Abstract
SARS-CoV-2 is a coronavirus causing a globalized outbreak called COVID-19. SARS-CoV-2 transmission is associated with inhalation of contaminated respiratory droplets and could causes severe complications. Until today several “waves” of infections have been observed despite implementation of strict health policies. Decisions for such sanitary measures are based on population health monitoring. Unfortunately, for COVID-19, a significant proportion of individuals are asymptomatic but play a role in the virus transmission.
To overcome these limitations, several strategies were developed including genome quantification in wastewater that could allow monitoring of the health status of population, since shedding of SARS-CoV-2 in patient stool is frequent. Wastewater-based epidemiology (WBE) was established and several countries implemented this approach to allow COVID-19 outbreak monitoring. In France, the OBEPINE project performed a quantitative analysis of SARS-CoV-2 in raw wastewater samples collected from major wastewater treatment plants (WWTP) since March 2020.
In the greater Paris area 1101 samples (507 for five WWTP and 594 for sewer) were collected. This 16 months monitoring allows us to observe the outbreak dynamics. Comparison of WBE indicators with health data lead to several important observation; the good level of correlation with incidence rates, the average 3 days lead time, and the sensitivity (WBE change when incidence is > to 7/100000 inhabitants). We also compared the local monitoring (city level) with the regional monitoring, to help cluster identification.
Moreover, variants of concern (VOC) emerged due to the selection pressure. We developed a specific RT-qPCR method targeting the deletion H69-V70 in the spike protein, using this deletion as a proxy of the B.1.1.7 presence in the wastewater. With this data we demonstrate the predominant role played by this strain in the third wave.
All these results allow a better description and understanding of the pandemic and highlight the role of such WBE indicators.
Keywords: SARS-CoV-2, COVID-19, Quantification, Wastewater, Variant monitoring, RT-qPCR, Epidemiology
Graphical abstract

1. Introduction
SARS-CoV-2 is a single-stranded positive RNA virus belonging to the Coronaviridae family, and the etiological agent of the coronavirus disease-19 (COVID-19) globalized outbreak since the end of 2019. SARS-CoV-2 transmission is associated with the inhalation of contaminated respiratory droplets via aerosols, although a possible contamination through inert surfaces was supposed (Bedrosian et al., 2021; World Health Organization, 2020). SARS-CoV-2 can cause severe complications in people suffering from comorbidity factors (such as diabetes, hypertension, obesity, acquired or iatrogenic immunosuppression) or elderly people (Yang et al., 2020). SARS-CoV-2 has infected more than 210 million people worldwide and caused 4,400,000 deaths at the August 20th, 2021 (Johns Hopkins, n.d.). Several “waves” of infections have been observed worldwide despite of the implementation of strict health policies to avoid hospital overload with patients requiring emergency care. Such sanitary measures range from mask wearing, cluster identification, limitation of human contact through lockdown or curfew. Decisions for these measures are based on population health monitoring toward COVID-19 testing.
Monitoring such an important outbreak is always a difficult task due to the intensive work for testing a sufficient and representative part of the population. Evaluating the prevalence of disease in the population generally relies on intensive testing of symptomatic patients, but in the case of COVID-19 the important proportion of individuals having moderate or no symptom (Mizumoto et al., 2020) but still actively participating in the transmission of the virus make the logical testing policies difficult to develop. For example, the screening strategy in France, during the pandemic, has been modified several times, depending on the increasing laboratory testing capacity, the willingness to test independently of the symptoms, or only after medical consultation. Moreover, an increase of the test number before holiday or special events or a decrease during a closed day is observed. Modification in the quantity of tests changes the representativeness of tested citizen and induces a shift in the collected data over time. New strategies could modify global health indicators and change the perception of the viral dynamics over time. Daily lethality or hospitalization rate could also change in accordance with the average age of the infected population (Lapidus et al., 2021) (less hospitalization when the age of the infected people is lower).
To overcome these limitations, several strategies were developed in France such as testing all the contacts of each SARS-CoV-2 positive case (“cluster approach”) by studying infected people's interactions during the previous days using survey or even smartphone tracing application (Ferretti et al., 2020; Sun and Viboud, 2020). Other indicators allow to monitor the outbreak dynamics, for example the proportion of COVID-19 positive tests by RT-qPCR or more recently by lateral flow assay, reporting the number of hospitalized patients (including intensive care unit) or daily lethal cases caused by the illness.
The viral dynamics monitoring based on solely clinical indicators is therefore incomplete, and small changes in the SARS-CoV-2 dynamics, crucial to identify for adapting health decisions, could be more difficult to detect in absence of stable indicators. That's why excretion of SARS-CoV-2 in infected patient stool whether they reported symptoms or not, has encountered an increasing interest. Wastewater-based epidemiology (WBE) allows epidemic monitoring, independently of the health policies.
Based on previous works showing that viral genome quantification could allow monitoring of the health status of the population (Prevost et al., 2015), SARS-CoV-2 genomes have been detected in raw wastewaters and various optimized isolation protocols have been now suggested to reliably quantify the viral genome (Barril et al., 2021; Pérez-Cataluña et al., 2021). Some works highlighted the correlation between the viral load in wastewater and the positivity of testing (Ahmed et al., 2020a; Medema et al., 2020; Randazzo et al., 2020; Wurtzer et al., 2020) and received a huge attention from health authorities and citizen. Several countries implemented WBE to allow COVID-19 outbreak monitoring. In France, since the beginning of the COVID 19 outbreaks (March 2020), the OBEPINE project performed a quantitative analysis of SARS-CoV-2 by RT-qPCR in raw wastewater samples collected from major wastewater treatment plant (WWTP) in France (Wurtzer et al., 2020; Bertrand et al., 2021), twice a week, but also from several major sewers linked to the WWTP. Starting with about 16 WWTP in March 2020, we are presently monitoring 168 WWTP covering the whole country, representing nearly 40% of the population.
