SUMMARY
The North American beaver is an exceptionally long-lived and cancer-resistant rodent species. Here, we report the evolutionary changes in its gene coding sequences, copy numbers, and expression. We identify changes that likely increase its ability to detoxify aldehydes, enhance tumor suppression and DNA repair, and alter lipid metabolism, potentially contributing to its longevity and cancer resistance. Hpgd, a tumor suppressor gene, is uniquely duplicated in beavers among rodents, and several genes associated with tumor suppression and longevity are under positive selection in beavers. Lipid metabolism genes show positive selection signals, changes in copy numbers, or altered gene expression in beavers. Aldh1a1, encoding an enzyme for aldehydes detoxification, is particularly notable due to its massive expansion in beavers, which enhances their cellular resistance to ethanol and capacity to metabolize diverse aldehyde substrates from lipid oxidation and their woody diet. We hypothesize that the amplification of Aldh1a1 may contribute to the longevity of beavers.
Graphical Abstract
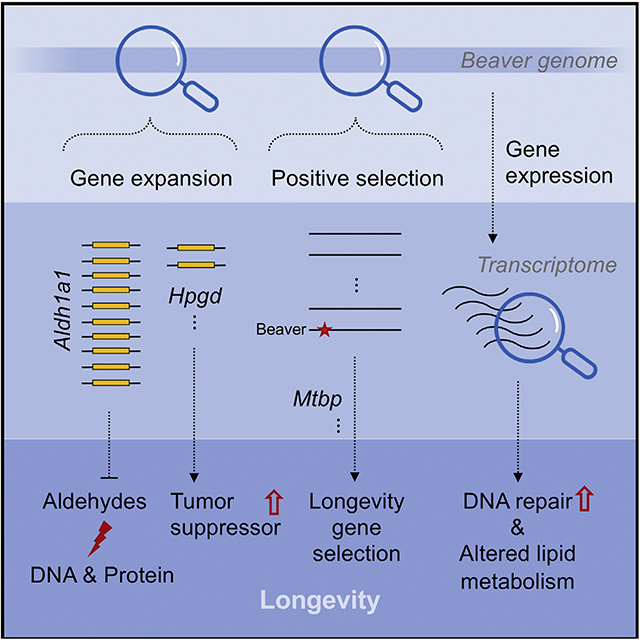
In brief
Zhang et al. examine the genome of North American beavers and find evolutionary changes that could contribute to beavers’ longevity. In particular, Aldh1a1, encoding an enzyme for aldehyde detoxification, is massively expanded in the beaver genome, protecting them against exposure to aldehydes from lipid oxidation and their woody diet.
INTRODUCTION
Between species, large animals tend to live longer. Recent studies of several long-lived mammalian species with large body sizes revealed diverse antitumor mechanisms. For example, elephants, the largest land mammals, were found to have enhanced Tp53 activity through gene copy-number expansion (~20 copies) (Abegglen et al., 2015), while bowhead whales, possibly the longest-lived mammals, show signals of natural selection in aging and cancer-associated genes (Keane et al., 2015). In this study, we focused on North American beavers (Castor canadensis), whose maximum lifespans (>24 years) and adult weights (from 24 to 71 lb) make them the longest lived and second largest rodent species.
The lifespans of rodent species are nearly 10 times more variable than those of other mammalian orders (Gorbunova et al., 2014). This heightened variability makes them excellent model organisms to study mechanisms of aging and longevity. Most notably, the naked mole rat, a rodent species with an extreme resistance to cancer, has a maximum lifespan of ~30 years, ~10 times the lifespan of mice (Gorbunova et al., 2014). Recent studies found unique amino acid changes in proteins involved in DNA repair and cell cycle (Kim et al., 2011), insulin β chain associated with insulin misfolding and diabetes (Fang et al., 2014), decreased expression of genes in insulin/Igf1 signaling in the liver (Kim et al., 2011), and an increased expression of genes involved in DNA repair and genome maintenance (MacRae et al., 2015). Beavers are another long-lived rodent species, and their cells are more resistant to malignant transformation than other rodent species (e.g., capybara, the largest rodent) and require similar “hits” as human cells for malignant transformation (Seluanov et al., 2018), making beavers a promising model organism to study human cancer. A recent study showed that Sirt6 in beavers can repair DNA double-strand breaks better than its ortholog in mice, and this increased genome maintenance contributes to the longevity of beavers (Tian et al., 2019). However, it remains unclear how beavers, the second largest rodents, evolved to have cancer resistance and longevity.
The beaver genome assembly was first released in 2017 (Lok et al., 2017). Before analyzing it, we re-sequenced and reassembled the beaver genome to improve the quality of its assembly in terms of both contiguity and completeness (Zhou et al., 2020). To find potential mechanisms of cancer resistance and longevity, using our improved beaver genome assembly, we explored three major types of signals of natural selection in beavers compared to other rodents: changes in gene copy numbers; positive selection in coding regions, which has affected protein structure, interaction, and function; and changes in gene expression. Through a systematic, comparative analysis, we identified a striking expansion of Aldh1a1, a stem cell marker gene, by ~10 copies in the beaver genome and also beaver-only expansion of several genes, including Hpgd, a tumor suppressor gene, and Cyp19a1, a gene involved in estrogen biosynthesis and linked to human longevity (Corbo et al., 2011). We also found signals of positive selection among beaver genes associated with lipid metabolism, oxidation reduction, and cancer suppression and increased expression of DNA repair genes in beavers compared to mice, which are consistent with observations in other long-lived species (MacRae et al., 2015).
RESULTS
Significant expansion of beaver Aldh1a1 and its functional consequences
Based on our gene annotation (Figure S1), we applied CAFE 3 (Han et al., 2013) to predict significant (false discovery rate [FDR] < 0.01) copy number increases in 12 beaver genes (Figure S2; see Method details and Data S2 for the gene list). Most of these significant increases are not specific to beavers. However, the expansion of one of them, Aldh1a1, is striking in beavers. We examined beaver Aldh1a1 in detail to explore whether its expansion could be important for the cancer resistance and/or longevity of beavers.
Expansion of beaver Aldh1a1
Compared with humans and chimpanzees, both of which have one copy of Aldh1a1, most rodent species have two copies of this gene (its close paralog in mouse is called Aldh1a7). This indicates an expansion in rodents, which very likely happened before their last common ancestor (Figure S2B). In contrast, 10 copies of Aldh1a1 were identified in the beaver genome, among which 2 copies are predicted as pseudogenes because of frameshift indels (Figure S3). One copy (ID: 266.2) has relatively lower annotation quality with two repeat fragments, which may indicate a mixture of two copies of Aldh1a1.
We next examined every copy of beaver Aldh1a1 and carried out qPCR to experimentally quantify duplication events. We compared synteny of the Aldh1a1 locus among beavers, humans, and mice (Figure 1A). Missing or unordered genes in a genomic region may indicate a poor assembly quality. In contrast, a perfect match of the order of corresponding genes indicates the good quality of genome assembly at this locus. After expansion, different gene copies evolved independently and accumulated differences in both sequence and gene structure. Using Apollo (Lee et al., 2013) and RNA sequencing (RNA-seq) data, we manually improved the gene annotation of seven functional copies of beaver Aldh1a1 (excluding two pseudogenes and the low-quality copy 266.2), which have different gene structures (Figure 1B). Bona fide gene copies likely encode homologous proteins with different substitutions. We picked 3 sites in the coding sequences of exon 4 (Figures 1C and S4A), which can distinguish the 7 copies from one another. The alignment of RNA-seq reads from different beavers indicates that those variable sites are genuine and that the different copies were transcribed simultaneously. While the RNA-seq read coverage at these variable sites was relatively low for the copy 266.5, we also analyzed another site, where the copy 266.5 is distinct from all of the other copies and with much higher RNA-seq reads coverage (Figure S4B). To further validate the expansion of Aldh1a1, we performed qPCR analysis, which showed that there are ~10 copies of Aldh1a1 in the beaver genome (Figure 1D).
Figure 1. Expansion of Aldh1a1 in the beaver genome.
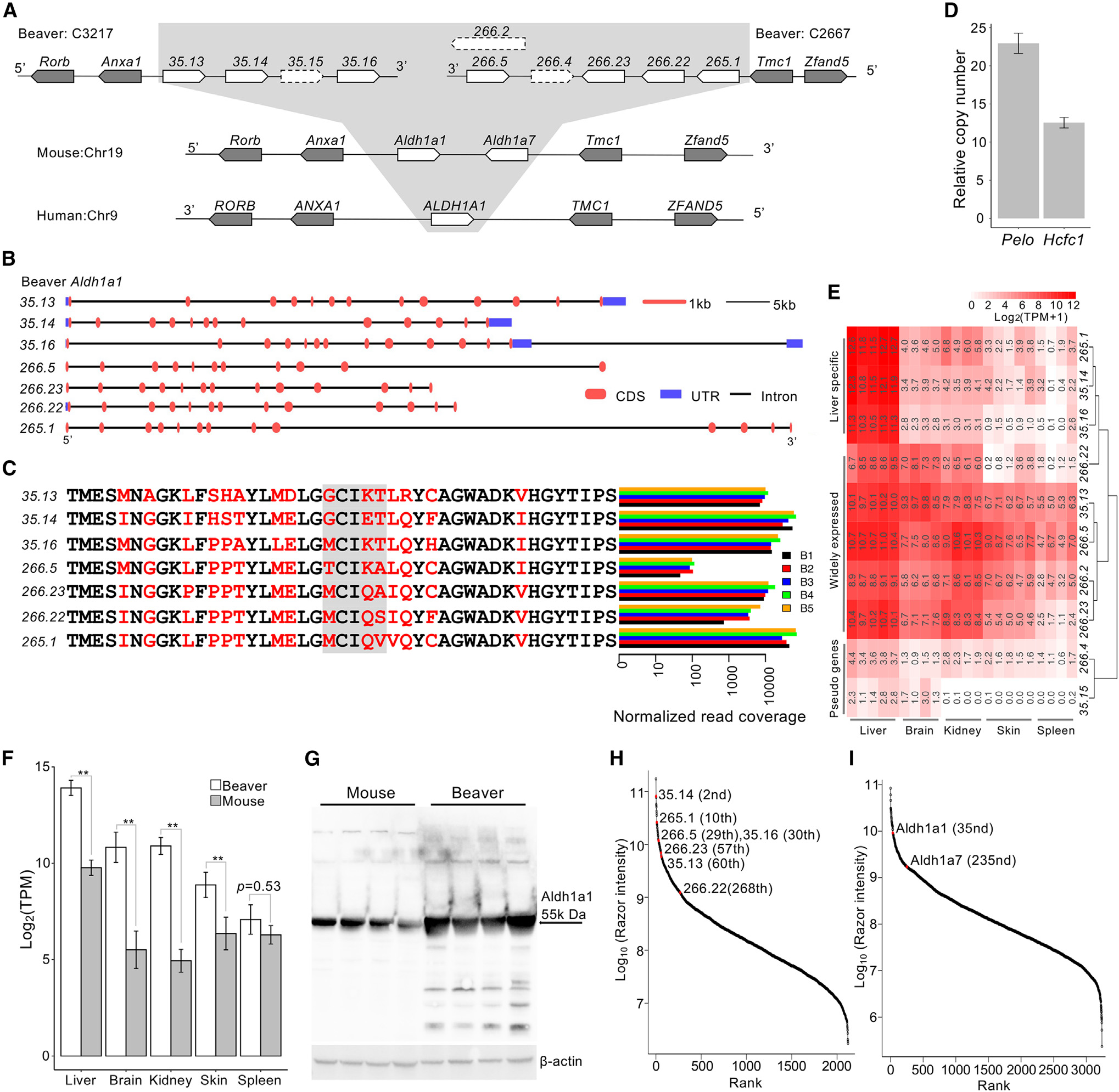
(A) The synteny of the Aldh1a1 locus in the genomes of the beaver, mouse, and human. We identified 10 copies of Aldh1a1 in the beaver genome, including 2 pseudogenes (35.15 and 266.4) and a low-quality copy (266.2).
(B) Gene structure of the 7 functional copies of Aldh1a1.
(C) RNA-seq reads coverage at genomic locations of different amino acid residues among 7 Aldh1a1 gene products. Variable sites were highlighted in red, and the gray box indicates the selected sites where we checked the coverage of RNA-seq reads. RNA-seq reads from the liver tissue of 5 beaver samples (B1–B5) were used in the plot (Table S1). Normalized read coverage is the read counts per 100 million mapped reads.
(D) Validation of the Aldh1a1 copy number by qPCR. Two single-copy genes were used as references: Hcfc1 on an autosome and Pelo on chromosome X. Three technical replicates for each of the 3 different amounts of DNA input. Data are shown as mean ± SD.
(E) Expression of different beaver Aldh1a1 copies across tissues. With an extremely high expression in liver alone, 3 copies show a liver-specific expression.
(F) Aldh1a1 expression comparison between beavers and mice. The double asterisk denotes adjusted p < 0.01 after Bonferroni correction. There is no difference in the spleen tissue (nominal p = 0.53). Five biological replicates for beaver liver and skin tissues; 4 biological replicates for beaver brain, kidney, and spleen tissues. Five biological replicates for each tissue of the mouse. Data are shown as mean ± SD.
(G) Western blot of Aldh1a1 protein from beaver and mouse liver extracts of 4 different samples of each species. Aldh1a1 protein quantity is statistically higher (p = 0.001 by 1-tailed Welch 2-sample t test) in the beaver liver than in the mouse liver.
(H) Relative abundance of Aldh1a1 copies among detected beaver liver proteins. The Aldh1a1 copies are highly abundant.
(I) Relative abundance of mouse Aldh1a1 and Aldh1a7 among all detected liver proteins. Relative protein rank is inversely correlated with abundance. The mean abundance of protein from 3 biological samples are shown in (H) and (I).
