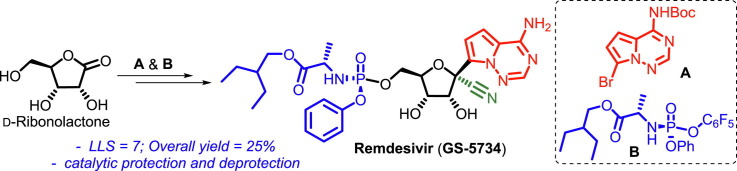
Graphical abstract
Keywords: Remdesivir, Nucleotide, Nucleoside, COVID-19, RdRp
Abstract
Remdesivir, the first drug approved by the FDA to treat COVID-19, is in high demand for patients infected with the SARS-CoV-2 virus. Herein, we report a facile approach minimizing the protecting group manipulations to afford remdesivir in good overall yield.
Introduction
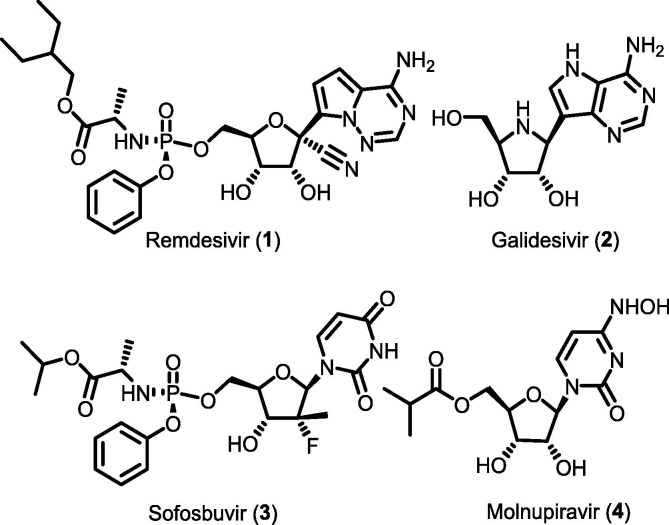
The COVID-19 pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has created a devastating health crisis around the globe [1], [2], [3], [4], [5], [6]. To combat this pandemic, effective and affordable treatments are still being urgently sought. In 2020, the FDA approved remdesivir (1, GS-5734) as the first drug for the treatment of COVID-19 [7], [8], and subsequently, related molecules such as galidesivir (2), sofosbuvir (3), and molnupiravir (4) were also found to be effective (Fig. 1 ) [9].
Fig. 1.
Structures of remdesivir and other related antiviral drugs.
Remdesivir (1), was initially developed by Gilead Sciences for the treatment of hepatitis C and Ebola virus infections [10], [11]. However, the clinical trials against Ebola were not successful [12]. Remdesivir is a prodrug with good cell permeability and hydrolyzes inside the cell to release a nucleotide triphosphate that targets the key RNA dependent RNA polymerase (RdRp) enzyme responsible for viral replication.
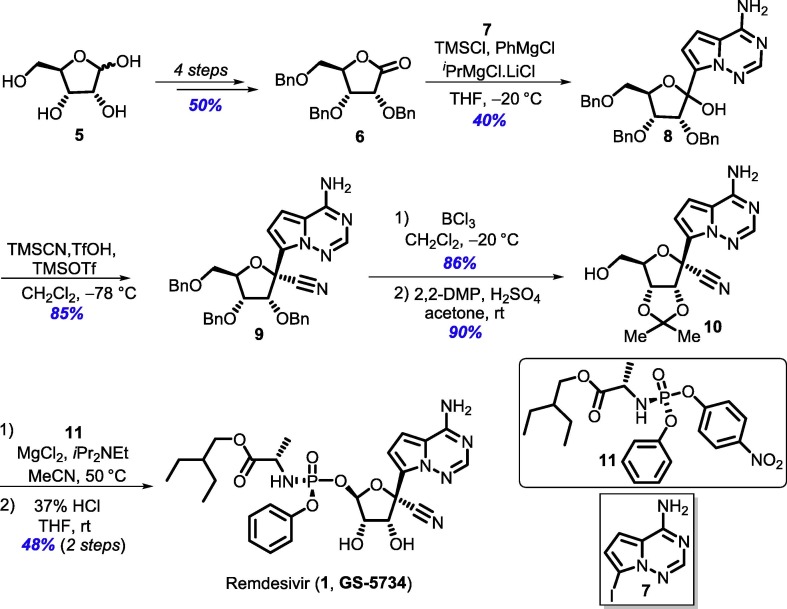
The synthetic route developed by Gilead Sciences for the commercial production of remdesivir [10], [11], involves protecting group manipulations of the sugar part during the synthesis, providing an opportunity for further improvements. Conversion of d-ribose (5) to 2,3,5-tri-O-benzyl-d-ribonolactone (6) followed by addition of modified nucleobase 4-amino-7-iodopyrrolo[2,1-f] [1–2,4]triazine (7) onto the lactone gave C-glycosylated product 8 in low yield (40%). After cyanation, all three benzyl groups were deprotected using BCl3 and the two secondary alcohols of the resulting triol were subsequently protected as isopropylidene ketal 10. Finally, introduction of phosphonate ester 11 to ketal 10 followed by hydrolysis of the ketal leads to the target compound remdesivir (1) (Scheme 1 ) [10]. Later, to improve the yield of the C-glycosylation reaction, expensive NdCl3 was used as an additive in stoichiometric amounts, which improved the yield to 69% [13], [14].
