Abstract
To investigate the roles of insulin receptor substrate 3 (IRS-3) and IRS-4 in the insulin-like growth factor 1 (IGF-1) signaling cascade, we introduced these proteins into 3T3 embryonic fibroblast cell lines prepared from wild-type (WT) and IRS-1 knockout (KO) mice by using a retroviral system. Following transduction of IRS-3 or IRS-4, the cells showed a significant decrease in IRS-2 mRNA and protein levels without any change in the IRS-1 protein level. In these cell lines, IGF-1 caused the rapid tyrosine phosphorylation of all four IRS proteins. However, IRS-3- or IRS-4-expressing cells also showed a marked decrease in IRS-1 and IRS-2 phosphorylation compared to the host cells. This decrease was accounted for in part by a decrease in the level of IRS-2 protein but occurred with no significant change in the IRS-1 protein level. IRS-3- or IRS-4-overexpressing cells showed an increase in basal phosphatidylinositol 3-kinase activity and basal Akt phosphorylation, while the IGF-1-stimulated levels correlated well with total tyrosine phosphorylation level of all IRS proteins in each cell line. IRS-3 expression in WT cells also caused an increase in IGF-1-induced mitogen-activated protein kinase phosphorylation and egr-1 expression (∼1.8- and ∼2.4-fold with respect to WT). In the IRS-1 KO cells, the impaired mitogenic response to IGF-1 was reconstituted with IRS-1 to supranormal levels and was returned to almost normal by IRS-2 or IRS-3 but was not improved by overexpression of IRS-4. These data suggest that IRS-3 and IRS-4 may act as negative regulators of the IGF-1 signaling pathway by suppressing the function of other IRS proteins at several steps.
Insulin and insulin-like growth factor 1 (IGF-1) initiate their diverse biological effects by binding to and activating their endogenous tyrosine kinase receptors (22, 44). The insulin receptor substrate (IRS) proteins are major substrates of both insulin receptor and IGF-1 receptor tyrosine kinases and are rapidly phosphorylated on their tyrosine residues following ligand stimulation (21). The resulting phosphotyrosine motifs in these substrates then bind proteins containing Src homology 2 (SH2) domains, notably phosphatidylinositol 3-kinase (PI 3-kinase) (5), growth factor receptor binding protein 2 (Grb-2) (36), and the protein tyrosine phosphatase SHP-2/Syp (38), thereby activating specific signaling cascades. In addition, depending on the cell type, IGF-1 and insulin receptor can phosphorylate other substrates, such as Shc (16, 28), and Gab1 (18), which link to one or another of these pathways. Together, these intermediate signals stimulate a variety of different downstream biological effects including mitogenesis, gene expression, glucose transport, and glycogen synthesis.
To date, four members of the IRS family (IRS-1, IRS-2, IRS-3, and IRS-4) have been identified (23, 24, 33, 40, 41). IRS-1 and IRS-2 are the best-characterized members and are very similar in their overall structure. Both are high-molecular-weight proteins consisting of a pleckstrin homology domain at the N terminus followed by a phosphotyrosine binding domain and a large C-terminal domain containing multiple potential tyrosine phosphorylation sites that can bind to specific SH2 domain-containing proteins (41). Experiments with mice lacking either IRS-1 or IRS-2, created using homologous recombinant gene-targeting techniques, have confirmed the importance of both of these IRS proteins to glucose homeostasis and growth (4, 42, 45). Deletion of IRS-1 leads to severe intrauterine growth retardation and peripheral insulin resistance, whereas deletion of IRS-2 results in insulin resistance and a defect in pancreatic β-cell development leading to diabetes. These in vivo data, as well as in vitro data (9), indicate that IRS-1 and IRS-2 are not fully interchangeable signaling intermediates for the biological effects of insulin and IGF-1.
IRS-3 and IRS-4 have the common overall architecture of the IRS family; however, IRS-3 is much smaller than the other IRS proteins and has fewer phosphorylation sites (23, 24, 33). Several in vivo and in vitro analyses have demonstrated that IRS-3 and IRS-4 can be phosphorylated by insulin and IGF-1, bind to SH2 domain-containing proteins including PI 3-kinase and Grb-2 (14, 30, 46), and promote some biological actions of insulin and IGF-1 (12, 43, 48). However, mice lacking either the IRS-3 or IRS-4 have recently been created, and, in contrast to the IRS-1- or IRS-2-deficient mice, IRS-3- and IRS-4-deficient mice have no apparent phenotype (15, 25), raising the question whether these proteins act as alternative substrates in the IGF-1 and insulin signaling pathway or play some other unique roles.
In the present study, we introduced IRS-3 and IRS-4 into normal wild-type and IRS-1-deficient embryonic fibroblast cells and investigated the impact of their expression on IGF-1 signaling and biological effects. The data obtained with these cells suggest that IRS-3 and IRS-4 may act as negative regulators of the IGF-1 signaling pathway by suppressing the function of other IRS proteins.
MATERIALS AND METHODS
Materials.
Human recombinant IGF-1 was obtained from Pepro Tec, Inc (Rocky Hill, N.J.). [γ-32P]ATP, [α-32P]dCTP, 125I-protein A, and [methyl-3H]thymidine were from New England Nuclear Inc. (Woburn, Mass.). Reagents for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting apparatus were from Bio-Rad Laboratories (Richmond, Calif.). Aluminum-backed silica gel thin-layer chromatographic plates were from Merck (Darmstadt, Germany). Protein A-Sepharose 6MB and the enhanced chemiluminescence Western blotting kit were from Boehringer Mannheim Co. (Indianapolis, Ind.). Taq DNA polymerase (AmpliTaq Gold) was from Applied Biosystems (Foster City, Calif.). ExpressHyb hybridization solution was from Clontech (Palo Alto, Calif.). All other common materials were from Sigma Chemical Co. (St. Louis, Mo.).
Antibodies.
Polyclonal antibodies to IRS-2 and p85α/β were generous gifts from M. F. White (Joslin Diabetes Center, Boston, Mass.). Polyclonal antibody to mouse IRS-4 was kindly provided by G. E. Lienhard (Dartmouth Medical School, Hanover, N.H.). Antibody to IRS-1 was prepared as described previously (40). The antibody against IRS-3 was prepared by immunizing rabbits with a glutathione S-transferase fusion protein containing amino acids 240 to 491 of rat IRS-3. Monoclonal antibody to phosphotyrosine (PY20) was purchased from Transduction Laboratories (San Diego, Calif.). Polyclonal antibodies to Akt, phosphospecific Akt (Ser 473), mitogen-activated protein (MAP) kinase, and phospho-specific MAP kinase (Tyr204) were from New England Biolabs, Inc. (Beverly, Mass.). Anti Grb-2 antibody, anti-Shc antibody, and anti-IGF-1 receptor antibody were from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.).
Generation of embryonic fibroblast cell lines.
Primary embryonic fibroblasts were obtained from 16.5-day fetuses of pregnant IRS-1+/− mice mated with IRS-1+/− males, as described previously (9). After the genotyping by PCR, IRS-1+/+ (wild-type; WT) and IRS-1−/− (IRS-1 knockout or KO) cells were passaged by the 3T3 protocol to establish permanent cell lines as described previously (9). Once established, the cell lines were maintained in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum at 37°C and 5% CO2. The cells were split every third day and never allowed to reach confluency, except as specified for experiments.
Plasmids and transfection.
Retrovirus expression vectors of human IRS-1 (pBABE-IRS-1) and mouse IRS-2 (pBABE-IRS-2) were prepared as described previously (9). Rat IRS-3 was generated by PCR from the rat genomic DNA sequence (33). The intron was removed by PCR overlap extension using the following two pairs of primers: 5′-CGTGGATCCGCGATGAAGCCTGCAGGTACG-3′ (sense) plus 5′-CTTGGGGGCTGAAACCCATGTTTGCTGGGCA-3′ (antisense) and 5′-TGCCCAGCAAACATGGGTTTCAGCCCCCAAG-3′ (sense) plus 5′-GCTGTCGACGTTCTAGAACTTGATGCTG-3′ (antisense) (19). BamHI and SalI sites were introduced at the 5′ and 3′ ends, respectively, of the coding sequence of IRS-3 by PCR. The subsequently generated coding sequence of rat IRS-3 was ligated into BamHI and SalI sites in pBABE-puro (pBABE-IRS-3). Mouse IRS-4 genomic DNA was screened from the BAC genomic DNA library (Genome Systems Inc. St. Louis, Mo.) by PCR with specific primers for mouse IRS-4 genomic DNA (13). The AvrII-ClaI fragment encompassing the entire exon 1 of mouse IRS-4 gene was blunt ended and ligated into the SnaBI site in pBABE-puro (pBABE-IRS-4). The nucleotide sequences of both IRS-3 and IRS-4 were determined to be identical to the published sequences (13, 33). ΦNX cells were transiently transfected by using the calcium precipitation technique with 20 μg of plasmid DNA per 10-cm-diameter dish. The cells were refed 12 to 16 h after transfection, and Polybrene (8 μg/ml)-supplemented virus-containing supernatant was transferred to the target cells 72 h after transfection. After an overnight infection period, the target cells were refed. Selection was begun by using 2 μg of puromycin per ml 48 h after infection. After the selection, the following three WT cell lines and five KO cell lines were generated: WT cell line infected with pBABE-puro (WT), pBABE-IRS-3 (WT-3), or pBABE-IRS-4 (WT-4) and KO cell line infected with empty pBABE-puro (KO), pBABE-IRS-1 (KO-1), pBABE-IRS-2 (KO-2), pBABE-IRS-3 (KO-1), or pBABE-IRS-4 (KO-4).
Immunoprecipitation and Western blot analysis.
