Abstract
Opioid misuse and mismanagement has been a public health crisis for several years. Pharmacogenomics (PGx) has been proposed as another tool to enhance opioid selection and optimization, with recent studies demonstrating successful implementation and outcomes. However, broad engagement with PGx for opioid management is presently limited. The purpose of this article is to highlight a series of barriers to PGx implementation within the specific context of opioid management. Areas of advancement needed for more robust pharmacogenomic engagement with opioids will be discussed, including clinical and economic research needs, education and training needs, policy and public health considerations, as well as legal and ethical issues. Continuing efforts to address these issues may help to further operationalize PGx toward improving opioid use.
Keywords: : opioids, pain, pain management, pharmacogenetics, pharmacogenomics
In 2017, the opioid crisis officially became a public health emergency as deeply troubling statistics were observed across the country [1]. While opioid prescribing has declined from a peak around 2010–2012, morphine milligram equivalents of opioids prescribed per person are roughly triple what they were 20 years ago [2]. Further, roughly 21–29% of those prescribed opioids for chronic pain misuse them and between 8 and 12% develop opioid use disorder [3].
One of the five points in the US Department of Health & Human Services Response to the Opioid Crisis is to advance the practice of pain management enabling access to high-quality, evidence-based pain care [4]. Resources have recently been developed, such as the CDC Guideline for Prescribing Opioids with Chronic Pain, to assist with improving prescribing practices [5]. In addition, an increased use of state-level prescription drug monitoring programs has been associated with opioid prescribing reductions [6], and an increase in the use of naloxone prescriptions has been recently observed [7]. Despite these positive advances in opioid management, 2017 data suggests there were 58 opioid prescriptions per every 100 Americans [8], and overdose deaths have accelerated during the COVID-19 pandemic [9].
The field of pharmacogenomics (PGx), or the science of the relationship between a person’s genetic attributes and drug response, has seen tremendous growth in the past few years. The first guidelines relating to opioid prescribing from the Clinical Pharmacogenetics Implementation Consortium (CPIC) were released nearly 10 years ago. Updated in late 2020 with more recent literature, these clinical guidelines now provide more precise recommendations for codeine, tramadol and hydrocodone prescribing as it pertains to different CYP2D6 phenotypes [10]. While there is potential for improved opioid management through PGx, as well as several practice-based examples of PGx-guided opioid optimization, these practices have yet to become mainstream [11,12]. Despite considerable progress, barriers remain to the widespread implementation of PGx within the context of opioid management [13,14].
The purpose of this article is to highlight example practices to illustrate the vision of opioid optimization through PGx while discussing barriers to PGx implementation in this context. Areas of advancement needed for more robust pharmacogenomic engagement with opioids will be discussed, including clinical and economic research needs, education and training needs, policy and public health considerations and legal and ethical issues. Each area of discussion will conclude with a specific belief or actionable step pertaining to how PGx can be advanced to further assist with opioid optimization.
Emerging practice models
While PGx is not widely observed as a key part of the clinical care process across the USA, several examples of practice models have been described in the literature where PGx information is routinely applied to patient care across inpatient and outpatient practice settings. The University of Florida began offering CYP2C19 testing in 2012 for percutaneous coronary intervention patients to assist with antiplatelet selection and optimization [15]. Further expansion to other drug-gene pairs, such as CYP2D6-opioids and CYP2D6/CYP2C19-antidepressants, followed over the next few years. Best Practice Advisories, a form of clinical decision support (CDS), were built into the electronic health record (EHR) for selected drug-gene pairs. A PGx outpatient consult service has also been implemented [16]. More specifically related to opioid optimization, recent literature more thoroughly describes implementation efforts specific to optimizing postsurgical pain management through PGx [11].
