Abstract
DNA double-strand breaks (DSBs) may be caused by normal metabolic processes or exogenous DNA damaging agents and can promote chromosomal rearrangements, including translocations, deletions, or chromosome loss. In mammalian cells, both homologous recombination and nonhomologous end joining (NHEJ) are important DSB repair pathways for the maintenance of genomic stability. Using a mouse embryonic stem cell system, we previously demonstrated that a DSB in one chromosome can be repaired by recombination with a homologous sequence on a heterologous chromosome, without any evidence of genome rearrangements (C. Richardson, M. E. Moynahan, and M. Jasin, Genes Dev., 12:3831–3842, 1998). To determine if genomic integrity would be compromised if homology were constrained, we have now examined interchromosomal recombination between truncated but overlapping gene sequences. Despite these constraints, recombinants were readily recovered when a DSB was introduced into one of the sequences. The overwhelming majority of recombinants showed no evidence of chromosomal rearrangements. Instead, events were initiated by homologous invasion of one chromosome end and completed by NHEJ to the other chromosome end, which remained highly preserved throughout the process. Thus, genomic integrity was maintained by a coupling of homologous and nonhomologous repair pathways. Interestingly, the recombination frequency, although not the structure of the recombinant repair products, was sensitive to the relative orientation of the gene sequences on the interacting chromosomes.
Genetic integrity relies on the faithful repair of DNA damage such as double-strand breaks (DSBs). Aberrantly repaired DSBs are expected to result in chromosomal rearrangements such as translocations, deletions, or chromosome loss. Multiple mechanisms have evolved to ensure proper repair of DSBs, details of which are now being elucidated (35). In mammalian cells, DSBs are repaired by both homology-dependent and homology-independent (nonhomologous) recombination, stimulating both pathways by 3 orders of magnitude or more (5, 27, 41, 42). These pathways have been considered mechanistically distinct since genetic analysis of DNA repair mutants demonstrates defects in either one process or the other (24, 28, 48, 49).
Although homologous recombination is a major DSB repair pathway, large fractions of mammalian genomes are composed of repetitive elements (44), raising the paradox that mammalian cells would seem to be at high risk for genome rearrangements; yet such rearrangements are not usually seen. One explanation for the generally nonmutagenic outcome of homologous repair in mammalian cells comes from the preferred use of sister chromatids as repair templates (23, 24, 37), as is also found in yeast (25). However, sequence repeats on nonhomologous chromosomes can also serve as homologous repair templates at a readily detectable frequency, albeit significantly reduced relative to sister chromatids (40), and repetitive Alu elements have been identified at or near recombinant breakpoints in cell lines with chromosomal translocations and other rearrangements (6, 22, 31). Thus, the role of repetitive sequences in interchromosomal DSB repair of mammalian cells remains unclear, but cells must limit, either actively or passively, the potential mutagenic outcomes of these events.
We previously used a mouse embryonic stem (ES) cell system to examine the repair of a single DSB by interchromosomal recombination within a reporter substrate. The overwhelming majority of events (97%) were determined to be gene conversions involving the transfer of a small amount of homologous sequence information from the unbroken chromosome into the broken chromosome (short-tract gene conversion [STGC]), with the remaining events (3%) involving the additional transfer of adjacent sequences (long-tract gene conversion [LTGC]) (40). The LTGC events were predicted to have been resolved within a region of fortuitous homology between the two chromosomes or by nonhomologous end joining (NHEJ). However, the structure of the LTGC events was not determined, and their small number would have precluded any definitive conclusions regarding the general nature of this repair class. Nevertheless, none of these events resulted in gross chromosomal alterations such as translocations, even though gene conversion associated with reciprocal exchange is predicted by some DSB repair models (47) and has been detected during yeast interchromosomal recombination (20).
Given that crossovers are predominantly associated with LTGC events in other systems (1, 15), we have now modified our recombination reporter substrates to favor the recovery of interchromosomal exchange events following homologous repair. The homology constraints thereby eliminate the recovery of frequent STGC events so as to analyze repair by alternative pathways. However, we find that although recombinants were readily obtained with these substrates, exchange events or other chromosomal rearrangements were extremely infrequent. Instead, the repair events were initiated by homologous invasion but NHEJ was used to complete the events, such that the newly synthesized strand arising from strand invasion was joined to the other end of the broken chromosome. These results demonstrate an important coupling of homologous and nonhomologous repair pathways for the maintenance of genomic integrity.
MATERIALS AND METHODS
DNA and cell line constructions.
The pim-1 allele was targeted by modifying the previously described p59 gene-targeting vector (52). A XhoI-RsrII fragment containing the promoter and 5′ coding region of S2neo (5′neo) (45) was modified to contain a 3′ XhoI site and inserted into the SalI site of p59 downstream of the hygromycin coding sequence (hyg) and into pim-1 exon 4. Plasmids with 5′neo in the opposite orientation as hyg (F5′) also had a SalI-XhoI fragment containing the polyadenylation signal of the bovine growth hormone (11) inserted for stabilization of the hyg mRNA. The Rb allele was targeted by modifying the previously described p129 gene-targeting vector (51). An XbaI-PstI fragment containing a hypoxanthine phosphoribosyltransferase gene (HPRT) and the 3′neo coding region from pMC1neo from the PstI site through the polyadenylation signal (3′neo) was inserted into Bluescript (Promega). From this, an XbaI fragment with the HPRT gene and 3′neo was inserted into an NheI site in intron 18 of the Rb locus in p129. Targeting constructs were cleaved away from the plasmid backbone prior to electroporation. The pΔnar plasmid was constructed from pMC1neo by deleting the NarI fragment from the 5′ portion of the neo gene (53). The homology fragment begins 136 bp 3′ of the neo ATG start codon and extends through the stop codon and polyA signal.
