Abstract
The general transcription factor TFIIE plays important roles in transcription initiation and in the transition to elongation. However, little is known about its function during these steps. Here we demonstrate for the first time that TFIIH-mediated phosphorylation of RNA polymerase II (Pol II) is essential for the transition to elongation. This phosphorylation occurs at serine position 5 (Ser-5) of the carboxy-terminal domain (CTD) heptapeptide sequence of the largest subunit of Pol II. In a human in vitro transcription system with a supercoiled template, this process was studied using a human TFIIE (hTFIIE) homolog from Caenorhabditis elegans (ceTFIIEα and ceTFIIEβ). ceTFIIEβ could partially replace hTFIIEβ, whereas ceTFIIEα could not replace hTFIIEα. We present the studies of TFIIE binding to general transcription factors and the effects of subunit substitution on CTD phosphorylation. As a result, ceTFIIEα did not bind tightly to hTFIIEβ, and ceTFIIEβ showed a similar profile for binding to its human counterpart and supported an intermediate level of CTD phosphorylation. Using antibodies against phosphorylated serine at either Ser-2 or Ser-5 of the CTD, we found that ceTFIIEβ induced Ser-5 phosphorylation very little but induced Ser-2 phosphorylation normally, in contrast to wild-type hTFIIE, which induced phosphorylation at both Ser-2 and Ser-5. In transcription transition assays using a linear template, ceTFIIEβ was markedly defective in its ability to support the transition to elongation. These observations provide evidence of TFIIE involvement in the transition and suggest that Ser-5 phosphorylation is essential for Pol II to be in the processive elongation form.
In eukaryotes, transcription of protein-encoding genes by RNA polymerase II (Pol II) is the first step in expression of those genes (for reviews, see references 4, 35, 44, and 51). Two sequential stages are now recognized in the establishment of Pol II processivity: transcription initiation and the transition from initiation to elongation. At initiation, five general transcription factors (TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) together with Pol II form the preinitiation complex (PIC) on the core promoter. Two models of PIC formation have been proposed on the basis of recent analyses. One model involves stepwise association of the general transcription factors and Pol II on promoter DNA, while the other model entails promoter sequences binding to a preassembled Pol II holoenzyme that contains most of the general transcription factors as well as SRB (suppressor of RNA polymerase B)- and Med-containing complex (reviewed in references 3 and 22). In vitro analyses of stepwise assembly of the PIC using purified factors have demonstrated that TFIIE joins the complex at a position near the transcription start site (between positions −14 and −2), after Pol II and TFIIF have joined the complex (25, 49). TFIIE then recruits TFIIH, and these two factors stabilize and activate the PIC, resulting in isomerization of double-stranded (ds) promoter DNA (promoter melting) upon transcription initiation. TFIIE and TFIIH are also involved in the transition from initiation to elongation, the stage during which they act to remove from the complex general transcription factors that have already completed their roles in the initiation step (promoter clearance) (reviewed in reference 35).
Human TFIIE (hTFIIE) consists of an α2β2 heterotetramer of 57-kDa α- and 34-kDa β-subunits (41). hTFIIEα is highly acidic (pI, 4.5) and possesses several putative structural motifs and characteristic sequences (40). The region essential for basal transcription is located within the N-terminal half of the molecule, in which all of the structural motifs reside (37, 38). The acidic region near the C terminus is the only region in the C-terminal half that has a stimulatory effect on basal transcription; this region binds directly to TFIIH. In contrast, hTFIIEβ is highly basic (pI, 9.5) and possesses several putative structural motifs and characteristic sequences different from those of hTFIIEα (36, 48, 57). The internal region of hTFIIEβ is essential for basal transcription. It has been found that TFIIEβ binds to single-stranded (ss) DNA through the basic region near its C terminus; the other general transcription factors, TFIIB and TFIIFβ (RAP30), bind to this region as well (42). In addition, we have recently determined the three-dimensional structure of the central core region in TFIIEβ that binds to dsDNA (43).
Human TFIIH consists of nine subunits and has three ATP-dependent catalytic activities: kinase activity that phosphorylates the carboxy-terminal domain (CTD) of the largest subunit of Pol II, DNA-dependent ATPase activity, and DNA helicase activity (reviewed in reference 59). TFIIE regulates these TFIIH activities, stimulating the CTD kinase and ATPase activities and repressing the helicase activity (8, 31, 39). At transcription initiation, TFIIE binds to Pol II, TFIIB, and TFIIF, recruits TFIIH into the PIC to stabilize and activate the PIC, and binds to stabilize the ssDNA region in promoter melting. Recent studies have provided support for this model. (i) Photo-crosslinking studies demonstrated that TFIIEβ binds directly to the core promoter region (between positions −14 and −2), where the promoter melts upon transcription initiation (49). (ii) Two-dimensional crystallography of yeast TFIIE (yTFIIE) with Pol II demonstrated that yTFIIE binds to the active center of Pol II, which is located near the transcription initiation site on the promoter (25). (iii) Short mismatched heteroduplex DNA around the transcription initiation site in topologically relaxed linear templates was shown to alleviate the requirement for TFIIE, TFIIH, and ATP (10, 20, 45, 61).
The above description summarizes our current understanding of the roles of TFIIE and TFIIH before and during transcription initiation. In contrast to initiation, extensive studies of the later stages of transcription have yet to be carried out. Functional involvement of TFIIE and TFIIH in the transition from initiation to elongation has been suggested by studies using a negatively supercoiled immunoglobulin heavy chain (IgH) promoter and the short mismatched heteroduplex linear promoter described above (10, 13, 19, 20, 46, 62). It has been suggested that TFIIE and TFIIH may suppress abortive initiation, which produces short transcripts (around 2 to 15 nucleotides) by releasing the general factors TFIID and TFIIB from the PIC, and may convert Pol II to its elongation-competent form (10, 21, 24). It has been demonstrated that this elongation-competent Pol II is hyperphosphorylated (34, 70). Since TFIIH is the only kinase that can phosphorylate the CTD in the in vitro reconstituted active Pol II complex, TFIIH has been a primary candidate for the biologically relevant CTD kinase (31, 39).
The CTD contains multiple repeats of the heptapeptide sequence YSPTSPS, which occurs 52 times in the largest subunit of human Pol II (7, 71). Several lines of evidence indicate that the integrity of the CTD is essential for basal and activated transcription. The unphosphorylated form of Pol II (Pol IIa) is preferentially recruited into a PIC reconstituted with purified general transcription factors (30). CTD phosphorylation may initially occur between transcription initiation and the transition from initiation to elongation, converting Pol IIa to the phosphorylated form (Pol IIo) (34, 35, 59). Recently, it has been demonstrated that CTD phosphorylation is also important for recruiting the mRNA processing enzymes to the nascent transcript, presumably reflecting the fact that mRNA processing (splicing, capping, and polyadenylation) occurs during and/or after transcription (6, 33).
To further investigate the mechanisms of transcription initiation and the transition to elongation and the TFIIE functions during these steps, we isolated TFIIE cDNAs from the nematode Caenorhabditis elegans (ceTFIIE cDNAs) and expressed two subunits of ceTFIIE in bacteria, both together and independently, based on the idea that the basic transcriptional mechanisms might be conserved among eukaryotic species. We compared the ceTFIIE subunits with respect to their abilities to substitute for their human counterparts in a human in vitro transcription system, their specificities of binding to the general transcription factors, their effects on CTD phosphorylation by TFIIH and, finally, their abilities to convert Pol II to the elongation form. Importantly, we demonstrated for the first time that TFIIE is directly involved in the transition from transcription initiation to elongation and suggested, through the use of transcription transition assays together with analyses of the sites of phosphorylation in the CTD heptapeptide repeat sequence, that TFIIE-induced phosphorylation of serine at position 5 (Ser-5) in the CTD heptapeptide repeat might be essential for this transition.
MATERIALS AND METHODS
Cloning of C. elegans TFIIE cDNAs.
The putative ceTFIIEβ coding sequence was identified using a TBLASTN homology search of the C. elegans translated expressed sequence tag (EST) databank (Sanger Centre, Cambridge, United Kingdom) to locate regions with significant homology to the hTFIIEβ amino acid sequence. Since there was an NdeI site approximately 25 bp upstream of the stop codon, four oligonucleotides were designed to perform two PCRs in order to amplify the N- and C-terminal halves independently. To amplify the N-terminal half of ceTFIIEβ, the oligonucleotide CEB1T (5′-CTGATCATATGGACCCGGAATTGTTAAGGC-3′) was designed to create an NdeI site (underlined) at the first methionine codon and to disrupt a BamHI site by changing the third nucleotide of the second aspartate codon (T to C, bold and underlined) and was used in conjunction with the oligonucleotide CEB2B (5′-GTCATTGTAGAAGACGAC-3′). To obtain the C-terminal half, the oligonucleotide CEB1B (5′-CTTGAGGATCCAGAAGTGTGTAATTAAAATC-3′) was designed to create a BamHI site (underlined) after the stop codon and was used in conjunction with the oligonucleotide CEB2T (5′-GTGGATTATATGAAGAAACG-3′). PCRs were performed using a C. elegans mixed-stage cDNA library (a kind gift from Yuji Kohara). After 50 cycles of PCR with an annealing temperature of 50°C, the PCR products (approximately 630 bp for the N-terminal portion and 640 bp for the C-terminal portion) were purified, blunt ended with the Klenow fragment of Escherichia coli DNA polymerase I, phosphorylated with T4 polynucleotide kinase, and subcloned into the SmaI site of pBluescript SK(−) (Stratagene). The nucleotide sequences of the cloned PCR products were confirmed using an ALFred DNA sequencer (Amersham Pharmacia Biotech).
Using the same strategy, several regions with high homology to the hTFIIEα amino acid sequence were identified. Since the extent of homology was lower than in the case of TFIIEβ, it was difficult to identify the entire putative coding region of ceTFIIEα from C. elegans genomic sequences, although the putative N and C termini were identified. The oligonucleotide CEA1T (5′-CAAGTCATATGTCATCTGGCCCAG-3′) was designed to create an NdeI site (underlined) at the first methionine codon, and the oligonucleotide CEA1B (5′-GAGCTGGATCCGAGACTTAATGAATAG-3′) was designed to create a BamHI site (underlined) after the stop codon. No product was obtained from the C. elegans mixed-stage cDNA library when these two oligonucleotides were used as the PCR primers. Several oligonucleotides which matched internal coding regions were synthesized and used in PCRs with CEA1B. The longest product was about 980 bp and was obtained with the oligonucleotide CEA2T (5′-CAACGTGGTGCGCTAC-3′), which matches a region approximately 100 amino acids internal from the N terminus. It was discovered by PCR and cDNA screening that the mixed-stage cDNA library was oligo-(dT)-primed and does not contain any clones which extend as far as the N-terminal region of the ceTFIIEα. Therefore, two different C. elegans embryonic cDNA libraries (kind gifts from Yuji Kohara and Hideyuki Okano) were used to obtain cDNA clones encoding the missing N-terminal region of ceTFIIEα. N-terminal cDNAs (600 bp) were obtained by PCR from both embryonic libraries using the N-terminal oligonucleotide CEA1T and the oligonucleotide CEA3B (5′-GGTGTCATTTGTTCGTTG-3′). After 50 cycles of PCR with an annealing temperature of 55°C, the PCR products were purified, blunt ended, and phosphorylated as described above and subcloned into the SmaI site of pBluescript SK(−) (Stratagene). The nucleotide sequences of the cloned PCR products were determined as described above.
