Abstract
Organoids offer self-organizing, three-dimensional tissue structures that recapitulate physiological processes in the convenience of a dish. The murine mammary gland is composed of two distinct epithelial cell compartments, serving different functions: the outer, contractile myoepithelial compartment and the inner, secretory luminal compartment. Here, we describe a method by which the cells comprising these compartments are isolated and then combined to investigate their individual lineage contributions to mammary gland morphogenesis and differentiation. The method is simple and efficient, and does not require sophisticated separation technologies such as fluorescence activated cell sorting. Instead, we harvest and enzymatically digest the tissue, seed the epithelium on adherent tissue culture dishes, and then use differential trypsinization to separate myoepithelial from luminal cells with ~90% purity. The cells are then plated in an extracellular matrix where they organize into bilayered, three-dimensional organoids that can be differentiated to produce milk over 10 days in culture. To test the effects of genetic mutations, cells can be harvested from wild type or genetically engineered mouse models, or they can be genetically manipulated prior to culture in three-dimensions. This technique can be used to generate mosaic organoids that allow investigation of gene function specifically in the luminal or myoepithelial compartment.
Keywords: mammary, breast, organoid, luminal, basal, myoepithelial, epithelial, 3D culture
Summary
The mammary gland is a bilayered structure, comprising outer myoepithelial and inner luminal epithelial cells. Presented is a protocol to prepare organoids using differential trypsinization. This efficient method allows researchers to separately manipulate these two cell types to explore questions concerning their roles in mammary gland form and function.
Introduction
The mammary gland (MG) is a tree-like, tubular epithelial structure embedded within an adipocyte rich stroma. The bilayered ductal epithelium comprises an outer, basal layer of contractile, myoepithelial cells (MyoECs) and an inner layer of luminal, secretory epithelial cells (LECs), encircling a central lumen 1. During lactation when the outer MyoECs contract to squeeze milk from the inner alveolar LECs, the mammary gland undergoes numerous changes that are under the control of growth factors (e.g. EGF and FGF) and hormones (e.g. progesterone, insulin and prolactin), that promote the differentiation of specialized structures, alveoli, which synthesize and secrete milk during lactation 1. The mammary epithelia can be experimentally manipulated using techniques in which either epithelial tissue fragments, cells or even a single basal cell are transplanted into host mammary fat pads, precleared of endogenous mammary parenchyma, and allowed to grow out to reconstitute an entire, functional epithelial tree 2–5. Transplantation is a powerful technique, but it is time-consuming and impossible if a mutation results in early embryonic lethality (< day E14) that prevents the rescue of transplantable mammary anlage. Furthermore, investigators frequently wish to research the roles of the two different compartments, which are derived from lineage-restricted progenitor cells. While Cre-lox technology allows differential genetic manipulation of MyoECs and LECs, again this is a time-consuming and expensive undertaking. Thus, since the 1950s, investigators have used in vitro mammary organoids as a relatively easy and efficient way to address questions concerning mammary tissue structure and function 6,7.
In early protocols describing the isolation and culture of primary mammary epithelial cells, investigators found that a basement membrane matrix (BME), composed of a plasma clot and chicken embryo extract, was required for MG fragments grown on a dish 6. In the following decades, extracellular matrices (ECMs, e.g. collagen and jellylike protein matrix secreted by Engelbreth-Holm-Swarm murine sarcoma cells) were developed that facilitate culture in three-dimensions (3D) and better mimic the in vivo environment 7–10. Culturing cells in 3D matrices revealed by multiple criteria (morphology, gene expression and hormone responsiveness) that such a microenvironment better models in vivo physiological processes 9–12. Research using primary murine cells identified key growth factors and morphogens necessary for the extended maintenance and differentiation of organoids 13. These studies have set the stage for the protocol presented here, and for the culture of human breast cells as 3D organoids, which is now a modern clinical tool, allowing for drug discovery and drug testing on patient samples 14. Overall, organoid culturing highlights the self-organization capacities of primary cells and their contributions to morphogenesis and differentiation.
Presented here is a protocol to culture murine epithelia that can be differentiated into milk-producing acini. A differential trypsinization technique is used to isolate the MyoECs and LECs that comprise the two, distinct MG cell compartments. These separated cell fractions can then be genetically manipulated to overexpress or knockdown gene function. Because lineage-intrinsic, self-organization is an innate property of mammary epithelial cells 15–17, recombining these cell fractions allows researchers to generate bilayered, mosaic organoids. We begin by enzymatically digesting the adipose tissue, and then incubating the mammary fragments on a tissue culture dish for 24 h (Figure 1). The tissue fragments settle on polystyrene dishes as bilayered fragments with their in vivo organization: outer myoepithelial layer surrounding inner luminal layers. This cellular organization allows for the isolation of the outer MyoECs by Trypsin-EDTA (0.5%) treatment for 3–6 minutes followed by a second round of Trypsin-EDTA (0.5%) treatment that detaches the remaining inner LECs (Figure 2). Thus, these cell types with differential trypsin sensitivity are isolated, and can subsequently be mixed together and plated in ECM (Figure 3). The cells undergo self-organization to form bilayered spheres, comprising an outer layer of MyoECs surrounding inner LECs. Lumen formation occurs as the cells grow in a medium containing a cocktail of growth factors (see recipes for Growth Medium) 13. After 5 days, organoids can be differentiated into milk-producing acini by switching to Alveologenesis Medium (see recipes and Figure 3F) and incubated for another 5 days. Alternatively, organoids will continue to expand and branch in Growth Medium for at least ten days. Organoids can be analyzed using immunofluorescence (Figure 3D–F) or released from the ECM using a recovery solution (see materials list) and analyzed via other methods (e.g. immunoblot, RT-qPCR).
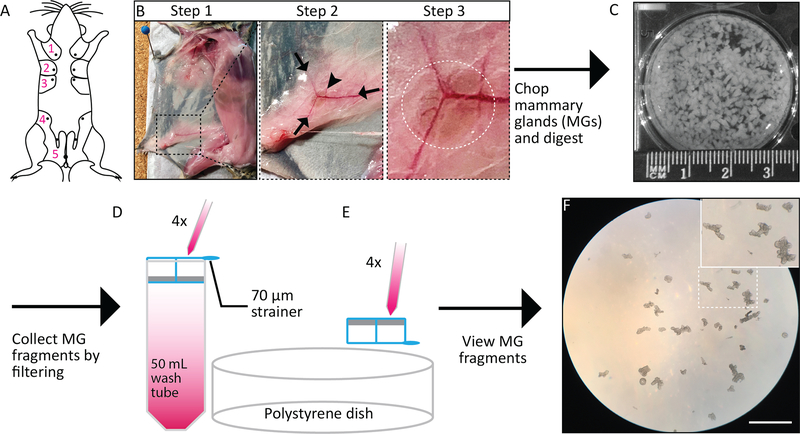
Figure 1: Mammary fragment isolation.
