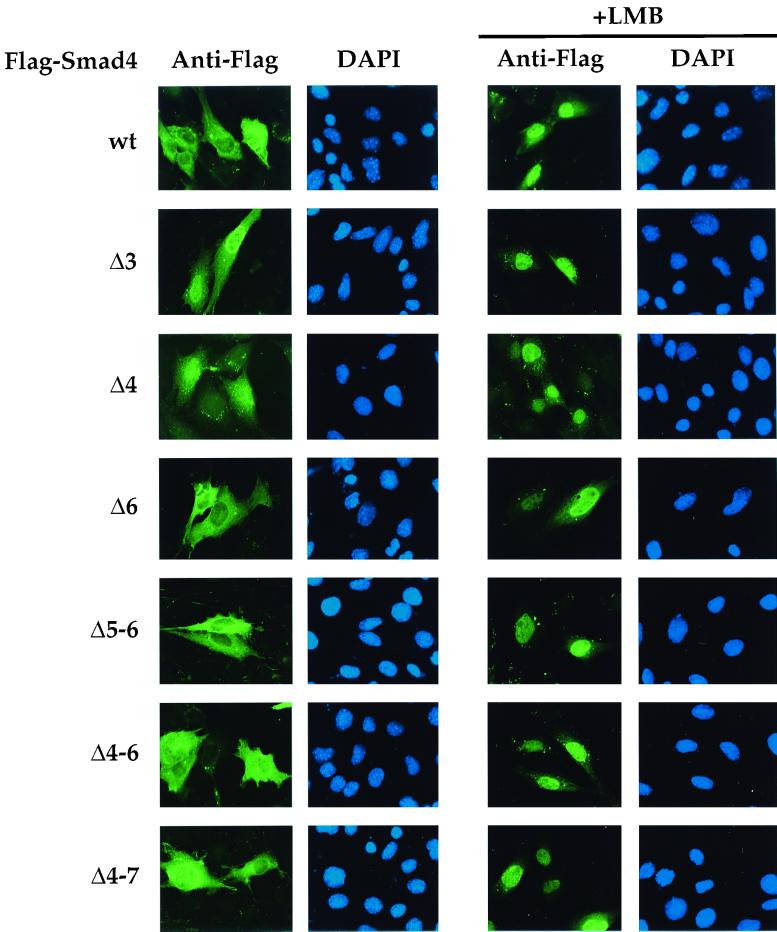
Flag-tagged derivatives of the Smad4 isoforms were transfected into NIH 3T3 cells, and their subcellular localization in the absence of TGF-β signaling was determined by indirect immunofluorescence (Fig. 4, left panels). Transfected Smad4 is localized both in the cytoplasm and in the nucleus, with cytoplasmic staining being stronger than nuclear staining in these cells (Fig. 4, left) (27). Deletion of exon 6 or exons 5 and 6 had no effect on the subcellular localization of Smad4 (Fig. 4, left; Smad4 Δ6 and Δ5-6). In contrast, deletion of exon 3 had a dramatic effect, in that Smad4 Δ3 was localized in the nuclei of all transfected cells (Fig. 4, left). Deletion of exon 4 gave a similar, though less emphatic, result, in that some strong staining is still seen outside the nucleus (Fig. 4, left, Smad4 Δ4). Smad4 isoforms that contained deletions of exon 4 in the context of larger deletions were more or less uniformly distributed throughout the cell (Fig. 4, left, Smad4 Δ4-6 and Δ4-7) (see below).
FIG. 4.
Subcellular localization of the alternatively spliced Smad4 variants in the absence and presence of LMB. NIH 3T3 cells were transfected with Flag-tagged Smad4 isoforms as indicated. (Left) The subcellular localization of the Smad4 isoforms in untreated cells was determined by indirect immunofluorescence using the anti-Flag antibody, and nuclei of the same cells were also stained with DAPI as indicated. (Right) Subcellular localization of the Smad4 isoforms in cells treated with LMB at a 20-ng/ml final concentration for 1 h. wt, wild type.
These results enabled us to make two predictions. Firstly, the nuclear localization of Smad4 Δ3 and, to a lesser extent, Smad4 Δ4 indicates that Smad4 must be capable of being constitutively transported to the nucleus in the absence of signaling, because the alternatively spliced Smad4 variants are relatively large proteins (greater than 50 kDa) that cannot freely diffuse across the nuclear membrane (12). Secondly, for Smad4 to be normally more cytoplasmic than nuclear, it must then be either actively exported from the nucleus or actively retained in the cytoplasm. This activity must require sequences encoded by exon 3 and also exon 4.
The best-understood nuclear export mechanism is that mediated by a short NES which requires the CRM1 protein (12). To test whether Smad4 undergoes CRM1-mediated nuclear export, we investigated whether the localization of wild-type Smad4 was sensitive to leptomycin B (LMB), a specific inhibitor of CRM1-mediated nuclear export (34). After treatment with LMB for 1 h, full-length Smad4 was indeed completely nuclear (Fig. 4, right panels, top). Thus, in untreated cells Smad4 must be constitutively imported into the nucleus and then immediately exported. When nuclear export is blocked by LMB treatment, Smad4 is trapped in the nucleus. We also investigated the behavior of all the Smad4 alternatively spliced variants upon LMB treatment. In all cases, they rapidly accumulated in the nucleus, except Smad4 Δ3, which was already nuclear (Fig. 4, right panels). This indicates that any cytoplasmic localization of the Smad4 isoforms is due to the activity of an NES, presumably located in exon 3, and that a strong NLS must be present in Smad4, outside the linker, in the MH1 domain or MH2 domain (see below).
