Golchoubian et al. show that the ER shaping protein REEP4 is a new player in nuclear pore complex (NPC) biogenesis. Their findings suggest that REEP4 cooperates with the key NPC assembly factor ELYS in late mitosis and that REEP4 may provide high-curvature ER membrane to the nascent NPC.
Abstract
Nuclear pore complexes (NPCs) are channels within the nuclear envelope that mediate nucleocytoplasmic transport. NPCs form within the closed nuclear envelope during interphase or assemble concomitantly with nuclear envelope reformation in late stages of mitosis. Both interphase and mitotic NPC biogenesis require coordination of protein complex assembly and membrane deformation. During early stages of mitotic NPC assembly, a seed for new NPCs is established on chromatin, yet the factors connecting the NPC seed to the membrane of the forming nuclear envelope are unknown. Here, we report that the reticulon homology domain protein REEP4 not only localizes to high-curvature membrane of the cytoplasmic endoplasmic reticulum but is also recruited to the inner nuclear membrane by the NPC biogenesis factor ELYS. This ELYS-recruited pool of REEP4 promotes NPC assembly and appears to be particularly important for NPC formation during mitosis. These findings suggest a role for REEP4 in coordinating nuclear envelope reformation with mitotic NPC biogenesis.
Introduction
Nuclear pore complexes (NPCs) are essential gateways for nucleocytoplasmic transport. NPCs are built from multiple copies of ∼30 different proteins called nucleoporins (Hampoelz et al., 2019) and form in nuclear pores. Such pores are openings within the nuclear envelope (NE) where the outer and inner nuclear membrane (ONM and INM) are fused, resulting in highly curved regions within the otherwise flat NE membrane. Early in mammalian open mitosis, the NE and NPCs disassemble. In consequence, soluble nucleoporins are released into the cytosol, and nuclear membrane proteins disperse in the cytoplasmic ER network (Kutay et al., 2021; Güttinger et al., 2009). During anaphase, a stepwise process reconstructs NPCs. This process depends on the nucleoporin ELYS, which binds to chromatin and recruits the Nup107-160 complex to establish a “seed” for new NPCs (Rasala et al., 2006; Franz et al., 2007; Schellhaus et al., 2016). At the same time, ER cisternae associate with chromatin through their resident INM proteins to initiate NE reassembly (Anderson et al., 2009). These ER cisternae are perforated with numerous openings called nanoholes or fenestrations, which are topologically equivalent to nuclear pores (Puhka et al., 2012; Otsuka et al., 2018). The association of ER nanoholes with ELYS-containing NPC seeds incorporates the seeds into the nascent nuclear membrane (Otsuka et al., 2018; Bilir et al., 2019), and NPC biogenesis is completed by sequential addition of the remaining nucleoporins (Dultz et al., 2008; Otsuka and Ellenberg, 2018).
The described mitotic pathway and a mechanistically distinct, ELYS-independent interphase pathway each build around half of the NPCs of a cell (Doucet et al., 2010; Dultz and Ellenberg, 2010; Otsuka et al., 2016; Otsuka and Ellenberg, 2018). During both pathways, high membrane curvature is induced and stabilized to create the nuclear pore. Factors that link the high-curvature pore membrane with proteinaceous NPC components are essential for integration of NPCs into the NE. The nucleoporins Nup133, Nup53 (also called Nup35), and Nup153 sense and potentially create membrane curvature. However, their curvature-sensing properties are required for NPC assembly only during interphase (Doucet et al., 2010; Vollmer et al., 2012, 2015). Additionally, the membrane-deforming proteins Reticulon4 and Tts1 contribute to NPC assembly into closed NEs (Dawson et al., 2009; Zhang and Oliferenko, 2014). For mitotic NPC assembly, however, factors that create high-curvature ER and coordinate ER remodeling with NPC formation are unknown.
Membrane shaping within the cytoplasmic ER depends on reticulon homology domain (RHD) proteins, namely Reticulon1–Reticulon4 and REEP1–REEP6 in mammals. RHD proteins bend membranes to establish tubules, curved edges of cisternae, and nanoholes within ER cisternae, and they preferentially localize to these regions of high membrane curvature (Voeltz et al., 2006; Shibata et al., 2010; Schroeder et al., 2019). The RHD proteins REEP3 and REEP4 mediate high curvature of ER specifically during mitosis (Kumar et al., 2019).
Here we report that REEP4 associates with the NE in addition to its expected localization to the cytoplasmic ER. We identify ELYS as a determinant of REEP4 targeting to the INM and show that REEP4 is required for normal levels of mitotically assembled NPCs. Our findings suggest that REEP4 links high-curvature ER with the ELYS-based NPC seed to promote NPC biogenesis during late mitosis.
Results
A fraction of REEP4 localizes to the INM
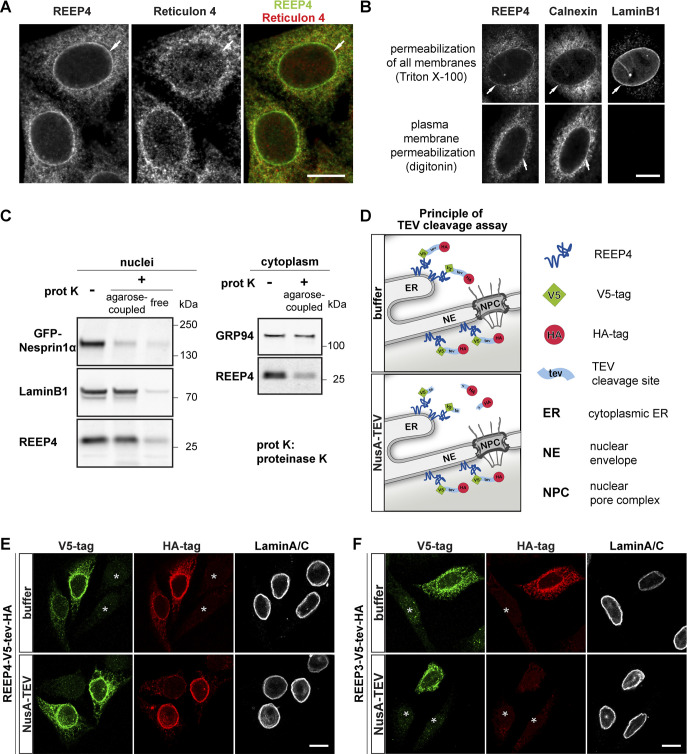
Previously studied RHD proteins, including REEP5/DP1 and Reticulon4, are excluded from the low-curvature NE (Voeltz et al., 2006; Shibata et al., 2010; Fig. 1 A). In contrast, endogenous REEP4, either untagged or chromosomally HA-tagged at the C-terminus, associates with the nuclear rim (Schlaitz et al., 2013; Fig. 1 A).
Figure 1.
A pool of REEP4 localizes to the INM. (A and B) HeLa cells with genomically HA-tagged REEP4 immunolabeled for HA. (A) Costained for Reticulon 4. (B) Costained for the ER marker Calnexin and the INM marker Lamin B1. Nuclear rim signal is detected for all three proteins after Triton X-100 treatment, but only for Calnexin after digitonin permeabilization. (C) HEK293T cells expressing the ONM marker GFP-Nesprin1α were fractionated into nuclei and cytoplasm. Left: Nuclei treated with agarose-coupled or unmodified (free) proteinase K and analyzed by immunoblotting. Right: In the cytoplasmic fraction, agarose-proteinase K degraded REEP4 but not the ER luminal protein GRP94, suggesting that ER integrity is maintained. (D) TEV cleavage assay: Purified NusA-TEV cleaves only cytoplasmic HA tags, while nucleoplasmic HA tags are protected. V5 tags mark cells expressing the reporter. (E and F) NusA-TEV cleavage assays were performed on HeLa cells expressing REEP4-V5-tev-HA (E) or REEP3-V5-tev-HA (F). Cells were immunostained for the V5 tag, the HA tag, and Lamin A/C to identify the nuclear rim. In A, B, E, and F, scale bars are 10 µm. In E and F, asterisks indicate untransfected cells.
We first asked whether REEP4 resides at the ONM or INM. The N-terminal RHD anchors REEP4 in the membrane, and the REEP4 C-terminus is exposed to the cytoplasm (Park et al., 2010; Brady et al., 2015; Kumar et al., 2019) or possibly the nucleoplasm. This structure allowed us to probe the topology of REEP4 at the NE using selective permeabilization and limited proteolysis. First, we permeabilized either all cellular membranes with the detergent Triton X-100 or only the plasma membrane with digitonin (Adam et al., 1990) and immunolabeled the REEP4 C-terminus. REEP4 at the nuclear rim was detected only in fully permeabilized cells (Fig. 1 B), demonstrating that nuclear REEP4 is inaccessible when NEs are intact. Next, we isolated cell nuclei and incubated them with either agarose-coupled or unmodified proteinase K. Agarose–proteinase K cannot enter intact nuclei and degrades only proteins of the ONM, such as Nesprin1α (Fig. 1 C). Unmodified proteinase K diffuses through nuclear pores and digests proteins of the ONM and also of the INM, such as Lamin B1. Endogenous REEP4 in the nuclear fraction resisted agarose–proteinase K treatment, similar to Lamin B1. Importantly, agarose–proteinase K degraded cytoplasmic REEP4, confirming that REEP4 is in principle susceptible to agarose–proteinase K digest (Fig. 1 C). Lastly, we adapted a tobacco etch virus (TEV)–based limited proteolysis assay (Theerthagiri et al., 2010; Ungricht et al., 2015). We tagged REEP4 with a V5 tag, a TEV protease cleavage site (tev), and an HA tag. Cells expressing REEP4-V5-tev-HA were semipermeabilized and incubated with purified NusA-TEV protease. NusA-TEV is restricted to the cytoplasm and cleaves the HA tag only from cytoplasmic REEP4-V5-tev-HA but not nucleoplasmic REEP4-V5-tev-HA (Fig. 1 D). Upon NusA-TEV treatment, HA signal from REEP4-V5-tev-HA disappeared in the cytoplasm but persisted at the nuclear rim, showing that REEP4 was protected from TEV cleavage within the nucleus (Fig. 1 E). The closely related REEP3 was not detected at the nuclear rim (Fig. 1 F). These results demonstrate that REEP4 associates with the INM or nuclear pore membrane. In the following, we will subsume both these domains under the term INM. REEP4 INM localization is in agreement with the findings of a proteomics study that detected REEP4 but not REEP3 enriched in the nuclear fraction (Itzhak et al., 2016). In fact, the localization of REEP4 to the NE appears to be unique among RHD proteins (Voeltz et al., 2006; Shibata et al., 2010; Schlaitz et al., 2013).
