Abstract
Background:
Duchenne muscular dystrophy (DMD) is an X-linked recessive disorder with early childhood onset characterized by profound loss of muscle strength and associated cardiomyopathy. DMD affects is most often caused by deletions involving single or multiple exons that disrupt the open reading frame of the DMD gene. Mutations causing loss or premature truncation of dystrophin result in dystrophin protein deficiency, which renders the plasma membrane of skeletal myofibers and cardiomyocytes weakened.
Aim of Review:
Genetic correction is in use to treat DMD, since several drugs have been already approved which partially restore dystrophin production through the use of antisense oligonucleotides. There are multiple ongoing clinical trials to evaluate the efficacy of treating DMD with micro-dystrophins delivered by adeno-associated viruses. Future approaches entail gene editing to target the single copy of the DMD gene on the X-chromosome. The primary, near-term goal is restoration of skeletal muscle dystrophin, and for some of these treatments, the efficacy in the heart is not fully known. Here, we discuss the anticipated cardiac outcomes of dystrophin-targeted therapies, and how this information informs genomic medicine for cardiomyopathies, especially in childhood.
Key Scientific Concepts of Review:
Many genetic treatment strategies are being implemented to treat DMD. Since most preclinical testing has focused on skeletal muscle, there is a gap in knowledge about the expected effects of these approaches on cardiac genetic correction and cardiomyopathy progression in DMD. Additional study is needed.
Keywords: heart failure, dilated cardiomyopathy, Duchenne muscular dystrophy, pediatrics, genome editing, gene therapy
INTRODUCTION
Duchenne muscular dystrophy (DMD) is an X-linked recessive neuromuscular disorder that affects skeletal and cardiac muscle [1, 2]. As an X-linked disorder, males present, usually before the age of 5 with falling, difficulty climbing stairs and raising up from the floor. Untreated, most DMD boys use a wheelchair for ambulation by 12 years of age [3]. Cardiomyopathy is also present, with detection of cardiac magnetic resonance changes around the age of ten. Respiratory musculature is also weakened and is often clinically apparent by mid-second decade. The cardiomyopathy of DMD progresses to a typical dilated cardiomyopathy (DCM) with both atrial and ventricular arrhythmias as the heart becomes more progressively dilated [4, 5]. The cardiomyopathy associated with DMD is amenable to traditional guideline-directed therapy of ACE inhibitors or angiotensin receptor blockade along with beta adrenergic blockade and mineralocorticoid receptor antagonism, and many patients receive these treatments early in life to slow progression of left ventricular dysfunction. Moreover, DMD patients are treated with glucocorticoid steroids, which also correlates with preservation of respiratory and cardiac muscle function [6–8]. Although glucocorticoid steroids, noninvasive respiratory treatments, and other supportive care have significantly extended the lifespan of DMD patients, the quality of life remains impaired by lack of skeletal muscle function. Because of the significant impairment of function along with a clear genetic understanding, there has been significant investment in devising and testing therapies to treat DMD. As might be expected, most have been developed and evaluated with skeletal muscle responses in mind.
DMD is caused by recessive mutations in the gene that encodes dystrophin, a 472 KDa cytoskeletal protein that stabilizes the sarcolemma of striated muscle cells [9]. Mutations in DMD can also cause Becker muscular dystrophy (BMD), a milder, less prevalent form of the disease characterized by delayed onset and slower progression. The dystrophin gene (DMD) spans approximately 2.4MB and is comprised of 79 exons on the X chromosome in the human genome [10]. The most common type of mutations responsible for DMD and BMD are exon deletions, followed by duplications, and then missense mutations, small deletions or insertions [3]. Importantly, mutational hotspots have been well-described and encompass exons 45-55 and exons 3-9 in the DMD gene [11]. Any mutation that prematurely terminates dystrophin protein production, resulting in complete or nearly complete abolishment of dystrophin protein production, tends to cause DMD. [12]. In contrast, BMD is associated with in-frame deletions, thereby leading to internally truncated dystrophin protein that appears to be stably expressed and partially functional, albeit mostly at lower levels than in healthy muscle. Importantly, it is widely reported that low levels of dystrophin restoration are expected to be clinically beneficial in DMD patients [13–15].
In mechanically active tissues such as the heart and skeletal muscle, dystrophin-deficient cardiomyocytes or skeletal myofibers are particularly susceptible to damage or death [16]. Genetic approaches to restore dystrophin expression or mitigate the secondary pathophysiological consequences that occur in the setting of DMD have emerged as viable therapeutic strategies to treat this disease [17–19]. The large size of the DMD gene has been problematic for viral gene therapy. However, because DMD is so well molecularly characterized and because it is encoded by a single copy on the X chromosome in males, this makes DMD an attractive target for genetic correction strategies.
1. Antisense Oligonucleotide-mediated Exon Skipping Therapy
Based on the observation that most DMD frameshifting mutations cluster in “hotspots” within the DMD gene, exon-skipping represents a feasible strategy to restore the open reading frame to generate internally truncated, partially functional dystrophin protein. It is estimated that up to 80% of DMD patients have genotypes that might be amenable to exon-skipping therapies [20]. Exon-skipping relies on antisense oligonucleotides (AONs), roughly 20-30 nucleotides, to manipulate pre-messenger RNA (mRNA) alternative splicing to circumvent mutated exons, effectively converting an out-of-frame DMD mutation to an in-frame BMD mutation. By transitioning an out of frame mutation to an in-frame mutation should, in principle, produce a dystrophin missing only a small internal component. However, the greatest challenge for AON-mediated approaches is their limited tissue penetration, especially in the heart. The relatively low level of dystrophin protein produced from AON-mediated therapies provides only partial correction. In order to improve stability and tissue penetration, AONs are chemically modified, for example, phosphorodiamidate morpholino oligomers and 2’-OMe-phosphorothioate. These chemical modifications have been designed, and in some cases selected, in order to minimize toxicity, improve stability, and increase bioavailability [21]. There are currently four AON therapies for DMD conditionally approved by the U.S. Food and Drug Administration (FDA).