A. Labeled schematic of a mouse’s 5 MGs with unlabeled, contralateral paired MGs. B. Images of mouse MGs with the #4 MG boxed and magnified to show how to identify the lymph node for removal. C. Image of chopped MGs in a 6-well low adhesion plate with a ruler showing the size of the tissue chunks (~0.1mm each). D-E. Schematic illustrating protocol steps 2.7. – 2.9. D. MG fragments are filtered through a 70 μm strainer and rinsed 4X. E. The strainer is then inverted over a 60 mm polystyrene tissue culture dish and fragments are released into the dish. F. Image showing the filtered tissue fragments collected on a 60 mm dish that are free of stroma. The arrows point to the smallest fragments that are collected on the 70 μm strainer. Scale bar = 100 μm.
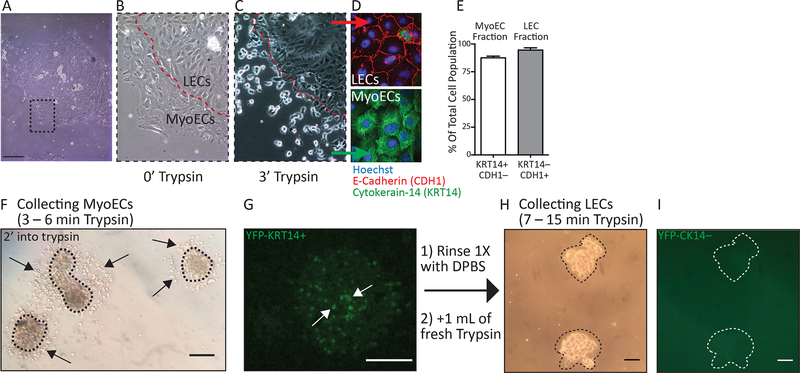
Figure 2: Differential trypsinization.
A. Brightfield image showing a tissue fragment adhered on a polystyrene dish, forming a pancake-like structure. B-C. The first differential trypsinization step detaches MyoECs that are clearly visible as bright, rounded cells after 3 minutes (3’). D. Immunofluorescent images of MyoECs, bottom, and LECs, top, using the Cytokeratin 14 (KRT14) cell marker for MyoECs, and E-Cadherin (CDH1) cell marker for LECs. E. The expression of KRT14 and CDH1 were used to quantify the yield and purity of the differentially trypsinized cell fractions. F-I. Representative phase-contrast and fluorescence (YFP) images of tissue fragments from Cytokeratin 14 (KRT14)-CreERT1; R26RYFP/+ MGs. Mice were injected with 75mg/kg tamoxifen 5 days prior to harvest. F. After 2 minutes incubation, detaching MyoECs (arrows) during the initial Trypsin-EDTA (0.5%). G. A sprinkling of KRT14-YFP-MyoECs (arrows) on top of a pancake of unlabeled LECs. H. Brightfield image of LECs after initial trypsinization and MyoEC detachment. I. After MyoEC detachment, KRT14-YFP-MyoECs are no longer visible as shown by the absence of YFP expression. Scale bars = 30 μm (A, brightfield) 100 μm (F, brightfield), 50 μm (G, fluorescence), 100 μm (H, I). Panels A-E of this Figure were modified from Macias et al. 19
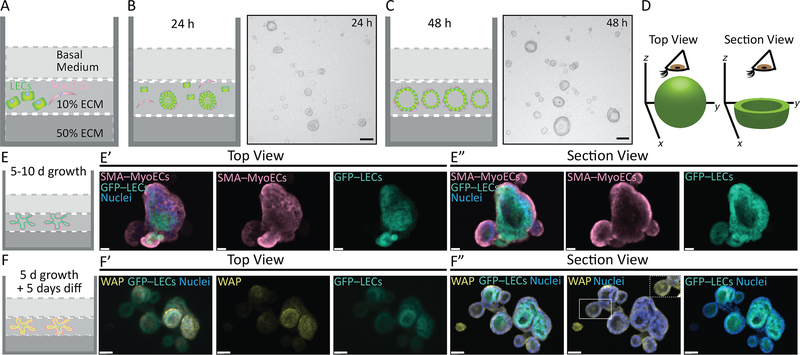
Figure 3: Three-dimensional organoid culture.
A. Schematic representation of single cells embedded in 10% ECM/90% Growth Medium and grown on a 50% ECM/50%DMEM base layer (protocol step 4.5). B- C. Illustrations and phase-contrast images showing the rapid self-organizing capacities of mammary organoids generated from differentially trypsinized and recombined MyoECs and LECs at 24 h (B) and 48 h (C). Images collected using a Keyance Biorevo BZ-9000 Digital Widefield microscope D. Schematic representation illustrating the top (left) or section (right) views used in E-F to show immunostained organoids. E. Schematic representation of a single well of an 8-well chamber slide containing mammary organoids that were grown for 5–10 days in Growth Medium. E’-E”. Top view (E’) and section view (E”) of immunostained organoids at day 10 of growth. MyoECs are marked with smooth actin muscle (SMA) in pseudocolor magenta. The LECs are from ACTb-EGFP mice and are shown in pseudocolor green. Nuclei are stained with Hoechst. F. Schematic representation of a single well of an 8-well chamber slide containing mammary organoids that were grown for 5 days in Growth Medium and 5 days in Alveologenesis Medium. F’-F”. Top view (F’) and section view (F”) of immunostained organoids at day 10 of growth. The MyoECs are unmarked. The LECs are from ACTb-EGFP mice and are shown in pseudocolor green. The milk protein, whey acidic protein (WAP), is shown in pseudocolor yellow in the LECs and coating the inside of the organoids’ lumens. Nuclei are stained with Hoechst. Images collected using a Spinning Disk Confocal Microscope and were reconstructed in 3D using Imaris (E’, E’) or bottom section ~30 slices (F’, F”). Scale bars = 100 μm (C), 20 μm (E), 40 μm (F).