During this outbreak, several variants of concern (VOC) emerged resulting from a constant and evolutionary selection pressure to which the virus is confronted (Huang and Wang, 2021). Several studies showed that multiples mutations give the variant new characteristics, such as increasing of infectivity and transmissibility or escaping from neutralizing antibodies induced by natural infection or vaccination (Harvey et al., 2021). We developed a specific RT-qPCR method targeting the del H69-V70 in the spike protein. This mutation is present on the B.1.1.7 strain (now called “alpha”) allowing us to use it as a proxy of the B.1.1.7 presence in the wastewater.
In the present paper we used more than one-year results of the Ile-de-France area monitoring, one of the first region implemented in the French WBE Network (called OBEPINE) and one of the first monitored in the world. Due to the previous interesting results, French research ministry provided a special grant to build the OBEPINE network around several research laboratories. With 1101 samples (507 for WWTP and 594 for sewer monitoring) collected since March 2020, comparison with available health data is performed and several important conclusions could be issued from this dataset, allowing a better understanding of the viral streams. We also describe the predominant role played by the B.1.1.7 strain in the third wave.
2. Material and methods
Five WWTP were sampled since March 5th, 2020 (see map in Supplementary Fig. 1 and WWTP information and Supplementary Table 1). Twenty-four-hour composite samples (according to NF T 90-90-523-2) were taken by automated samplers from March 5th, 2020 without interruption until today (data presented until June 6th 2021). Sampling was based on flow rate, started at 7:00 AM and finish at J + 1, 7:00 AM. Samples were taken by suction and collected in a refrigerated polyethylene tank at 5 °C (+ or −3 °C). The final collected volume was between 8.7 and 14 L. Then samples were carefully homogenized and subsampled in 100 mL polyethylene bottle, transported to the laboratory at 4 °C and processed in less than 24 h after sampling. The map of the watershed, connected at the several WWTP is indicated in Supplementary Fig. 1 panel A.
During the same period, samples were taken from the sewer network system of the city of Paris, to assess the epidemic at a more local scale. From May 11th 2020, sewer effluent samples were grab samples (n = 6, twice a week), and after the January 18th 2021 24 h-composite samples were taken at 16 localizations, once a week as indicated in Supplementary Fig. 1, panel B, until June the 6th. This 24-hour integrated sampling helps to avoid dispersion of results due to the non-homogenized wastewater in sewer observed when comparing grab sample with the 24 h composite sample (data not shown).
2.1. Concentration methods
The sample processing was done as previously described (Wurtzer et al., 2020). Briefly, samples were homogenized, then 11 mL were centrifugated at 200,000 ×g for 1 h at +4 °C using a XPN80 Coulter Beckman ultracentrifuge equipped with a swing rotor (SW41Ti). Pellets were resuspended in 200 μL of Dubelcco's phosphate buffer saline 1× w/o calcium w/o magnesium pH 7.4 (X0515-100, Dutscher, France). The pellet was then homogenized and pretreated for dissociating viruses and organic matter, and then removing organic compounds that may reduce extraction efficiency, according to the manufacturer's recommendation. Briefly, 600 μL of CD1 reagent of QIAsymphony PowerFecal Pro kit (#938036, QIAGEN) were added to the sample and vortexed for 10 min in a bead beater. Then 150 μL of CD2 reagent of QIAsymphony PowerFecal Pro kit (#938036, QIAGEN) were added and incubated for 5 min on ice before pelleting precipitated organic matter by centrifugation at 16,000 ×g for 1 min. Supernatant was collected and total nucleic acids were purified using of QIAsymphony PowerFecal Pro kit (#938036, QIAGEN) on a QIAsymphony automated extractor (QIAGEN) according to a slightly modified manufacturer's protocol allowing to handle 900 μL-sample volume and including an additional washing of beads. Extracted nucleic acids were filtered through OneStep PCR inhibitor removal kit (Zymoresearch) according the manufacturer's instructions.
2.2. Validation of the concentration methods
For evaluating the performance of the process, 200 mL of raw wastewater sample were seeded with 106 genome units of adenovirus 41 (ATCC VR-9300) and 107 genome units of bovine coronavirus (BCoV), kindly provided by Dr. Delphine Perrotte (LABEO, France). Twelve 11-mL subsamples were independently processed, and recovery rates were estimated based on the quantification of adenovirus and BCoV in raw wastewater compared to the seeded quantities (Supplementary Fig. 3). The repeatability of the measurement was assessed by quantifying spiked viruses (purified suspensions of adenovirus 41 and BCoV) and endogenous viruses (Pepper Mild Mottle virus (PMMoV), CrAssphages, F specific RNA phages genotype 2 (FRNAPH G2) and SARS-CoV-2) on 12 independent samples. Raw SARS-CoV-2 positive wastewater samples were collected during a low prevalence period of COVID-19 for demonstrating the robustness and repeatability (with a CV% near to 40% for mean concentration of 3100 GU/L)of analysis on samples with low viral content.
2.3. Molecular detection method
The RT-qPCR primers and PCR conditions used herein have been previously described (E sarbeco set, del69-70 set) (Table 1 ) (Wurtzer et al., 2021). The amplification was done using Fast virus 1-step Master mix 4× (Lifetechnologies) with oligonucleotide concentrations recommended previously. Detection and quantification were performed by RT-qPCR using a standard curve based on a plasmid carrying a full-length amplicon for gene E. We cloned the E amplicon (112 bp, 26269–26381 according to the Wuhan strain, EPI ISL 402124) using a vector pCR 2.1 TOPO TA (kit #451641, ThermoFisher scientific). Standard curve used for quantifying gene E is provided in Supplementary data (Supplementary Fig. 2). Gene E-results were confirmed by quantification of a region located within the gene encoding for the viral RNA-dependent RNA polymerase (RdRp, a WHO recommended assay). For quantification a plasmid was constructed caring the RdRp amplified sequence, using the plasmid pCR 2.1 TOPO TA (kit #451641, ThermoFisher scientific) with the RdRp amplicon cloned: 106 bp, set IP4: 26269 – 26381 Additionally, 5.000 copies of an internal positive control (IPC) was added in the raw sample to evaluate the presence of residual inhibitors and excluding false negative results. The IPC consists in a plasmid containing beta-acting gene flanked by enterovirus-specific primers (Wurtzer et al., 2014). Amplification reaction and fluorescence detection were performed on Viaa7 Real Time PCR system (Lifetechnologies). The detection limit of gene E assay was estimated to be around 103 genome units per liter of raw wastewater based on a mean recovery rate for BCoV about 75%.