For stimulation of IGF-1-mediated responses, cells were serum deprived overnight in medium containing 0.1% bovine serum albumin (BSA) and then, unless noted otherwise, treated for the indicated times with IGF-1 at a final concentration of 10 nM in DMEM supplemented with 0.1% BSA. Protein extracts were prepared by using buffer A (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 2 mM Na3 VO4, 20 mM Na4P2O2, 100 mM NaF, 1% NP-40, 2 mM phenylmethylsulfonyl fluoride, 20 μg of aprotinin per ml, 10 μg of leupeptin per ml) for 30 min at 4°C, and insoluble protein was removed by centrifugation at 13,400 × g in a microcentrifuge. The protein content was determined by the method of Bradford. The extract was then resolved directly in SDS-polyacrylamide gels after boiling in Laemmli SDS sample buffer or subjected to immunoprecipitation with the indicated antibodies. For immunoprecipitation, 500 μg of cellular protein was incubated with the indicated antibodies for 12 h at 4°C. Immunocomplexes were collected and washed with buffer A three times and resuspended in SDS sample buffer. Proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The blots were blocked with 3% BSA in TBS buffer (10 mM Tris [pH 7.5], 150 mM NaCl), incubated with antibodies in TBS containing 2% BSA, and then incubated with either secondary antibodies conjugated to horseradish peroxidase or 125I-protein A. The immunoreactive bands were visualized by either enhanced chemiluminescence or a Molecular Dynamics PhosphorImager.
PI 3-kinase assay.
Quiescent cells were stimulated for 10 min with 10 nM IGF-1 in DMEM containing 0.1% BSA. After three washes with ice-cold phosphate-buffered saline (PBS), cells were lysed in PI-3 kinase buffer (20 mM Tris [pH 7.4], 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 2 mM Na3 VO4, 10% glycerol, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml) for 10 min at 4°C and cleared by centrifugation at 13,400 × g at 4°C, and the protein content of the supernatant was determined. A 250-μg portion of cellular protein was subjected to immunoprecipitation for 2 h at 4°C. The resulting immunocomplexes were washed three times with PBS containing 1% NP-40, three times with 500 mM LiCl–100 mM Tris (pH 7.5), and twice with reaction buffer (10 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA). The pellets were resuspended sequentially in 50 μl of reaction buffer, 10 μl of 100 mM MgCl2, and 10 μl of PI (2 μg/μl) and sonicated in 10 mM Tris (pH 7.5) containing 1 mM EDTA. The phosphorylation reaction was started by adding 5 μl of 65 μM ATP containing 3 μCi of [γ-32P]ATP. After 15 min at room temperature, the reaction was stopped with 20 μl of 8 N HCl and then 160 μl of CHCl3-methanol (1:1). The samples were briefly centrifuged, and the lower (organic) phase was spotted on thin-layer silica gel chromatography plates. The plates were developed in CHCl3-methanol-H2O-NH4OH (120:94:23:2.4), dried, visualized, and quantified on a Molecular Dynamics PhosphorImager.
RT-PCR analysis.
Total RNA was isolated from serum-starved cells with TRIzol reagent (GIBCO-BRL, Gaithersburg, Md.). Isolated RNA was treated with DNase I as recommended by the manufacturer (GIBCO-BRL). First-strand cDNA was synthesized from 5 μg of total RNA by using a first-strand cDNA synthesis kit (GIBCO-BRL) with a random primer. In control reactions, water replaced the reverse transcriptase. Aliquots of the reverse transcription RT and control reaction mixtures were amplified by PCR using AmpliTaq Gold and the following pairs of primers: 5′-AGCGAGCTCGAGCATGGCGAGCCCTC-3′ and 5′-ATCGTCGACTCGAGATCTCCGAGTCA-3′ for mouse IRS-1, 5′-AAGGCCAGCACCTTACCTCG-3′ and 5′-AGCCATGGTGGCCCTGGGCAG-3′ for human IRS-1, 5′-CTCTGACTATATGAACCTG-3′ and 5′-ACCTTCTGGCTTTGGAGGTG-3′ for mouse IRS-2, 5′-GGCCCCACAGTCTCCTCCGG-3′ and 5′-GCCTCTTGGGGACTGAAAC-3′ for mouse and rat IRS-3, and 5′-CCCTTCTACAAAGATGTGTGGC-3′ and 5′-TCTCCAGAAACAGCTCATGC-3′ for mouse IRS-4. The PCR products were separated on a 2% agarose gel and visualized by ethidium bromide staining.
Northern blot analysis.
Northern blot analysis was performed by standard techniques in denaturing formamide-containing agarose gels (31). Total RNA was isolated by using TRIzol reagent. A 10-μg sample of total RNA was subjected to electrophoresis in 1% agarose gels. Ethidium bromide staining of the gels confirmed equal loading and integrity of the RNA. After being transferred to a nylon membrane, the blots were hybridized in ExpressHyb hybridization solution with [α-32P]dCTP-labeled probes. After incubation for 2 h at 65°C, the blots were washed twice in 1× SSC buffer (150 mM NaCl, 15 mM sodium citrate)–0.1% SDS for 20 min each at room temperature and once for 30 min in 0.1× SSC–0.1% SDS at 50°C. The membranes were air dried and subjected to autoradiography.
[methyl-3H]thymidine incorporation into DNA.
Cells were plated at a density of 2 × 105 per well in 24-well dishes. After 1 day, the medium was changed for 48 h to DMEM with 0.1% BSA. The cells were then stimulated with IGF-1 for 15 h and pulsed with 1 μCi of [methyl-3H]thymidine per well for 1 h at 37°C. After two washes with ice-cold PBS, the cells were incubated in ice-cold 10% trichloroacetic acid (TCA) for 1 h. TCA-precipitated DNA was washed once with ice-cold 10% TCA, lysed for 30 min in 0.1 N NaOH–0.1% SDS solution, and then counted for incorporated radioactivity. All assays were performed in duplicate.
Statistical analysis.
Data are expressed as mean ± standard error of the mean (SEM). Differences between two groups were evaluated by an unpaired Student t test. P < 0.05 was defined as indicating the presence of a statistically significant difference.
RESULTS
Expression of endogenous and retroviral introduced IRSs in embryonic fibroblast cells.
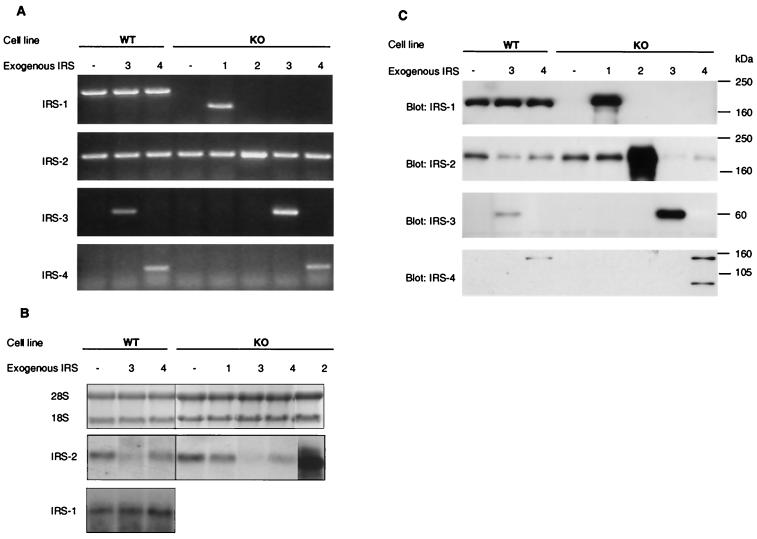
To analyze the impact of IRS-3 or IRS-4 expression on the IGF-1 signaling pathway, immortalized embryonic fibroblast cell lines prepared from normal (IRS-1+/+) mice (WT) and IRS-1−/− knockout mice (KO) by infection with pBABE retrovirus containing rat IRS-3 or mouse IRS-4. KO cells were also infected with pBABE containing either human IRS-1 or mouse IRS-2 to reconstitute the impaired signaling caused by IRS-1 deficiency. Expression of the four potential endogenous and exogenous IRS genes was examined by RT-PCR using sets of specific primers for each IRS coding sequence. Because the primer pairs for IRS-1, IRS-2, and IRS-4 were designed within one exon, the RNA samples were also amplified by PCR without RT to rule out contamination of the RNA samples by genomic DNA. A fragment amplified by mouse IRS-1 primers was detectable in all WT cell lines infected with either empty pBABE-puro (WT), pBABE-IRS-3 (WT-3), or pBABE-IRS-4 virus (WT-4) (Fig. 1A). A smaller fragment, which was amplified by the primers for human IRS-1, was detected from only IRS-1 virus-infected KO cells (KO-1 cells). A fragment amplified by the IRS-2 primers appeared in all cell lines (Fig. 1A). By contrast, amplified IRS-3 and IRS-4 fragments were detected from only the cells transfected with either IRS-3 (WT-3 and KO-3) or IRS-4 (WT-4 and KO-4), respectively (Fig. 1A). No visible fragment was detected from any PCR samples without RT (data not shown).
FIG. 1.
Expression of the four IRS proteins in various mouse embryonic fibroblast cell lines. (A) RT-PCR for IRS-1, IRS-2, IRS-3, and IRS-4 mRNA was performed on total RNA prepared from the three WT cell lines (transfected with empty pBABE [WT], rat IRS-3 [WT-3], or mouse IRS-4 [WT-4] viruses) and the five IRS-1 KO cell lines (transfected with empty pBABE [KO], human IRS-1 [KO-1], mouse IRS-2 [KO-2], rat IRS-3 [KO-3], or mouse IRS-4 [KO-4] viruses). The sizes of the bands specific for the IRS proteins were 922 bp (mouse IRS-1), 498 bp (human IRS-1), 339 bp (IRS-2), 431 bp (IRS-3), and 294 bp (IRS-4). (B) Northern blot analysis of IRS-1 and IRS-2 mRNA. Total RNA (10 μg) prepared from serum-starved cells were separated on formaldehyde-containing agarose gels and transferred to nylon membranes. Equal loading and integrity of the RNA were confirmed by ethidium bromide staining of agarose gels (upper panel). Blots were hybridized with a specific probe for either IRS-2 (middle panel) or IRS-1 (lower panel) and visualized by autoradiography. (C) Equal amounts of cell extracts from serum-starved cells were separated by SDS-PAGE and analyzed by immunoblotting using the indicated antibodies. The experiments shown are representative of multiple experiments.