Gene expression of expanded beaver Aldh1a1 copies
All of the predicted beaver Aldh1a1 copies have regular gene structures with introns, and only two of them were identified as pseudogenes by GeneWise (Birney et al., 2004) (see Method details and Figure S3C). At least seven copies of Aldh1a1 are of high annotation quality and transcribed in parallel in several different beavers (Figure 1C). Based on their expression patterns in different tissues, beaver Aldh1a1 copies can be clustered into three groups (Figure 1E). As expected, the two pseudogenes show very low expression in all of the tissues. While some copies are expressed in multiple tissues, others show particularly high expression levels in liver, which is the organ most important to metabolize aldehydes. To understand the overall transcriptional activity of Aldh1a1 copies in beavers, we compared expression levels of Aldh1a1 copies in liver, brain, kidney, skin, and spleen between beavers and mice using RNA-seq data (see Method details). The overall expression of Aldh1a1 was significantly higher in beaver liver, brain, kidney, and skin than that in the same mouse organs, respectively (Figure 1F). Although significantly expanded in beavers, Aldh1a1 was not differentially expressed in the spleen tissue between beavers and mice. We also examined the protein levels of Aldh1a1 with the western blot. Using cytosolic extracts from beaver and mouse liver cells, we found that there is more Aldh1a1 in beaver liver cells than in mouse liver cells (Figure 1G). To measure the protein quantity of each Aldh1a1 copy, we performed the label-free quantitation of beaver and mouse liver proteins by mass spectrometry. Of ~2,000 proteins measured in beaver livers, the protein levels of the 7 best annotated Aldh1a1 copies ranked among the highest (all in the top 300 and 6 of them in the top 60) (Figure 1H). These results suggest that the expanded Aldh1a1 copies play important roles in beavers, especially in their livers. Aldh1a1 and Aldh1a7 are also abundant among ~3,000 measured proteins in the mouse livers (Figure 1I).
Divergent evolution of beaver Aldh1a1 copies after expansion
Expanded beaver Aldh1a1 copies have shown differences in their gene structures (Figure 1B) and coding sequences (Figure S4). In addition, they show differences in the expression pattern across beaver tissues; several copies are highly expressed in the beaver livers (Figure 1E). To further check whether they have different functions, we modeled their three-dimensional (3D) structures with SwissModel (Waterhouse et al., 2018) and the structure of the sheep Aldh1a1, 5ABM.pdb (Koch et al., 2015), as the template, which show >80% identity with beaver Aldh1a1 copies (Figure S5A). While beaver Aldh1a1 copies have distinct 3D structures (Figure S5B), much clearer structural differences were revealed by measuring the solvent accessible volume of the substrate entry channel (Figure S5C). Beaver Aldh1a1 copies 35.14, 265.1, 35.16, and 266.22 showed a higher degree of accessibility as if “gates” were more open to the external solvent area and the copy 266.5 was even more extreme. More detailed models with implicit hydrogens added were shown in Figure S5D. The absence of Voronoi spheres for some copies could be a result of either a narrow outer gate or a shallow substrate entry channel.
Aldh1a1 expansion among other conserved Aldh genes
Aldh1a1 belongs to the Aldh super gene family: there are 19 human and 21 mouse Aldh genes, respectively. To investigate whether the expansion of beaver Aldh1a1 is a compensation for the loss of other Aldh genes or a result of natural selection on its own, we manually checked all Aldh genes in the beaver genome (Figure S6A). Except for Aldh1a1, beavers have the same set of Aldh genes as mice and humans, with regular gene structures and open reading frames (Figures S6B and S6C). The conservation of other Aldh genes suggests that natural selection likely drove the fixation of multiple copies of Aldh1a1 in beavers.
Enhanced tolerance of alcohol and aldehydes of beaver cells
Aldh gene products can protect organisms against damage from oxidative stress by processing toxic aldehydes generated as a result of lipid peroxidation (Singh et al., 2013). Both alcohol and endogenous aldehydes can lead to DNA damage and increase mutations in stem cells (Garaycoechea et al., 2018). We next tested the resistance of beaver cells to ethanol and aldehydes. After treatment with 18% ethanol for 7 h, both beaver and mouse lung fibroblasts showed significantly decreased cell viability (Figure 2A). However, the reduction of cell viability was significantly lower for beaver lung fibroblasts than mouse cells (p = 0.002). Higher expression of Aldh1a1 in beaver lung fibroblasts was validated by quantitative real-time PCR (Figure 2B). This result suggests that higher Aldh1a1 levels in beaver may lead to higher alcohol resistance. We also tested Aldh1a1 activity for three types of endogenous aldehydes: all-trans-retinal (RET), malonaldehyde (MDA), and 4-hydroxynonenal (HNE). Beaver and mouse liver extracts did not differ in their ability to process RET; however, beaver extract showed much stronger activity on MDA and HNE (Figure 2C). This result clearly shows that the beaver liver possesses higher Aldh1a1 activity than the mouse liver.
Figure 2. Functional characterization of Aldh1a1.
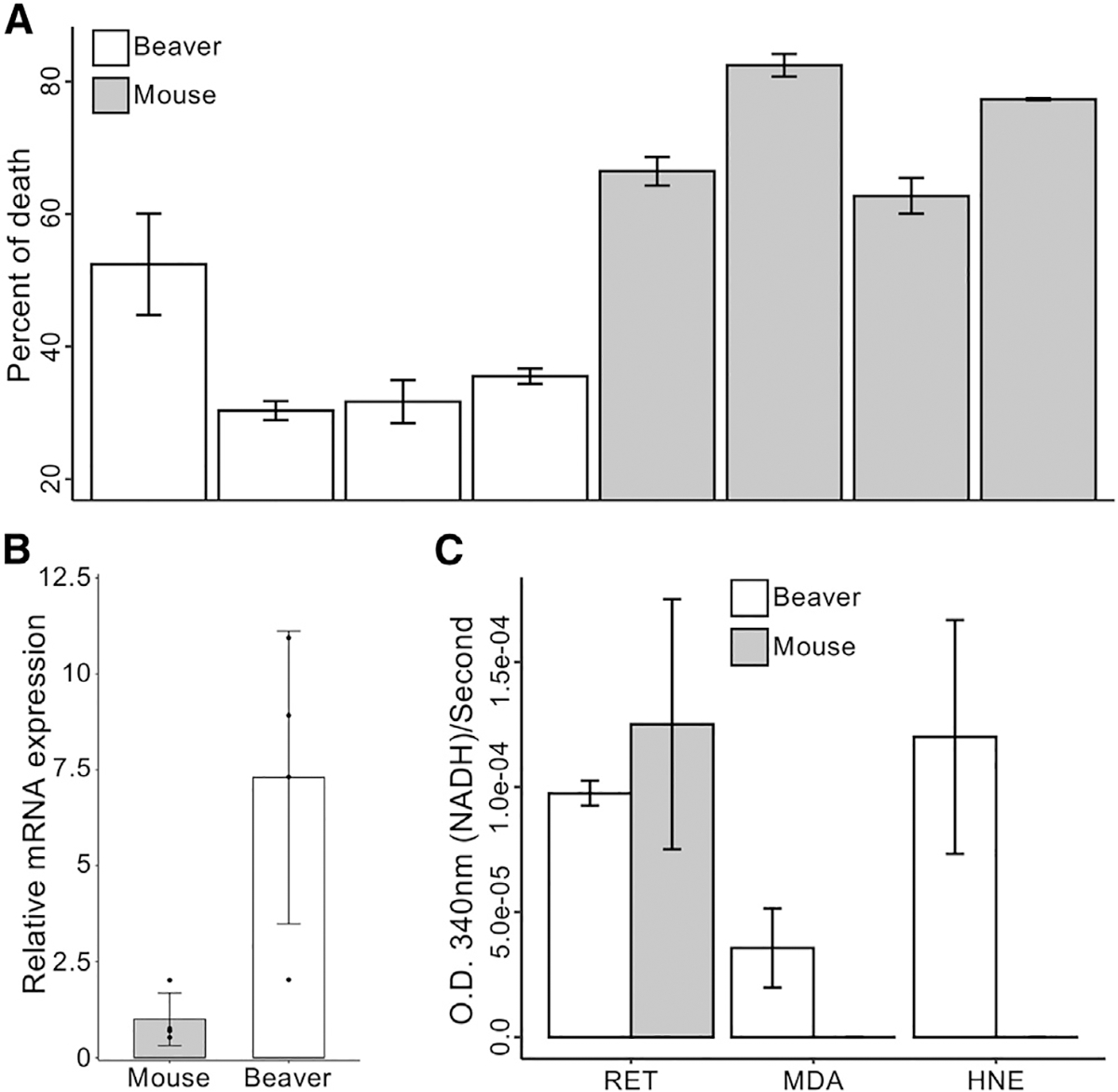
(A) Cell viability. Beaver lung fibroblasts have a lower percentage of death in the presence of high concentrations of ethanol, normalized by corresponding controls (p = 0.002 by mixed-effects model; see Method details).
(B) Expression of Aldh1a1 in lung fibroblasts measured by qPCR with β-actin normalization. The expression of Aldh1a1 in beaver cells is higher than in mouse cells (p = 0.02, 1-tailed t test).
(C) Aldehyde metabolic activity. Enhanced Aldh1a1 activity of beaver in aldehyde metabolism. Activities are normalized by corresponding controls without any addition (see Method details). HNE, 4-hydroxynonenal; MDA, malonaldehyde; RET, all-trans-retinal.
Lung fibroblasts were used in experiments of (A) and (B). Cytosolic extracts from hepatocytes were used for the experiment of (C). Four different biological samples of each species were used in experiments of (A)–(C), with 3 technical replicates of each sample for (A) and (C), and 2 technical replicates of each sample for (B). Data are shown as mean ± SD.
Beaver-only gene expansion
In addition to significant gene expansion identified by CAFE 3 (Han et al., 2013), beaver-specific expansion of highly conserved genes could be biologically relevant. We identified 12 gene candidates for potential beaver-specific expansion and successfully validated 5 of them by qPCR (see Method details): Hpgd (15-hydroxyprostaglandin dehydrogenase), Fitm1 (Fat storage inducing transmembrane protein 1), Cyp19a1 (Cytochrome P450 family 19 subfamily A member 1), Pla2g4c (Phospholipase A2 Group IVC), and Cenpt (Centromere protein T) (Figures 3A and S7–S10). There are two copies of Hpgd, a well-known tumor suppressor gene, in the beaver genome. The Hpgd loci in beaver, mouse, and human genomes show good synteny (Figure 3B), indicating a good assembly quality at this region. The beaver-specific expansion of Hpgd was validated by real-time qPCR (Figure 3A). We aligned beaver and mouse Hpgd sequences together (Figure 3C). While the two copies of beaver Hpgd have a high degree of sequence similarity, there are several amino acid residue differences. RNA-seq reads from individual beavers covered variable sites in both Hpgd copies, indicating that both copies are transcribed (and likely functional) and that sequence differences were not due to DNA sequencing errors but instead divergent evolution after the expansion (Figure 3D). Hpgd is a tumor suppressor of many cancer types, including cancers of the liver (Lu et al., 2014), colon (Myung et al., 2006), lung (Ding et al., 2005), and breast (Wu et al., 2017). Our RNA-seq data analysis showed that the expression of Hpgd in both liver and skin was significantly higher in beavers than in mice (Figure 3E). The expansion of Hpgd and its higher expression likely contribute to the cancer resistance of beavers. Among those expanded genes, the extra copies of Cenpt and Fitm1 were predicted to be pseudogenes and still expressed at levels similar to those of the functional copies (Data S2), suggesting potential functionality. Studies have shown that some pseudogenes (e.g., PTENP1 in humans [Poliseno et al., 2010] and Lethe in mice [Rapicavoli et al., 2013]) are expressed and functional (Cheetham et al., 2020).
Figure 3. Beaver-only gene expansion, including tumor suppressor Hpgd.
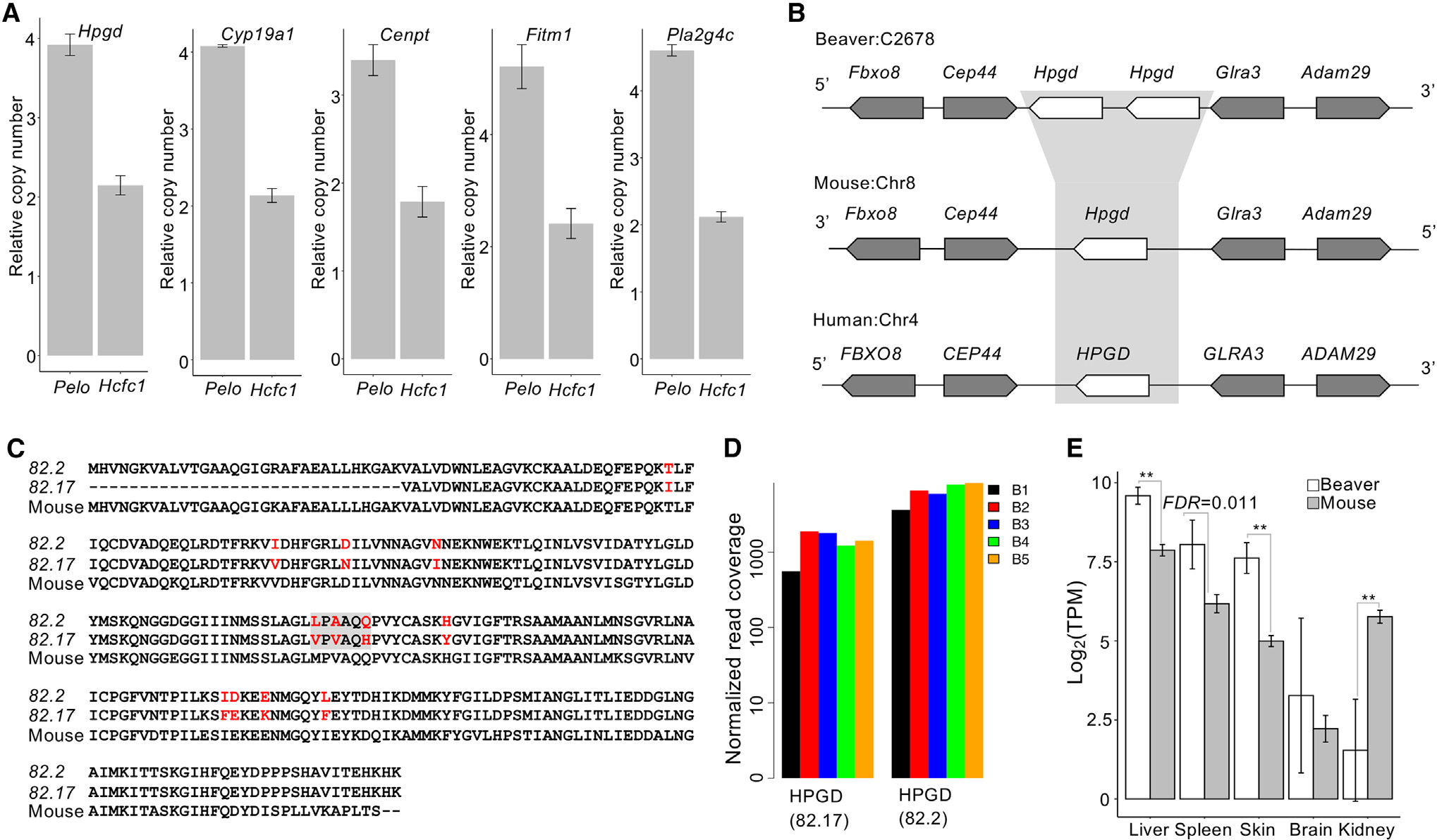
(A) qPCR validation of the copy number of beaver-only expanded genes. The copy numbers relative to the reference gene on chromosome X (i.e., Pelo) and on an autosome (i.e., Hcfc1) are shown for each candidate gene. Three technical replicates for each of the 3 different amounts of DNA input. Data are shown as mean ± SD.