In the absence of TGF-β signaling, therefore, a nuclear export mechanism is responsible for the substantially cytoplasmic localization of Smad4 in NIH 3T3 cells.
Smad4 contains a canonical NES.
Prototypic NESs are short hydrophobic sequences, rich in leucine residues (Fig. 5A) (12, 33). Human Smad4 contains a putative leucine-rich NES in exon 3 (Fig. 5A). This sequence is absolutely conserved in mouse, rat, and chick Smad4; Xenopus Smad4α; and Drosophila Medea. Significantly, it is absent in Xenopus Smad4β, which is known to be constitutively nuclear (18, 31). The R-Smads do not contain such sequences.
To test whether this putative NES was functional, we mutated two of the critical leucine residues (Leu-146 and Leu-148) to alanines (Fig. 5A). The resulting Smad4-NES mutant was exclusively localized in the nucleus (Fig. 5B), indicating that exon 3 of Smad4 contains a functional NES that is absolutely required for nuclear export.
In addition, deletion of exon 4, either alone or in the context of a larger deletion, was shown above to at least partially abolish the nuclear export activity in Smad4, indicating that sequences in exon 4, outside the canonical NES, are also required for full NES function. Deleting exon 4 alone had a bigger effect than deleting it with exons 5 and 6, or with exons 5, 6, and 7 (Fig. 4). This suggests that the important residues are at the exon 3-exon 4 boundary, which are different in Smad4 Δ4 from those in Smad4 Δ4-6 or Smad4 Δ4-7. The most likely explanation is that the requirement for the residues in exon 4 is structural, providing the NES in exon 3 with an appropriate protein context to adopt the correct conformation ftk;2or CRM1 binding, as previously proposed for other NESs (12).
We have, therefore, identified a functional NES encoded by exon 3 of Smad4, which is necessary, though not sufficient, to mediate nuclear export.
Smad4 contains a functional NLS in its MH1 domain.
The rapid accumulation of Smad4 in the nucleus in the presence of LMB and the exclusively nuclear localization of some of the Smad4 isoforms indicate that Smad4 must be actively transported to the nucleus in the absence of TGF-β signaling. The best-understood nuclear import mechanism is mediated by a classical NLS and requires the importin-α/β heterodimer (12). NLSs are characterized by clusters of basic residues, often followed by a hydrophobic aliphatic residue and an acidic residue (12). Two types of NLSs have been identified: those exemplified by the simian virus 40 large-T-antigen NLS, which is a cluster of basic amino acids, and bipartite NLSs, such as those in nucleoplasmin and other histone binding proteins, N1 and N2, which are defined as two basic amino acids and a spacer of any 10 amino acids followed by a cluster of basic amino acids (7) (Fig. 5A). Smad4 contains a sequence reminiscent of a bipartite NLS in its MH1 domain (Fig. 5A). This motif is conserved in all Smad4s, and the basic cluster is conserved in all the R-Smads (48). To test whether this sequence is a functional NLS in Smad4, we generated a mutant in which five lysine residues in this putative motif were mutated to alanines (Fig. 5A). The resulting Smad4-NLS mutant is completely excluded from the nucleus in transfected NIH 3T3 cells. This is not dramatically different from wild-type Smad4, which is substantially cytoplasmic anyway due to its strong NES activity (Fig. 5B). Therefore, to provide stronger evidence for the activity of the NLS, we combined the NLS mutation with the NES mutation. We reasoned that the Smad4-NES mutant was exclusively nuclear because the activity of the NLS transported it into the nucleus, and in the absence of NES function, the mutant was retained there. Mutating the NLS in the context of the mutated NES gave rise to a protein that was uniformly distributed throughout the cell (Fig. 5B). Given that this subcellular localization is completely different from that seen with the Smad4-NES mutant, which is exclusively nuclear, we conclude that this NLS plays a major role in the nuclear import of Smad4. However, the fact that the Smad4-NLS–NES mutant is not entirely cytoplasmic suggests that some additional NLS activity resides elsewhere in Smad4 (see Discussion).
Thus, Smad4 has a functional NLS in its MH1 domain, required for rapid transport of Smad4 to the nucleus, even in the absence of TGF-β signaling. In unstimulated cells, this activity is counteracted by the activity of the NES, which exports Smad4 back into the cytoplasm. Smad4 is therefore shuttling between the nucleus and cytoplasm in unstimulated cells.
Smad4 can associate with activated Smad2 in the nucleus.
Upon TGF-β signaling, activated Smad2 and Smad3 are assumed to form complexes with Smad4 in the cytoplasm which translocate to the nucleus (29). However, activated Smad2 can translocate to the nucleus in the absence of Smad4 in the Smad4-deficient lines SW480.7, MDA-MB468, and BxPC3 (reference 27 and unpublished data). To address whether activated Smad2 can form complexes with a Smad4 that is already nuclear prior to TGF-β signaling, we analyzed the transcriptional activity of the Smad4-NES mutant, which is constitutively nuclear. The transcriptional activity of the Smad4-NES mutant was identical to that of wild-type Smad4, in the context of both a Fast-1–Smad2–Smad4 complex and a Mixer-Smad2-Smad4 complex (10) (Fig. 5C). This indicates that Smad4 can efficiently form complexes with activated Smad2 in the nucleus. Equivalent results were obtained with Smad4 Δ3, which is also constitutively nuclear (Fig. 3C). In addition, no increase in basal activity of the reporters was seen in the absence of TGF-β signaling in cells expressing the Smad4-NES mutant, indicating that the nuclear Smad4 in unstimulated cells is not transcriptionally active.