Materials List.
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
|---|---|---|---|
| BD Insulin syringe 0.5 mL | Thermo Fisher Scientific | 14-826-79 | |
| Pentobarbital | Millipore Sigma | P3761 | |
| Trypsin EDTA 0.05% | Thermo Fisher Scientific | 25300-062 | |
| Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12) | Thermo Fisher Scientific | 11330-057 | |
| Antibiotic-Antimycotic (100X) | Thermo Fisher Scientific | 15240062 | Pen/Strep also works |
| DMEM/F12, no phenol red | Thermo Fisher Scientific | 11039-021 | |
| Dulbecco’s phosphate-buffered saline (DPBS) | Thermo Fisher Scientific | 14190-250 | Without Mg2+/Ca2+ |
| Matrigel™ Growth Factor Reduced (GFR); Phenol Red-Free; 10 mL | Fisher Scientific | CB-40230C | Lot: 8204010, stock concentration 8.9 mg/mL |
| DNase (Deoxyribonuclease I) | Worthington Biochemical | LS002007 | |
| Millicell EZ SLIDE 8-well glass, sterile | Millipore Sigma | PEZGS0816 | These chamber slides are great for gasket removal but other brands can work well (e.g. Lab Tek II). |
| Class 3 Collagenase | Worthington Biochemical | LS004206 | |
| Class 2 Dispase (Roche) | Millipore Sigma | 4942078001 | |
| Fetal Bovine Serum | VWR | 97068-085 | 100% US Origin, premium grade, Lot: 059B18 |
| MillexGV Filter Unit 0.22μm | Millipore Sigma | SLGV033RS | |
| Gentamicin | Thermo Fisher Scientific | 15710064 | |
| Insulin | Millipore Sigma | I6634-100mg | |
| EGF | Fisher Scientific | AF-100-15-100ug | |
| N-2 Supplement (100x) | Thermo Fisher Scientific | 17502048 | |
| B27 supplement without vitamin A (50x) | Thermo Fisher Scientific | 12587010 | |
| Nrg1 | R&D | 5898-NR-050 | |
| R-spondin | Peprotech | 120-38 | |
| Rho inhibitor Y-27632 | Tocris | 1254 | |
| 70μM nylon cell strainer (Corning) | Fisher Scientific | 08-771-2 | |
| 15 ml High-Clarity Polypropylene Conical Tube (BD Falcon) | Fisher Scientific | 352096 | |
| 50 ml High-Clarity polypropylene conical tube (BD Falcon) | Fisher Scientific | 352098 | |
| 35 mm TC-treated Easy-Grip Style Cell Culture Dish (BD Falcon) | Fisher Scientific | 353001 | |
| 60 mm TC-treated Easy-Grip Style Cell Culture Dish (BD Falcon) | Fisher Scientific | 353004 | |
| Corning® Costar® Ultra-Low Attachment 6-well | Fisher Scientific | CLS3471 | |
| Ovine Pituitary Prolactin | National Hormone and Peptide Program | Purchased from Dr. Parlow at Harbor-UCLA Research and Education Institute | |
| Dexamethasone | Millipore Sigma | D4902-25MG | |
| Fluoromount-G (Southern Biotech) | Fisher Scientific | 0100-01 | Referred to as mounting media in text |
| Corning Cell Recovery solution | Fisher Scientific | 354253 | Follow the guidelines for use – Extraction of Three-Dimensional Structures from Corning® Matrigel® Matrix |
| Paraformaldahyde | Millipore Sigma | PX0055-3 | |
| Sterile Filtered Donkey Serum | Equitech-Bio Inc. | SD30-0500 | |
| Triton X-100 | Millipore Sigma | x100-500ML | Laboratory grade |
| NaCl | Fisher Scientific | S671-3 | |
| KCl | Fisher Scientific | P217-500 | |
| NaH2PO4 | Fisher Scientific | S468-500 | |
| KH2PO4 | Fisher Scientific | P285-500 | |
| Sterile Filtered Donkey Serum | Equitech-Bio Inc. | SD30-0500 | |
| Sodium Hydroxide | Fisher Scientific | S318-500 | |
| Glycine | Fisher Scientific | BP381-5 | |
| 24 well ultra–low attachment plate (Corning) | Fisher Scientific | CLS3473-24EA | |
| KRT14–CreERtam | The Jackson Laboratory | 5107 | |
| R26R-EYFP | The Jackson Laboratory | 6148 | |
| B6 ACTb-EGFP mice | The Jackson Laboratory | 003291 | |
| Mouse anti-SMA | Millipore Sigma | A2547 | Use at 1:500, Lot: 128M4881V, stock 5.2 mg/mL, RRID:AB_476701 |
| Donkey anti-Mouse 647 | Jackson ImmunoResearch | 715-606-150 | Use at 1:1000, Lot: 140554, stock 1.4 mg/mL |
| Goat anti-WAP | Santa Cruz Biotech | SC-14832 | Use at 1:250, Lot: J1011, stock 200 μg/mL, RRID:AB_677601 |
| Donkey anti-Goat 647 | Thermo Fisher Scientific | A21447 | Use at 1:500, Lot: 1608641, stock 2 mg/mL, RRID:AB_2535864 |
| Hoechst 33342 | AnaSpec | AS-83218 | Use 1:2000, stock is 20mM |
Protocol
All methods described here have been approved by the Institutional Animal Care and Use Committee (IACUC) of the University of California, Santa Cruz.
1. Day 1: mammary gland digestion
1.1. Prepare to harvest the MGs from mature female mice (10–14 weeks of age).
1.1.1. Perform the harvesting on an open bench under aseptic conditions.
1.1.2. Sterilize all surgical supplies, cork boards and pins (autoclave and soak in 70% alcohol for 20 minutes prior to surgery).
1.1.3. Because euthanasia is the outcome, anesthetize animals with sodium pentobarbital (2X anesthetic dose of 0.06mg/g body weight) delivered via intraperitoneal injection with a 0.5 mL insulin syringe.
1.1.4. Monitor the level of anesthesia by pinching the animal’s toes for reflex response and commence the protocol only after the animal is fully anesthetized.
1.1.5. Place the animal on its back, pin appendages to the corkboard and wipe down its abdomen and chest with ethanol.
1.2. To harvest the #2, 3, 4, and 5 MGs from one mouse (i.e. 8 MGs) (Figure 1A), identify the midline between the two hind legs and make a small incision (1cm) on the abdominal skin with sharp scissors, then extend the cut up to the neck 18.