Eteplirsen was approved by the FDA in 2016, representing the first exon-skipping drug to treat DMD [22]. Eteplirsen is a morpholino AON that is administered intravenously once weekly [23–25]. In 2019, golodirsen was approved by the FDA to treat exon 53-skippable DMD mutations [26]. Golodirsen is also a morpholino AON that is administered systemically once weekly [27]. In August of 2020, viltolarsen was also approved by the FDA to treat DMD patients with mutations that are genotypically eligible for exon 53 skipping [28]. It is estimated that approximately 8% of the DMD population is amenable to AON-mediated skipping of exon 53. Most recently, casimersen was approved to treat exon 45-skippable DMD patients. These AONs were approved by the FDA based on their ability to increase dystrophin expression in skeletal muscle. These AON compounds received conditional regulatory approvals based on documented dystrophin protein production in skeletal muscle, which is ascertained from skeletal muscle biopsies. Given the younger age of treatment, coupled with modest protein production produced from these methods, it may take 3-5 years or more, to detect functional improvements in walking duration, compared to historical controls. Additional clinical trials are underway to comprehensively assess potential clinical benefits for the FDA-approved AONs, as well as to test additional AONs that target different DMD exons, such as exon 45 [3].
It remains unknown at this time whether AON-exon skipping exerts benefit for cardiac function in DMD patients, and this possibility warrants further investigation. Targeting the heart with AONs has been historically challenging and is thought to be due to increased endosomal entrapment in cardiomyocytes compared to skeletal muscle fibers [29]. Additionally, disproportionate improvement in skeletal muscle dystrophin restoration without concomitant cardiac muscle dystrophin production might exacerbate cardiomyopathy owing increased skeletal muscle function, which would thereby increase cardiac demand. Because of the mechanical role of dystrophin at the membrane, including cardiomyocyte plasma membrane, increased cardiac contractile demand could exacerbate cardiomyocyte degeneration, thus creating a faster timeline for cardiac dysfunction in DMD. The degree to which this may occur is not well known in humans, and additional studies are needed in boys who have been receiving these therapies.
2. Gene Replacement Therapy
AON-mediated exon skipping is suitable for specific DMD mutations. At this point in time, DMD patients with exon 51-, 53-, or 45-skippable mutations can be treated with AONs, which leaves the considerable majority of DMD patients without viable therapy. Gene replacement therapy represents an alternative approach to restore dystrophin expression. Complete replacement of the DMD gene is precluded due to the large size of the coding region required to produce full length dystrophin protein (~11.4 kb) [3]. Based on limited reports of BMD patients with milder clinical outcomes and specific large internal deletions, it is predicted that micro-dystrophin constructs will be efficacious. Backed by studies in preclinical models like mice and dogs, these smaller, internally truncated dystrophins have been shown to be physiologically protective for skeletal muscle function and in some settings cardiac muscle [30–32]. How well these preclinical models reflect what occurs in humans is not known since the cardiomyopathy in dog models is variable and generally less robust in mouse models compared to human DMD-associated cardiomyopathy. Micro-dystrophins retain the capacity to fit into AAV vectors, and micro-dystrophins include the N- and C-termini as well as a small number of central spectrin repeat rod domains central for its function in skeletal muscle. Patients with BMD, the phenotypically milder form of muscular dystrophy, that harbor massive in-frame intragenic deletions in DMD have pointed to which regions of dystrophin are essential [33]. These insights have led to the design and development of minimally functional, truncated forms of dystrophin, referred to as micro-dystrophin or mini-dystrophins, depending on size [34]. Importantly, the micro-dystrophin cDNA constructs are within the packaging limit of AAV vectors (~4.6 kb), which are necessary for in vivo delivery to cardiac and skeletal muscles [35]. The constructs encode varying lengths of truncated dystrophin protein (from 228 KDa to 108 KDa) but contain the minimal amino-terminal actin-binding domain 1 (ABD1), several spectrin-like repeats, a few hinge regions and the carboxy-terminal cysteine rich domain [10]. Based on promising results from preclinical experimental studies in small rodent and canine models of DMD [36–38], multiple clinical trials are ongoing for AAV-mediated systemic delivery of micro-dystrophins. The clinical trials currently underway differ in study design, recruitment age, AAV serotype, promoter to direct expression of dystrophin, the precise details of the dystrophin construct, and immune suppression. Each trial requires one single injection of AAV micro-dystrophin to treat the patients.
Clinical trial NCT037669116 is using an AAV vector, AAVrh74, which is most similar to AAV8, and encodes a micro-dystrophin construct driven by the cardiac and skeletal muscle specific MHCK7 enhancer-promoter hybrid [34]. Strong cardiac and skeletal muscle expression is achieved with the MHCK7 promoter because it contains regulatory nucleotide sequences derived from the M-type creatine kinase promoter and alpha-myosin heavy chain enhancer. In this randomized, double-blind, placebo-controlled phase II clinical trial, safety and efficacy of micro-dystrophin delivery is being tested in DMD males between 4 and 7 years of age. Inclusion criteria include the use of a stable oral corticosteroid dose for at least 12 weeks. A second non-randomized phase I/IIa clinical trial (NCT03375164) is assessing this same micro-dystrophin construct in two different age cohorts of males. Cohort A will comprise very young DMD patients between 3 months and 3 years of age and no previous treatment with corticosteroids, whereas Cohort B will comprise patients between 4 and 7 years of age and a stable dose of oral corticosteroids for at least 12 weeks before screening. In September 2020, results of NCT03375164 were reported in JAMA Neurology [39]. Results from this trial indicate that, upon a single dose of 2.0 x 1014 viral particles per kilogram body weight rAAVrh74.MHCK7.micro-dystrophin administered intravenously, no serious adverse events occurred. Furthermore, robust expression of micro-dystrophin was detected in at least 80% of all skeletal muscle fibers examined, with mostly correct localization at the sarcolemma. Finally, there were notable functional improvements, as well as reduced CK levels that were maintained for one year. A more recent study using skeletal muscle magnetic resonance imaging reported decreased fat infiltration in a cohort of DMD patients who received rAAVrh74.MHCK7.micro-dystrophin therapy, compared to an age-matched natural history cohort [40].