Scheme 1.
Gilead route for the synthesis of remdesivir (1, GS-5734).
In view of the importance of this being the only FDA-approved drug for COVID-19 mitigation, we reasoned any improvement on the synthetic route would have a large impact on supplies. The opportunities we realized include (a) protecting group manipulations, viz, replacing the benzyl groups with those which could be introduced and removed by catalytic processes avoiding stoichiometric reagents, (b) avoiding the use of NdCl3, and (c) achieving anomeric selectivity with better yields.
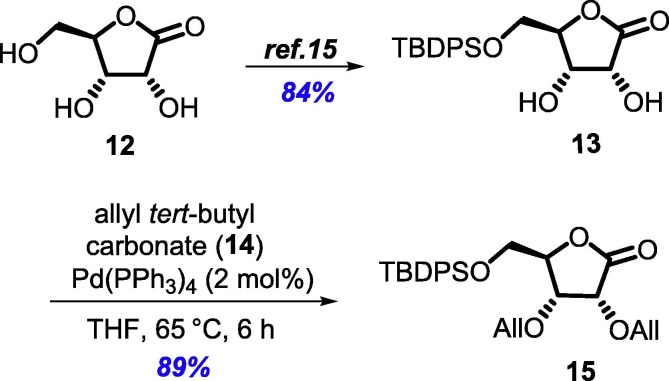
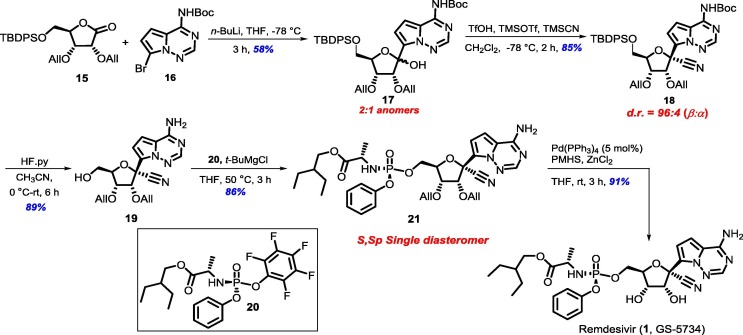
We planned our synthesis from d-ribonolactone (12), which is in the correct oxidation form to introduce the heterocyclic fragment at the anomeric carbon. It was also assumed that a bulky protecting group at the C-5 hydroxyl would enable better selectivity at the anomeric position. Thus, the primary hydroxyl group of commercially available d-ribonolactone (12, can also be synthesized from d-ribose in one step) was silylated with tert-butyldiphenyl silyl (TBDPS) group to afford the corresponding silyl ether 13 in 84% yield [15]. Further protection of the remaining two secondary alcohols using allyl tert-butyl carbonate (14) in the presence of 2 mol% Pd(PPh3)4 furnished diallylated ribonolactone 15 in 89% yield (Scheme 2 ) [16].
Scheme 2.
Synthesis of 5-O-TBDPS-2,3-O-diallyl ribonolactone (15).
Next, lactone 15 was subjected to C-glycosylation [10], [11], [13], [14], [17] with the protected nucleobase tert-butyl(7-bromopyrrolo[2,1-f] [1–2,4] triazin-4-yl) carbamate (16) which in presence of n-BuLi provided an anomeric mixture (2:1) of C-glycosylated product 17 in 58% yield. The cyanation [10], [11], [13], [18] of 17 under standard conditions gave cyano-glycoside 18 in 85% yield with excellent selectivity in favor of the desired isomer (d.r. = 96:4, β:α). To introduce the desired phosphonate on the primary hydroxyl group, deprotection of the silyl group and Boc group was achieved under HF.pyridine conditions in one-pot to afford amino-alcohol 19 in 89% yield [19]. The next task was to carry out the P-chiral phosphorylation, a key step in the synthesis of remdesivir [20]. Coupling of 19 with the known chiral pentafluoro-phosphoramidate (20) [21] in the presence of t-BuMgCl gave phosphoramidate ester 21 in 86% yield as a single diastereoisomer. Finally, removal of the diallyl groups from 21 was accomplished by Pd-catalyzed reductive deallylation, using a method developed by our group [22], to obtain the target molecule remdesivir (1) in 91% yield, whose spectral data were in good agreement with the reported data (Scheme 3 ).
