Abstract
Simple Summary
Non-coding RNAs have been recently attained attention because of their contribution in the pathogenesis of brain tumors. These transcripts have been shown to be dysregulated in pituitary gland tumors as well as meningiomas. In these two types of brain tumors, dysregulation of non-coding RNAs has been associated with some clinical features and response to therapeutic options. Different types of non-coding RNAs have been shown to interact with each other to promote progression of brain tumors. Further research is needed to find the possible application of non-coding RNAs as biomarkers for pituitary gland tumors as well as meningiomas, particularly in patients’ follow-up.
Abstract
Long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs) are non-coding transcripts which are involved in the pathogenesis of pituitary gland tumors. LncRNAs that participate in the pathogenesis of pituitary gland tumors mainly serve as sponges for miRNAs. CLRN1-AS1/miR-217, XIST/miR-424-5p, H19/miR-93a, LINC00473/miR-502-3p, SNHG7/miR-449a, MEG8/miR-454-3p, MEG3/miR-23b-3p, MEG3/miR-376B-3P, SNHG6/miR-944, PCAT6/miR-139-3p, lncRNA-m433s1/miR-433, TUG1/miR-187-3p, SNHG1/miR-187-3p, SNHG1/miR-302, SNHG1/miR-372, SNHG1/miR-373, and SNHG1/miR-520 are identified lncRNA/miRNA pairs that are involved in this process. Hsa_circ_0001368 and circOMA1 are two examples of circRNAs that contribute to the pathogenesis of pituitary gland tumors. Meanwhile, SNHG1, LINC00702, LINC00460, and MEG3 have been found to partake in the pathogenesis of meningioma. In the current review, we describe the role of non-coding RNAs in two types of brain tumors, i.e., pituitary tumors and meningioma.
Keywords: lncRNA, miRNA, circRNA, pituitary gland cancer, meningioma
1. Introduction
Non-coding RNAs comprise heterogeneous types of transcripts in terms of functions, size, evolutionary conservation, and expression level. The integrated application of large-scale sequencing methods and bioinformatics tools has facilitated the annotation of non-coding RNAs; thus, they are not considered as either junk portions of the genomes or byproducts of massive transcription. Soon after the completion of the Human Genome Project, several non-coding RNAs were detected in mammals [1]. With the advent of high-throughput sequencing strategies, the expression profile of non-coding RNAs has been more precisely identified [2]. The ENCODE project has stated that approximately 80% of the human genome is transcribed into non-coding RNAs [3]. These transcripts take part in manifold biological processes, controlling physiological and developmental events. Most notably, they have been recognized as tumor suppressors and oncogenes in numerous types of cancers [4]. Three classes of non-coding RNAs, i.e., long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and, more recently, circular RNAs (circRNAs), have attained much attention in this field. LncRNAs are transcripts with more than 200 nucleotides and several shared features with mRNAs, yet lack detectable open reading frames [5]. They regulate the expression of genes by serving as signals, decoys, scaffolds, guide RNAs, or enhancer RNAs [5]. On the other hand, miRNAs are short-sized RNAs that mainly regulate the expression of genes at the post-transcriptional level through binding with 3′ UTR of transcripts and induction of gene silencing or mRNA degradation [6]. CircRNAs are produced mostly through the back-splicing of exons in precursor mRNAs. These covalently closed RNA transcripts function as miRNA sponges, serve as scaffolds for proteins, or modulate transcriptional and splicing events. Occasionally, circRNAs are used as templates for the synthesis of polypeptides. Abnormal expression of circRNAs has been reported in diverse malignancies. In addition to sustaining cell growth and proliferation, they promote tumor invasiveness and facilitate bypass of cell senescence and death [7]. In the current review, we describe the role of these classes of non-coding RNAs in two types of brain tumors, i.e., pituitary tumors and meningioma.
2. Pituitary Gland Tumors
Pituitary adenomas comprise a heterogeneous group of tumors historically classified based on their size into micro- and macroadenomas considering a threshold of 1 cm. More recently, immunohistochemical analyses and electron microscopy have been used to classify these tumors further [8]. These tumors can also be categorized as functional or nonfunctional, based on their hormonal activity. They might be identified either as an incidental finding in radiology or being associated with symptoms, particularly visual disturbance, based on their size, growth rate, or hormone secretion [9]. The overall prevalence of pituitary adenomas is estimated to be around 16% [10]. Recent studies have shown dysregulation of lncRNAs, miRNAs, and circRNAs in pituitary gland tumors. For instance, RPSAP52, an antisense lncRNA from the HMGA2 locus, has been found to be overexpressed in both gonadotroph- and prolactin-producing adenomas of this gland, where its expression has been correlated with the expression of HMGA2. Contrariwise, the expression of RPSAP52 has been variable among somatotroph adenomas. RPSAP52 has been shown to act as a molecular sponge for miR-15a, miR-15b, and miR-16 to increase the expression of HMGA2. In addition, RPSAP52 could enhance the expression of HMGA1. RPSAP52 has been shown to enhance cell growth through increasing G1/S transition [11]. XIST is another upregulated lncRNA in pituitary tumors whose expression is increased parallel with upregulation of bFGF and downregulation of miR-424-5p. Functionally, XIST acts as a sponge for miR-424-5p to increase bFGF levels. Silencing of XIST or bFGF or overexpression of miR-424-5p has been shown to inhibit proliferation, migration, and invasiveness of pituitary neuroendocrine tumor cells [12].