NCT03368742 is a randomized, controlled, open-label phase I/II study involving the use of AAV9 and a micro-dystrophin construct driven by a muscle specific creatine kinase 8 (CK8) promoter [34]. The recruitment age for this trial is between 4 and 17 years of age, and this trial is assessing the safety and tolerability in DMD. Inclusion criteria for this trial includes the use of oral corticosteroids for at least 12 weeks. Another trial, initiated in 2017, is conducting a multicenter, open-label, non-randomized phase I clinical trial to evaluate the safety and tolerability of their micro-dystrophin construct driven by a minimized murine muscle-specific creatine kinase (CK) promoter and delivered by a single infusion of AAV9 vectors [41]. In February 2020, the phase III, randomized, double-blind, placebo-controlled trial was initiated to assess ambulatory function and monitor safety of the AAV9 micro-dystrophin investigational therapy (NCT04281485).
In contrast to AON exon-skipping therapies, viral gene delivery has the potential to directly target the myocardium with high efficiency; AAV9 and rAAVrh74 possess high cardiac tissue tropism, however preexisting neutralizing antibodies in potential recipients may limit the available number of treatable patients [42]. The ongoing clinical trials of micro-dystrophin include young DMD boys. Therefore, these studies can potentially address the degree to which micro-dystrophin can prevent or mitigate the cardiomyopathy in DMD patients. Fundamentally, these clinical trials for neuromuscular disease represent the first gene therapy trials for cardiomyopathy in children. Despite the noted progress, some challenges remain. It should be noted that corticosteroids are used at high doses in most of these clinical trials to prevent adverse immune responses to the AAV vectors. Corticosteroid use in DMD patients has been shown to improve muscle function and strength, so this could represent a potential confounding factor in these studies. Furthermore, despite compelling results from micro-dystrophin studies in small animal models of DMD (e.g., mdx mice), these animal models often experience a milder skeletal and cardiac muscle phenotype than humans, so a functional benefit in DMD patients may be more difficult to achieve. Furthermore, micro-dystrophins are approximately 40% shorter than the average length of truncated dystrophins in BMD patients [43], so micro-dystrophin therapies in DMD patients may fall short of the mild BMD phenotype. Additionally, not all myofibers or cardiomyocytes are transduced by these AAVs, and their duration in the human myofiber and cardiomyocyte setting is not known. There is ongoing degeneration in BMD skeletal muscle, and the micro-dystrophin vectors are not expected to transduce satellite cells, the stem cells of muscle. There is less known about the dynamics of AAV-mediated transgene expression in DMD hearts, and cardiac biopsies may be required to determine if expression is durably maintained long-term. Nevertheless, micro-dystrophins as a gene replacement approach in dystrophin-deficient DMD patients seems to remain a promising therapy.
3. Therapeutic Potential of CRISPR-Mediated Genome Editing
Somatic cell genome editing with CRISPR (clustered regularly interspaced short palindromic repeat)-Cas9 technology is evolving at a rapid pace and has the potential to revolutionize the treatment or prevention of human disease, particularly monogenetic, X-linked recessive disorders like DMD [44]. Notably, this approach is aimed at correcting the somatic cells and not the germline to avoid ethical concerns with germline gene editing. CRISPR-Cas9 represents a tool that can be programmed to produce precisely targeted modifications in the genome and epigenome [45]. Similar to exon-skipping and micro-dystrophin gene replacement strategies, CRISPR-Cas9-based approaches aim to restore the production by creating internally truncated, partially functional dystrophin protein.
A number of elegant preclinical studies have demonstrated the immense power of harnessing this tool to permanently correct certain DMD mutations in vitro and in vivo. In essence, this is accomplished by designing guide RNAs (gRNAs) that hybridize to specific complementary regions in the DMD gene, which enables the Cas9 endonuclease to make precise, double-stranded DNA breaks [46]. This in turn leads to editing of the gene via activation endogenous DNA repair pathways, namely nonhomologous end joining (NHEJ) or homology directed repair (HDR), depending on the cell type and absence/presence of a DNA template. The field has predominantly focused attention on NHEJ-based CRISPR-Cas9 editing because with current tools, HDR is a more efficient event in dividing cells. The end result is restoration of the DMD reading frame, such that a single exon or multiple exons is/are excluded from the mature mRNA transcript, thereby leading to the translation of truncated, partially active dystrophin protein. Like AON-mediated correction, CRISPR-Cas9 mediated genetic correction generates a dystrophin protein with more of its internal components, compared to micro-dystrophin gene therapy.
