Graphical abstract
Keywords: Carmofur, SARS-CoV-2 main protease, Structural modification
Abstract
A coherent account of the reaction mechanistic details, structural modifications, and inhibition potentials of antineoplastic drug carmofur and its modified analogs to inhibition of SARS-CoV-2 main protease (Mpro) is reported. The survey is performed by integrating the density functional based tight binding (DFTB3) with density functional theory (DFT) calculations. The inhibition process commences with nucleophilic attack from the sulfur atom on the carbonyl group, yielding a C-S bond formation, followed by a bond formation of the H-O9 by 2.07 Å, which results in a transition state contains a ring of six atoms. We found that although the direct addition of sulfhydryl group hydrogen to the N3 position is likely to happen, the proper position of the hydrogen to O9 decreases its accessibility. The thermodynamic stability of the complex was calculated to be highly sensitive to the substituent on the N11 position. Compounds with CH2NH2 and CH2F at N11 positions of carmofur revealed high thermodynamic stability to complexation with Mpro but induced no change in substrate-binding pocket comparable to carmofur. Replacing the N11 of carmofur with carbon (C-carmofur) was effective in terms of complexation stability at CH2CH2CH2F and CH2CH2CH2OH substitutions and occupation of S1 subsite by these structures in addition to the S2 subsite. Based on the resulted data, increasing the length of the carbon chain at introduced substitutions in N-carmofur almost decreases the complexation stability while in C-carmofur the trend is reversed. Throughout these information outputs, it was suggested that compounds d, e, i′, and k′ might be novel and more efficacious drug candidates instead of carmofur. We believe that our characterization of mechanistic details and structural modification on Mpro/carmofur complex will significantly intensify researchers' understanding of this system, and consequently help them to take advantage of results into practice and design various valuable derivatives for inhibition of SARS-CoV-2 main protease.
1. Introduction
COVID-19 as a highly contagious pathogenic has rapidly spread worldwide in December 2019 from Wuhan city in Hubei Province of China [1]. While the number of reported confirmed cases worldwide continues to rise, efforts are underway to overcome this challenge. So far, no known effective drug is available to treat or alleviate the disease symptoms [2]. The development of new generation of antiviral drugs is urgently needed. Since, the process of new drug design is highly time‐consuming, risky, and costly, drug repurposing can be used as an alternative strategy, determining the new indications of existing drugs for another disease [3]. Reducing the risk of adverse side effects, drug interactions, and drug development time and expenditure are advantages of this method [4]. Considering the matter of time, the computational approaches for drug repurposing provide the best possible chance of selecting the most effective drug among the broad list of approved drugs for the life‐threatening emergency condition of COVID‐19 [5].
Carmofur, a derivative of 5-fluorouracil, is an antineoplastic drug containing an electrophilic carbonyl reactive group that inhibits human acid ceramidase (AC) through covalent modification of its catalytic cysteine [6]. Raised AC levels are linked to several malignancies, including breast [7], prostate [8], colorectal [9], melanoma [10], and brain [11]. The ability of carmofur to inhibit AC led to its anticancer activity. Since the 1980s, carmofur has been used to treat colorectal cancer [12]. It has also been shown clinical benefits against breast [13], gastric [14], and bladder cancers [15].
Similar to SARS and MERS, the coronavirus genome encodes four non-structural proteins including Spike (S‐protein), RNA-dependent RNA polymerase (RdRp), the main protease (MPro), and papain-like protease (PLpro), which recognized as promising targets for developing drugs and treatment against the recent coronavirus epidemic [16], [17], [18], [19]. A recent study shows that carmofur inhibits the SARS-CoV-2 main protease (Mpro) [20]. The X-ray crystal structure of the Mpro/carmofur complex shows that the carbonyl reactive group of carmofur is covalently bound to the Sγ atom of Cys145, while its fatty acid tail is inserted into the hydrophobic S2 subsite. Based on these observations, the authors suggested that the sulfhydryl group of Cys145 attacks the electrophilic carbonyl group of carmofur, resulting in covalent modification of Cys145 and release of the 5-FU moiety. Considering this, the present study is designed to elucidate the detailed molecular mechanism of inhibition of MPro by carmofur along with structural modification of the drug to inhibit SARS-CoV-2 main protease in a more effective affinity. We hope that the obtained results will help in the repurposing of already approved drugs to combat the recent dangerous coronavirus epidemic.