(B) Synteny at the Hpgd locus.
(C) Alignment of beaver and mouse Hpgd protein sequences. Variable sites are highlighted in red, and the gray box indicates the selected sites where we checked the coverage of RNA-seq reads.
(D) Coverage of RNA-seq reads at the selected sites in individual beavers. RNA-seq reads from the liver tissue of 5 beaver samples (B1–B5) were used in the plot (Table S1). Normalized read coverage is the read counts per 100 million mapped reads.
(E) Expression of Hpgd in tissues of both beavers and mice (**adjusted p < 0.01, Bonferroni correction). Five biological replicates for beaver liver and skin tissues; 4 biological replicates for beaver brain, kidney, and spleen tissues. Five biological replicates for each tissue of mouse. Data are shown as mean ± SD.
Beaver genes under positive selection and their association with tumor suppression and longevity
We identified 21 beaver genes putatively under positive selection (FDR < 0.01; Table 1; Data S1), using the branch site model (Zhang et al., 2005) implemented in the PosiGene (Sahm et al., 2017) pipeline, followed by manual curation (see Method details). Although not enriched at the pathway level, several top genes under positive selection are known to be associated with lipid metabolism (e.g., Erlin2, Fabp3, Cilp2), tumor suppression (e.g., Vwa5a, Fabp3), and oxidation reduction process (e.g., Hsd17b1, Fabp3, Cox15, Cyb5a, Aoc1). For several of them, past studies have shown their potential association with aging/longevity. Different alleles of Hsd17b1 were found to be significantly associated with human longevity in females (Scarabino et al., 2015). A single knockout of Mtbp in mice led to an extension of lifespan (Grieb et al., 2016). Knockdown of Mrpl37 increased the lifespan of Caenorhabditis elegans by 41% on average (Houtkooper et al., 2013). Long-lived bats have a greater abundance of Fabp3 in muscle mitochondria than that of short-lived mice, and thus its regulation of lipids may influence mitochondrial function (Pollard et al., 2019). FABP3 also acts as a tumor suppressor in human embryonic cancer cells and breast cancer (Song et al., 2012). In addition, Ptx3 is an inflammatory protein that protects the organism against pathogens and controls autoimmunity and is involved in tissue remodeling and cancer development (Doni et al., 2019).
Table 1.
Putative positive selection genes in beaver (FDR < 0.01)
| Gene | Gene name | No. seq.a | No. sitesb | FDR |
|---|---|---|---|---|
|
| ||||
| Mtbp | MDM2 Binding Protein | 12 | 4 | 6.58E–4 |
| Ptx3 | Pentraxin 3 | 16 | 6 | 1.56E–3 |
| Tbxa2r | Thromboxane A2 Receptor | 15 | 6 | 2.75E–3 |
| Hsd17b1 | Hydroxysteroid 17-Beta Dehydrogenase 1 | 14 | 6 | 3.32E–3 |
| Erlin2 | ER Lipid Raft Associated 2 | 18 | 3 | 3.86E–3 |
| Fabp3 | Fatty Acid Binding Protein 3 | 17 | 7 | 3.86E–3 |
| Adam19 | ADAM Metallopeptidase Domain 19 | 18 | 6 | 4.38E–3 |
| Cilp2 | Cartilage Intermediate Layer Protein 2 | 14 | 30 | 5.07E–3 |
| Chst12 | Carbohydrate Sulfotransferase 12 | 18 | 5 | 5.07E–3 |
| Urb2 | URB2 Ribosome Biogenesis Homolog | 16 | 4 | 5.07E–3 |
| Mrpl37 | Mitochondrial Ribosomal Protein L37 | 18 | 6 | 5.07E–3 |
| Depdc7 | DEP Domain Containing 7 | 17 | 7 | 5.61E–3 |
| Neu2 | Neuraminidase 2 | 12 | 3 | 6.07E–3 |
| Vwa5a | Von Willebrand Factor A Domain Containing 5A | 10 | 16 | 6.07E–3 |
| Cox15 | Cytochrome C Oxidase Assembly Homolog COX15 | 18 | 4 | 6.64E–3 |
| Gdf2 | Growth Differentiation Factor 2 | 17 | 9 | 7.31E–3 |
| Sit1 | Signaling Threshold Regulating Transmembrane Adaptor 1 | 10 | 4 | 7.31E–3 |
| Aoc1 | Amine Oxidase Copper Containing 1 | 17 | 9 | 8.02E–3 |
| Cyb5a | Cytochrome B5 Type A | 18 | 1 | 8.52E–3 |
| Scn4b | Sodium Voltage-Gated Channel Beta Subunit 4 | 16 | 6 | 8.64E–3 |
| Il23a | Interleukin 23 Subunit Alpha | 16 | 4 | 1.00E–2 |
No. seq., the number of available orthologous sequences from a subset of our selected 18 species to build the high-quality alignment for detecting selection signals.
No. sites, the number of amino acid residues that show selection signal in beavers.
Mtbp (Mdm2 binding protein) interacts with and enhances the stability of oncoprotein Mdm2 (mouse double minute 2), which promotes the degradation of Tp53 (Figure S11). Sites with positive selection signals were identified in exons encoding the mid-domain and the C-domain of Mtbp (Figure 4A). In addition to codon changes (i.e., selection signals identified by PosiGene), there are indels in beaver Mtbp, compared with the ortholog from other rodent species. In particular, there is a 15-bp insertion in exon 18 of beaver Mtbp that resembles an insertion at a similar location in Mtbp of the long-lived naked mole rat. We explore potential functional effects of changes in codons and indels using PROVEAN (Choi and Chan, 2015), PolyPehn2 (Adzhubei et al., 2013), and CADD (Rentzsch et al., 2019) by considering changes from those in the human genome (human sequences are the consensus sequence at those loci) to those in the beaver genome (Figure 4B). Two sites under positive selection are consistently predicted to be deleterious, suggesting that they could result in decreased Mtbp function and hence decrease the stability of Mdm2, leading to higher Tp53 activity. We also checked the top 3 sites under positive selection identified in the multiple sequence alignment of Mtbp from 62 mammals (Figures S11B–S11D) and examined in the gnomAD (Genome Aggregation Database) (Karczewski et al., 2020) the frequency of the human allele identical to the beaver nucleotide. We examined a few other sequence changes at the positive selection sites in other mammals and found that only changes in beaver Mtbp were predicted to be deleterious (Figure S11D), which may contribute to its cancer resistance. Haploinsufficiency of Mtbp in mice delays spontaneous cancer development and extends lifespan by enhancing Tp53 function (Grieb et al., 2016). Our hypothesis on the decreased function of Mtbp, due to deleterious mutations, is supported by the lower expression of Mtbp in beavers than in mice (Figure 4C).
Figure 4. Mtbp is under positive selection.
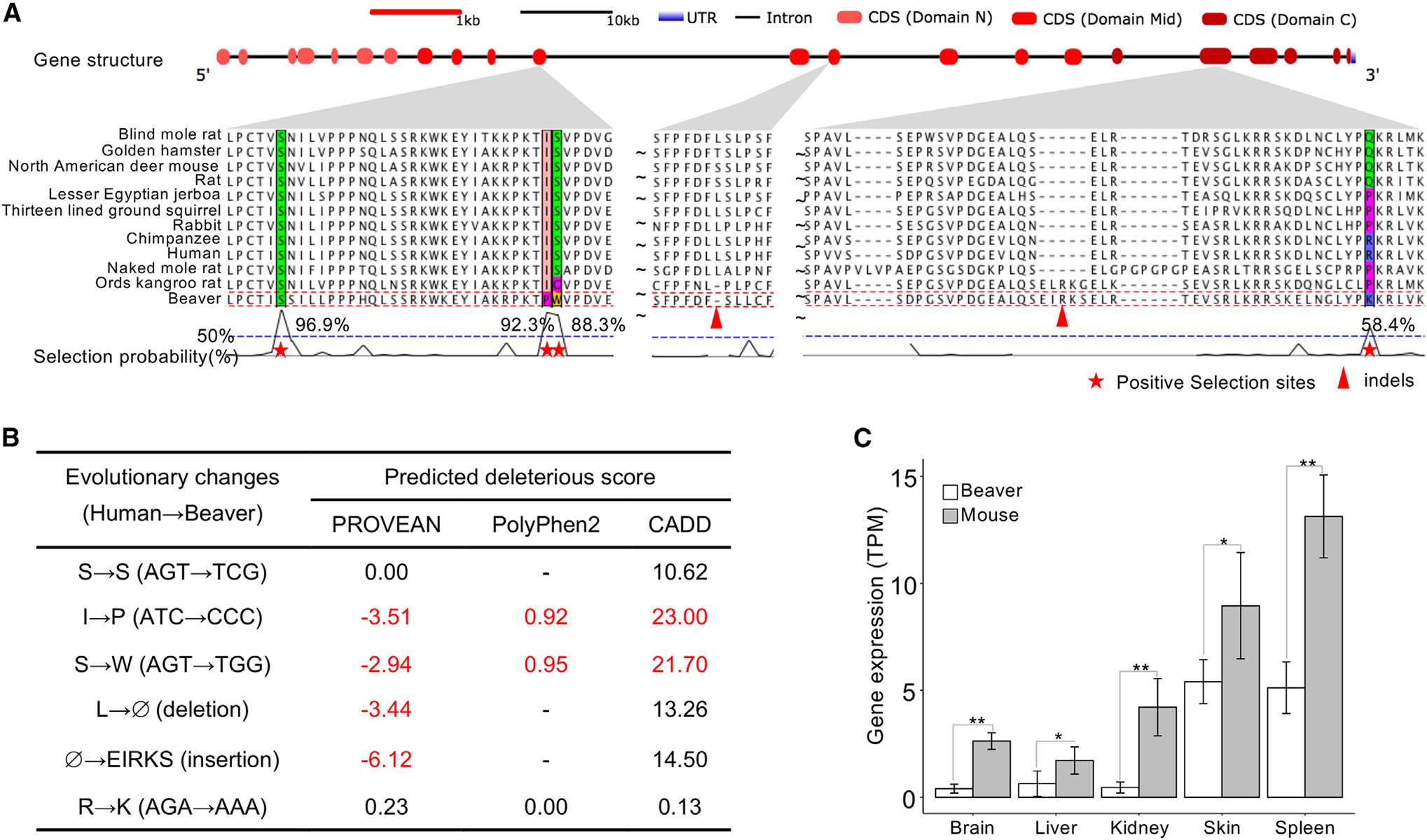
(A) Positive selection signals in Mtbp gene. We observed nucleotide positions and indels under positive selection in the coding sequences of 3 exons, corresponding to the mid-domains and the C domains of Mtbp.
(B) Nucleotide sequence changes between the beaver and human genomes. We predicted the deleteriousness of these changes based on their human genome annotation, using the following metrics and thresholds: PROVEAN score < ‒2.5, 0.85 < PolyPhen2 score < 1.0, and CADD score > 20. In addition, the change of S→W (AGT→TGG) is a splicing variant, 1 bp away from the splicing junction.
(C) Mtbp gene expression in tissues of beavers and mice. *FDR < 0.01; **adjusted p < 0.01 (Bonferroni correction). Five biological replicates for beaver liver and skin tissues; 4 biological replicates for beaver brain, kidney, and spleen tissues. Five biological replicates for each tissue of mouse. Data are shown as mean ± SD.
Enhanced expression of DNA repair genes and changes in genes involved in lipid metabolism
Among 12,090 genes with one-to-one orthologs between beavers and mice, we detected 3,267 (1,657 upregulated and 1,610 downregulated), 2,813 (1,593 upregulated and 1,220 downregulated), 3,681 (1,908 upregulated and 1,773 downregulated), 3,270 (1,465 upregulated and 1,805 downregulated), 3,270 (1,465 upregulated and 1,805 downregulated), and 3,451 (1,754 upregulated and 1,697 downregulated) differentially expressed genes (fold change > 2.5 and adjusted p < 0.01 after Bonferroni correction) in liver, brain, kidney, skin, and spleen, respectively, between beavers and mice (see Method details). Except for the spleen, upregulated genes in beavers were enriched in DNA repair and cell-cycle pathways, while downregulated ones in beavers were enriched in metabolism of lipids, post-translational protein modification, and transport of small molecules pathways (Figure 5; Data S2).
Figure 5. REACTOM pathways enriched with genes differentially expressed in liver between beavers and mice.