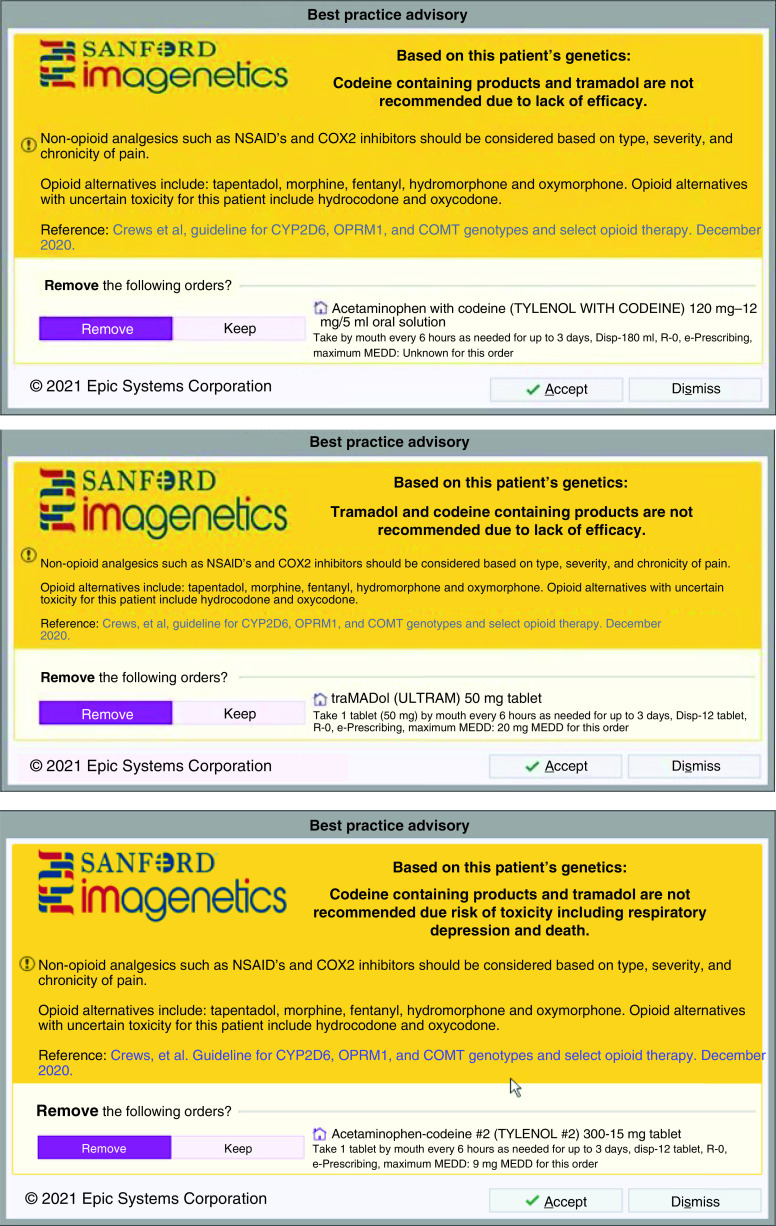
Similar to the University of Florida, Sanford Health’s precision medicine program, Imagenetics, also utilizes CDS tools. Clinical PGx pharmacists at Sanford provide a retrospective review once PGx test results are signed out into the EHR. Pharmacists review for drug–gene interactions and evaluate for any medications that are relevant enzyme inhibitors or inducers. A chart note is written with any recommendations. Depending on patient-specific variables, recommendations may be limited to monitoring suggestions, or no interventions may be recommended at all. After initial review, because PGx results are incorporated as a discrete value, CDS is the main approach to alert prescribers to any drug–gene interactions. CDS alerts include a description of the interaction, a link to associated CPIC guidelines, and relevant recommendations. PGx CDS is branded with ‘Sanford Imagenetics’ to inform providers about the nature of the alert [12]. Specific to opioid optimization, language includes considering nonopioid medications if appropriate, including opioid alternatives not dependent on CYP2D6, and lists opioid alternatives with uncertain toxicity (i.e., hydrocodone and oxycodone) [10]. Additionally, the alert has the option to keep or remove the order for the opioid prescribed. Figure 1 provides examples of CDS tools.
Figure 1. . Clinical decision support examples.
© 2021 Epic Systems Corporation.
Community pharmacy-focused examples exist as well [17–19], with multiple publications suggesting community pharmacy as a good fit for clinical PGx management [20,21]. However, in a similar fashion to other clinical settings, implementation is not widespread. Increasing reliance on community pharmacy models may help in areas where hospitals do not have the infrastructure to support in-house PGx testing. In this approach, smaller community hospitals could collect PGx samples, send off to an off-site PGx laboratory, with results communicated back to the community pharmacist to manage follow-up with any/all involved providers. In this way, expansion to different health systems in a variety of geographies could be more easily accomplished. One additional potential positive influencer for increased uptake of PGx in community pharmacy settings may be the widespread availability of direct-to-consumer PGx testing that is available for sale in many community pharmacies. Such availability may create a need for community pharmacists to counsel patients on direct-to-consumer PGx testing [22].
These emerging practice models have shown promise for leveraging PGx in a variety of clinical settings to optimize medication use in general, with more specific and positive demonstration that pertains to opioid management. Evidence is not isolated to a single study or case report. Instead, replication in different healthcare environments implies that there is potential for PGx guidance to positively impact opioid selection in many settings. We believe that replicating these emerging models of successful PGx implementation should be strongly considered to serve more patients.
Research driving clinical practice
CYP2D6 plays a role in the metabolism of many opioids; CYP2D6 polymorphisms can affect clinical efficacy and safety with opioid use. Additionally, CYP2B6 and CYP3A are other CYP pathways involved in opioid metabolism but lack evidence-provoking associated clinical recommendations, providing an opportunity for more research. CPIC guidelines provide therapeutic recommendations for CYP2D6 with codeine and tramadol. The data for CYP2D6 is not as strong for hydrocodone, oxycodone and methadone [10]. Therefore, some of the most commonly used opioids do not have PGx therapeutic recommendations at this time. Further research needs to explore the implications of CYP2D6 polymorphisms on these medications to translate into evidence-based guideline therapeutic recommendations. Additionally, the impact of OPRM1 and COMT need to be explored to further define their role. While CYP2D6 is commonly included in PGx panel testing, OPRM1 and COMT generally are not. Currently, there is no standardization for OPRM1, and COMT genotype to phenotype association, and most of the platforms that do include OPRM1 and COMT only test OPRM1 rs1799971 and COMT rs4680 [10]. Another genetic consideration is ABCB1 which encodes for P-glycoprotein and may alter the pharmacokinetics of opioids. The clinical implications are unclear at this point but exist as another target for further research [23].