2. Model and computational details
The three-dimensional structure of the SARS-CoV-2 main protease (PDB ID: 7BUY) was downloaded from RCSB PDB database (http://www.rcsb.org/pdb). Before calculation, the protein was edited by cutting the Arg40 to Met49 residues, Leu141 to Cys145 residues, and His163 to Glu166 residues from the crystal structure. The starting configuration consisted of the crystal structure of the Mpro/carmofur complex, which has been fully optimized using the density functional based tight binding (DFTB3) method [21] by third-order parametrization for organic and biological systems (3OB)-3–1 parameter [22], [23], [24]. Vibrational frequencies were computed to obtain the Gibbs free energy for all the Mpro/carmofur analog complexes. The DFTB + program [25] was used for this part of the calculations. B3LYP hybrid functional (20% HF exchange) with Grimme’s DFT-D3 dispersion correction [26] were used for all geometry optimization related to transition state structures. The basis set was set to 6–311 + G(d,p) [27] for all atoms during the calculations. The Berny algorithm was used to transition states optimization. Vibrational frequencies are computed by using the same method/basis set as the geometries optimized. The first-order saddle point (transition state) was confirmed by only one imaginary frequency on computed vibrational frequencies. Intrinsic reaction coordinate (IRC) calculation was computed to confirmed transition structure connection to minima [28], [29]. SMD/Water variation was considered to consider the solvent effects on all the calculations. All the optimization and frequency calculations in this part of the study were executed with the ORCA program package [30]. In the present study, the free energy for each compound in solution was computed through the following formula:
where ΔG1atm→1M = 1.89 kcal/mol is the change of free-energy for the contraction of 1 mol of an ideal gas from 1 atm to the 1 M solution-phase standard state. The atom numbering used throughout the study is illustrated in Fig. 1 .
Fig. 1.

The molecular structure of carmofur with atom numbering.
3. Results and discussion
The main purpose of the present study is to modify the carmofur molecular structure to develop novel carmofur analogs that inhibit SARS-CoV-2 main protease and therefore cause the virus to malfunction. The crystal structure of Mpro in complex with carmofur shows that the hydrophobic sequence tail of carmofur is inserted into the S2 subsite, while the carbonyl moiety (C O) is linked to the Sγ atom of Cys145 through a 1.8 Å covalent bond [20]. Moreover, a hydrogen bond between the O12 of the inhibitor and the NH2 hydrogen of Gly143 further stabilized the complex. The carmofur only occupies the S2 subsite of the Mpro [20]. Considering that the carbonyl group of carmofur is easy to be attacked by sulfhydryl group [31], we investigated the mechanism of Mpro/carmofur complexation according to Scheme 1 .