Table 1.
Primers and double dye probes for the specific amplification.
| Name | Sequence 5′-3′ | Target | Optimal concentration | Reference |
|---|---|---|---|---|
| E_Sarbeco_F | ACAGGTACGTTAATAGTTAATAGCGT | SARS-CoV-2 | 400 nM | (Corman et al., 2020) |
| E_Sarbeco_R | ATATTGCAGCAGTACGCACACA | 400 nM | ||
| E_Sarbeco_P | FAM-ACACTAGCCATCCTTACTGCGCTTCG-BHQ1 | 200 nM | ||
| DEL69-70_F186 | TACTTGGTTCCATGCTATCT | SARS-CoV-2 | 300 nM | (Wurtzer et al., 2021) |
| DEL69-70_R340 | ACTGGGTCTTCGAATCTAA | 600 nM | ||
| DEL69-70_Ps307 | 6-FAM-AGAAGTCTAACATAATAAGAGGCTGGA-BHQ1 | 200 nM | ||
| CPQ64-F | TGTATAGATGCTGCTGCAACTGTACTC | CrAssPhage | 500 nM | (Stachler et al., 2017) |
| CPQ64-R | CGTTGTTTTCATCTTTATCTTGTCCAT | 500 nM | ||
| CPQ64-P | FAM-CTGAAATTGTTCATAAGCAA-MGB ECLIPSE | 200 nM | ||
| PMMV-FP1 | GAGTGGTTTGACCTTAACGTTTGA | PMMoV | 500 nM | (Haramoto et al., 2013) |
| PMMV-RP1 | TTGTCGGTTGCAATGCAAGT | 500 nM | ||
| PMMV-Probe | FAM-CCTACCGAAGCAAATG-MGB ECLISPE | 200 nM | ||
| AdV_F_F102 | GCGCACTTTGTAAGARTA | Adenovirus type F | 600 nM | (Prevost et al., 2015) |
| AdV_F_R231 | CACCGATACGTACTTCAG | 900 nM | ||
| AdV_F_Ps160 | FAM-CACGATGTAACCACAGACAGG-BHQ1 | 200 nM | ||
| Bovine_coronavirus_F | GGACCCAAGTAGCGATGAG | BCoV | 400 nM | (Kishimoto et al., 2017) |
| Bovine_coronavirus_R | GACCTTCCTGAGCCTTCAATA | 400 nM | ||
| Bovine_coronavirus_P | FAM-ATTCCGACTAGGTTTCCGCCTGG-BHQ1 | 200 nM | ||
| GII Fwd | TGCAAACCTAACTCGGAATGG | FRNAPH G2 | 500 nM | (Ogorzaly and Gantzer, 2006) |
| GII Rev | AGGAGAGAACGCAGGCCTCTA | 500 nM | ||
| GII Pr | FAM-TCCCTCTATTTCCTC-NFQMGB | 200 nM |
A relative quantification method was performed for estimating the Alpha variant carrying del69-70 in the spike. To achieve this goal, the ∆Cq between the del69-70 target (Alpha variant signature) and the gene E (Total SARS-CoV-2 genomes) is calculated for each sample:
Then the difference between the ∆Cq of the samples and the ∆Cq of a calibrator based on B.1.1.7 RNA is calculated, giving the ∆∆Cq value:
The normalized del69-70 positive target proportion in the sample is then equal to:
With similar PCR efficiencies for gene E and del69-70 mutation (Supplementary data Fig. 3), no correction according to PCR efficiencies was necessary (Schmittgen and Livak, 2008).
2.4. Mathematical and statistical methods used
To allow regional dynamics representation, we calculated the daily average concentration (one to five WWTP were analyzed each day). To avoid outlier effects, we used a LOWESS smoothing method on 10 adjacent points (GraphPad Prism v9.0). Proportion of variants, using the ∆Cq was used on gene E to allow evaluation of the viral genome concentration for B.1.1.7 then the same LOWESS smoothing procedure was applied.
Another, advanced model was also introduced to better take into account the dynamics of the presence of the viral genome and have better properties in real time: the wastewater indicator (WWI) is calculated from log-transformed flow-adjusted RT-qPCR analyses on wastewater 24 h-composite samples, by Kalman smoother (Courbariaux et al., 2021) followed by a normalizer (Cluzel et al., 2021). The quantification results from RT-qPCR are flow-adjusted using the following formula:
where V 0, t is the total volume at the inlet of the treatment plant on day t, V t is the mean volume of the WWTP over a year of history and C 0, t is the RT-qPCR quantification result. Then, flow-adjusted data is processed through Kalman smoothing.
Kalman smoother is a mathematical model based on the hypothesis that the viral load in wastewater of the day depends on the day before. The model estimates daily viral load of the WWTP from the first to the latest sampling, in a way that allowing to calculate an indicator robust to outliers and censured data. If a measurement is flagged as a censored value, we consider that it can take every value below the limit of quantification. The smoother output is the estimated process that maximizes the likelihood of the observations.