Construction of ceTFIIE expression vectors.
Plasmids containing either the N- or C-terminal portion of the ceTFIIEβ (ceTFIIEβ cDNA) open reading frame were digested with either NdeI and PstI (N-terminal clone; 0.54 kb) or PstI and BamHI (C-terminal clone; 0.33 kb). These fragments were subcloned into the pET3a and 6HisT-pET11d vectors to construct expression plasmids containing the entire coding region of ceTFIIEβ cDNA, expressing nontagged ceTFIIEβ (ceTFIIEβ) and six-histidine-tagged ceTFIIEβ (6H-ceTFIIEβ), respectively (17). Similarly, plasmids containing either the N- or C-terminal portion of the open reading frame of ceTFIIEα (ceTFIIEα cDNA) were digested with NdeI and SacI (N-terminal clone; 0.41 kb) or with SacI and BamHI (C-terminal clone; 0.89 kb). These fragments were subcloned into the pET3a and 6HisT-pET11d vectors to construct nontagged (ceTFIIEα) and six-histidine-tagged (6H-ceTFIIEα) ceTFIIEα expression plasmids containing the full coding region of ceTFIIEα cDNA (17).
Construction of coexpression plasmids encoding two TFIIE subunits was performed essentially as described previously (15). Both C. elegans and human TFIIEα expression plasmids (in pET11d) with six-histidine tags at the N termini of the inserted DNAs (40) were digested with XbaI and BamHI, and the resulting fragments containing TFIIEα cDNAs were blunt ended as described above. C. elegans and human TFIIEβ expression plasmids (in pET3a) (57) were digested with XbaI, blunt ended as described above, and then treated with calf intestine alkaline phosphatase. Finally, TFIIEα cDNA fragments were subcloned into the (blunted) XbaI sites of the TFIIEβ expression plasmids to place both cDNAs in tandem in the same orientation. Four different chimeric 6H-TFIIE coexpression plasmids (ceTFIIEα-ceTFIIEβ, ceTFIIEα-hTFIIEβ, hTFIIEα-ceTFIIEβ, and hTFIIEα-hTFIIEβ) were constructed in this manner.
The HA-pET11d vector was constructed as described elsewhere (42). Hemagglutinin (HA)-tagged ceTFIIEβ (HA-ceTFIIEβ) and ceTFIIEα (HA-ceTFIIEα) expression plasmids were prepared by subcloning the NdeI-BamHI fragments of either ceTFIIEβ or ceTFIIEα cDNA into the same sites of the HA-pET11d vector.
Glutathione S-transferase (GST) fusion constructs were made in pGEX-2TL(+) as described previously (42). Expression plasmids containing GST-fused human TFIIH subunits were prepared as follows. cDNA encoding the TFIIH subunits XPB (ERCC3), p52, p44, Cdk7 (MO15), and cyclin H was digested with NdeI and BamHI and subcloned into pGEX-2TL(+), digested with the same restriction enzyme. XPD (ERCC2) cDNA was digested with NdeI and HindIII and subcloned into pGEX-2TL(+) digested with the same restriction enzyme. p62 cDNA was digested with NdeI and SspI and subcloned into pGEX-2TL(+) digested with NdeI and SmaI. p34 cDNA was digested with NdeI and EcoRI and subcloned into pGEX-2TL(+) digested with the same restriction enzymes. Finally, MAT1 cDNA was digested with NcoI and BamHI and subcloned into pGEX-2TL(+) digested with the same restriction enzymes.
Expression and purification of recombinant proteins.
Recombinant proteins were expressed in E. coli BL21(DE3)pLysS by induction with isopropyl-β-d-thiogalactopyranoside (IPTG) (55). For general purification, soluble bacterial lysates were used. For miniscale preparations, lysates (1 ml) representing 50 to 100 ml of culture were mixed directly with 1 ml of buffer B (20 mM Tris-HCl [pH 7.9 at 4°C], 0.5 mM EDTA, 10% [vol/vol] glycerol, 1 mM phenylethylsulfonyl fluoride [PMSF], 2-μg/ml antipain, 2-μg/ml aprotinin, 1-μg/ml leupeptin, 0.8-μg/ml pepstatin, 10 mM 2-mercaptoethanol) containing 500 mM NaCl (BB500) and 100 μl of Ni-nitrilotriacetic acid (NTA) agarose resin (Qiagen) and incubated for 4 h at 4°C. The resin samples were washed twice with 1 ml of BB500, twice with 1 ml of buffer D (20 mM Tris-HCl [pH 7.9 at 4°C], 20% [vol/vol] glycerol, 1 mM PMSF, 10 mM 2-mercaptoethanol) containing 500 mM KCl (BD500), and twice with 500 μl of BD500 containing 20 mM imidazole-HCl (pH 7.9). Bound proteins were eluted twice with 300 μl of BD500 containing 100 mM imidazole-HCl (pH 7.9). Typical preparations were >80% pure as judged by Coomassie blue staining of a sodium dodecyl sulfate (SDS)-polyacrylamide gel.
For large-scale preparations of 1 to 4 liters of IPTG-induced bacterial cultures, lysates were mixed with 10 ml of BB500 and purified on a Ni-NTA column (1-ml column volume; Qiagen) as described previously (55). After washing with 2 column volumes of the same buffer (BB500), 10 column volumes of BD500, and finally 2 column volumes of BD500 containing 20 mM imidazole-HCl (pH 7.9), expressed proteins (>95% pure, judging by Coomassie blue staining of an SDS-polyacrylamide gel) were eluted with 2 column volumes of BD500 containing 100 mM imidazole-HCl (pH 7.9). Large-scale preparations were carried out for four different coexpressed chimeric forms of TFIIE. Purification of recombinant human TATA-binding protein (TBP), TFIIB, TFIIF, and TFIIE has been described in detail elsewhere (32, 38, 42, 60).
HA-tagged and GST fusion proteins were expressed in E. coli BL21(DE3)pLysS by IPTG induction. Cells were harvested from 50 to 100 ml of culture, resuspended in 1 ml of BB500, and sonicated. Soluble lysates were separated from insoluble debris by ultracentrifugation at 20,000 × g using a 50.2 Ti rotor (Coulter-Beckman) and stored at −80°C until use.
In vitro transcription assays.
The general transcription factor TFIIH was purified either from HeLa nuclear extracts or from cytoplasmic S100 fractions as previously described (39). All other general transcription factors (TBP, TFIIB, TFIIF, and TFIIE) were purified essentially as follows: the recombinant proteins were expressed in E. coli, solubilized by sonication, and purified on a Ni-NTA agarose column. Pol II was purified to near-homogeneity from HeLa nuclear pellets by DE52, A25, P11, and high-performance liquid chromatography-DEAE 5PW columns. In vitro transcription was carried out as described previously (38). The plasmid pML(C2AT)Δ-50, which contains the adenovirus type 2 major late (AdML) promoter and gives a 390-nucleotide (nt) transcript, was used as a template for basal transcription assays (53). Autoradiography was performed at −80°C with Fuji RX-U X-ray film. The incorporation of [α-32P]CTP into transcripts was quantified using a Fuji BAS2500 Bio-Imaging analyzer.
Generation of antibodies against C. elegans TFIIE subunits.
Both 6H-ceTFIIE subunits were expressed independently in E. coli, solubilized by sonication, and purified on a Ni-NTA agarose column. Since 6H-ceTFIIEβ was mostly soluble (>80% in soluble lysate) and 6H-ceTFIIEα was mostly insoluble (>90% in pellet), 6H-ceTFIIEβ was purified from bacterial lysates and 6H-ceTFIIEα was purified from bacterial pellets after solubilization with 4 M guanidine-HCl (pH 7.5). Two milligrams of each purified protein was subjected to SDS-polyacrylamide gel electrophoresis (PAGE), and the appropriate bands were excised from the gel after Coomassie blue staining.
To raise rabbit polyclonal antibodies against ceTFIIEα, 500 μg of 6H-ceTFIIEα was mixed with complete Freund's adjuvant (Difco) and injected intramuscularly into each of two rabbits. Two weeks after the first injection, a second injection of 250 μg of 6H-ceTFIIEα mixed with incomplete Freund's adjuvant (Difco) was given by two methods, intramuscularly and subcutaneously. A third injection, identical to the second, was given after a further 2 weeks. Blood was collected 8 days after the third injection. The antibody generated recognized both natural and recombinant ceTFIIEα in solution and on Western blots.
Polyclonal antibodies against ceTFIIEβ were raised in rats. One hundred micrograms of 6H-ceTFIIEβ was mixed with complete Freund's adjuvant (Difco) and injected both subcutaneously and intraperitoneally into each of five rats. Two weeks after the first injection, a second injection of 100 μg of 6H-ceTFIIEβ mixed with incomplete Freund's adjuvant (Difco) was given in the same manner. Third and fourth injections, identical to the second, were given at two-week intervals. Blood was collected 8 days after the fourth injection. The antibody generated recognized both natural and recombinant ceTFIIEβ in solution and on Western blots.
Preparation of C. elegans embryonic nuclear extracts.
The Bristol N2 wild-type strain of C. elegans was grown in liquid culture essentially as described previously (56). Liquid cultures were started by seeding two 9-cm plates of N2 into 1 liter of S medium (10 mM potassium citrate [pH 6], 50 mM potassium phosphate [pH 6], 50 μM EDTA, 5 μg of cholesterol/ml, 3 mM CaCl2, 3 mM MgSO4, 25 μM FeSO4, 10 μM MnCl2, 10 μM ZnSO4, 1 μM CuSO4) in a 2-liter flask with a culture paste of E. coli OP50 from a 2-liter culture. Worms were grown at 22°C with shaking at 350 rpm for 4 days, and growth was monitored until most worms were gravid hermaphrodites. Growth synchronization of C. elegans was then carried out essentially as described previously (28), except that the culture volume was 6 liters. Final recovery of embryos was 2.8 g. To prepare embryonic nuclear extracts, the embryos were harvested and homogenized as described previously (28).
Coimmunoprecipitation and depletion of C. elegans TFIIE from the nuclear extract.
Rat polyclonal antisera against ceTFIIEβ (0.5 μl) and 6 μl (packed volume) of protein G-Sepharose 4FF (Amersham Pharmacia Biotech) were incubated in buffer C (20 mM Tris-HCl [pH 7.9 at 4°C], 0.5 mM EDTA, 20% [vol/vol] glycerol, 0.5 mM PMSF, 10 mM 2-mercaptoethanol, 0.002% [vol/vol] Nonidet P-40) containing 100 mM KCl (BC100) and 200 μg of bovine serum albumin (BSA)/ml for 2 h at 4°C with rotation. The protein G-Sepharose beads were precipitated and washed twice with 500 μl of buffer C containing 1 M KCl (BC1000) and twice with 500 μl of BC500. One hundred and fifty microliters of C. elegans nuclear extract (2.1 mg of protein/ml) pre-equilibrated with BC500 was then incubated with the prepared anti-ceTFIIEβ antibody-protein G beads in a 500-μl reaction volume for 4 h at 4°C with rotation. This step was repeated three times, and the resulting supernatant was used as a ceTFIIE-depleted nuclear extract. To check for complete depletion of ceTFIIE, the beads were washed twice with 500 μl of BC500 and twice with 500 μl of BC100 and boiled in SDS sample buffer, and the proteins released from the beads were analyzed by SDS–10% PAGE.