Endogenous Smad4 shuttles between the cytoplasm and the nucleus.
Our analysis above suggests that transfected Smad4 shuttles continuously between the nucleus and cytoplasm in the absence of TGF-β signaling due to the combined activities of nuclear import and export signals in Smad4. It was essential to prove that endogenous Smad4 also exhibited this behavior and to investigate whether it was specific for Smad4. The TGF-β-responsive keratinocyte cell line HaCaT was used for this analysis, as its cellular architecture facilitates the visualization of endogenous Smads in both unstimulated and stimulated cells. The Smads were detected with monoclonal antibodies specific for Smad4 (exon 5) or for Smad2 and -3, which do not cross-react with any other proteins on Western blots (data not shown) (see Fig. 7).
FIG. 7.
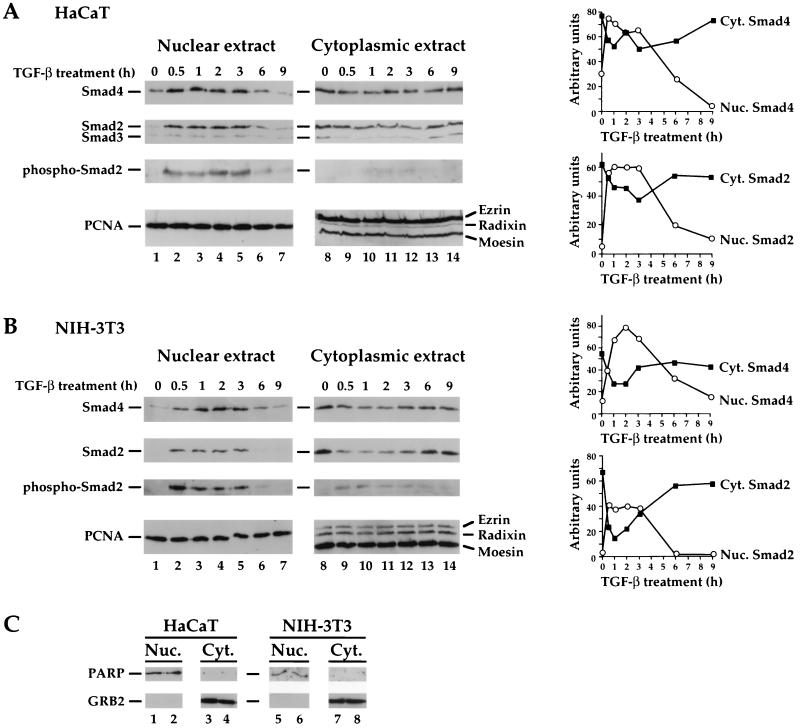
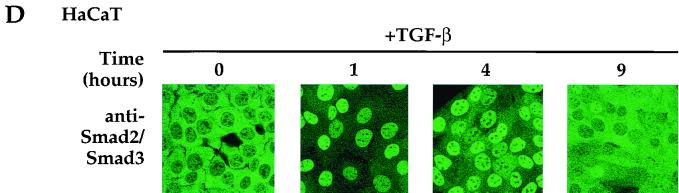
Prolonged TGF-β signaling leads to export of Smad2 and Smad4 from the nucleus to the cytoplasm. (A and B) Nuclear and cytoplasmic extracts were made from HaCaT cells (A) or NIH 3T3 cells (B) that were treated with 20 μg of cycloheximide per ml and were either uninduced or induced with TGF-β (2 ng/ml) for the times shown. They were analyzed by Western blotting with antibodies against Smad4 or Smad2 and -3 (as in Fig. 6); phosphorylated Smad2; the ERM proteins ezrin, radixin, and moesin; or PCNA. Nuclear and cytoplasmic extracts were from the same cells. Cytoplasmic extracts were concentrated to the same volume as nuclear extracts, and equal volumes were loaded on the SDS-polyacrylamide gel. The Western blots were quantitated, and the results are presented graphically (right panels). (C) Western blotting of the nuclear and cytoplasmic extracts from two time points with antibodies against the cytoplasmic protein GRB2 and the nuclear protein poly(ADP-ribose) polymerase (PARP) indicates that the extracts are virtually free from cross-contamination. (D) TGF-β-dependent nuclear import of Smad2 and Smad3 and export after prolonged signaling in HaCaT cells as detected by immunofluorescence. Cells were treated with 20 μg of cycloheximide per ml and were either uninduced or induced with TGF-β (2 ng/ml) for 1, 4, or 9 h and processed for immunofluorescence as described for Fig. 6. Cells were examined by confocal laser scanning microscopy using a Zeiss LSM 510 confocal microscope. Nuc., nuclear; Cyt., cytoplasmic.