The mathematical writing of the Kalman smoother's underlying model is as follows:
where t is the time index (ranging from 1 to n days), X t ∈ ℝ is the logarithm of the real concentration in wastewater at time t, X = (X t)t∈{1,…,n} is the vector of log-transformed real concentrations (to be recovered) and Y t ∈ ℝ is the logarithm transformation of the estimated concentration in wastewater measured by RT-qPCR at time t, C t, previously defined (Y t = log (C t)). Y t is generally only partially observed. We note the set of t at which Y t is observed. is the vector of measurements. Y ∗ is an accessory latent variable corresponding to a non-censored version of Y. I is the identity matrix. η ∈ ℝ, δ ∈ ℝ, κ ∈ ℝ + and τ ∈ ℝ + are parameters (to be estimated). ℓ is the threshold below which censorship applies (i.e. the LOQ of the laboratory). O t ∈ {0, 1} is, for any , the indicator variable of the event “Y t ∗ is an outlier”. . stands for the Bernoulli distribution of parameter p and for the Uniform distribution on the interval [a, b]. p is a meta-parameter designating the a priori probability of being an outlier (we take p = 2% here). a and b have to be chosen, respectively minimum of Y minus 2 standard deviation of Y, and maximum of Y + 2 standard deviations of Y. The parameters η ∈ ℝ, δ ∈ ℝ, κ ∈ ℝ + and τ ∈ ℝ + of maximum likelihood are estimated by numerical optimization through Nelder-Mead, as explained in (Courbariaux et al., 2021). At time n, the developed smoother gives the law of X t for t ∈ {1, …, n} knowing , as well as the probability for each Y t to be an outlier. We note the produced reconstitution . As several laboratories with varying quantification protocols are providing RT-qPCR results, an additional step of normalization was needed to enhance the comparability of results between WWTPs. We then perform the following normalization:
where WWI t is the WWI value at time t, is the previously defined reconstitution, C m represents a quantification threshold of 1000 GU/L and C M is the maximum concentration historically recorded by the reference laboratory on plants with average daily flows similar to that of the plant of interest. Flow bins were selected in order to obtain a distribution that is as uniform as possible. The factor of 150 is an arbitrary coefficient. WWI is a unitless value.
2.5. Correlation computations and lag estimations
As we want to calculate the correlation between clinical and wastewater data over a period where they are supposed to be similar and thus where the WWI is supposed to mainly capture a majority of people also likely to be diagnosed, we decided to focus on the period corresponding to the second and third waves of the epidemic in France. To avoid being biased by the movements of individuals during the 2020 summer vacations, we consider the start date of the second wave as September the 1st, 2020, from which the majority of holidaymakers returned to their residence city. We consider that the last point of the interval of interest is the date from which the signal undergoes a new growth phase following the decay of the second peak of the epidemic.
We then drag the subpart of the incidence rate curve over a ±30-day window until we find the time lag that yields the best correlation with the WWI. We use Pearson cross-correlation as a measure of similarity between the two signals. The cross-correlation calculation is performed between the WWI and the log transformation of the incidence rate. Since correlation is sensitive to outliers especially when sample size is small, we subsampled the incidence signal using 50% of the available data so as to avoid certain special patterns resulting in an unnaturally high correlation. The time lag resulting to the highest positive correlation is recorded. A positive lag value indicates that the WWI is ahead of the studied epidemic signal. A negative lag value indicates that the WWI is lagging behind it. Performing the subsampling 1000 times gives confidence intervals for correlation and lag values.
Incidence data was collected at the regional scale (i.e. Parisian area) or at the departmental scale (i.e. subdivision of the Parisian area). It must be noted that the departmental scale doesn't exactly correspond to the WWTP watershed (except for Paris). Two stations (MAV and SEM) were representative to the same department.
3. Results
3.1. Method performance evaluation
The combination of ultracentrifugation method coupled with the RTq PCR, used in this project, shows a good repeatability (with a CV% near to 40% for concentration around 10^3 UG/L), but also an important recovery rate, around 80% for a bovine coronavirus (Supplementary Fig. 3), by minimizing the treatment step of the sample and using automatized extraction procedure. This allows a good consistence of data through the 16-month project.
3.2. Monitoring SARS-CoV-2 viral load in wastewaters from the Greater Paris area
Viral load was quantified from samples collected from 5 WWTP (Fig. 1 ) of Greater Paris area from March 5th, 2020 to June 6th, 2021. Time course analysis of the average viral load is depicted in Fig. 2 . Strikingly SARS-CoV-2 concentration in raw wastewater closely matched the rate of incidence (i.e. the number of newly infected patients over 7 rolling days) on the same area (r2 = 0.81 with average value and r2 = 0.87 with Lowess regression, Spearman r = 0.9248, p < 0.0001 for regional Lowess vs incidence).
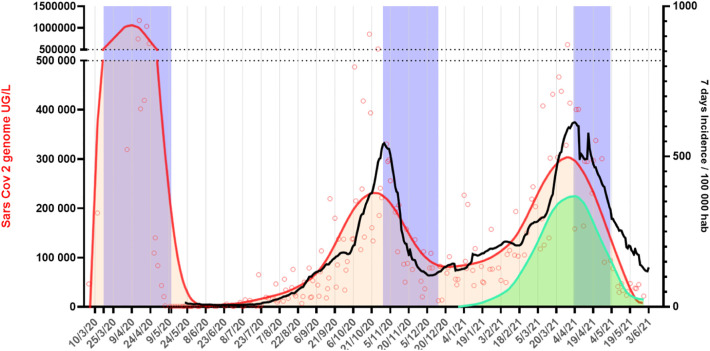
Fig. 1.
Average viral load in several WWTP from Ile de France global. Open red circles represent average concentration (1 to 5 WWTP) per day. Red line is the lowess curve resulting from the smoothing of the concentration values. Incidence data for the whole region (black curve) 7 day cumulated. Estimated concentration of genomes containing the del-69-70 mutation (green). Blue area represents the lockdown periods. Concentration ordinate are split in two to allow first wave representation.