A series of reports in 2016 and 2017 demonstrated the feasibility of this strategy in mdx mouse models of DMD [47–50]. These studies used AAV9 vectors to locally or systemically deliver the CRISPR-Cas9 gene editing machinery to diseased mice, which resulted in significantly increased dystrophin protein production, close to the full length size, as well functional improvements in both cardiac and skeletal muscle tissues. Additional CRISPR-based gene editing studies for DMD have been carried out in human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) with similar success [51–53]. In a 2018, gene editing was used to generate dystrophin expression in a large animal model of DMD, a canine model. This was achieved using AAV9 vectors carrying a single gRNA and Cas9 driven by a CK promoter which was delivered intramuscularly or systemically to the deltaE50-MD dog model of DMD, which harbors a mutation that corresponds to a mutational “hotspot” in humans [54]. Importantly, robust expression of dystrophin was observed in cardiac muscle as well as various skeletal muscles upon treatment. While improvements in muscle histology were also observed after viral treatment, the study was not sufficiently powered to assess muscle or heart functional response to gene editing. In a more recent study, CRISPR-Cas9-based adenine base and prime editing was used to correct exon 51 deletions in mice and also in human induced pluripotent stem cells (hiPSCs) [55]. The hiPSCs were subsequently differentiated into cardiomyocytes, demonstrating correction of dystrophin production, along with functional improvement in cellular features of cardiomyopathy [55]. Further experiments in larger animal models, as well as optimization of in vivo delivery and minimization of off-target effects, are necessary before these potential therapies can progress to human clinical trials.
The nuclease activity of Cas9 can be rendered catalytically inactive by specific point mutations (referred to as “dCas9”) and fused effectors that activate or repress gene expression [45]. This approach, although still in its infancy, has the potential to completely reprogram the transcriptional landscape in a cell [56]. An immediately apparent advantage of the dCas9 system lies in its ability to modify the genome without creating double-stranded DNA breaks, thus avoiding the concern of off-target mutagenesis. The application of CRISPR-dCas9 to modulate endogenous gene expression has already been tested in mouse models by designing gRNAs targeting cis-regulatory regions (e.g., enhancers, promoters, and silencers) in the genome [57–59]. Systemic administration of AAV9 vectors carrying gRNAs targeting the Mef2d gene in transgenic mice expressing dCas9 fused to the VPR transcriptional activator resulted in not only significant up-regulation MEF2D protein in the postnatal heart, but also the expected functional effects such as pathological gene expression, cardiac hypertrophy, and fibrosis [60]. This approach is very powerful since this method can be used to regulate the expression of any human gene, even those too large for AAV vectors. While upregulation of dystrophin is not a viable strategy to treat DMD, transcriptional upregulation of compensating proteins could be. A study showed that transcriptional upregulation of Lama1 via dCas9 fused to transcriptional activator VP64 was effective in a mouse model of Lama2-mediated congenital muscular dystrophy, leading to significant functional improvements and thwarting disease progression [61]. This study supports the concept that CRISPR-dCas9-mediated transcriptional modulation of modifier genes, i.e., a mutation-independent approach, may represent an alternative avenue to treat DMD. Indeed, this approach was also used to significantly upregulate the dystrophin homolog, utrophin, in human DMD myoblasts [62]. The dCas9 nuclease is capable of more than activation endogenous gene expression. It can also be engineered to repress gene expression by fusion to transcriptional effectors (KRAB) or epigenetic modifiers (LSD1) [45]. Ultimately, the CRISPR-based genome editing toolkit may be used to therapeutically modulate the expression of genes in the heart that are expected to improve myocardial performance or delay adverse ventricular remodeling.
Despite exciting and rapid progress in the field of CRISPR-based gene editing, numerous obstacles remain before this technology could be leveraged as a safe and effective therapy in the clinical setting. One of the major challenges is optimization of delivering the gene editing machinery to target tissues, especially the heart. Factors such as dosage, time, and frequency of intervention must be addressed. Toxicity has been observed in the human DMD trials, seen as complement activation, and may be indicating upper limits of AAV dosing. Correspondingly, insufficient dosing is less likely to result in meaningful functional benefit. The timing of genetic intervention is another important consideration, especially for clinical trials, where there is a need to more urgently identify clinical improvement. Gene editing, like gene therapy, may only require a single treatment. However, despite AAV9 and a few other AAV serotypes having high cardiac and skeletal muscle tropism, many people in the population, including DMD patients, have neutralizing antibodies (nAbs) against various AAVs, which could reduce the effectiveness of the therapy [42]. For gene editing, off-target mutations remain a serious concern, so gRNA safety must be comprehensively evaluated prior to use in humans. Furthermore, once administered to a patient, it may be desirable to ablate the Cas9 nuclease to eliminate its activity and potential side effect profile. Finally, one key advantage of CRISPR-Cas9-based gene editing, in contrast to gene replacement approaches with micro-dystrophin, is the ability to manipulate endogenous loci, which are subject to natural regulation of the gene rather than artificially through elements in the vector. This detail may be particularly relevant for upregulation of proteins without toxic overexpression.
4. Concluding Remarks
Genetic correction strategies are becoming a reality for treating genetic diseases and even some non-genetic disorders [63, 64]. DMD has emerged as a prime target for genetic correction, given its early onset, severe symptoms and well understood genetic defects. Multiple AON based compounds have been approved for use in the US, but these treat only a fraction of DMD patients and are comparatively inefficient producing only 1-5% of normal dystrophin content. The large size of the dystrophin gene significantly slowed advances in gene therapy, but even this concern has been met through the use of micro-dystrophins. The ongoing clinical trials with micro-dystrophins also represent the first gene therapy for cardiomyopathy in children. The future looks to genetic correction strategies. In theory, genetic correction should restore expression of more of the internal components of dystrophin, which may have important functional consequences to cardiac function. In each case, it will be essential to monitor the effects of these treatments on the heart.
Fig. 1.