Scheme 1.
The different reaction pathways for inhibition of Mpro by carmofur.
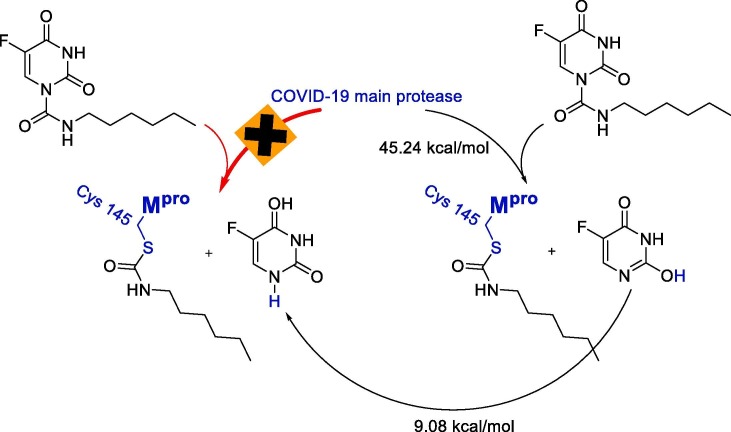
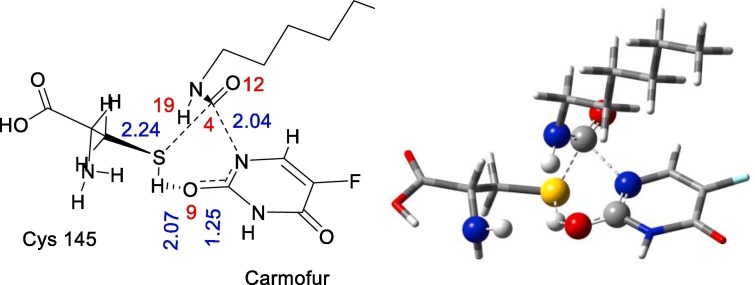
The reaction pathway leading to products Mpro/carmofur complex and 5-fluoro-2-hydroxypyrimidin-4(3H)-one, overcomes an energy barrier of as high as 45.24 kcal/mol through the transition state shown in Fig. 2 . The transition state contains a ring of six atoms, in which the sulfhydryl group acts as an entering group while 5-fluoro-2-hydroxypyrimidin-4(3H)-one leaves the carbonyl group. Nucleophilic attack from the sulfur atom on the carbonyl group leads to the bond formation of C10-S by 2.24 Å. Moreover, the process involves the changing of the C10-N3 bond from 1.47 to 2.04 Å and a bond formation of the SH-O9 by 2.07 Å. With the formation of the SH-O9 bond, the C10-N3 bond length increases from 1.47 to 2.04 Å, and hence the C4-N3 bond length decreases from 1.38 to 1.35 Å. In addition to the mentioned bond formations, the transition state is stabilized by two hydrogen bonds of H19-O9 (2.10 Å) and the hydrogen of the NH2 group at Cys145 and O9 (2.34 Å). This indicates that the formation of 5-fluoropyrimidine-2,4(1H,3H)-dione is unlikely through the direct addition of sulfhydryl group hydrogen to the N3 position as proposed by Jin, Z. et al [20], while it can be formed in the next steps by a tautomerization reaction with an energy barrier of as high as 9.08 kcal/mol [32] (see Scheme 1).
Fig. 2.
Key parameters for the transition state structure located for Mpro/carmofur complexation using the B3LYP-D3BJ/6–311 + G(d,p) level of calculations. The interatomic distances are given in Å.
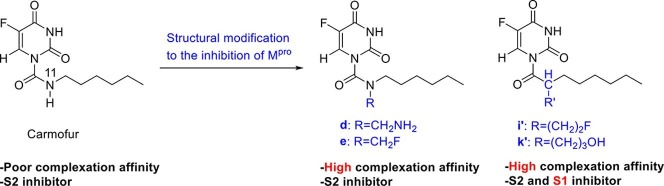
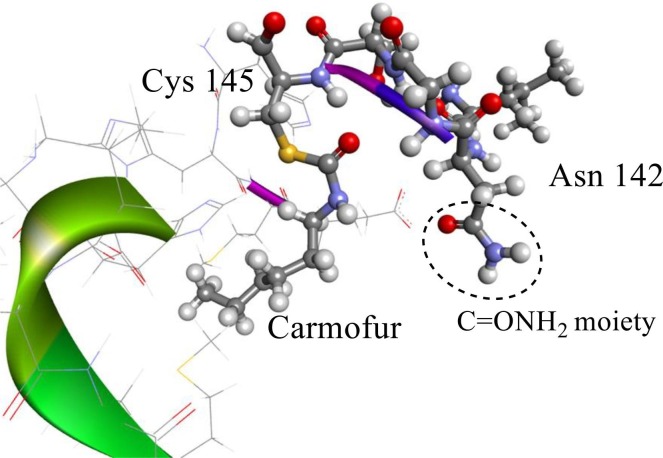
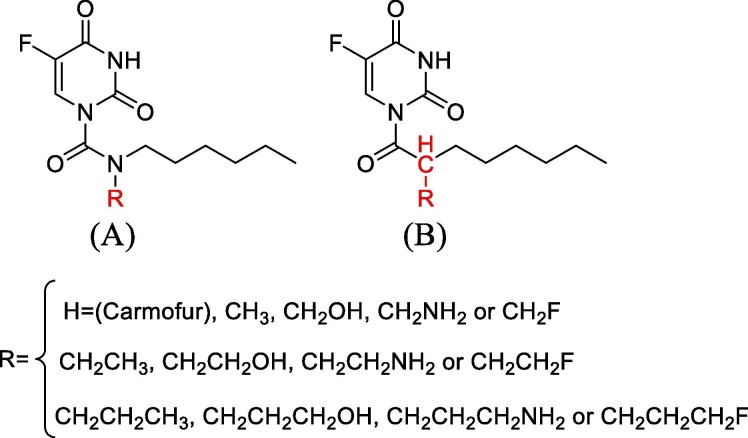
To apply the structural modification found in this study to future drug discovery research, especially for drug candidates containing carboxamides, this work aimed to stabilized the Mpro/inhibitor complexation along with the inhibitor structural flexibility to block different sites on Mpro. Because the C ONH2 moiety in Asn142 can increase the stability of the complex by creating a hydrogen bond (see Fig. 3 ), we first modified carmofur by introducing substitutions at the N11 position of this inhibitor (see Fig. 4 , A).