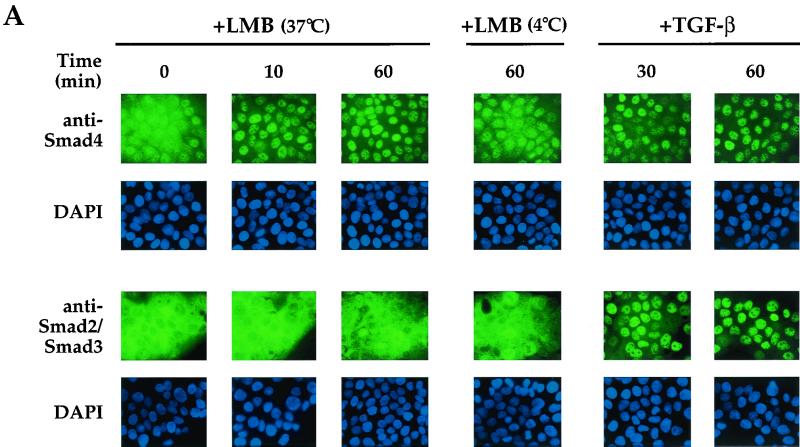
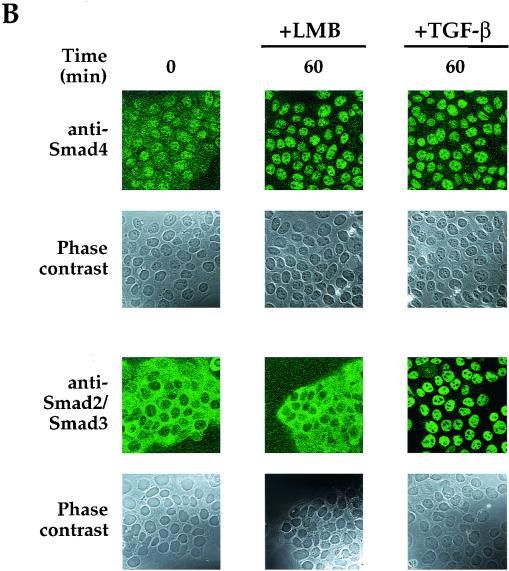
We asked whether endogenous Smad4 accumulated in the nucleus when cells were treated with LMB. If so, this would indicate that the Smad4 NLS was functional in the absence of TGF-β signaling and that Smad4 was normally actively transported from the nucleus via a CRM1-dependent mechanism. In the absence of LMB, and in the absence of TGF-β, Smad4 was distributed throughout the HaCaT cells (Fig. 6A, left panels) with clear nuclear staining seen in addition to cytoplasmic staining. This is also very obvious in the confocal section shown in Fig. 6B. In contrast, in the same conditions, Smad2 and Smad3 were almost exclusively cytoplasmic (Fig. 6). After 10 min of LMB treatment, endogenous Smad4 was largely concentrated in the nuclei, whereas Smad2 and Smad3 remained cytoplasmic (Fig. 6A), demonstrating that only Smad4 is efficiently imported into the nucleus and accumulates there when nuclear export is blocked by LMB. This striking difference between Smad4 and Smad2 and -3 was also clearly seen after longer incubations with LMB. Smad4 was exclusively nuclear after 60 min of treatment with LMB, while Smad2 and Smad3 were completely unaffected by this treatment (Fig. 6). When the LMB treatment was performed at 4°C, little relocalization of Smad4 was observed (Fig. 6A), indicating that import of Smad4 to the nucleus is an active process. As a control for the immunostaining, the behavior of Smad4 and Smad2 and -3 was tested in response to TGF-β signaling. As expected, 30 min after TGF-β stimulation, all the Smads were beginning to accumulate in the nucleus, and this was complete by 60 min (Fig. 6A, right panels, and 6B).
FIG. 6.
Endogenous Smad4 rapidly shuttles between the nucleus and the cytoplasm in unstimulated cells. (A) HaCaT cells were treated with LMB (20 ng/ml) for the times shown at either 37 or 4°C or with 2 ng of TGF-β per ml and then processed for immunofluorescence using either an anti-Smad4 monoclonal antibody or an anti-Smad2 and -Smad3 monoclonal antibody. Nuclei of the same cells were also stained with DAPI. Fluorescence was visualized using a Zeiss Axioplan upright fluorescence microscope. (B) Fields from a subset of the samples shown in panel A were examined by confocal laser scanning microscopy using a Zeiss LSM 510 confocal microscope. Phase-contrast images of the same fields are also shown.
Thus, in unstimulated HaCaT cells, Smad4 is continuously shuttling between the cytoplasm and the nucleus. This results in some nuclear localization of Smad4 in the absence of TGF-β signaling. Smad2 and Smad3, in contrast, do not shuttle between the cytoplasm and nucleus in these conditions and are clearly regulated independently of Smad4 in unstimulated cells.
Prolonged TGF-β signaling leads to export of Smad2, Smad3, and Smad4 from the nucleus to the cytoplasm.