Some complexities exist with CYP2D6 when compared with some other CYP450 enzymes. Copy number variants (CNVs) are common with CYP2D6 [24]. CYP2D6 CNVs include duplication or deletion and potential rearrangement with CYP2D7 [25]. Many laboratory tests lack the ability to determine the exact number of CNV present, which can complicate interpretation and cause confusion among providers and patients, particularly if different results are returned from different laboratories upon repeated testing [26]. In some cases, CYP2D6 results may be reported out as indeterminate when uncertain function alleles are identified. Utilizing PGx to guide opioid prescribing is not nearly as beneficial for patients with such results at this time, but advances in research and technology will hopefully provide a clearer picture of CYP2D6 analysis in the future.
These and other scientific considerations have led to the development of clinical practice guidelines from the CPIC. Originally published in February 2012 under the name ‘Clinical Pharmacogenetic Implementation Consortium (CPIC) Guidelines for Codeine Therapy in the Context of Cytochrome P450 2D6 (CYP2D6) Genotype’, these guidelines were further updated in April 2014, April 2017, October 2019, and most recently under a revised name in December 2020 [10]. Future research must continue as implementation work progresses to perpetually revise and strengthen treatment guidelines over time as new evidence emerges.
The need for more studies related to pain and PGx is evidenced by current investigations in the field. A Depression and Opioid Pragmatic Trial in Pharmacogenomics (ADOPT-PGx), is an NIH-funded pragmatic trial examining if available pharmacogenetic data results in better pain control in patients when compared with usual care. The study is enrolling patients with acute post-surgical pain and chronic pain [27]. Medstar Health Research Institute is enrolling patients in its PGx Applied to Chronic Pain Treatment in primary care trial which is an open-label, prospective, randomized trial. Patients on opioid therapy will be randomized to PGx-guided care versus usual care [28]. A search of open clinical trials suggests many related studies also exist. These studies are moving the field forward, however much more research is needed especially looking beyond the role of solely CYP2D6 in treatment-guided opioid therapy.
Proper utilization of CYP2D6 information to help guide opioid prescribing should include, among many other variables, consideration of phenoconversion. Medications that are strong CYP2D6 inhibitors can cause a CYP2D6 normal metabolizer patient to respond as a poor metabolizer (activity score of 0). Moderate CYP2D6 inhibitors also can influence patient response. Currently, PGx-related CDS may not account for potential phenoconversion. Incorporating CYP2D6 inhibitors that a patient is on into an algorithm to provide recommendations will be a crucial step in optimizing PGx opioid CDS.
It is important to note that PGx guidelines alone provide only one variable in the highly complex topic of opioid-driven pain management. More robust opioid selection and dosing algorithms may become more helpful when taking into account PGx as well as many other patient-specific factors. Because pain is subjective, there is potential for more variation of outcomes compared with more quantitatively monitored conditions, such as warfarin/anticoagulation (warfarindosing.org). Even within the context of PGx alone, there are multiple factors to consider related to opioid metabolism. Demographics (age, gender, ethnicity, race), environment, other disease states/conditions and medications and indications for opioid use should be considered. Researching these interwoven issues may help to provide more clarity on opioid optimization. We believe that research must expand to further guide opioid-focused PGx implementation efforts in a data-driven manner.
Clinician education & CDS
The difficulties of clinically implementing PGx have been described for years in a variety of literature sources, with education being regularly referenced as a specific barrier [29,30]. As the science of PGx is newer, many clinicians may not have received education or training in their undergraduate, graduate or residency training experiences. Specifically, some physicians may feel unprepared to use genetic information in their practice [14], with similar findings observed for pharmacists and other healthcare professionals [31,32]. This lack of knowledge continues to be documented in recent literature, despite generally favorable feelings toward PGx [33]. Although it has been ensured that education and training programs involve PGx in the classroom [34], continuing education materials may be helpful for practicing clinicians with a limited understanding of PGx concepts [30]. Strong examples exist in different settings for healthcare professional education, such as what has been described for the Mayo Clinic, but challenges of broad education remain [35].
One global pharmacy and medical education survey suggests that PGx education has improved in recent years [36]. However, the breadth and depth of PGx training vary across educational programs [37,38] despite the presence of consensus-based pharmacist competencies and guidelines for PGx knowledge attainment and clinical application [39]. Guidelines from the Center for the Advancement of Pharmacy Education and Accreditation Council for Pharmacy Education, continuing education, certificate programs and degree programs are currently being offered to educate practicing pharmacists [39,40].
One method to reduce the breadth and depth of educational needs on the topic is through CDS infrastructure [13]. Active CDS that guides prescribing practices based on genetic data has been noted as a key component of successful implementation [30]. Although interpreting the CDS tools may still require some baseline level of knowledge about PGx, these tools may be one way to facilitate greater engagement without substantial education and may help to overcome educational barriers [41]. Rather than needing to thoroughly understand clinical decisions that should be made in the context of a drug–gene interaction, support tools can provide more specific guidance to the clinician. However, CDS tools have been developed in markedly different ways across different electronic health records, and diversity among them has created difficulty in assessing the effectiveness of interventions [42]. Further, CDS functionality within EHRs has been identified as a barrier to effective CDS integration [43], as has the relative prioritization of PGx CDS [44].