Scheme 3.
Total synthesis of remdesivir (1, GS-5734).
In conclusion, a short synthetic approach to access remdesivir has been developed in seven longest linear steps (LLS) with 25% overall yield starting from d-ribonolactone (12). The present strategy using a silyl/allyl protected sugar 15 has advantages including (i) avoiding the use of hazardous BCl3 for benzyl deprotection, (ii) improvement in the coupling yield without any additive, (iii) use of catalytic methods for protection and deprotection of the allyl groups. We believe that the present approach is scalable and can be used for the synthesis of novel analogues in addition to remdesivir as anti-viral agents.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Srivari Chandrasekhar reports financial support was provided by Science and Engineering Research Board.
Acknowledgments
Acknowledgments
We thank the Council of Scientific and Industrial Research (CSIR), Ministry of Science and Technology, Government of India, for research funding (COVID-19 Mission Mode Project, HCP 0029). K.K.P thanks the University Grants Commission (UGC), New Delhi, for a research fellowship. P.G. thanks the CSIR, New Delhi, for a research fellowship. S.C. thanks the Science and Engineering Research Board (SERB), Government of India for the J C Bose fellowship (SB/S2/JCB-002/2015). We also thank Dr. B. Jagadeesh, Head, Centre for NMR, CSIR-IICT, for NMR analytical support.
CSIR-IICT Manuscript Communication Number: IICT/Pubs./2021/279.
Footnotes
This work is dedicated to the memory of Dr. Surendar Reddy Bathula (Principal Scientist, CSIR-IICT), who lost the battle with COVID-19 infection after a fierce fight.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tetlet.2021.153590.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., de F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wit E.D., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Proc. Natl. Acad. Sci. USA. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kupferschmidt K., Cohen J. Science. 2020;367:610–611. doi: 10.1126/science.367.6478.610. [DOI] [PubMed] [Google Scholar]
- 5.Habibzadeh P., Stoneman E.K. Int. J. Occup. Environ. Med. 2020;11:65–71. doi: 10.15171/ijoem.2020.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J., Xing F., Liu J., Yip C.C.-Y., Poon R.W.-S., Tsoi H.-W., Lo S.K.-F., Chan K.-H., Cheng V.C.-C., Chen H., Hui C.K.-M., Yuen K.-Y. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., de Castilla D.L., Finberg R.W., Engl N. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.X., Nicastri E., Oda R. N. Engl J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. ACS Cent Sci. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Namchuk M.N. ACS Infect. Dis. 2021;7:1298–1302. doi: 10.1021/acsinfecdis.0c00874. [DOI] [PubMed] [Google Scholar]
- 10.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Tongeren S.A.V., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.-S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 12.Mulangu S., Dodd L.E., Davey R.T., Mbaya O.T., Proschan M., Mukadi D., Manzo M.L., Nzolo D., Oloma A.T., Ibanda A., Ali R., Coulibaly S., Levine A.C., Grais R., Diaz J., Lane H.C., Muyembe-Tamfum J.-J. N. Engl J. Med. 2019;381:2293–2303. [Google Scholar]
- 13.Vieira T., Stevens A.C., Chtchemelinine A., Gao D., Badalov P., Heumann L. Org. Process Res. Dev. 2020;24:2113–2121. doi: 10.1021/acs.oprd.0c00172. [DOI] [PubMed] [Google Scholar]
- 14.Xue F., Zhou X., Zhou R., Zhou X., Xiao D., Gu E., Guo X., Xiang J., Wang K., Yang L., Zhong W., Qin Y. Org. Process Res. Dev. 2020;24:1772–1777. doi: 10.1021/acs.oprd.0c00310. [DOI] [PubMed] [Google Scholar]
- 15.(a) Drew M.G.B., Mann J., Thomas A. J. Chem. Soc. Perkin Trans. 1986;1:2279–2285. [Google Scholar]; (b) Williams J.D., Kamath V.P., Morris P.E., Townsend L.B. Org. Synth. 2005;82:75–79. [Google Scholar]
- 16.Haight A.R., Stoner E.J., Peterson M.J., Grover V.K. J. Org. Chem. 2003;68:8092–8096. doi: 10.1021/jo0301907. [DOI] [PubMed] [Google Scholar]
- 17.(a) T. Butler, A. Cho, C.U. Kim, O.L. Saunders, L. Zhang, U.S. Patent 2009041447 Apr. 22 (2009); (b) T.V. Keutz, J.D. Williams, C.O. Kappe, Org. Process Res. Dev.24 (2020) 2362−2368; (c) T. Butler, A. Cho, B.R. Graetz, C.U. Kim, S.E. Metobo, O.L. Saunders, A.W. Waltman, J. Xu, L. Zhang, U.S. Patent 20100459508 Sep. 20 (2010); (d) S.E. Metobo, J. Xu, O.L. Saunders, T. Butler, E. Aktoudianakis, A. Cho, C.U. Kim, Tetrahedron Lett. 53(2012) 484−486; (e) Y. Xie, T. Hu, Y. Zhang, D. Wei, W. Zheng, F. Zhu, G. Tian, H.A. Aisa, J. Shen, J. Org. Chem. 86(2021) 5065-5072.