REEP4 is proximal to and partially colocalizes with the nucleoporin ELYS
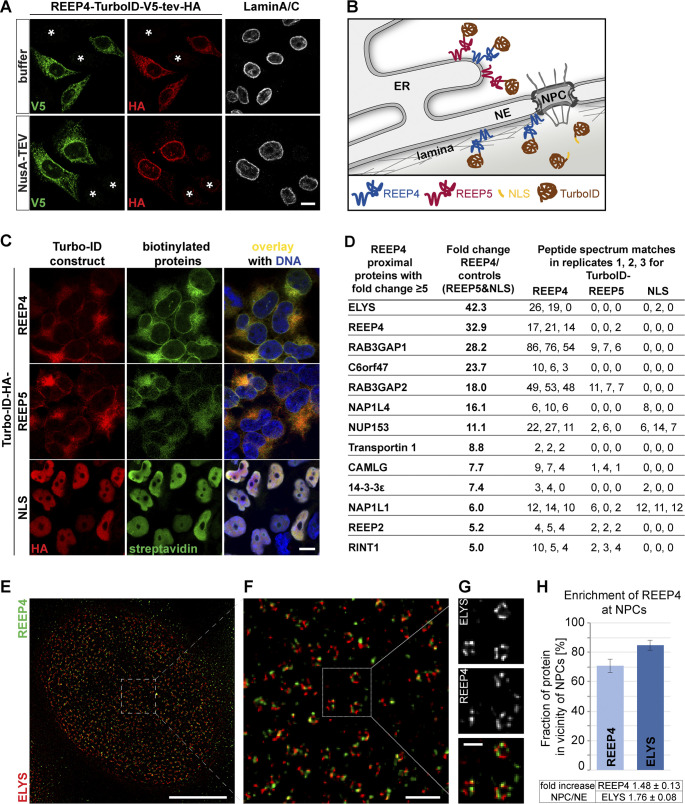
To characterize the immediate molecular environment of REEP4, we used proximity-dependent biotin identification (BioID; Roux et al., 2012). We inducibly expressed REEP4 fused to the biotin ligase TurboID (Branon et al., 2018), which biotinylates REEP4-proximal proteins in the cell. To control for nuclear and cytoplasmic background, we analyzed TurboID fusions of a nuclear localization sequence (NLS) and the RHD protein REEP5 (Fig. 2, A–C). Proteins captured on a streptavidin matrix were identified by LC-MS/MS, and the fold enrichment in the REEP4 sample over controls was calculated based on the spectral count information (Fig. 2 D; Mellacheruvu et al., 2013). The resulting list of REEP4-proximal proteins contained the known REEP4 interaction partners Rab3GAP1, Rab3GAP2, and 14-3-3 (Tinti et al., 2012), validating our approach (Fig. 2 D; Table S1). The most strongly enriched REEP4-proximal protein was the nucleoporin ELYS. We compared REEP4 and ELYS localization patterns by stimulated emission depletion (STED) microscopy and found them to frequently reside on the same ring-like structures, consistent with an association of both proteins with nuclear pores (Fig. 2, E–H).
Figure 2.
REEP4 BioID experiment identifies ELYS. (A) NusA-TEV cleavage assay on HeLa cells expressing REEP4-TurboID-V5-tev-HA reveals that REEP4-TurboID-V5-tev-HA localizes to peripheral ER and INM like endogenous REEP4. Asterisks indicate untransfected cells. (B) Scheme for REEP4-BioID with controls. (C) HEK cells were induced for expression of the indicated TurboID constructs for 24 h and treated with biotin for 1 h before fixation. TurboID constructs were detected with an HA antibody, and biotinylated proteins with fluorescently labeled streptavidin. (D) Proteins identified as proximal to REEP4 with fivefold or larger enrichment in REEP4 over control samples. See Table S1 for full list, raw MS datasets, and enrichment of proteins in REEP4 sample over single control samples. (E–G) REEP4-HA cells immunolabeled for the HA tag and ELYS and imaged by STED microscopy. (H) Quantification of REEP4 fraction in the vicinity of NPCs; ELYS included as a control. Scale bars are 10 µm (A and C), 5 µm (E), 500 nm (F), and 200 nm (G).
ELYS promotes REEP4 targeting to the INM
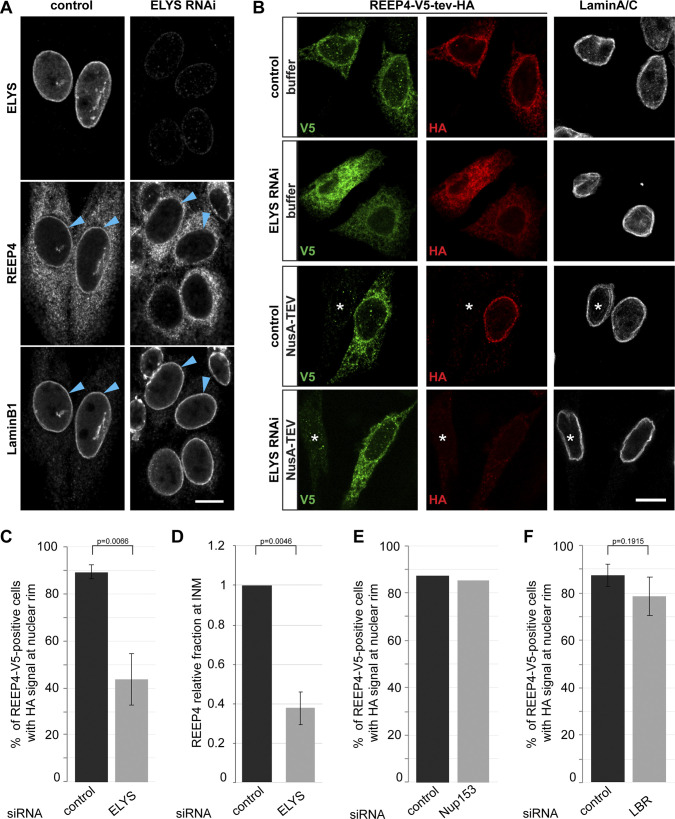
The accumulation of proteins at the INM depends on retention partners (Ungricht et al., 2015; Boni et al., 2015). We therefore tested whether ELYS retains REEP4 at the INM. After ELYS depletion, REEP4 signal at the nuclear rim was markedly reduced (Fig. 3 A). Based on TEV cleavage assays (Fig. 1 D), REEP4 was present at the INM in 89% of control cells but only 44% of ELYS-depleted cells (Fig. 3, B and C; and Fig. S1, A and B). Furthermore, the fraction of REEP4 that localized to the INM dropped by 60% upon ELYS depletion (Fig. 3 D). Depletion of Nup153, also found proximal to REEP4 by BioID (Fig. 2 D), did not reduce nuclear REEP4 (Fig. 3 E and Fig. S1, A and C). In ELYS RNAi cells, the INM proteins SUN2, Emerin, and Lap2β localized normally, and Lamin A/C targeted normally but showed elevated levels at the nuclear rim for unknown reasons (Fig. S1, E and F). These observations exclude a general role for ELYS in establishing the INM. The INM protein LBR mistargeted after ELYS RNAi, as reported previously (Fig. S1 E, far right column; Mimura et al., 2016). Yet, association of REEP4 with the INM was unaffected by LBR depletion (Fig. 3 F and Fig. S1, A and D), ruling out that REEP4 mislocalization in ELYS RNAi resulted from a lack of LBR at the INM. Finally, after ELYS depletion, the independent marker HA-tev-V5-Lap2β was not reduced at the nuclear rim, indicating that NusA-TEV protease did not aberrantly enter nuclei (Fig. S1 F). In conclusion, ELYS appears to directly mediate nuclear targeting of REEP4.
Figure 3.
ELYS promotes REEP4 INM targeting. (A) REEP4-HA cells treated with control or ELYS-targeting siRNA; labeled for the HA tag, ELYS, and Lamin B1; and imaged by confocal microscopy. Arrowheads indicate the nuclear rim. (B–D) NusA-TEV assay of HeLa cells expressing REEP4-V5-tev-HA, treated with control or ELYS siRNA, and stained for V5 tag, HA tag, and Lamin A/C. (B) Representative images. Asterisks indicate untransfected cells. (C) Quantification of cells with HA staining at the nuclear rim after NusA-TEV treatment (n = 5). (D) For an estimate of changes to the fraction of REEP4 at the nuclear rim, HA signal intensity (i.e., INM pool) was divided by V5 signal intensity (i.e., total pool of REEP4), and the mean value was normalized to the HA/V5 ratio in the respective control. n = 5 with 25 cells analyzed per condition. (E and F) Fraction of REEP4-V5-tev-HA–expressing, NusA-TEV–treated cells with HA staining at the nuclear rim in control cells or after depletion of Nup153 (E; average of two experiments with very similar outcomes) or LBR (F; n = 3). In A and B, scale bar is 10 µm. In C, E, and F, ≥35 cells were analyzed per condition in a blinded manner. In C, D, and F, error bars are SEM. P values were obtained using two-tailed, paired t tests. See Fig. S1, A–D, for analysis of ELYS, Nup153, and LBR depletions and expression of REEP4-V5-tev-HA.
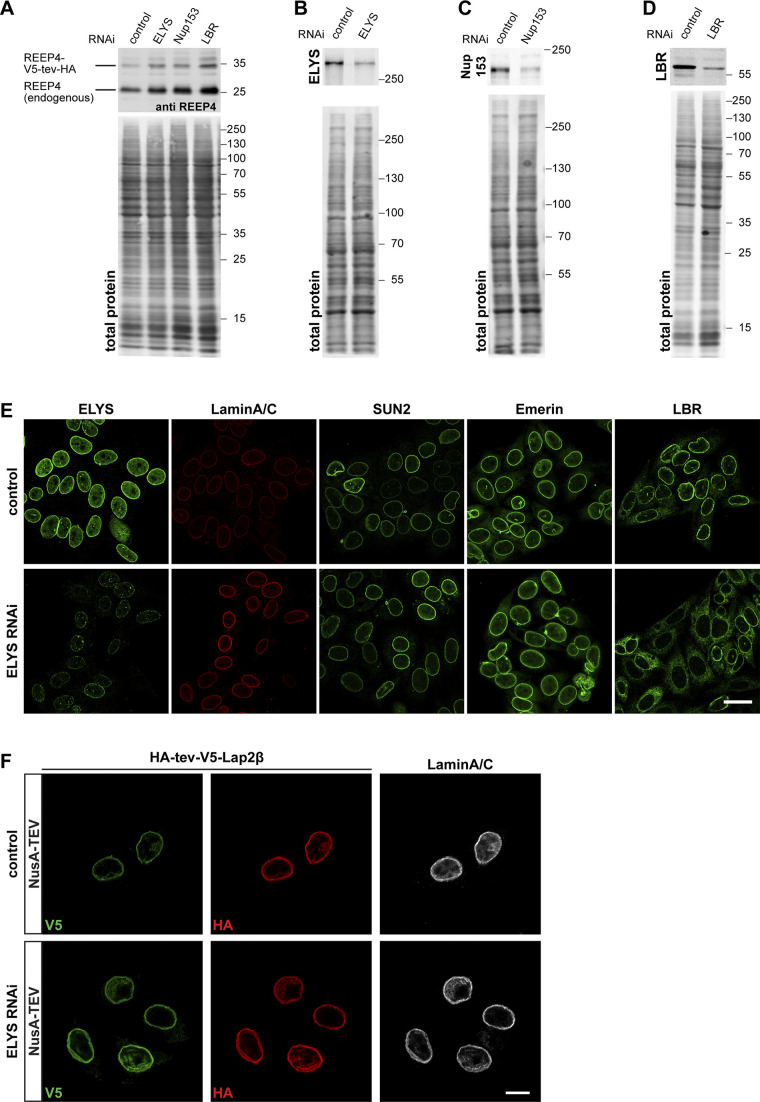
Figure S1.
Role of ELYS in REEP4 INM targeting. Related to Fig. 3. (A) Cells transfected with a construct for expression of REEP4-V5-tev-HA and with control, ELYS-, Nup153-, or LBR-targeting siRNA were lysed in Laemmli buffer and analyzed by SDS-PAGE and immunoblotting using an antibody against REEP4, which detects both endogenous REEP4 and REEP4-V5-tev-HA. Expression of REEP4-V5-tev-HA was not reduced after either of the siRNA treatments. (B–D) Lysates of cells transfected for the TEV assays with nontargeting control and ELYS- (B), Nup153- (C), or LBR- (D) targeting siRNA were analyzed by SDS-PAGE and immunoblotting with antibodies against the respective depleted proteins. (B and C) ELYS (n = 4) and Nup153 (n = 2) protein levels were on average reduced by 70%. (D) LBR protein levels were on average reduced by 90% (n = 3). (A–D) Images below the immunoblots show total protein stain of the membrane to visualize the amounts of loaded protein in the different samples. The total protein amounts in the respective lanes were used for normalization before determining depletion efficiency. (E) HeLa cells were treated with control or ELYS siRNA and immunostained for ELYS and the INM proteins Lamin A/C, SUN2, Emerin, and LBR. After ELYS-specific RNAi, ELYS was markedly reduced at the nuclear rim (far left column) but localization of the INM proteins Lamin A/C, SUN2, and Emerin was not impaired. Lamin A/C staining appeared increased after ELYS depletion for unknown reasons. Note that the two left columns show the same cells for control and ELYS RNAi, respectively, that were labeled for both ELYS and Lamin A/C. LBR targeting to the INM was impaired after ELYS depletion (far right column), as previously described by Mimura et al. (2016). Scale bar is 20 µm. (F) Cells expressing the INM protein Lap2β tagged N-terminally with HA-tev-V5 were treated with control or ELYS-targeting siRNAs and subjected to NusA-TEV treatment. Only the NusA-TEV treatment condition is shown; buffer-treated cells show an identical pattern. At least 30 cells were analyzed per condition in two different experiments. All control cells with V5-labeling, indicating successful transfection, also showed HA-staining of corresponding intensity. Among 68 V5-positive ELYS RNAi cells, one cell was lacking clear HA staining, but all other cells showed HA staining that corresponded to V5 intensity, suggesting that ELYS depletion did not lead to increased permeability of NPCs for NusA-TEV protease. Scale bar is 10 µm.