Coimmunoprecipitated ceTFIIE subunits were detected by Western blotting with either anti-ceTFIIEα rabbit antiserum (1:3,000 dilution) or anti-TFIIEβ rat polyclonal antiserum (1:3,000 dilution) after transfer to an Immobilon-P polyvinylidene difluoride membrane (Millipore) as described previously (38). Signals were detected using the enhanced chemiluminescence detection system (Amersham Pharmacia Biotech) and RX-U film (Fuji Film) after incubation of the immunoblots with horseradish peroxidase-linked secondary antibodies against rabbit or rat IgG as appropriate.
Primer extension reaction.
In vitro transcription reactions were performed essentially as described previously (28) except that the reaction temperature was 24°C, 200 ng of the AdML promoter pMLH1 (14) was used as a supercoiled DNA template, and the reaction mixture volume was 50 μl. C. elegans embryonic nuclear extract (42 μg of total protein) or ceTFIIE-depleted nuclear extract was used for each reaction. Transcription was stopped by the addition of 75 μl of 450 mM sodium acetate (pH 5.3)–10 mM EDTA–0.5% SDS–yeast tRNA (50 μg/ml). Primer extension reactions were carried out as described previously (29). A synthetic oligonucleotide (5′-CTGACAATCTTAGCGCAGAAGTCATG-3′) was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase and used as a primer. The products (92 nt) were analyzed on 12% denaturing polyacrylamide-urea gels. Autoradiography was performed at −80°C with Fuji RX-U X-ray film.
GST-pull down assay.
GST fusion proteins were used for protein interaction assays. Each protein to be tested (200 ng) was incubated with lysates containing 400 ng of GST fusion proteins together with 5 μl (packed volume) of glutathione-Sepharose (Amersham Pharmacia Biotech) in a 500-μl reaction mixture in BC100 containing 200 μg of BSA/ml for 4 h at 4°C with rotation. The glutathione-Sepharose resin was then washed twice with 500 μl of buffer C containing 200 mM KCl (BC200) and once with 500 μl of BC100 and boiled in SDS sample buffer. The proteins released from the resin were analyzed by SDS-PAGE and Western blotting as described above.
Kinase assay.
Kinase assays were carried out as described elsewhere (39) using the general transcription factors together with Pol II and a DNA fragment containing AdML promoter sequences from −39 to +29. Phosphorylation reactions were carried out at 30°C for 1 h and stopped by the addition of 75 μl of phosphorylation stop solution (10 mM EDTA, 0.1% Nonidet P-40, 0.05% SDS). Phosphorylated proteins were precipitated with trichloroacetic acid, analyzed by SDS–5% PAGE (5.5% acrylamide), and detected by autoradiography performed at −80°C with Fuji RX-U X-ray film. The extent of 32P-phosphorylation of the CTD of the Pol II largest subunit was quantified using a Fuji BAS2500 Bio-Imaging analyzer.
Transcription transition assay.
To measure the effects of various TFIIE constructs on the transition from initiation to elongation, the pML(C2AT)100 insert was constructed by PCR using the 5′ oligonucleotide ML100-1T (5′-GACTATCTAGAGTGTTCCTGAAGGGGG-3′) to create an XbaI site (underlined) at the 5′ end of the AdML core promoter and the 3′ oligonucleotide ML100-1B (5′-CGATCTCCCGGGAAATATAGAAGAAGGAG-3′) to create a SmaI site (underlined) at the 3′ end of the short (97 bp) G-less cassette. The product of a PCR using these promers and pML(C2AT)Δ-50 as a template was subcloned into the SmaI site of pBluescript SK(−) (Stratagene) to yield the pML(C2AT)100 transcription template, which gives a short 107-nt transcript. To provide a linear template, pML(C2AT)100 was digested with SmaI. PICs for use in the transcription transition assays were performed in a 15-μl reaction mixture containing either no TFIIE or 15 ng of one of the four different TFIIE proteins, together with all other general transcription factors, Pol II, and 100 ng of the pML(C2AT)100 template (linear or supercoiled). These mixtures were incubated for 45 min at 28°C under the in vitro transcription conditions, except that no nucleoside triphosphates were added. Transcription was then initiated by addition of 15 μl of reaction mixture containing 6 μCi of [α-32P]CTP (400 Ci/mmol; Amersham Pharmacia Biotech), 50 μM ATP, 50 μM UTP, 12.5 μM CTP, 40 μM 3′-O-methyl GTP, and 0.15 U of RNase T1 (Amersham Pharmacia Biotech). After transcription, reactions were stopped by heat treatment for 3 min at 68°C and treated with 4 U of calf intestine alkaline phosphatase for 20 min at 37°C to reduce the background signal due to nonincorporated [α-32P]CTP. The reactions were stopped, and transcripts were then ethanol precipitated and analyzed on 10% denaturing polyacrylamide-urea gels. Autoradiography was performed at −80°C with Fuji RX-U X-ray film. The incorporation of [α-32P]CTP into transcripts was quantified using a Fuji BAS2500 Bio-Imaging analyzer.
Transcription initiation reaction.
Initiation reactions were performed essentially as described previously (13, 20). Each reaction (15 μl) contained 40 mM Hepes-KOH (pH 8.4), 12 mM Tris-HCl (pH 7.9 at 4°C), 60 mM KCl, 4 mM MgCl2, 0.3 mM EDTA, 12% (vol/vol) glycerol, 0.3 mM PMSF, 6 mM 2-mercaptoethanol, and 150 μg of BSA/ml. By preincubation for 45 min at 28°C, the preinitiation complex was formed with 10 ng of the 158-bp AflIII-ScaI fragment of pMLH1 (containing the AdML promoter sequence from −111 to +47) (14) together with Pol II and all general transcription factors, except that TBP was used instead of TFIID and hTFIIE was replaced with various combinations of chimeric TFIIE. Transcription initiation was then carried out for 45 min at 28°C by addition of 5 μl of reaction mixture containing 60 μM ATP and 10 μCi [α-32P]CTP (800 Ci/mmol; NEN) in the same buffer composition. Reactions were stopped by heat treatment for 3 min at 68°C and treated with 10 U of calf intestine alkaline phosphatase for 20 min at 37°C. After inactivation for 10 min at 75°C, transcripts were analyzed on 20% denaturing polyacrylamide-urea gels. Autoradiography was performed at −80°C with Fuji RX-U X-ray film.
Nucleotide sequence accession numbers.
The EMBL accession numbers for the ceTFIIEα and ceTFIIEβ cDNA sequences are Y08816 and Y08815, respectively.
RESULTS
Isolation of C. elegans TFIIE cDNAs.
In order to examine the relationship between TFIIE structure and its role in transcription, we have been isolating human TFIIE homologs from different species and studying their structural and functional similarities to their human counterparts. We isolated both subunits of a TFIIE homolog from Xenopus laevis (xTFIIE), and other groups isolated homologs from Drosophila melanogaster (dTFIIE) and yeast Saccharomyces cerevisiae (yTFIIE) (11, 36, 37, 68). When the functional exchangeabilities of these TFIIE homologs with human TFIIE were tested, xTFIIE and dTFIIE were functionally exchangeable in the human in vitro reconstituted transcription system but yTFIIE was not. In amino acid sequence comparison with their human counterparts, xTFIIE has 79% identity in xTFIIEα and 84% in xTFIIEβ and dTFIIE has 46% identity in dTFIIEα and 59% in dTFIIEβ, whereas yTFIIE has only 22% identity in yTFIIEα and 23% in yTFIIEβ. Judging from these data, the border of a functional exchangeability may evolutionarily lie between Drosophila and yeast, indicating that an hTFIIE homolog from C. elegans will be of use in the examination of structure-function relationships in TFIIE.
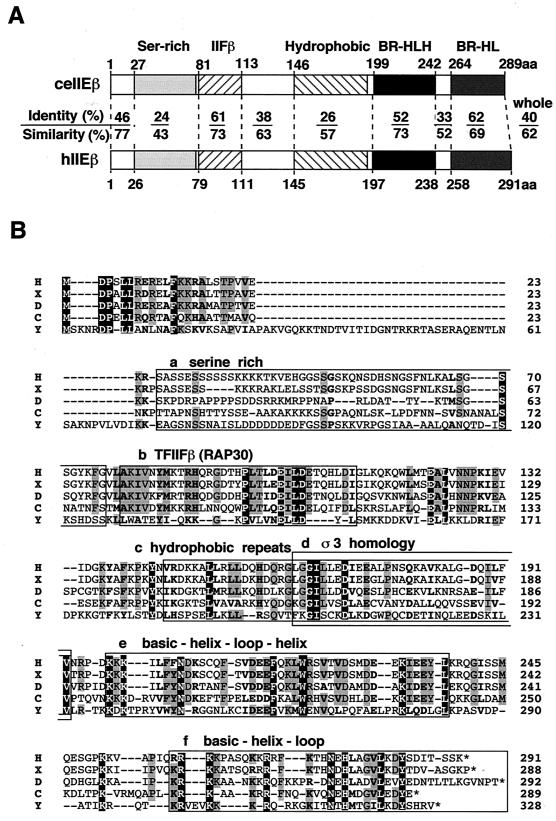
Here we report the isolation of a putative C. elegans TFIIEβ (ceTFIIEβ) that perfectly matched the open reading frame predicted from the sequence of C. elegans genomic DNA (chromosome II). The ceTFIIEβ cDNA was about 0.9 kb in length and encoded a highly basic 289-amino-acid protein (pI, 9.3) with a calculated molecular weight of 33.0 kDa. As shown in Fig. 1A, comparison of the predicted amino acid sequence of ceTFIIEβ with that of hTFIIEβ (57) revealed relatively high conservation throughout the entire sequence (40% identity and 62% similarity), except for the region corresponding to the serine-rich sequence (residues 27 to 78; 24% identity and 43% similarity). The region corresponding to the C-terminal 7 amino acid residues of hTFIIEβ was missing in ceTFIIEβ. These results are consistent with our earlier conclusions, drawn from the basal transcription activities of hTFIIEβ deletion mutants, that the N-terminal 50 amino acid residues are dispensable, and the C-terminal 14 residues are stimulatory but not essential, for transcription (42). Figure 1B shows an alignment of the amino acid sequences of TFIIEβ homologs from five different species. The lack of sequence conservation in the serine-rich region was more obvious when this region was compared among five species. On the whole, aromatic residues and many hydrophobic residues were well conserved. In addition, charge-carrying acidic and basic residues (especially in the TFIIFβ [RAP30] homology region) were well conserved, as were the basic region-helix-loop-helix (BR-HLH) and the BR-HL motifs.
FIG. 1.