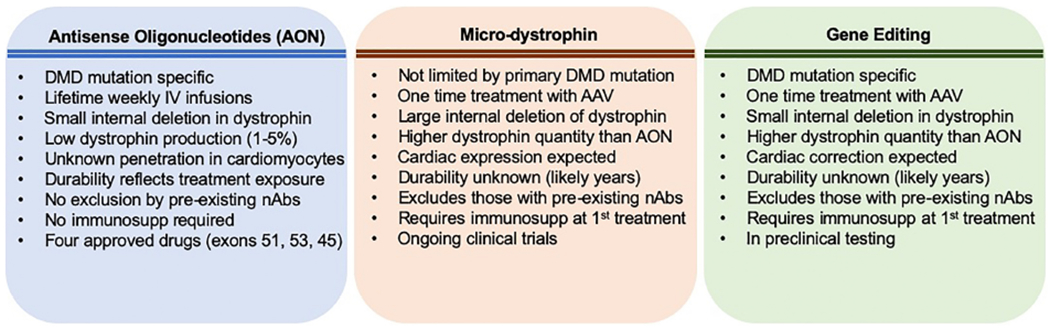
Comparison of major approaches for genetic correction for DMD. Four different antisense oligonucleotides (AON) have been approved for use in the US. These target specific DMD mutations. Adeno-associated viral gene therapy is being investigated in ongoing clinical trials and aims to produce micro-dystrophin in skeletal and cardiac muscle. Gene editing is also contemplated for DMD; gene editing has been tested in preclinical models, including a large animal model for DMD.
Highlights.
Antisense oligonucleotides are approved to treat DMD
Gene therapy to express micro-dystrophin is in clinical trial
Gene editing for DMD is being assessed preclinically
Each of these genetic correction strategies may differentially affect the heart
Acknowledgments
Supported by: NIH, Department of Defense, and the American Heart Association, Leducq Fondation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
Dr. McNally consults for Amgen, Avidity, AstraZeneca, Cytokinetics, 4D Molecular Therapeutics, Janssen, Pfizer, PepGen, Tenaya Therapeutics and Invitae. She is the founder of Ikaika Therapeutics. These activities are unrelated to the content of this work.
JRJ has no conflicts to disclose.
REFERENCES
- [1].Ryder S, Leadley RM, Armstrong N, Westwood M, de Kock S, Butt T, Jain M, Kleijnen J, The burden, epidemiology, costs and treatment for Duchenne muscular dystrophy: an evidence review, Orphanet J Rare Dis 12(1) (2017) 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mah JK, Korngut L, Dykeman J, Day L, Pringsheim T, Jette N, A systematic review and meta-analysis on the epidemiology of Duchenne and Becker muscular dystrophy, Neuromuscul Disord 24(6) (2014) 482–91. [DOI] [PubMed] [Google Scholar]
- [3].Duan D, Goemans N, Takeda S, Mercuri E, Aartsma-Rus A, Duchenne muscular dystrophy, Nat Rev Dis Primers 7(1) (2021) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Buddhe S, Cripe L, Friedland-Little J, Kertesz N, Eghtesady P, Finder J, Hor K, Judge DP, Kinnett K, McNally EM, Raman S, Thompson WR, Wagner KR, Olson AK, Cardiac Management of the Patient With Duchenne Muscular Dystrophy, Pediatrics 142(Suppl 2) (2018) S72–s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].McNally EM, Kaltman JR, Benson DW, Canter CE, Cripe LH, Duan D, Finder JD, Groh WJ, Hoffman EP, Judge DP, Kertesz N, Kinnett K, Kirsch R, Metzger JM, Pearson GD, Rafael-Fortney JA, Raman SV, Spurney CF, Targum SL, Wagner KR, Markham LW, Contemporary cardiac issues in Duchenne muscular dystrophy. Working Group of the National Heart, Lung, and Blood Institute in collaboration with Parent Project Muscular Dystrophy, Circulation 131(18) (2015) 1590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Griggs RC, Miller JP, Greenberg CR, Fehlings DL, Pestronk A, Mendell JR, Moxley RT 3rd, King W, Kissel JT, Cwik V, Vanasse M, Florence JM, Pandya S, Dubow JS, Meyer JM, Efficacy and safety of deflazacort vs prednisone and placebo for Duchenne muscular dystrophy, Neurology 87(20) (2016) 2123–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Manzur AY, Kuntzer T, Pike M, Swan A, Glucocorticoid corticosteroids for Duchenne muscular dystrophy, Cochrane Database Syst Rev (2) (2004) CD003725. [DOI] [PubMed] [Google Scholar]
- [8].Bylo M, Farewell R, Coppenrath VA, Yogaratnam D, A Review of Deflazacort for Patients With Duchenne Muscular Dystrophy, Ann Pharmacother 54(8) (2020) 788–794. [DOI] [PubMed] [Google Scholar]
- [9].Hoffman EP, Brown RH Jr., Kunkel LM, Dystrophin: the protein product of the Duchenne muscular dystrophy locus, Cell 51(6) (1987) 919–28. [DOI] [PubMed] [Google Scholar]
- [10].Gao QQ, McNally EM, The Dystrophin Complex: Structure, Function, and Implications for Therapy, Compr Physiol 5(3) (2015) 1223–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Flanigan KM, Dunn DM, von Niederhausern A, Soltanzadeh P, Gappmaier E, Howard MT, Sampson JB, Mendell JR, Wall C, King WM, Pestronk A, Florence JM, Connolly AM, Mathews KD, Stephan CM, Laubenthal KS, Wong BL, Morehart PJ, Meyer A, Finkel RS, Bonnemann CG, Medne L, Day JW, Dalton JC, Margolis MK, Hinton VJ, C. United Dystrophinopathy Project, Weiss RB, Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort, Hum Mutat 30(12) (2009) 1657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Garcia-Rodriguez R, Hiller M, Jimenez-Gracia L, van der Pal Z, Balog J, Adamzek K, Aartsma-Rus A, Spitali P, Premature termination codons in the DMD gene cause reduced local mRNA synthesis, Proc Natl Acad Sci U S A 117(28) (2020) 16456–16464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Neri M, Torelli S, Brown S, Ugo I, Sabatelli P, Merlini L, Spitali P, Rimessi P, Gualandi F, Sewry C, Ferlini A, Muntoni F, Dystrophin levels as low as 30% are sufficient to avoid muscular dystrophy in the human, Neuromuscul Disord 17(11-12) (2007) 913–8. [DOI] [PubMed] [Google Scholar]