Finally, we wanted to confirm the subcellular localization of the endogenous Smads in unstimulated cells by subcellular fractionation, comparing NIH 3T3 cells which were used for the transfection studies with HaCaT cells that were used for the studies of the endogenous Smads. We also wanted to investigate the fate of the Smads after prolonged TGF-β signaling. Nuclear and cytoplasmic extracts were made from HaCaT or NIH 3T3 cells that had been preincubated with the protein synthesis inhibitor cycloheximide (20 μg/ml) to prevent any Smad synthesis and then incubated with TGF-β for different times. Control experiments indicated that this amount of cycloheximide was sufficient to inhibit protein synthesis by 93% in both cell types (see Materials and Methods). The Smads were visualized by Western blotting with the specific monoclonal antibodies described above. To control for protein loading, the cytoplasmic extracts were blotted with an antibody that recognizes the exclusively cytoplasmic ezrin, radixin, and moesin (ERM proteins) (42), and the nuclear extracts were blotted with an antibody against PCNA (Fig. 7A, bottom panels). Control experiments indicated that there was virtually no cross-contamination of nuclei with cytoplasm or vice versa in either cell type (Fig. 7C).
In HaCaT cells, in the absence of TGF-β signaling, some Smad4 was detected in the nuclear fraction as predicted from the observation that Smad4 is undergoing continuous nucleocytoplasmic shuttling (Fig. 7A, lane 1). In the same conditions, Smad2 and Smad3 are predominantly cytoplasmic (Fig. 7A, lanes 1 and 8). This is in precise agreement with the immunofluorescence data shown in Fig. 6. After a 30-min treatment with TGF-β, increased levels of Smad4 are seen in the nuclear fraction, together with Smad2 and Smad3 (lane 2). As the levels of the Smads increase in the nuclear extracts, they correspondingly fall in the cytoplasmic extracts (Fig. 7A, lanes 2 to 5 and 9 to 12). The nuclear Smad2 is clearly phosphorylated, as seen by its detection with a polyclonal antibody directed against Smad2 phosphorylated at the C-terminal “SSXS” motif (29) which has previously been well characterized (11, 28). A low level of Smad2 in the cytoplasm is also phosphorylated. After 6 h of TGF-β treatment, the levels of bulk Smad2, phosphorylated Smad2, Smad3, and Smad4 in the nuclear fraction were very low (lane 6), and by 9 h, very little of any of the Smads was detected in the nuclear extracts (lane 7). This could be due to Smad degradation (28) or to export of the Smads to the cytoplasm. The latter mechanism appears to be the dominant one. After prolonged TGF-β induction (6 and 9 h), levels of Smad2, Smad3, and Smad4 in the cytoplasmic extracts increase again to approximately the levels seen in unstimulated cells (Fig. 7A, lanes 13 and 14). This is not due to new protein synthesis, as the cells had been incubated with cycloheximide prior to the initial treatment with TGF-β. The fact that the anti-phosphorylated Smad2 antibody does not detect any Smad2 in the cytoplasm after prolonged TGF-β treatment indicates that the Smad2 accumulating back in the cytoplasm is dephosphorylated.
The same experiment was performed in NIH 3T3 cells, the cell line used for the transfection studies described above (Fig. 7B). A major difference between these cells and HaCaT cells is that NIH 3T3 cells have extremely low levels of Smad3 relative to Smad2. The Smads exhibit essentially the same behavior in NIH 3T3 cells as in HaCaT cells. Some Smad4 was nuclear prior to signaling, although this was considerably less than in HaCaT cells, in agreement with the immunofluorescence data (Fig. 4 and 6); Smad 2 was exclusively cytoplasmic. Both Smad2 and Smad4 accumulated in the nucleus 30 min after stimulation with TGF-β and were depleted from the cytoplasm. After 6 h, Smad2 became dephosphorylated, and coincidentally, both Smad2 and Smad4 accumulated back in the cytoplasm (Fig. 7B).
The export of Smad2 and Smad3 back to the cytoplasm after prolonged TGF-β signaling was confirmed by immunofluorescence (Fig. 7D). HaCaT cells were treated with cycloheximide and then TGF-β for the times shown. It is clear in these confocal images that after 1 h of TGF-β stimulation Smad2 and Smad3 are exclusively nuclear. After 4 h, Smad2 and Smad3 begin to reappear in the cytoplasm, and after 9 h, this is almost complete.
DISCUSSION
The Smad4 mRNA is alternatively spliced.
We have described six alternatively spliced variants of Smad4, some of which are expressed at substantial levels (at least at the mRNA level) relative to wild-type Smad4 and are widely expressed in many different cell types. The six alternatively spliced Smad4 mRNAs can be translated into proteins that interact with Smad2 and Smad3 in a ligand-inducible manner and are incorporated into a DNA-binding transcription factor complex with activated Smad2 and Fast-1. However, they differ in their transcriptional activity and in their subcellular localization in unstimulated cells. This diversity in Smad4 could contribute to determining how different cell types respond to TGF-β ligands, possibly dictating the transcriptional activation potential of Smad-transcription factor complexes. Although we have raised antibodies that detect the specific spliced Smad4 isoforms when overexpressed, they are not avid enough to detect the endogenous proteins. The production of better antibodies is required to enable us to address the in vivo role of the alternatively spliced variants.
Smad4 is continuously shuttling between the cytoplasm and nucleus in the absence of TGF-β signaling.