Sequence analysis of C. elegans TFIIEβ. (A) Comparison of ceTFIIEβ (ceIIEβ) with hTFIIEβ (hIIEβ). The serine-rich sequence (Ser-rich), a region similar to the small subunit of TFIIF, TFIIFβ (IIFβ), a hydrophobic region (Hydrophobic), a BR-HLH and a BR-HL are indicated (42). In the middle panel, identity (above) and similarity (below) between ceIIEβ and hIIEβ over each structural motif and characteristic sequence region are indicated as percentages. The numbers presented above and below the diagram indicate the amino acid residues that delimit each structure. (B) Sequence alignment of TFIIEβ from five different species. Amino acid sequences of TFIIEβ from human (H), X. laevis (X), D. melanogaster (D), C. elegans (C), and yeast S. cerevisiae (Y) were aligned. Completely identical residues are shaded in black, and residues identical in four species are shaded in gray. Conserved similar residues are shown in bold type. Identical and similar amino acids were assigned as described previously (36). Putative structural motifs and characteristic sequences are shown as described for Fig. 1A and boxed (boxes a to f), except that a ς3 homology region (box d, ς3 homology) is additionally indicated. A hyphen indicates a gap.
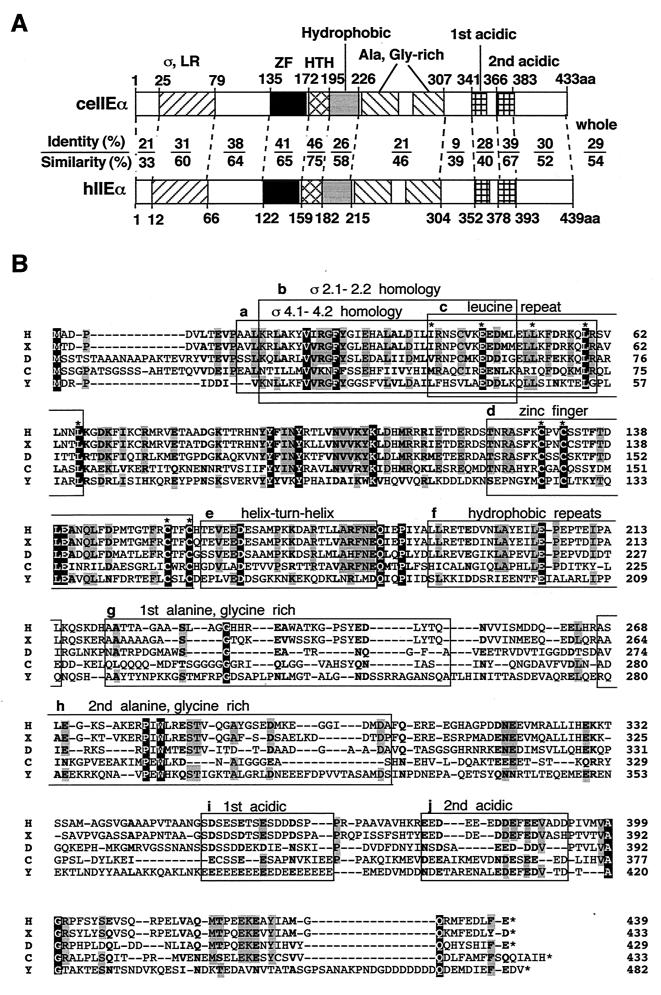
We then isolated a putative C. elegans TFIIEα (ceTFIIEα) using the same strategies as for ceTFIIEβ. The ceTFIIEα cDNA was 1.5 kb in length and was identical to the open reading frame predicted from the genomic sequence (chromosome IV). It included a stop codon (TAA) located 12 bases upstream of the translation start codon (ATG) and nucleotide sequences flanking this translation start codon which matched the Kozak sequence (8 of 10 nt) and encoded a highly acidic 433-amino-acid protein (pI, 4.8) with a calculated molecular weight of 49.1 kDa. Comparison of C. elegans and human (40) TFIIEα sequences revealed 29% identity and 54% similarity over the entire sequence (Fig. 2A). These values were approximately 10% lower than the corresponding values for TFIIEβ. However, the region between residues 25 and 195 of ceTFIIEα, which may correspond to a region necessary for transcription in hTFIIEα (38), showed 37% identity and 64% similarity to hTFIIEα, which represents a level of sequence conservation similar to that observed for TFIIEβ. The second acidic region (residues 366 to 383), which corresponds to the TFIIH binding region of hTFIIEα (residues 378 to 393), also showed strong sequence conservation (39% identity and 67% similarity), although the rest of the C-terminal half of ceTFIIEα (residues 196 to 365 and 384 to 433) showed only 22% identity and 46% similarity to the human homolog (Fig. 2A). Fig. 2B shows an alignment of the amino acid sequences of TFIIEα from five different species; regions of higher sequence conservation, which may indicate functional domains, were more obvious in this wider alignment. As was observed for TFIIEβ, many aromatic residues, most of which are located in the N-terminal half and very near the C terminus (Tyr-414, Phe-422, and Phe-426 in ceTFIIEα), were strongly conserved. Hydrophobic residues, most of which are located in the N terminus and especially in the leucine repeat region, were also well conserved. In addition, the positions of four cysteine residues in the zinc finger motif were perfectly conserved, including the internal spacing (21 amino acid residues).
FIG. 2.
Sequence analysis of C. elegans TFIIEα. (A) Comparison of ceTFIIEα (ceIIEα) with hTFIIEα (hIIEα). The region with ς subdomain homology and a leucine repeat (ς, LR), a zinc finger motif (ZF), a helix-turn-helix motif (HTH), a hydrophobic region (Hydrophobic), two regions rich in alanine and glycine residues (Ala, Gly-rich), and the first and second acidic regions (2nd acidic) are indicated (38). Identity (above) and similarity (below) between ceIIEα and hIIEα over each structural motif and characteristic sequence region are indicated as shown in Fig. 1A. (B) Sequence alignment of TFIIEα from five different species. Amino acid sequences of TFIIEα from four species were aligned as shown in Fig. 1B. Identical and similar amino acids were assigned as described previously (37). Putative structural motifs and characteristic sequences are shown as described for panel A and boxed (boxes a to j), except that two overlapping ς subdomain homology regions (box a, ς 2.1–2.2 homology, and box b, ς 4.1–4.2 homology) and two alanine, glycine-rich regions (box g, 1st alanine, glycine-rich, and box h, 2nd alanine, glycine-rich) are additionally indicated. A hyphen indicates a gap.
Recombinant C. elegans TFIIE is identical to the natural form.
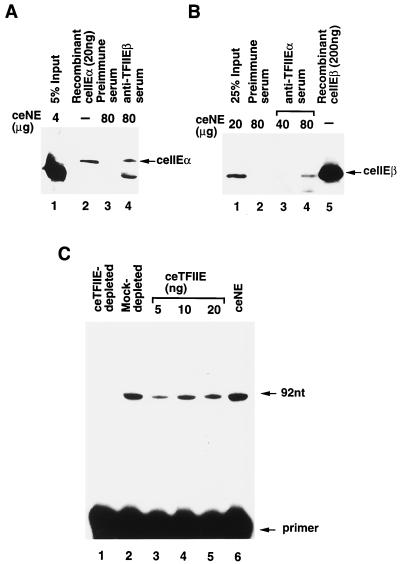
In order to confirm that we had isolated bona fide ceTFIIE cDNAs, both ceTFIIEβ and ceTFIIEα were expressed independently in bacteria with N-terminal six-histidine tags and purified on a Ni-NTA agarose column. Polyclonal antibodies were raised against recombinant ceTFIIE subunits by immunizing experimental animals with specific bands excised from SDS-PAGE gels. Recombinant and natural ceTFIIE subunits were compared with respect to migration on SDS-PAGE, antibody recognition, and function in primer extension assays (Fig. 3). Each subunit of natural ceTFIIE was immunoprecipitated from C. elegans embryonic nuclear extracts, and the precipitates were examined to determine whether the other subunit was coimmunoprecipitated. Rat anti-ceTFIIEβ antiserum was used to coimmunoprecipitate natural ceTFIIEα, which produced a band of the same size as recombinant ceTFIIEα on Western blots probed with rabbit anti-ceTFIIEα antiserum (Fig. 3A, lane 4 versus lane 2). Natural ceTFIIEβ was similarly coimmunoprecipitated with rabbit anti-ceTFIIEα antiserum and produced a band of the same size as recombinant ceTFIIEβ on Western blots probed with rat anti-ceTFIIEβ antiserum (Fig. 3B, lanes 3 and 4 versus lane 5).
FIG. 3.
Identification of natural C. elegans TFIIE. (A) Coimmunoprecipitation of natural ceTFIIEα with ceTFIIEβ. To demonstrate association of natural ceTFIIEα with ceTFIIEβ, 80 μg of C. elegans embryonic nuclear extract was incubated with anti-ceTFIIEβ antibody–protein G-Sepharose, and natural ceTFIIEβ was precipitated. After SDS-PAGE on a 10% polyacrylamide gel, coprecipitated ceTFIIEα was detected with anti-ceTFIIEα rabbit antibody after Western blotting. Lane 1, 5% input of embryonic nuclear extract (4 μg); lane 2, recombinant ceTFIIEα (20 ng); lane 3, nuclear extract treated with preimmune serum (80 μg); lane 4, nuclear extract treated with anti-ceTFIIEβ serum (80 μg). An arrow indicates the position of ceTFIIEα (ceIIEα). (B) Coimmunoprecipitation of natural ceTFIIEβ with ceTFIIEα. The same strategy as detailed for panel A was employed to study the association of natural ceTFIIEβ with ceTFIIEα. Eighty micrograms of nuclear extract was incubated with anti-ceTFIIEα antibody–protein G-Sepharose, and natural ceTFIIEα was precipitated. Coprecipitated ceTFIIEβ was detected by anti-ceTFIIEβ rabbit antibody after Western blotting. Lane 1, 25% input of embryonic nuclear extract (20 μg); lane 2, nuclear extract treated with preimmune serum (80 μg); lanes 3 and 4, increasing amounts of nuclear extract treated with anti-ceTFIIEβ serum (40 and 80 μg, respectively); lane 5, recombinant ceTFIIEβ (200 ng). An arrow indicates the position of ceTFIIEβ (ceIIEβ). (C) Transcription complementation assay of ceTFIIE. ceTFIIE was depleted from nuclear extracts by treatment with anti-ceTFIIEβ antibody–protein G-Sepharose. In the same way, mock-depleted nuclear extracts were prepared by treatment with preimmune IgG–protein G-Sepharose. Complementation of natural ceTFIIE was studied by adding increasing amounts of purified recombinant ceTFIIE and carrying out primer extension reactions. Lane 1, ceTFIIE-depleted nuclear extracts (42 μg); lane 2, mock-depleted nuclear extracts (42 μg); lanes 3 to 5, ceTFIIE-depleted nuclear extracts (42 μg) with increasing amounts of recombinant ceTFIIE (5, 10, and 20 ng, respectively); lane 6, C. elegans nuclear extracts (42 μg). Arrows indicate the positions of the reverse transcript (92 nt) and the primer.