To quantitate the expression levels of IRS-1 and IRS-2 mRNA, total RNA prepared from serum-starved cells was subjected to Northern blot analysis. The level of IRS-2 mRNA in KO cells was slightly higher than that in WT cells (121% ± 5.4% of control), whereas IRS-3-expressing cells (WT-3 and KO-3) showed a marked decrease in IRS-2 mRNA levels compared to their host cells (52% ± 5.4% and 11% ± 7.8% in WT-3 and KO-3, respectively) (Fig. 1B). IRS-4 expression also caused an significant decrease in IRS-2 mRNA expression in KO-4 cells (41% ± 7.8%) and produced a tendency toward reduced IRS-2 mRNA levels in WT-4 cells. In contrast, endogenous IRS-1 gene expression in WT cell lines was not changed by IRS-3 or IRS-4 expression (Fig. 1B).
To confirm the IRS protein expression level, cell lysates were prepared from serum-starved cells and analyzed by Western blotting using the respective anti-IRS protein antibodies. IRS-1 protein was detected in all WT cell lines, and the protein amount was not altered by IRS-3 or IRS-4 expression (Fig. 1C). KO-1 cells overexpressed IRS-1 twofold compared to WT cells. As expected from the results of RT-PCR, IRS-2 protein was detected in all cell lines (Fig. 1C). KO cells exhibited a ∼20% increase in IRS-2 protein expression compared to that in WT cells (122% ± 5.4% with respect to WT cells), and this was not altered by IRS-1 overexpression in KO-1 cells. KO cells transfected with IRS-2 revealed a fivefold overexpression of IRS-2 compared with the IRS-1 KO cells. Both IRS-3 and IRS-4 proteins could be detected only in KO and WT cells infected with expression retroviruses and migrated at 60 and 155 kDa, respectively (Fig. 1C). The expression level of these proteins was about threefold higher in KO cell lines than in WT cell lines in which these cDNAs had been introduced (Fig. 1C). By Western blot analysis with an IRS-4 specific antibody, an additional (85-kDa) protein was detected in both WT-4 and KO-4 cells. This protein was also detected by the phosphotyrosine (PY) antibody (see Fig. 2) and could bind with p85 (see Fig. 3), suggesting that this protein is degraded IRS-4. Surprisingly, following expression of either IRS-3 or IRS-4, both WT and KO cell lines showed a significant decrease in IRS-2 protein expression compared with their host cells (∼61% ± 7.2% and ∼45% ± 6.0% reduction in WT-3 and WT-4 cells and ∼78% ± 11.2% and ∼60% ± 9.3% reduction in KO-3 and KO-4 cells, respectively). There was no significant change in the amount of other signaling molecules, including IGF-1 receptor, p85 subunit of PI 3-kinase, Akt, Grb-2, and p44/42 MAP kinase (data not shown).
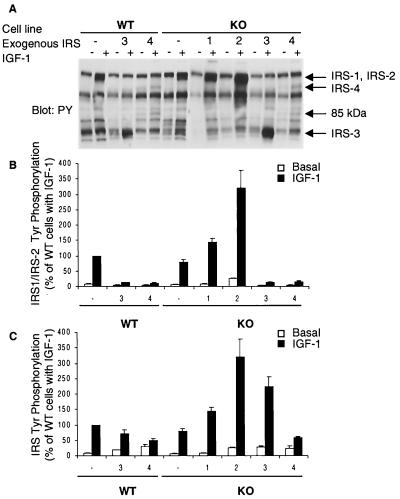
FIG. 2.
IGF-1-induced tyrosine phosphorylation of IRS proteins. (A) Serum-starved cells were stimulated with 10 nM IGF-1 for 3 min at 37°C. Equal amounts of proteins in cell lysates were separated by SDS-PAGE and immunoblotted with PY-specific antibody. Arrows indicate the migration of proteins immunoreactive with each IRS protein-specific antibody. The 85-kDa protein was detectable in WT-4 and KO-4 cells. The experiment shown is representative of multiple experiments. (B) The relative intensity of the PY band corresponding to IRS-1 and IRS-2 in each lane was measured by densitometric analysis. The phosphorylation level is expressed as a percentage of the intensity observed in IGF-1-induced WT cells and is presented as mean and SEM of four independent experiments. (C) Total tyrosine phosphorylation level of the four IRS proteins. Relative intensities of the area corresponding to IRS-1, IRS-2, IRS-3, and IRS-4 in each lane were measured separately by densitometric analysis. The sum total intensities of four IRS proteins were calculated and expressed as percentages of the intensity seen in IGF-1-induced WT cells.
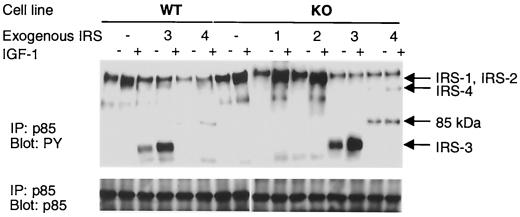
FIG. 3.
Association of IRS proteins with p85 subunit PI 3-kinase. Serum-starved cells were stimulated with 10 nM IGF-1 for 3 min at 37°C. The protein concentration of cell lysates was determined, and equal amounts of proteins were immunoprecipitated (IP) with anti-p85 antibody and analyzed by immunoblotting with anti-PY antibody (upper panel) or anti-p85 antibody (lower panel). The result shown is representative of multiple experiments. Arrows indicate the migration of proteins immunoreactive with each IRS protein-specific antibody. The 85-kDa protein detectable in WT-4 and KO-4 cells probably represents a degradation fragment of IRS-4.
IRS protein tyrosine phosphorylation and its association with p85 PI 3-kinase.
To examine the tyrosine phosphorylation of IRS proteins, serum-deprived cells were stimulated with 10 nM IGF-1 for 3 min and cell lysates were analyzed by Western blotting with antiphosphotyrosine (PY) antibody. In WT and KO cells, IGF-1 caused a rapid tyrosine phosphorylation of the 175- to 185-kDa protein, which corresponded to phosphorylated endogenous IRS-1 and/or IRS-2 (Fig. 2A). The intensity of this band in IGF-1-treated KO cell was ∼20% lower than that in WT cells (Fig. 2B). A stronger phosphotyrosine signal appeared at 175 to 185-kDa in IGF-1-treated KO-1 and KO-2 cells, corresponding to phosphorylated endogenous and exogenous IRS-1 and IRS-2, and the intensity was increased by 1.5- and 3-fold from that in KO cells, respectively (Fig. 2A and B). In both WT-3 and KO-3 cells, IGF-1 led to an increase in the intensity of a band corresponding IRS-3 (60 kDa) (Fig. 2A and B). In addition, in these IRS-3-expressing cells, the signal of phosphorylated 175- to 185-kDa protein (representing IRS-1 and/or IRS-2) showed a marked decrease (∼87%) from that in control cells (Fig. 2A and B). IGF-1 caused phosphorylation on two proteins (155 and 85 kDa) in both WT-4 and KO-4 cells (Fig. 2A and B). More importantly, these cells also showed a ∼85% decrease in 175- to 185-kDa protein phosphorylation. This decrease of IRS-1 and IRS-2 phosphorylation seen in IRS-3- and IRS-4 expressing cells could be explained only in part by a decrease in the amount of IRS-2 protein, since WT-3 and WT-4 cells express equal amounts of IRS-1 compared with WT cells. The sum of the intensities of the bands corresponding to all phosphorylated IRS proteins in IGF-1-treated WT-3 and WT-4 cells was about 70 and 50% of the intensity in WT cells, respectively (Fig. 2C). Total IRS protein phosphorylation in KO-3 cells was significantly higher (∼125%) than that in control cells, whereas the level in KO-4 cell was comparable to that in WT-4 cells.
An important component of the IRS-1- and IRS-2-mediated response to IGF-1 is docking of the p85 regulatory subunit of PI 3-kinase to tyrosine-phosphorylated IRS proteins and activation of the p110 catalytic subunit. Thus interaction could be detected in all cells by immunoprecipitation with anti-p85 antibody following Western blotting with anti-PY antibody. In WT, KO, KO-1, and KO-2 cells treated with IGF-1, a strong signal was detected at 175 to 185 kDa, corresponding to phosphorylated IRS-1 and IRS-2, while a fainter signals were seen in WT-3 and WT-4 cells (Fig. 3). A strong signal at 60 kDa was also detected in both WT-3 and KO-3 cells in the basal state, and IGF-1 stimulation caused a marked increase in the intensity of this band (Fig. 3). We detected 155- and 85-kDa PY proteins in only WT-4 and KO-4 cells (Fig. 3). These results confirm that tyrosine-phosphorylated IRS-3 and IRS-4 bind the p85 subunit of PI 3-kinase in these cells. Furthermore, the levels of p85 subunit in these cells were similar, indicating IRS-3 and IRS-4 expression did not affect the p85 protein level (Fig. 3).