REEP4 may target to the forming NE through a direct interaction with ELYS during nuclear reformation in anaphase. However, REEP4 may also accumulate at the nuclear membrane during interphase by passive diffusion through NPC peripheral channels and retention at the INM. Thus, the mechanism of ELYS-mediated targeting of REEP4 to the INM requires future clarification.
REEP4 is required for normal NPC levels
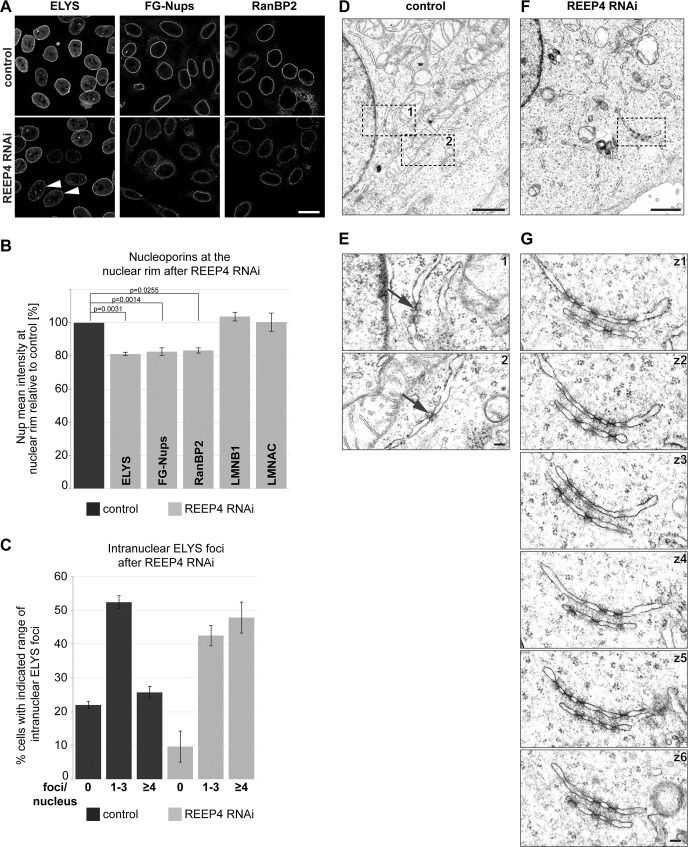
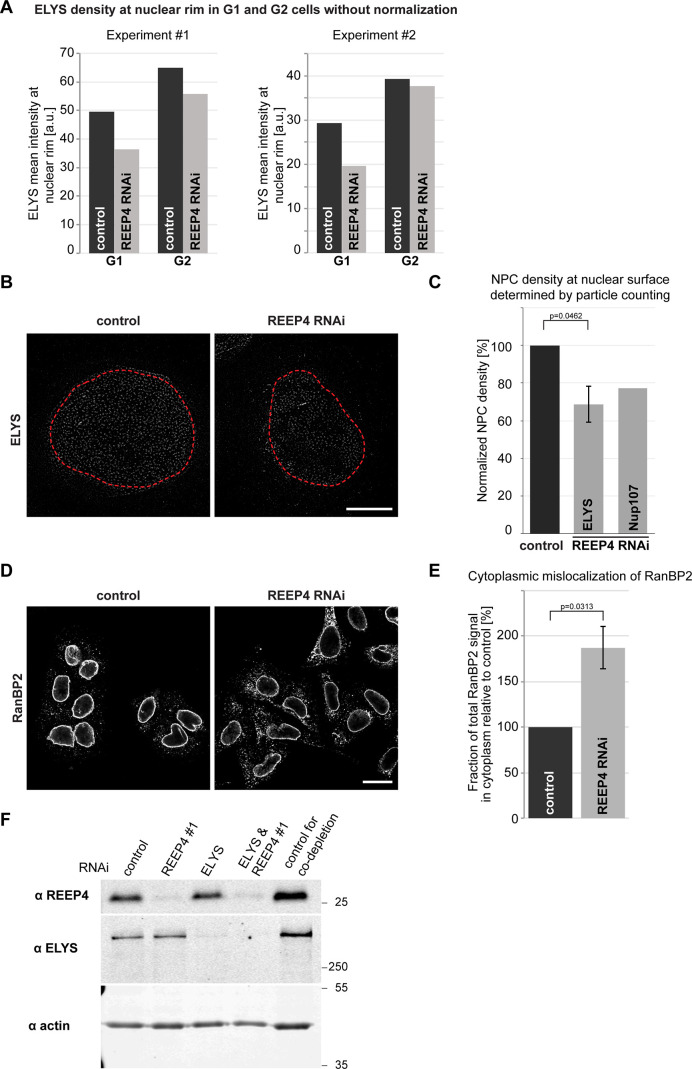
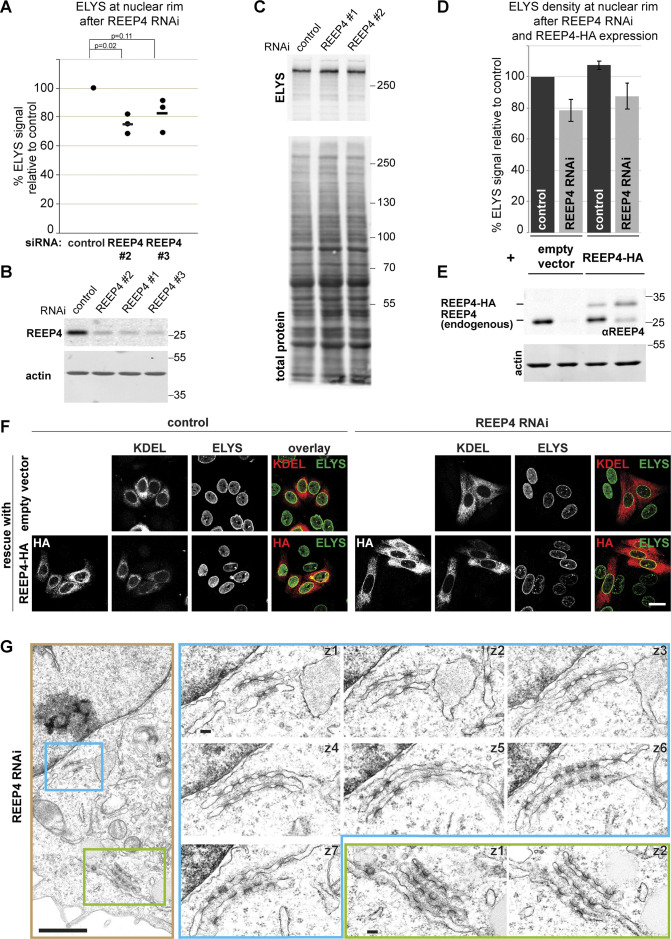
ELYS is a critical determinant of NPC biogenesis (Rasala et al., 2006; Franz et al., 2007), prompting us to examine NPCs after REEP4 depletion. The levels of three nucleoporins belonging to different NPC subcomplexes were decreased by ∼20%, while the levels of Lamin A/C or Lamin B1 at the nuclear rim were unaltered after REEP4 RNAi (Fig. 4, A and B). The number of NPCs per area counted from STED images of early interphase cells was reduced to a similar extent (Fig. S3, B and C). Different REEP4 siRNAs reduced ELYS signal at the NE and expression of exogenous REEP4-HA elevated ELYS signal at the NE (Fig. S2, A–F). Thus, REEP4 is required for normal NPC density.
Figure 4.
REEP4 is required for normal NPC densities. (A–C) HeLa cells treated with control or REEP4 siRNA, immunolabeled for indicated nucleoporins, and imaged by confocal microscopy. (A) Representative images. Arrowheads indicate cells with increased nucleoplasmic ELYS foci. (B) Mean intensities of the respective nuclear rim proteins in control and depleted cells. Lamin B1 (LMNB1), Lamin A/C (LMNAC), n = 3; ELYS, RanBP2, n = 4; FG-Nups, n = 6. At least 100 cells were analyzed per condition. Error bars are SEM. Two-sided, paired t tests were performed on the raw data and yielded the indicated P values. (C) Percentage of cells with zero, one to three, or four or more intranuclear ELYS foci in control and REEP4 RNAi cells. n = 3; ≥55 cells were analyzed per condition in a blinded manner; error bars are SEM. (D–G) Transmission EM analysis of control and REEP4 RNAi cells. (D and F) Overview images. (E) Enlarged views of the regions marked as 1 and 2 in D. Arrows indicate examples of rare NPC intermediates in the ER of control cells. (G) Images of consecutive serial z-sections (z1–z6) of the region outlined in F, showing clustered NPC intermediates in stacked ER cisternae in the cell periphery in REEP4 RNAi cells. See Fig. S2 G for another example. Scale bars: 20 µm (A), 1 µm (D and F), 100 nm (E and G).
Figure S3.
Further characterization of the role of REEP4 in mitotic NPC assembly. Related to Fig. 5. (A) Two examples for ELYS signal mean intensity changes in G1 versus G2 after REEP4 RNAi. These are two of the eight experiments that were incorporated into Fig. 5 B but showing the raw data without normalization to the G1 or G2 controls. (B and C) Quantification of NPC numbers per surface area in control and REEP4-depleted HeLa cells in G1 cells (identified by low CENP-F signal) expressing endogenously GFP-tagged Nup107 (Otsuka et al., 2016) and immunostained for GFP or ELYS. (B) Representative images. Red outline marks mask generated for particle counting in the two nuclei. Scale bar is 5 µm. (C) NPC densities were determined by particle counting of ELYS- or Nup107-positive NPC-sized structures at the nuclear surface in control and REEP4-depleted cells and normalized to control conditions. At least eight cells were analyzed per condition in each experiment. ELYS: n = 4; error bars are SEM. A two-sided t test for unequal variance was performed on the normalized data. Nup107: average of two experiments with very similar outcomes is shown. (D and E) Quantification of the cytoplasmic pool of the nucleoporin RanBP2 in control and REEP4-depleted HeLa G1 cells. (D) Representative images. Scale bar is 20 µm. (E) Mean signal intensity of RanBP2 in the cytoplasm in REEP4 RNAi cells relative to control. At least 80 cells were analyzed per condition in each experiment. (C and E) Shown are the means from three experiments. Error bars are SEM. A one-sided, unequal variance t test was performed on normalized data. (F) Western blot analysis of REEP4 and ELYS depletion in experiments shown in Fig. 5, C and D. ELYS was depleted by ∼85% and REEP4 by ∼90% in both single- and double-depletion experiments (average of two experiments).
Figure S2.