Our identification of the alternatively spliced isoforms of Smad4 has enabled us to uncover a novel mechanism for Smad4 regulation. From the experiments described here, we conclude that in the absence of TGF-β signaling Smad4 shuttles between the cytoplasm and the nucleus. The strongest evidence for this is provided by the experiments with LMB. After inhibiting the nuclear export machinery with LMB, endogenous Smad4 has substantially accumulated in the nucleus after 10 min and is completely nuclear after 60 min. We propose that this behavior is regulated by the combined activities of an NES that we have identified in the Smad4 linker and an NLS in its MH1 domain. We have demonstrated the functionality of the NES and NLS in Smad4 by making point mutations in these motifs in the context of full-length Smad4 and showing that this inhibits their activity. In the case of the NLS, the inhibition was not complete, suggesting that another NLS exists elsewhere in Smad4 or alternatively that the mutant Smad4 forms complexes with endogenous Smads or with other proteins that contain strong NLSs. We are currently investigating this further. Consistent with this idea, we have shown that the NLS that we have identified in the MH1 domain is not sufficient to direct an otherwise cytoplasmic protein to the nucleus (our unpublished data).
The NLS that we have identified in human Smad4 is conserved in all Smad4s (see below). The NES, however, is present in only a subset of Smad4s; Xenopus Smad4β and Caenorhabditis elegans SMA-4 and DAF-3 do not contain an NES. We would thus expect all the Smad4s containing the NES to shuttle between the cytoplasm and the nucleus in the absence of signaling and expect those that do not contain an NES to be constitutively nuclear. Indeed, consistent with this view, Xenopus Smad4β has been shown to be constitutively nuclear (31) and DAF-3 is thought to have a nuclear role in the absence of signaling by the C. elegans TGF-β ligand, DAF-7 (35).
Taken together, our data suggest that the distribution of Smad4 between the cytoplasm and the nucleus will be governed by the relative strengths of nuclear import over export in different cell types. For example, in HaCaT cells, a substantial proportion of the endogenous Smad4 is nuclear in unstimulated cells, which we demonstrate by immunofluorescence and by subcellular fractionation. In NIH 3T3 cells, in contrast, Smad4 is predominantly cytoplasmic in similar conditions.
Although this mode of regulation is novel for Smad4, the retention of molecules in the cytoplasm through a mechanism of active export from the nucleus is not without precedent and has been recently demonstrated for such disparate proteins as the target of the Hedgehog (Hh) signaling pathway, the 155-kDa transcription factor Cubitus interruptus (Ci155); cyclin B1; the NF-κB inhibitor IκBα; and the yeast AP-1-like transcription factor yap1p (1, 23).
Mechanism of accumulation of Smad4 in the nucleus upon TGF-β signaling.
Smad4 accumulates in the nucleus within 30 min of stimulation of cells with TGF-β, presumably due to formation of complexes with the R-Smads Smad2 and Smad3. In principle, this could be achieved either by increasing the rate of nuclear import or by reducing the rate of nuclear export. We favor the latter mechanism, since nuclear import of Smad4 is extremely rapid, even in the absence of signal. We do not yet understand the mechanism whereby formation of complexes with R-Smads inhibits nuclear export, although obvious possibilities are masking of the NES either through complex formation per se or through binding of the Smad complexes to DNA.
The R-Smads contain an NLS but not an NES.
The continuous nucleocytoplasmic shuttling that we have demonstrated for Smad4 prior to TGF-β signaling is not a property of the R-Smads, as they do not contain an NES and are completely insensitive to the effects of LMB. This suggests that they are retained in the cytoplasm in the absence of TGF-β by a different mechanism, possibly through interaction with other cytoplasmic proteins, such as microtubules (8) and/or molecules such as the SARA (41). The receptor-regulated Smads, however, do have NLSs related to the one that we have identified in Smad4 (48). We propose that, in the absence of signal, the NLS in Smad2 and Smad3 is masked and that phosphorylation of these Smads by the activated type I receptor unmasks the NLS, resulting in translocation of the Smads to the nucleus (48).
It has been assumed that formation of complexes composed of an activated R-Smad and Smad4 occurs in the cytoplasm, before translocation to the nucleus (29). However, here we demonstrate that TGF-β-inducible transcriptional activity of Smad4 is the same whether the Smad4 is completely nuclear prior to stimulation or predominantly cytoplasmic, suggesting that complexes of activated Smad2 or -3 and Smad4 can form in the nucleus. It will be important to determine directly where complex formation normally occurs.
The fate of the Smads after prolonged TGF-β signaling.
In this study, we have also addressed the fate of the Smads after prolonged TGF-β signaling. We conclude that, 6 to 9 h after TGF-β stimulation, the bulk of Smad2, Smad3, and Smad4 is exported from the nucleus to the cytoplasm. Since the kinetics of this correlate with the kinetics of R-Smad dephosphorylation, we favor a model whereby dephosphorylation of the R-Smads leads to dissociation of Smad4 from the complexes and export of the Smads to the cytoplasm. Smad4 is presumably exported via a CRM1-dependent mechanism. The R-Smads, however, which do not contain a recognizable NES and are completely insensitive to the effects of LMB, must be exported from the nucleus by a distinct mechanism.