ES cell line E14TG2a (21) was grown in standard media supplemented with leukemia inhibitory factor at 105 U/ml (GIBCO/Life Technologies). For gene targeting, 1.6 × 107 cells were electroporated at 250 V/960 μF with 100 μg of targeting construct. Selection medium containing hygromycin (110 μg/ml) was added 48 h after transfection of the pim-1 targeting construct. Targeted clones were identified by Southern blots of genomic DNA cleaved with HincII using a genomic HincII-BstXI pim-1 fragment as a probe (52). Selection medium containing hypoxanthine-aminopterin-thymidine was added 20 h after transfection of the Rb targeting construct. Targeted clones were identified by Southern blots of genomic DNA cleaved with PstI, using a genomic PstI-PvuII Rb fragment as a probe (51). Independently derived cell lines for both F5′/3′ (lines F12 and G12) and R5′/3′ (lines B3 and C11) were identified and used in subsequent experiments. Southern blotting was used to confirm that each line contains a single targeted integration event.
DSB induction and DNA analysis.
Electroporations were performed as above with 50 μg of the circular pCBASce I-SceI expression vector (40), 25 μg of circular pΔnar plasmid, or both, as indicated. The number of surviving cells was determined 20 h after electroporation, at which time G418 was added (200 μg/ml). G418R colonies were scored and expanded 12 days later. DNA extractions, Southern blot analysis, and PCR for amplification of repair junctions were performed as described previously (39). The PCR primers were from intron 4 of pim-1 and hyg (39). PCR products were cloned with the TA cloning system (Invitrogen) and sequenced by the Sloan-Kettering Institute core facility.
FISH.
All 21 clones from class IV and 3 clones from class III were analyzed by fluorescence in situ hybridization (FISH). Chromosome metaphase spreads from individual clones were made as previously described (40). Spreads were hybridized to whole chromosome mouse chromosome 14 (chr.14) fluorescein isothiocyanate and chr.17 Cy3 probes (Vysis-Cambio), counterstained with 4′,6′-diamidino-2-phenylindole (DAPI), and visualized by confocal microscopy at the Sloan-Kettering Institute core facility.
RESULTS
Interchromosomal recombination induced by a DSB.
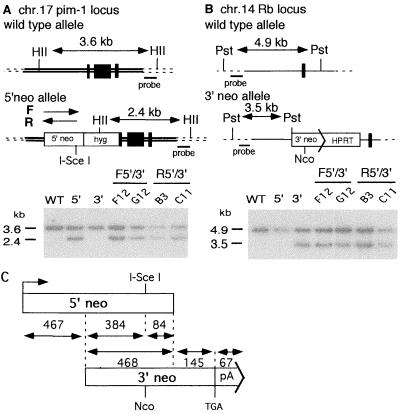
To determine if DSB repair events in which homology is constrained are associated with chromosomal rearrangements, we inserted truncated neomycin phosphotransferase (neo) gene substrates into loci on two heterologous chromosomes in mouse ES cells. The truncated neo gene inserted at the pim-1 locus of chr.17 contains a 5′neo sequence in which the 18-bp recognition site for the rare-cutting I-SceI endonuclease has been incorporated (Fig. 1A). The truncated neo gene inserted at the Rb locus of chr.14 contains a 3′neo sequence which does not have this site (Fig. 1B). Since the neo sequences are truncated, each is nonfunctional. The substrate design provides 468 bp of homology between the neo sequences, which are identical except at the I-SceI site (Fig. 1C).
FIG. 1.
Interchromosomal recombination substrates. The 5′neo allele (A) was constructed by targeting 5′neo and the hyg gene to the pim-1 locus on chr.17. The forward orientation (F allele) of the 5′neo sequence is present in the F5′/3′ cell lines, and the reverse orientation (R allele) is present in the R5′/3′ cell lines, as indicated by the arrow orientation. The 3′neo allele (B) was constructed by targeting 3′neo and the HPRT gene to the Rb locus on chr.14. Wild-type (top) and targeted (bottom) alleles are diagrammed for each locus, with confirmatory Southern analyses presented using HincII and PstI for the 5′neo and the 3′neo alleles, respectively, and probing with sequences as indicated. An I-SceI site disrupts the 5′neo gene at the NcoI site. Two independently derived clones were analyzed for both F5′/3′ and R5′/3′, as indicated (F5′/3′, F12 and G12; R5′/3′, B3 and C11). Black bars, pim-1 (A) or Rb (B) exons. HII, HincII; Pst, PstI; Nco, NcoI. (C) The truncated 5′neo and 3′neo sequences. Distances are indicated in bp. 5′neo contains a promoter, and 3′neo contains the neo gene stop codon and a polyadenylation signal. There is 468 bp of overlap between the 5′neo and 3′neo sequences.
Reconstruction of a functional neo+ gene in these cell lines is dependent on homologous recombination between the two chromosomes at the neo sequences. To determine the effect of the relative orientation of the neo sequences on recombination, independently derived cell lines termed F5′/3′ (clones F12 and G12) and R5′/3′ (clones B3 and C11) were constructed. F5′/3′ has 5′neo in the same orientation as 3′neo relative to the centromere (F, forward), and R5′/3′ has the neo sequences in reverse orientation (R, reverse). As previously demonstrated with related substrates (39, 40), interchromosomal homologous recombination in the absence of a DSB is rare. No neo+ colonies were derived from any of the parental cell lines, nor were recombinants detected following electroporation of the cell lines with the pΔnar control plasmid that contains 3′neo (Fig. 1C) and can therefore correct the truncation mutation in 5′neo by recombination (frequency, <8 × 10−8 [Table 1 and data not shown]).