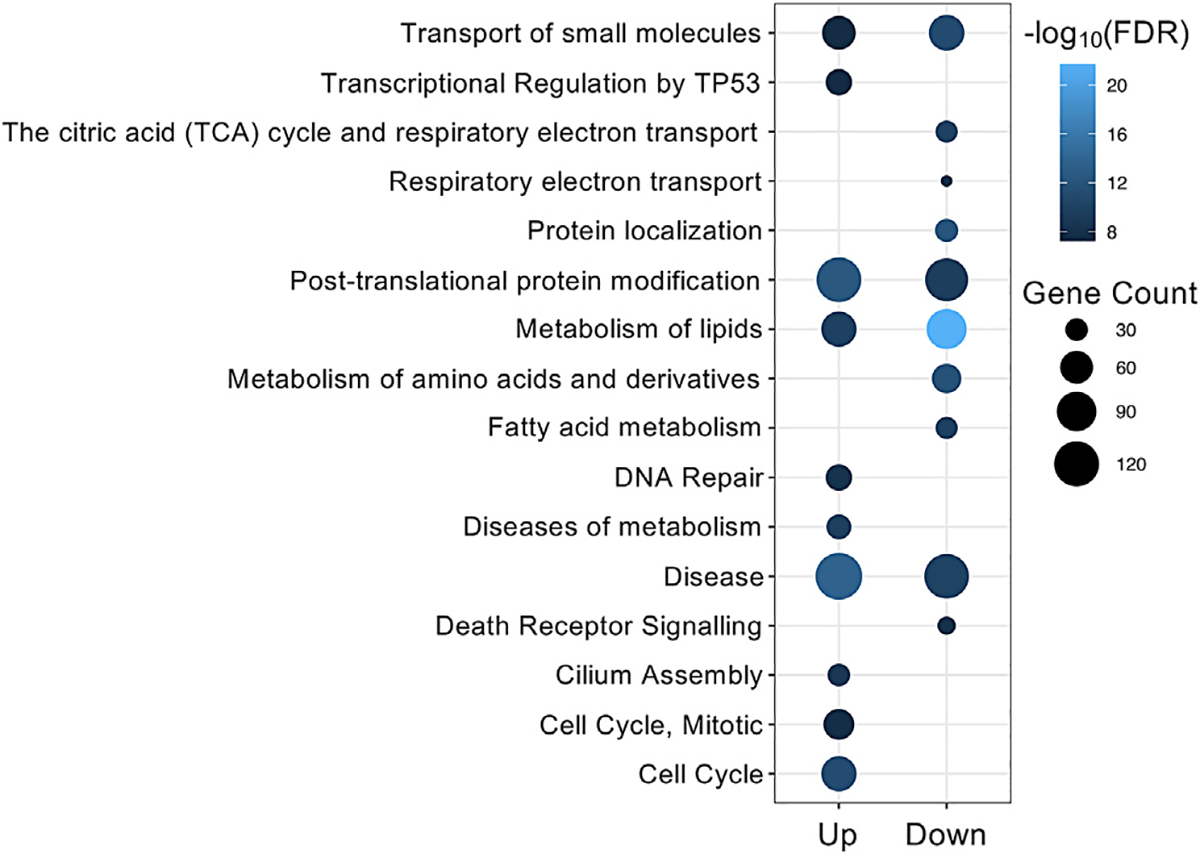
Up (Down) refers to gene upregulation (downregulation) in beavers. See Data S2 for the full pathway enrichment analysis results.
Among differentially expressed genes, Igf2 (insulin-like growth factor 2) showed much higher liver expression in beavers than in mice (fold change = 58.87, adjusted p = 1.43E–76 after Bonferroni correction), and the gene of its binding protein Igf2bp2 also shows significant upregulation in beaver livers (adjusted p = 8.63E–5 after Bonferroni correction) (Figure S12). The expression of Igf2 significantly decreases after birth in the liver of most mammals, including mice and rats. Interestingly, Igf2 and Igf2bp2 show relatively higher expression levels in the livers of naked mole rats and Damaraland mole rats (Fang et al., 2014; Ma and Gladyshev, 2017), both long-lived rodents.
DISCUSSION
Aldh gene products process aldehydes generated as a result of lipid peroxidation, thus protecting the organism from the consequence of oxidative stress (Singh et al., 2013). A recent study showed that both alcohol and endogenous aldehydes damage chromosomes and increase the mutation rate of stem cells (Garaycoechea et al., 2018). The activation of Aldh1a1 was found with the ability to reduce cell oxidative stress damage (Calleja et al., 2021). We found that beaver cells exhibited better tolerance to ethanol than mouse cells and showed strikingly enhanced capabilities for metabolizing MDA and HNE (Figure 2). While MDA is the most mutagenic aldehyde product of lipid peroxidation, making up 70% of the total aldehydes produced (Townsend et al., 2001), HNE is the most toxic (Ayala et al., 2014). The upregulation of Aldh occurs in mammals in response to lipid peroxidation (Vassalli, 2019), which is a major source of endogenous aldehydes. Polyunsaturated fatty acids (PUFAs) are more susceptible to oxidation than monounsaturated fatty acids, and beavers have a high proportion of PUFAs, which is unusual among mammals (Domaradzki et al., 2019; Martysiak-Zurowska et al., 2009; Zalewski et al., 2009). For example, beaver tail fat contains >80% UFAs (Zalewski et al., 2009). High levels of PUFAs will result in increased lipid peroxidation, which is reversely correlated with the lifespan of diverse species, including mammals (Hulbert et al., 2014). Compared to Aldh2 and Aldh3a1, Aldh1a1 is the most important enzyme to oxidize aldehydes formed by lipid peroxidation in murine hepatocytes (Makia et al., 2011). Thus, the long lifespan of beavers indicates the presence of a potential protective mechanism against lipid peroxidation, and Aldh1a1 expansion could be a key factor.
In addition, one major difference between beavers and other rodents (and also most other mammals) is their woody diet. They eat tree barks and twigs (e.g., aspen, maple, willow, birch, black alder), stems, buds, and some soft plants (e.g., grass, leaves). Woody plants produce a large variety of volatile organic compounds (VOCs) as antiherbivore defense mechanisms (Dyer et al., 2018; Iason et al., 2012; Pallardy, 2008). The beaver diet of woody plants may have provided evolutionary pressure that selected for the expansion of Aldh1a1. A study of VOCs in the tree barks of six coniferous and nine deciduous tree species found an abundance of aldehydes, including hexanal, heptanal, and benzaldehyde (Ozgenc et al., 2017). Similarly, a recent metabolomics analysis of sugar maple sap revealed additional aldehydes with complex structures such as sinapaldehyde and 4-hydroxybenzaldehyde, among several other natural products such as polyphenols (Garcia et al., 2020). This suggests that beavers are also exposed to high degrees of aldehydes and alcohol through their woody diet. The exposure of beavers to aldehydes through both diet and PUFA oxidation may entail the increased expression of Aldh1a1 to cope with this challenge. Gene expansion is one effective way to increase gene dosage rapidly. From this point of view, it is very likely that Aldh1a1 expansion in beavers is a result of natural selection against exposure to aldehydes. Since Aldh1a1 expansion significantly improves their tolerance against oxidative stress, we hypothesize that as a result it may contribute to beavers’ longevity.
In addition to measuring their overall expression level and metabolic activity on aldehydes, we explored whether different Aldh1a1 copies have evolved different gene regulation and functionalities after expansion. Several copies showed a high expression in the liver, the organ most important for aldehyde metabolism (Figure 1E). Interestingly, the expansion of beaver Aldh1a1 does not lead to universally higher expression in beavers than in mice (e.g., spleen; Figure 1F). Such results indicate tissue-specific regulation of expanded Aldh1a1 copies. Furthermore, our structural modeling revealed potential differences in their protein structures, suggesting that they may recognize diverse aldehyde substrates (Figure S5). This is consistent with the idea that function diversification of the Aldhs arose from gene duplication events, and the substrate entry channel dictates the specificity of each Aldh (Vasiliou et al., 2000). Different substrate entry channels of beaver Aldh1a1 copies (Figure S5) indicate their different functionalities, which may respond to the organ-specific toxicity of aldehydes.
While no difference in Aldh1a1 activity on RET was observed between beavers and mice, Aldh1a1 in beavers showed much higher activities on MDA and HNE than in mice, supporting the idea that the extra copies of Aldh1a1 may confer a greater ability to oxidize diverse aldehyde substrates. Increased Aldh1 activity is expected to increase an organism’s resistance to oxidative stress. Enhanced tolerance for oxidative stress has been found in long-lived fruit flies compared to those with a normal lifespan (Deepashree et al., 2019). Human centenarians have been found to have less oxidative stress damage compared to controls (Belenguer-Varea et al., 2020). Linked to multiple age-related diseases, the oxidative stress is likely one of the major contributors to aging (Liguori et al., 2018). Lipoxidation increases with age (Mitchell et al., 2007), while reactive aldehyde, a known carcinogen, interferes with DNA replication, causes DNA damage, and induces the formation of DNA adducts (Langevin et al., 2011). Aldh1a1 expansion can increase cellular protection against these toxins. Our finding of better toleration of ethanol by beaver cells is consistent with a previous human study, which showed that low Aldh1a1 activity may account for alcohol sensitivity in some White populations (Marchitti et al., 2008). Stem cell exhaustion is one of hallmarks of the aging process. Aldh1a1 is also a marker gene associated with the stemness of cells and is expressed higher in stem cells (Li et al., 2017). Aldh1a1 expansion may also contribute to stem cell maintenance in beavers. This hypothesis is consistent with results from other studies, which found that UFAs can maintain cancer cell stemness (Mukherjee et al., 2017) and the positive association between Aldh1a1 expression and lipid unsaturation level (Li et al., 2017). Thus, Aldh1a1 expansion may affect the aging process of beavers by increasing both resilience to oxidative stress and the stemness of beaver cells. Mice deficient in both Aldh1a1 and Aldh3a1 had fewer hematopoietic stem cells, more reactive oxygen species, and increased sensitivity to DNA damage (Gasparetto et al., 2012), all of which are early aging phenotypes.
Several studies have shown the association of Aldh1a1 expression with the aging process. The expression of Aldh1a1 dramatically increases with age in mouse hematopoietic stem cells (Levi et al., 2009). It is significantly lower in CD4+ T cells from aged mice than those from young mice, and its low expression may affect the migration of T cells in aged mice (Park et al., 2014). However, how and in which tissues the expression of Aldh1a1 is significantly changed with age is not clear. To explore this, we used human gene expression data in diverse tissues from GTEx (Genotype-Tissue Expression project) (Carithers et al., 2015) and studied Aldh1a1 expression patterns during aging in the presence of several potential covariates (see Method details). In several tissues, the expression of Aldh1a1 significantly changes with age (FDR < 0.01) (Figure 6). For example, it significantly increases with age in adipose tissue, which may be associated with an increase in the percentage of body fat among older people (St-Onge and Gallagher, 2010). It also significantly decreases with age in several human brain regions. This may be associated with an increase in some brain disorders among older individuals, as patients with Parkinson’s disease tend to have a decreased expression of Aldh1a1 in corresponding brain regions (Galter et al., 2003). It is believed that aldehyde dehydrogenases (ALDHs) play a significant role in neuroprotection (Marchitti et al., 2007). For example, ALDH1A1 has been found to be a marker of astrocytic differentiation during brain development (Adam et al., 2012). Thus, Aldh1a1 expansion may also protect beavers from age-related brain impairment.
Figure 6. Age-associated expression of ALDH1A1 in humans.
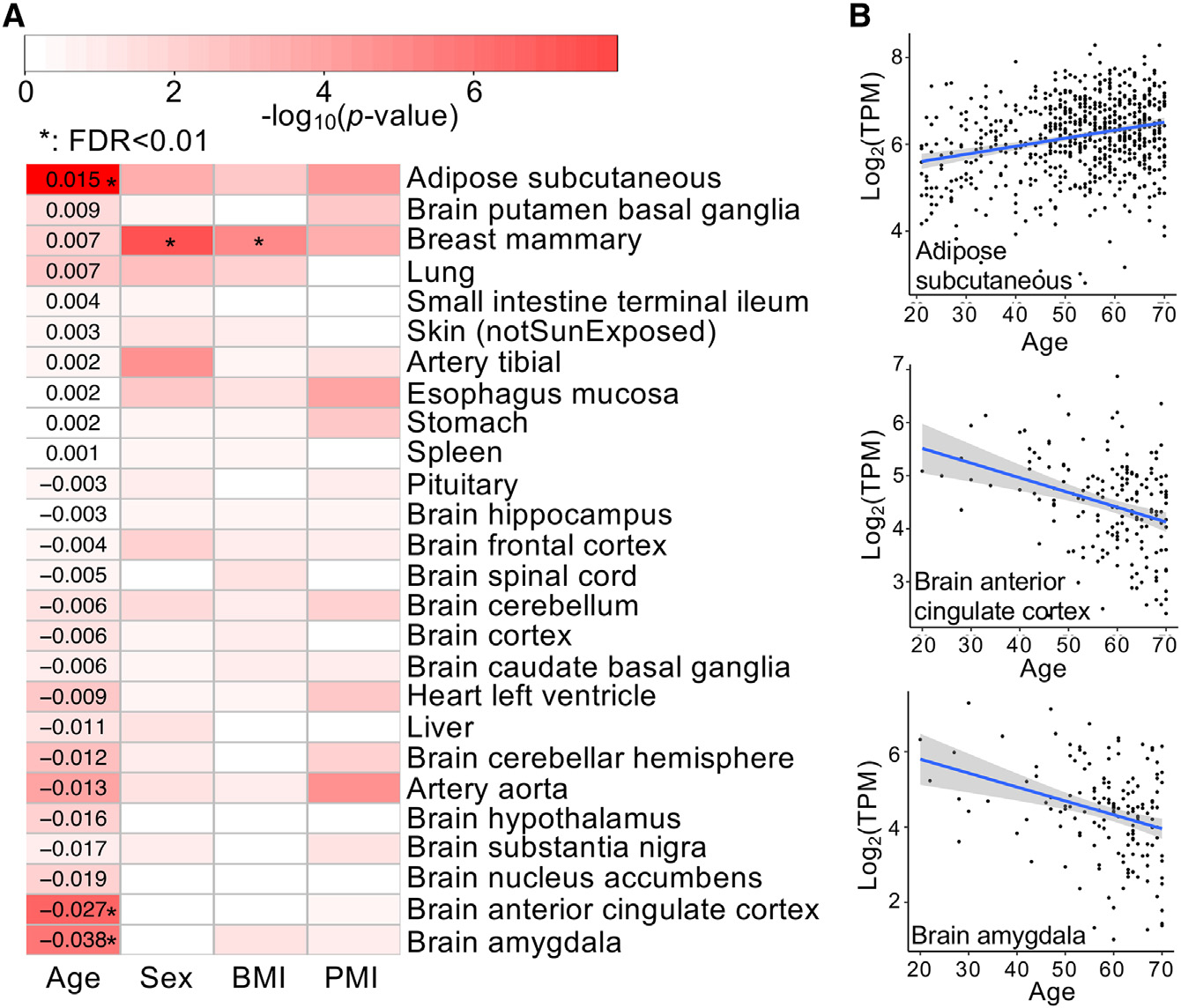
(A) Correlation of ALDH1A1 expression with age, sex, body mass index (BMI), and post-mortem interval (PMI, minutes between death and sample collection). Tissues are ordered according to the regression coefficients on the age, as the digits show, from the highest positive coefficient to the lowest negative coefficient. The color represents p values (on a −log10 scale), and a significant correlation (FDR < 0.01) is denoted by an asterisk.
(B) Significant correlation between ALDH1A1 expression and age in 3 human tissues. The gray-shaded area around the regression line indicates 95% confidence interval.