- 18.(a) Xie Y., Hu T., Zhang Y., Wei D., Zheng W., Zhu F., Tian G., Aisa H.A., Shen J. J. Org. Chem. 2021;86:5065–5072. doi: 10.1021/acs.joc.0c02986. [DOI] [PubMed] [Google Scholar]; (b) Savi C.D., Hughes D.L., Kvaerno L. Org. Process. Res. Dev. 2020;24:940–976. doi: 10.1021/acs.oprd.0c00233. [DOI] [PubMed] [Google Scholar]; (c) Vargas D.F., Larghi E.L., Kaufman T.S. ACS Omega. 2021;6:19356–19363. doi: 10.1021/acsomega.1c03082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Mishra B., Manmode S., Walke G., Chakraborty S., Neralkar M., Hotha S. Org. Biomol. Chem. 2021;19:1315–1328. doi: 10.1039/d0ob02176h. [DOI] [PubMed] [Google Scholar]; (b) Sugawara A., Takada H., Hirose T., Kimishima A., Yamada T., Toda M., Kojima T., Matsumaru T., Sunazuka T. Org. Lett. 2021;23:1758–1763. doi: 10.1021/acs.orglett.1c00183. [DOI] [PubMed] [Google Scholar]; (c) Nagamalla S., Johnson D.K., Sathyamoorthi S. Med. Chem. Res. 2021;30:1348–1357. doi: 10.1007/s00044-021-02724-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Liu G., Zhang Z., Fu S., Liu B. Org. Lett. 2021;23:290–295. doi: 10.1021/acs.orglett.0c03748. [DOI] [PubMed] [Google Scholar]; (e) Matsuura S., Niu C.-H., Cohen J.S. JCS Chem. Comm. 1976:451–452. [Google Scholar]
- 20.(a) Ross B.S., Reddy P.G., Zhang H.-R., Rachakonda S., Sofia M.J. J. Org. Chem. 2011;76:8311–8319. doi: 10.1021/jo201492m. [DOI] [PubMed] [Google Scholar]; (b) Wang M., Zhang L., Huo X., Zhang Z., Yuan Q., Li P., Chen J., Zou Y., Wu Z., Zhang W. Angew. Chem., Int. Ed. 2020;59:20814–20819. doi: 10.1002/anie.202011527. [DOI] [PubMed] [Google Scholar]; (c) Gannedi V., Villuri B.K., Reddy S.N., Ku C.-C., Wong C.-H., Hung S.-C. J. Org. Chem. 2021;86:4977–4985. doi: 10.1021/acs.joc.0c02888. [DOI] [PubMed] [Google Scholar]; (d) Wang M., Zhang L., Huo X., Zhang Z., Zhang W. Curr. Protocols. 2021 doi: 10.1002/cpz1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) B.K. Chun, M.O. Clarke, E. Doerffler, H.C. Hui, R. Jordan, R.L. Mackman, A.S. Ray, D. Siegel, Patent WO2016/69826 (2016); (b) M.O. Clarke, R. Jordan, R.L. Mackman, A.S. Ray, D. Siegel, Patent WO2017/184668 (2017); (c) O. Moukha-Chafiq, M.J. Suto, J. Liu, R. Boohaker, Patent WO2020/247633, (2020).
- 22.Chandrasekhar S., Reddy C.R., Rao R.J. Tetrahedron. 2001;57:3435–3438. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.