Interaction of the IRS proteins with p85 could also be detected by Western blot analysis of anti-PY precipitates with anti-p85 antibody. In all cell lines, IGF-1 treatment led to a significant increase in the amount of precipitated p85 (Fig. 4A and B). The increased p85 precipitation following IGF-1 stimulation is likely to be associated with the phosphorylated IRS proteins, since the IRS proteins were major PY proteins which showed IGF-1-induced increases in coimmunoprecipitation with p85 (Fig. 3). However, IRS-4 binding to p85 in WT-4 and KO-4 cells was very weak (Fig. 3); thus, p85 might also be associated with other PY proteins in these cells. The amount of p85 coprecipitated in anti-PY precipitates in IGF-1 stimulated cells correlated with the level of total tyrosine phosphorylation of all four IRS proteins, as shown in Fig. 2, in most cell types (Fig. 4B). The exception was in the IRS-2- or IRS-3-expressing cell lines (KO-2, WT-3, and KO-3). These cells showed relatively less p85 precipitation with respect to the phosphorylation levels of IRS proteins. We have confirmed that all the phosphorylated IRS proteins were precipitated almost completely with anti-PY antibody by immunoblotting of the supernatants in the immunoprecipitation tubes (data not shown). Therefore, this reduced p85 binding in IRS-2- or IRS-3-overexpressing cells was not due to a lower efficiency of immunoprecipitation in these cell lines.
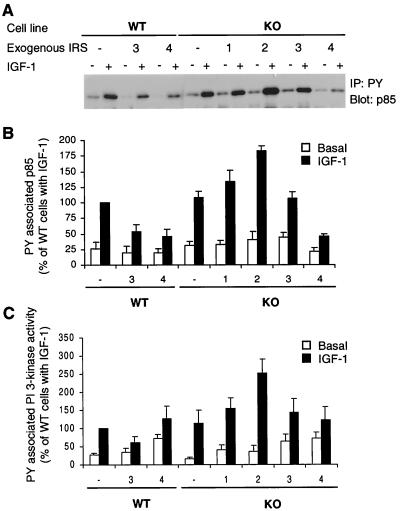
FIG. 4.
PY-containing protein-associated p85 and PI 3-kinase activity. (A) Serum-starved cells were stimulated with 10 nM IGF-1 for 10 min at 37°C. Cell lysates were immunoprecipitated (IP) with anti-PY antibody and immunoblotted with anti-p85 antibody. The experiment shown is representative of multiple experiments. (B) Quantitation of four independent experiments. The results are expressed as the percentage of the level present in IGF-1-induced WT cells and are presented as the mean and SEM of four experiments. (C) IGF-1-stimulated activation of PI 3-kinase. Quiescent cells were stimulated with 10 nM IGF-1 for 10 min at 37°C, and in vitro kinase assays of cell lysates were performed as described in Materials and Methods using anti-PY immunoprecipitates. Phosphorylated PI was quantified on a Molecular Dynamics PhosphorImager. Results are expressed as percentages of IGF-1-induced WT cells and are the mean and SEM of three independent experiments.
PI 3-kinase activity in PY antibody-precipitated samples paralleled the amount of p85 precipitated by anti-PY antibody, and no significant difference was found between these two values except in IRS-4-expressing cells (Fig. 4B). The relative value of PI 3-kinase activity in samples precipitated from WT-4 and KO-4 cells was significantly higher than that of p85 association under both basal and IGF-1-stimulated conditions, suggesting that IRS-4 may also associate with other proteins that can activate PI 3-kinase.
Inhibition of IRS-1- and IRS-2-mediated IGF-1 signaling by IRS-3 or IRS-4 expression.
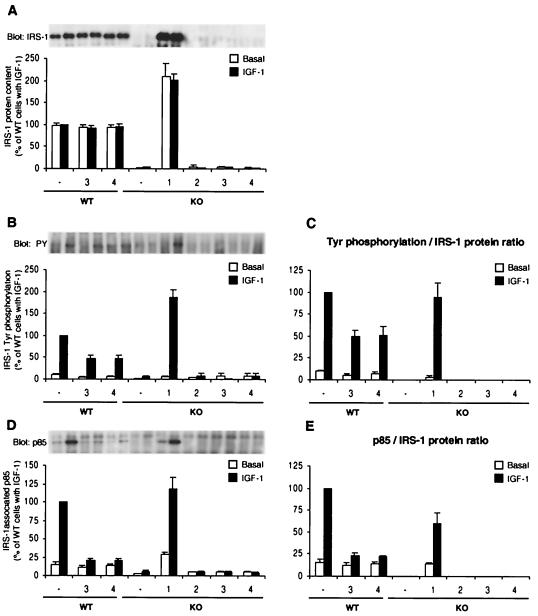
To investigate how IRS-3 or IRS-4 expression affects IRS-1- and IRS-2-mediated IGF-1 signaling pathways, we individually analyzed IRS-1 and IRS-2 tyrosine phosphorylation and p85 binding by immunoprecipitation with IRS-1- or IRS-2-specific antibodies followed by Western blotting. There was no significant difference in the IRS-1 protein content between WT, WT-3, and WT-4 cells (Fig. 5A). Nevertheless, both WT-3 and WT-4 cells showed a ∼50% decrease in IGF-1-stimulated IRS-1 tyrosine phosphorylation compared to WT cells (Fig. 5B and C). The level of IGF-1-induced p85 association per IRS-1 protein in WT-3 and WT-4 cells was also reduced to ∼15% of that in control cells, a decrease that was significantly greater than the decrease in IRS-1 tyrosine phosphorylation (Fig. 5D and E). KO-1 cells showed a ∼90% increase in tyrosine phosphorylation compared to that in WT cells (Fig. 5B), and this increase was comparable to the increase in the amount of IRS-1 (Fig. 5C). These cells also showed a ∼30% decrease in p85 association per IRS-1 protein (Fig. 5E).
FIG. 5.
Tyrosine phosphorylation of IRS-1 and its association with p85 PI 3-kinase. Serum-starved cells were stimulated with 10 nM IGF-1 for 3 min at 37°C. (A, B, and D) The protein concentration of cell lysates was determined, and equal amounts of proteins were immunoprecipitated with anti-IRS-1 antibody and immunoblotted with anti-IRS-1 antibody (A), anti-PY antibody (B), or anti-p85 antibody (D). Blots were visualized by autoradiography (upper panels of A, B, and D) and quantified (lower panels of A, B, and D). Quantitative results, expressed as percentages of IGF-1-induced WT cells, represent the mean and SEM of three independent experiments. (C) The levels of tyrosine phosphorylation per IRS-1 protein in IRS-1-positive cells (WT, WT-3, WT-4, and KO-1 cells) were calculated by dividing the relative phosphorylation level of IRS-1 measured in panel B by the relative amount of IRS-1 protein shown in panel A. (E) The levels of p85 association per IRS-1 were calculated by dividing the amount of IRS-1-associated p85 in panel D by the amount of IRS-1 protein in panel A.
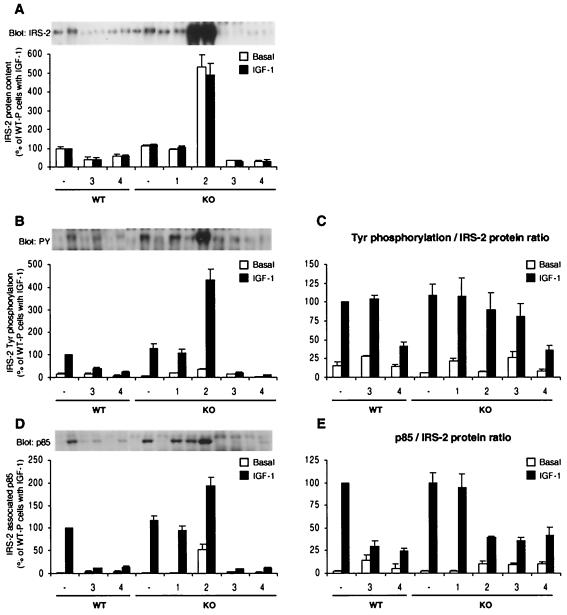
Immunoprecipitation with anti-IRS-2 antibody followed by IRS-2 immunoblotting reproduced the results of the Western blot analysis using whole-cell lysates (Fig. 6A). Thus, WT-3, WT-4, KO-3, and KO-4 cells showed a marked decrease in IRS-2 content (∼62, 40, 70, and 70%, respectively, from their control cells), while KO-2 cells exhibited a fivefold overexpression of IRS-2 compared to KO cells (Fig. 6A). The intensity of IGF-1-stimulated tyrosine-phosphorylated IRS-2 correlated in general with the level of IRS-2 protein (Fig. 6C). The exception was in the two IRS-4-expressing cell lines, WT-4 and KO-4, which showed ∼59 and ∼64% reductions in IRS-2 phosphorylation, respectively (Fig. 6C). The IRS-2 phosphorylation level per IRS-2 protein in IRS-3-expressing KO-3 cells also tended to be lower than that in control, although the decrease was not statistically significant (Fig. 6C). Interestingly, p85 association with IRS-2, adjusted for changes in IRS-2 protein content, was significantly reduced in WT-4 and KO-4 cells, as well as in WT-3 and KO-3 cells (Fig. 6D and E). These results suggest that expression of either IRS-3 or IRS-4 can cause inhibition of IRS-1- and IRS-2-mediated signaling in at least three steps: (i) decreasing the amount of IRS-2 protein, (ii) decreasing the phosphorylation of IRS-1 and IRS-2, and (iii) decreasing the association of p85 with IRS-1 or IRS-2.
FIG. 6.
Tyrosine phosphorylation of IRS-2 and its association with p85 PI 3-kinase. Cell lysates prepared as described in the legend to Fig. 5 were immunoprecipitated with anti-IRS-2 antibody and immunoblotted with anti-IRS-2 antibody (A), anti-PY antibody (B), or anti-p85 antibody (D). The results are expressed as in Fig. 5. All of the results, expressed as percentages of IGF-1-induced WT cells, are presented as the mean and SEM of three independent experiments.
Impact of IRS-3 or IRS-4 expression on Akt phosphorylation.