Compared to other mammals, the adipose tissue and muscles of beavers show different lipid composition, with the highest proportion of PUFAs (Domaradzki et al., 2019; Martysiak-Zurowska et al., 2009; Zalewski et al., 2009), which may indicate an unusual method of lipid metabolism and antilipoxidation in beavers. Consistent with the unusual lipid composition in beavers, we found that genes differentially expressed between mice and beavers are also enriched in lipid metabolism (Figure 5). Several genes expanded in the beaver genome, including Aldh1a1, Hpgd, Fitm1, Cyp19a1, and Pla2g4c, are also associated with lipid metabolism. Aldh1a1-knockout mice show decreased accumulation of both subcutaneous and visceral fat pads (Ziouzenkova et al., 2007), indicating that Aldh1a1 expansion may contribute to the accumulation of protective layers of fat that beavers evolved to provide insulation in cold water. Furthermore, Fabp3, one of the top positively selected genes in the beaver, has also been found to be associated with intramuscular fat deposition and body weight in chickens (Ye et al., 2010) and pigs (Gerbens et al., 1999). In mice, Fabp3 is essential for fatty acid oxidation in brown adipose tissue and plays a central role in cold tolerance (Vergnes et al., 2011).
Among the genes duplicated specifically in beavers, Hpgd is highly expressed in regulatory T cells, which prevents autoimmunity and maintains adipose tissue homeostasis (Schmidleithner et al., 2019). Fitm1 plays an important role in lipid droplet accumulation. Cyp19a1 is involved in the synthesis of cholesterol, steroids, and other lipids. It also catalyzes estrogen biosynthesis, which enhances healthy aging and human longevity (Horstman et al., 2012). A significant association between Cyp19a1 polymorphisms and longevity was observed in humans (Corbo et al., 2011). As a member of the phospholipase A2 enzyme family, Pla2g4c plays a role in hydrolyzing glycerol phospholipids to produce lysophospholipids and free fatty acids. Although not enriched on the pathway level, among the top 10 putative positive selection genes, 3 are associated with lipid metabolism. Erlin2 is a regulator of the cytosolic lipid of cancer cells (Wang et al., 2012). Fabp3 plays a role in the metabolism and transport of long-chain PUFAs, and its role in lipid regulation may be important for mitochondrial function (Pollard et al., 2019). Genetic variant association studies indicate that Cilp2 regulates lipid species in both mice and humans (Jha et al., 2018). All of these genetic and phenotypic features indicate a natural selection on lipid metabolism in beavers. Genes involved in lipid composition have been found under stronger selective pressure in long-lived species (Jobson et al., 2010). The evolution of lipid metabolism is connected to mammalian longevity (Li and de Magalhães, 2013), and the important role of lipid metabolism in aging and lifespan regulation has been observed in different species, including humans (Johnson and Stolzing, 2019). Altered regulation of genes associated with lipid metabolism in the beaver genome likely contributes to its longevity. Although how altered lipid metabolism affects the longevity of beavers needs further exploration, several unusual fatty acids, such as branched-chain (BCFA) and odd-chain (OCFA), which are typical for ruminants, were detected in beaver muscles. BCFA and OCFA have shown positive biological effects on anticancer activity (Domaradzki et al., 2019).
Like other long-lived rodent species, beavers are cancer resistant (Tian et al., 2018). The malignant transformation of beaver cells needs both inactivation of Tp53, Rb1, and Pp2ca and constitutive activation of telomerase and HRas (Seluanov et al., 2018). Such requirements for beaver cells are more stringent than for mouse cells and identical to those for human cells, which makes beavers a promising model to study human cancer. Several findings from this study may shed some light on the cancer resistance of beavers. Hpgd, a known tumor suppressor for many types of cancer, has been expanded uniquely in beavers among rodents. The expansion of Aldh1a1 can better protect beaver cells against exposure to aldehyde from both their woody diet and lipid peroxidation, which can damage DNA and proteins, by metabolizing reactive aldehyde into harmless acetic acid. The increased expression of genes in DNA repair pathway can also protect beavers from cancer.
STAR⋆METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zhengdong D. Zhang (zhengdong.zhang@einsteinmed.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Most data are listed in the Key resources table. Duplicated genes, gene sets enriched with genes differentially expressed between beavers and mice, and proteome quantity data are provided in the Data S2. The raw mass spectrometry proteomics data have been deposited into the ProteomeXchange Consortium (Deutsch et al., 2017) via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD028546.
This paper does not report original code.
Any additional data that support the findings of this study are available from the corresponding author upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Anti-ALDH1A1 | Invitrogen | Cat#PA5-95937; RRID:AB_2807739 |
| 4-hydroxy-nonenal | Cayman Chemical Company | 32100 |
| Malondialdehyde tetrabutylammonium salt | Millipore Sigma | 36357-100MG |
| All trans-Retinal, powder, ≥ 98% | Sigma-Aldrich | R2500-100MG |
| Ethanol | Millipore-Sigma | 1009831011 |
| S-Trap mini columns (100 – 300 μg) | Protifi | CO2-mini-80 |
| C 18 Tips | Thermo Fisher Scientific | 87784 |
| Triethylammonium bicarbonate buffer | Sigma-Aldrich | T7408-500ML |
| DTT,1,4-Dithiothreitol | Millipore-Sigma | 11583786001 |
| Iodoacetamide | Sigma-Aldrich | I1149-5G |
| Thermo Scientific Pierce MS Grade Trypsin | Thermo Fisher Scientific | 90058 |
| β-Nicotinamide adenine dinucleotide sodium salt | Sigma-Aldrich | N0632-5G |
|
| ||
| Critical commercial assays | ||
|
| ||
| Cell Proliferation Reagent WST-1 | Millipore-Sigma | Cat.#5015944001 |
|
| ||
| Deposited data | ||
|
| ||
| Proteomics | This paper | ProteomeXchang: PXD028546 |
| RNA-Seq | This paper | NCBI: SRS6617922 |
| RNA-Seq | This paper | NCBI: SRS6617923 |
| RNA-Seq | This paper | NCBI: SRS6617924 |
| RNA-Seq | This paper | NCBI: SRS6617925 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644691 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644699 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644707 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644715 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644723 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644690 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644698 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644706 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644714 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644722 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644695 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644703 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644711 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644719 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644727 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644694 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644702 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644710 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644718 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644726 |
| RNA-Seq | This paper | NCBI: SRR15970260 |
| RNA-Seq | This paper | NCBI: SRR15970261 |
| RNA-Seq | This paper | NCBI: SRR15970262 |
| RNA-Seq | This paper | NCBI: SRR15970263 |
| RNA-Seq | This paper | NCBI: SRR15970264 |
| RNA-Seq | This paper | NCBI: SRR15970265 |
| RNA-Seq | This paper | NCBI: SRR15970266 |
| RNA-Seq | This paper | NCBI: SRR15970267 |
| RNA-Seq | This paper | NCBI: SRR15970268 |
| RNA-Seq | This paper | NCBI: SRR15970269 |
| RNA-Seq | This paper | NCBI: SRR15970270 |
| RNA-Seq | This paper | NCBI: SRR15970271 |
| RNA-Seq | This paper | NCBI: SRR15970272 |
| RNA-Seq | This paper | NCBI: SRR15970273 |
| RNA-Seq | This paper | NCBI: SRR15970274 |
| RNA-Seq | This paper | NCBI: SRR15970275 |
| RNA-Seq | This paper | NCBI: SRR15970276 |
| RNA-Seq | This paper | NCBI: SRR15970277 |
| RNA-Seq | This paper | NCBI: SRR15970278 |
| RNA-Seq | This paper | NCBI: SRR15970279 |
| RNA-Seq | This paper | NCBI: SRR15970280 |
| RNA-Seq | This paper | NCBI: SRR15970281 |
| RNA-Seq | This paper | NCBI: SRR15970282 |
| RNA-Seq | This paper | NCBI: SRR15970283 |
| RNA-Seq | This paper | NCBI: SRR15970284 |
| RNA-Seq | This paper | NCBI: SRR15970285 |
| RNA-Seq | This paper | NCBI: SRR15970286 |
|
| ||
| Software and algorithms | ||
|
| ||
| Trinity | Haas et al., 2013 | https://github.com/trinityrnaseq/trinityrnaseq/wiki |
| BUSCO | Simão et al., 2015 | https://busco.ezlab.org/ |
| RepBase | Bao et al., 2015 | https://www.girinst.org/repbase/ |
| RepeatMasker | Tarailo-Graovac and Chen, 2009 | https://www.repeatmasker.org/ |
| Maker2 | Holt and Yandell, 2011 | https://www.yandell-lab.org/software/maker.html |
| MUSCLE | Edgar, 2004 | https://www.drive5.com/muscle/ |
| RAxML | Stamatakis, 2014 | https://github.com/stamatak/standard-RAxML |
| PosiGene | Sahm et al., 2017 | https://github.com/gengit/PosiGene |
| OrthoDB | Zdobnov et al., 2017 | https://www.orthodb.org/v9/index.html |
| CAFÉ 3 | Han et al., 2013 | https://github.com/hahnlab/CAFE |
| GeneWise | Birney et al., 2004 | ftp://ftp.ebi.ac.uk/pub/software/unix/wise2/ |
| DeSeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Kallisto | Bray et al., 2016 | https://pachterlab.github.io/kallisto/ |
| SwissModel | Waterhouse et al., 2018 | https://swissmodel.expasy.org/ |
|
| ||
| Experimental models: cell lines | ||
|
| ||
| Beaver/Mouse lung fibroblast cells | This paper | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals and ethics statement
Compliance with ethical regulations and all animal experiments were approved by University Committee on Animal Resources of University of Rochester. The beavers and mice were caught in the wild. Mice were females and about 3–4 months old. Beavers were all young adults, while their exact ages are not known. Among the beaver samples (Table S1), B1, B4, B5, and B7 were from males, B8 from a female, while the sex of the animals is unknown for B2, B3, and B6.
METHOD DETAILS
Genome annotation and phylogeny
On the basis of our highly improved beaver genome assembly (Zhou et al., 2020), we did gene annotation using the Maker2 pipeline (Holt and Yandell, 2011). We predicted 26,515 beaver genes with support from either transcriptomes or protein sequences (Figure S1A). Among these genes, 20,670 (78%) were found with functional domains by InterProScan (v5.25) (Jones et al., 2014). A genome is considered well annotated if more than 90% genes have an annotation edit distance (AED) score, which measures the goodness of fit of each gene to the evidence supporting it, lower than 0.5, and over 50% proteins contain a recognizable domain (Campbell et al., 2014). In our beaver genome annotation, ~90.7% gene models have AED scores lower than 0.5 (Figure S1C) and 78% gene products contain domains. The number of predicted 26,515 protein coding genes in the beaver genome could be an overestimation due to potentially redundantly predicted genes (or gene fragments) in tens of thousands of shorter scaffolds fragments that currently cannot be assembled into longer ones. For our analysis, we only used high-quality predicted beaver genes: ~17k high-quality orthologs for gene expansion identification, ~16k high-quality orthologs between beavers and mice for gene expression comparison, and ~9k high-quality orthologs with good alignment of coding sequences from at least 10 species for discovery of genes under positive selection.
A total of 14 rodent species, 2 rabbit species, and human and chimpanzee (as outgroup species), are included in our analysis. With 5,087 single copy genes across them, we generated their phylogeny and estimated that beaver and its evolutionally closest species in our set, Ord’s Kangroo rat, separated about 46 million years ago (Figure S1B).
De novo transcriptome assembly
RNA-seq data from brain and liver (each with two replicates) were assembled using Trinity (Haas et al., 2013). We collected 16,816 high quality beaver transcripts, by requiring each transcript to meet the following conditions: (1) proper start and stop codons, (2) a correct reading frame (codons in triplets), (3) a gene length similar to the mouse ortholog (±20%), (4) a good alignment with the mouse ortholog (in peptide sequence), (5) the best candidate among all beaver Trinity assembled sequences. Such transcriptome assemblies were used as evidence support in our gene annotation pipeline (Figure S1A).
Training for ab initio gene prediction
Augustus was trained by running BUSCO (Simão et al., 2015) with the ‘–long’ parameter, which performs a full optimization of training for Augustus gene finding. SNAP was trained following the previously described pipeline (Campbell et al., 2014) with three iterations.
Gene structure and function annotation
Maker2 (Holt and Yandell, 2011) was used for gene structure prediction (see Figure S1A for the pipeline). Repeating elements were first masked by RepeatMasker (v4.07) (Tarailo-Graovac and Chen, 2009), with RepBase repeat libraries (20170127) (Bao et al., 2015) and beaver-specific repeating elements constructed by RepeatModeler (version 1.0.10) following the instruction given by: http://weatherby.genetics.utah.edu/MAKER/wiki/index.php/Repeat_Library_Construction-Basic. RepeatRunner was then used to further identify more divergent transposable protein elements provided by Maker2.
With the repeat-masked assembly, genes were predicted by ab initio gene predictors (i.e., SNAP (Korf, 2004) and Augustus (Stanke and Waack, 2003)) and evidence-based gene calling (i.e., using transcript assembly and protein sequences). For beaver gene transcripts, we used our 16,816 assembled transcripts (see above) and the 9,805 full length open reading frames from the published beaver genome (Lok et al., 2017). For protein evidence, about 66.7 thousands of reviewed mammalian protein sequences from Swiss-Prot (The UniProt Consortium, 2017) were used for homolog-based gene prediction. These transcript and protein sequences were used to train the ab initio gene predictors (see above), polish the predicted gene models, and evaluate each predicted gene model. Finally, we predicted 26,515 beaver genes with evidence support from either transcript or protein sequences.
For each gene, Maker2 calculates an annotation edit distance (AED) score, which measures the goodness of fit of each gene to the evidence supporting it. A genome is usually considered well annotated if more than 90% genes have AED scores < 0.5 and over 50% proteins contain a recognizable domain (Campbell et al., 2014). In our beaver genome annotation, ~90.7% gene models have AED scores lower than 0.5 (Figure S1C) and 78% of predicted gene products contain known protein domains by InterProScan (v5.25) (Jones et al., 2014).
Orthology and phylogeny analyses
To identify gene families across species, we used OrthoDB (release 9) (Zdobnov et al., 2017), which covers more than 600 eukaryotic species with functional annotation from more than 100 sources. In our analysis we selected 14 rodent and rabbit species from OrthoDB and mapped beaver and naked mole rat protein sequences to OrthoDB. We chose human and chimpanzee as out-group species. We identified ~21,000 gene families, among which 5,087 gene families have a single copy across all 18 species.