Fig. 2.
SARS-CoV-2 viral concentration in 5 WWTP from Ile-de-France. Average daily (red open circles) and smoothed (Lowess) (red line) viral RNA concentrations are depicted as well as incidence at the level of the department (black line) The calculated WWI is represented in blue. Gray surface indicates lockdown periods. Each graph is a different WWTP.
Although the first lockdown (March 17th) had a dramatic effect on virus concentration in wastewater (Wurtzer et al., 2020) which resulted in the absence of detectable viral genome in samples from the beginning of May 2020, re-circulation of the virus was detected as early as the end of June 2020. This was followed by a slow but steady increase during the summer of 2020 that culminated at the end of October, when a curfew followed by a 2nd lockdown was decided (October 30). After the first lockdown the number of COVID-19 positive patients remained stable in the same geographic area and began to slowly increase at the beginning of July 2020. We must note that only symptomatic patients were individually tested due to the low number of available tests before August 2020. The number of patients in ICU only increased after July 12th.
During the so-called second wave the increase in viral RNA concentration was slower than the one observed during the first wave which may likely reflect the effect of the health prevention measures deployed (face covering, social distancing etc.). This increase could be split in two times periods (1) from June to September when virus circulation slowly increased and (2) from September to the end of October when viral circulation markedly increased. Viral RNA concentration level reached a few millions genome equivalents per liter in October 2020, underlying the second wave in the greater Paris population. A curfew period followed by lockdown was applied after October 30 at the end of a school holiday in the area. Effects were rapidly observed but the decrease stopped at the end of December 2020, one month after the lockdown ended. Viral concentration, once again started a slow but steady increase. The third wave had the same aspect with a slow start followed by a logarithmic increasing of the viral concentration after mid-February 2021. The increase halted when the third lockdown was put in place in April 3.
3.3. Local monitoring of SARS-CoV-2 viral load in WWTP
Whereas we provided convincing evidences that the average viral load in 5 WWTP was in good agreement with other epidemiological markers in the greater Paris area — a wide area of approximately 8 million inhabitants — we wondered whether local survey could provide additional information on the local distribution of SARS-CoV-2 carriers.
We first noticed that viral dynamics were strikingly similar from one WWTP to another (Fig. 2). Health related data from the administrative area related to watershed (French department) were gathered and compared to the level of viral genomes quantified in wastewaters. If the global dynamics of all WWTP were similar, small local differences could be observed. For example, at Seine center (SEC) wastewater treatment plant, the increase in viral load for the second wave was visually delayed. In addition, the highest viral concentrations were lower in SEC WWTP during the second wave, compared to other WWTP in the region. This difference may not strictly reflect differences in the circulation of the virus. Indeed, the SEC collection area, the center of Paris, is structured by a unitary network (pluvial and wastewater) which could partly explain the difference observed. On the other hand, the samples have been frozen during this period and we cannot exclude that freezing/thawing may have an impact on procedures that require virus concentration before genome extraction. Several experiments in the lab confirm the effect of sample frozen of virus recovery when using the ultracentrifugation methods (data not shown).
3.4. At the city sewer scale
After the relevant results at the regional and departmental scale, we also monitored smaller watershed by collecting samples in Parisian sewers, to allow for a more precise description of the viral dynamics and allow for the detection of clusters. Viral concentrations in samples collected from the sewer system of the city of Paris are plotted in Fig. 3 . Results showed that the median of data taken from the sewer visually correlated with the city trends, emphasizing the possibility to refine the virus spreading in the area. Monitoring of individual sampling points showed some specific dynamics before the start of the second wave, demonstrating the possibility to detect local cluster using sewer monitoring (lower panel). For example, an important concentration was locally detected during week 22 (October 14, 2020), at the beginning of the second wave, probably due to local clusters. Same trend could be easily observed before the second lockdown.
Fig. 3.

Upper panel median value of the sewer measurement (open red circles), red line show the dynamics of the concentration (Lowess). Black line represents the incidence of the 7-days cases in the area. In gray lockdown period. Lower panel: heat map of each sample point SARS CoV 2 concentration, white line represent beginning and ending of the lockdown.
3.5. Correlation of genome concentration with epidemiological data and time lag
Correlations (Spearman) of the relationship between smoothed average concentration of RNA genome and number of COVID 19 positives patients were performed for each WWTP and for the regional global trends (see Fig. 4 panel A, B, C, D and E). The relationship looks quite similar at the WWTP level and at the regional level, underlining that the COVID-19 outbreaks dynamic and characteristics of stool excretion, as well as hydrological sewer network, were quite similar in all the area when the number of patients was important. These results underlined the population movement inside the greater Paris area (i.e. people could live and work in area served by different WWTP). These movements could have homogenized the virus spreading between areas. Coefficients of correlation were quite similar for all WWTP (Fig. 4 panel F), with r2 equal to 0.80, 0.82, 0.81, 0.77, for regional, MAV, SEM, and SEV respectively. Two stations show a lower correlation SEC with a r2 = 0.62 and STV with a r2 of 0.56. For each WWTP, an incidence of nearly 6.8 inhabitant/100,000 for 1 rolling week was associated with the detection of viral genome in the connected WWTP.
Fig. 4.
Relationship between log of the concentration of SARS-CoV-2 genome and the log number of incidence number. Panel A regional scale, in blue average of the 5 WWTP, in red Lowess of the same data. Panel B, in blue SEM WWTP, in red MAV. panel C relationship between SEV and departmental incidence number. Panel D relation between STV and the departmental incidence number panel E, relationship for SEC and Paris incidence panel F, linear regression of all WWTP and the regional average.