- [14].Gentil C, Le Guiner C, Falcone S, Hogrel JY, Peccate C, Lorain S, Benkhelifa-Ziyyat S, Guigand L, Montus M, Servais L, Voit T, Pietri-Rouxel F, Dystrophin Threshold Level Necessary for Normalization of Neuronal Nitric Oxide Synthase, Inducible Nitric Oxide Synthase, and Ryanodine Receptor-Calcium Release Channel Type 1 Nitrosylation in Golden Retriever Muscular Dystrophy Dystrophinopathy, Hum Gene Ther 27(9) (2016) 712–26. [DOI] [PubMed] [Google Scholar]
- [15].Godfrey C, Muses S, McClorey G, Wells KE, Coursindel T, Terry RL, Betts C, Hammond S, O’Donovan L, Hildyard J, El Andaloussi S, Gait MJ, Wood MJ, Wells DJ, How much dystrophin is enough: the physiological consequences of different levels of dystrophin in the mdx mouse, Hum Mol Genet 24(15) (2015) 4225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lammerding J, Lee RT, Torn apart: membrane rupture in muscular dystrophies and associated cardiomyopathies, J Clin Invest 117(7) (2007) 1749–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chemello F, Bassel-Duby R, Olson EN, Correction of muscular dystrophies by CRISPR gene editing, J Clin Invest 130(6) (2020) 2766–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Olson EN, Toward the correction of muscular dystrophy by gene editing, Proceedings of the National Academy of Sciences (2021) 202004840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Min YL, Bassel-Duby R, Olson EN, CRISPR Correction of Duchenne Muscular Dystrophy, Annu Rev Med 70 (2019) 239–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, Kosma K, Dawkins H, Lamont L, Roy AJ, Chamova T, Guergueltcheva V, Chan S, Korngut L, Campbell C, Dai Y, Wang J, Barisic N, Brabec P, Lahdetie J, Walter MC, Schreiber-Katz O, Karcagi V, Garami M, Viswanathan V, Bayat F, Buccella F, Kimura E, Koeks Z, van den Bergen JC, Rodrigues M, Roxburgh R, Lusakowska A, Kostera-Pruszczyk A, Zimowski J, Santos R, Neagu E, Artemieva S, Rasic VM, Vojinovic D, Posada M, Bloetzer C, Jeannet PY, Joncourt F, Diaz-Manera J, Gallardo E, Karaduman AA, Topaloglu H, El Sherif R, Stringer A, Shatillo AV, Martin AS, Peay HL, Bellgard MI, Kirschner J, Flanigan KM, Straub V, Bushby K, Verschuuren J, Aartsma-Rus A, Beroud C, Lochmuller H, The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations, Hum Mutat 36(4) (2015) 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nguyen Q, Yokota T, Antisense oligonucleotides for the treatment of cardiomyopathy in Duchenne muscular dystrophy, Am J Transl Res 11(3) (2019) 1202–1218. [PMC free article] [PubMed] [Google Scholar]
- [22].Aartsma-Rus A, Krieg AM, FDA Approves Eteplirsen for Duchenne Muscular Dystrophy: The Next Chapter in the Eteplirsen Saga, Nucleic Acid Ther 27(1) (2017) 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Alfano LN, Charleston JS, Connolly AM, Cripe L, Donoghue C, Dracker R, Dworzak J, Eliopoulos H, Frank DE, Lewis S, Lucas K, Lynch J, Milici AJ, Flynt A, Naughton E, Rodino-Klapac LR, Sahenk Z, Schnell FJ, Young GD, Mendell JR, Lowes LP, Long-term treatment with eteplirsen in nonambulatory patients with Duchenne muscular dystrophy, Medicine (Baltimore) 98(26) (2019) e15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lim KR, Maruyama R, Yokota T, Eteplirsen in the treatment of Duchenne muscular dystrophy, Drug Des Devel Ther 11 (2017) 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mendell JR, Goemans N, Lowes LP, Alfano LN, Berry K, Shao J, Kaye EM, Mercuri E, Eteplirsen Study G, Telethon Foundation DMDIN, Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy, Ann Neurol 79(2) (2016) 257–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Heo YA, Golodirsen: First Approval, Drugs 80(3) (2020) 329–333. [DOI] [PubMed] [Google Scholar]
- [27].Frank DE, Schnell FJ, Akana C, El-Husayni SH, Desjardins CA, Morgan J, Charleston JS, Sardone V, Domingos J, Dickson G, Straub V, Guglieri M, Mercuri E, Servais L, Muntoni F, S.-N.S. Group, Increased dystrophin production with golodirsen in patients with Duchenne muscular dystrophy, Neurology 94(21) (2020) e2270–e2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Clemens PR, Rao VK, Connolly AM, Harper AD, Mah JK, Smith EC, McDonald CM, Zaidman CM, Morgenroth LP, Osaki H, Satou Y, Yamashita T, Hoffman EP, C.D. Investigators, Safety, Tolerability, and Efficacy of Viltolarsen in Boys With Duchenne Muscular Dystrophy Amenable to Exon 53 Skipping: A Phase 2 Randomized Clinical Trial, JAMA Neurol 77(8) (2020) 982–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Godfrey C, Desviat LR, Smedsrod B, Pietri-Rouxel F, Denti MA, Disterer P, Lorain S, Nogales-Gadea G, Sardone V, Anwar R, El Andaloussi S, Lehto T, Khoo B, Brolin C, van Roon-Mom WM, Goyenvalle A, Aartsma-Rus A, Arechavala-Gomeza V, Delivery is key: lessons learnt from developing splice-switching antisense therapies, EMBO Mol Med 9(5) (2017) 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Banks GB, Gregorevic P, Allen JM, Finn EE, Chamberlain JS, Functional capacity of dystrophins carrying deletions in the N-terminal actin-binding domain, Hum Mol Genet 16(17) (2007) 2105–13. [DOI] [PubMed] [Google Scholar]