Single-copy gene families were used for phylogenetic analysis. Briefly, orthologs from each family were first aligned by MUSCLE (Edgar, 2004). Poorly aligned regions was removed by TrimAI (Capella-Gutiérrez et al., 2009). Trimmed alignments were then concatenated and used to generate the phylogenetic tree by RAxML (Stamatakis, 2014). The best substitution model for the full data matrix was determined by the Akaike information criterion in MrModeltest software (Nylander, 2004). The best-scoring maximum likelihood (ML) tree was inferred using a novel rapid bootstrap algorithm combined with ML searches following 1000 RAxML runs (using the ‘f-a’ option) (Stamatakis, 2014). The divergence times for the species analyzed were estimated by Reltime (Tamura et al., 2012). Diverged about 46 million years ago, Ords Kangroo rat is evolutionarily closest to beaver (Figure S1B).
RNA extraction and sequencing
Initially, we generated RNA-seq data from brain and liver tissues, with two replicates each, and used the assembled transcriptomes as evidences in our gene annotation. For those samples, the total RNA was isolated from frozen tissues using the mRNA-Seq Sample Prep Kit Illumina (San Diego, CA. USA) in accordance with the manufacturer’s instructions, and the mRNA integrity was checked by the agarose gel analysis. Polyadenylated RNA was then isolated using a poly-dT bead procedure and followed by reverse transcription. Short-insert ‘paired-end’ libraries were prepared using the Illumina TruSeq Sample Preparation Kit v2, and the sequencing was performed on the Illumina HiSeq2000 platform. The raw data were processed by NGS QC Toolkit (v.2.3.3)(Patel and Jain, 2012) to remove low-quality reads.
RNA-Seq data of 5 tissues (brain, liver, kidney, skin and spleen) from 4 or 5 samples, depending on the tissue type, were generated to identify differentially expressed genes between beavers and mice. For each of those tissues, total RNA from 4 or 5 young adult beavers and 5 young adult wild mice, 3–4 months old, were extracted using a PureLink RNA Mini Kit (ThermoFisher). A DNase I on column digestion was performed to remove genomic DNA contamination. The RNA samples were processed with the Illumina TruSeq stranded total RNA RiboZero Gold kit and then subjected to Illumina NovaSeq 6000 paired-end 150bp sequencing at University of Rochester Genomics Research Center.
Identification of significant gene expansion in beaver
With the phylogeny that we built and the gene counts from OrthoDB (Zdobnov et al., 2017), we predicted gene family expansion using CAFE 3 (Han et al., 2013). CAFE first calculated an error model for gene family size estimation as a part of genome assembly and annotation. For our gene counts, it estimated ~4.8% of the gene families had incorrect gene numbers assigned to them. It corrected this error before calculating ancestral family sizes and then estimated a more accurate gene family evolution rate. Finally, it calculated the probability of observing the sizes of each gene family of those species by Monte Carlo re-sampling procedure. Families with large variance in size, especially observed in closely related species, will tend to have a lower P-value. For families with low P-values (0.01 as the default), a P-value for the transition between parent and child nodes for each branch in the phylogeny was also calculated to identify where the large change of family size takes place.
We identified 84 candidate gene families with significant expansion (FDR < 0.01) in the beaver lineage. Assembly and annotation artifacts and pseudogenes can reduce the accuracy of gene expansion prediction (Keane et al., 2015). To improve the prediction reliability, we iteratively removed the gene copy with the lowest average sequence similarity to other beaver genes in the family, if it was below 70%. We also removed any gene copy whose expression was low (TPM < 0.5 across all tissues). With these criteria, we reduced our candidate expanded gene families to 12 genes (Figure S2C).
Genes expanded only in the beaver
To find beaver-specific expansion of conserved genes, we require identical gene copy numbers in all other species included in our study and a higher copy number only in the beaver genome. Initially, we identified 121 gene candidates. After we removed gene copies with low sequence similarity between gene products (identity < 0.7) and low expression (TPM < 0.5 across all tissues), 61 genes remained. We then manually checked them to select most reliable candidates with the following qualifications: (1) gene predictions with matching synteny in their genomic neighborhood among beaver, mouse, and human and (2) genes copies with different coding sequences supported by RNA-seq reads (from the same beaver). 12 beaver-specific gene expansions met these stringent criteria and were further checked by qPCR. We verified more copy numbers for five of them when compared to reference genes (Figures 3, S2A, and S2D).
Pseudogene identification
GeneWise (Birney et al., 2004) was used to identify pseudogenes (Figure S3). For a predicted beaver gene, we extracted the genomic sequence from its locus with both upstream and downstream 5-kb regions. Using its mouse (or human) ortholog as the reference, we then scanned the gene sequence by GeneWise and checked the presence of frameshift indels that can ‘pseudogenize’ the gene. We also checked if the predicted gene can be a processed pseudogene, which can be generated through mRNA retrotransposition. If a predicted beaver gene has no introns, we checked its orthologs in other rodents to determine whether it is a processed pseudogene or a single-exon gene.
Positive selection
Branch-site likelihood method (Zhang et al., 2005) implemented in PosiGene (Sahm et al., 2017) was used to identify genes under positive selection in the beaver genome. Only 9,749 genes with alignment of ortholog coding sequence from at least 10 species were considered. In addition to the quality control procedures implemented in PosiGene (Sahm et al., 2017), we also manually checked all ~150 genes potentially under positive selection (p < 0.05) and removed ones with poor local sequence alignments as potential false positives. In case, the poor local sequence alignment is due to relatively low quality of beaver gene annotation, Apollo (Lee et al., 2013) was used to improve the gene annotation based on evidence support from ortholog protein sequence and RNA-seq reads. We then used the improved coding sequences as the input for PosiGene. Finally, we ran PosiGene three times at different starting points and identified 21 beaver genes under positive selection with FDR < 0.01 consistently among these independent runs. The coding sequences of other species were download from NCBI as of January of 2020.
Reverse transcription quantitative PCR (RT-qPCR)
Total RNA from mouse and beaver lung fibroblasts were extracted with a PureLink RNA Mini Kit (Thermo Fisher Scientific), with DNase I on column digestion to remove genomic DNA contamination. Reverse transcription was performed using an iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed using a CFX Connect Real-Time PCR Detection System (Bio-Rad).
Primers were designed based on the conserved regions of beaver and mouse Aldh1a1 gene, covering the consensus sequences of all seven copies of the gene in beavers and two copies in mice. Designed to cross exons to prevent the amplification of genomic DNA, the following primer sequences were used: AAGGCCCTCAGATTGACAAGG (Aldh1a1 forward) and AAAGTAGCCTTTGTTCCCCCA (Aldh1a1 reverse). Results were normalized by β-actin.
qPCR validation of gene expansion
Candidate primers were designed by Primer-Blast (Ye et al., 2012) in the exons of selected genes. Specificity of the primers was first checked by Primer-Blast against all of the coding sequences in the beaver genome. Then genome-wide specificity was checked by MFEprimer (Qu et al., 2012). Specificity of primers was further inspected by gel electrophoresis and the melting curve analysis. Amplification efficiency was checked by a standard curve analysis for each candidate primers with different amount of DNA input to make sure that the finally used primers have equal amplification efficiency. Quantitative PCR was used to quantify gene copy number, with three replicates for each of the three different amount of DNA input. Two genes – Hcfc1 on an autosome and Pelo on chromosome X – with no predicted expansion in beaver and no annotated expansion in other rodents (according to orthoDB (Zdobnov et al., 2017)) were used as references. The copy number of each target gene relative to its corresponding reference genes was calculated by ΔCt. And the genomic DNA of a male beaver, different from ones whose samples were used for the genome sequencing and assembly, was used for the experiment. Primers for target and reference genes are listed in Table S2.
Western blot
Liver cytosolic extracts were quantitated using the BCA assay (Thermo). 30 ug of each extract was resolved through 4%–20% Criterion Tris-Glycine (TGX) Stain-Free SDS-PAGE (Biorad) and transferred to nitrocellulose. Prior to transfer, total proteins were imaged using a Biorad Gel Documentation System to control for protein loading. Rabbit polyclonal anti-Aldh1a1 (Invitrogen cat# PA5–95937) was used as the primary to detect Aldh1a1 isoforms. Protein loading was also checked by performing western blot using anti-beta Actin (Abcam, ab8227).
Liver proteomics of beaver and mouse
Data dependent analysis (DDA), otherwise known as ‘shotgun’ or random proteome analysis, was combined with label free quantitation (LFQ) to analyze beaver and mouse liver proteomes by mass spectrometry (MS). Liver tissues from three wild caught beavers and three mice were flash frozen in liquid nitrogen and stored at −80C. Approximately half of a gram of tissue was pulverized to a fine powder under liquid nitrogen using a stainless-steel pulverizer chilled in liquid nitrogen. Approximately 5 mg of liver tissue was mixed with 250 ul of 50 mM TEAB PH 7.6; 5% SDS and mixed by pipetting. Samples were sonicated in a chilled cup-horn Q800R3 Sonicator System (Qsonica; Newtown, CT) for a total of 15 minutes at 30% output- 30 × 30 s pulses with 30 s in between pulses. Next, samples reduced with 5 mM DTT and alkylated with 10 mM iodoacetamide. Samples were processed using the standard S-trap mini column method (Protifi; Farmingdale, NY). Prior to MS, peptides were mixed with common internal Retention Time standards (CiRT) peptide mix (50 fmol CiRT/2ug total tryptic peptides) followed by analysis by MS on a Orbitrap Tribrid Fusion Lumos instrument (Thermo) equipped with an EASY-Spray HPLC Column (500 mm × 75 um 2 um 100A P/N ES803A, Nano-Trap Pep Map C18 100A; Thermo). Buffer A was 0.1% formic acid and buffer B was 100% acetonitrile (ACN) with 0.1% FA. Flow rate was 300nl/min and runs were 150 minutes: 0–120 minutes, 5% B to 35% B; then from 120–120.5 minutes, 35%–80% B; followed by a 9-minute 80% B wash until 130 min. From 130–130.5 minutes B was decreased to 5% and the column was re-equilibrated for the remaining 20 minutes at 5% B. The instrument was run in data dependent analysis (DDA) mode. MS2 fragmentation was with HCD (30% energy fixed) and dynamic exclusion was operative after a single time and lasted for 30 s. Additional instrument parameters may be found in the Thermo RAW file. Raw files were analyzed directly with the MSFragger/Philosopher pipeline (da Veiga Leprevost et al., 2020; Kong et al., 2017) and included Peptide and Protein Prophet modules (Ma et al., 2012) for additional quality control. Label-free quantitation at the level of MS1 was performed along with the ‘match-between-runs’ function for alignment of chromatographic peaks between separate runs. MaxLFQ with a minimum of two ions was implemented and normalization was selected. The beaver complete proteome database derived from an earlier build of the beaver genome (Lok et al., 2017) was used as the search for searching and ciRT peptides, contaminants. The Aldh1a1 copy 266.23 was added along with the reverse complement of all to determine false discovery rate (FDR). Search and refinement iterations converged in their stringency to keep the FDR below 1% for all peptide assignments. CiRT peptides were also analyzed for consistency of retention times (RTs) between runs. Additional details are available in MSFragger Log files. In total 2,121 beaver proteins and 3,239 mouse proteins are quantified with sufficient data for accurate quantitation (Data S2).
Model of protein structure of Aldh1a1 copies
In an effort to understand the substrate specificity and potential mechanistic/kinetic differences between beaver Aldh1a1 copies, each copy was modeled using SwissModel (Waterhouse et al., 2018), with the reported structure of sheep Aldh1a1, 5ABM.pdb, (Koch et al., 2015) as the template. The sheep enzyme is > 80% conserved with all of the beaver alleles, suggesting the models are reasonably accurate. UCSF Chimera (Pettersen et al., 2004) was used to view the structural models of beaver Aldh1a1 alleles. In order to more accurately compare differences in the characteristics of the substrate channels of each allele, the BetaCavityWeb web server (Kim et al., 2015) was used to measure the solvent accessible volume of the channel. In order to have a more realistic appraisal, implicit hydrogens were added to the models with the Dock prep function in UCSF Chimera (Pettersen et al., 2004).
Ethanol treatment of cells
The effects of ethanol on cellular functions were studied with cultured primary beaver and mouse lung fibroblasts. We used two different beaver and mouse cell lines, low PD cells stabilized in culture. Cells were seed in complete medium containing 15% FBS in 96 well plate for 24 hours, then incubated in medium containing 300 mM ethanol for seven hours. Then WST-1 assay was performed by adding WST-1 reagent (cell proliferation reagent, Roche) directly to the culture wells, incubating for 4 hours at 37°C and 5% CO, shaking thoroughly for 1 minute on a shaker and then measuring the absorbance at 430–480 nm with TECAN spark 20M spectrophotometric reader. The stable tetrazolium salt WST-1 was cleaved to a soluble formazan by a complex cellular mechanism that occurs primarily at the cell surface. This bio-reduction is largely dependent on the glycolytic production of NAD(P)H in viable cells. Therefore, the amount of formazan dye formed directly correlates to the number of metabolically active cells in the culture.
To examine whether beaver and mouse lung fibroblast cells show significantly different rate of cell death under the ethanol treatment, relative to corresponding controls, a linear mixed effects model was used with species as the fixed effect and individuals (n = 4 for each species with 3 replicates each individual) as random effects: lmer(cell death rate ~species+(1|individual)), which was implemented with the R package lme4 (Bates et al., 2015).
Liver aldehyde dehydrogenase activity
In addition to the cytosolic Aldh1a1, a mitochondrial enzyme, Aldh2, also plays an important role in the aldehyde metabolism. A previous study had shown that in murine liver Aldh1a1 plays a more important role in the metabolism of endogenous aldehydes than Aldh2 (Makia et al., 2011). To exclude influence from Aldh2, we tested aldehyde dehydrogenase activity using cytosolic protein extracts from liver cells. Crude cytosolic protein extracts were prepared from wild beaver or mouse (C57/Bl6) livers using phosphate buffer similar to a previously described method (Makia et al., 2011). Specifically, tissue was resuspended in K-Phos Buffer (50 mM potassium phosphate pH7.4, 250 mM sucrose, 1 mM EDTA) at 1g/3.0ml. Tissues were dounce homogenized in ice using a Teflon pestle followed by ten passages through a 27Ga needle to lyse the cells. Samples were centrifuged (Thermo/Sorvall Legend Micro21R) at 1,000 rpm (100 3 g) for 10 minutes at 4°C to remove any remaining intact cells. Supernatant was transferred to a clean tube and centrifuged at 14,000 rpm (18,800 3 g) for 20 minutes at 4°C. We found that the supernatants/extracts prepared this way and rapidly aliquotted and frozen with liquid nitrogen did not lose significant activity after one freeze/thaw cycle. Just prior to assaying, a 100 μL aliquot of the supernatant was thawed on ice and passaged through a 0.5 mL Zeba desalting spin column (Thermo; 7,000 mwco) equilibrated in the same K-Phos buffer to remove small molecules that might compete with added substrates. Aldehyde dehydrogenase was measured in 384-well transparent microplates in 50 μL volume consisting of 10 μL extract in the same K-Phos buffer supplemented with 1 mM NAD+. Aldehyde substrates all-trans-retinal (RET), malonaldehyde tetrabutylammonium salt (MAD), and 4-hydroxynonenal (NHE) were added at indicated concentrations from DMSO stocks. Final DMSO in assay was 4%. Dehydrogenase activity was measured as the change in absorbance at 340 nm (reduction of NAD+ to NADH) using a Tecan Spark 20M plate reader pre-equilibrated at 37°C. Rates were determined after a few minutes of lag during which time the plate temperature was adjusting to 37°C. NAD+, all-trans-retinal, and malonaldehyde tetrabutylammonium salt, were purchased from Sigma. 4-hydroxynonenal was purchased from Cayman Chemical. Aldh1a1-class of specific activity was determined by subtracting any trace amount of NAD+ to NADH conversion that occurred in the absence of added aldehyde substrate.