There are several unique demonstration projects that each suggest operationalizing CDS for PGx is viable in a variety of settings [45,46]. The genomic prescribing system is one example of an online, secure, custom interface for PGx CDS that has been operational for several years, which leverages color-coded indicators of PGx risk for ease of understanding [47]. In the first 43 months since inception of the genomic prescribing system program, 100% of red lights were clicked, 79% of yellow lights were clicked and 43% of green lights were clicked, demonstrating strong and sustained utilization by a variety of clinicians. Even sites with limited EHR infrastructure solutions have been suggested to support CDS [48]. Further, publications that highlight strategies for effective CDS integration have been published in recent years that can provide guidance to organizations that are looking to add CDS [47,49–51].
To be most useful, CDS technologies must take into account several clinical variables. As pain management is inherently complex and patient-specific, it is difficult to assemble a comprehensive CDS tool. However, the PGx complexities of opioid management are largely straightforward due to their focus on CYP2D6 polymorphisms at this time. Therefore, PGx-focused CDS for pain management may be appropriate as a confirmatory step after the provider or team has identified a preferred pain management strategy. It may also be appropriate to consider related drug–drug interactions and phenoconversion at this confirmatory stage as well. However, drug–drug interactions within the context of opioid use should not be a novel consideration. Provider-focused educational efforts are still important when CDS is involved [52], but less education may be required to more narrowly support pharmacogenetics of opioid use when CDS is already in place. Given the relatively specific and limited necessary guidance, CDS tools for opioid use may offer more viable implementation. Consistent clinical consults have similarly been described as a pathway toward PGx-optimized opioid use [53].
Just as no clinician is expected to be a profound expert of every content area in the field of medicine, it may be more practical for most clinicians to possess a more minimal level of competence and fluency with PGx as a broad science, while a smaller set of specialists take on the role of specific content-level expert [34]. Similar to other specialty services, PGx consult services have demonstrated utility in the hospital setting [54]. Referrals to ambulatory clinics also exist in similar ways [12,16,50,55]. These approaches may be beneficial when the front-line provider is not finding success in pain management and when PGx is a suspected contributing factor. We believe that basic and general PGx knowledge is necessary for all clinicians, that CDS is not a substitute for education, and that PGx specialists should be identified and available to assist with complex PGx decisions.
Economic, health policy & public health considerations
Widespread implementation of PGx use in clinical decision making, as with all healthcare practices, requires evidence of both clinical and economic benefits. Specifically related to economic considerations, the implementation of PGx treatment algorithms must be justified by virtue of cost savings for health payers and to the healthcare system as a whole. The general economic and societal cost of opioid misuse is well-documented [56]. In recognition of this, payers may consider reimbursement incentives to support the evaluation of a patient’s relevant PGx data prior to initiating outpatient opioid treatment as a way to better ensure therapeutic optimization.
Recent increases in the prescribing of codeine and tramadol creates both concern and opportunity for intervention [57,58], as codeine and tramadol are two agents with some of the strongest evidence for PGx-guided therapeutic optimization [10]. There may be an opportunity to provide prior authorization criteria to enforce the administration of PGx testing and the documentation of a CYP2D6 activity score, which could support the selection of alternate agents in the case of CYP2D6 poor or ultra-rapid metabolizers. As the evidence evolves over time, as PGx testing results become more broadly available, and as the cost of PGx testing continues to decrease, prior authorization criteria could be adjusted accordingly. Balancing pharmacoeconomic considerations with health outcomes is difficult but necessary in a healthcare environment with scarce resources.
It is important to clarify that the implementation of prior authorization criteria should not be done in such a way as to further limit or delay the critical access to necessary pain medications, but as a way to support engagement of PGx testing in appropriate circumstances. For instance, prospective PGx testing could be ordered prior to elective surgeries or other situations where opioid needs could be reasonably anticipated. When such foresight is impractical, a prior authorization could serve as a warning that prescribing beyond an initial quantity may require PGx testing to better ensure a beneficial safety and efficacy balance exists for the patient. Similarly, to reduce impediments to opioid access, systems must be in place to support a robust and easy-to-access, yet secure, framework which allows each patient’s PGx data available. The use of systems such as prescription drug monitoring programs has been discussed as one potential way to document and store a patient’s relevant PGx phenotypes and make them available at the point of prescribing or dispensing [59]. Expanded integration of EHRs both between health-systems and outpatient pharmacies, or the implementation of other data sharing strategies would be substantially beneficial in ensuring continuity of PGx-guided decision making for patients across healthcare settings.
Availability of PGx information at the point of dispensing for community pharmacists, coupled with expansion in the scope of practice to allow for therapeutic substitution, may reduce or eliminate delays that may arise through a prior authorization-based process. In lieu of scope expansion, implementation of collaborative agreements between physicians and pharmacists that allow pharmacists to make substitutions in accordance with prior authorization criteria could reduce delays at the point of dispensing. However, a collaborative practice may be less effective in implementation, as multiple collaborative agreements may need to be in place to allow for therapeutic substitution across their entire patient population. Efforts should be made to advance health policy in such a way that empowers healthcare professionals to ensure opioid optimization has taken place while mitigating safety issues and supporting efficacy.