It was recently proposed that Smad signaling was terminated through the destruction of Smad2 (28), based on the ability of proteasome inhibitors such as MG-132 and lactacystin to stabilize phosphorylated Smad2 and the clear demonstration that at least a fraction of activated nuclear Smad2 is ubiquitinated and thus targeted for destruction via the proteasome. Since we do not detect any substantial depletion of the R-Smads or Smad4 after prolonged TGF-β signaling, we suggest that perhaps only a small proportion of activated Smad2 is degraded by the proteasome. In addition, other components of the TGF-β signaling pathway are regulated by proteolysis, in particular the activated receptors and the corepressors Ski and SnoN (16, 39, 50). It is possible that the gross stabilization of phosphorylated Smad2 in the presence of proteasome inhibitors (28) is a combination of direct stabilization of Smad2 and some indirect effects such as stabilizing the type I receptor kinase or stabilizing phosphorylated Smad2 in complexes with molecules such as Ski and SnoN. These corepressors associate preferentially with phosphorylated Smad2 and Smad3 and are then normally targeted for degradation by the proteasome (16); proteasome inhibitors stabilize the Smad-Ski and Smad-SnoN complexes (39). It will be important to determine how stabilization of these other components by the proteasome inhibitors affects the stabilization of activated Smad2.
The role of nuclear Smad4 prior to signaling.
The fact that Smad4 undergoes continuous nucleocytoplasmic shuttling prior to TGF-β signaling, while the R-Smads are actively retained in the cytoplasm, suggests that the nuclear Smad4 may have an important role in unstimulated cells. Indeed, there are readily detectable levels of Smad4 in the nucleus in unstimulated HaCaT and NIH 3T3 cells, but virtually no Smad2 or Smad3. This nuclear Smad4 is clearly transcriptionally inactive, since we detect no increase in basal transcription even when Smad4 is entirely targeted to the nucleus by mutating the NES. A role for such nuclear Smad4 was recently proposed (38). Smad4 was shown to form complexes with the nuclear proto-oncoprotein SnoN in the absence of a signal, and it was suggested that the role of these complexes was to bind to promoter sequences and repress the transcription of target genes prior to TGF-β signaling. Upon stimulation, SnoN is rapidly degraded, allowing Smad4 to form complexes with the activated R-Smads to activate transcription of target genes (38). This is an attractive mechanism which would ensure very tight regulation of transcription in response to TGF-β signaling.
ACKNOWLEDGMENTS
We thank Rik Derynck, Steve Goodbourne, Jon Graff, Tim Hunt, Peter ten Dijke, and Malcolm Whitman for plasmids and antibodies and Minoru Yoshida for LMB. We are very grateful to Christine Tran Quang for invaluable help with the immunofluorescence experiments and to Mike Howell for the experiment in Fig. 7C. We thank Stéphane Germain, Mike Howell, Gareth Inman, Helen McNeill, Peter ten Dijke, and Rebecca Randall for helpful discussions and comments on the manuscript.
The work was supported by the Imperial Cancer Research Fund, a TMR Research Network grant (ERBFMRXCT980216) to C.S.H., and an MRC Research Fellowship to F.J.N.
REFERENCES
- 1.Chen C H, von Kessler D P, Park W, Wang B, Ma Y, Beachy P A. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell. 1999;98:305–316. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Rubock M J, Whitman M. A transcriptional partner for MAD proteins in TGF-β signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 4.de Caestecker M P, Hemmati P, Larisch-Bloch S, Ajmera R, Roberts A B, Lechleider R J. Characterization of functional domains within Smad4/DPC4. J Biol Chem. 1997;272:13690–13696. doi: 10.1074/jbc.272.21.13690. [DOI] [PubMed] [Google Scholar]
- 5.de Caestecker M P, Yahata T, Wang D, Parks W T, Huang S, Hill C S, Shioda T, Roberts A B, Lechleider R J. The Smad4 activation domain (SAD) is a proline-rich, p300-dependent transcriptional activation domain. J Biol Chem. 2000;275:2115–2122. doi: 10.1074/jbc.275.3.2115. [DOI] [PubMed] [Google Scholar]
- 6.de Winter J P, Roelen B A, ten Dijke P, van der Burg B, van den Eijnden-van Raaij A J. DPC4 (SMAD4) mediates transforming growth factor-β1 (TGF-β1) induced growth inhibition and transcriptional response in breast tumour cells. Oncogene. 1997;14:1891–1899. doi: 10.1038/sj.onc.1201017. [DOI] [PubMed] [Google Scholar]
- 7.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 8.Dong C, Li Z, Alvarez R, Jr, Feng X H, Goldschmidt-Clermont P J. Microtubule binding to Smads may regulate TGFβ activity. Mol Cell. 2000;5:27–34. doi: 10.1016/s1097-2765(00)80400-1. [DOI] [PubMed] [Google Scholar]
- 9.Enoch T, Zinn K, Maniatis T. Activation of the human β-interferon gene requires an interferon-inducible factor. Mol Cell Biol. 1986;6:801–810. doi: 10.1128/mcb.6.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Germain S, Howell M, Esslemont G M, Hill C S. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 2000;14:435–451. [PMC free article] [PubMed] [Google Scholar]
- 11.Gorelik L, Flavell R A. Abrogation of TGFβ signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity. 2000;12:171–181. doi: 10.1016/s1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- 12.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 13.Hahn S A, Schutte M, Hoque A T, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, Kern S E. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 14.Hata A, Lo R S, Wotton D, Lagna G, Massagué J. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature. 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- 15.Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massagué J. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 16.Heldin C H, ten Dijke P. SMAD destruction turns off signalling. Nat Cell Biol. 1999;1:E195–E197. doi: 10.1038/70223. [DOI] [PubMed] [Google Scholar]