Gene expression comparison between beavers and mice
Transcriptomic analysis was done for 5 tissues (liver, brain, kidney, skin and spleen), with 4 or 5 replicates, between wild beavers (young adults) and wild mice (young adults, 3–4 months old). Distribution of beaver and mouse samples on the first two principal components was shown in Figure S12B. To remove the potential normalization bias due to uneven mapping of reads to non-orthologous genes between beaver and mouse genes, only orthologous genes between these two species were included for read mapping and downstream analysis. Gene expression levels were measured using Kallisto (Bray et al., 2016). Different lengths of orthologous genes could lead to bias in differential expression analysis between species. Taking the same approach as a previous study (Cooper et al., 2020), we used DESeq2 (Love et al., 2014) and included lengths of orthologous genes as a normalization factor to identify genes differentially expressed between beavers and mice.
ALDH1A1 expression during human aging
Using data from the GTEx Project (phs000424.v8.p2) (Carithers et al., 2015), we studied gene expression changes of ALDH1A1 during aging. In the GTEx Project, many samples were taken from post-mortem individuals. Studies have demonstrated that expression of some genes may change in certain tissues after death and show significant association with post-mortem interval (PMI, in minutes between death and sample collection) (Ferreira et al., 2018). To reduce artifacts from PMIs, we ignored tissues where the expression of ALDH1A1 shows significant association with PMI (p < 0.01). For the remaining tissues, we used robust liner regression (Greco et al., 2019) to assess the association between gene expression and age with sex, body mass index, and PMI as covariates.
QUANTIFICATION AND STATISTICAL ANALYSIS
A linear mixed model was used to measure the effects of ethanol on cellular functions. Robust linear regression was used to analzye gene expression pattern of ALDH1A1 in human tissues during aging. Differentially expressed genes were identified using DESeq2 (Love et al., 2014). Details can be found in Method details. Numbers of biological and technical replicates are indicated in the figure legends.
Supplementary Material
Highlights.
Aldh1a1 expansion protects beavers from their high exposure to aldehydes
Hpgd, a tumor suppressor, shows beaver-specific expansion among rodents
Mtbp, an antilongevity gene, shows deleterious mutations and low expression in beavers
Beavers show enhanced DNA repair and altered lipid metabolism, compared with mice
ACKNOWLEDGMENTS
This work was supported by NIH grant P01AG047200 to V.G., A.S., V.N.G., J.V., and Z.D.Z.
Footnotes
DECLARATION OF INTERESTS
J.V. is a founder of Singulomics. All of the other authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109965.
REFERENCES
- Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, Campbell MS, Kiso WK, Schmitt DL, Waddell PJ, Bhaskara S, et al. (2015). Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA 314, 1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam SA, Schnell O, Pöschl J, Eigenbrod S, Kretzschmar HA, Tonn JC, and Schüller U (2012). ALDH1A1 is a marker of astrocytic differentiation during brain development and correlates with better survival in glioblastoma patients. Brain Pathol. 22, 788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I, Jordan DM, and Sunyaev SR (2013). Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. Chapter 7, Unit 7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala A, Muñoz MF, and Argüelles S (2014). Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev 2014, 360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, and Kohany O (2015). Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, and Walker S (2015). Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 67, 1–48. [Google Scholar]
- Belenguer-Varea A, Tarazona-Santabalbina FJ, Avellana-Zaragoza JA, Martinez-Reig M, Mas-Bargues C, and Ingles M (2020). Oxidative stress and exceptional human longevity: systematic review. Free Radic. Biol. Med. 149, 51–63. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, and Durbin R (2004). GeneWise and Genomewise. Genome Res. 14, 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Calleja LF, Yoval-Sánchez B, Hernández-Esquivel L, Gallardo-Pérez JC, Sosa-Garrocho M, Marín-Hernández Á, Jasso-Chávez R, Macías-Silva M, and Salud Rodríguez-Zavala J (2021). Activation of ALDH1A1 by omeprazole reduces cell oxidative stress damage. FEBS J. 288, 4064–4080. [DOI] [PubMed] [Google Scholar]
- Campbell MS, Holt C, Moore B, and Yandell M (2014). Genome Annotation and Curation Using MAKER and MAKER-P. Curr. Protoc. Bioinformatics 48, 4.11.1–4.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, and Gabaldón T (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET, et al. ; GTEx Consortium (2015). A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreserv. Biobank. 13, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham SW, Faulkner GJ, and Dinger ME (2020). Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat. Rev. Genet. 21, 191–201. [DOI] [PubMed] [Google Scholar]
- Choi Y, and Chan AP (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K, Saxena A, Sharma V, Neufeld S, Tran M, Gutierrez H, Erberich J, Birmingham A, Cobb J, and Hiller M (2020). Interspecies transcriptome analyses identify genes that control the development and evolution of limb skeletal proportion. FASEB J. 34, 1. [Google Scholar]
- Corbo RM, Ulizzi L, Positano L, and Scacchi R (2011). Association of CYP19 and ESR1 pleiotropic genes with human longevity. J. Gerontol. A Biol. Sci. Med. Sci. 66, 51–55. [DOI] [PubMed] [Google Scholar]
- da Veiga Leprevost F, Haynes SE, Avtonomov DM, Chang HY, Shanmugam AK, Mellacheruvu D, Kong AT, and Nesvizhskii AI (2020). Philosopher: a versatile toolkit for shotgun proteomics data analysis. Nat. Methods 17, 869–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepashree S, Niveditha S, Shivanandappa T, and Ramesh SR (2019). Oxidative stress resistance as a factor in aging: evidence from an extended longevity phenotype of Drosophila melanogaster. Biogerontology 20, 497–513. [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, Campbell DS, Bernal-Llinares M, Okuda S, Kawano S, et al. (2017). The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45 (D1), D1100–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tong M, Liu S, Moscow JA, and Tai HH (2005). NAD+-linked 15-hydroxyprostaglandin dehydrogenase (15-PGDH) behaves as a tumor suppressor in lung cancer. Carcinogenesis 26, 65–72. [DOI] [PubMed] [Google Scholar]
- Domaradzki P, Florek M, Skałecki P, Litwińczuk A, Kędzierska-Matysek M, Wolanciuk A, and Tajchman K (2019). Fatty acid composition, cholesterol content and lipid oxidation indices of intramuscular fat from skeletal muscles of beaver (Castor fiber L.). Meat Sci. 150, 131–140. [DOI] [PubMed] [Google Scholar]
- Doni A, Stravalaci M, Inforzato A, Magrini E, Mantovani A, Garlanda C, and Bottazzi B (2019). The Long Pentraxin PTX3 as a Link Between Innate Immunity, Tissue Remodeling, and Cancer. Front. Immunol 10, 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer LA, Philbin CS, Ochsenrider KM, Richards LA, Massad TJ, Smilanich AM, Forister ML, Parchman TL, Galland LM, and Hurtado PJ (2018). Modern approaches to study plant–insect interactions in chemical ecology. Nat. Rev. Chem. 2, 50–64. [Google Scholar]
- Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Seim I, Huang Z, Gerashchenko MV, Xiong Z, Turanov AA, Zhu Y, Lobanov AV, Fan D, Yim SH, et al. (2014). Adaptations to a subterranean environment and longevity revealed by the analysis of mole rat genomes. Cell Rep. 8, 1354–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PG, Muñoz-Aguirre M, Reverter F, SáGodinho CP, Sousa A, Amadoz A, Sodaei R, Hidalgo MR, Pervouchine D, Carbonell-Caballero J, et al. (2018). The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat. Commun. 9, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galter D, Buervenich S, Carmine A, Anvret M, and Olson L (2003). ALDH1 mRNA: presence in human dopamine neurons and decreases in substantia nigra in Parkinson’s disease and in the ventral tegmental area in schizophrenia. Neurobiol. Dis. 14, 637–647. [DOI] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Mulderrig L, Louzada S, Yang F, Guilbaud G, Park N, Roerink S, Nik-Zainal S, et al. (2018). Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature 553, 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia EJ, McDowell T, Ketola C, Jennings M, Miller JD, and Renaud JB (2020). Metabolomics reveals chemical changes in Acer saccharum sap over a maple syrup production season. PLoS ONE 15, e0235787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparetto M, Sekulovic S, Brocker C, Tang P, Zakaryan A, Xiang P, Kuchenbauer F, Wen M, Kasaian K, Witty MF, et al. (2012). Aldehyde dehydrogenases are regulators of hematopoietic stem cell numbers and B-cell development. Exp. Hematol. 40, 318, 29.e2. [DOI] [PubMed] [Google Scholar]
- Gerbens F, van Erp AJ, Harders FL, Verburg FJ, Meuwissen TH, Veerkamp JH, and te Pas MF (1999). Effect of genetic variants of the heart fatty acid-binding protein gene on intramuscular fat and performance traits in pigs. J. Anim. Sci. 77, 846–852. [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Zhang Z, Gladyshev VN, and Vijg J (2014). Comparative genetics of longevity and cancer: insights from long-lived rodents. Nat. Rev. Genet. 15, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco L, Luta G, Krzywinski M, and Altman N (2019). Analyzing outliers: robust methods to the rescue. Nat. Methods 16, 275–276. [DOI] [PubMed] [Google Scholar]
- Grieb BC, Boyd K, Mitra R, and Eischen CM (2016). Haploinsufficiency of the Myc regulator Mtbp extends survival and delays tumor development in aging mice. Aging (Albany NY) 8, 2590–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Thomas GW, Lugo-Martinez J, and Hahn MW (2013). Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 30, 1987–1997. [DOI] [PubMed] [Google Scholar]
- Holt C, and Yandell M (2011). MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstman AM, Dillon EL, Urban RJ, and Sheffield-Moore M (2012). The role of androgens and estrogens on healthy aging and longevity. J. Gerontol. A Biol. Sci. Med. Sci. 67, 1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, and Auwerx J (2013). Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert AJ, Kelly MA, and Abbott SK (2014). Polyunsaturated fats, membrane lipids and animal longevity. J. Comp. Physiol. B 184, 149–166. [DOI] [PubMed] [Google Scholar]
- Iason GR, Dicke M, and Hartley SE (2012). The Ecology of Plant Secondary Metabolites: From Genes to Global Processes (Cambridge University Press; ). [Google Scholar]
- Jha P, McDevitt MT, Gupta R, Quiros PM, Williams EG, Gariani K, Sleiman MB, Diserens L, Jochem A, Ulbrich A, et al. (2018). Systems Analyses Reveal Physiological Roles and Genetic Regulators of Liver Lipid Species. Cell Syst. 6, 722–733.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson RW, Nabholz B, and Galtier N (2010). An evolutionary genome scan for longevity-related natural selection in mammals. Mol. Biol. Evol. 27, 840–847. [DOI] [PubMed] [Google Scholar]
- Johnson AA, and Stolzing A (2019). The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell 18, e13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. (2020). Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. Nature 581, 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M, Semeiks J, Webb AE, Li YI, Quesada V, Craig T, Madsen LB, van Dam S, Brawand D, Marques PI, et al. (2015). Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10, 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, Han L, Marino SM, Sun X, Turanov AA, Yang P, et al. (2011). Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Cho Y, Lee M, Laskowski RA, Ryu SE, Sugihara K, and Kim DS (2015). BetaCavityWeb: a webserver for molecular voids and channels. Nucleic Acids Res. 43 (W1), W413–W418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MF, Harteis S, Blank ID, Pestel G, Tietze LF, Ochsenfeld C, Schneider S, and Sieber SA (2015). Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues. Angew. Chem. Int. Ed. Engl. 54, 13550–13554. [DOI] [PubMed] [Google Scholar]
- Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, and Nesvizhskii AI (2017). MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 14, 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I (2004). Gene finding in novel genomes. BMC Bioinformatics 5, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, and Patel KJ (2011). Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475, 53–58. [DOI] [PubMed] [Google Scholar]
- Lee E, Helt GA, Reese JT, Munoz-Torres MC, Childers CP, Buels RM, Stein L, Holmes IH, Elsik CG, and Lewis SE (2013). Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 14, R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi BP, Yilmaz OH, Duester G, and Morrison SJ (2009). Aldehyde dehydrogenase 1a1 is dispensable for stem cell function in the mouse hematopoietic and nervous systems. Blood 113, 1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, and de Magalhães JP (2013). Accelerated protein evolution analysis reveals genes and pathways associated with the evolution of mammalian longevity. Age (Dordr.) 35, 301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Condello S, Thomes-Pepin J, Ma X, Xia Y, Hurley TD, Matei D, and Cheng JX (2017). Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell 20, 303–314.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori I, Russo G, Curcio F, Bulli G, Aran L, Della-Morte D, Gargiulo G, Testa G, Cacciatore F, Bonaduce D, and Abete P (2018). Oxidative stress, aging, and diseases. Clin. Interv. Aging 13, 757–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok S, Paton TA, Wang Z, Kaur G, Walker S, Yuen RK, Sung WW, Whitney J, Buchanan JA, Trost B, et al. (2017). De Novo Genome and Transcriptome Assembly of the Canadian Beaver (Castor canadensis). G3 (Bethesda) 7, 755–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Han C, and Wu T (2014). 15-PGDH inhibits hepatocellular carcinoma growth through 15-keto-PGE2/PPARγ-mediated activation of p21WAF1/Cip1. Oncogene 33, 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, and Gladyshev VN (2017). Molecular signatures of longevity: insights from cross-species comparative studies. Semin. Cell Dev. Biol. 70, 190–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Vitek O, and Nesvizhskii AI (2012). A statistical model-building perspective to identification of MS/MS spectra with PeptideProphet. BMC Bioinformatics 13 (Suppl 16), S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae SL, Croken MM, Calder RB, Aliper A, Milholland B, White RR, Zhavoronkov A, Gladyshev VN, Seluanov A, Gorbunova V, et al. (2015). DNA repair in species with extreme lifespan differences. Aging (Albany NY) 7, 1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makia NL, Bojang P, Falkner KC, Conklin DJ, and Prough RA (2011). Murine hepatic aldehyde dehydrogenase 1a1 is a major contributor to oxidation of aldehydes formed by lipid peroxidation. Chem. Biol. Interact. 191, 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchitti SA, Deitrich RA, and Vasiliou V (2007). Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol. Rev 59, 125–150. [DOI] [PubMed] [Google Scholar]