1.3. Follow by making small cuts laterally towards the legs and arms to allow for the release of the skin using a cotton swab, pull the skin away and stretch it tight before pinning it down on one side (Figure 1B, Step 1) 18. Remove the MGs by cutting under them and remove the lymph nodes from the #4 glands (Figure 1B, Steps 2, 3) 18. Repeat the procedure on the other side of the body.
1.4. Collect the MG tissue in 50 mL of 4 °C Dulbecco’s Modified Eagle’s Media (DMEM)/Nutrient Mixture F12 (F12) supplemented with 5% fetal bovine serum (FBS) and 1X Antibiotic-Antimycotic (Anti-Anti) 18.
1.5. Chop the glands in a 35 mm dish or on a ceramic plate using a razor blade or tissue chopper. Rotate the plate every 5 manual chops or every round on the tissue chopper until the tissue pieces can fit through a 1 mL micropipette tip with ease (~0.1mm/fragment) (Figure 1C).
1.6. Digest the MGs in Digestion Medium (see Table 1) for 14 h in a 6-well low adhesion dish at 37 °C, 5% CO2.
Table 1:
Recipes for reagents
| 10 mL | Digestion Medium | |
| Amount | Reagent | Notes |
| 9.45 mL | DMEM/F12 | |
| 100 μL | Antibiotic-Antimycotic (100X) | |
| 0.04 g | Class 3 Collagenase | |
| 0.04 g | Class 2 Dispase | |
| 50 μL | Gentamicin | Final Concentration: 500 μg |
| 2.5 mL | Fetal Bovine Serum | Final concentration: 5% (v/v) |
| Pass through 0.22μm filter | ||
| 50 mL | Maintenance Medium | |
| Amount | Reagent | Notes |
| 49.47 mL | DMEM/F12 | |
| 0.5 mL | Antibiotic-Antimycotic (100X) | |
| 2.5 mL | Fetal Bovine Serum | Final concentration: 5% (v/v) |
| 25 μL | Insulin | Final concentration: 250 μg |
| 5 μL | EGF | Final concentration: 500 ng |
| 10 mL | Growth Medium | |
| Amount | Reagent | Notes |
| 9.6455 mL | DMEM/F12, no phenol red | |
| 100 μL | N-2 Supplement (100x) | |
| 200 μL | B27 supplement without vitamin A (50x) | |
| 10 μL | Nrg1 | Stock: 100μg/mL |
| 42.5 μL | R-spondin | Stock: 10 μg/mL |
| 1 μL | Rho inhibitor Y-27632 | Stock: 10 μM |
| 1 μL | EGF | Stock: 0.1 μg/μL |
| 10 mL | Alveologenesis Medium | |
| Amount | Reagent | Notes |
| 9.6355 mL | DMEM/F12, no phenol red | |
| 100 μL | N-2 Supplement (100x) | |
| 200 μL | B27 supplement without vitamin A (50x) | |
| 10 μL | Nrg1 | Stock: 100μg/mL |
| 42.5 μL | R-spondin | Stock: 10 μg/mL |
| 1 μL | Rho inhibitor Y-27632 | Stock: 10 μM |
| 5 μL | Ovine Pituitary Prolactin | Final concentration: 1 μg/mL |
| 1 μL | Dexamethasone | Final concentration: 5 μg/mL |
| 5 μL | Insulin | Final concentration: 5 μg/mL |
| 1 L | 10X DPBS | |
| Amount | Reagent | Notes |
| 80 g | NaCl | |
| 2 g | KCl | |
| 14.4 g | NaH2PO4 | |
| 2.4 g | KH2PO4 | |
| 1 L | di H2O | |
| Fill to 800 mL before adding dry reagents and dissolve. Fill volume to 1 L. Adjust pH to 7.4. Autoclave to sterilize. | ||
| 1 L | 1X DPBS | |
| Amount | Reagent | Notes |
| 100 mL | 10X PBS | |
| 900 mL | di H2O | |
| 1 L | PBST | |
| Amount | Reagent | Notes |
| 100 mL | 10X PBS | |
| 2.5 mL | Triton X-100 | |
| 250 mL | 4% Paraformaldehyde | |
| Amount | Reagent | Notes |
| 10 g | Paraformaldehyde | |
| 200 mL | di H20 | water must be at 60 °C |
| 25 mL | 10X DPBS | |
| 50 μL | 10 N Sodium Hydroxide | |
| Pass through a 0.45 μm filter to sterilize and assure pH is 7.4 | ||
| 10 mL | 1 % Donkey Serum | |
| Amount | Reagent | Notes |
| 100 μL | Sterile Filtered Donkey Serum | |
| 9.9 mL | 1X DPBS | |
| 10 mL | 0.2% Glycine | |
| Amount | Reagent | Notes |
| 0.02 g | Glycine | |
| 10 mL | 1X DPBS | |
2. Day 2: isolation of mammary epithelial tissue fragments
2.1. After 14 h of digestion, gently mix by pipetting the digested tissue 10X using a 1 mL micropipette to break down any remaining stroma or adipose tissue, ensuring that neither bubbles nor excess mechanical force are generated.
Note: If there is incomplete digestion after 14 h, this could be due to the accumulation of sheared DNA. In this circumstance, add 1 μL of 1 mg/mL Deoxyribonuclease I (DNaseI) per 2 mL Digestion Medium. Incubate for another 30 min at 37 °C, 5% CO2.
2.2. Collect tissue in a 15 mL tube and rinse the well used for digestion with 2–3 mL of tissue culture grade 1X Dulbecco’s Phosphate Buffered Saline (DPBS) free of Ca2+ and Mg2+. Centrifuge at 600 × g for 10 min.
2.4. Evacuate supernatant containing lipid layer and medium, and then resuspend the pellet in 5 mL DPBS and switch to a new 15 mL tube. Centrifuge at 600 × g for 10 min.
2.5. During the centrifugation time place a 70 μm nylon cell strainer in a 50 mL tube and pre-wet the strainer using 10 mL of 37 °C DMEM/F12.
2.6. Resuspend tissue fragments from protocol step 2.4 using 5 mL DPBS and pass the suspension through a pre-wet 70 μm nylon cell strainer to remove stromal cells and single cells (Figure 1D).