TABLE 1.
Induction of interchromosomal recombination by a DSB
| Cell line | DNA | Recombination frequency (10−6)a |
|---|---|---|
| F5′ | pCBASce | <0.084 |
| pΔnar | <0.084 | |
| pCBASce + pΔnar | 260 ± 92 | |
| F5′/3′ | pCBASce | 3.9 ± 2.3 |
| pΔnar | <0.053 | |
| pCBASce + pΔnar | 150 ± 24 | |
| R5′/3′ | pCBASce | 0.42 ± 0.19 |
| pΔnar | <0.048 | |
| pCBASce + pΔnar | 100 ± 12 |
Recombination frequency was calculated as the total number of G418R clones divided by the total number of cells surviving electroporation and was derived from at least three experiments for each cell line. Two independently derived cell lines for each F5′/3′ and R5′/3′ were tested (Fig. 1).
To determine if a DSB would promote interchromosomal recombination between the truncated neo genes, the I-SceI endonuclease was expressed in cells from the expression vector pCBASce (40) so as to introduce a DSB in 5′neo. Following electroporation of pCBASce alone, neo+ colonies were readily obtained from both the F5′/3′ and R5′/3′ cell lines but not the F5′ cell line (Table 1), indicating that recombination between the two neo sequences is required to create a functional neo+ gene. The R5′/3′ cell lines gave neo+ recombinants at a frequency of 0.42 × 10−6, and the F5′/3′ cell lines gave recombinants at a frequency of 3.9 × 10−6. The frequency of DSB-promoted interchromosomal recombination in the F5′/3′ cell lines is similar to that seen in cell lines containing two full-length neo genes, regardless of relative substrate orientation (FN and RN lines, average frequencies of 3.8 × 10−6 and 3.2 × 10−6, respectively [data not shown and reference 40]). This indicates that the homology constraint for generating a functional product does not by itself reduce the recovery of recombinants, as would have been predicted if recombination between the neo sequences led to a large class of nonselectable or lethal repair events. However, the ninefold reduction in recombinants in the R5′/3′ cell lines implies that the relative orientation of the truncated neo repeats may significantly affect the recovery of recombinants.
As a control, cell lines were electroporated with both pCBASce and pΔnar to detect DSB-promoted gene targeting events. Gene targeting was significantly more frequent than interchromosomal recombination, as previously seen (39, 40), and was similar for the four cell lines, 1.5 × 10−4 and 1.0 × 10−4 for the F5′/3′ and R5′/3′ cell lines, respectively. The similar frequency of gene targeting again indicates that the orientation of 5′neo does not by itself affect the overall ability to recombine. Rather, the observed ninefold difference in interchromosomal recombination appears to be related to the relative orientation of the truncated neo sequences on the two chromosomes.
Interchromosomal recombination with variable products.
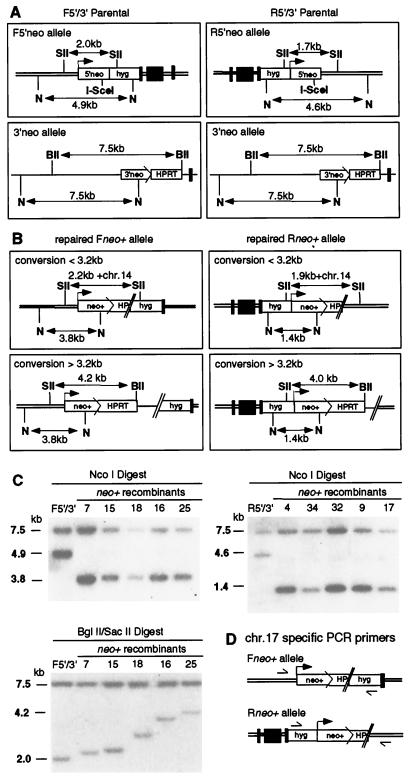
To characterize the interchromosomal recombination products, we performed Southern blotting on genomic DNA from 45 neo+ clones derived from the F5′/3′ cell line and 35 neo+ clones derived from the R5′/3′ cell line. Representative repair events are shown in Fig. 2, and classification of repair events is summarized in Table 2. DSB-promoted recombination restoring a functional, full-length neo+ gene should convert the I-SceI site in 5′neo to an NcoI site, and this was verified for each of the neo+ clones. The NcoI fragment for the 5′neo allele (4.9 kb, F5′; 4.6 kb, R5′) was altered in each of the neo+ clones as expected if chr.17 contained the recombinant neo+ allele (3.8 kb, Fneo+; 1.4 kb, Rneo+) (Fig. 2C and data not shown). By contrast, the unbroken 3′neo allele on chr.14 remained the parental size (7.5 kb) in each of the clones.
FIG. 2.
DSB repair products. Restriction maps of parental (A) and repaired neo+ alleles (B). NcoI digestion is diagnostic for homologous repair of the I-SceI-generated DSB in either 5′neo allele. BglII/SacII digestion is used to characterize the type of recombination event. BglII/SacII fragments of variable size (B) indicate gene conversion tracts incorporating variable lengths of chr.14 sequences (+chr.14) and NHEJ at variable positions of chr.17. BII, BglII; N, NcoI; SII, SacII. (C) Southern blot analysis of representative clones. Note that the 3′neo allele is unchanged from the parental allele in each of the recombinant clones. The probe was a 5′neo gene fragment. (D) Position of PCR primers derived from chr.17 sequences that flank the repair junctions.