Characterization of the REEP4 depletion phenotype. Related to Fig. 4. (A) HeLa cells were transfected with control siRNA or REEP4 siRNAs #2 and #3 (REEP4 siRNA #1 is used in all other experiments), fixed, immunolabeled for ELYS, and imaged by confocal microscopy. Mean intensities at the nuclear rim were measured in control and REEP4 RNAi cells from the microscopy images. At least 100 cells were analyzed per condition; shown are the normalized mean intensities obtained in three experiments (dots). The horizontal line indicates the mean of the three experiments. Welch’s t test was performed on normalized data. For REEP4 siRNA #3, the P value is larger than 0.05; however, in each single experiment, ELYS levels are reduced compared with the control. We therefore conclude that ELYS reduction at the nuclear rim is a specific effect of REEP4 depletion. (B and C) Whole lysates of HeLa cells depleted of REEP4 using the indicated siRNAs were analyzed by SDS-PAGE and Western blotting and probed for REEP4 (B) or ELYS (C). To indicate amounts of loaded proteins, either actin (B) or total protein (C) was visualized. (D–F) Empty HA-tagging vector or RNAi-resistant, HA-tagged wild-type REEP4 was expressed in control cells or REEP4 RNAi cells, fixed, immunolabeled for ELYS, and imaged by confocal microscopy. (D) ELYS mean intensity at the nuclear rim was measured in transfected cells identified by expression of cotransfected RFP-KDEL (this transfection marker was used because the empty HA-tagging vector control does not yield an HA signal in transfected cells). Results of eight experiments are shown; ≥100 cells were analyzed per condition. Error bars are SEM. (E) Western blot analysis of lysates from cells used for experiment described in D. The lanes correspond to the bars of the graph directly above. Endogenous REEP4 and REEP4-HA were detected with anti-REEP4 antibody; actin served as a loading control. (F) Example images from experiment analyzed in E and D. In cells depleted of endogenous REEP4 and expressing REEP4-HA, ELYS levels at the nuclear rim are restored. However, in the entire population, REEP4-HA is not expressed to the same levels as endogenous REEP4 (see E) and only partially rescues the REEP4 RNAi phenotype (see D). Scale bar is 20 µm. (G) Additional example for transmission EM analysis of REEP4 RNAi phenotype as in Fig. 4, F and G. Left: Overview image. Right: Images of consecutive serial z-sections of the regions outlined in blue and green in the overview image. Scale bars: overview 1 µm, serial sections 100 nm.
Further, after REEP4 RNAi, ELYS-positive nuclear foci increased (Fig. 4, A and C), and clusters of NPC intermediates appeared in the cytoplasmic ER (Fig. 4, D–G; and Fig. S2 G). The ER-based cytoplasmic structures containing NPC intermediates likely correspond to the previously described annulate lamellae (Weberruss and Antonin, 2016), which seem expanded after REEP depletion. These observations suggest that nucleoporins not incorporated into NPCs after REEP4 RNAi accumulate in aberrant nucleoplasmic or cytoplasmic regions. Together, our findings indicate that REEP4 is required for NPC formation.
Inactivation of the membrane-shaping protein Atlastin disrupts the ER network and impairs the targeting of INM proteins and NPC biogenesis (Pawar et al., 2017). After REEP4 RNAi, however, ER morphology is unaltered (Schlaitz et al., 2013; Kumar et al., 2019), and the accumulation of membrane-associated Lamin B1 at the INM is not affected (Fig. 4 B). Thus, it is improbable that REEP4 RNAi reduces NPC levels due to disruption of ER morphology.
REEP4 is required for normal levels of NPCs assembled during mitosis
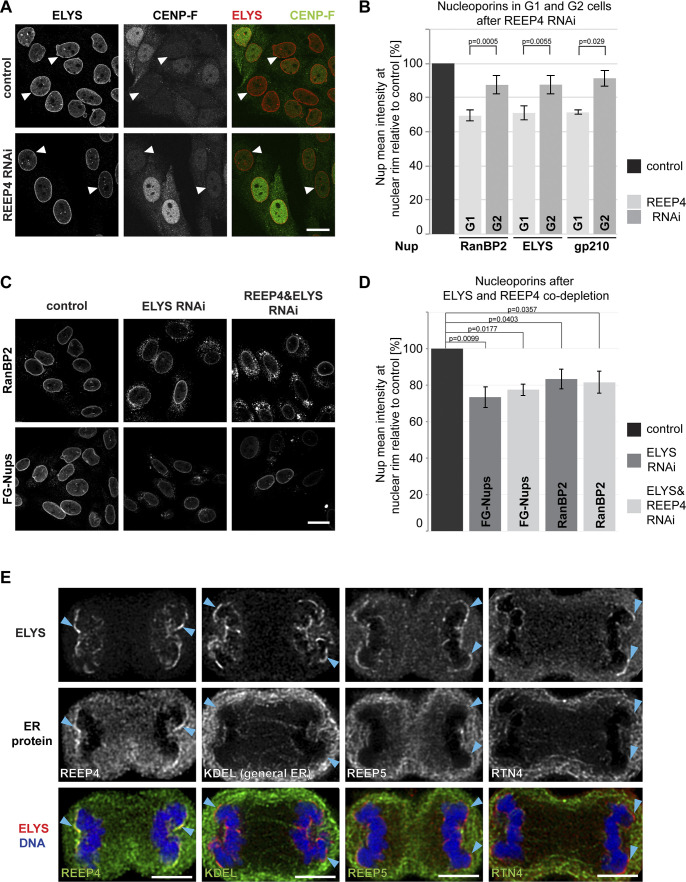
REEP4 (together with REEP3) shapes the ER during mitosis and ELYS initiates NPC assembly in anaphase. Thus, REEP4 and ELYS might collaborate in mitotic NPC biogenesis. To analyze the role of REEP4 in mitotic NPC formation, we determined NPC levels in early interphase (G1) cells, in which the NPCs present have largely arisen through mitotic assembly, and in late interphase (G2) cells, whose NPCs have originated to nearly equal parts during mitosis and interphase. If REEP4 contributed specifically to mitotic NPC assembly, its depletion should cause a larger reduction of nucleoporin levels in G1 than in G2 cells. We classified cells as G1 or G2 based on expression of the cell cycle–regulated protein CENP-F (Liao et al., 1995) and measured intensities of the nucleoporins ELYS, RanBP2, and gp210 in control and REEP4 RNAi cells. All three markers were reduced more strongly in G1 than in G2 cells following REEP4 depletion (Fig. 5, A and B; and Fig. S3 A), indicating that cells in early interphase had a lower density of NPCs. Counting NPCs from STED images confirmed their decrease in G1 cells (Fig. S3, B and C). The nucleoporin RanBP2 markedly mislocalized to the cytoplasm in REEP4-depleted G1 cells (Fig. S3, D and E). Together, these findings support a role of REEP4 in mitotic NPC assembly.
Figure 5.
REEP4 promotes mitotic NPC formation. (A–C) HeLa cells treated with control siRNA or depleted of REEP4 (A and B) or ELYS and ELYS/REEP4 (C and D), immunolabeled for indicated nucleoporins and CENP-F (A and B), and imaged by confocal microscopy. (A and C) Representative images. (A) Arrowheads indicate cells with low CENP-F signal. (B and D) Mean intensities of the respective nucleoporins at the nuclear rim. (B) Mean intensities were measured in either G1 cells (low CENP-F) or G2 cells (high CENP-F signal). Nucleoporin levels shown are relative to the respective control (G1 or G2). ELYS, n = 8; RanBP2, n = 10; gp210, n = 4; ≥21 cells analyzed per G1 or G2 condition. (D) n = 4. At least 100 cells were analyzed per condition. See Fig. S3 F for analysis of ELYS and REEP4 depletion efficiency. (B and D) Error bars are SEM. Two-sided, paired t tests were performed on the raw data. (E) REEP4-HA cells in anaphase, immunolabeled for ELYS and the HA tag (to detect REEP4), KDEL (general ER), REEP5, or RTN4 (both markers of high curvature ER), and imaged by confocal microscopy. Arrowheads indicate chromosomal noncore regions with accumulated ELYS. Scale bars: 20 µm (A and C), 5 µm (E).
To further probe whether REEP4 and ELYS act in the same NPC biogenesis pathway, we codepleted the proteins and measured NPC levels. Both ELYS single and REEP4/ELYS codepletion caused an ∼20% reduction in nucleoporin densities (Fig. 5, C and D; and Fig. S3 F), supporting the idea that both proteins act in the mitotic NPC assembly pathway. Given that REEP4 is lost from the nuclear rim after ELYS depletion (Fig. 3), this result also indicates that NE-localized REEP4 is the pool that is critical for NPC biogenesis. Depletion of ELYS caused an ∼40% reduction in NPC densities in U2OS and MCF10 cells (Doucet et al., 2010; Jevtić et al., 2019). The reduction of 20% we observed may be due to compensatory mechanisms in HeLa cells or high toxicity of ELYS RNAi and a resulting disappearance of well-depleted cells.
Endogenous REEP4 partially colocalized with ELYS at noncore regions of anaphase chromosomes (Fig. 5 E), consistent with a joint role in NPC biogenesis during late mitosis. Overall, these results suggest that REEP4 collaborates with ELYS in mitotic NPC assembly.
Conclusions
We report here that REEP4, a protein creating high-curvature ER specifically during mitosis, is recruited to the INM by the NPC biogenesis factor ELYS. Our findings suggest that the two proteins jointly promote NPC formation in late mitosis. We speculate that REEP4 aids the association of high-curvature ER, possibly nanoholes, with chromatin-bound ELYS to coordinate NE reformation with NPC biogenesis. ER nanoholes are abundant in mitotic ER and have been implicated in NPC formation (Puhka et al., 2012; Otsuka et al., 2018; Bilir et al., 2019). Live superresolution microscopy or correlative light and electron microscopy may in the future resolve whether REEP4 tethers nanoholes to the NPC seed. The contributions of the REEP4 membrane-shaping motifs, i.e., the RHD and an amphipathic helix, and whether REEP4 and ELYS directly interact to promote NPC biogenesis, remain to be elucidated.
REEP4 is at the INM of nearly 90% of control cells; thus, REEP4 remains at the INM after mitotic NPC formation. REEP4 may be stored in the nucleus for the next mitosis when it promotes an increase of ER membrane curvature (Puhka et al., 2012; Kumar et al., 2019) or it may play still undefined roles in the nucleus during interphase.
During metaphase, REEP4 controls microtubule-dependent clearing of ER from chromatin and establishes high-curvature morphology of ER (Schlaitz et al., 2013; Kumar et al., 2019). Together with REEP4’s role in mitotic NPC assembly described here, these findings define REEP4 as a central regulator of ER and NE dynamics during mitosis.
Materials and methods
Cell culture, siRNAs, transfections, and generation of HeLa with endogenously HA-tagged REEP4
HeLa cells (ATCC CCL-2, authenticated by University of California Berkeley tissue culture facility) and HEK293T cells (ATCC CRL-11268) were cultured in DMEM, supplemented with 10% FBS, at 37°C in a humidified 5% CO2 incubator. Cells were checked for mycoplasma contamination approximately every 6 mo and were always negative. For plasmid transfections or siRNA/plasmid cotransfections, 1.8 × 105 cells were seeded in 1 well of a 24-well plate and transfected the next day with 0.4 µg plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. 4–6 h after transfection, cells were split onto coverslips and incubated for a total of 72 h with siRNAs/plasmids before fixation. siRNAs were transfected twice, first concomitant with cell seeding, and second, the next day when they had adhered, using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s instructions. 4–6 h after the second transfection, the cells were split onto coverslips and incubated for a total of 72 h before fixation.
To establish a HeLa cell line with REEP4 C-terminally HA-tagged at the endogenous locus, we used a CRISPR/Cas12-based gene-editing approach (Fueller et al., 2020) with the primers M1-REEP4 and M2-REEP4 and plasmid pMacTagP27 (Fueller et al., 2020). We analyzed REEP4-HA localization in a pool of cells as well as in two different clones obtained from single cells by light microscopy, and all yielded identical results. Single-cell clones were characterized by Western blotting, and the gene-edited locus was analyzed by sequencing a PCR product obtained from genomic DNA with primers targeting exon 7 of REEP4 and a region within the tagging cassette. siRNAs used were from Thermo Fisher Scientific and are specified in Table 1. Primer sequences are specified in Table 2. The images shown are from a clone in which only size-shifted, i.e., tagged, REEP4 was detected by Western blot and for which sequencing revealed that all alleles were homogeneously tagged but otherwise unaltered.