Fig. 3.
The crystal structure of Mpro/carmofur complex labeled Cys145 and Asn142.
Fig. 4.
Modification of carmofur analogs by introducing substitutions at the N11 position (N-Carmofur-(CH2)nX) and or by altering the N11 to carbon and introducing substitutions at the new position of C11 (C-Carmofur-(CH2)nX).
Although increasing the length of the substitutions brings the hydrogen donor closer to the oxygen lone pair electrons of the C ONH2 moiety, but according to results in Table 1 , the most complexation stability is seen at n = 1. While introducing NH2 and OH into the N-Carmofur (g and k) exhibited a modest increase in relative Gibbs’s free energy of the complex, introducing NH2 and F into the N11 position at n = 1 (N-Carmofur-(CH2)2NH2 and N-Carmofur-(CH2)3OH) made the complex highly thermodynamically stable (see d and e in Table 1). The d and e structures with −12.73 and −10.12 kcal/mol respectively are both stable than the Mpro/carmofur complex. The d inhibitor is stabilized by both a 2.04 Å hydrogen bond between the NH2 hydrogen of introduced substitution and the oxygen lone pair electrons of C ONH2 moiety in Asn142, and also hydrophobic interactions (see Fig. 5 ). In the same manner, e structure also forms a hydrogen bond of 2.30 Å between the CH2F hydrogen of introduced substitution and the oxygen lone pair electrons of C ONH2 moiety. Hydrogen bonding in e structure can occur due to the high electronegativity of the fluoride and so leaving the carbon relatively electron-poor. These data indicate that compounds d and e might inhibit Mpro by a relatively high negative binding energy. Besides, the results show that H at n = 1, F at n = 2, and H, NH2, and F at n = 3 are not preferable for Mpro inhibition (see b, I, j, l, and m in Table 1).
Table 1.
The relative free energies for carmofur (black row) and its analogs in complex with Mpro. The relative free energies are given in kcal/mol.
| n/X | N-Carmofur-(CH2)nX | C-Carmofur-(CH2)nX | ||
|---|---|---|---|---|
| n = 0, X = H | a | −7.17 | a′ | −2.60 |
| n = 1, X = H | b | −6.12 | b′ | −6.40 |
| n = 1, X = OH | c | −7.01 | c′ | −2.28 |
| n = 1, X = NH2 | d | −12.73 | d′ | −5.75 |
| n = 1, X = F | e | −10.12 | e′ | −3.63 |
| n = 2, X = H | f | −9.46 | f′ | −7.50 |
| n = 2, X = OH | g | −7.58 | g′ | −7.51 |
| n = 2, X = NH2 | h | −8.53 | h′ | −5.64 |
| n = 2, X = F | i | −4.93 | i′ | −13.15 |
| n = 3, X = H | j | −5.12 | j′ | −5.98 |
| n = 3, X = OH | k | −8.57 | k′ | −11.77 |
| n = 3, X = NH2 | l | −5.60 | l′ | −5.99 |
| n = 3, X = F | m | −5.78 | m′ | −4.64 |
Fig. 5.
Overall structure of Mpro in complex with d, e, i′, and k′ structures.
Next, we modified carmofur by altering the N13 to carbon and introducing substitutions at the new position of C11 (see Fig. 4, B). Replacing nitrogen with carbon without any other substitution modifications drastically reduces the Mpro/carmofur complexation stability (see a′ at Table 1). Also, unlike the previous case, all of the n = 1 compounds are highly unstable than the Mpro/carmofur complex, a. Nevertheless, compounds i′ and k′ are more thermodynamically stable than a and d. The i′ analog form a hydrogen bond of 2.34 Å between the CH2F hydrogen of its modified substitution and the oxygen lone pair electrons of C ONH2 moiety. In addition to this hydrogen bond, the i′ inhibitor is also stabilized by hydrophobic interactions of modified substitution moiety (CH2CH2F) with Met165 of S1 subsite (see Fig. 5). Altogether, this structure with complexation energy of −13.15 kcal/mol is −5.98 kcal/mol more stable than a. On the other hand k′ by form a strong hydrogen bond of 1.95 Å between the OH hydrogen of its modified substitution and the oxygen lone pair electrons of C ONH2 moiety and also hydrophobic interactions with Met165, His163 and His164 of S1 subsite is −4.60 kcal/mol more stable than a. Therefore, our results show that the mechanism of C-carmofur modifications is different from that of the N-carmofur compounds. In addition to the greater thermodynamic stability of i′ and k′ than N-Carmofur modifications, the largest conformational differences occur in the substrate-binding pocket, d and e analogs only occupy the S2 subsite, whereas C-Carmofur-(CH2)2F and C-Carmofur-(CH2)3OH occupy S1 subsite in addition to the S2 subsite. These findings indicate the structural elaboration potential of carmofur and will be helpful for the design of more potent derivatives against the Mpro.