Using the WWI developed during the project, we also estimated the correlation between WWI and the incidence rate for each WWTP, for the 2nd and 3th wave (Fig. 5 panel A). To avoid being biased by the movements of individuals during the 2020 summer vacations, we considered the start date of the second wave as September the 1st, 2020, from which most holidaymakers returned to their primary residences. We considered the end date as the last day before the incidence rate increase. Likewise, the first day of the third wave is the incidence rate's turning point after winter holidays in early January. Since correlation is sensitive to outliers, especially when sample size is small, we subsampled the incidence signal using 50% of the available data to avoid certain special patterns resulting in an unnaturally high correlation. We performed this subsampling experiment 1.000 times. In accordance with the simple correlation coefficient we found a very positive relationship between the WWI and the incidence, except for the 2nd wave of the station, SEC and STV, in accordance with the previous correlation approach.
Fig. 5.
Panel A (upper), correlation between the WWI for each WWTP and 2nd and 3th wave. Panel B (lower) positive or negative lag (in day) between the WWI. Lag was calculated by maximizing the correlation between the two datasets. For both panel, correlation was calculated 1000 time using a subsample of 50% of the values. LSM mean STV.
This specific behavior is probably due to the difference between the sewer network and the regional incidence data, for example STV is more rural than other WWTPs and the frozen sample for SEC, as explains before, could have play a role.
We also monitored the time between WWI and population incidence (Fig. 5 panel B). The observed time lag between the incidence rate and the wastewater signal was observed by finding the configurations that maximize the correlation between the two signals at the pandemic's second and third waves in France. The highest positive correlation is recorded at each experiment, giving a distribution of correlation and time lags for each WWTP and each wave. Once again to avoid outliers effect, a 50% subsampling experiment was performed 1.000 times. Positive lag mean that the wastewater signal was observed before the incidence one, and negative number indicate that wastewater follow the incidence rate (i.e. change after). An average lag of 3 days is observed, with some stations with a better prediction advance. Wastewater could be used as an early warning system in such situation. The station with average correlation (see above) also shows a bad predictive behavior, with a negative delay.
3.6. Variant monitoring
Based on a specific del69-70RT-qPCR applied on all the samples starting from December 15th, 2020, we have observed the relative proportion of this mutation of the spike protein among the total quantity of SARS-CoV-2-genome. We have extrapolated the quantity of genomes sharing this mutation and using it as a proxy of the alpha variant (B.1.1.7 viral strain) (see Fig. 1). The raise of this viral strain could be one of the explanations of the 3rd wave, in accordance with some modeling data based on a higher infectious rate for this strain (Di Domenico and Colizza, 2021).
The evolution of del69-70 mutation was put into the perspective of the evolution of alpha variants reported among people diagnosed in all Paris Hospital (“AP-HP”) centers including hospitalized, non-hospitalized patients and healthcare workers (see Fig. 6 ). The diagnosis of alpha variants was performed by both specific-mutation screening and sequencing during the months of January and February and only by specific-mutation screening from the beginning of March. A very good superposition can be observed between the variants dynamic curve issued from WWI and that of issued from the AP-HP patients data (see Fig. 6).
Fig. 6.

Evolution of the average concentration of del 69-70 in the Greater Paris area (open blue circle) and representation of the dynamics by Lowess smoothing (blue line). In black 7 days rolling average sequence of variants in the Paris city (sub population of the Greater Paris, data SPF). In purple data from the Parisian hospital (data AP-HP).
4. Discussion
The monitoring of wastewater treatment since the start of the COVID-19 epidemic (wastewater monitoring starting from March 5th, 2020) allowed us to have a whole picture of the virus spreading dynamic during the entire outbreak. Our measurement methodology, based on direct ultracentrifugation, without any preparation step such as clarification, allow us to have a high recovery rate (>90% for bovine coronavirus surrogate, see Supplementary Fig. 3) and a low dispersion of results (less than 40 CV% BCoV for low concentration around 10^3 UG/L). Due to the low number of sample manipulation steps, repeatability is quite similar for all the viruses in the sample (i.e. spiked or naturally present, naked or enveloped virus). This method, without any clarification step considers the so called free viruses as well as viruses adsorbed on particular organic matters (Ahmed et al., 2020b).
Since WBE is independent form health monitoring strategies, these results could be one of the most reliable indicators of the three waves in the greater Paris area. Indeed, other health indicators have changed during the outbreak event for several reasons such as test availability, testing policy, change in the population impacted by the viruses and WBE is the only indicator that has not been modified since the beginning of the outbreak. Beyond incidence measurement based on test, other indicators such as number of deaths could also have changed due to the improvement of ICU treatment or because of the different ages of the infected individuals in the population. Recent data underline the shift in the age classes infected by the virus (Lapidus et al., 2021; O'Driscoll et al., 2021). In addition, the more fragile patients (older and or with comorbidity factors) were likely most protected during the second wave (better information, compliance with “preventive measures”, mask was mandatory in many situation).
RNA concentration, could allow description of “the real dynamic” of the virus spreading into the population, allowing for a direct comparison between the three waves. Our data showed that in the Greater Paris area, the first wave showed more highly concentrated samples (i.e. samples with about 1 million UG/L) than the 2nd and 3rd waves (see Fig. 3). Quantitative assessment of SARS-CoV-2 genomes in wastewaters demonstrates the impact of prevention measures such as lockdown or curfew which can be once again observed in this project. Some general modeling papers have postulated the decrease of the wave intensity (Di Domenico et al., 2021) in accordance with our data.
Good reliance of the viral dynamic in population monitored by WBE is confirmed by the visually and statistically good correlation between concentrations and incidence data (see Fig. 1, Fig. 2, Fig. 4, Fig. 5). The observation of results between successive waves shows that the sensitivity of such indicator and a regression method allowed us to calculate the incidence threshold when viral genome raises in wastewater. In accordance with the work of Weidhaas and colleagues (Weidhaas et al., 2021), we observed vRNA in WW when incidence reaches 7/100,000 inhabitants for 7 days. Once again, these results showed that WBE could be used as an early warning system, even if the advance of the WW indicators could be sensitive to some specific effect that still needs to be better understood. These effects, could be linked to the sewer network, sample conservation, or other effect yet to be discovered.