Table 1. siRNAs used in this study.
| Name | Catalogue number; si RNAi ID | Sequence (5′ to 3′) |
|---|---|---|
| REEP4 siRNA#1 | AM16708; 32438 | GGCUGUGAAGACCAAGAACTT |
| REEP4 siRNA#2 | 4392420; s37272 | GCAGAGAUCGUUACAGACATT |
| REEP4 siRNA#3 | 4392420; s37270 | CAAGAACAUUCGUGAAUAUTT |
| ELYS siRNA | AM16708; 108720 | GGUCUCCUCAACGACUUAATT |
| LBR siRNA | 4392420; s8101 | GCCUCUUAUUGAUGGAAGATT |
| Nup153 siRNA | 4392420; s19375 | GCAUCGCCGAAGAUAGAUUTT |
| Negative control siRNA | AM4611 | Not disclosed by manufacturer |
Table 2. Primers and oligonucleotides used in this study.
| Name | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| M1-REEP4 | CTCATGCTGTGCCTCCCCTTTCCCCAGGGCACCTCGCGCTCCCTGAAGGTTCGGACGAGGAAAAAGACTGTGCCCTCAGACGTGGACAGCTCAGGTGGAGGAGGTAGTG | Generation of amplicon for HA-tagging of REEP4 at the endogenous locus with CRISPR/Cas12 (Fueller et al., 2020) |
| M2-REEP4 | ATAGCCCTGGAGCCCTGCAGGGCACGAGGTAAGAAGGGGGCAGATGCAGCAGACCAAAAAATGGACAGCTAGGGTCTGCTGATCTACAAGAGTAGAAATTAGCTAGCTGCATCGGTACC | Generation of amplicon for HA-tagging of REEP4 at the endogenous locus with CRISPR/Cas12 (Fueller et al., 2020) |
| RGP09 | AGATACTGAGGCAGTCCCCC | Forward primer for sequencing of gene-edited REEP4 locus |
| RGP10 | GACTTTCCACACCGTCGACAT | Reverse primer for sequencing of gene-edited REEP4 locus |
| oAS363 | GCCCTGAAAATAAAGATTCTCTGTTGTGGGTCCACTACCAGATCCGGAGCTGTCCACGTCTGAGGGCAC | Generation of REEP4-V5-tev-HA (see text) |
| oAS365 | TCTTTATTTTCAGGGCAGTGGTAGTGGCAGCGGATACCCATACGATGTTCCAGATTAC | Generation of REEP4-V5-tev-HA (see text) |
| oAS416 | TCCGAGCAGGGGATTGGGGATAGGCTTGCCTCCACTACCAGATCCGGAGC | Generation of REEP4-V5-tev-HA (see text) |
| oAS417 | AATCCCCTGCTCGGACTGGATAGCACCCCCACAACAGAGAATCTTTATTTTC | Generation of REEP4-V5-tev-HA (see text) |
| oAS421 | TCCGGATCTGGTAGTGGAGGCAAGCCTATCCCCAATCCCCTGCTCGGACTGGATAGCACCCCCACAACAGAGAATCTTTATTTTCAGGGC | Generation of REEP3-V5-tev-HA (see text) |
| oAS422 | GCCCTGAAAATAAAGATTCTCTGTTGTGGGGGTGCTATCCAGTCCGAGCAGGGGATTGGGGATAGGCTTGCCTCCACTACCAGATCCGGA | Generation of REEP3-V5-tev-HA (see text) |
| oAS423 | CACTACCAGATCCGGAAAAATACACTTGTGGTCGTTTCTTC | Generation of REEP3-V5-tev-HA (see text) |
| oAS424 | AATCTTTATTTTCAGGGCAGTGGTAGTGGCAGCGGATACCCATACGATGTTCCAGATTAC | Generation of REEP3-V5-tev-HA (see text) |
| oAS492 | AGTGGTAGTGGCAGCGGACCCACAACAGAGAATCTTTATTTTCAGGGCGGCAAGCCTATCCCCAATCCCCTGCTCGGACTGGATAGCACCTCCGGATCTGGTAGTGGA | Generation of HA-tev-V5-Lap2β (see text) |
| oAS493 | TCCACTACCAGATCCGGAGGTGCTATCCAGTCCGAGCAGGGGATTGGGGATAGGCTTGCCGCCCTGAAAATAAAGATTCTCTGTTGTGGGTCCGCTGCCACTACCACT | Generation of HA-tev-V5-Lap2β (see text) |
| oAS494 | GCTGCCACTACCACTAGCGTAATCTGGAACATCGTATGGGTACATGGTGGCGACCGGTAGC | Generation of HA-tev-V5-Lap2β (see text) |
| oAS495 | CGGATCTGGTAGTGGAATGCCGGAGTTCCTAGAGGAC | Generation of HA-tev-V5-Lap2β (see text) |
| AB20 | GGCAGCGGATCCGGGTCTGGCTCCGGATCAAAAGACAATACTGTGCCTCTGAAGC | Amplification of TurboID sequence |
| AB21 | CTATGCGTAATCCGGTACATCGTAAGGGTAGCTAGCCTTTTCGGCAGAC | Amplification of TurboID sequence |
| AB03 | ATGTACCGGATTACGCATAGTGTACAAGTAAAGCGGCCGC | Amplification/linearization of REEP4-HA–containing vector |
| AB15 | CCAGACCCGGATCCGCTGCCGCTGTCCACGTCTGAGGG | Amplification/linearization of REEP4-HA–containing vector |
| AB22 | CGGACTCAGATCTCGAGCTCATGTCTGCGGCCATGAG | Amplification of REEP5 sequence |
| AB23 | CCAGACCCGGATCCGCTGCCGGTGCTCTTCTTTTCTTCACCC | Amplification of REEP5 sequence |
| AB24 | ATGTACCGGATTACGCATAGTGTACAAGTAAAGCGGCCGC | Amplification of REEP4-HA vector backbone (excluding REEP4-HA) |
| AB25 | GAGCTCGAGATCTGAGTCCG | Amplification of REEP4-HA vector backbone (excluding REEP4-HA) |
| AB26 | AGAAATCAGTCTGCGGTCTGC | Amplification of vector backbone (from 3xHA-miniturbo-NLS; Addgene) |
| AB27 | TGCGTAGTCTGGGACGTC | Amplification of vector backbone (from 3xHA-miniturbo-NLS; Addgene) |
| AB28 | ATGACGTCCCAGACTACGCAAAAGACAATACTGTGCCTCTGAAGC | Amplification of TurboID-NLS |
| AB29 | CTTTTCGGCAGACCGCAG | Amplification of TurboID-NLS |
| AB08 | CTAGAGGGCCCGTTTAAACC | Amplification/linearization of pcDNA5/FRT/TO vector |
| AB09 | CAAGCTTAAGTTTAAACGCTAGAGTCC | Amplification/linearization of pcDNA5/FRT/TO vector |
| AB10 | AGCGTTTAAACTTAAGCTTGATGGTGTCCTGGATGATCTGTC | Amplification of REEP4-TurboID-HA |
| AB11 | GGTTTAAACGGGCCCTCTAGCTATGCGTAATCCGGTACATCG | Amplification of REEP4- and REEP5-TurboID-HA |
| AB30 | AGCGTTTAAACTTAAGCTTGATGTCTGCGGCCATGAG | Amplification of REEP5-TurboID-HA |
| AB32 | GGTTTAAACGGGCCCTCTAGCCTCTAGATGCATGCTCGAGTC | Amplification of HA-TurboID-3xNLS |
| AB33 | AGCGTTTAAACTTAAGCTTGTACCCCTATGACGTCCCAGACTACGCAAAAGACAATAC | Amplification of HA-TurboID-3xNLS |
Molecular cloning
Plasmids encoding human REEP3-HA and REEP4-HA have been described previously (Schlaitz et al., 2013; Kumar et al., 2019) and are derived from pEGFP-N1 (Clontech), in which the EGFP sequence was replaced by a sequence encoding the HA tag. REEP4-HA was amplified with primers oAS363 and oAS365, which targeted the region between REEP4 and HA sequence and contained the sequences for a 3x(serine-glycine) linker and the TEV cleavage site (tev) in their overhangs. The resulting PCR product was purified and ligated using Gibson assembly (NEB) to generate REEP4-tev-HA. The V5 tag sequence was inserted into REEP4-tev-HA 5′ of the TEV cleavage site sequence by amplifying the plasmid with the primers oAS416 and oAS417, which contained the V5 sequences in their overhangs, and ligating the PCR product by Gibson assembly (NEB). The REEP3-V5-tev-HA construct was generated by amplification of the REEP3-HA plasmid using primers oAS423 and oAS424 and Gibson assembly of this PCR product with an annealed oligonucleotide generated from oligonucleotides oAS421 and oAS422.
For generating HA-tev-V5-Lap2β, pEGFP-Lap2β was used as the host plasmid (P30463; Euroscarf; Beaudouin et al., 2002). pEGFP-Lap2β was amplified without the GFP tag but with overhangs encoding the HA tag using oligonucleotides oAS494 and oAS495 and the annealed oligonucleotides oAS492 and oAS493, encoding linker, TEV cleavage site, V5 tag, and a second linker, were combined with the PCR product using Gibson assembly.
The plasmids encoding the TurboID enzyme (ID107172) and 3xHA-miniTurbo-ID-NLS (ID107173) were obtained from Addgene (Branon et al., 2018). The TurboID-HA sequence was amplified. REEP4-TurboID-HA was generated by PCR-based linearization of the REEP4-HA plasmid, excluding the HA tag sequence, and ligation with the TurboID-HA sequence. REEP5-TurboID-HA was generated by (1) amplifying the REEP4-HA plasmid backbone excluding the REEP4-HA sequence, (2) amplifying the REEP5 sequence (from HA-REEP5; Schlaitz et al., 2013), and (3) combination of these two amplicons with the Turbo-ID-HA-sequence. In plasmid ID107173, the sequence encoding miniTurbo-ID was replaced by TurboID to yield the HA-TurboID-3xNLS construct. The resulting sequences were transferred into pcDNA5/FRT/TO vectors. Cloning was done using Gibson assembly or circular polymerase extension cloning (Quan and Tian, 2009). Primers used were AB3, AB20-AB32, AB8-AB11, and AB15 as detailed in Table 2. GFP-Nesprin1α was a gift from Andreas Merdes (Université Paul Sabatier/CNRS, Toulouse, France; Espigat-Georger et al., 2016) and is based on pEGFP-C2 (Clontech).
Immunofluorescence and light microscopy
For standard immunofluorescence, cells were grown on coverslips or in 18-well glass-bottom µ dishes (Ibidi) and fixed with 4% formaldehyde. Primary and secondary antibodies used are specified in Table 3 and Table 4. DNA was labeled with Hoechst 33342 (Merck) or SytoxGreen (Thermo Fisher Scientific).