2.7. Collect the tissue fragments on the cell strainer. Rinse 4X with 10mL of 37 °C DMEM/F12 (Figure 1D).
Caution: Incomplete rinsing will result in cultures contaminated with non-epithelial cells.
2.8. Release the tissue fragments by holding the strainer tab with your gloved fingers, inverting the strainer over a 60 mm tissue culture dish and passing 4X, 1 mL aliquots of Maintenance Medium (see Table 1) through the bottom of the strainer (Figure 1E).
2.9. Check the strainer for remnant tissue fragments, which will be visible by the naked eye, and rinse the strainer 1 more time with 1 mL of Maintenance Medium if any fragments are still adhering to the strainer. Rinsed tissue fragments should now be free of stromal cells.
2.9. Quickly examine under an inverted microscope (4X or 10X objective) the 60 mm dish containing the MG fragments from protocol step 2.9. (Figure 1F). A typical preparation of 8 MGs yields ~500 fragments. Look for single cells or fat droplets and follow the note below if there are contaminating cells.
Note: If there are contaminating cells, re-perform the filtration step by collecting the medium and fragments from the 60 mm dish using a 5 mL pipette and filtering the fragment again through a fresh 70 μm strainer, repeating protocol steps 2.6., 2.8. – 2.10.
2.10. Incubate 24 h at 37 °C, 5% CO2, allowing the tissue fragment to adhere and generate bilayered fragments (Figure 2A).
Note: If the fragments have not settled by 24 h, continue to incubate until adhered. If the fragments do not adhere well, the separation of the cell compartments will not work. If researchers are concerned about adhesion, the tissue culture plates can be treated to promote fragment attachment (e.g. poly-L-lysine).
3. Day 3: differential trypsinization of myoepithelial and luminal epithelial cells
3.1. To separate MyoECs from LECs, evacuate the media from the dish, rinse 1x with 1 mL DPBS and add 1 mL of fresh Trypsin-EDTA (0.5%), and carefully monitor digestion under an inverted microscope (10X or 20X objective) (Figure 2B, C, F). The detachment of the outer MyoEC layer will require 3–6 min, depending on Trypsin-EDTA (0.5%) strength.
Note: Please see Figure 2 for representative images showing this process. Under brightfield illumination, the MyoECs appear rounded up and have a brighter appearance in comparison to the LECs, which remain adhered in the center and appear darker.
3.2. Collect the MyoEC fraction in a 15 mL tube containing 2 mL 10% FBS/DPBS. Without disturbing the LECs, gently rinse, the 60 mm dish with 2 mL of DPBS and then dispose of the DPBS (Figure 2H–I).
Note: The usual recovery for MyoECs is within a range of (3.5 × 106 – 1.5 × 106) depending on the size of the MGs.
3.3. To remove the LEC fraction, add 1 mL of Trypsin-EDTA (0.5%) to the dish again and incubate 7–15 min, monitoring carefully to prevent over-digestion. Quench the Trypsin-EDTA (0.5%) on the dish with 2 mL 10% FBS/DPBS. Collect the LEC fraction in a new 15 mL tube
Note: The usual recovery for LECs is within a range (2E+6 – 4.2E+6) depending on the size of the MGs. Routinely, the purity of both fractions as assayed by immunohistochemistry is ~90% 19 (Figure 2E).
3.4. Centrifuge each fraction for 5 min at 300 × g to remove the Trypsin-EDTA (0.5%). Resuspend the pellet in 250 μL of Maintenance Medium and count each cell population using a hemocytometer or automated cell counter. Place cells on ice while counting.
Note: If genetic manipulation of cell fractions is desired, primary cells can be grown on low adhesion dishes and infected with lentivirus 20.
4. Day 3: combine and embed cell fractions in an Extracellular Matrix
NOTE: Once the MyoEC and LEC fractions have been collected and counted, they can be combined. The typical MyoEC/LEC ratio is 1:3 (Figure 3A)19. Different studies can be performed. For example to perform mosaic studies, fractions can be generated from wild type (WT) and mutant (Mut) mice and combined (MyoEC/LEC: WT/WT; WT/Mut; Mut/WT; Mut/Mut) 21, or fractions can be combined using different ratios of MyoECs/LECs 19.
4.1. Based on cell counts, calculate the number of wells (8-well chamber, see list of materials) that need to be prepared (12,000 cells/well (e.g. 3000 MyoECs:9000 LECs).
4.2. Establish the base layer for 3D culture by adding 90 μL of 50% ECM (50% ECM/50% DMEM/F12, no phenol red – see the table of materials) to each well. Ensure there are no bubbles and the wells are coated evenly. To solidify the base layer, incubate the slides at 37 °C, 5% CO2 for 30 min.
Note: The ECM needs to stay ice-cold until this step; otherwise it will polymerize prematurely and lead to uneven base coating and polymerization. Using a base ECM enhances organoid growth in a single plane, which aids image capture. We also obtain faster organoid growth and use less ECM 22.
4.3. During the polymerization time, prepare cell mixes. Pellet the MyoEC and LEC fractions at 300 × g for 5 min and resuspend each cell fraction in 10% ECM/90% Growth Medium so each well has 100 μL (see Table 1 for how to make Growth Medium).
Note: For preparation ease, replicate wells using the same cell mixes can be combined and prepared in 1 tube (e.g. 4 wells of the same cell mix can be prepared in 400 μL of 10% ECM/90% Growth Medium).
4.4. Add 100 μL of each cell mix in 10% ECM/90% Growth Medium to each well and allow organoids to settle for 20 min at 37 °C, 5% CO2.
4.5. Once the cells have settled, gently add 100 μL of Growth Medium by gently pipetting down the chamber wall of each well. Incubate slides at 37 °C, 5% CO2 (see Figure 3A for the total composition of each well).
Note: The Rho Kinase inhibitor, R-spondin, and Nrg1 are factors that have been identified as important for the long-term culture of organoids, grown from both primary murine mammary cells and human breast cancer cells 13,14. In addition, the stem cell factors B27 and N2 extend the time that organoids can be cultured.
4.6. Image cells every 24 h to track their growth and gently change Growth Medium every 2–3 days using 100 μL/well (Figure 3B–E).
Note: Use extreme care when changing the medium. Tilt the chamber slides to collect medium to one corner of the wells. Only remove ≤100 μL of medium and replenish with care so as to leave the ECM layer undisturbed.
4.7. If researchers are interested in investigating lactation/alveologenesis, switch to Alveologenesis Medium (see Table 1) on day 5 and continue to change medium every 2–3 days until day 10 (Figure 3F) or beyond by passaging using recovery solution (see the list of materials) 13.