This long-time monitoring period confirmed that the concentration of SARS-CoV-2 fell below our detection level when lockdown was stopped (May 11th, 2020), and SARS-CoV-2 genomes were not detected until June 20th. At that time, there was no epidemiological evidence of virus circulation. Moreover, similar trends are observed among all the WWTP at the Great Paris scale and show the homogeneity of the WWTP viral dynamics. This effect suggests a homogenization of the infected population, according to the population important movements in the area during worktime.
The link between SARS-CoV-2 genome concentration in wastewater and incidence data seem to be stable among several WWTP in the same region, allowing, in the future, to describe the “typical excretion pattern” via stool and have information about the hospitalization rate evolutions in the following days. Effect of vaccination on viral fecal shedding should be described to change the model accordingly.
For important urban area linked to huge WWTP treating important volume of wastewater, deciphering the area with infected patients or identification of clusters could be difficult. In this case, WBE results, only allow observation of the whole area. To overcome this limitation, it is could be helpful to use sewer monitoring. In this project we monitored WWTPs at a regional scale but also in the Parisian city center sewers, to work at the local level. Using 24 h-composite sampling method allow a more precise localization of cluster. Results show a good correlation between the sewer dynamics and the regional trend (see Fig. 3). Moreover, some specific “hot spots” especially before the epidemic waves (see heatmap Fig. 3) was observed, probably related to clusters of infected people. This type of data could allow regional authorities to make specific health-related decisions. A global scheme of monitoring could be installed and for example, an increasing concentration in WWTP could lead to a more local analysis via seers and identification of clusters. Our results prove the very good relationship between city sewage system viral RNA concentration and the WWTP dynamic. Other authors have also suggested this type of relation (Ahmed et al., 2021).
Lastly variant monitoring at the sewer network could allow a fine description of the variant spreading and could help to explain the cause of the incidence increase. For example, the 3rd wave is mainly resulted from variants carrying the delta 69-70 mutation in the spike protein, one of the signatures of the newly called “alpha variant” or B.1.1.7. WBE could allow detection of such type of dynamics, and to understand the role of new variants and functional mutations. If the 3th wave is due to this variant, the outbreak dynamics is still comparable to the two first waves. Few study are available regarding the enteric replication and fecal shedding dynamics, moreover virus variants could have a different one compared to the original Wuhan strain. Moreover, vaccination of the population could impact all these enteric parameters.
5. Conclusion
During this COVID 19 events, measurement of WW has allowed several research groups to have the whole picture of the outbreak dynamics. Wastewater-based epidemiology is a new discipline which still needs some refinement for a better understanding of the results. WBE allows a monitoring of the whole population, including asymptomatic people. The pioneer Ile-de-France project, included in the OBEPINE national monitoring based on 1101 analyzed samples over 16 months in the Greater Paris area, allowed to describe some aspects of this indicator. The good correlation between incidence and viral RNA concentration, the relative stability of this correlation over several WWTP and the ability to discover hot spots of concentration in the sewer network when 24 h composite samples were used, make WBE an easy and helpful tool to deploy at regional and city scale. WBE results are complementary to other indicators and give other type of information. Utilization of a mathematically calculated WWI allows us to describe the short advance in some specific WWTP.
Some limits of the WBE should be overcome, and analytical noise is important, in relation with sampling strategies. In fact, the “nuggets effect” (i.e. non-homogenous wastewater) and non-perfectly diluted polluted wastewater introduce noise in the measurement. Moreover, change in the inhibitors concentration should be monitored. That's why utilization of smoothing technics such as Lowess are mandatory but the need of a better mathematical methodology is needed for expressing the results and allowing fast interpretation of the daily data and in development (Cluzel et al., submitted). Furthermore, a mathematical indicator should help for comparison at the national level; indeed, when different laboratories are involved the relationship between incidence and viral concentration could be different.
The following are the supplementary data related to this article.
Supplementary figures
WWTP characteristics.
Funding
Analyses were carried on the OBEPINE Research grant (Ministry of Higher Education, Research and Innovation), as well as CNRS, Eau de Paris and Sorbonne University Grant.
CRediT authorship contribution statement
S. Wurtzer: Conceptualization, Data curation, Formal analysis, Funding acquisition, Writing – review & editing. P. Waldman: Investigation, Methodology. M. Levert: Investigation, Methodology. N. Cluzel: Visualization. J.L. Almayrac: Data curation, Validation. C. Charpentier: Data curation. S. Masnada: Data curation. M. Gillon-Ritz: Data curation, Validation. J.M. Mouchel: Conceptualization, Data curation, Project administration, Formal analysis. Y. Maday: Conceptualization, Data curation, Formal analysis, Project administration, Funding acquisition, Writing – review & editing. M. Boni: Conceptualization, Data curation, Project administration, Funding acquisition, Formal analysis. V. Marechal: Conceptualization, Data curation, Formal analysis, Project administration, Funding acquisition, Writing – review & editing. L. Moulin: Conceptualization, Data curation, Formal analysis, Project administration, Funding acquisition, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge all the sampling teams involved and several laboratory members for technical discussion. Thanks to Alban Robin for helpful support.