Table 3. Primary antibodies used in this study.
| Target (clone) | Source/reference and catalogue number | Host species | Dilutions used |
|---|---|---|---|
| HA tag (6E2) | CST, 2367 | Mouse | IF: 1/100 |
| GFP | Roche, 11814460001 | Mouse | WB: 1/5,000; IF: 1/500 |
| HA tag (3F10) | Sigma-Aldrich, 11867423001 | Rat | WB: 1/5,000 |
| Reticulon4 | Abcam, ab47085 | Rabbit | IF: 1/250 |
| Calnexin | Abcam, ab22595 | Rabbit | IF: 1/250 |
| REEP4 | Kumar et al., 2019 | Rabbit | WB: 1/1,000 |
| Lamin B1 | Abcam, ab16048 | Rabbit | IF: 1/500 |
| ELYS | Sigma-Aldrich, HPA031658 | Rabbit | IF: 1/200; WB: 1/1,000 |
| FG Nups (Mab414) | BioLegend, 902901 | Mouse | IF: 1/2,000 |
| RanBP2 | Pichler et al., 2002 | Goat | IF: 1/4,000 |
| Lamin A/C | Santa Cruz, sc-6215 | Goat | IF: 1/100 |
| V5 tag | CST, 13202 | Rabbit | IF: 1/1,000 |
| GRP94 (9G10) | Stressgen, SPA-850 | Rat | WB: 1/1,000 |
| SUN2 | Abcam, ab124916 | Rabbit | IF: 1/100 |
| Emerin | Proteintech, 10351-AP | Rabbit | IF: 1/250 |
| LBR | Abcam, ab32535 | Rabbit | IF: 1/100; WB: 1/1,000 |
| Nup153 | Abcam, ab84872 | Rabbit | WB: 1/1,000 |
| actin | Abcam, ab8224 | Mouse | WB: 1/1,000 |
| CENP-F | BD Transduction Laboratories, 610768 | Mouse | IF: 1/100 |
| gp210 | Bethyl Laboratories | Rabbit | IF: 1/100 |
| KDEL | Enzo, ADI-SPA-827 | Mouse | IF: 1/100 |
| Lamin B1 (CoraLite 647 conjugate) | Proteintech, CL647-66095 | Mouse | IF: 1/100 |
IF, immunofluorescence; WB, Western blotting.
Table 4. Secondary antibodies and reagents for detection of biotinylated proteins used in this study.
| Conjugation | Target species | Host species | Source and catalogue number | Dilutions used |
|---|---|---|---|---|
| IRDye 800CW | Rabbit | Goat | Licor, 926-32211 | WB: 1/10,000 |
| IRDye 680D | Mouse | Goat | Licor, 926-68070 | WB: 1/10,000 |
| Alexa Fluor 680 | Rat | Goat | Thermo Fisher Scientific, A-21096 | WB: 1/10,000 |
| Alexa Fluor 488 | Rabbit | Donkey | Thermo Fisher Scientific, A21206 | IF: 1/500 |
| Alexa Fluor 488 | Mouse | Donkey | Thermo Fisher Scientific, A21202 | IF: 1/500 |
| Alexa Fluor 647 | Rabbit | Donkey | Thermo Fisher Scientific, A31573 | IF: 1/500 |
| Alexa Fluor 647 | Mouse | Donkey | Thermo Fisher Scientific, A31571 | IF: 1/500 |
| Alexa Fluor 568 | Rabbit | Donkey | Thermo Fisher Scientific, A10042 | IF: 1/500 |
| Alexa Fluor 568 | Mouse | Donkey | Thermo Fisher Scientific, A10037 | IF: 1/500 |
| STAR 635P | Rabbit | Goat | Abberior, ST635P-1002 | IF (STED): 1/200 |
| ATTO 594 | Mouse | Goat | Sigma-Aldrich, 76085 | IF (STED): 1/200 |
| Alexa Fluor 647–streptavidin | NA | NA | Thermo Fisher Scientific, S21374 | IF: 1/2,000 |
| DyLight800-streptavidin | NA | NA | Thermo Fisher Scientific, 21851 | WB: 1/15,000 |
IF, immunofluorescence; NA, not applicable; WB, Western blotting.
For selective permeabilization of the plasma membrane, coverslips were treated with 40 µg/ml digitonin (Merck) in PBS for 5 min on ice. For staining of digitonin-treated coverslips, 5% normal donkey serum for blocking and antibody dilutions were prepared in PBS. For immunofluorescence following TEV assays, cells grown on coverslips were fixed in 4% formaldehyde, permeabilized with 0.1% Triton X-100 (Merck) in PBS and blocked with 5% normal donkey serum (Abcam) in PBS/0.1% Tween. Antibodies were diluted in PBS/0.1% Tween. For immunofluorescence of nucleoporins, coverslips were blocked and permeabilized by incubation with immunofluorescence buffer (1% BSA, 0.02% SDS, and 0.1% Triton X-100 in PBS). All antibody incubation and wash steps were performed using this buffer (Gomez-Cavazos and Hetzer, 2015). Primary antibody incubations with anti-NPC antibodies were performed overnight at 4°C. For all experiments described above, coverslips were mounted with ProLong Diamond (Thermo Fisher Scientific) directly after the staining procedure and left to cure overnight.
For visualization of NPCs by STED microscopy, cells were grown in eight-well µ-slides with polymer bottom (Ibidi). To improve labeling efficiency and resolution, osmotic swelling was performed as follows: cells were washed with PBS and incubated with deionized water for 5 min at room temperature before fixation (Jaiswal et al., 2019). Blocking and immunolabeling were performed as described above, and cells were kept in PBS and imaged within 24 h.
STED imaging was performed on a Leica TCS SP8 3X STED system (Leica Microsystems) with an HC PL APO 93×/1.30-NA STED glycerol objective lens at room temperature. STED images were acquired using the Leica Application Suite X software (Leica), with a white light laser at 635 or 594 nm and a 775-nm STED laser. A pinhole of 0.75 airy units was used, and HyD detectors (Leica Microsystems) were used for signal detection of STED samples. The bottom surfaces of NEs were acquired with <20 nm pixel size using z-stacks of three to four slices with 70–100-nm distance.
Confocal imaging was performed at room temperature using an HCX PL APO 63×/1.40-NA oil objective lens on the TCS SP8 system with Leica Application Suite X software (Leica) and HyD detectors or a 63×/1.40-NA Plan-Apochromat oil objective lens on an LSM 780 system (Zeiss) equipped with argon laser (488 nm), 561-nm DPSS laser, and 633-nm HeNe laser. At the LSM780, images were acquired with HyD detectors, a pinhole of 1 airy units was used, and acquisition was performed with Zen software (Zeiss).
Western blotting
Proteins separated by SDS-PAGE were transferred onto a nitrocellulose membrane (GE Healthcare Amersham) by Western blotting. Total protein on Western blots was detected with Revert700 Total Protein Stain (LI-COR Biosciences). For normalization of Western blot data, either total protein or actin staining was used. Western blot signals were quantified using Image Studio Lite (LI-COR Biosciences), and Western blot images were cropped and their levels adjusted in Photoshop (Adobe).
TEV cleavage assay
Cells were subjected to TEV assays 48 h after transient transfection of the respective TEV assay reporters. Cells were semipermeabilized by a 10-min incubation on ice with freshly prepared permeabilization buffer (20 mM Hepes, pH 7.5, 110 mM CH3COOK, 5 mM (CH3COO)2Mg, 250 mM sucrose, and 0.5 mM EGTA) containing 0.0025% digitonin (from 5% stocks in water, prepared the same day), washed three times with permeabilization buffer without digitonin, and incubated with purified NusA-TEV protease (Theerthagiri et al., 2010) for 15 min at 30°C. Immediately following incubation with NusA-TEV, coverslips were fixed with 4% formaldehyde in PBS and processed for immunofluorescence. The TEV assay procedure was adapted from Ungricht et al. (2015). Purified NusA-TEV protease was a gift of Rosemarie Ungricht and Ulrike Kutay (ETH Zurich, Zurich, Switzerland).
Image analysis and quantifications
Fiji was used for all microscopy image analysis (Schindelin et al., 2012). Microscopy images from NusA-TEV–treated samples and of ELYS intranuclear clusters that were qualitatively analyzed (Fig. 3, C, E, and F; and Fig. 4 C) were classified using the Blind Analysis Tools plugin developed by Astha Jaiswal and Holger Lorenz (https://imagej.net/Blind_Analysis_Tools).
To quantify nuclear rim signal either for HA in the TEV assays (Fig. 3 D) or for nucleoporin densities (Fig. 4 B; Fig. 5, B and D; and Fig. S2, A and D), background subtraction of the raw images was performed using a rolling ball algorithm with a diameter of 150 pixels (corresponding to the average cell size). Next, a mask was generated for the nuclear rim using either the staining of the nucleoporin itself or the staining of Lamin A/C or Lamin B1 in a separate channel. Segmentation was performed by applying a combination of the Gaussian filter, the Make Binary tool, iterative erosion, and iterative dilation operations. The segmentation was corrected manually for out-of-focus regions, and the final mask covered ≥30% of the whole rim. Mean nucleoporin fluorescence intensities were measured in the resulting rim masks. We analyzed three experiments in a blind manner and confirmed that blind analysis did not alter the quantification outcomes; therefore, the remaining experiments were analyzed without blinding.
For quantification of V5 mean intensity for Fig. 3 D, a mask was generated using the V5 signal. To determine the change in nuclear rim-localized REEP4 as a fraction of total expressed REEP4, nuclear rim mean intensity from the HA channel (corresponding to REEP4 at the INM; obtained as with nucleoporin mean intensities above) was divided by the mean intensity from the V5 channel (corresponding to the entire REEP4-V5-tev-HA pool) in control or ELYS RNAi cells after NusA-TEV treatment. This assay did not allow an absolute quantification of the nuclear rim pool of REEP4. However, by comparing the HA/V5 ratios in control and ELYS RNAi conditions, we could estimate the reduction of the REEP4 pool at the INM.
For quantification of CENP-F signal (Fig. 5 B), CENP-F mean intensity was measured inside the nuclear rim mask that was generated as described above. For each condition (control siRNA or REEP4 siRNA with quantification of ELYS, RanBP2, or gp210), the 20% of the cell population with the lowest CENP-F signal were classified as G1 cells, the 20% with the highest CENP-F signal were classified as G2 cells, and the corresponding nucleoporin mean intensities for these populations were determined.
STED images were deconvolved using the Decon Express function of Huygens Professional Software (Scientific Volume Imaging). For quantification of NPC numbers based on particle counting in STED images (Fig. S3, B and C), HeLa cells expressing endogenously GFP-tagged Nup107 (Otsuka et al., 2016) immunolabeled for GFP or for ELYS were used. A maximum projection of the deconvolved z-stack slices was generated in Fiji. Images were analyzed blindly using the Blind Analysis Tools plugin and an in-house macro based on the following steps. The channel with nucleoporin labeling was used to generate a mask of the NE surface to determine its area. In the same channel but with a different, manually adjusted threshold, particles containing nucleoporin signal and representing putative NPCs were detected after despeckling, smoothing, and watershedding of the images. Particles with the following parameters were selected and counted as NPCs: area 0.016–0.3 µm2, circularity 0.5–1, and roundness 0.5–1. Single NPCs in our images had a diameter of 140–150 nm, probably due to the location of epitopes for antibody binding and the added distance due to primary and secondary antibodies; thus, the area of one NPC was roughly 0.016 µm2 in our images. Particles that were >0.016 µm2 therefore likely represented clusters of NPCs that were not separated by the previously applied watershedding. These larger particles were divided by 0.016 and rounded to the next integer to determine the absolute number of NPCs. Finally, NPC number per surface area was calculated, and the resulting mean densities were normalized to controls.
To analyze the enrichment of REEP4 in the vicinity of NPCs from STED images (Fig. 2 H), NPC masks were obtained as described above based on ELYS signal. The diameter of these NPC masks was expanded by three pixels, and the raw integrated densities of REEP4 and ELYS were measured in these new dilated masks as well as on the entire nuclear surface. The NPC-associated signal was divided by the total NE signal for each protein to obtain the fraction of the total protein at NPCs. To calculate enrichment of the proteins in the NPC region over the entire NE surface, the value for the NPC fraction of each protein was divided by the fraction of the area of the NE occupied by the NPC masks.
For quantification of cytoplasmic mislocalization of RanBP2 in REEP4-depleted cells (Fig. S3, D and E), cells were first immunolabeled for RanBP2 and afterward for calnexin and CENP-F. Cells with weak CENP-F signal were preferentially imaged. RanBP2 and calnexin channels were used to generate masks of the nuclear rim and the whole cell, respectively. A new mask was generated by subtracting the nuclear region from the whole cell mask, and RanBP2 signal intensity was measured in this new mask to determine the amount of cytoplasmically localized RanBP2. The mean of the mean signal intensities in the cytoplasm was normalized to the respective control values.