TABLE 2.
Repair classes of DSB-induced interchromosomal recombination
| Repair class | No. of clones
|
|
|---|---|---|
| F5′/3′ | R5′/3′ | |
| Gene conversion without an exchange | ||
| I (conversion tract, <0.2 kb)a | 0 | 0 |
| II (conversion tract, <1.0 kb) | 29 | 24 |
| III (conversion tract, <3.2 kb) | 4 | 1 |
| IV (conversion tract, >3.2 kb) | 12 | 9 |
| Translocation | 0 | 1b |
| Total | 45 | 35 |
Gene conversion tracts of less than 229 bp do not produce a functional neo+ gene, and thus these products are not recovered.
The translocation was between chr.17 and an unidentified chromosome(s) (see Fig. 4B).
We next wanted to verify that a full-length neo+ gene was created and to determine how the overall structures of the two chromosomal loci were altered by recombination. Genomic DNA was cleaved with the enzymes BglII and SacII, which have sites flanking the neo alleles. As seen for the NcoI digest, the chr.14 3′neo allele was unchanged in each of the clones (Fig. 2C). By contrast, the chr.17 allele containing the neo+ gene gave fragments of varying size. For the Fneo+ allele, the size ranged from 2.2 to 4.2 kb (Fig. 2C and data not shown); for the Rneo+ allele, the size ranged from 1.9 to 4.0 kb (data not shown). The smallest fragments, 2.2 kb for Fneo+ and 1.9 kb for Rneo+, are as expected for a full-length neo+ gene (Fig. 2B). The largest fragments, 4.2 kb for Fneo+ and 4.0 kb for Rneo+, are the sizes expected if the entire HPRT gene along with the end of the neo coding region on chr.14 had been incorporated into chr.17 (Fig. 2B).
Because the chr.14 3′neo allele was unchanged in each of the clones, none of the products were consistent with a gene conversion event associated with reciprocal exchange. Rather, the variability of the chr.17 allele suggested that the predominant event was a noncrossover gene conversion in which one broken end of the 5′neo allele on chr.17 invaded the 3′neo allele on chr.14 to initiate repair synthesis. NHEJ of the newly synthesized strands to the noninvading end of chr.17 could be used to complete the repair event. Variability could result from either the incorporation of variable amounts of chr.14 sequences to the chr.17 break site and/or NHEJ of the newly synthesized strands to variable positions along chr.17.
To fully characterize the gene conversion events, PCR was performed with primers from chr.17 that were expected to flank the neo+ gene (Fig. 2D). PCR was performed on each of the clones with BglII/SacII fragments of less than 4.2 kb (Fneo+/3′ clones) or 4.0 kb (Rneo+/3′ clones), which were expected to have conversion tracts of less than 3.2 kb. As with the Southern analysis, PCR products of variable size were obtained. Conversion tracts encompassing the sequence incorporated from chr.14 that was joined to chr.17 during repair ranged from just over 0.2 to approximately 3.2 kb (Table 2). As expected, tracts of less than 0.2 kb (class I) were not obtained, as they would have been insufficient in length to have incorporated the 3′ end of the neo coding region to produce a functional neo+ gene (Fig. 1C). The majority of recombinants had conversion tracts that extended more than 0.2 kb but less than 1 kb (class II). The two parental cell lines gave similar results, with 64% (29 of 45) of Fneo+/3′ clones and 69% (24 of 35) of Rneo+/3′ clones having tract lengths in this range. A few of the clones (4 of 45 from Fneo+/3′; 1 of 35 from Rneo+/3′) had gene conversion tract lengths between 1 and 3.2 kb (class III). The sizes of the conversion tracts correlated well with those predicted by Southern blotting (data not shown).
Homologous recombination associated with NHEJ events.
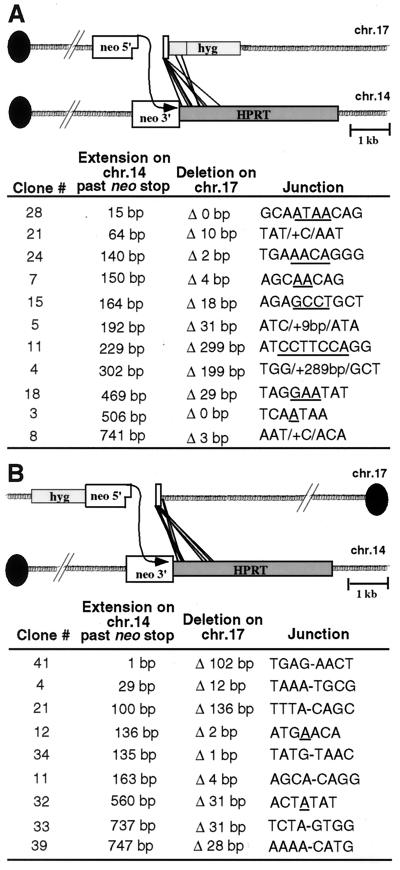
To conclusively determine if NHEJ was used to complete the repair events, as well as to precisely determine the length of the conversion tracts, junctions from class II neo+ clones were determined by sequencing of the PCR products. A total of 11 Fneo+ and 9 Rneo+ junctions were analyzed. Sequence analysis confirmed that the neo+ phenotype in each of the clones was due to the incorporation of at least 229 bp from the 3′ end of the neo coding region on chr.14 to create a full-length neo+ gene (Fig. 3). Clones with larger fragments had incorporated more sequence from chr.14. In one clone, 1 additional bp was incorporated beyond the neo gene stop codon for a total gene conversion tract of 230 bp (Rneo+/3′ clone 41). Apparently, the neo gene polyadenylation site is unnecessary for expression since it was not incorporated in this or three other clones (Fneo+/3′ clones 28 and 21; Rneo+/3′ clone 4). As expected, the longest gene conversion tract in these class II clones was less than 1 kb (Fneo+/3′ clone 8; Rneo+/3′ clones 33 and 39).