Editor: Warish Ahmed
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O’Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barril P.A., Pianciola L.A., Mazzeo M., Ousset M.J., Jaureguiberry M.V., Alessandrello M., Sánchez G., Oteiza J.M. Evaluation of viral concentration methods for SARS-CoV-2 recovery from wastewaters. Sci. Total Environ. 2021;756 doi: 10.1016/j.scitotenv.2020.144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedrosian N., Mitchell E., Rohm E., Rothe M., Kelly C., String G., Lantagne D. A systematic review of surface contamination, stability, and disinfection data on SARS-CoV-2 (Through July 10, 2020) Environ. Sci. Technol. 2021;55:4162–4173. doi: 10.1021/acs.est.0c05651. [DOI] [PubMed] [Google Scholar]
- Bertrand I., Challant J., Jeulin H., Hartard C., Mathieu L., Lopez S., Schvoerer E., Courtois S., Gantzer C. Epidemiological surveillance of SARS-CoV-2 by genome quantification in wastewater applied to a city in the northeast of France: comparison of ultrafiltration- and protein precipitation-based methods. Int. J. Hyg. Environ. Health. 2021;233 doi: 10.1016/j.ijheh.2021.113692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluzel N., Courbariaux M., Wang S., Moulin L., Wurtzer S., Bertrand I., Laurent K., Montfort P., OBEPINE consortium. Le Guyader S., Boni M., Mouchel J.-M., Marechal V., Nuel G., Maday Y. 2021. Mathematical modeling and adequate environmental sampling plans are essential for the public health assessment of COVID-19 pandemics: Development of a monitoring indicator for SARS-CoV-2 in wastewater. Submitted. [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courbariaux M., Cluzel N., Wang S., Maréchal V., Moulin L., Wurtzer S., Consortium O., Mouchel J.-M., Maday Y., Nuel G. 2021. An Autoregressive Model for a Censored Data Denoising Method Robust to Outliers With Application to the Ob\’epine SARS-Cov-2 Monitoring. arXiv:2108.02115 [stat] [Google Scholar]
- Di Domenico L., Colizza V. INSERM; 2021. Epidemic Scenarios of Delta Variant in France in the Summer 2021 (No. 31) [Google Scholar]
- Di Domenico L., Sabbatini C.E., Pullano G., Lévy-Bruhl D., Colizza V. Impact of January 2021 curfew measures on SARS-CoV-2 B.1.1.7 circulation in France. Eurosurveillance. 2021;26:2100272. doi: 10.2807/1560-7917.ES.2021.26.15.2100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti L., Wymant C., Kendall M., Zhao L., Nurtay A., Abeler-Dörner L., Parker M., Bonsall D., Fraser C. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368:eabb6936. doi: 10.1126/science.abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Kishida N., Konno Y., Katayama H., Asami M., Akiba M. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol. 2013;79:7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., COVID-19 Genomics UK (COG-UK) Consortium. Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.-W., Wang S.-F. SARS-CoV-2 entry related viral and host genetic variations: implications on COVID-19 severity, immune escape, and infectivity. IJMS. 2021;22:3060. doi: 10.3390/ijms22063060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns Hopkins, n.d.Johns Hopkins , n.d. Coronavirus Resource Center.
- Kishimoto M., Tsuchiaka S., Rahpaya S.S., Hasebe A., Otsu K., Sugimura S., Kobayashi S., Komatsu N., Nagai M., Omatsu T., Naoi Y., Sano K., Okazaki-Terashima S., Oba M., Katayama Y., Sato R., Asai T., Mizutani T. Development of a one-run real-time PCR detection system for pathogens associated with bovine respiratory disease complex. J. Vet. Med. Sci. 2017;79:517–523. doi: 10.1292/jvms.16-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus N., Paireau J., Levy-Bruhl D., de Lamballerie X., Severi G., Touvier M., Zins M., Cauchemez S., Carrat F. Do not neglect SARS-CoV-2 hospitalization and fatality risks in the middle-aged adult population. Infect. Dis. Now. 2021;51:380–382. doi: 10.1016/j.idnow.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. 2020:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the diamond princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M., Ribeiro Dos Santos G., Wang L., Cummings D.A.T., Azman A.S., Paireau J., Fontanet A., Cauchemez S., Salje H. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590:140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- Ogorzaly L., Gantzer C. Development of real-time RT-PCR methods for specific detection of F-specific RNA bacteriophage genogroups: application to urban raw wastewater. J. Virol. Methods. 2006;138:131–139. doi: 10.1016/j.jviromet.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Cuevas-Ferrando E., Randazzo W., Falcó I., Allende A., Sánchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost B., Lucas F.S., Goncalves A., Richard F., Moulin L., Wurtzer S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015;79:42–50. doi: 10.1016/j.envint.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Stachler E., Kelty C., Sivaganesan M., Li X., Bibby K., Shanks O.C. Quantitative CrAssphage PCR assays for human fecal pollution measurement. Environ. Sci. Technol. 2017;51:9146–9154. doi: 10.1021/acs.est.7b02703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Viboud C. Impact of contact tracing on SARS-CoV-2 transmission. Lancet Infect. Dis. 2020;20:876–877. doi: 10.1016/S1473-3099(20)30357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., Torgersen K., Laan J.V., LaCross N. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Transmission of SARS-CoV-2: Implications for Infection Prevention Precautions (No. WHO/2019-nCoV/Sci_Brief/Transmission_modes/2020.3) [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Maday Y., Teyssou R., Richard E., Almayrac J.-L., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, greater Paris, France, 5 March to 23 April 2020. Eurosurveillance. 2020;25:2000776. doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Prevost B., Lucas F.S., Moulin L. Detection of enterovirus in environmental waters: a new optimized method compared to commercial real-time RT-qPCR kits. J. Virol. Methods. 2014;209:47–54. doi: 10.1016/j.jviromet.2014.08.016. [DOI] [PubMed] [Google Scholar]
- Wurtzer S., Waldman P., Levert M., Mouchel J.-M., Gorgé O., Boni M., Maday Y., OBEPINE Consortium, Marechal V., Moulin L. 2021. Monitoring the Propagation of SARS CoV2 Variants by Tracking Identified Mutation in Wastewater Using Specific RT-qPCR (preprint) [DOI] [Google Scholar]
- Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q., Ji R., Wang H., Wang Y., Zhou Y. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
WWTP characteristics.