Implementation of prior authorization criteria would necessitate patient and provider access to PGx phenotypes. As testing costs decline over time and as more patients have either direct-to-consumer or provider-ordered PGx information on file, the lack of PGx data may diminish over time. However, PGx testing coverage and other related economic variables may create a foundational barrier to the relatively limited number of patients who currently receive PGx testing. This may be corrected by adopting public health recommendations that encourage preemptive PGx testing for patients, similar to existing guidelines for preventative health screenings and vaccinations. Proactive PGx testing may be more easily justified from a cost perspective, as a patient’s genetic information will not change over time, and if results are obtained early and are accessible, the cost of the test is theoretically spread across multiple pharmacotherapeutic decisions that span a patient’s entire life. The use of a robust data storage and sharing system, as described above, would limit (or ideally, eliminate) the need for repeat genetic testing and optimize the cost–effectiveness for the testing procedure. We believe that continued economic analyses will help to guide policy decisions for PGx implementation and that existing PGx data must be shared among healthcare professionals to ensure that previously obtained PGx information is not wasted but is used to optimize patient care.
Ethical clarity & consensus
PGx is an area of medicine replete with complex ethical, social, and legal concerns. Some of these complicated issues include the implications of the high-cost therapeutics and the inequitable access to them; ownership of genetic information and how it is obtained, stored, and used; and potential discrimination and social justice ramifications of genetic-based stratification of patients [60]. But lacking widely accepted explicit professional and ethical norms, attempts to merely balance costs/benefits seem ill-suited to address the wide-ranging PGx related moral issues.
Novel therapeutics and diagnostics entail novel and potentially astronomical research and development costs, irrespective of potential efficacy. The cost of development, as well as the search for return on investment, may lead to unintentional but foreseeable disparities in access to potentially life-saving therapeutics. Historically disadvantaged communities will be the most adversely impacted by limited access to novel therapies, particularly genetic-based therapies. While the overall cost of genetic testing has declined in recent years, direct test costs, insurance coverage, and co-pays still represent additional substantial barriers to equitable access to PGx on top of existing structural and systemic healthcare disparities.
Ownership of genetic data is also a serious, long-standing concern [61]. Certain specific protections against unconsented, harmful genetic information disclosure exist, including the Genetic Information Nondiscrimination Act [62] and the Patient Protection and Affordable Care Act [63]. These protections should, theoretically, shield patients and research participants and encourage wider acceptance. However, serious concerns persist, specifically regarding disclosures that adversely impact the patient in unexpected ways [64–66].
Consider the following example: a primary care provider is considering sending a patient's sample to an outside laboratory for genetic analysis with the hope of developing a targeted, individualized treatment plan. Of the four laboratories available for this genetic analysis, each has very different approaches to handling the patent’s genetic data. Neither Laboratory 1 or 2 permit nor make accommodations for individual patients to opt-out of their samples being used in subsequent research, either by the laboratory or, as is more often the case, by third-party researchers. Laboratories 1 and 2 operate under an ethically tenuous implied consent approach to subsequent research; they presume that when samples are sent in for genetic analysis, then consent for any subsequent research activities is provided. Laboratory 3 allows for patients to opt-out of subsequent genetic testing. Still, the process is arduous on the patient, and the process is only detailed in the fine print of the sample submission process, which the patient may or may not have access to before the sample is submitted. Thus, while an opt-out process is beneficial to the patient’s ability to control their data, the burden of exercising this benefit is disproportionately on the clinician who obtains informed consent from the patient and on the patient herself. At last, Laboratory 4 takes a very different approach to issues of consent for research. Laboratory 4 provides documents and training for clinicians to utilize when gaining informed consent from the patient, which includes details about future research participation and the ability to opt-out without adverse consequences to the patient.
Many additional questions exist related to data ownership and research. For instance, who owns the data yielded from genetic tests? Does the patient have the opportunity to remove their data from research projects or business activities? Are patients owed compensation if their data results in financial windfall? Should an individual have the opportunity to monetize their genetic data? No current consensus exists on these questions, but answering these questions may help with confidence in PGx, specifically if the target patient population is historically underserved or marginalized.
Finally, many are concerned that the categorization and sub-categorization of groups of people intrinsic to the clinical application of PGx may further exacerbate existing social inequalities and discrimination. Shields worried about the framing of PGx data in relation to race. Framing new PGx data ‘in terms of ‘racial differences’ in allele frequencies relevant to disease risk or drug response continue[s] a long and painful history of comparative racial science in the US’ [67]. The intersection of amorphous psychosocial categories and ‘socially charged phenotypes’ may result in unanticipated, socially problematic consequences that have the potential to outstrip the expected benefits of PGx.
Although conceptually straightforward and widely employed in public health ethical analyses, a utilitarian cost-benefit approach falters in the face of multifaceted intersections of ethical ambiguity. PGx requires more normative sophistication precisely because of the intersectionality of patients whom PGx intends to benefit. We believe that addressing the resilient, morally ambiguous concerns associated with personalized medicine and PGx will require persistent dialogue between professionals and within professions.