FIG. 3.
Sequence analysis of repair junctions from class II neo+ clones. PCR products from the neo+ allele of the Fneo+/3′ (A) and Rneo+/3′ cell lines (B) were cloned and sequenced. Extension on chr.14 indicates the length of newly synthesized DNA incorporated at one end of the chr.17 break site beyond the neo stop codon. Thus, the total extension includes an additional 229 bp to the number indicated. Deletion on chr.17 indicates the number of base pairs deleted from the nonconverted end of the I-SceI break site prior to NHEJ. The sequences at the junctions of chr.14 and chr.17 are separated by a hyphen when there is no sequence overlap, underlined if microhomology is present, or indicated by the base pairs that were inserted at the junction.
Sequencing demonstrated that completion of the repair events occurred by NHEJ, in most cases with minimal deletion from the chr.17 end (Fig. 3). In some clones, the 4-base 3′ overhang of the I-SceI site was maintained (Fneo+/3′ clones 28 and 3) or bases from the overhang were the only ones deleted (Fneo+/3′ clones 24, 7, and 8; Rneo+/3′ clones 12, 34, and 11). Deletions were generally ≤31 bp, although larger deletions were found in four clones, with up to 299 bp deleted (Fneo+/3′ clone 11). Microhomology was observed at approximately half of the junctions, and in three clones nucleotide addition was observed (Fneo+/3′ clones 21, 5, and 4). Overall, junctions of the 21 sequenced clones were similar to those obtained from NHEJ at a DSB within a single chromosome (27, 36) and at translocation breakpoints (13, 39, 54). Although the frequency of neo+ clones differed between the Fneo+/3′ and Rneo+/3′ cell lines, no difference was found in the recombinant products in either the conversion tract length involving chr.14 or the deletion from chr.17 (Fig. 3). Fewer Rneo+/3′ clones appeared to use microhomology in joining the two ends, although more clones must be examined to determine if this is significant.
Rare translocation during interchromosomal gene conversion.
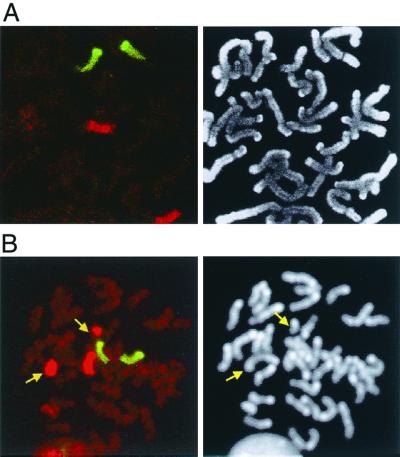
The remaining clones containing the entire HPRT gene (Fig. 2; Table 2) were obtained at similar frequencies from the F5′/3′ (27%; 12 of 45) and R5′/3′ (29%; 10 of 35) parental cell lines. These could have arisen either by gene conversion involving tracts of greater than 3.2 kb or by nonreciprocal translocation. To distinguish between these possibilities, FISH was performed on each of the 22 clones by using whole chromosome probes to mouse chr.17 and chr.14. Most of these clones (21 of 22) had not undergone a translocation or any other gross chromosomal rearrangements, including nonreciprocal translocations or large duplications (Fig. 4A and data not shown). Thus, these clones (class IV) arose from a gene conversion event that extended more than 3.2 kb but less than a cytologically observable distance (i.e., 1 Mb). The extent of the conversion is currently being mapped.
FIG. 4.
Visualization of a rare translocation by FISH using whole chromosome mouse chr.14 fluorescein isothiocyanate (green) and chr.17 Cy3 (red) probes (left panel) and DAPI (right panel). (A) Representative class IV clone having two normal chrs.14 and chrs.17. (B) Reciprocal translocation clone (Rneo+/3′ clone 17) having two normal chrs.14, one normal untargeted chr.17, and two hybrid translocation chromosomes involving chr.17 and an unidentified chromosome (red/unstained), as indicated by the arrows.
By Southern blot analysis, the remaining neo+ recombinant which was derived from the R5′/3′ parental cell line appeared to be similar to the class IV clones. However, FISH analysis indicated that this clone had undergone a reciprocal translocation involving chr.17 in the region of the DSB and an unidentified chromosome (Fig. 4B). (We cannot, however, rule out the possibility that a second unidentified chromosome was involved in the translocation.) The frequency of this event was 1.2 × 10−8. This contrasts with our previous results in which the repair of two chromosomal DSBs led to reciprocal translocations at a frequency of 10−4 (39). Therefore, it is likely that in this one clone the translocation involved another chromosome which had fortuitously undergone a DSB. Because this was the only clone that exhibited an unusual structure, we do not know if the event was specific to repair of the DSB in the R5′/3′ cell line.