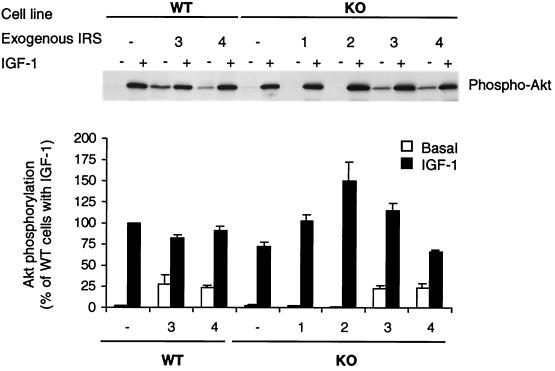
Activation of PI 3-kinase leads to the formation of phosphatidylinositol-3,-4-bisphosphate [PI(3,4)P2] and/or PI(3,4,5)P3, thereby recruiting Akt (PKB/Rac) to membranes and promoting its phosphorylation by other membrane-associated phospholipid-dependent kinases (1, 2). Ser473 phosphorylation plays an important role in the activation of Akt; therefore, we examined the activity of the Akt pathway by Western blotting with Akt phosphoserine 473-specific antibody. IGF-1 treatment caused a great increase in Ser phosphorylation of Akt in all cell types, and the levels of phosphorylation in cells treated with IGF-1 roughly paralleled the increase in PI 3-kinase activity (Fig. 7). Interestingly, IRS-3- or IRS-4-overexpressing cells also showed significant increases in phosphorylation of Akt in the absence of IGF-1 (i.e., in the basal state), resulting in poor stimulation.
FIG. 7.
Effect of IRS protein overexpression on Akt phosphorylation in embryonic fibroblast cell lines. After overnight serum starvation, cells were stimulated with 10 nM IGF-1 for 10 min at 37°C. Cell lysates were subjected to SDS-PAGE and analyzed by Western blotting with anti-phospho-Akt antibody (Ser473). Blots were visualized by autoradiography (upper panel) and quantified by scanning densitometry (lower panel). Quantitative results, expressed as percentages of IGF-1-induced WT cells, are presented as the mean and SEM of three independent experiments.
Impact of IRS-3 or IRS-4 expression on MAP kinase signaling.
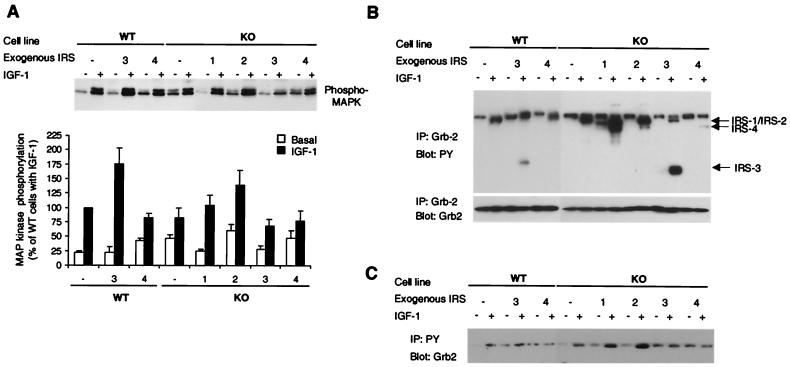
The MAP kinase cascade is another major signaling cascade activated by insulin or IGF-1 stimulation. Tyrosine-phosphorylated IRS proteins, as well as tyrosine-phosphorylated Shc, can bind to Grb-2, which links signaling via Ras to a cascade of serine/threonine kinases, Raf, MAP kinase kinase, and MAP kinase (32). We examined the activation of this pathway by Western blotting with a phospho-p44/p42 MAP kinase-specific antibody. Elevated basal phosphorylation of p44/p42 MAP kinase was observed in WT-4, KO, KO-2, and KO-4 cells (95, 107, 200, and 105% increase from basal phosphorylation of WT cells, respectively) (Fig. 8A). Following IGF-1 stimulation, all cell lines except KO-4 showed a significant increase in MAP kinase phosphorylation from their basal states (Fig. 8A). WT-3 cells showed the greatest phosphorylation (1.8-fold increase from WT cells).
FIG. 8.
(A) Effect of IRS protein overexpression on MAP kinase phosphorylation in embryonic fibroblast cell lines. After overnight serum starvation, cells were stimulated with 10 nM IGF-1 for 10 min at 37°C. Cell lysates were subjected to SDS-PAGE and analyzed by Western blotting with anti-phospho-p44/42 MAP kinase antibody (Tyr204). Blots were visualized by autoradiography (upper panel) and quantified by scanning densitometry (lower panel). Quantitative results are expressed as percentages of the level present in IGF-1-induced WT cells and are presented as the mean and SEM of three independent experiments. (B) Association of IRS proteins with Grb-2. Serum-starved cells were stimulated with 10 nM IGF-1 for 3 min at 37°C. Protein samples were immunoprecipitated (IP) with anti-Grb-2 antibody and analyzed by immunoblotting with anti-PY (upper panel) or anti-Grb-2 antibody (lower panel). Arrows indicate the migration of proteins immunoreactive with each IRS protein-specific antibody. (C) Grb-2 binding with PY proteins. Serum-starved cells were stimulated with 10 nM IGF-1 for 3 min at 37°C. Protein samples were immunoprecipitated with anti-PY antibody and analyzed by immunoblotting with anti-Grb-2 antibody.
The association of Grb-2 with IRS proteins and Shc was assessed by coimmunoprecipitation with either anti-PY antibody or anti-Grb-2 antibody. IGF-1-induced Grb-2 binding to IRS proteins was confirmed by immunoprecipitation with anti-Grb-2-specific antibody following Western blotting with anti-PY antibody (Fig. 8B). In the precipitates from IGF-1-stimulated cells, all four tyrosine-phosphorylated IRS proteins were clearly detected, although the signal of phosphorylated IRS-4 was faint, especially in WT-4 cells. By contrast, phosphorylated Shc, another receptor substrate which could link the IGF-1 signal to Grb-2, was not detectable in Grb-2 antibody precipitates blotted with anti-PY antibody (Fig. 8B) or anti-Shc specific antibody (data not shown). This suggests that IRS proteins, but not Shc, are the main receptor substrates which bind to Grb-2 in these cell lines.
IGF-1 caused a significant increase in Grb-2 precipitation by anti-PY antibody in both WT and KO control cells (Fig. 8C). IRS-1 or IRS-2 overexpression in KO cells caused a significant elevation of IGF-1-induced Grb-2 association compared to that in control cells, suggesting that Grb-2 can bind to tyrosine-phosphorylated IRS-1 or IRS-2 in these cells. IRS-3 expression in WT cells also caused a slight increase in the amount of precipitated Grb-2 in the basal state, and the IGF-1-induced Grb-2 association was similar to that in WT cells. In contrast to WT-3 cells, KO-3 cells showed about a threefold increase in Grb-2 precipitation in the basal state and did not show an IGF-1-induced increase in Grb-2 coprecipitation. Both of the IRS-4 expressing cells, WT-4 and KO-4, showed no IGF-1-induced increase in Grb-2 precipitation. This PY-associated Grb-2 protein levels did not parallel MAP kinase phosphorylation levels.
IGF-1-mediated immediate-early gene induction in IRS-3- or IRS-4 expressed cells.
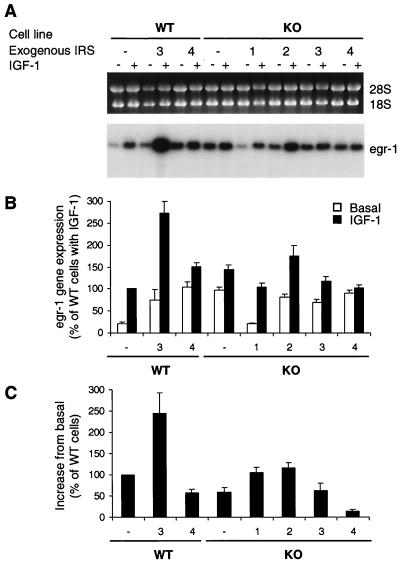
IGF-1 stimulates immediate-early gene induction and mitogenesis in 3T3 cells, and both of these biological effects are impaired by IRS-1 deficiency (9). Previous studies in our laboratory have shown that in IRS-1−/− cells early gene induction could be reconstituted by either IRS-1 or IRS-2 whereas mitogenesis could be reconstituted by IRS-1 but was not fully reconstituted by IRS-2 (9). Therefore, we evaluated the role of IRS-3 and IRS-4 in these IGF-1-mediated biological responses. IGF-1-mediated immediate-early gene induction was examined by Northern blot analysis using a probe against the early growth response 1 (egr-1) cDNA (Fig. 9A). Compared to WT cells, KO cells showed significantly higher basal egr-1 gene expression (about a fourfold increase from WT cells) (Fig. 9B). IRS-3 or IRS-4 overexpression in WT cells caused a significant increase in expression of this gene in the basal state (about three- and fourfold above that in WT cells, respectively). The elevated basal egr-1 expression in KO cells was decreased to the WT level by IRS-1 overexpression. IRS-2, IRS-3, or IRS-4 overexpression in KO cells did not cause any change in basal egr-1 gene expression compared to that in nontransfected KO cells. Since each cell line exhibited a somewhat different level of egr-1 mRNA expression in the basal state, we evaluated the IGF-1-induced gene expression as a function of the increase in egr-1 mRNA expression from basal in each cell type (Fig. 9C). Compared to WT cells, KO cells showed a ∼32% decrease in IGF-1-induced egr-1 gene expression, and this impairment was fully recovered by exogenous IRS-1 (KO-1 cells) or IRS-2 (KO-2) expression (Fig. 9C). Interestingly, IRS-3 expression in WT cells, but not in KO cells, was associated with an increase in egr-1 gene expression (220% of that in WT cells), while IRS-4 expression led to an impaired response in both WT and KO cells. The level of egr-1 gene expression roughly paralleled the level of MAP kinase phosphorylation in each cell line.