- [31].Howard ZM, Dorn LE, Lowe J, Gertzen MD, Ciccone P, Rastogi N, Odom GL, Accornero F, Chamberlain JS, Rafael-Fortney JA, Micro-dystrophin gene therapy prevents heart failure in an improved Duchenne muscular dystrophy cardiomyopathy mouse model, JCI Insight 6(7) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yue Y, Pan X, Hakim CH, Kodippili K, Zhang K, Shin JH, Yang HT, McDonald T, Duan D, Safe and bodywide muscle transduction in young adult Duchenne muscular dystrophy dogs with adeno-associated virus, Hum Mol Genet 24(20) (2015) 5880–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, Zubrzycka-Gaarn EE, Bulman DE, Harris JB, Davies KE, Very mild muscular dystrophy associated with the deletion of 46% of dystrophin, Nature 343(6254) (1990) 180–2. [DOI] [PubMed] [Google Scholar]
- [34].Duan D, Systemic AAV Micro-dystrophin Gene Therapy for Duchenne Muscular Dystrophy, Mol Ther 26(10) (2018) 2337–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gregorevic P, Blankinship MJ, Allen JM, Crawford RW, Meuse L, Miller DG, Russell DW, Chamberlain JS, Systemic delivery of genes to striated muscles using adeno-associated viral vectors, Nat Med 10(8) (2004) 828–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gregorevic P, Blankinship MJ, Allen JM, Chamberlain JS, Systemic microdystrophin gene delivery improves skeletal muscle structure and function in old dystrophic mdx mice, Mol Ther 16(4) (2008) 657–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yue Y, Li Z, Harper SQ, Davisson RL, Chamberlain JS, Duan D, Microdystrophin gene therapy of cardiomyopathy restores dystrophin-glycoprotein complex and improves sarcolemma integrity in the mdx mouse heart, Circulation 108(13) (2003) 1626–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Le Guiner C, Servais L, Montus M, Larcher T, Fraysse B, Moullec S, Allais M, Francois V, Dutilleul M, Malerba A, Koo T, Thibaut JL, Matot B, Devaux M, Le Duff J, Deschamps JY, Barthelemy I, Blot S, Testault I, Wahbi K, Ederhy S, Martin S, Veron P, Georger C, Athanasopoulos T, Masurier C, Mingozzi F, Carlier P, Gjata B, Hogrel JY, Adjali O, Mavilio F, Voit T, Moullier P, Dickson G, Long-term microdystrophin gene therapy is effective in a canine model of Duchenne muscular dystrophy, Nat Commun 8 (2017) 16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mendell JR, Sahenk Z, Lehman K, Nease C, Lowes LP, Miller NF, Iammarino MA, Alfano LN, Nicholl A, Al-Zaidy S, Lewis S, Church K, Shell R, Cripe LH, Potter RA, Griffin DA, Pozsgai E, Dugar A, Hogan M, Rodino-Klapac LR, Assessment of Systemic Delivery of rAAVrh74.MHCK7.micro-dystrophin in Children With Duchenne Muscular Dystrophy: A Nonrandomized Controlled Trial, JAMA Neurol 77(9) (2020) 1122–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Willcocks RJ, Forbes SC, Walter GA, Sweeney L, Rodino-Klapac LR, Mendell JR, Vandenborne K, Assessment of rAAVrh.74.MHCK7.micro-dystrophin Gene Therapy Using Magnetic Resonance Imaging in Children With Duchenne Muscular Dystrophy, JAMA Netw Open 4(1) (2021) e2031851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shieh PB, Emerging Strategies in the Treatment of Duchenne Muscular Dystrophy, Neurotherapeutics 15(4) (2018) 840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Leborgne C, Latournerie V, Boutin S, Desgue D, Quere A, Pignot E, Collaud F, Charles S, Simon Sola M, Masat E, Jouen F, Boyer O, Masurier C, Mingozzi F, Veron P, Prevalence and long-term monitoring of humoral immunity against adeno-associated virus in Duchenne Muscular Dystrophy patients, Cell Immunol 342 (2019) 103780. [DOI] [PubMed] [Google Scholar]
- [43].Mercuri E, Muntoni F, Muscular dystrophies, Lancet 381(9869) (2013) 845–60. [DOI] [PubMed] [Google Scholar]
- [44].Doudna JA, The promise and challenge of therapeutic genome editing, Nature 578(7794) (2020) 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Adli M, The CRISPR tool kit for genome editing and beyond, Nat Commun 9(1) (2018) 1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, Genome engineering using the CRISPR-Cas9 system, Nat Protoc 8(11) (2013) 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA, In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy, Science 351(6271) (2016) 403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, Cong L, Zhang F, Vandenberghe LH, Church GM, Wagers AJ, In vivo gene editing in dystrophic mouse muscle and muscle stem cells, Science 351(6271) (2016) 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN, Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy, Science 351(6271) (2016) 400–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS, Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy, Nat