DISCUSSION
These results clearly indicate that following a DSB, a sequence on a heterologous chromosome can serve as a repair template for homologous recombination in mammalian cells, even when homology is constrained. All but one of the clones (79 of 80) we examined had undergone interchromosomal recombination between the neo sequences without exchange of flanking markers or other genome rearrangement. Instead, repair was initiated by gene conversion and completed by NHEJ. Thus, repair of a single DSB by interchromosomal gene conversion, whether a fully homologous event (40) or a compound event involving NHEJ as in this report, rarely compromises genomic integrity. The results presented here contrast with the repair of two DSBs in which chromosomal rearrangements (translocations) were readily recovered (39). In that case, translocations did not arise by gene conversion but rather by joining of the ends of two different chromosomes by NHEJ or single-strand annealing, suggesting that gene conversion has a higher fidelity for maintaining genomic integrity than these other repair pathways.
The importance of NHEJ and homologous repair for the maintenance of genomic integrity in mammalian cells is emphasized by the observations that cell mutants in either repair pathway exhibit a high frequency of chromosomal aberrations (7, 10, 12, 16, 24, 26, 29, 32, 37, 48, 50). The importance of these two repair pathways is evident in both embryonic and adult cell types, although recent studies suggest that there may be differences between these stages in the contribution of various repair pathways and proteins. For example, ES cells deficient in the homologous repair protein Rad54 are sensitive to ionizing radiation (8), a potent inducer of DSBs, although this sensitivity seems to decrease through development to the adult mouse (9). By contrast, adult mice mutant for the NHEJ repair protein DNA-PKcs are hypersensitive to ionizing radiation, although DNA-PKcs−/− ES cells do not display this phenotype (3, 12). Nevertheless, mutation of other NHEJ proteins Ku70 and Ku80 leads to ionizing radiation sensitivity in both ES and adult mouse cells (17, 18, 34), and therefore, it is likely that the Ku protein participates in the NHEJ events that we report here.
NHEJ and homologous repair have also been proposed to have different contributions to repair during different stages of the cell cycle, i.e., G0/G1 and S/G2. Based on previous work, we expect that the overwhelming majority of repair events at the chr.17 break site are intrachromosomal, involving either NHEJ of the two broken ends or homologous repair from the sister chromatid, which are not selected for in this system (23, 27). This is supported by the frequency of gene targeting, which is 40- to 240-fold higher than interchromosomal events. What governs the use of a homologous sequence on a heterologous chromosome for repair of a DSB is unclear, but considering the nuclear volume, it is possible that random collision plays a role in homologous partner choice.
The results presented here provide convincing evidence that NHEJ and homologous repair are not completely separable and that coupling of the two pathways can preserve genomic integrity for the repair of a single DSB. Coupling of the two pathways has previously been predicted in some gene targeting events (see, e.g., references 2, 38, and 41). This report provides direct evidence for such events and detailed analysis of the junctions. The structure of the recombinant products demonstrates that repair of the DSB was initiated by invasion of one chr.17 end into the homologous sequence on chr.14, priming DNA replication which extended into heterologous sequences. Sequence analysis demonstrated that NHEJ was used to join the newly replicated strands to the other chr.17 end, which in some cases was preserved to such an extent as to maintain the overhang of the break site. The consistent recovery of clones that maintain the other chr.17 end indicates that this end is maintained close to the repair complex even though it does not participate in the homologous invasion step. This coupled repair mechanism can also account for previously observed infrequent LTGC events from allelic and interchromosomal recombination with related substrates (33, 40), although this has not been verified.
Although similar models for the initiation of recombination have been proposed for yeast (19) and Drosophila DSB repair (14), the coupling of NHEJ and homologous repair appears to occur more readily in mammalian cells and possibly plant cells (38), presumably due to an overall greater contribution of NHEJ to DSB repair in higher eukaryotes. As a result of this process, the heterologous sequences that are replicated during repair synthesis become duplicated. In most clones we found that the duplication was a few kilobases or less. In none of the clones did replication extend to the end of the chromosome, as has been detected in yeast (30). However, in mammalian cells replication to the end of the chromosome may lead to inviable progeny, resulting in either unbalanced genetic information (as in the F5′/3′ cell line) or an acentric product (as in the R5′/3′ cell line). Similar constraints on product recovery might also exist if gene conversion with a reciprocal exchange were exclusive to the S/G2 phase of the cell cycle and recombinant chromosomes always segregated from each other.
Although overall genome integrity is maintained in the coupled repair products we observed, the resulting duplication of sequences 3′ to the break site is likely to be deleterious in some cases. Alterations of the ALL1 locus in leukemic cells have been found which involve partial tandem duplications of the ALL1 gene at or near Alu repeats (43, 46). These duplications mechanistically could have arisen similarly to the events described here (Fig. 5). Thus, a DSB within or near a repetitive element could initiate strand invasion into the identical element on the homologue in G0/G1 (33) or sister chromatid in S/G2 (23) and prime DNA synthesis. Following repair synthesis, the repair event would resolve by NHEJ (Fig. 5). Alternatively, repair synthesis could continue into a downstream repetitive element of the same class, so that the event is resolved by annealing of the newly synthesized strand with the complementary end of the broken chromosome (not shown). The advantage of this model is that it allows for invasion to occur within an identical Alu element or other sequence but has no constraints on the completion of the repair event, since it can occur by either NHEJ or homologous annealing.