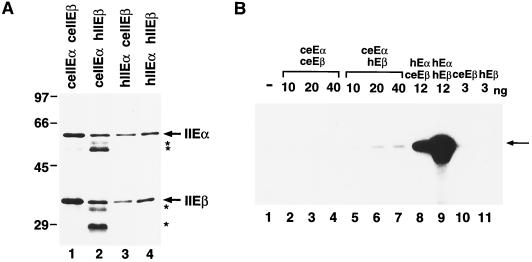
We next tested whether recombinant ceTFIIE could functionally replace natural ceTFIIE in transcription using primer extension analysis (Fig. 3C). Before doing this, we prepared soluble recombinant ceTFIIE. Since it was reported that three subunits of human replication protein A, which were almost insoluble when expressed independently, could be expressed in a soluble form by subcloning their cDNAs in tandem in bacterial expression vectors (15), we applied this method to ceTFIIEα and hTFIIEα by adding a six-histidine tag at the N terminus and coexpressing them with either ceTFIIEβ or hTFIIEβ in bacteria. The four different subunit combinations of TFIIE were then readily purified on a Ni-NTA agarose column. Figure 4A shows SDS-PAGE analysis of these chimeric proteins. As expected, six-histidine-tagged ceTFIIEα (6H-ceTFIIEα) formed a soluble complex with either ceTFIIEβ (Fig. 4A, lane 1) or hTFIIEβ (lane 2), although several degraded polypeptides were observed in the case of purified TFIIE with ceTFIIEα and hTFIIEβ (ceTFIIEα-hTFIIEβ) (lane 2). 6H-ceTFIIEα migrated slightly faster than 6H-hTFIIEα (58 kDa versus 59 kDa) (Fig. 4A, lanes 1 and 2 versus lanes 3 and 4). Finally, natural ceTFIIE was depleted from C. elegans embryonic nuclear extracts using rat anti-ceTFIIEβ antiserum. As shown in Fig. 3C, almost no transcription activity was observed in this depleted extract (lane 1), whereas mock depletion with rat preimmune serum did not alter the transcription activity of the nuclear extract (lane 2 versus lane 6). Addition of increasing amounts of recombinant ceTFIIE restored the transcription activity of ceTFIIE-depleted embryonic nuclear extract (Fig. 3C, lanes 3 to 5). These results indicate that recombinant ceTFIIE was identical to natural ceTFIIE.
FIG. 4.
Characterization of recombinant C. elegans TFIIE. (A) SDS-PAGE analysis of four different recombinant TFIIE proteins, with subunits from either human or C. elegans. Recombinant TFIIE subunits were coexpressed in four different combinations from the coexpression plasmids described in Materials and Methods, purified, and analyzed by SDS–10% PAGE (lanes 1 to 4). The sizes of molecular mass markers are indicated on the left (in kilodaltons). The approximate positions of TFIIEα (IIEα) and TFIIEβ (IIEβ) are indicated by arrows on the right. Asterisks indicate degradation products derived from ceTFIIEα and hTFIIEβ in lane 2. (B) Basal transcription activities of chimeric TFIIE. In vitro transcription assays were carried out with increasing amounts of chimeric TFIIE proteins. Lane 1, no TFIIE (−); lanes 2 to 4, 10, 20, and 40 ng of TFIIE made up of 6H-ceTFIIEα and nontagged ceTFIIEβ (ceEαceEβ); lanes 5 to 7, 10, 20, and 40 ng of TFIIE made up of 6H-ceTFIIEα and nontagged hTFIIEβ (ceEαhEβ); lane 8, 12 ng of TFIIE made up of 6H-hTFIIEα and nontagged ceTFIIEβ (hEαceEβ); lane 9, 12 ng of TFIIE made up of 6H-hTFIIEα and nontagged hTFIIEβ (hEαhEβ); lanes 10 and 11, 3 ng of 6H-ceTFIIEβ (ceEβ) and 6H-hTFIIEβ (hEβ), respectively. The arrow indicates the position of the specific transcript (390 nt).
Transcriptional exchangeabilities of ceTFIIE subunits with their human counterparts.
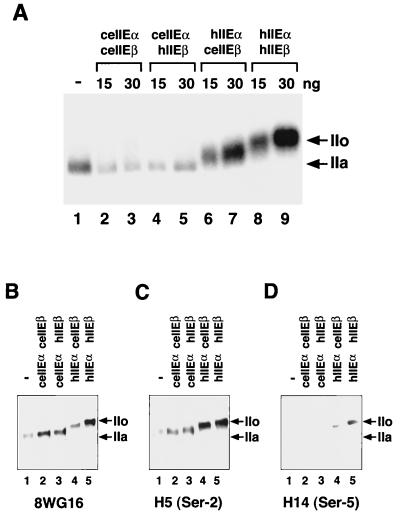
We then tested the abilities of ceTFIIE subunits to functionally replace their human counterparts using four different subunit combinations of TFIIE in a human in vitro transcription system with a supercoiled template (Fig. 4B). Although ceTFIIEβ, when complexed with hTFIIEα, showed approximately 30% of wild-type hTFIIE activity (lanes 8 and 9), ceTFIIEα showed less than 5% of the wild-type activity when complexed with hTFIIEβ (lanes 5 to 7). Moreover, wild-type ceTFIIE showed almost no activity regardless of the amount added (lanes 2 to 4). These results demonstrate that ceTFIIEβ can partially replace hTFIIEβ but ceTFIIEα cannot replace hTFIIEα in a human in vitro transcription system with a supercoiled template.
C. elegans and human TFIIEβ show similar binding specificities to human general transcription factors.
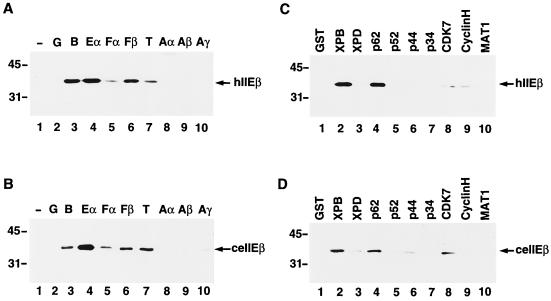
In an attempt to determine why ceTFIIEβ was only partially able to replace its human counterpart, the binding specificities of ceTFIIEβ and hTFIIEβ for human general transcription factors were compared using GST-pull down assays (Fig. 5). Both C. elegans and human TFIIEβ bound strongly to TFIIB, TFIIEα, and TFIIEβ (RAP30), weakly to TFIIFα (RAP74) and TBP, and very weakly to TFIIAγ (Fig. 5A and B). The human TFIIH subunits were similarly tested (Fig. 5C and D); both C. elegans and human TFIIEβ bound to XPB, p62, and Cdk7, albeit relatively weakly (Fig. 5C and D, lanes 2, 4, and 8). The only difference detected between ceTFIIEβ and hTFIIEβ was that hTFIIEβ bound weakly to cyclin H (Fig. 5C, lane 9), whereas ceTFIIEβ did not bind to cyclin H but did bind weakly to p44 instead (Fig. 5D, lane 6). In addition, since we observed that human Pol II bound predominantly to TFIIB and hTFIIEβ, the binding of human Pol II to ceTFIIEβ was also tested; weaker but significant binding (about half-efficiency relative to that of hTFIIEβ) was observed (Y. Ohkuma, data not shown). Judging from these results, there was not much difference between ceTFIIEβ and hTFIIEβ in binding to the human general transcription factors. It therefore remains difficult to explain the partial exchangeability of ceTFIIEβ with hTFIIEβ simply from these binding results.
FIG. 5.
Binding assays using C. elegans TFIIEβ. (A) Binding of hTFIIEβ to the various general transcription factors. All of the human general transcription factors (400 ng each) except for hTFIIEβ, the TBP activation factors of TFIID, and the TFIIH subunits were fused to GST and expressed in bacteria. GST-pull down assays were carried out with 200 ng of 6H-hTFIIEβ. After SDS-PAGE on a 10% polyacrylamide gel and Western blotting, bound 6H-hTFIIEβ was detected with anti-hTFIIEβ antibody. Lane 1, control bacterial lysate (no GST protein) (−); lane 2, GST alone (no fusion protein) (G); lane 3, GST-TFIIB (B); lane 4, GST-hTFIIEα (Eα); lane 5, GST-TFIIFα (Fα); lane 6, GST-TFIIFβ (Fβ); lane 7, GST-TBP (T); lane 8, GST-TFIIAα (Aα); lane 9, GST-TFIIAβ (Aβ); lane 10, GST-TFIIAγ (Aγ). An arrow indicates the position of 6H-hTFIIEβ (hIIEβ). (B) Binding of ceTFIIEβ to the various general transcription factors. GST-pull down assays were carried out as described for panel A, except that HA-ceTFIIEβ was used instead of 6H-hTFIIEβ. After Western blotting, bound HA-ceTFIIEβ was detected with anti-HA monoclonal antibody (12CA5). An arrow indicates the position of HA-ceTFIIEβ (ceIIEβ). (C) Binding of hTFIIEβ to the TFIIH subunits. The assay was done as described for panel A. Four hundred nanograms of each GST-fused TFIIH subunit, with GST alone (lane 1) as a control, were used to examine binding to hTFIIEβ. An arrow indicates the position of 6H-hTFIIEβ (hIIEβ). (D) Binding of ceTFIIEβ to the TFIIH subunits. GST-pull down assays were carried out as described for panel C, except that HA-ceTFIIEβ was used instead of 6H-hTFIIEβ. An arrow indicates the position of HA-ceTFIIEβ (ceIIEβ). Molecular mass markers are shown to the left.
C. elegans TFIIEα does not bind efficiently to human TFIIEβ.
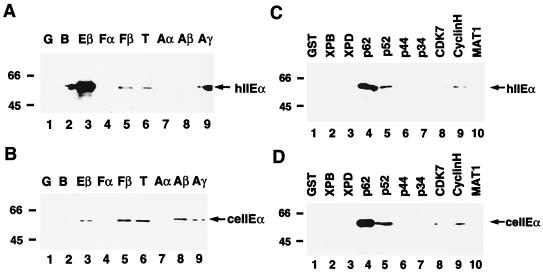
In contrast to ceTFIIEβ, ceTFIIEα was unable to replace hTFIIEα in a human in vitro transcription system (Fig. 4B). To address this issue, the binding specificities of ceTFIIEα and hTFIIEα for various human general transcription factors were analyzed (Fig. 6). Of the general transcription factors, hTFIIEα bound most strongly to hTFIIEβ (Fig. 6A, lane 3). However, ceTFIIEα did not bind well to hTFIIEβ (about 10% of the efficiency of hTFIIEα) (Fig. 6A and B, compare lanes 3). In addition, ceTFIIEα bound more strongly to TFIIAβ than hTFIIEα did (Fig. 6A and B, lanes 8). Binding to TFIIB, TFIIFβ (RAP30), and TBP was similar for C. elegans and human TFIIEα (Fig. 6A and B, lanes 2, 5, and 6), and both also bound predominantly to p62 and weakly to p52 among the nine TFIIH subunits (Fig. 6C and D, lanes 4 and 5). These results indicate that a different binding affinity to hTFIIEβ might be a main reason for the inability of ceTFIIEα to substitute functionally for hTFIIEα.
FIG. 6.