4. Conclusions
In addition to the characterization of mechanistic details and specify the exact geometry of transition state involved in Mpro/carmofur complexation, we suggested several carmofur analogs and evaluated their complexation stability and intermolecular interactions with M pro. Compounds d, e, i′, and k′ were highly stable to Mpro and their Gibs free energy was much stronger than that of carmofur. Although these four compounds interact relatively identically with Mpro, compounds i′ and k′ occupy S1 subsite in addition to the S2 subsite. We propose that all d, e, i′, and k′ compounds might serve as drug candidates that could be developed for use as efficacious Mpro inhibitors drugs with much stronger binding energy than carmofur.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Niloofar Hemati: Project administration. Saba Hadidi: Conceptualization, Formal analysis, Investigation, Resources, Software, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. Farshad Shiri: Formal analysis, Investigation, Methodology. Mohammad Hosein Farzaei: Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Carlos W.G., Dela Cruz C.S., Cao B., Pasnick S., Jamil S. Novel Wuhan (2019-nCoV) coronavirus. Am J Respir Crit Care Med. 2020:P7–P8. doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]
- 2.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci. Trends. 2020;14:69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 3.Mohamed K., Yazdanpanah N., Saghazadeh A., Rezaei N. Computational drug discovery and repurposing for the treatment of Covid-19: a systematic review. Bioorg. Chem. 2021;106:104490. doi: 10.1016/j.bioorg.2020.104490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrawal P. Advantages and challenges in drug re-profiling. J. Pharmacovigil. 2015;2:2. [Google Scholar]
- 5.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020;60:3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dementiev A., Joachimiak A., Nguyen H.a., Gorelik A., Illes K., Shabani S., Gelsomino M., Ahn E.-Y., Nagar B., Doan N. Molecular mechanism of inhibition of acid ceramidase by carmofur. J. Med. Chem. 2019;62(2):987–992. doi: 10.1021/acs.jmedchem.8b01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sänger N., Ruckhäberle E., Györffy B., Engels K., Heinrich T., Fehm T., Graf A., Holtrich U., Becker S., Karn T. Acid ceramidase is associated with an improved prognosis in both DCIS and invasive breast cancer. Mol. Oncol. 2015;9:58–67. doi: 10.1016/j.molonc.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seelan R.S., Qian C., Yokomizo A., Bostwick D.G., Smith D.I., Liu W. Human acid ceramidase is overexpressed but not mutated in prostate cancer. Genes Chromosom. Cancer. 2000;29(2):137–146. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1018>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 9.Klobučar M., Grbčić P., Pavelić S.K., Jonjić N., Visentin S., Sedić M. Acid ceramidase inhibition sensitizes human colon cancer cells to oxaliplatin through downregulation of transglutaminase 2 and β1 integrin/FAK− mediated signalling. Biochem. Biophys. Res. Commun. 2018;503(2):843–848. doi: 10.1016/j.bbrc.2018.06.085. [DOI] [PubMed] [Google Scholar]
- 10.Realini N., Palese F., Pizzirani D., Pontis S., Basit A., Bach A., Ganesan A., Piomelli D. Acid ceramidase in melanoma: expression, localization, and effects of pharmacological inhibition. J. Biol. Chem. 2016;291(5):2422–2434. doi: 10.1074/jbc.M115.666909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doan N.B., Alhajala H., Al-Gizawiy M.M., Mueller W.M., Rand S.D., Connelly J.M., Cochran E.J., Chitambar C.R., Clark P., Kuo J. Acid ceramidase and its inhibitors: a de novo drug target and a new class of drugs for killing glioblastoma cancer stem cells with high efficiency. Oncotarget. 2017;8 doi: 10.18632/oncotarget.22637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakamoto J., Hamada C., Rahman M., Kodaira S., Ito K., Nakazato H., Ohashi Y., Yasutomi M. An individual patient data meta-analysis of adjuvant therapy with carmofur in patients with curatively resected colon cancer. Jpn. J. Clin. Oncol. 2005;35:536–544. doi: 10.1093/jjco/hyi147. [DOI] [PubMed] [Google Scholar]