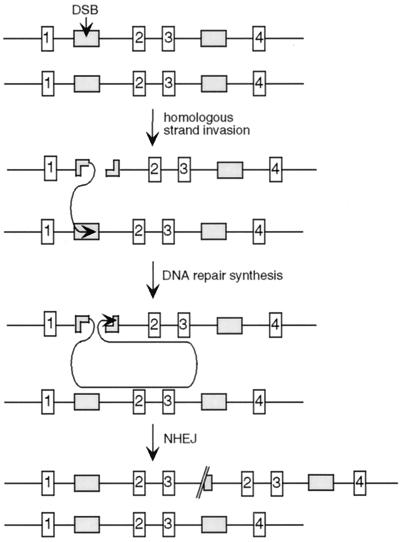
FIG. 5.
Model for partial tandem gene duplications. Events can be initiated by a DSB within a repetitive element (gray box) on one chromosome, followed by invasion of one end into the same element on the homologue or sister chromatid. Repair synthesis extends downstream to duplicate sequences, shown here as exons 2 and 3 (white bars). Completion of the repair event occurs by NHEJ of the newly synthesized strands at or near the end of the broken chromosome. The result is a partial tandem gene duplication on one chromosome or chromatid, while the other chromosome or chromatid remains unaltered.
It is unclear what minimal length of homology is required to promote interchromosomal homologous recombination in mammalian cells and what effect the length of homology has on crossing over. As little as 68 bp of homology is sufficient for homologous invasion to occur at a detectable frequency during DSB-promoted gene targeting in ES cells, although in this case recombination occurs at a significantly lower frequency than when >200 bp of homology is used (C. Richardson, J. Winderbaum, and M. Jasin, unpublished results). The majority of dispersed repetitive elements, SINEs (<200 bp in the mouse or 300 bp in humans) or small truncated LINEs (as small as 300 bp) (44), are within the size range of the homologous repeat used in this study. However, LINEs can be longer than this repeat unit. Full-length LI elements are 7 kb, although the majority are truncated to smaller units of a few kilobases or less (44). It is unclear whether interchromosomal recombination between repeats as long as several kilobases would give rise to repair products different than those reported here.
Surprisingly, we observed a ninefold higher frequency of interchromosomal recombination in the F5′/3′ cell lines in which the neo sequences are in the same orientation relative to the centromere, compared with the R5′/3′ cell lines in which the neo sequences are in the opposite orientation, even though the overall structure of the recombinants was very similar for the two cell lines. Cell lines with the other two configurations of the truncated neo sequences gave similar results (data not shown); for example, a cell line with the neo repeats in the same relative orientation but opposite to F5′/3′ gave similar recombination frequencies as the F5′/3′ cell line (C. Richardson and M. Jasin, unpublished results). Unless there is loss of a major class of repair product from the R5′/3′ cell lines, these results suggest an unexpected sensing of the relative orientation of the neo sequences on the two interacting chromosomes during these compound repair events. Rates of interchromosomal Cre/loxP recombination in yeast have been shown to be affected by centromere clustering (4), although thus far there has not been a study of this in mammalian cells. It will be interesting to determine if this orientation effect will be generally observed in mammalian cells and, if so, to determine the factors responsible for this phenomenon.
ACKNOWLEDGMENTS
We thank Hein te Riele (Amsterdam) for materials, Diane Tabarini in the core sequencing facility, and Katia Manova and Scott Kerns in the core microscopy facility.
C.R. is a Vrushalli Ranadive Special Fellow of the Leukemia and Lymphoma Society (formerly the Leukemia Society of America). This work was supported by an NSF grant (MCB-9728333) to M.J.
REFERENCES
- 1.Aguilera A, Klein H L. Yeast intrachromosomal recombination: long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics. 1989;123:683–694. doi: 10.1093/genetics/123.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belmaaza A, Chartrand P. One-sided invasion events in homologous recombination at double-strand breaks. Mutat Res. 1994;314:199–208. doi: 10.1016/0921-8777(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 3.Biedermann K A, Sun J R, Giaccia A J, Tosto L M, Brown J M. scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc Natl Acad Sci USA. 1991;88:1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess S M, Kleckner N. Collisions between yeast chromosomal loci in vivo are governed by three layers of organization. Genes Dev. 1999;13:1871–1883. doi: 10.1101/gad.13.14.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choulika A, Perrin A, Dujon B, Nicolas J-F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1963–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper D N, Krawczak M, Antonarkis S E. The nature and mechanisms of human gene mutation. In: Vogelstein B, Kinzler K W, editors. The genetic basis of human cancer. New York, N.Y: McGraw-Hill; 1998. pp. 65–94. [Google Scholar]
- 7.Difilippantonio M J, Zhu J, Chen H T, Meffre E, Nussenzweig M C, Max E E, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Essers J, Hendriks R W, Swagemakers S M, Troelstra C, de Wit J, Bootsma D, Hoeijmakers J H, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 9.Essers J, van Steeg H, de Wit J, Swagemakers S M, Vermeij M, Hoeijmakers J H, Kanaar R. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank K M, Sekiguchi J M, Seidl K J, Swat W, Rathbun G A, Cheng H L, Davidson L, Kangaloo L, Alt F W. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 11.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 12.Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver D T, Alt F W. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 13.Gillert E, Leis T, Repp R, Reichel M, Hosch A, Breitenlohner I, Angermuller S, Borkhardt A, Harbott J, Lampert F, Griesinger F, Greil J, Fey G H, Marschalek R. A DNA damage repair mechanism is involved in the origin of chromosomal translocations t(4;11) in primary leukemic cells. Oncogene. 1999;18:4663–4671. doi: 10.1038/sj.onc.1202842. [DOI] [PubMed] [Google Scholar]