- 17.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 18.Howell M, Itoh F, Pierreux C E, Valgeirsdottir S, Itoh S, ten Dijke P, Hill C S. Xenopus Smad4β is the co-Smad component of developmentally-regulated transcription factor complexes responsible for induction of early mesodermal genes. Dev Biol. 1999;214:354–369. doi: 10.1006/dbio.1999.9430. [DOI] [PubMed] [Google Scholar]
- 19.Huang H-C, Murtaugh L C, Vize P D, Whitman M. Identification of a potential regulator of early transcriptional responses to mesoderm inducers in the frog embryo. EMBO J. 1995;14:5965–5973. doi: 10.1002/j.1460-2075.1995.tb00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen J, Serup P, Karlsen C, Nielsen T F, Madsen O D. mRNA profiling of rat islet tumors reveals nkx 6.1 as a β-cell-specific homeodomain transcription factor. J Biol Chem. 1996;271:18749–18758. doi: 10.1074/jbc.271.31.18749. [DOI] [PubMed] [Google Scholar]
- 21.Johnson C, Van Antwerp D, Hope T J. An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IκBα. EMBO J. 1999;18:6682–6693. doi: 10.1093/emboj/18.23.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonk L J, Itoh S, Heldin C H, ten Dijke P, Kruijer W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem. 1998;273:21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- 23.Kaffman A, O'Shea E K. Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- 24.Kageyama H, Seki N, Yamada S, Sakiyama S, Nakagawara A. DPC4 splice variants in neuroblastoma. Cancer Lett. 1998;122:187–193. doi: 10.1016/s0304-3835(97)00389-3. [DOI] [PubMed] [Google Scholar]
- 25.Kretzschmar M, Doody J, Massagué J. Opposing BMP and EGF signalling pathways converge on the TGF-β family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 26.Kretzschmar M, Doody J, Timokhina I, Massagué J. A mechanism of repression of TGFβ/Smad signaling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Pouponnot C, Massagué J. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo R S, Massagué J. Ubiquitin-dependent degradation of TGF-β-activated Smad2. Nat Cell Biol. 1999;1:472–478. doi: 10.1038/70258. [DOI] [PubMed] [Google Scholar]
- 29.Massagué J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 30.Massagué J, Wotton D. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuyama N, Hanafusa H, Kusakabe M, Shibuya H, Nishida E. Identification of two Smad4 proteins in Xenopus. Their common and distinct properties. J Biol Chem. 1999;274:12163–12170. doi: 10.1074/jbc.274.17.12163. [DOI] [PubMed] [Google Scholar]
- 32.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin C H, Miyazono K, ten Dijke P. TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 34.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 35.Patterson G I, Koweek A, Wong A, Liu Y, Ruvkun G. The DAF-3 Smad protein antagonizes TGF-β-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes Dev. 1997;11:2679–2690. doi: 10.1101/gad.11.20.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schutte M, Hruban R H, Hedrick L, Cho K R, Nadasdy G M, Weinstein C L, Bova G S, Isaacs W B, Cairns P, Nawroz H, Sidransky D, Casero R A, Jr, Meltzer P S, Hahn S A, Kern S E. DPC4 gene in various tumor types. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 37.Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 38.Stroschein S L, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-β signaling by the SnoN oncoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 39.Sun Y, Liu X, Ng-Eaton E, Lodish H F, Weinberg R A. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor β signaling. Proc Natl Acad Sci USA. 1999;96:12442–12447. doi: 10.1073/pnas.96.22.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ten Dijke P, Miyazono K, Heldin C H. Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem Sci. 2000;25:64–70. doi: 10.1016/s0968-0004(99)01519-4. [DOI] [PubMed] [Google Scholar]
- 41.Tsukazaki T, Chiang T A, Davison A F, Attisano L, Wrana J L. SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 42.Tsukita S, Yonemura S. Cortical actin organization: lessons from ERM (ezrin/radixin/moesin) proteins. J Biol Chem. 1999;274:34507–34510. doi: 10.1074/jbc.274.49.34507. [DOI] [PubMed] [Google Scholar]
- 43.van Hengel J, Vanhoenacker P, Staes K, van Roy F. Nuclear localization of the p120(ctn) Armadillo-like catenin is counteracted by a nuclear export signal and by E-cadherin expression. Proc Natl Acad Sci USA. 1999;96:7980–7985. doi: 10.1073/pnas.96.14.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitman M. Smads and early developmental signaling by the TGFβ superfamily. Genes Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- 45.Wieser R, Wrana J L, Massagué J. GS domain mutations that constitutively activate TβR-I, the downstream signaling component in the TGF-β receptor complex. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong C, Rougier-Chapman E M, Frederick J P, Datto M B, Liberati N T, Li J M, Wang X F. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor β. Mol Cell Biol. 1999;19:1821–1830. doi: 10.1128/mcb.19.3.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu R-Y, Zhang Y, Feng X-H, Derynck R. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol Cell Biol. 1997;17:2521–2528. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Z, Liu X, Lodish H F. Importin β mediates nuclear translocation of Smad3. J Biol Chem. 2000;275:23425–23428. doi: 10.1074/jbc.C000345200. [DOI] [PubMed] [Google Scholar]
- 49.Yagi K, Goto D, Hamamoto T, Takenoshita S, Kato M, Miyazono K. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J Biol Chem. 1999;274:703–709. doi: 10.1074/jbc.274.2.703. [DOI] [PubMed] [Google Scholar]
- 50.Zhu H, Kavsak P, Abdollah S, Wrana J L, Thomsen G H. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]