All processing of microscopy data, statistical testing, and generation of graphs was performed in Excel (Microsoft). For the statistical tests used, data distribution was assumed to be normal but this was not formally tested. Images were cropped, levels adjusted, and channels combined using Photoshop (Adobe).
Proteinase K assay
HEK293T cells were lysed with a Dounce homogenizer in S250 buffer (10 mM Hepes, pH 7.5, 250 mM sucrose, 50 mM KCl, and 2.5 mM MgCl2) supplemented with 40 µM MG132 and 1 mM DTT and spun for 10 min at 1,000 g to obtain cytoplasmic (supernatant) and crude nuclear (pellet) fractions. The fractions were incubated with buffer only, unmodified proteinase K (Sigma-Aldrich), or agarose-coupled proteinase K (Sigma-Aldrich) in lysis buffer for 30 min at room temperature. Reactions were stopped by addition of PMSF (Sigma-Aldrich). Samples were supplemented with Laemmli buffer and analyzed by SDS-PAGE and immunoblotting.
EM
HeLa cells were seeded on coverslips and cultured until they reached a confluency of ∼80–90%. Cells were rinsed with PBS three times and then prefixed with 2.5% glutaraldehyde, 1.6% formaldehyde, and 2% sucrose in 50 mM cacodylate buffer for 30 min at room temperature. The fixative was washed out with 50 mM cacodylate buffer. After postfixation with 2% OsO4 for 1 h at 4°C, in the dark, cells were rinsed four times with distilled water and incubated overnight at 4°C in 0.5% uranyl acetate (in water). On the following day, coverslips were again rinsed four times with water, and cells were dehydrated stepwise with 40, 60, 70, 80, 90, 95, and 100% ethanol. Coverslips were immediately placed on capsules filled with Spurr-resin (Sigma-Aldrich) and polymerized at 60°C for ∼48 h. Polymerized blocks with embedded cells were sectioned using a Reichert Ultracut S Microtome (Leica Instruments) to a thickness of 80 nm. Poststaining with 3% uranyl acetate and lead citrate was performed. Serial sections on slot grids were imaged on a JE-1400 (Jeol), operating at 80 kV, equipped with a 4K × 4K digital camera (F416; TVIPS). Micrographs were adjusted in brightness and contrast with ImageJ.
Generation of stable cell lines for the inducible expression of TurboID constructs
For the generation of stable cell lines, Flp-In HEK293 cells (Gossen et al., 1995) were used and maintained in DMEM supplemented with 10% vol/vol FBS, 1 mM sodium pyruvate, 2 mM GlutaMAX, and 1% (vol/vol) penicillin/streptomycin at 37°C under 5% CO2. 10 µg/ml blasticidin was added to select for cells that express the Tet repressor and 100 µg/ml zeocin to select for cells that have no gene of interest inserted downstream of the Tet promoter. Mycoplasma testing was performed before experiments and showed no contamination. Cotransfection of the Flp-recombinase plasmid and the pcDNA5/FRT/TO vector (Thermo Fisher Scientific) containing the gene of interest (REEP4-TurboID-HA, REEP5-TurboID-HA, or HA-TurboID-3xNLS) was performed with Lipofectamine 2000 (Invitrogen) in a 1:3 ratio in cells growing in the medium described above but without antibiotics. After 48 h, selection of positive clones was started by changing the medium to DMEM supplemented with 10% vol/vol FBS, 1 mM sodium pyruvate, 2 mM GlutaMAX, 1% (vol/vol) penicillin/streptomycin, 100 µg/ml hygromycin, and 10 µg/ml blasticidin. Selection medium was changed every 2–4 d until colonies appeared after ∼8–10 d. Individual colonies were isolated and grown to confluency in 24-well plate wells (with 50 µg/ml hygromycin). Clones were tested for successful insertion of the construct by inducing its expression with 100 ng/ml doxycycline for 24 h.
BioID experiment
Before large scale BioID experiments, we determined concentrations of doxycycline for induction of all TurboID constructs to a level similar to endogenous REEP4. The doxycycline concentrations used were REEP4-TurboID, 4 ng/ml; REEP5-TurboID, 0.35 ng/ml; and TurboID-3xNLS, 2 ng/ml. 48 h before harvest, cells were seeded in 15-cm dishes at a density of ∼20%, and they were induced with doxycycline 24 h before cell harvest. For biotinylation of proteins, biotin was added to a final concentration of 500 µM 1 h before cell harvest. Cells were harvested by scraping, washed in PBS, and snap frozen in liquid nitrogen.
Cells were lysed at room temperature by resuspension in 1 ml lysis buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.4% SDS, 5 mM EDTA, 1 mM DTT, 1 mM PMSF [Sigma-Aldrich], complete protease inhibitors [Roche], 40 µM MG132 [Sigma-Aldrich], and PhosStop phosphatase inhibitors [Roche]) and passed 10 times through a 20G needle. Subsequently, cell lysates were sonicated in a BioRuptur (Diagenode) for 10 intervals of 30 s at high power, 30 s off to break protein aggregates and homogenize the lysate. 200 µl of 10% Triton X-100 was added to resolubilize precipitated SDS, and the samples were sonicated once more. 2.13 ml of 50 mM Tris, pH 7.5 (containing the protease and phosphatase inhibitors specified above), was added to reduce the salt concentration of the lysate to 150 mM, and lysates were distributed to two tubes. Lysates were cleared by centrifugation at 15,000 g for 10 min, and 3 ml of supernatants were combined with 50 µl of equilibrated streptavidin-coated beads (17-5113-01; GE) and incubated for 3 h at 4°C. After incubation, the beads were pelleted at 2,500 g for 2 min at 4°C and washed twice for 5 min each in a volume of 500 µl with the following buffers: wash 1, adjusted lysis buffer (42.5 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.12% SDS, 1.5 mM EDTA, 0.3 mM DTT, 0.3 mM PMSF, 1× complete protease inhibitor, 40 µM MG132, and 1× PhosSTOP), 4°C; wash 2, 2% SDS, room temperature; wash 3, 50 mM Hepes, pH 7.4, 1 mM EDTA, 500 mM NaCl, 1% Triton X-100, and 0.1% sodium deoxycholate, room temperature; and wash 4, 50 mM Tris-HCl, pH 7.5, 50 mM NaCl, and 0.1% Triton X-100 (procedure adapted from Schopp and Béthune [2018]).
To evaluate the success of the pulldown, 10% of beads were boiled at 95°C for 10 min with sample buffer containing 2 mM biotin and analyzed by immunoblotting. The remaining beads were washed once in urea buffer (8 M urea and 100 mM Tris-HCl, pH 8.8), resuspended in 50 µl urea buffer, and stored at −80°C until mass spectrometry analysis.
Mass spectrometry analysis of BioID samples
On-bead tryptic proteolysis
Streptavidin agarose beads were washed extensively with 8 M urea in 0.1 M Tris/HCl, pH 8.5, followed by reduction and alkylation steps. Finally, the beads were washed with 50 mM ammonium bicarbonate and incubated with trypsin overnight in 50 mM ammonium bicarbonate at 37°C. The resulting peptides were collected via centrifugation at 1,000 g for 5 min. The beads were then rinsed with 50 mM ammonium bicarbonate, and this second tryptic fraction was pooled with the first one. After acidification, tryptic peptides were desalted by stage tipping using Empore C18 47-mm disks and then dried in Speed-Vacuum. They were then resuspended in 5% formic acid and 5% acetonitrile for LC-MS/MS analysis.
Data acquisition
Peptides were analyzed by C18 nanoflow reversed-phase HPLC (Dionex Ultimate 3000 RSLC nano; Thermo Fisher Scientific) combined with orbitrap mass spectrometer (Q Exactive HF Orbitrap; Thermo Fisher Scientific). Samples were separated in an in-house-packed 75-µm internal diameter × 35-cm C18 column (Reprosil-Gold C18, 3 µm, 200 Å; Dr. Maisch) using 70-min linear gradients of 5–25%, 25–40%, and 40–95% acetonitrile in 0.1% formic acid with 300 nL/min flow in 90-min total run time. The scan sequence began with an MS1 spectrum (Orbitrap analysis; resolution 120,000; mass range 400–1,500 m/z; automatic gain control [AGC] target 16; and maximum injection time 60 ms). Up to 20 of the most intense ions per cycle were fragmented and analyzed in the orbitrap with data-dependent acquisition. MS2 analysis consisted of collision-induced dissociation (higher-energy collisional dissociation; resolution 30,000; AGC 15; normalized collision energy 27; and maximum injection time 60 ms). The isolation window for MS/MS was 2.0 m/z.
Data processing
The datasets were searched against the human Swissprot/Uniprot database. Proteome Discoverer 1.4 software was used to identify proteins. Carbamidomethylation of cysteine was used as fixed modification, and acetylation (protein N-termini) and oxidation of methionine were used as variable modifications. A maximum of two missed cleavages were allowed for the tryptic peptides. The precursor mass tolerance was set to 10 ppm, and both peptide and protein false discovery rates were set to 0.01. The other parameters were used with default settings. The database search was performed against the human Uniprot database (release 2016) with taxonomy Homo sapiens using Mascot and Sequest.
Analysis of mass spectrometry data
Fold-change (FC) calculation of identified proteins (sample of interest/control) was done using REPRINT (Resource for Evaluation of Protein Interaction Networks; Mellacheruvu et al., 2013; https://reprint-apms.org). Peptide spectrum matches (PSMs) for protein i identified with bait j were normalized to the sum of PSMs of all proteins identified by bait j. With Nj = ΣPSMi,j, the normalized PSMs for protein i identified with bait j, Ti,j, are calculated as Ti,j = PSMi,j/Nj. The PSMs for protein i were also normalized for each control sample x in the same way, and normalized PSMs for protein i obtained in control sample x, Ci,x, are calculated as Ci,x = PSMi,x/Nx. The averaged normalized count across all n controls was then computed as Ci = 1/nΣCi,x. The FC for every protein i was then calculated by the formula FCi,j = (Ti,j + α)/(Ci + α). α represents a small background factor to prevent division by 0 (Mellacheruvu et al., 2013). In our final analysis, we removed known contaminants documented in the REPRINT database. We considered proteins as top hits (listed in Fig. 2 D) that showed an FC equal to or larger than five and were detected in at least two of three replicates with an average of at least two peptide spectrum matches. The entire dataset is available in Table S1.
Online supplemental material
Fig. S1 (related to Fig. 3) shows the role of ELYS in REEP4 INM targeting. Fig. S2 (related to Fig. 4) shows characterization of the REEP4 depletion phenotype. Fig. S3 (related to Fig. 5) shows further characterization of the role of REEP4 in mitotic NPC assembly. Table S1 lists full datasets for MS analyses of BioID purification samples and REPRINT outputs.
Supplementary Material
lists full datasets for MS analyses of BioID purification samples and REPRINT outputs.
Acknowledgments
We are indebted to Ulrike Kutay and Rosemarie Ungricht for advice on the TEV assay, the generous gift of purified NusA-TEV protease, and thoughtful discussions. We gratefully acknowledge Frauke Melchior (ZMBH, Heidelberg University, Heidelberg, Germany) for anti-RanBP2 antibody and manifold support. We thank Elmar Schiebel for assistance with EM experiments and advice throughout this study. Thanks to Jan Ellenberg (EMBL Heidelberg, Heidelberg, Germany) for the GFP-Nup107 HeLa cell line, Andreas Merdes for the GFP-Nesprin1α plasmid, and Julien Béthune for advice on the BioID procedure. We are grateful to Holger Lorenz, Astha Jaiswal, Christian Hörth (ZMBH Imaging Facility), and Ulrike Engel (Nikon Imaging Center Heidelberg University) for expert light microscopy support, and to members of the Knop, Lemberg, Melchior, Schiebel, and Schuck laboratories for discussions, reagents, and advice. We thank Rebecca Heald and Sebastian Schuck for comments on the manuscript.