Note: At this point, organoids can be isolated from the ECM by incubating at 4 °C in 400 μl of 4 °C recovery solution (see materials list for protocol and reagent info).
5. Day 5 or 10: fix and immunostain organoids
5.1. Remove medium carefully by gently pipetting off the media (a bulb pipette works best). Rinse each well using 200 μL 1X Dulbecco’s Phosphate Buffered Saline (DPBS, see recipes).
5.2. Fix organoids using cold (4 °C), 4% (w/v) paraformaldehyde (PFA, see recipes) for 10 min at room temperature.
Caution: PFA is considered hazardous. Wear personal protective equipment (lab coat, gloves and safety glasses). This step should be performed inside a fume hood.
Note: The ECM is dissolved by PFA treatment. Incomplete ECM removal can lead to background staining when organoids are analyzed by immunofluorescence.
5.3. Remove the 4% PFA and add 200 μL of 0.2% (w/v) glycine/DPBS (see Table 1) to each well. Incubate slides at room temperature for 30 min or 4 °C overnight on a rocking surface set to a slow setting.
Note: Organoids can be stored for 1–3 days in DPBS at 4 °C prior to the next step.
5.4. Permeabilize the organoids using DPBS + 0.25% Triton X-100 (PBST, see Table 1) for 10 min at room temperature.
5.5. Block organoids using 5% donkey serum (DS) in DPBS for 1 h on a rocking surface.
Note: This step can be performed overnight at 4 °C on a rocking surface.
5.7. Prepare primary antibodies in 1% DS/DPBS, use 125–200 μL for each well. Perform immunostaining by incubating the organoids in primary antibody overnight at 4 °C on a rocking surface.
6. Day 11: complete immunofluorescence
6.1. Wash each well 2X with 200 μL PBST for 5 min. Add secondary antibody in 1% DS/DPBS using 125–200 μL for each well. Incubate at room temperature on a rocking surface for 45 min.
6.2. Wash each well 2X using 200 μL DPBS per well.
6.3. Stain nuclei using Hoechst in DPBS (1:2000 in DPBS) for 5 min at room temperature.
6.4. Evacuate all liquid left on the well by gently suctioning with a vacuum.
6.5. Carefully remove the chambers and gasket, place 1 dot (~30 μL) of mounting media (see table of materials) on each well and coverslip, taking care to remove bubbles. Allow the slide to dry in dark space for 1–2 days. Seal with clear nail polish. Image organoids on a confocal microscope (Figure 3 E–F).
Representative Results:
The protocol presented here describes a method for investigating specific lineage contributions of mammary epithelial cells by making use of mosaic organoids. To obtain primary murine cells for organoids, the mammary gland epithelium must first be isolated from the surrounding adipocyte rich stroma (Figure 1). This process is described briefly here and can also be visualized in a previously published paper 18. To obtain sufficient cells, it is recommended that #2, 3, 4, and 5 MGs are removed (Figure 1A). An important step and key to isolating a pure population of epithelial cells is removal from the #4 MGs of the lymph nodes, which are rich in immune cells that will contaminate the preparation (Figure 1A, B). The MGs are minced to generate fragments that are ~0.1mm in size (Figure 1B). The tissue fragments are then enzymatically digested, a process occurring in the presence of collagenase, to release epithelia from stroma, and in the absence of trypsin, to prevent the digestion of proteins such as cadherins that maintain cell-cell contacts. The digested tissue is then centrifuged to remove lipids and filtered through a cell strainer and washed (Figure 1C). Epithelial fragments, adhering to the strainer, are released by inverting the filter and washing the membrane, which transfers the epithelial fragments onto a polystyrene dish (Figure 1D). These fragments appear as small, branched structures (Figure 1E).
The purified epithelial fragments are incubated for 24 h. They settle down onto the dish and adhere, forming flat, pancake-like structures with an outer layer of MyoECs encircling inner LECs (Figure 2A–B). In Figure 2C, the edge of such a pancake-like structure from a wild type animal is observed. Trypsin treatment differentially detaches the MyoECs, which de-adhere first and appear as bright, rounded cells that encircle the core of remaining cuboidal LECs (Figure 2C, F). The detachment of the MyoECs is carefully monitored using brightfield microscopy and occurs within 3–6 minutes. Once the MECs are collected, LECs are subsequently detached through a second trypsin treatment that takes longer, 7–15 minutes. The time required for cell detachment depends on the trypsin concentration and freshness. The overall purity of the two cell compartments is ~90%, as assayed by counting cells that are KRT14-positive and E-Cadherin (CDH1)-negative in the MyoEC fraction and cells that are KRT14-negative and E-Cadherin-positive in the LEC fraction (Figure 2D–E) 19. We discovered that some of the MyoECs are removed from the top of the pancake-like structure as well as from the outer edges. This is observed by using tissue fragments collected from mice labeled with an inducible, fluorescent basal marker (Cytokeratin 14 (KRT14)-CreERT1; R26RYFP/+) and injected with 75mg/kg tamoxifen, 5 days prior to harvest. In Figure 2F, G the detachment of MyoECs from around the edges of the pancake structure is readily apparent within the first 2 min of trypsin treatment (Figure 2F). In addition, YFP-KRT14-positive cells are observed on top of the structure, where they rounded up after trypsin treatment and are removed by the rinse/collection step (Figure 2G). The unlabeled core of LECs (Figure 2H), which contains few or no YFP-KRT14-positive cells, (Figure 2I) are subsequently detached in the second round of trypsin treatment.