Conclusion & future perspective
Areas of advancement needed for more robust pharmacogenomic engagement include clinical and economic research needs, education and training needs, policy and public health considerations and legal and ethical issues. Leaning on published models of implementation successes may help to facilitate the expansion of PGx-driven opioid optimization. While PGx represents only one variable in the highly complex field of pain management, continuing efforts to address these issues may help to further operationalize PGx as a strategy for improving opioid use. Further research on the role of pharmacogenomic guidance within the opioid management process, advances in PGx education, and the development of health policy and responsible data sharing will help to clarify next steps in PGx-focused opioid optimization to improve pain management.
Executive summary.
Opioid misuse is a public health crisis with troubling trends & statistics
The US Department of Health & Human Services Response to the Opioid Crisis includes five points, one of which is to advance the practice of pain management enabling access to high-quality, evidence-based pain care.
Clinical guidelines from the Clinical Pharmacogenetics Implementation Consortium have been recently updated and include guidance related to the intersection of pharmacogenomics (PGx) and opioid use.
Barriers remain to widespread implementation of PGx within the context of opioid management.
Emerging practice models exist in hospitals, ambulatory care and community pharmacies where PGx-informed pain management is practiced.
Existence of emerging practice models with associated replication in different healthcare environments implies that there is potential for PGx guidance to positively impact opioid selection in many settings.
Ongoing research, education & clinical decision support
Current research and practice guidelines suggest strongest levels of evidence for therapeutic recommendations for CYP2D6 with codeine and tramadol.
Additional trials are ongoing to explore PGx-guided opioid management improves outcomes as compared with usual care.
Though strong examples of PGx-focused education for healthcare professionals exist, broad education across health professions is inconsistent.
Clinical decision support (CDS) tools may help to guide clinicians as PGx is implemented. However, as pain management is highly complex, creation of a comprehensive CDS tool is difficult.
Economic, health policy, public health & ethical/legal considerations
Costs associated with opioid use and misuse are problematic, and balancing pharmacoeconomic considerations with health outcomes is difficult but necessary.
PGx data sharing is necessary to ensure that previously obtained PGx information is not wasted.
Continued economic analyses may help to guide policy decisions for PGx implementation.
Disclosure issues surrounding PGx data should be carefully balanced to ensure appropriate access without misuse.
Footnotes
Author contributions
All authors have contributed substantially based on collaborative drafting of the design, drafting, revisions and final approval of the materials.
Financial & competing interests disclosure
D Bright has conducted funded research with commercial pharmacogenetics companies and has a patent pending. N Petry is a co-investigator for RFA-HG-17-008 (PI: J Peterson/site PI: RA Wilke), an NIH/NHGRI Administrative Supplement: A Depression and Opioid Pragmatic Trial (ADOPT-PGx), which looks at gene-based dosing vs. standard of care for post-operative opioids. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.HHS acting secretary declares public health emergency to address national opioid crisis (2017). https://www.hhs.gov/about/news/2017/10/26/hhs-acting-secretary-declares-public-health-emergency-address-national-opioid-crisis.html
- 2.Prescribing practices (2019). https://www.cdc.gov/drugoverdose/data/prescribing/prescribing-practices.html
- 3.Opioid overdose crisis (2021). https://www.drugabuse.gov/drug-topics/opioids/opioid-overdose-crisis
- 4.Strategy to combat opioid abuse, misuse, and overdose. https://www.hhs.gov/opioids/sites/default/files/2018-09/opioid-fivepoint-strategy-20180917-508compliant.pdf
- 5.Dowell D, Haegerich TM, Chou R. CDC Guideline for prescribing opioids for chronic pain – United States. MMWR Recomm. Rep. 2016 65(No. RR-1), 1–49 (2016). [DOI] [PubMed] [Google Scholar]
- 6.State successes. https://www.cdc.gov/drugoverdose/policy/successes.html
- 7.Life-saving naloxone from pharmacies (2019). https://www.cdc.gov/vitalsigns/naloxone/index.html?deliveryName=DM8022
- 8.Prescribing practices (2019). https://www.cdc.gov/drugoverdose/data/prescribing/prescribing-practices.html
- 9.Overdose deaths accelerating during COVID-19 (2020). https://www.cdc.gov/media/releases/2020/p1218-overdose-deaths-covid-19.html
- 10.Crews KR, Monte AA, Huddart R et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid therapy. Clin. Pharmacol. Ther. (2021) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Referenced repeatedly, the Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2D6, OPRM1and COMT genotypes help to inform opioid optimization.
- 11.Thomas CD, Parvataneni HK, Gray CF et al. A hybrid implementation-effectiveness randomized trial of CYP2D6-guided postoperative pain management. Genet. Med. 23(4), 621–628 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petry N, Baye JF, Aifaoui A et al. Implementation of wide-scale pharmacogenetic testing in primary care. Pharmacogenomics 20(12), 903–913 (2019). [DOI] [PubMed] [Google Scholar]; • Describes an example of an outpatient pharmacogenomics (PGx) practice implementation.
- 13.Klein ME, Parved MM, Shin J-G. Clinical implementation of pharmacogenomics for personalized precision medicine: barriers and solutions. J. Pharm. Sci. 106, 2368–2379 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Owusu-Obeng A, Fei K, Levy KD et al. Physician-reported benefits and barriers to clinical implementation of genomic medicine: a multi-site IGNITE-Network survey. J. Pers. Med. 8, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavallari LH, Weitzel KW, Elsey AR et al. Institutional profile: University of Florida Health personalized medicine program. Pharmacogenomics 18(5), 421–426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes an example of an inpatient PGx practice implementation.