FIG. 9.
Effect of IRS protein overexpression on IGF-1-induced immediate-early gene expression. (A) After overnight serum starvation, cells were stimulated with 10 nM IGF-1 for 30 min at 37°C. RNA samples extracted from serum-starved or IGF-1-stimulated cells were separated on formaldehyde-containing agarose gels and transferred to nylon membranes. Equal loading and integrity of the RNA were confirmed by ethidium bromide staining of agarose gels (upper panel). Blots were hybridized with a specific probe for the egr-1 gene as described in the text and visualized by autoradiography (lower panel). (B and C) The blots were quantified on a Molecular Dynamics PhosphorImager. Results are expressed as percentages of IGF-1-induced WT cells (B) and the increases from basal values by IGF-1-induction from the basal condition (C). Data represent the mean and SEM of three independent experiments.
IGF-1-mediated mitogenesis in IRS-3- or IRS-4-expressing cells.
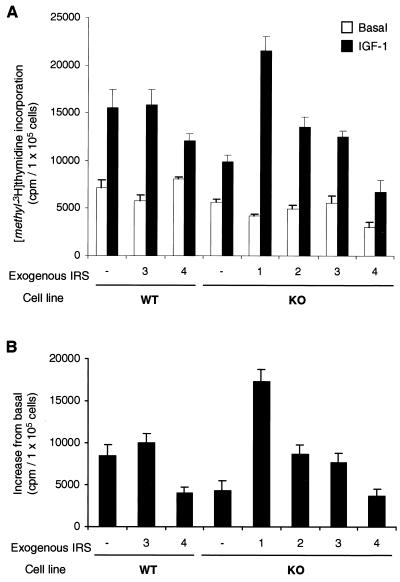
As previously noted (9), mitogenesis, as measured by [methyl-3H]thymidine incorporation into DNA, was impaired in IRS-1 KO cells compared to WT cells (Fig. 10). In the present study, there was a ∼55% decrease in IGF-1-induced DNA synthesis in the KO cells (Fig. 10B). Overexpression of IRS-1 reversed the decrease in [methyl-3H]thymidine incorporation to supranormal level (200%), while overexpression of IRS-2 brought this response back to a level similar to that in WT cells. IRS-3 overexpression had no effect on IGF-1-stimulated DNA synthesis in WT-3 cells and produced an effect similar to IRS-2 in KO-3 cells. By contrast, IRS-4 expression caused a ∼55% decrease in IGF-1-induced DNA synthesis in WT-4 cells. The reduced DNA synthesis in IRS-1 KO cells was not improved by overexpression of IRS-4.
FIG. 10.
Effect of IRS protein overexpression on IGF-1-mediated mitogenesis in embryonic fibroblast cell lines. Quiescent cells were cultured in the presence or absence of 10 nM IGF-1 for 15 h and pulsed with 1 μCi of [methyl-3H]thymidine per 2 × 105 cells for 1 h at 37°C. Radioactivity in trichloroacetate-precipitable DNA was measured with a scintillation counter. Results represent the mean and SEM of four independent experiments and are expressed as net cpm per 105 cells in the presence or absence IGF-1 (A) or the increase due to the presence of IGF-1 in each cell line (B).
DISCUSSION
A variety of studies have demonstrated the importance of the IGF-1 receptor-mediated signals in embryonic growth and development (6, 10, 34). The IRS proteins are major substrates of both insulin receptor and IGF-1 receptor tyrosine kinases. The four known IRS proteins share similar overall architecture, including conservation of numerous tyrosine phosphorylation sites that bind SH2 domain-containing proteins (14, 24, 40, 41). Disruption of the IRS-1 gene causes severe intrauterine growth retardation; however, mice deficient for other IRS proteins show little or no impairment in growth (15, 25, 45), suggesting that the four IRS proteins play different roles in growth and development.
Using embryonic fibroblast cell lines created from IRS-1 KO and normal WT mouse embryos, we previously demonstrated some aspects of differential IGF-1 signaling that occurs via IRS-1 and IRS-2 (9). In the present study, we have used these cells to evaluate the impact of IRS-3 and IRS-4 on IGF-1 signaling and the interaction of these substrates on the IRS-1- and IRS-2-mediated signaling pathway. We found that both IRS-3 and IRS-4 are phosphorylated upon IGF-1 stimulation and that both bind the p85 subunit of PI 3-kinase and Grb-2. Interestingly, however, both IRS-3 and IRS-4 have some inhibitory effects on IRS-1- and IRS-2-mediated IGF-1 signaling. These effects occur at three steps in the signaling pathway. Thus, both IRS-3 and IRS-4 expression cause a decrease in IRS-2 protein production. IRS-3 expression also decreases IGF-1-stimulated tyrosine phosphorylation of IRS-1, while IRS-4 expression decreases the phosphorylation of both IRS-1 and IRS-2. Finally, both IRS-3 and IRS-4 can decrease the association between p85 and IRS-1 or IRS-2.
The ability of either IRS-3 or IRS-4 expression to cause a decrease in IRS-2 protein occurs without any change in the amount of IRS-1 or other signaling components measured in WT and KO cell lines. This decrease of IRS-2 protein could be due to the reduction in the protein synthesis, since IRS-3- and IRS-4-expressing cells showed a decreased IRS-2 mRNA level under serum-starved condition, although the decrease found in WT-4 cells is not statistically significant. This effect may also be cell specific, since IRS-3-deficient mice show no significant change in IRS-2 protein content in adipose tissues (25). Despite the finding that IRS-1-deficient fibroblasts have a slight increase (∼20%) in IRS-2 mRNA and protein content, IRS-1 overexpression does not cause a decrease in endogenous IRS-2 protein, indicating that this effect of IRS-3 and IRS-4 is relatively specific.
Both IRS-3 and IRS-4 have been clearly tyrosine-phosphorylated by IGF-1 stimulation, confirming that both of them are potential IGF-1 substrates. However, it may be difficult to compare directly the tyrosine phosphorylation levels of IRS-3 with those of other IRS proteins since IRS-3 has fewer tyrosine phosphorylation motifs and this protein gives a wide band on the gel. The different IRS proteins may transfer to the blotting membrane with different efficiencies and may react with their respective antibodies with different affinities.
All the IRS-3- and IRS-4-overexpressing cells show a drastic decrease in tyrosine-phosphorylated IRS-1 and IRS-2. This is not simply due to the reduced IRS-2 protein content, since tyrosine phosphorylation is reduced even when normalized for protein content. This suggests a competition by IRS-3 and IRS-4 for either the IGF-1 receptor kinase domain itself or the NPXY motif in the juxtramembrane domain of the IGF-1 receptor that binds the IRS substrates (46). Indeed, the affinity of each IRS protein for the insulin or IGF-1 receptor may not be same. In a previous study, we found that IRS-3 binds more strongly than IRS-1 to immobilized peptides containing a phosphorylated NPXY motif (37). A study using the yeast two-hybrid system also revealed that interaction of IRS-3 with the insulin receptor is comparable to that of IRS-2 and stronger than that of IRS-1 (46). Therefore, it is possible that IRS-3 and IRS-4 inhibit the phosphorylation of other IRS proteins by competing for binding to the NPXY motif in the IGF-1 receptor. To confirm this hypothesis, we have examined the IRS protein–IGF-1 receptor binding by coimmunoprecipitation analysis of the IRS protein–IGF-1 receptor complexes. However we could not detect the receptor in the precipitates with IRS-1- or IRS-2-specific antibody, even in WT cells, suggesting that the IGF-1 receptor–IRS protein complexes are not abundant or sufficiently stable to be detected by coimmunoprecipitation (data not shown). The subcellular distribution has also been shown to be different for the IRS proteins. IRS-1 and IRS-2 are located mainly in the low-density microsome fraction while IRS-3 is associated more with the plasma membrane fraction in rat adipocytes (3). This different localization in the cell may also contribute to the efficiency of interaction between the IGF-1 receptor and the various IRS proteins.
Numerous SH2 domain-containing proteins bind to the tyrosine-phosphorylated forms of the IRS proteins, including PI 3-kinase, Grb-2, and SHP-2 (14, 30, 39–41). PI 3-kinase is a critical link to the signaling of insulin and IGF-1 and can bind to all four IRS proteins. This kinase is a heterodimeric enzyme composed of a catalytic subunit (p110) associated with one of several SH2 domain-containing regulatory subunits (p50α, AS53/p55α, p55PIK, p85α, and p85β) that can bind to tyrosine-phosphorylated YXXM motifs on signaling proteins such as the IRS proteins. Immunoprecipitation analysis of our cell lines confirms that p85 can bind to all four IRS proteins. In this study, we also find that p85 binding to IRS-1 or IRS-2 is impaired by IRS-3 or IRS-4 overexpression. The decrease of IRS-1 and IRS-2 tyrosine phosphorylation, especially of the tyrosine residues in YXXM motifs, probably contributes to the impaired p85 binding. It has also been shown that during insulin stimulation, p85 associated with IRS-3 more rapidly than with IRS-1 and IRS-2 in rat adipocytes (37) and with IRS-3 more strongly than with IRS-2 in adipocytes from IRS-1-deficient mice (20, 37). Thus, in cells that express IRS-3, IRS-3 may be the dominant protein in binding with p85, by competing with IRS-2 and IRS-1.
Despite these various interactions, reconstitution of IRS-1 KO cells by IRS-1 is more effective than by IRS-2 in recovering the impaired mitogenic responses caused by IRS-1 disruption. In previous studies, we found that the reduced DNA synthesis in IRS-1 KO cells was recovered by IRS-1 reconstitution but not by overexpression of IRS-2 (9). However, in the present study, IRS-2-overexpressing KO cells (KO-2 cells) showed an elevation in IGF-1-induced DNA synthesis to the WT level. The difference between these studies is likely to be due to the different levels of expression of IRS-2 (about fivefold above the endogenous level in the present study and only twofold above the endogenous level in the previous study). Other reports have shown that IRS-2 can support insulin-induced mitogenic responses, although the response mediated by IRS-2 is also weaker than that by IRS-1 in 32D cells (43). Therefore, it is likely that larger amounts of IRS-2 can mediate the mitogenic signal in KO-2 cells. Indeed, in the present study, IRS-1 overexpression reconstituted the mitogenic signal to a supranormal level, probably due to supranormal expression of IRS-1 protein.