The MyoEC and LEC fractions are collected, combined and embedded into 10% ECM that is plated onto a 50% ECM base; this allows for better optical resolution of the organoids that grow primarily along the base layer (Figure 3A). After 24 h, cells have assembled into aggregated structures that largely lack a lumen (Figure 3B). After 48 H, nascent organoids form as the central lumens hollows and appears as a lighter internal space (Figure 3C). After 10 days, the organoids are large, branched structures with well-developed lumens. Mosaic organoids that were generated from MyoECs, harvested from wild type mice, and LECs, harvested from ACTb-EGFP mice, were fixed in situ, immunostained with an antibody direct against the basal marker, alpha-smooth muscle actin (SMA) and stained with the Hoechst DNA dye to show nuclei. The top and section views show different sets of images collected as a Z-stack and reconstructed into a 3-D view (Figure 3D). The top view reveals the branched morphology of the organoids (Figure 3E’). The section view shows the bilayered epithelial structure and open lumen of these organoids (Figure 3E”). These organoids can also be differentiated at Day 5 using Alveologenesis Medium and incubated for an additional 5 days (Figure 3F). The organoids grow larger, have more branches and contain milk. Differentiated organoids were generated as described above and immunostained with an antibody directed against the milk marker, whey acidic protein (WAP) (Figure 3F). WAP is a soluble protein, secreted into milk, and much of this liquid is lost when the cells are fixed and immunostained in situ. Therefore, in the top and section views, WAP staining is visible intracellularly in secreting cells and extracellularly in milk that is trapped at the cell surface during fixation (Figure 3F), although in section view a small organoid appears to contain liquid milk (Figure 3F” boxed overlay).
Discussion
Here, a method is presented detailing how researchers can generate 3D organoid cultures using primary MG cells. The difference between this and many protocols is that here we detail a method to separate the two, distinct MG cell compartments: outer basal MyoECs and inner LECs. Our method employs a two-step, Trypsin-EDTA (0.5%) treatment that we call differential trypsinization 19. This procedure allows researchers to isolate basal and luminal cells without using sophisticated flow cytometry and thus can be used for studying MGs harvested from a wide variety of mammalian species that may not have the well-characterized biomarkers required for FACS. The ability to segregate the two cell subpopulations enables researchers to genetically modify the isolated cells independently, or recombine cells from animals harboring genetic mutations or labels, and thus generate mosaic organoids in 3D culture. A limitation with the current protocol is that the stromal compartment is not included in the culturing conditions. However, new methods are being developed to co-culture stromal components with organoids generated from either primary cells or cell lines to better recapitulate in vivo ECM 23–26, and these methods may be adapted to this protocol. In addition, it is important to note that while this protocol achieves a great enrichment of the MyoEC and LEC fractions (~90% purification), the fractions do not represent pure cell lineages.
The success of this protocol relies on a number of key steps. First, it is important to gently but thoroughly digest the MG tissue. Over digestion of the tissue will lead to cell death and lower recovery of epithelial cells. Under digestion will result in stromal and adipose cell contamination, which will interfere with later analyses (e.g. immunofluorescence, protein and mRNA measurements). Second, it is important to thoroughly rinse the MG tissue to remove contaminating cells in protocol step 2.8. In protocol step 2.9, the MG tissue fragments are released into a 60 mm dish. Researchers should monitor the released fragments immediately, before they adhere to the dish. If fat droplets or single cells are observed, protocol steps 2.6., 2.8–2.11. must be repeated. To do this, the medium and tissue fragments are collected from the dish, placed into a new 70 μm strainer, washed 4X with 37 °C DMEM/F12 and then released into a new 60 mm dish. Third, it is essential to watch the first Trypsin-EDTA (0.5%) incubation closely because the MyoECs can detach within the first 3 minutes, but they can also adhere for up to 6 minutes. There have even been instances when the TrypsinEDTA (0.5%) was suboptimal and this incubation proceeded for 10 minutes with successful purification of MyoECs. However, past 10 min of trypsinization resulted in the co-collection of MyoECs and LECs. It is also important that the dish is undisturbed during the first incubation because this will lead to contamination of the MyoEC fraction with LECs. The reverse is also true; if MyoECs are not detached well from the dish, they will contaminate the LEC fraction. If researchers are using reporter mice that label MyoECs or LECs exclusively, it is easier to visualize the separation under a fluorescence microscope (Figure 2F–I). Finally, if researchers plan on fixing organoids for immunofluorescence analyses, the pH (7.4) and temperature (4 °C) of the 4% PFA is important for successful dissociation of the ECM. If the organoids are collected for other modes of analyses (e.g. protein and mRNA measurements), it is important that the recovery solution be at 4 °C. If the ECM is not dissolving, incubation with the recovery solution can be extended by ten min (i.e. 30 min total incubation). However, longer incubation periods will lead to loss of 3D structure and cell death. The recovery protocol (listed in the material list) specifies the use of wide-bore tips and this is important for maintaining the 3D structure of the organoids as well as the integrity of the cells.
In addition to these four key steps, there are two factors that influence the success of the protocol. First, organoid growth can be limited by genetic mutations that reduce cell proliferation and therefore reduce organoid growth in ECM. If only a few organoids are obtained, the subsequent fixation step frequently results in their loss. To address this, the number of cells embedded within the ECM should be increased, while retaining the ratio of MyoECs:LECs (protocol step 4.1. – 4.2.). Second, once the cells are transferred into an ECM it is important to watch their growth daily and be vigilant about media changes (every 2–3 days). This protocol specifies phenol-red free reagents for better visualization, but the same success and growth is achieved using phenol-red positive reagents. The days when medium changes occur prior to fixation (protocol step 4.6.) should be performed with extreme care to reduce cell loss. The 10% ECM top layer is delicate; therefore washes or medium changes should be performed by pipetting fluid down the chamber walls to minimize mechanical disturbances.
Differentiation of the organoids into milk-producing acini requires treatment with differentiation supplements: hydrocortisone or dexamethasone, insulin and prolactin. In this protocol, dexamethasone is recommended. In addition, while prolactin is commercially available, the prolactin used in this protocol was obtained from the National Hormone and Peptide Program. Again, it is very important to leave the organoids undisturbed when changing the Alveologenesis Medium. Differentiation requires a minimum of 5 days and this can be extended another 3–5 days, but the base layer of ECM degrades after 10–12 days. Differentiated organoids are filled with milk and their lumens appear darker.
Presented here, is an efficient technique that can be used to address compartment-specific, lineage contributions to mammary epithelial morphogenesis and differentiation. With this technique, researchers can generate mosaic organoids comprising differentially genetically manipulated MyoECs and LECs 21, or MyoECs and LECs obtained from mice harboring different genetic mutations. This allows researchers to better understand the contributions of lineage-specific cell compartments to organ morphogenesis and the acquisition of specialized functions such as milk production.
Acknowledgments
We thank Ben Abrams for technical assistance and core support from the University of California, Santa Cruz (UCSC) Institute for the Biology of Stem Cells (IBSC). We thank Susan Strome and Bill Saxton for the use of their Solamere Spinning Disk Confocal Microscope. This work was supported in part by grants to UCSC from the Howard Hughes Medical Institute through the James H. Gilliam Fellowships for Advanced Study program (S.R.), from the NIH (NIH GM058903) for the initiative for maximizing student development (H.M.) and from the National Science Foundation for a graduate research fellowship (O.C. DGE 1339067) and by a grant (A180370) from the UC-Cancer Research Coordinating Committee (LH).