- 16.Arwood MJ, Dietrich EA, Duong BQ et al. Design and early implementation successes and challenges of a pharmacogenetics consult clinic. J. Clin. Med. 9(7), 2274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrari SP, Greco AJ, Michaels NM et al. Implementation of a pharmacogenomics service in a community pharmacy. J. Am. Pharm. Assoc. 54, 172–180 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Bright DR, Kisor DF, Smith A, Conaway M, Yu M. Implementation of a pharmacogenetic management service for post-myocardial infarction care in a community pharmacy. Per. Med. 12(4), 319–325 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Breaux S, Desrosiers FAD, Niera M, Sinha S, Nislow C. Pharmacogenomics at the point of care: a community pharmacy project in British Columbia. J. Pers. Med. 11, 11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padgett L, O'Connor S, Roederer M, McLeod H, Ferrari S. Pharmacogenomics in a community pharmacy: ACT now. J. Am. Pharm. Assoc. 51(2), 189–193 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Owen JA. Integrating pharmacogenomics into pharmacy practice via medication therapy management. J. Am. Pharm. Assoc. 51(6), e64–e74 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Gammal RS, Mayes J, Caudle KE. Ready or not, here it comes: direct-to-consumer pharmacogenomic testing and its implications for community pharmacists. J. Am. Pharm. Assoc. 59, 646–650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes direct-to-consumer PGx testing, which has the potential to drastically change clinical workflow and practices that involve PGx data.
- 23.Parchure AK, Peng YB. The impact of opioid analgesics and the pharmacogenomics of ABCB1 in opioid dependence and pharmacotherapies: a short review. Open Pain J. 13, 7–21 (2020). [Google Scholar]
- 24.Beoris M, Amos Wilson J, Garces JA, Lukowiak AA. CYP2D6 copy number distribution in the US population. Pharmacogenet. Genomics 26, 96–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Botton MR, Scott ER, Scott SA. Sequencing the CYP2D6 gene: from variant allele discovery to clinical pharmacogenetic testing. Pharmacogenomics 18(7), 673–685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarvis JP, Peter AP, Shaman JA. Consequences of CYP2D6 copy-number variation for pharmacogenomics in psychiatry. Front. Psychiatry 10, 432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.A depression and opioid pragmatic trial in pharmacogenetics (2021). https://clinicaltrials.gov/ct2/show/NCT04445792
- 28.Pharmacogenomics applied to chronic pain treatment in primary care (2020). https://ichgcp.net/clinical-trials-registry/NCT04685304
- 29.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature 526, 343–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shuldiner AR, Relling MV, Peterson JF et al. The pharmacogenomics research network translational pharmacogenetics program: overcoming challenges of real-world implementation. Clin. Pharmacol. Ther. 94(2), 207–210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calzone K, Jenkins J, Culp S, Bonham VL, Badzek L. National nursing workforce survey of nursing attitudes, knowledge and practice in genomics. Per. Med. 10(7), 719–728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roederer MW, Riper MV, Valgus J, Knafl G, McLeod H. Knowledge, attitudes and education of pharmacists regarding pharmacogenetic testing. Per. Med. 9(1), 19–27 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Frigon M-P, Blackburn M-E, Dubois-Bouchard C, Gagnon A-L, Tardif S, Tremblay K. Pharmacogenetic testing in primary care practice: opinions of physicians, pharmacists and patients. Pharmacogenomics 20(8), 589–598 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Haga SB. Integrating pharmacogenetic testing into primary care. Expert Rev. Precis. Med. Drug Dev. 2(6), 327–336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giri J, Curry TB, Formea CM, Nicholson WT, Rohrer Vitek CR. Education and knowledge in pharmacogenomics: still a challenge? Clin. Pharmacol. Ther. 103(5), 752–755 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Kuželički NK, Zitnik IP, Gurwitz D et al. Pharmacogenomics education in medical and pharmacy schools: conclusions of a global survey. Pharmacogenomics 20(9), 643–657 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Haga SB, Moaddeb J. Pharmacogenomics courses in pharmacy school curricula. Pharmacogenomics 20(9), 625–630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shatnawi A, Khanfar NM, Latif DA, Shear M. A comparative study of the depth, breadth, and perception of pharmacogenomics instruction in a subgroup of US pharmacy curricula. Curr. Pharm. Teach. Learn. 11(5), 476–484 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Weitzel KW, Aquilante CL, Johnson S, Kisor DF, Empey PE. Educational strategies to enable expansion of pharmacogenomics-based care. Am. J. Health Syst. Pharm. 73(23), 1986–1998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes pharmacist competencies and educational standards to support training of future clinicians.