Binding assays using C. elegans TFIIEα. (A) Binding of hTFIIEα to the various general transcription factors. All of the human general transcription factors (400 ng each) except for hTFIIEα, the TBP activation factors of TFIID, and the TFIIH subunits were fused to GST and expressed in bacteria. GST-pull down assays were carried out as described for Fig. 5A using 200 ng of 6H-hTFIIEα. After Western blotting, bound hTFIIEα was detected with anti-hTFIIEα antibody. Lane 1, GST alone (G); lane 2, GST-TFIIB (B); lane 3, GST-hTFIIEβ (Eβ); lane 4, GST-TFIIFα (Fα); lane 5, GST-TFIIFβ (Fβ); lane 6, GST-TBP (T); lane 7, GST-TFIIAα (Aα); lane 8, GST-TFIIAβ (Aβ); lane 9, GST-TFIIAγ (Aγ). Arrows indicate the position of 6H-hTFIIEα (hIIEα). (B) Binding of ceTFIIEα to the various general transcription factors. GST-pull down assays were carried out as described for panel A using 200 ng of HA-ceTFIIEα. After Western blotting, bound HA-ceTFIIEα was detected with anti-HA monoclonal antibody (12CA5). An arrow indicates the position of HA-ceTFIIEα (ceIIEα). (C) Binding of hTFIIEα to the TFIIH subunits. Assays were carried out as described for panel A. Four hundred nanograms of each GST-fused TFIIH subunit, with GST alone (lane 1) as a control, were used to examine binding to hTFIIEα. An arrow indicates the position of 6H-hTFIIEα (hIIEα). (D) Binding of ceTFIIEα to the TFIIH subunits. Assays were carried out as described for panel C, except that HA-ceTFIIEα was used instead of 6H-hTFIIEα. An arrow indicates the position of HA-ceTFIIEα (ceIIEα). Molecular mass markers are indicated to the left.
The intermediate shift in the Pol II phosphorylation state induced by C. elegans TFIIEβ may be caused by defective phosphorylation at serine-5 of the CTD heptapeptide sequence.
In light of accumulated evidence suggesting a tight connection between CTD phosphorylation and transcription and our observation that ceTFIIEβ is partially able to replace its human counterpart in transcription, we investigated the effects of four different chimeric forms of TFIIE on CTD phosphorylation during PIC formation (Fig. 7A). Wild-type hTFIIE fully stimulated CTD phosphorylation, causing the largest subunit of Pol II to shift completely from the IIa to the IIo form (lanes 8 and 9). In contrast, chimeric TFIIE containing hTFIIEα and ceTFIIEβ (hTFIIEα-ceTFIIEβ) caused an intermediate shift of Pol II to a point between the IIa and IIo forms (lanes 6 and 7), while neither wild-type ceTFIIE nor TFIIE containing ceTFIIEα and hTFIIEβ (ceTFIIEα-hTFIIEβ) produced any significant Pol II shift upon phosphorylation reaction (lanes 2 to 5).
FIG. 7.
Effects of ceTFIIE subunits on CTD phosphorylation by TFIIH during preinitiation complex formation. Kinase assays (25 μl) were carried out as described in Materials and Methods. TFIIE proteins were prepared and purified as described for Fig. 4A and in Materials and Methods. (A) CTD phosphorylation of intact Pol II during PIC formation. Lane 1, kinase reaction without TFIIE (−); lanes 2 and 3, ceTFIIE (ceIIEαceIIEβ); lanes 4 and 5, chimeric TFIIE made up of ceTFIIEα and hTFIIEβ (ceIIEαhIIEβ); lanes 6 and 7, chimeric TFIIE made up of hTFIIEα and ceTFIIEβ (hIIEαceIIEβ); lanes 8 and 9, hTFIIE (hIIEαhIIEβ). For lanes 2, 4, 6, and 8, 15 ng of each TFIIE was added. For lanes 3, 5, 7, and 9, 30 ng of each TFIIE was added. Phosphorylated proteins were analyzed on a 5.5% acrylamide-SDS gel and detected by autoradiography. Arrows indicate the positions of the phosphorylated (IIo) and unphosphorylated (IIa) forms of the largest subunit of Pol II. (B) Detection of the largest subunit of Pol II after treatment with kinases. The kinase reaction was carried out essentially as described for panel A, except that nonisotopic ATP was used instead of [γ-32P]ATP and a monoclonal antibody (8WG16) against the CTD heptapeptide was used to detect the largest subunit of Pol II. (C) Detection of phosphorylated Ser-2 in the CTD of the largest subunit of Pol II. Reactions were carried out as described for panel B, and phospho-Ser-2 in the CTD was detected using the monoclonal antibody H5. (D) Detection of phosphorylated Ser-5 in the CTD of the largest subunit of Pol II. Reactions were carried out as described for panel B, and phospho-Ser-5 in the CTD was detected using the monoclonal antibody H14.
Recently, CTD phosphorylation sites have been reported from studies using a GST-CTD fusion protein, CTD heptapeptide repeat peptides, and a free form of intact Pol II; for example, TFIIH phosphorylates Ser-5, and Cdk8-cyclin C of the SRB- and Med-containing complex NAT phosphorylates Ser-2 and Ser-5 (58, 64). Since TFIIH is the only CTD kinase to exist in the reconstituted active PIC at transcription, we thought that phosphorylation of the CTD heptapeptide repeats (YSPTSPS; the first Tyr [Y] is here designated Tyr-1) of Pol II in the active PIC must correspond to the conformational change of Pol II to be processive. To identify the site(s) of CTD phosphorylation, we carried out phosphorylation reactions in the active PIC using intact Pol II and analyzed the largest subunit of Pol II by Western blotting with the following monoclonal antibodies: 8WG16, which preferentially detects the hypophosphorylated form of the CTD; H5, which detects phosphorylated Ser-2; and H14, which detects phosphorylated Ser-5 (Fig. 7B to D) (47, 63). Using this method, we recently observed that TFIIH phosphorylated both Ser-2 and Ser-5, but Ser-5 phosphorylation was dependent almost entirely on TFIIE both in solution and in the PIC (69). In the present study, both wild-type TFIIE and chimeric TFIIE (hTFIIEα-ceTFIIEβ) actively phosphorylated Pol II, although the shift induced by hTFIIEα-ceTFIIEβ was relatively lower than that induced by hTFIIE (Fig. 7B, lane 4 versus lane 5). In contrast, neither ceTFIIE nor another chimeric TFIIE (ceTFIIEα-hTFIIEβ) produced any significant shift (Fig. 7B, lanes 1 to 3). When the phosphorylation sites were analyzed, active chimeric TFIIE (hTFIIEα-ceTFIIEβ) and wild-type hTFIIE phosphorylated Ser-2 to a similar extent (Fig. 7C, lanes 4 and 5). However, intriguingly, this hTFIIEα-ceTFIIEβ induced Ser-5 phosphorylation very little, in clear contrast to hTFIIE, which strongly induced Ser-5 phosphorylation (Fig. 7D, lane 4 versus lane 5).
C. elegans TFIIEβ shows a severe defect in its ability to support transcription at the transition from initiation to elongation on a linear DNA template.
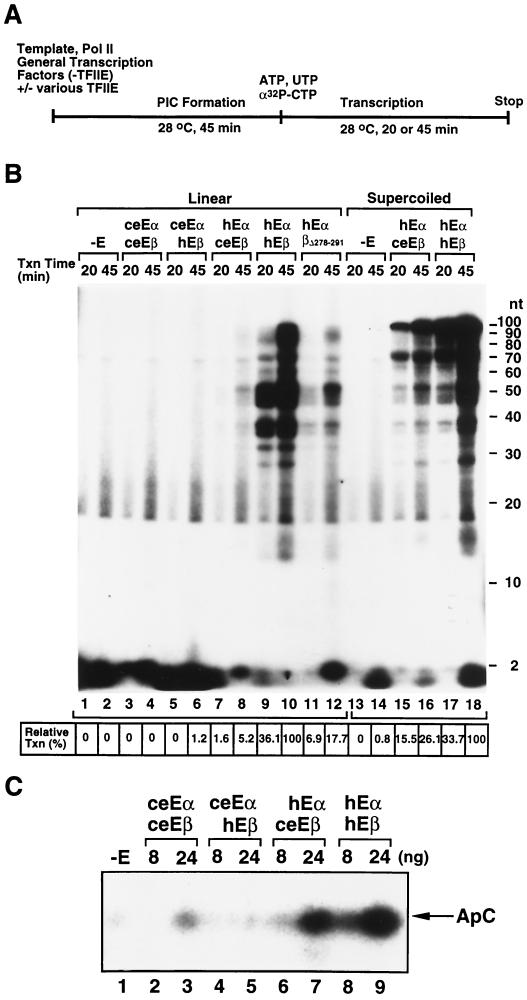
Since ceTFIIEβ showed a partial transcriptional exchangeability with hTFIIEβ on a supercoiled template and failed to induce Ser-5 phosphorylation well in the CTD of Pol II, we thought this defect of Ser-5 phosphorylation was a potential reason for the partial exchangeability. Thus, we characterized its function in transcription by focusing on the transition step to elongation. As shown in Fig. 8A, the PICs were preformed by incubation of Pol II with general transcription factors (with or without various forms of TFIIE) and either a linear or supercoiled AdML template [pML(C2AT)100]. Transcription was started by addition of nucleoside triphosphates at a low concentration to limit Pol II processivity so as to allow study of the transition stage. Although transcription occurred at a very low level (0.8% of transcription in the presence of wild-type hTFIIE; Fig. 8B, lane 14 versus lane 18) even in the absence of TFIIE on the supercoiled template, hTFIIE was stringently required for efficient transcription on both linear and supercoiled templates (lanes 9, 10, 17, and 18). Chimeric TFIIE (hTFIIEα-ceTFIIEβ) showed 26% of wild-type hTFIIE transcription activity on the supercoiled template (Fig. 8B, lane 16 versus lane 18), a level consistent with the results shown in Fig. 4B. However, hTFIIEα-ceTFIIEβ showed only 5% of wild-type hTFIIE transcription on the linear template (Fig. 8B, lane 8 versus lane 10). The reason why we did not observe shorter (abortive) transcripts as well as unincorporated [α-32P]CTP so much may be that we treated samples with calf intestine alkaline phosphatase as described previously (21) and ethanol precipitated to reduce the background. The hTFIIEβ Δ278–291 mutant showed similarly reduced transcription (about 20% of the wild-type level) on both templates (Fig. 8B, lanes 11 and 12) (Ohkuma, data not shown; 42). This is clearly different from the transcription activity of hTFIIEα-ceTFIIEβ. To examine whether this difference between linear and supercoiled templates reflects the difference at the transition stage to elongation, we carried out transcription initiation assays to see the first phosphodiester bond formation (Fig. 8C). The activity of hTFIIEα-ceTFIIEβ was also about 30% of that of wild-type hTFIIE at initiation (lanes 6 and 7 versus lanes 8 and 9). These results strongly indicate that hTFIIEα-ceTFIIEβ has a severe defect in the transition activity from transcription initiation to elongation and, in other words, TFIIE is directly involved in this stage.
FIG. 8.