Commun 8 (2017) 14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang Y, Long C, Li H, McAnally JR, Baskin KK, Shelton JM, Bassel-Duby R, Olson EN, CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice, Sci Adv 3(4) (2017) e1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Min YL, Chemello F, Li H, Rodriguez-Caycedo C, Sanchez-Ortiz E, Mireault AA, McAnally JR, Shelton JM, Zhang Y, Bassel-Duby R, Olson EN, Correction of Three Prominent Mutations in Mouse and Human Models of Duchenne Muscular Dystrophy by Single-Cut Genome Editing, Mol Ther 28(9) (2020) 2044–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Min YL, Li H, Rodriguez-Caycedo C, Mireault AA, Huang J, Shelton JM, McAnally JR, Amoasii L, Mammen PPA, Bassel-Duby R, Olson EN, CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells, Sci Adv 5(3) (2019) eaav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, Caballero D, Harron R, Stathopoulou TR, Massey C, Shelton JM, Bassel-Duby R, Piercy RJ, Olson EN, Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy, Science 362(6410) (2018) 86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chemello F, Chai AC, Li H, Rodriguez-Caycedo C, Sanchez-Ortiz E, Atmanli A, Mireault AA, Liu N, Bassel-Duby R, Olson EN, Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing, Sci Adv 7(18) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Thakore PI, Black JB, Hilton IB, Gersbach CA, Editing the epigenome: technologies for programmable transcription and epigenetic modulation, Nat Methods 13(2) (2016) 127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Matharu N, Rattanasopha S, Tamura S, Maliskova L, Wang Y, Bernard A, Hardin A, Eckalbar WL, Vaisse C, Ahituv N, CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency, Science 363(6424) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lau CH, Ho JW, Lo PK, Tin C, Targeted Transgene Activation in the Brain Tissue by Systemic Delivery of Engineered AAV1 Expressing CRISPRa, Mol Ther Nucleic Acids 16 (2019) 637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhou H, Liu J, Zhou C, Gao N, Rao Z, Li H, Hu X, Li C, Yao X, Shen X, Sun Y, Wei Y, Liu F, Ying W, Zhang J, Tang C, Zhang X, Xu H, Shi L, Cheng L, Huang P, Yang H, In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice, Nat Neurosci 21(3) (2018) 440–446. [DOI] [PubMed] [Google Scholar]
- [60].Schoger E, Carroll KJ, Iyer LM, McAnally JR, Tan W, Liu N, Noack C, Shomroni O, Salinas G, Gross J, Herzog N, Doroudgar S, Bassel-Duby R, Zimmermann WH, Zelarayan LC, CRISPR-Mediated Activation of Endogenous Gene Expression in the Postnatal Heart, Circ Res 126(1) (2020) 6–24. [DOI] [PubMed] [Google Scholar]
- [61].Kemaladewi DU, Bassi PS, Erwood S, Al-Basha D, Gawlik KI, Lindsay K, Hyatt E, Kember R, Place KM, Marks RM, Durbeej M, Prescott SA, Ivakine EA, Cohn RD, A mutation-independent approach for muscular dystrophy via upregulation of a modifier gene, Nature 572(7767) (2019) 125–130. [DOI] [PubMed] [Google Scholar]
- [62].Wojtal D, Kemaladewi DU, Malam Z, Abdullah S, Wong TW, Hyatt E, Baghestani Z, Pereira S, Stavropoulos J, Mouly V, Mamchaoui K, Muntoni F, Voit T, Gonorazky HD, Dowling JJ, Wilson MD, Mendoza-Londono R, Ivakine EA, Cohn RD, Spell Checking Nature: Versatility of CRISPR/Cas9 for Developing Treatments for Inherited Disorders, Am J Hum Genet 98(1) (2016) 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Saha K, Sontheimer EJ, Brooks PJ, Dwinell MR, Gersbach CA, Liu DR, Murray SA, Tsai SQ, Wilson RC, Anderson DG, Asokan A, Banfield JF, Bankiewicz KS, Bao G, Bulte JWM, Bursac N, Campbell JM, Carlson DF, Chaikof EL, Chen ZY, Cheng RH, Clark KJ, Curiel DT, Dahlman JE, Deverman BE, Dickinson ME, Doudna JA, Ekker SC, Emborg ME, Feng G, Freedman BS, Gamm DM, Gao G, Ghiran IC, Glazer PM, Gong S, Heaney JD, Hennebold JD, Hinson JT, Khvorova A, Kiani S, Lagor WR, Lam KS, Leong KW, Levine JE, Lewis JA, Lutz CM, Ly DH, Maragh S, McCray PB Jr., McDevitt TC, Mirochnitchenko O, Morizane R, Murthy N, Prather RS, Ronald JA, Roy S, Roy S, Sabbisetti V, Saltzman WM, Santangelo PJ, Segal DJ, Shimoyama M, Skala MC, Tarantal AF, Tilton JC, Truskey GA, Vandsburger M, Watts JK, Wells KD, Wolfe SA, Xu Q, Xue W, Yi G, Zhou J, Consortium S, The NIH Somatic Cell Genome Editing program, Nature 592(7853) (2021) 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Green ED, Gunter C, Biesecker LG, Di Francesco V, Easter CL, Feingold EA, Felsenfeld AL, Kaufman DJ, Ostrander EA, Pavan WJ, Phillippy AM, Wise AL, Dayal JG, Kish BJ, Mandich A, Wellington CR, Wetterstrand KA, Bates SA, Leja D, Vasquez S, Gahl WA, Graham BJ, Kastner DL, Liu P, Rodriguez LL, Solomon BD, Bonham VL, Brody LC, Hutter CM, Manolio TA, Strategic vision for improving human health at The Forefront of Genomics, Nature 586(7831) (2020) 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]