- 14.Gloor G B, Lankenau D H. Gene conversion in mitotically dividing cells: a view from Drosophila. Trends Genet. 1998;14:43–46. doi: 10.1016/s0168-9525(97)01365-6. [DOI] [PubMed] [Google Scholar]
- 15.Godwin A R, Liskay R M. The effects of insertions on mammalian intrachromosomal recombination. Genetics. 1994;136:607–617. doi: 10.1093/genetics/136.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber M R. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 17.Gu Y, Jin S, Gao Y, Weaver D T, Alt F W. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu Y, Seidl K J, Rathbun G A, Zhu C, Manis J P, van der Stoep N, Davidson L, Cheng H L, Sekiguchi J M, Frank K, Stanhope-Baker P, Schlissel M S, Roth D B, Alt F W. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 19.Haber J E. DNA recombination: the replication connection. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 20.Haber J E, Leung W Y. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci USA. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 22.Jeffs A R, Benjies S M, Smith T L, Sowerby S J, Morris C. The BCR gene recombines preferentially with Alu elements in complex BCR-ABL translocations of chronic myeloid leukemia. Hum Mol Genet. 1998;7:767–776. doi: 10.1093/hmg/7.5.767. [DOI] [PubMed] [Google Scholar]
- 23.Johnson R D, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson R D, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 25.Kadyk L C, Hartwell L H. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karanjawala Z E, Grawunder U, Hsieh C L, Lieber M R. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr Biol. 1999;9:1501–1504. doi: 10.1016/s0960-9822(00)80123-2. [DOI] [PubMed] [Google Scholar]
- 27.Liang F, Han M, Romanienko P J, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang F, Romanienko P J, Weaver D T, Jeggo P A, Jasin M. Chromosomal double-strand break repair in Ku80 deficient cells. Proc Natl Acad Sci USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu N, Lamerdin J E, Tebbs R S, Schild D, Tucker J D, Shen M R, Brookman K W, Siciliano M J, Walter C A, Fan W, Narayana L S, Zhou Z-Q, Adamson A W, Sorensen K J, Chen D J, Jones N J, Thompson L H. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 30.Malkova A, Ivanov E L, Haber J E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris C, Jeffs A, Smith T, McDonald M, Board P, Kennedy M, Fitzgerald P. BCR gene recombines with genomically distinct sites on band 11q13 in complex BCR-ABL translocations of chronic myeloid leukemia. Oncogene. 1996;12:677–685. [PubMed] [Google Scholar]
- 32.Moynahan M E, Chiu J W, Koller B H, Jasin M. Brca1 controls homology-directed repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 33.Moynahan M E, Jasin M. Loss of heterozygosity induced by a chromosomal double-strand break. Proc Natl Acad Sci USA. 1997;94:8988–8993. doi: 10.1073/pnas.94.17.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nussenzweig A, Sokol K, Burgman P, Li L, Li G C. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc Natl Acad Sci USA. 1997;94:13588–13593. doi: 10.1073/pnas.94.25.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paques F, Haber J E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips J W, Morgan W F. Illegitimate recombination induced by DNA double-strand breaks in a mammalian chromosome. Mol Cell Biol. 1994;14:5794–5803. doi: 10.1128/mcb.14.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce A J, Johnson R D, Thompson L H, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puchta H. Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Plant J. 1998;13:331–339. doi: 10.1093/genetics/152.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 40.Richardson C, Moynahan M E, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sargent R G, Brenneman M A, Wilson J H. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schichman S A, Caligiuri M A, Strout M P, Carter S L, Gu Y, Canaani E, Bloomfield C D, Croce C M. ALL-1 tandem duplication in acute myeloid leukemia with a normal karyotype involves homologous recombination between Alu elements. Cancer Res. 1994;54:4277–4280. [PubMed] [Google Scholar]
- 44.Schmid C W. Alu: structure, origin, evolution, significance, and function of one-tenth of human DNA. Prog Nucleic Acid Res. 1996;53:283–319. doi: 10.1016/s0079-6603(08)60148-8. [DOI] [PubMed] [Google Scholar]
- 45.Smih F, Rouet P, Romanienko P J, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strout M P, Marcucci G, Bloomfield C D, Caligiuri M A. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc Natl Acad Sci USA. 1998;95:2390–2395. doi: 10.1073/pnas.95.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 48.Taccioli G E, Rathbun G, Oltz E, Stamato T, Jeggo P A, Alt F W. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 49.Takata M, Sasaki M S, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tebbs R S, Zhao Y, Tucker J D, Scheerer J B, Siciliano M J, Hwang M, Liu N, Legerski R J, Thompson L H. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc Natl Acad Sci USA. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.te Riele H, Maandag E R, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Natl Acad Sci USA. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.te Riele H, Maandag E R, Clarke A, Hooper M, Berns A. Consecutive inactivation of both alleles of the pim-1 proto-oncogene by homologous recombination in embryonic stem cells. Nature. 1990;348:649–651. doi: 10.1038/348649a0. [DOI] [PubMed] [Google Scholar]
- 53.Thomas K R, Capecchi M R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 54.Wang P, Zhou R, Zou Y, Jackson-Cook C, Povirk L. Highly conservative reciprocal translocations formed by apparent joining of exchanged DNA double-strand break ends. Proc Natl Acad Sci USA. 1997;94:12018–12023. doi: 10.1073/pnas.94.22.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]