Involvement of TFIIE in the transcription transition step. (A) Schematic representation of the transcription transition assay. Pol II and the general transcription factors were preincubated on either linear or supercoiled AdML template pML(C2AT)100. Transcription was initiated by the addition of nucleotides. Both templates give a 107-nt transcript. (B) Effects of ceTFIIE subunits on the transcription transition. Transcription was carried out for 20 min (odd-numbered lanes) or 45 min (even-numbered lanes) in the presence of the various TFIIE proteins listed on the top or in the absence of TFIIE. To the right of the panel, the sizes of markers are shown in nucleotides. Lane 1, reaction without TFIIE (−E); lanes 2 and 3, ceTFIIE (ceEαceEβ); lanes 4 and 5, chimeric TFIIE made up of ceTFIIEα and hTFIIEβ (ceEαhEβ); lanes 6 and 7, chimeric TFIIE made up of hTFIIEα and ceTFIIEβ (hEαceEβ); lanes 8 and 9, hTFIIE (hEαhEβ). Lanes 1 to 12, transcription on a linear template; lanes 13 to 18, transcription on a supercoiled template. Transcripts longer than 35 nt were considered to be elongating transcripts, and their amounts were measured by a Fuji-BAS2500 phosphoimager. Transcription by wild-type TFIIE on the two different templates was defined as 100% (lanes 10 and 18). Relative transcription activities (%) are presented in the bottom panel. (C) Effects of ceTFIIE subunits on transcription initiation. The PIC was preformed as described for panel A on the AflIII-ScaI fragment of pMLH1 (containing the AdML promoter sequence from −111 to +47) (14). Transcription initiation was then carried out for 45 min at 28°C by addition of [α-32P]CTP in the presence of the four different TFIIE proteins or in the absence of TFIIE. The position of the dinucleotide transcript (adenylyl cytidine [ApC]) is indicated by an arrow. Lane 1, reaction without TFIIE (−E); lanes 2, 4, 6, and 8, 8 ng of each TFIIE protein was added; lanes 3, 5, 7, and 9, 24 ng of each TFIIE protein was added. Abbreviations for TFIIE proteins were the same as those used in panel B.
DISCUSSION
In this study, we isolated and characterized both subunits of C. elegans TFIIE (ceTFIIE) for the purpose of elucidating TFIIE functions in transcription. Since the essential transcription mechanisms are conserved between human and C. elegans, these ceTFIIE subunits were nice tools, as if they were two useful human TFIIE subunit mutants, for studying TFIIE functions by looking at the functional defects of those subunits in the human in vitro transcription system. We showed that only the smaller subunit, ceTFIIEβ, could partially replace its human counterpart in the human system. By means of further functional studies, we found that ceTFIIEα did not bind well to hTFIIEβ, which explains the inability of ceTFIIEα to substitute for its human counterpart. We also found that in the human in vitro system, ceTFIIEβ was severely defective in its ability to support the transition from initiation to elongation. Importantly, CTD phosphorylation in the presence of chimeric TFIIE consisting of hTFIIEα and ceTFIIEβ (hTFIIEα- ceTFIIEβ) resulted in an intermediate form of the largest subunit of Pol II because hTFIIEα-ceTFIIEβ did not efficiently support phosphorylation of Ser-5 in the CTD repeat sequence. Although many studies have suggested that CTD phosphorylation is important for Pol II processivity, this is, to our knowledge, the first report to suggest that Ser-5 phosphorylation is somehow related to transcription transition efficiency and that this might be the basis of TFIIE involvement in this step.
Differences between TFIIEα and TFIIEβ.
Both ceTFIIE subunits were expressed in bacteria and tested for the ability to functionally replace their human homologs in a human in vitro transcription system. By transcription analysis, we demonstrated that ceTFIIEβ could partially substitute for hTFIIEβ in transcription on a supercoiled template but that ceTFIIEα could not replace hTFIIEα. In order to examine this functional difference, we compared the binding of ceTFIIE and hTFIIE subunits to human general transcription factors (Fig. 5 and 6). The only differences observed between human and C. elegans TFIIEβ was the latter's weak binding to the TFIIH subunits cyclin H and p44 (Fig. 5C and D). In contrast, ceTFIIEα, as opposed to hTFIIEα, failed to bind strongly to hTFIIEβ (Fig. 6B, lane 3). This weak binding of ceTFIIEα to hTFIIEβ is probably the main reason for the inability of ceTFIIEα to functionally replace hTFIIEα, while the partial ability of ceTFIIEβ to replace hTFIIEβ cannot be simply explained by these binding results. Further analyses revealed that ceTFIIEβ was unable to fully support Ser-5 phosphorylation in the CTD of Pol II or the transition to transcription elongation (Fig. 7 and 8). These results, together with those of protein-DNA cross-linking experiments (49) and our recent structural study of the hTFIIEβ core dsDNA-binding domain (43), strongly suggest the following model for the different functional roles of each subunit. TFIIEβ may play a fundamental role: it is located inside the PIC and makes contact with the promoter region at the point where the dsDNA starts to open (between positions −14 and −2 with the transcription start site defined as +1). TFIIEα, on the other hand, appears to have evolved in a more species-specific fashion and plays an antenna-like role in receiving signals for transcriptional regulation; it is located on the outside of the PIC and makes contact with regulatory factors and SRB- and Med-containing complex in addition to recruiting TFIIH into the PIC.
Stimulation of TFIIH-mediated CTD phosphorylation by TFIIE coincides with induction of Ser-5 phosphorylation in the CTD heptapeptide sequence.
It is now widely recognized that the CTD of Pol II is hyperphosphorylated during active transcription elongation (34, 70) and that only hypophosphorylated Pol II can form the PIC to initiate transcription (30, 66). These observations clearly indicate that CTD phosphorylation occurs between PIC formation and the transition from transcription initiation to elongation. Since TFIIH is the only factor located inside the PIC that possesses a CTD kinase activity in an in vitro reconstituted transcription system and is regulated by TFIIE (31, 39), we believe that these two molecules play central roles in the formation of processive Pol II complexes.
Since ceTFIIEβ was partially able to replace hTFIIEβ in transcription on the supercoiled AdML template (Fig. 4B and 8B), its effect on CTD phosphorylation during PIC formation was examined (Fig. 7). An intermediate form of the Pol II largest subunit with electrophoretic mobility between those of the IIa and IIo forms was produced in the presence of chimeric TFIIE (hTFIIEα-ceTFIIEβ) (Fig. 7A, lanes 6 and 7). Ser-2 in the CTD repeat motif was phosphorylated by TFIIH even in the absence of TFIIE (Fig. 7C, lane 1) but was stimulated in the presence of four different forms of TFIIE (Fig. 7C, lanes 2 to 5), whereas Ser-5 phosphorylation was strongly induced by wild-type hTFIIE (Fig. 7D, lane 5) and weakly by chimeric TFIIE (hTFIIEα-ceTFIIEβ) (Fig. 7D, lane 4). These results indicate that ceTFIIEβ showed only partial transcriptional complementation of its human counterpart because of a defect in supporting Ser-5 phosphorylation.
The transcription transition activity of TFIIE coincides with Ser-5 phosphorylation in the CTD.
Using two different forms of the AdML template, we found that chimeric TFIIE (hTFIIEα-ceTFIIEβ) showed 26% transcription activity (relative to wild-type hTFIIE) on the supercoiled template (Fig. 8B, lane 16 versus lane 18) but, significantly, only 5% of wild-type activity on the linear template (lane 8 versus lane 10). In a transcription initiation assay, hTFIIEα-ceTFIIEβ also showed approximately 30% of the activity of wild-type hTFIIE (Fig. 8C, lanes 6 and 7 versus lanes 8 and 9). These results clearly indicate that hTFIIEα-ceTFIIEβ was markedly defective in supporting the transition to elongation. This, to our knowledge, is the first indication that induction of Ser-5 phosphorylation in the CTD by TFIIE may play a role in the transition from transcription initiation to elongation. On the contrary, Ser-2 phosphorylation was observed even in the absence of TFIIE (Fig. 7C, lane 2). This evokes two possibilities: one is that there is no effect of Ser-2 phosphorylation on transcription, and the other is that Ser-2 phosphorylation is essential but requires, in addition, Ser-5 phosphorylation to change Pol II to a fully processive transcription complex.
A long-lasting issue has been whether CTD phosphorylation is essential for Pol II transcription. More recently, two lines of studies, which support the importance of CTD phosphorylation in transcription, have been reported. (i) Reinberg and colleagues have demonstrated a link between transcription and CTD phosphorylation by showing that Cdk8 (the human SRB10 homolog) targets not only the CTD of Pol II but also cyclin H of TFIIH and that Cdk8 inactivates both transcription and CTD kinase activities of TFIIH by phosphorylating cyclin H (1). (ii) Two novel studies using the chromatin immunoprecipitation method have demonstrated that Ser-5, but not Ser-2, phosphorylation was dependent on transcription and that Ser-5-phosphorylated Pol II was primarily localized at promoter regions (23, 54). In addition, two studies on human immunodeficiency virus type 1 gene transcription have reported that the transcriptional activator of this gene, Tat, stimulates both Ser-5 phosphorylation of the CTD of Pol II by Cdk9 and Pol II processivity for transcription elongation (12, 73). These results all support the idea that Ser-5 phosphorylation makes Pol II processive in transcription. They also support our observation that Ser-5 phosphorylation is a prerequisite for the transcription transition step and further confirm the importance of CTD phosphorylation of Pol II at Ser-5. Of course, further evidence will be required to establish a causal link between CTD phosphorylation and transcription, because it is still possible that something else, e.g., the helicase activity of TFIIH, provides the essential function for the transition step and that Ser-5 phosphorylation is just a consequence of an inactive Pol II complex. Therefore, the current focus of our research is to pursue the possibility that Ser-5 phosphorylation is directly involved in this transcription transition.
Recent studies have also shown that pre-mRNAs are targeted for capping through binding of the guanylyltransferase component of the capping apparatus to the phosphorylated CTD of Pol II (6, 33, 72). Mammalian guanylyltransferase binds synthetic CTD peptides containing phosphoserine at either Ser-2 or Ser-5 of the YSPTSPS repeat (16). However, only CTD peptides containing phospho-Ser-5 stimulate guanylyltransferase activity, enhancing enzyme affinity for GTP and increasing the yield of enzyme-GMP intermediate. CTD peptides containing phospho-Ser-2, on the other hand, have no effect on guanylyltransferase activity. Importantly, Rodriguez et al. have proved genetically that only Ser-5-phosphorylating Kin28 of yeast Cdk7 is able to recruit the capping enzyme guanylyltransferase (Ceg1) and the polyadenylation factor Pta1 to Pol II and that other CTD kinases, such as SRB10 of yeast Cdk8, are not able to carry out this function (50). In summary, our present results together with the recent genetic and biochemical data clearly suggest that TFIIE and TFIIH play important roles via Ser-5 phosphorylation during the transition from transcription initiation to elongation as well as in subsequent RNA processing steps.
ACKNOWLEDGMENTS
We thank Tetsuro Kokubo and Kiyoe Ura for critical reading of the manuscript, Takehiro Kobayashi and Kiyoji Tanaka for human XPB (ERCC3) and XPD (ERCC2) cDNA clones, Koji Hisatake for the human p52 cDNA clone, Jean-Marc Egly for human p52 and MAT1 cDNA clones, Charles J. Sherr for the mouse Cdk7 (MO15) cDNA clone, David O. Morgan for the human cyclin H cDNA clone, Yuji Kohara for the C. elegans embryonic and mixed-stage cDNA libraries, Hideyuki Okano for the C. elegans embryonic cDNA library, and Katsuyuki Tamai for raising antibodies. We also thank Masayuki Yokoi, Toshihiko Oka, and Tomoko Okamoto for technical assistance and Robert G. Roeder and our colleagues for helpful discussion.