Footnotes
Disclosures
The authors have nothing to disclose.
References
- 1.Macias H & Hinck L Mammary gland development. Wiley Interdisciplinary Reviews in Developmental Biology 1 (4), 533–557, doi: 10.1002/wdev.35, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniel CW, De Ome KB, Young JT, Blair PB & Faulkin LJ Jr. The in vivo life span of normal and preneoplastic mouse mammary glands: a serial transplantation study. Proceedings of the National Academy of Science U S A. 61 (1), doi 10.1073/pnas.61.1.53, 53–60 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ip MM & Asch BB Methods in Mammary Gland Biology and Breast Cancer Research. (Kluwer Academic/Plenum Publishers, 2000). [Google Scholar]
- 4.Shackleton M et al. Generation of a functional mammary gland from a single stem cell. Nature. 439 (7072), 84–88, doi: 10.1038/nature04372, (2006). [DOI] [PubMed] [Google Scholar]
- 5.Stingl J et al. Purification and unique properties of mammary epithelial stem cells. Nature. 439 (7079), 993–997, doi: 10.1038/nature04496, (2006). [DOI] [PubMed] [Google Scholar]
- 6.Lasfargues EY Cultivation and behavior in vitro of the normal mammary epithelium of the adult mouse. II. Observations on the secretory activity. Experimental Cell Research. 13 (3), 553–562, doi: 10.1016/0014-4827(57)90085, (1957). [DOI] [PubMed] [Google Scholar]
- 7.Simian M & Bissell MJ Organoids: A historical perspective of thinking in three dimensions. Journal of Cell Biology 216 (1), 31–40, doi: 10.1083/jcb.201610056, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orkin RW et al. A murine tumor producing a matrix of basement membrane. Journal of Experimental Medicine. 145 (1), 204–220, doi: 10.1084/jem.145.1.204, (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee EY, Parry G & Bissell MJ Modulation of secreted proteins of mouse mammary epithelial cells by the collagenous substrata. Journal of Cell Biology. 98 (1), 146–155, doi: 10.1083/jcb.98.1.146, (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee EY, Lee WH, Kaetzel CS, Parry G & Bissell MJ Interaction of mouse mammary epithelial cells with collagen substrata: regulation of casein gene expression and secretion. Proceedings of the National Academy of Science U S A. 82 (5), 1419–1423, doi: 10.1073/pnas.82.5.1419, (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bissell MJ & Barcellos-Hoff MH The influence of extracellular matrix on gene expression: is structure the message? Journal of Cell Science. Supplement. 8 327–343 (1987). [DOI] [PubMed] [Google Scholar]
- 12.Petersen OW, Ronnov-Jessen L, Howlett AR & Bissell MJ Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proceedings of the National Academy of Science U S A. 89 (19), 9064–9068, doi: 10.1073/pnas.89.19.9064, (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarde T et al. Wnt and Neuregulin1/ErbB signalling extends 3D culture of hormone responsive mammary organoids. Nat Commununications. 7 13207, doi: 10.1038/ncomms13207, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sachs N et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell. 172 (1–2), 373–386 e310, doi: 10.1016/j.cell.2017.11.010, (2018). [DOI] [PubMed] [Google Scholar]
- 15.Daniel CW, Strickland P & Friedmann Y Expression and functional role of E- and P-cadherins in mouse mammary ductal morphogenesis and growth. Developmental Biology 169 (2), 511–519, doi: 10.1006/dbio.1995.1165, (1995). [DOI] [PubMed] [Google Scholar]
- 16.Runswick SK, O’Hare MJ, Jones L, Streuli CH & Garrod DR Desmosomal adhesion regulates epithelial morphogenesis and cell positioning. Nature Cell Biology. 3 (9), 823–830, doi: 10.1038/ncb0901-823, (2001). [DOI] [PubMed] [Google Scholar]
- 17.Chanson L et al. Self-organization is a dynamic and lineage-intrinsic property of mammary epithelial cells. Proceedings of the National Academy of Science U S A. 108 (8), 3264–3269, doi: 10.1073/pnas.1019556108, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honvo-Houeto E & Truchet S Indirect Immunofluorescence on Frozen Sections of Mouse Mammary Gland. Journal of Visualized Experiments. (106), doi: 10.3791/53179, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macias H et al. SLIT/ROBO1 signaling suppresses mammary branching morphogenesis by limiting basal cell number. Developmental Cell. 20 (6), 827–840, doi: 10.1016/j.devcel.2011.05.012, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welm BE, Dijkgraaf GJ, Bledau AS, Welm AL & Werb Z Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell. 2 (1), 90–102, doi: 10.1016/j.stem.2007.10.002, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith P et al. VANGL2 regulates luminal epithelial organization and cell turnover in the mammary gland. Scientific Reports. 9 (1), 7079, doi: 10.1038/s41598-019-43444-8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee GY, Kenny PA, Lee EH & Bissell MJ Three-dimensional culture models of normal and malignant breast epithelial cells. Nature Methods. 4 (4), 359–365, doi: 10.1038/nmeth1015, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell JJ, Davidenko N, Caffarel MM, Cameron RE & Watson CJ A multifunctional 3D co-culture system for studies of mammary tissue morphogenesis and stem cell biology. PLoS One. 6 (9), e25661, doi: 10.1371/journal.pone.0025661, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Labarge MA, Garbe JC & Stampfer MR Processing of human reduction mammoplasty and mastectomy tissues for cell culture. Journal of Visualized Experiments. (71), doi: 10.3791/50011, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marlow R & Dontu G Modeling the breast cancer bone metastatic niche in complex three-dimensional cocultures. Methods in Molecular Biology. 1293 213–220, doi: 10.1007/978-1-4939-2519-3_12, (2015). [DOI] [PubMed] [Google Scholar]
- 26.Koledova Z & Lu PA 3D Fibroblast-Epithelium Co-culture Model for Understanding Microenvironmental Role in Branching Morphogenesis of the Mammary Gland. Methods in Molecular Biology. 1501 217–231, doi: 10.1007/978-1-4939-6475-8_10, (2017). [DOI] [PubMed] [Google Scholar]