This work was funded by the German Research Foundation (project no. SCHL1876/2-1, A-L. Schlaitz) and a Chica and Heinz Schaller Foundation Short Term-Fellowship (to A. Schlaitz).
The authors declare no competing financial interests.
Author contributions: Conceptualization, Writing - original draft: A-L. Schlaitz; Funding acquisition, Supervision: A-L. Schlaitz, N. Ozlu; Investigation, Formal analysis: B. Golchoubian, A. Brunner, H. Bragulat-Teixidor, A. Neuner, B.A. Akarlar, A-L. Schlaitz; Methodology, Visualization, Writing - review & editing: B. Golchoubian, A. Brunner, H. Bragulat-Teixidor, A. Neuner.
References
- Adam, S.A., Marr R.S., and Gerace L.. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807–816. 10.1083/jcb.111.3.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, D.J., Vargas J.D., Hsiao J.P., and Hetzer M.W.. 2009. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J. Cell Biol. 186:183–191. 10.1083/jcb.200901106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin, J., Gerlich D., Daigle N., Eils R., and Ellenberg J.. 2002. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 108:83–96. 10.1016/S0092-8674(01)00627-4 [DOI] [PubMed] [Google Scholar]
- Bilir, Ş., Kojidani T., Mori C., Osakada H., Kobayashi S., Koujin T., Hiraoka Y., and Haraguchi T.. 2019. Roles of Nup133, Nup153 and membrane fenestrations in assembly of the nuclear pore complex at the end of mitosis. Genes Cells. 24:338–353. 10.1111/gtc.12677 [DOI] [PubMed] [Google Scholar]
- Boni, A., Politi A.Z., Strnad P., Xiang W., Hossain M.J., and Ellenberg J.. 2015. Live imaging and modeling of inner nuclear membrane targeting reveals its molecular requirements in mammalian cells. J. Cell Biol. 209:705–720. 10.1083/jcb.201409133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, J.P., Claridge J.K., Smith P.G., and Schnell J.R.. 2015. A conserved amphipathic helix is required for membrane tubule formation by Yop1p. Proc. Natl. Acad. Sci. USA. 112:E639–E648. 10.1073/pnas.1415882112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon, T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., Feldman J.L., Perrimon N., and Ting A.Y.. 2018. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36:880–887. 10.1038/nbt.4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, T.R., Lazarus M.D., Hetzer M.W., and Wente S.R.. 2009. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J. Cell Biol. 184:659–675. 10.1083/jcb.200806174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet, C.M., Talamas J.A., and Hetzer M.W.. 2010. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 141:1030–1041. 10.1016/j.cell.2010.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz, E., and Ellenberg J.. 2010. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J. Cell Biol. 191:15–22. 10.1083/jcb.201007076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz, E., Zanin E., Wurzenberger C., Braun M., Rabut G., Sironi L., and Ellenberg J.. 2008. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180:857–865. 10.1083/jcb.200707026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espigat-Georger, A., Dyachuk V., Chemin C., Emorine L., and Merdes A.. 2016. Nuclear alignment in myotubes requires centrosome proteins recruited by nesprin-1. J. Cell Sci. 129:4227–4237. 10.1242/jcs.191767 [DOI] [PubMed] [Google Scholar]
- Franz, C., Walczak R., Yavuz S., Santarella R., Gentzel M., Askjaer P., Galy V., Hetzer M., Mattaj I.W., and Antonin W.. 2007. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 8:165–172. 10.1038/sj.embor.7400889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fueller, J., Herbst K., Meurer M., Gubicza K., Kurtulmus B., Knopf J.D., Kirrmaier D., Buchmuller B.C., Pereira G., Lemberg M.K., and Knop M.. 2020. CRISPR-Cas12a-assisted PCR tagging of mammalian genes. J. Cell Biol. 219:e201910210. 10.1083/jcb.201910210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cavazos, J.S., and Hetzer M.W.. 2015. The nucleoporin gp210/Nup210 controls muscle differentiation by regulating nuclear envelope/ER homeostasis. J. Cell Biol. 208:671–681. 10.1083/jcb.201410047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen, M., Freundlieb S., Bender G., Müller G., Hillen W., and Bujard H.. 1995. Transcriptional activation by tetracyclines in mammalian cells. Science. 268:1766–1769. 10.1126/science.7792603 [DOI] [PubMed] [Google Scholar]
- Güttinger, S., Laurell E., and Kutay U.. 2009. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10:178–191. 10.1038/nrm2641 [DOI] [PubMed] [Google Scholar]
- Hampoelz, B., Andrés-Pons A., Kastritis P., and Beck M.. 2019. Structure and Assembly of the Nuclear Pore Complex. Annu. Rev. Biophys. 48:515–536. 10.1146/annurev-biophys-052118-115308 [DOI] [PubMed] [Google Scholar]
- Itzhak, D.N., Tyanova S., Cox J., and Borner G.H.. 2016. Global, quantitative and dynamic mapping of protein subcellular localization. eLife. 5:e16950. 10.7554/eLife.16950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal, A., Hoerth C.H., Zúñiga Pereira A.M., and Lorenz H.. 2019. Improved spatial resolution by induced live cell and organelle swelling in hypotonic solutions. Sci. Rep. 9:12911. 10.1038/s41598-019-49408-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtić, P., Schibler A.C., Wesley C.C., Pegoraro G., Misteli T., and Levy D.L.. 2019. The nucleoporin ELYS regulates nuclear size by controlling NPC number and nuclear import capacity. EMBO Rep. 20:e47283. 10.15252/embr.201847283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D., Golchoubian B., Belevich I., Jokitalo E., and Schlaitz A.-L.. 2019. REEP3 and REEP4 determine the tubular morphology of the endoplasmic reticulum during mitosis. Mol. Biol. Cell. 30:1377–1389. 10.1091/mbc.E18-11-0698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay, U., Jühlen R., and Antonin W.. 2021. Mitotic disassembly and reassembly of nuclear pore complexes. Trends Cell Biol. 31:1019–1033. 10.1016/j.tcb.2021.06.011 [DOI] [PubMed] [Google Scholar]
- Liao, H., Winkfein R.J., Mack G., Rattner J.B., and Yen T.J.. 1995. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell Biol. 130:507–518. 10.1083/jcb.130.3.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu, D., Wright Z., Couzens A.L., Lambert J.-P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y., et al. 2013. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 10:730–736. 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura, Y., Takagi M., Clever M., and Imamoto N.. 2016. ELYS regulates the localization of LBR by modulating its phosphorylation state. J. Cell Sci. 129:4200–4212. 10.1242/jcs.190678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka, S., and Ellenberg J.. 2018. Mechanisms of nuclear pore complex assembly - two different ways of building one molecular machine. FEBS Lett. 592:475–488. 10.1002/1873-3468.12905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka, S., Bui K.H., Schorb M., Hossain M.J., Politi A.Z., Koch B., Eltsov M., Beck M., and Ellenberg J.. 2016. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. eLife. 5:e19071. 10.7554/eLife.19071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka, S., Steyer A.M., Schorb M., Hériché J.-K., Hossain M.J., Sethi S., Kueblbeck M., Schwab Y., Beck M., and Ellenberg J.. 2018. Postmitotic nuclear pore assembly proceeds by radial dilation of small membrane openings. Nat. Struct. Mol. Biol. 25:21–28. 10.1038/s41594-017-0001-9 [DOI] [PubMed] [Google Scholar]
- Park, S.H., Zhu P.-P., Parker R.L., and Blackstone C.. 2010. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J. Clin. Invest. 120:1097–1110. 10.1172/JCI40979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar, S., Ungricht R., Tiefenboeck P., Leroux J.-C., and Kutay U.. 2017. Efficient protein targeting to the inner nuclear membrane requires Atlastin-dependent maintenance of ER topology. eLife. 6:e28202. 10.7554/eLife.28202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler, A., Gast A., Seeler J.S., Dejean A., and Melchior F.. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 108:109–120. 10.1016/S0092-8674(01)00633-X [DOI] [PubMed] [Google Scholar]
- Puhka, M., Joensuu M., Vihinen H., Belevich I., and Jokitalo E.. 2012. Progressive sheet-to-tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol. Biol. Cell. 23:2424–2432. 10.1091/mbc.e10-12-0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan, J., and Tian J.. 2009. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 4:e6441. 10.1371/journal.pone.0006441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala, B.A., Orjalo A.V., Shen Z., Briggs S., and Forbes D.J.. 2006. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA. 103:17801–17806. 10.1073/pnas.0608484103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, K.J., Kim D.I., Raida M., and Burke B.. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196:801–810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellhaus, A.K., De Magistris P., and Antonin W.. 2016. Nuclear Reformation at the End of Mitosis. J. Mol. Biol. 428(10, 10 Pt A):1962–1985. 10.1016/j.jmb.2015.09.016 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaitz, A.-L., Thompson J., Wong C.C.L., Yates J.R. III, and Heald R.. 2013. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev. Cell. 26:315–323. 10.1016/j.devcel.2013.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopp, I.M., and Béthune J.. 2018. Split-BioID - Proteomic Analysis of Context-specific Protein Complexes in Their Native Cellular Environment. J. Vis. Exp. (134):57479. 10.3791/57479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, L.K., Barentine A.E.S., Merta H., Schweighofer S., Zhang Y., Baddeley D., Bewersdorf J., and Bahmanyar S.. 2019. Dynamic nanoscale morphology of the ER surveyed by STED microscopy. J. Cell Biol. 218:83–96. 10.1083/jcb.201809107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, Y., Shemesh T., Prinz W.A., Palazzo A.F., Kozlov M.M., and Rapoport T.A.. 2010. Mechanisms determining the morphology of the peripheral ER. Cell. 143:774–788. 10.1016/j.cell.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerthagiri, G., Eisenhardt N., Schwarz H., and Antonin W.. 2010. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J. Cell Biol. 189:1129–1142. 10.1083/jcb.200912045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinti, M., Johnson C., Toth R., Ferrier D.E.K., and Mackintosh C.. 2012. Evolution of signal multiplexing by 14-3-3-binding 2R-ohnologue protein families in the vertebrates. Open Biol. 2:120103. 10.1098/rsob.120103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht, R., Klann M., Horvath P., and Kutay U.. 2015. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J. Cell Biol. 209:687–703. 10.1083/jcb.201409127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz, G.K., Prinz W.A., Shibata Y., Rist J.M., and Rapoport T.A.. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586. 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Vollmer, B., Schooley A., Sachdev R., Eisenhardt N., Schneider A.M., Sieverding C., Madlung J., Gerken U., Macek B., and Antonin W.. 2012. Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J. 31:4072–4084. 10.1038/emboj.2012.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer, B., Lorenz M., Moreno-Andrés D., Bodenhöfer M., De Magistris P., Astrinidis S.A., Schooley A., Flötenmeyer M., Leptihn S., and Antonin W.. 2015. Nup153 Recruits the Nup107-160 Complex to the Inner Nuclear Membrane for Interphasic Nuclear Pore Complex Assembly. Dev. Cell. 33:717–728. 10.1016/j.devcel.2015.04.027 [DOI] [PubMed] [Google Scholar]
- Weberruss, M., and Antonin W.. 2016. Perforating the nuclear boundary - how nuclear pore complexes assemble. J. Cell Sci. 129:4439–4447. 10.1242/jcs.194753 [DOI] [PubMed] [Google Scholar]
- Zhang, D., and Oliferenko S.. 2014. Tts1, the fission yeast homologue of the TMEM33 family, functions in NE remodeling during mitosis. Mol. Biol. Cell. 25:2970–2983. 10.1091/mbc.e13-12-0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists full datasets for MS analyses of BioID purification samples and REPRINT outputs.