- 40.Guy JW, Patel I, Oestreich JH. Clinical application and educational training for pharmacogenomics. Pharmacy 6, 163 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunnenberger HM, Crews KR, Hoffman JM et al. Preemptive clinical pharmacogenetics interpretation: current programs in five United States medical centers. Annu. Rev. Pharmacol. Toxicol. 55, 89–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herr TM, Pterson JF, Rasmussen LV, Caraballo PJ, Peissig PL, Starren JB. Pharmacogenomic clinical decision support design and multi-site process outcomes analysis in the eMERGE network. J. Am. Med. Inform. Assoc. 26(2), 143–148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zebrowski AM, Ellis DE, Barg FK et al. Qualitative study of system level factors related to genomic implementation. Genet. Med. 21(7), 1534–1540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chase DA, Baron S, Ash JS. Clinical decision support and primary care acceptance of genomic medicine. Stud. Health Technol. Inform. 245, 700–703 (2017). [PubMed] [Google Scholar]
- 45.Herr TM, Bielinski SJ, Bottinger E et al. Practical considerations in genomic decision support: the eMERGE experience. J. Pathol. Inform. 6, 50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dolin RH, Boxwala A, Shalaby J. A pharmacogenomics clinical decision support service based on FHIR and CDS hooks. Methods Inf. Med. 57, e115–e123 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Danahey K, Borden BA, Furner B et al. Simplifying the use of pharmacogenomics in clinical practice: building the genomic prescribing system. J. Biomed. Inform. 75, 110–121 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Krebs K, Milani L. Translating pharmacogenomics into clinical decisions: do not let the perfect be the enemy of the good. Human Genomics 13, 39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hicks JK, Dunnenberger HM, Gumpper KF, Haidar CE, Hoffman JM. Integrating pharmacogenomics into electronic health records with clinical decision support. Am. J. Health Syst. Pharm. 73(23), 1967–1976 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes implementation considerations for integrating pharmacogenomics into electronic health records with clinical decision support.
- 50.Hicks JK, Stowe D, Willner MA, Wai M, Daly T, Gordon SM. Implementation of clinical pharmacogenomics within a large health system: from electronic health record decision support to consultation services. Pharmacotherapy 36(8), 940–948 (2016). [DOI] [PubMed] [Google Scholar]
- 51.O'Donnell PH. Incorporating preemptive pharmacogenomic testing into the clinical setting. Clin. Adv. Hematol. Oncol. 18(9), 526–529 (2020). [PubMed] [Google Scholar]
- 52.Roden DM, Van Driest SL, Mosley JD et al. Benefit of pre-emptive pharmacogenetic information on clinical outcome. Clin. Pharmacol. Ther. 103(5), 787–794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith DM, Weitzel KW, Elsey AR et al. CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genet. Med. 21(8), 1842–1850 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Owusu-Obeng A, Weitzel KW, Hatton RC et al. Emerging roles for pharmacists in clinical implementation of pharmacogenomics. Pharmacotherapy 34(10), 1102–1112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunnenberger HM, Biszewski M, Bell GC et al. Implementation of a multidisciplinary pharmacogenomics clinic in a community health system. Am. J. Health Syst. Pharm. 73(23), 1956–1966 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Oderda GM, Lake J, Rüdell K, Roland CL, Masters ET. Economic burden of prescription opioid misuse and abuse: a systematic review. J. Pain Palliat. Care Pharmacother. 29(4), 388–400 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Bigal LM, Bibeau K, Dumbar S. Patterns in opioid prescription in the United States by region and prescribers over a 4-year period. J. Opioid Manag. 15(6), 499–506 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Bigal LM, Bibeau K, Dunbar S. Tramadol prescription over a 4-year period in the USA. Curr. Pain Headache Rep. 23(10), 76 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Bright DR, Petry N, Roath E, Reckow E, Chavour S. Barriers, solutions, and effect of using pharmacogenomics data to support opioid prescribing. J. Manag. Care Spec. Pharm. 26(12), 1597–1602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lipton P. Pharmacogenetics: the ethical issues. Pharmacogenomics J. 3, 14–16 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Ormond KE, Cho MK. Translating personalized medicine using new genetic technologies in clinical practice: the ethical issues. Per. Med. 11(2), 211–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.U.S. Dept. of Labor, Employee Benefits Security Administration. The Genetic Information Nondiscrimination Act of 2008 (GINA). HR 493, 110th Cong., 2nd Sess. DC, USA : (2009). [Google Scholar]
- 63.Compilation of Patient Protection and Affordable Care Act: as Amended through November 1, 2010 Including Patient Protection and Affordable Care Act Health-Related Portions of the Health Care and Education Reconciliation Act of 2010. HR 3590, 111th Cong., 2nd Sess. US Government Printing Office, DC, USA: (2010). [Google Scholar]
- 64.Hudson KL, Holohan MK, Collins FS. Keeping pace with the times – the Genetic Information Nondiscrimination Act of 2008. N. Engl. J. Med. 358(25), 2661–2663 (2008). [DOI] [PubMed] [Google Scholar]
- 65.McGuire AL, Majumder MA. Two cheers for GINA? Genome Med. 1(1), 6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Slaughter LM. The Genetic Information Nondiscrimination Act: why your personal genetics are still vulnerable to discrimination. Surg. Clin. North Am. 88(4), 723–738 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Shields AE. Ethical concerns related to developing pharmacogenomic treatment strategies for addiction. Addict. Sci. Clin. Pract. 6(1), 32–43 (2011). [PMC free article] [PubMed] [Google Scholar]