- Marchitti SA, Brocker C, Stagos D, and Vasiliou V (2008). Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin. Drug Metab. Toxicol. 4, 697–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martysiak-Zurowska D, Zalewski K, and Kamieniarz R (2009). Unusual odd-chain and trans-octadecenoic fatty acids in tissues of feral European beaver (Castorfiber), Eurasian badger (Melesmeles) and raccoon dog (Nyctereutesprocyonoides). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 153, 145–148. [DOI] [PubMed] [Google Scholar]
- Mitchell TW, Buffenstein R, and Hulbert AJ (2007). Membrane phospholipid composition may contribute to exceptional longevity of the naked molerat (Heterocephalus glaber): a comparative study using shotgun lipidomics. Exp. Gerontol. 42, 1053–1062. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Kenny HA, and Lengyel E (2017). Unsaturated Fatty Acids Maintain Cancer Cell Stemness. Cell Stem Cell 20, 291–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung SJ, Rerko RM, Yan M, Platzer P, Guda K, Dotson A, Lawrence E, Dannenberg AJ, Lovgren AK, Luo G, et al. (2006). 15-Hydroxyprostaglandin dehydrogenase is an in vivo suppressor of colon tumorigenesis. Proc. Natl. Acad. Sci. USA 103, 12098–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA (2004). MrModeltest v2 (Evolutionary Biology Centre, Uppsala University; ). [Google Scholar]
- Ozgenc O, Durmaz S, Celik G, Korkmaz B, and Yaylı N (2017). Comparative phytochemical analysis of volatile organic compounds by SPME-GC-FID/MS from six coniferous and nine deciduous tree bark species grown in Turkey. S. Afr. J. Bot. 113, 23–28. [Google Scholar]
- Pallardy S (2008). Physiology of Woody Plants, Third Edition (Academic Express Elsevier; ). [Google Scholar]
- Park J, Miyakawa T, Shiokawa A, Nakajima-Adachi H, Tanokura M, and Hachimura S (2014). Attenuation of migration properties of CD4+ T cells from aged mice correlates with decrease in chemokine receptor expression, response to retinoic acid, and RALDH expression compared to young mice. Biosci. Biotechnol. Biochem. 78, 976–980. [DOI] [PubMed] [Google Scholar]
- Patel RK, and Jain M (2012). NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7, e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, and Pandolfi PP (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard AK, Ingram TL, Ortori CA, Shephard F, Brown M, Liddell S, Barrett DA, and Chakrabarti L (2019). A comparison of the mitochondrial proteome and lipidome in the mouse and long-lived Pipistrelle bats. Aging (Albany NY) 11, 1664–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu W, Zhou Y, Zhang Y, Lu Y, Wang X, Zhao D, Yang Y, and Zhang C (2012). MFEprimer-2.0: a fast thermodynamics-based program for checking PCR primer specificity. Nucleic Acids Res. 40, W205–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, and Chang HY (2013). A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2, e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch P, Witten D, Cooper GM, Shendure J, and Kircher M (2019). CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47 (D1), D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahm A, Bens M, Platzer M, and Szafranski K (2017). PosiGene: automated and easy-to-use pipeline for genome-wide detection of positively selected genes. Nucleic Acids Res. 45, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarabino D, Scacchi R, Pinto A, and Corbo RM (2015). Genetic Basis of the Relationship Between Reproduction and Longevity: A Study on Common Variants of Three Genes in Steroid Hormone Metabolism–CYP17, HSD17B1, and COMT. Rejuvenation Res. 18, 464–472. [DOI] [PubMed] [Google Scholar]
- Schmidleithner L, Thabet Y, Schönfeld E, Köhne M, Sommer D, Abdullah Z, Sadlon T, Osei-Sarpong C, Subbaramaiah K, Copperi F, et al. (2019). Enzymatic Activity of HPGD in Treg Cells Suppresses Tconv Cells to Maintain Adipose Tissue Homeostasis and Prevent Metabolic Dysfunction. Immunity 50, 1232–1248.e14. [DOI] [PubMed] [Google Scholar]
- Seluanov A, Gladyshev VN, Vijg J, and Gorbunova V (2018). Mechanisms of cancer resistance in long-lived mammals. Nat. Rev. Cancer 18, 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, and Zdobnov EM (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. [DOI] [PubMed] [Google Scholar]
- Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, Thompson DC, and Vasiliou V (2013). Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med. 56, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song GX, Shen YH, Liu YQ, Sun W, Miao LP, Zhou LJ, Liu HL, Yang R, Kong XQ, Cao KJ, et al. (2012). Overexpression of FABP3 promotes apoptosis through inducing mitochondrial impairment in embryonic cancer cells. J. Cell. Biochem. 113, 3701–3708. [DOI] [PubMed] [Google Scholar]
- St-Onge MP, and Gallagher D (2010). Body composition changes with aging: the cause or the result of alterations in metabolic rate and macronutrient oxidation? Nutrition 26, 152–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, and Waack S (2003). Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 (Suppl 2), ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, and Kumar S (2012). Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 109, 19333–19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, and Chen N (2009). Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics Chapter 4, Unit 4.10. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45 (D1), D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Doerig K, Park R, Can Ran Qin A, Hwang C, Neary A, Gilbert M, Seluanov A, and Gorbunova V (2018). Evolution of telomere maintenance and tumour suppressor mechanisms across mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Firsanov D, Zhang Z, Cheng Y, Luo L, Tombline G, Tan R, Simon M, Henderson S, Steffan J, et al. (2019). SIRT6 Is Responsible for More Efficient DNA Double-Strand Break Repair in Long-Lived Species. Cell 177, 622–638.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend AJ, Leone-Kabler S, Haynes RL, Wu Y, Szweda L, and Bunting KD (2001). Selective protection by stably transfected human ALDH3A1 (but not human ALDH1A1) against toxicity of aliphatic aldehydes in V79 cells. Chem. Biol. Interact. 130–132, 261–273. [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Pappa A, and Petersen DR (2000). Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem. Biol. Interact. 129, 1–19. [DOI] [PubMed] [Google Scholar]
- Vassalli G (2019). Aldehyde Dehydrogenases: Not Just Markers, but Functional Regulators of Stem Cells. Stem Cells Int. 2019, 3904645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes L, Chin R, Young SG, and Reue K (2011). Heart-type fatty acid-binding protein is essential for efficient brown adipose tissue fatty acid oxidation and cold tolerance. J. Biol. Chem. 286, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhang X, Lee JS, Wang X, Yang ZQ, and Zhang K (2012). Endoplasmic reticulum factor ERLIN2 regulates cytosolic lipid content in cancer cells. Biochem. J. 446, 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Liu T, Yang P, Liu X, Liu F, Wang Y, Xiong H, Yu S, Huang X, and Zhuang L (2017). Association of 15-hydroxyprostaglandin dehydrogenate and poor prognosis of obese breast cancer patients. Oncotarget 8, 22842–22853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye MH, Chen JL, Zhao GP, Zheng MQ, and Wen J (2010). Associations of A-FABP and H-FABP markers with the content of intramuscular fat in Beijing-You chicken. Anim. Biotechnol. 21, 14–24. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, and Madden TL (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalewski K, Martysiak-Zurowska D, Chylinska-Ptak M, and Nitkiewicz B (2009). Characterization of Fatty Acid Composition in the European Beaver (Castor fiber L.). Pol. J. Environ. Stud. 18, 493–499. [Google Scholar]
- Zdobnov EM, Tegenfeldt F, Kuznetsov D, Waterhouse RM, Simão FA, Ioannidis P, Seppey M, Loetscher A, and Kriventseva EV (2017). OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 45 (D1), D744–D749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, and Yang Z (2005). Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Oreskovic E, Zhang Q, Lu Q, Gilman A, Lin YS, He J, Zheng Z, Lu JY, Lee J, et al. (2021). Transposon-triggered innate immune response confers cancer resistance to the blind mole rat. Nat. Immunol. 22, 1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Dou Q, Fan G, Zhang Q, Sanderford M, Kaya A, Johnson J, Karlsson EK, Tian X, Mikhalchenko A, et al. (2020). Beaver and Naked Mole Rat Genomes Reveal Common Paths to Longevity. Cell Rep. 32, 107949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, Hamilton JA, Tang G, Dolnikowski GG, Vogel S, et al. (2007). Retinaldehyde represses adipogenesis and diet-induced obesity. Nat. Med. 13, 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Most data are listed in the Key resources table. Duplicated genes, gene sets enriched with genes differentially expressed between beavers and mice, and proteome quantity data are provided in the Data S2. The raw mass spectrometry proteomics data have been deposited into the ProteomeXchange Consortium (Deutsch et al., 2017) via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD028546.
This paper does not report original code.
Any additional data that support the findings of this study are available from the corresponding author upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Anti-ALDH1A1 | Invitrogen | Cat#PA5-95937; RRID:AB_2807739 |
| 4-hydroxy-nonenal | Cayman Chemical Company | 32100 |
| Malondialdehyde tetrabutylammonium salt | Millipore Sigma | 36357-100MG |
| All trans-Retinal, powder, ≥ 98% | Sigma-Aldrich | R2500-100MG |
| Ethanol | Millipore-Sigma | 1009831011 |
| S-Trap mini columns (100 – 300 μg) | Protifi | CO2-mini-80 |
| C 18 Tips | Thermo Fisher Scientific | 87784 |
| Triethylammonium bicarbonate buffer | Sigma-Aldrich | T7408-500ML |
| DTT,1,4-Dithiothreitol | Millipore-Sigma | 11583786001 |
| Iodoacetamide | Sigma-Aldrich | I1149-5G |
| Thermo Scientific Pierce MS Grade Trypsin | Thermo Fisher Scientific | 90058 |
| β-Nicotinamide adenine dinucleotide sodium salt | Sigma-Aldrich | N0632-5G |
|
| ||
| Critical commercial assays | ||
|
| ||
| Cell Proliferation Reagent WST-1 | Millipore-Sigma | Cat.#5015944001 |
|
| ||
| Deposited data | ||
|
| ||
| Proteomics | This paper | ProteomeXchang: PXD028546 |
| RNA-Seq | This paper | NCBI: SRS6617922 |
| RNA-Seq | This paper | NCBI: SRS6617923 |
| RNA-Seq | This paper | NCBI: SRS6617924 |
| RNA-Seq | This paper | NCBI: SRS6617925 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644691 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644699 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644707 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644715 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644723 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644690 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644698 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644706 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644714 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644722 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644695 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644703 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644711 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644719 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644727 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644694 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644702 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644710 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644718 |
| RNA-Seq | (Zhao et al., 2021) | NCBI: SRX11644726 |
| RNA-Seq | This paper | NCBI: SRR15970260 |
| RNA-Seq | This paper | NCBI: SRR15970261 |
| RNA-Seq | This paper | NCBI: SRR15970262 |
| RNA-Seq | This paper | NCBI: SRR15970263 |
| RNA-Seq | This paper | NCBI: SRR15970264 |
| RNA-Seq | This paper | NCBI: SRR15970265 |
| RNA-Seq | This paper | NCBI: SRR15970266 |
| RNA-Seq | This paper | NCBI: SRR15970267 |
| RNA-Seq | This paper | NCBI: SRR15970268 |
| RNA-Seq | This paper | NCBI: SRR15970269 |
| RNA-Seq | This paper | NCBI: SRR15970270 |
| RNA-Seq | This paper | NCBI: SRR15970271 |
| RNA-Seq | This paper | NCBI: SRR15970272 |
| RNA-Seq | This paper | NCBI: SRR15970273 |
| RNA-Seq | This paper | NCBI: SRR15970274 |
| RNA-Seq | This paper | NCBI: SRR15970275 |
| RNA-Seq | This paper | NCBI: SRR15970276 |
| RNA-Seq | This paper | NCBI: SRR15970277 |
| RNA-Seq | This paper | NCBI: SRR15970278 |
| RNA-Seq | This paper | NCBI: SRR15970279 |
| RNA-Seq | This paper | NCBI: SRR15970280 |
| RNA-Seq | This paper | NCBI: SRR15970281 |
| RNA-Seq | This paper | NCBI: SRR15970282 |
| RNA-Seq | This paper | NCBI: SRR15970283 |
| RNA-Seq | This paper | NCBI: SRR15970284 |
| RNA-Seq | This paper | NCBI: SRR15970285 |
| RNA-Seq | This paper | NCBI: SRR15970286 |
|
| ||
| Software and algorithms | ||
|
| ||
| Trinity | Haas et al., 2013 | https://github.com/trinityrnaseq/trinityrnaseq/wiki |
| BUSCO | Simão et al., 2015 | https://busco.ezlab.org/ |
| RepBase | Bao et al., 2015 | https://www.girinst.org/repbase/ |
| RepeatMasker | Tarailo-Graovac and Chen, 2009 | https://www.repeatmasker.org/ |
| Maker2 | Holt and Yandell, 2011 | https://www.yandell-lab.org/software/maker.html |
| MUSCLE | Edgar, 2004 | https://www.drive5.com/muscle/ |
| RAxML | Stamatakis, 2014 | https://github.com/stamatak/standard-RAxML |
| PosiGene | Sahm et al., 2017 | https://github.com/gengit/PosiGene |
| OrthoDB | Zdobnov et al., 2017 | https://www.orthodb.org/v9/index.html |
| CAFÉ 3 | Han et al., 2013 | https://github.com/hahnlab/CAFE |
| GeneWise | Birney et al., 2004 | ftp://ftp.ebi.ac.uk/pub/software/unix/wise2/ |
| DeSeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Kallisto | Bray et al., 2016 | https://pachterlab.github.io/kallisto/ |
| SwissModel | Waterhouse et al., 2018 | https://swissmodel.expasy.org/ |
|
| ||
| Experimental models: cell lines | ||
|
| ||
| Beaver/Mouse lung fibroblast cells | This paper | N/A |