IRS-3 seems to be able to mediate the mitogenic signal. Thus, both of the IRS-3-expressing cell lines exhibit normal or increased IGF-1-induced DNA synthesis despite the drastic impairment of IRS-1- and IRS-2-mediated signaling. In contrast, IRS-4 expression in WT cells causes a significant reduction in IGF-1-induced DNA synthesis. IRS-4 is a mediator of the mitogenic signal in 32D cells (14, 43). However, there is some controversy whether IRS-4 can mediate mitogenic signals as well as IRS-1 can (14, 43). In this study, IRS-4 overexpression in WT cells caused a significant decrease in IGF-1-induced mitogenesis. This decrease is probably due to the inhibition of the IGF-1 signal through IRS-1, which is thought to be a most effective mediator for the mitogenic signal. IRS-4 expression in KO cells showed a similar mitogenic response to that of IRS-1 in KO cells (Fig. 10). The IGF-1 signal mediated by IRS-2 was dramatically inhibited in KO-4 cells (Fig. 6), suggesting that the IGF-1-induced mitogenic response in KO-4 cells might be mediated by IRS-4 protein, at least in part. The inability to reconstitute the mitogenic signal in KO-4 cells may be because the expression level of IRS-4 in KO cells was lower than that of other IRS proteins during reconstitution.
Induction of expression of immediate-early genes, such as egr-1, constitutes one of the first steps in the expression of growth-regulatory proteins (8). Although our previous report shows that egr-1 gene expression is impaired by IRS-1 deficiency (9), the KO cells we used in the present study (which differ from those used in the previous study) exhibit somewhat elevated basal and IGF-1-stimulated egr-1 expression compared to WT cells. The reason for this difference is unknown; however, it has been shown that the egr-1 gene induction can be mediated by signals from the MAP kinase cascades, including ERK1/2, JNK, and p38 MAP kinase (7, 17, 29). Although egr-1 gene expression is usually rapid and transient (7, 29), one recent study has shown a sustained level of expression of egr-1 gene in atherosclerotic vascular lesions (26), suggesting that the basal expression of this gene can be continuously elevated under some conditions. In KO cells, the elevation in egr-1 gene expression tends to correlate with elevated basal p44/42 MAP kinase phosphorylation. It is also possible that the elevated basal expression in KO cells is due to the lack of IRS-1 protein. Indeed, reconstitution of IRS-1 in KO cells may have caused the decrease in basal egr-1 gene expression, as well as p44/42 MAP kinase phosphorylation, to the levels in WT cells, while overexpression of IRS-4 in WT cells caused an elevation in both basal egr-1 gene and basal p44/42 MAP kinase phosphorylation. Whatever the cause, the effects of IRS-4 expression on egr-1 gene expression are mainly on its basal level.
More importantly, IRS-3 expression causes an increase in IGF-1-stimulated MAP kinase phosphorylation and egr-1 gene induction in WT cells but not in IRS-1-deficient cells. Activation of MAP kinase may depend on the interaction of Grb-2 with the guanine nucleotide exchange factor SOS, thus activating the small GTP binding protein Ras (11) and the MAP kinase cascade (32). Rat IRS-3, which was used in this study, has been shown to bind with Grb-2 in rat adipose tissue (30); however, the association of IRS-3 with Grb-2 does not explain why only WT-3 cells show an elevation in MAP kinase phosphorylation, since KO-3 cells have threefold more IRS-3 proteins than do WT-3 cells. IRS-3 has been shown to bind the PY phosphatase SHP-2 with a greater affinity than the other IRS proteins do (25, 30). Since this phosphatase is necessary for full activation of the MAP kinase cascade in response to insulin and IGF-1 (27, 35, 47), it is possible that the association of IRS-3 with SHP-2 contributes to the activation of MAP kinase observed in these experiments.
In conclusion, IRS-3 and IRS-4 are phosphorylated in response to IGF-1 and can bind to p85 and activate PI 3-kinase in embryonic cell lines. IRS-3 expression causes a significant elevation of IGF-1-stimulated MAP kinase phosphorylation and immediate-early gene induction in the presence of IRS-1, suggesting that IRS-3 can act in cooperation with IRS-1 in mediating gene expression in this cell line. Moreover, IRS-3 or IRS-4 expression causes an impairment of IRS-1- and IRS-2-mediated signaling in at least three steps in the signaling pathway: (i) decreasing the IRS-2 mRNA and protein amount, (ii) decreasing the IGF-1-stimulated tyrosine phosphorylation of IRS-1 and IRS-2, and (iii) decreasing the p85 binding to IRS-1 and IRS-2. These results indicate that IRS-3 and IRS-4 may act as negative regulators of some aspects of the signaling pathway by suppressing the function of other IRS proteins and that the biological responses to IGF-1 in individual cells or tissues may be regulated by the combination of all four IRS proteins.
ACKNOWLEDGMENTS
We are grateful to M. F. White and G. E. Lienhard for providing regents used in this study. We thank T.-L. Azar and J. Konigsberg for excellent secretarial assistance.
This work was supported by NIH grants DK 33201 (C.R.K.) and DK 55545 (C.R.K.), as well as Joslin DERC grant DK 36836. C.R.K. is the recipient of an ADA mentor-based grant.
REFERENCES
- 1.Alessi D R, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings B A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 2.Alessi D R, Cohen P. Mechanism of activation and function of protein kinase B. Curr Opin Genet Dev. 1998;8:55–62. doi: 10.1016/s0959-437x(98)80062-2. [DOI] [PubMed] [Google Scholar]
- 3.Anai M, Ono H, Funaki M, Fukushima Y, Inukai K, Ogihara T, Sakoda H, Onishi Y, Yazaki Y, Kikuchi M, Oka Y, Asano T. Different subcellular distribution and regulation of expression of insulin receptor substrate (IRS)-3 from those of IRS-1 and IRS-2. J Biol Chem. 1998;273:29686–29692. doi: 10.1074/jbc.273.45.29686. [DOI] [PubMed] [Google Scholar]
- 4.Araki E, Lipes M A, Patti M E, Brüning J C, Haag III B L, Johnson R S, Kahn C R. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature. 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 5.Backer J M, Myers M G, Jr, Shoelson S E, Chin D J, Sun X J, Miralpeix M, Hu P, Margolis B, Skolnik E Y, Schlessinger J, et al. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992;11:3469–3479. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker J, Liu J P, Robertson E J, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- 7.Barroso I, Santisteban P. Insulin-induced early growth response gene (Egr-1) mediates a short term repression of rat malic enzyme gene transcription. J Biol Chem. 1999;274:17997–18004. doi: 10.1074/jbc.274.25.17997. [DOI] [PubMed] [Google Scholar]
- 8.Biesiada E, Razandi M, Levin E R. Egr-1 activates basic fibroblast growth factor transcription. Mechanistic implications for astrocyte proliferation. J Biol Chem. 1996;271:18576–18581. doi: 10.1074/jbc.271.31.18576. [DOI] [PubMed] [Google Scholar]
- 9.Brüning J C, Winnay J, Cheatham B, Kahn C R. Differential signaling by insulin receptor substrate 1 (IRS-1) and IRS-2 in IRS-1-deficient cells. Mol Cell Biol. 1997;17:1513–1521. doi: 10.1128/mcb.17.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang L M, Myers M G, Jr, Backer J M, Shoelson S E, White M F, Birnbaum M J, Kahn C R. Insulin-stimulated oocyte maturation requires insulin receptor substrate 1 and interaction with the SH2 domains of phosphatidylinositol 3-kinase. Mol Cell Biol. 1993;13:6653–6660. doi: 10.1128/mcb.13.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper J A, Kashishian A S. In vivo binding properties of SH2 domains from STPase-activating protein and phosphatidylinositol 3-kinase. Mol Cell Biol. 1993;13:1737–1745. doi: 10.1128/mcb.13.3.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Fea K, Roth R A. Protein kinase C modulation of insulin receptor substrate-1 tyrosine phosphorylation requires serine 612. Biochemistry. 1997;36:12939–12947. doi: 10.1021/bi971157f. [DOI] [PubMed] [Google Scholar]
- 13.Fantin V R, Lavan B E, Wang Q, Jenkins N A, Gilbert D J, Copeland N G, Keller S R, Lienhard G E. Cloning, tissue expression, and chromosomal location of the mouse insulin receptor substrate 4 gene. Endocrinology. 1999;140:1329–1337. doi: 10.1210/endo.140.3.6578. [DOI] [PubMed] [Google Scholar]
- 14.Fantin V R, Sparling J D, Slot J W, Keller S R, Lienhard G E, Lavan B E. Characterization of insulin receptor substrate 4 in human embryonic kidney 293 cells. J Biol Chem. 1998;273:10726–10732. doi: 10.1074/jbc.273.17.10726. [DOI] [PubMed] [Google Scholar]