- 13.Morimoto K., Koh M. Postoperative adjuvant use of carmofur for early breast cancer. Osaka City Med. J. 2003;49:77–83. [PubMed] [Google Scholar]
- 14.Gröhn P., Heinonen E., Kumpulainen E., Länsimies H., Lantto A., Salmi R., Pyrhönen S., Numminen S. Oral carmofur in advanced gastrointestinal cancer. Am. J. Clin. Oncol. 1990;13:477–479. doi: 10.1097/00000421-199012000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Nishio S., Kishimoto T., Maekawa M., Kawakita J., Morikawa Y., Funai K., Hayahara N., Yuki K., Nishijima T., Yasumoto R. Study on effectiveness of carmofur (Mifurol) in urogenital carcinoma, especially bladder cancer, as a post-operative adjuvant chemotherapeutic agent, Hinyokika kiyo. Acta Urol. Japonica. 1987;33:295–303. [PubMed] [Google Scholar]
- 16.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 17.Luan B., Huynh T., Cheng X., Lan G., Wang H.-R. Targeting Proteases for Treating COVID-19. J. Proteome Res. 2020;19:4316–4326. doi: 10.1021/acs.jproteome.0c00430. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y., Yang C., Xu X.-F., Xu W., Liu S.-W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., El Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti–COVID-19 drug design. Sci. Adv. 2020;6(42) doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 21.Gaus M., Cui Q., Elstner M. DFTB3: extension of the self-consistent-charge density-functional tight-binding method (SCC-DFTB) J. Chem. Theory Comput. 2011;7:931–948. doi: 10.1021/ct100684s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubillus M., Kubař T., Gaus M., Řezáč J., Elstner M. Parameterization of the DFTB3 Method for Br, Ca, Cl, F, I, K, and Na in Organic and Biological Systems. J. Chem. Theory Comput. 2015;11(1):332–342. doi: 10.1021/ct5009137. [DOI] [PubMed] [Google Scholar]
- 23.Gaus M., Lu X., Elstner M., Cui Q. Parameterization of DFTB3/3OB for sulfur and phosphorus for chemical and biological applications. J. Chem. Theory Comput. 2014;10(4):1518–1537. doi: 10.1021/ct401002w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaus M., Goez A., Elstner M. Parametrization and Benchmark of DFTB3 for Organic Molecules. J. Chem. Theory Comput. 2013;9(1):338–354. doi: 10.1021/ct300849w. [DOI] [PubMed] [Google Scholar]
- 25.Hourahine B., Aradi B., Blum V., Bonafé F., Buccheri A., Camacho C., Cevallos C., Deshaye M.Y., Dumitrică T., Dominguez A. DFTB+, a software package for efficient approximate density functional theory based atomistic simulations. J. Chem. Phys. 2020;152(12):124101. doi: 10.1063/1.5143190. [DOI] [PubMed] [Google Scholar]
- 26.Grimme S., Antony J., Ehrlich S., Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010;132:154104. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard B.P., Altarawy D., Didier B., Gibson T.D., Windus T.L. New basis set exchange: an open, up-to-date resource for the molecular sciences community. J. Chem. Inf. Model. 2019;59:4814–4820. doi: 10.1021/acs.jcim.9b00725. [DOI] [PubMed] [Google Scholar]
- 28.Fukui K. The path of chemical reactions-the IRC approach. Acc. Chem. Res. 1981;14:363–368. [Google Scholar]
- 29.Fukui K. Formulation of the reaction coordinate. J. Phys. Chem. 1970;74:4161–4163. [Google Scholar]
- 30.Neese F. The ORCA program system. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2012;2(1):73–78. [Google Scholar]
- 31.Lienhard G.E., Jencks W.P. Thiol addition to the carbonyl group. Equilibria and kinetics1. J. Am. Chem. Soc. 1966;88:3982–3995. doi: 10.1021/ja00969a017. [DOI] [PubMed] [Google Scholar]
- 32.Rastogi V.K., Palafox M.A. Vibrational spectra, tautomerism and thermodynamics of anticarcinogenic drug: 5-Fluorouracil. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011;79(5):970–977. doi: 10.1016/j.saa.2011.04.008. [DOI] [PubMed] [Google Scholar]