This work was supported in part by grants from the Ministry of Education, Science and Culture of Japan (F.H. and Y.O.), the Core Research for Evolutional Science and Technology (CREST) (F.H. and Y.O.), the Biodesign Research Program of the Institute of Physical and Chemical Research (RIKEN) (F.H.), the Terumo Life Science Foundation (Y.O.), and the Yamanouchi Foundation for Research on Metabolic Disorders (Y.O.).
REFERENCES
- 1.Akoulitchev S, Chuikow S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomei M S, Halden N F, Cullen C R, Corden J L. Genetic analysis of the repetitive carboxy-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1988;8:330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björklund S, Kim Y-J. Mediator of transcriptional regulation. Trends Biochem Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 4.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 5.Chiang C-M, Ge H, Wang Z, Hoffmann A, Roeder R G. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerase II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho E J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 8.Drapkin R, Reardon J T, Ansali A, Huang J-C, Zawel L, Ahn K, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 9.Dvir A, Garrett K P, Chalut C, Egly J-M, Conaway J W, Conaway R C. A role for ATP and TFIIH in activation of the RNA polymerase II preinitiation complex prior to transcription initiation. J Biol Chem. 1996;271:7245–7248. doi: 10.1074/jbc.271.13.7245. [DOI] [PubMed] [Google Scholar]
- 10.Dvir A, Conaway R C, Conaway J W. A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc Natl Acad Sci USA. 1997;94:9006–9010. doi: 10.1073/pnas.94.17.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feaver W J, Henry N L, Bushnell D A, Sayre M H, Brickner J H, Gileadi O, Kornberg R D. Yeast TFIIE. Cloning, expression, and homology to vertebrate proteins. J Biol Chem. 1994;269:27549–27553. [PubMed] [Google Scholar]
- 12.Garber M E, Mayall T P, Suess E M, Meisenhelder J, Thompson N E, Jones K A. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 Tat-P-TEFb complex to TAR RNA. Mol Cell Biol. 2000;20:6958–6969. doi: 10.1128/mcb.20.18.6958-6969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 14.Hawley D K, Roeder R G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987;262:3452–3461. [PubMed] [Google Scholar]
- 15.Henricksen L A, Umbricht C B, Wold M S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 16.Ho C K, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann A, Roeder R G. Purification of His-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991;19:6337. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann A, Roeder R G. Cloning and characterization of human TAF20/15. Multiple interactions suggest a central role in TFIID complex formation. J Biol Chem. 1996;271:18194–18202. doi: 10.1074/jbc.271.30.18194. [DOI] [PubMed] [Google Scholar]
- 19.Holstege F C P, Tantin D, Carey M, van der Vliet P C, Timmers H T M. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holstege F C P, van der Vliet P C, Timmers H T M. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 21.Holstege F C P, Fiedler U, Timmers H T M. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koleske A J, Young R A. The RNA polymerase II holoenzyme and its implication for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 23.Komarnitsky P, Cho E-J, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar K P, Akoulitchev S, Reinberg D. Promoter-proximal stalling results from the inability to recruit transcription factor IIH to the transcription complex and is a regulated event. Proc Natl Acad Sci USA. 1998;95:9767–9772. doi: 10.1073/pnas.95.17.9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leuther K K, Bushnell D A, Kornberg R D. Two-dimensional crystallography of TFIIB- and IIE-RNA polymerase II complexes: implications for start site selection and initiation complex formation. Cell. 1996;85:773–779. doi: 10.1016/s0092-8674(00)81242-8. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Kornberg R D. Interplay of positive and negative effectors in function of the C-terminal repeat domain of RNA polymerase II. Proc Natl Acad Sci USA. 1994;91:2362–2366. doi: 10.1073/pnas.91.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao S-M, Taylor I C A, Kingston R E, Young R A. RNA polymerase II carboxy-terminal domain contributes to the response to multiple acidic activators in vitro. Genes Dev. 1991;5:2431–2440. doi: 10.1101/gad.5.12b.2431. [DOI] [PubMed] [Google Scholar]
- 28.Lichtsteiner S, Tjian R. Synergistic activation of transcription by UNC-86 and MEC-3 in Caenorhabditis elegans embryo extracts. EMBO J. 1995;14:3937–3945. doi: 10.1002/j.1460-2075.1995.tb00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lillie J W, Green M, Green M R. An adenovirus E1a protein region required for transformation and transcriptional repression. Cell. 1986;46:1043–1051. doi: 10.1016/0092-8674(86)90704-x. [DOI] [PubMed] [Google Scholar]
- 30.Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu H, Zawel L, Fisher L, Egly J-M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 32.Malik S, Hisatake K, Sumimoto H, Horikoshi M, Roeder R G. Sequence of general transcription factor TFIIB and relationships to other initiation factors. Proc Natl Acad Sci USA. 1991;88:9553–9557. doi: 10.1073/pnas.88.21.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 34.O'Brien T, Hardin S, Greenleaf A, Lis J. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 35.Ohkuma Y. Multiple functions of general transcription factors TFIIE and TFIIH in transcription: possible points of regulation by trans-acting factors. J Biochem (Tokyo) 1997;122:481–489. doi: 10.1093/oxfordjournals.jbchem.a021777. [DOI] [PubMed] [Google Scholar]
- 36.Ohkuma Y, Hashimoto S, Roeder R G, Horikoshi M. Structural conservation of putative functional motifs between Xenopus and human TFIIE-β. Nucleic Acids Res. 1992;20:4363. doi: 10.1093/nar/20.16.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohkuma Y, Hashimoto S, Roeder R G, Horikoshi M. Identification of two large subdomains in TFIIE-α on the basis of homology between Xenopus and human sequences. Nucleic Acids Res. 1992;20:5838. doi: 10.1093/nar/20.21.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohkuma Y, Hashimoto S, Wang C K, Horikoshi M, Roeder R G. Analysis of the role of TFIIE in basal transcription and TFIIH-mediated carboxy-terminal domain phosphorylation through structure-function studies of TFIIE-α. Mol Cell Biol. 1995;15:4856–4866. doi: 10.1128/mcb.15.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohkuma Y, Roeder R G. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature. 1994;368:160–163. doi: 10.1038/368160a0. [DOI] [PubMed] [Google Scholar]
- 40.Ohkuma Y, Sumimoto H, Hoffmann A, Shimasaki S, Horikoshi M, Roeder R G. Structural motifs and potential ς homologies in the large subunit of human general transcription factor TFIIE. Nature. 1991;354:398–401. doi: 10.1038/354398a0. [DOI] [PubMed] [Google Scholar]
- 41.Ohkuma Y, Sumimoto H, Horikoshi M, Roeder R G. Factors involved in specific transcription by mammalian RNA polymerase II: purification and characterization of general transcription factor TFIIE. Proc Natl Acad Sci USA. 1990;87:9163–9167. doi: 10.1073/pnas.87.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto T, Yamamoto S, Watanabe Y, Ohta T, Hanaoka F, Roeder R G, Ohkuma Y. Analysis of the role of TFIIE in transcriptional regulation through structure-function studies of the TFIIEβ subunit. J Biol Chem. 1998;273:19866–19876. doi: 10.1074/jbc.273.31.19866. [DOI] [PubMed] [Google Scholar]
- 43.Okuda M, Watanabe Y, Okamura H, Hanaoka F, Ohkuma Y, Nishimura Y. Structure of the central core domain of TFIIEβ with a novel double-stranded DNA-binding surface. EMBO J. 2000;19:1346–1356. doi: 10.1093/emboj/19.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 45.Pan G, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 46.Parvin J D, Sharp P. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 47.Patturajan M, Schulte R J, Sefton B M, Berezney R, Vincent M, Bensaude O, Warren S L, Corden J L. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- 48.Peterson M G, Inostroza J, Maxon M E, Flores O, Admon A, Reinberg D, Tjian R. Structure and function of the recombinant subunits of human TFIIE. Nature. 1991;354:369–373. doi: 10.1038/354369a0. [DOI] [PubMed] [Google Scholar]
- 49.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. Localization of subunits of transcription factors IIE and IIF immediately upstream of the transcriptional initiation site of the adenovirus major late promoter. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez C R, Cho E-J, Keogh M-C, Moore C L, Greenleaf A L, Buratowski S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol. 2000;20:104–112. doi: 10.1128/mcb.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 52.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 53.Sawadogo M, Roeder R G. Factors involved in specific transcription by human RNA polymerase II: analysis by rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schroeder S C, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Studier W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 56.Sulston J, Hodgkin J. Methods. In: Wood W B, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1988. pp. 587–606. [Google Scholar]
- 57.Sumimoto H, Ohkuma Y, Sinn E, Kato H, Shimasaki S, Horikoshi M, Roeder R G. Conserved sequence motifs in the small subunit of human general transcription factor TFIIE. Nature. 1991;354:401–404. doi: 10.1038/354401a0. [DOI] [PubMed] [Google Scholar]
- 58.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 59.Svejstrup J Q, Vichi P, Egly J-M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 60.Takada R, Nakatani Y, Hoffmann A, Kokubo T, Hasegawa S, Roeder R G, Horikoshi M. Identification of human TFIID components and direct interaction between a 250-kDa polypeptide and the TATA box-binding protein (TFIIDτ) Proc Natl Acad Sci USA. 1992;89:11809–11813. doi: 10.1073/pnas.89.24.11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tantin D, Carey M. A heteroduplex template circumvents the energetic requirement for ATP during activated transcription by RNA polymerase II. J Biol Chem. 1994;269:17397–17400. [PubMed] [Google Scholar]
- 62.Timmers H T M. Transcription initiation by RNA polymerase II does not require hydrolysis of the β-γ phosphoanhydride bond of ATP. EMBO J. 1994;13:391–399. doi: 10.1002/j.1460-2075.1994.tb06273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson N E, Steinberg T H, Aronson D B, Burgess R R. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- 64.Trigon S, Serizawa H, Conaway J W, Conaway R C, Jackson S P, Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J Biol Chem. 1998;273:6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- 65.Tyree C M, George C P, Lira-De Vito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 66.Usheva A, Maldonado E, Golding A, Lu H, Houbavi D, Reinberg D, Aloni Y. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992;69:871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- 67.Van Dyke M W, Sawadogo M, Roeder R G. Stability of transcription complex on class II genes. Mol Cell Biol. 1989;9:342–344. doi: 10.1128/mcb.9.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X, Hansen S K, Ratts R, Zhou S, Snook A J, Zehring W. Drosophila TFIIE: purification, cloning, and functional reconstitution. Proc Natl Acad Sci USA. 1997;94:433–438. doi: 10.1073/pnas.94.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watanabe Y, Fujimoto H, Watanabe T, Maekawa T, Masutani C, Hanaoka F, Ohkuma Y. Modulation of TFIIH-associated kinase activity by complex formation and its relationship with CTD phosphorylation of RNA polymerase II. Genes Cells. 2000;5:407–423. doi: 10.1046/j.1365-2443.2000.00336.x. [DOI] [PubMed] [Google Scholar]
- 70.Weeks J R, Hardin S E, Shen J, Lee J M, Greenleaf A L. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlation with gene activity and transcript processing. Genes Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- 71.Young R A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 72.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou M, Halanski M A, Radonovich M F, Kashanchi F, Peng J, Price D H, Brady J N. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxy-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]