- 15.Fantin V R, Wang G E, Lienhard G E, Keller S R. Mice lacking insulin receptor substrate 4 exhibit mild defects in growth, reproduction, and homostasis. Am J Physiol. 2000;278:E127–E133. doi: 10.1152/ajpendo.2000.278.1.E127. [DOI] [PubMed] [Google Scholar]
- 16.Giorgetti S, Pelicci P G, Pelicci G, Van Obberghen E. Involvement of Src-homology/collagen (SHC) proteins in signaling through the insulin receptor and the insulin-like-growth-factor-I receptor. Eur J Biochem. 1994;223:195–202. doi: 10.1111/j.1432-1033.1994.tb18983.x. [DOI] [PubMed] [Google Scholar]
- 17.Hodge C, Liao J, Stofega M, Guan K, Carter-Su C, Schwartz J. Growth hormone stimulates phosphorylation and activation of elk-1 and expression of c-fos, egr-1 and junB through activation of extracellular signal-regulated kinases 1 and 2. J Biol Chem. 1998;273:31327–31336. doi: 10.1074/jbc.273.47.31327. [DOI] [PubMed] [Google Scholar]
- 18.Holgado-Madruga M, Emlet D R, Moscatello D K, Godwin A K, Wong A J. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature. 1996;379:560–563. doi: 10.1038/379560a0. [DOI] [PubMed] [Google Scholar]
- 19.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 20.Kaburagi Y, Satoh S, Tamemoto H, Yamamoto-Honda R, Tobe K, Veki K, Yamauchi T, Kono-Sugita E, Sekihara H, Aizawa S, Cushman S W, Akanuma Y, Yazaki Y, Kadowaki T. Role of insulin receptor substrate-1 and pp60 in the regulation of insulin-induced glucose transport and GLUT4 translocation in primary adipocytes. J Biol Chem. 1997;272:25839–25844. doi: 10.1074/jbc.272.41.25839. [DOI] [PubMed] [Google Scholar]
- 21.Kahn C R, White M F, Shoelson S E, Backer J M, Araki E, Cheatham B, Csermely P, Folli F, Goldstein B J, Huertas P, Rothenberg P L, Saad M J A, Siddle K, Sun X J, Wilden P A, Yamada K, Kahn S A. The insulin receptor and its substrate: molecular determinants of early events in insulin action. Recent Prog Horm Res. 1993;48:291–339. doi: 10.1016/b978-0-12-571148-7.50015-4. [DOI] [PubMed] [Google Scholar]
- 22.Kasuga M, Karlsson F A, Kahn C R. Insulin stimulates the phosphorylation of the 95,000-dalton subunit of its own receptor. Science. 1982;215:185–187. doi: 10.1126/science.7031900. [DOI] [PubMed] [Google Scholar]
- 23.Lavan B E, Fantin V R, Chang E T, Lane W S, Keller S R, Lienhard G E. A novel 160 kDa phosphotyrosine protein in insulin-treated embryonic kidney cells is a new member of the insulin receptor substrate family. J Biol Chem. 1997;272:21403–21407. doi: 10.1074/jbc.272.34.21403. [DOI] [PubMed] [Google Scholar]
- 24.Lavan B E, Lane W S, Lienhard G E. The 60-kDa phosphotyrosine protein in insulin-treated adipocytes is a new member of the insulin receptor substrate family. J Biol Chem. 1997;272:11439–11443. doi: 10.1074/jbc.272.17.11439. [DOI] [PubMed] [Google Scholar]
- 25.Liu S C, Wang Q, Lienhard G E, Keller S R. Insulin receptor substrate 3 is not essential for growth or glucose homeostasis. J Biol Chem. 1999;274:18093–18099. doi: 10.1074/jbc.274.25.18093. [DOI] [PubMed] [Google Scholar]
- 26.McCaffrey T A, Fu C, Du B, Eksinar S, Kent K C, Bush H, Jr, Kreiger K, Rosengart T, Cybulsky M I, Silverman E S, Collins T. High-level expression of Egr-1 and Egr-1-inducible genes in mouse and human atherosclerosis. J Clin Investig. 2000;105:653–662. doi: 10.1172/JCI8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noguchi T, Matozaki T, Horita K, Fujioka Y, Kasuga M. Role of SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in insulin-stimulated ras activation. Mol Cell Biol. 1994;14:6674–6682. doi: 10.1128/mcb.14.10.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pronk G J, McGlade J, Pelicci G, Pawson T, Bos J L. Insulin-induced phosphorylation of the 46- and 52-kDa Shc proteins. J Biol Chem. 1993;268:5748–5753. [PubMed] [Google Scholar]
- 29.Rolli M, Kotlyarov A, Sakamoto K M, Gaestel M, Neininger A. Stress-induced stimulation of early growth response gene-1 by p38/stress-activated protein kinase 2 is mediated by a cAMP-responsive promoter element in a MAPKAP kinase 2-independent manner. J Biol Chem. 1999;274:19559–19564. doi: 10.1074/jbc.274.28.19559. [DOI] [PubMed] [Google Scholar]
- 30.Ross S A, Lienhard G E, Lavan B E. Association of insulin receptor substrate 3 with SH2 domain-containing proteins in rat adipocytes. Biochem Biophys Res Commun. 1998;247:487–492. doi: 10.1006/bbrc.1998.8821. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 7.3–7.87. [Google Scholar]
- 32.Schaeffer H J, Weber M J. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sciacchitano S, Taylor S I. Cloning, tissue expression, and chromosomal localization of the mouse IRS-3 gene. Endocrinology. 1997;138:4931–4940. doi: 10.1210/endo.138.11.5518. [DOI] [PubMed] [Google Scholar]
- 34.Sell C, Dumenil G, Deveaud C, Miura M, Coppola D, DeAngelis T, Rubin R, Efstratiadis A, Baserga R. Effect of a null mutation of the insulin-like growth factor I receptor gene on growth and transformation of mouse embryo fibroblasts. Mol Cell Biol. 1994;14:3604–3612. doi: 10.1128/mcb.14.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Z Q, Lu W, Feng G S. The shp-2 tyrosine phosphatase has opposite effects in mediating the activation of extracellular signal-regulated and c-Jun NH2-terminal mitogen-activated protein kinases. J Biol Chem. 1998;273:4904–4908. doi: 10.1074/jbc.273.9.4904. [DOI] [PubMed] [Google Scholar]
- 36.Skolnik E Y, Lee C H, Batzer A G, Vicentini L M, Zhou M, Daly R J, Myers M G, Jr, Backer J M, Ullrich A, White M F, Schlessinger J. The SH2/SH3 domain-containing protein GRB2 interacts with tyrosine-phosphorylated IRS-1 and Shc: implications for insulin control of ras signalling. EMBO J. 1993;12:1929–1936. doi: 10.1002/j.1460-2075.1993.tb05842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith-Hall J, Pons S, Patti M E, Burks D J, Yenush L, Sun X J, Kahn C R, White M F. The 60-kDa insulin receptor substrate functions like an IRS-protein (pp60IRS3) in adipose cells. Biochemistry. 1997;36:8304–8310. doi: 10.1021/bi9630974. [DOI] [PubMed] [Google Scholar]
- 38.Sun X J, Crimmins D L, Myers M G, Jr, Miralpeix M, White M F. Pleiotropic insulin signals are engaged by multisite phosphorylation of IRS-1. Mol Cell Biol. 1993;13:7418–7428. doi: 10.1128/mcb.13.12.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun X J, Pons S, Wang L M, Zhang Y, Yenush L, Burks D, Myers M G, Jr, Glasheen E, Copeland N G, Jenkins N A, Pierce J H, White M F. The IRS-2 gene on murine chromosome 8 encodes a unique signaling adapter for insulin and cytokine action. Mol Endocrinol. 1997;11:251–262. doi: 10.1210/mend.11.2.9885. [DOI] [PubMed] [Google Scholar]
- 40.Sun X J, Rothenberg P L, Kahn C R, Backer J M, Araki E, Wilden P A, Cahill D A, Goldstein B J, White M F. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- 41.Sun X J, Wang L M, Zhang Y, Yenush L, Myers M G, Jr, Glasheen E M, Lane W S, Pierce J H, White M F. Role of IRS-2 in insulin and cytokine signalling. Nature. 1995;377:173–177. doi: 10.1038/377173a0. [DOI] [PubMed] [Google Scholar]
- 42.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S, Sekihara H, Yoshioka S, Horikoshi H, Furuta Y, Ikawa Y, Kasuga M, Yazaki Y, Aizawa S. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 43.Uchida T, Myers M G, Jr, White M F. IRS-4 mediates protein kinase B signaling during insulin stimulation without promoting antiapoptosis. Mol Cell Biol. 2000;20:126–138. doi: 10.1128/mcb.20.1.126-138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ullrich A, Gray A, Tam A W, Yang Feng T, Tsubokawa M, Collins C, Henzel W J, Le Bon T, Kathuria S, Chen E, Jacobs S, Francke U, Ramachandran J, Fujita-Yamaguchi Y. Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 1986;5:2503–2512. doi: 10.1002/j.1460-2075.1986.tb04528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Withers D J, Gutierrez J S, Towery H, Burks D J, Ren J M, Previs S, Zhang Y, Bernal D, Pons S, Shulman G I, Bonner-Weir S, White M F. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 46.Xu P, Jacobs A R, Taylor S I. Interaction of insulin receptor substrate 3 with insulin receptor, insulin receptor-related receptor, insulin-like growth factor-1 receptor, and downstream signaling proteins. J Biol Chem. 1999;274:15262–15270. doi: 10.1074/jbc.274.21.15262. [DOI] [PubMed] [Google Scholar]
- 47.Yamauchi K, Milarski K L, Saltiel A R, Pessin J E. Protein-tyrosine-phosphatase SHPTP2 is a required positive effector for insulin downstream signaling. Proc Natl Acad Sci USA. 1995;92:664–668. doi: 10.1073/pnas.92.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou L, Chen H, Xu P, Cong L N, Sciacchitano S, Li Y, Graham D, Jacobs A R, Taylor S I, Quon M J. Action of insulin receptor substrate-3 (IRS-3) and IRS-4 to stimulate translocation of GLUT4 in rat adipose cells. Mol Endocrinol. 1999;13:505–514. doi: 10.1210/mend.13.3.0242. [DOI] [PubMed] [Google Scholar]