2.1. LncRNAs
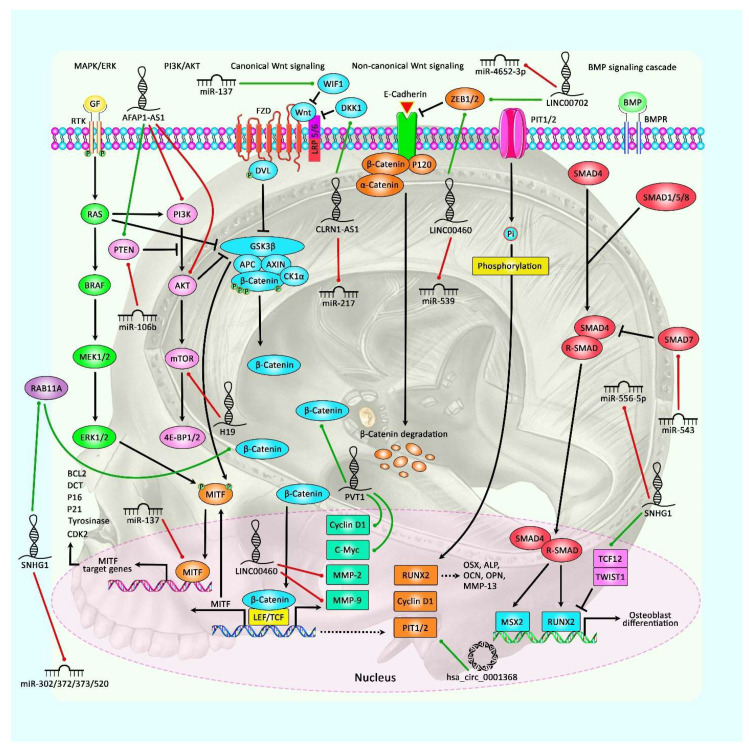
Several lncRNAs have been found to act as suppressors of tumorigenesis processes in pituitary adenomas. Among these, lncRNAs is clarin 1 antisense RNA 1 (CLRN1-AS1), which is underexpressed in prolactinomas. This lncRNA has a role in the inactivation of Wnt/β-catenin signaling. Being mainly located in the cytoplasm, CLRN1-AS1 has been shown to suppress cell proliferation, promote apoptosis, and inhibit autophagy. In addition, CLRN1-AS1 sponges miR-217 to increase the expression of the dickkopf WNT signaling pathway inhibitor 1 (DKK1). The FOXP1 transcription factor could suppress the expression of CLRN1-AS1 [13]. Additionally, H19 has been found to be commonly downregulated in primary pituitary adenoma samples. Downregulation of H19 has been correlated with tumor progression. Overexpression of H19 could suppress the proliferation of pituitary adenoma cells in vitro and their growth in vivo. Functionally, H19 could regulate tumorigenesis via suppressing the activity of mTORC1 but not mTORC2. In fact, H19 blocks mTORC1-mediated phosphorylation of 4E-BP1 without changing the activity of S6K1. H19 also interacts with 4E-BP1 and inhibits 4E-BP1 binding to Raptor. Notably, H19 could suppress pituitary tumors more effectively than cabergoline [14]. Moreover, H19 has been found to have a synergistic effect with dopamine agonists in prolactinoma. From a mechanistical point of view, H19 increases the expression of ATG7 through sequestering miR-93a [15]. Table 1 shows the list of lncRNAs participating in the pathogenesis of pituitary gland tumors. Figure 1 represents the role of several ncRNAs in regulating the MAPK/ERK, PI3K/AKT, Wnt/β-Catenin, and BMP cascades in pituitary gland tumors and meningiomas.
Table 1.
LncRNAs and pituitary gland tumors (ANTs: adjacent non-tumor samples).
| lncRNA | Accession Number/Location | Expression Pattern | Clinical Samples/Animal Model | Assessed Cell Lines | Targets/Regulators | Signaling Pathways | Description | Reference |
|---|---|---|---|---|---|---|---|---|
| RPSAP52 | ENSG00000241749/12q14.3 | ↑ | 12 gonadotroph tumors and 3 normal samples | HT-29, HCT-116, BCPAP, GH3, ATt-20 | HMGA2↑, HMGA1↑, miR-15a↓, miR-15b↓, miR-16↓ | - | Microarray analysis and RT-PCR confirmed RPSAP52 upregulation, which enhances cell cycle progression and cell growth. | [11] |
| CLRN1-AS1 | ENSG00000239265/3q25.1 | ↓ | 42 pairs of pituitary prolactinoma and ANTs, male BALB/C athymic nude mice | 293T | miR-217↑, DKK1↓/FOXP1↑ | Wnt/β-catenin | This lncRNA has the potential to limit cell proliferation and autophagy but enhance apoptosis rate. Additionally, FOXP1 diminishes CLRN1-AS1 transcription. CLRN1-AS1 downregulation was assessed by qRT-PCR. | [13] |
| XIST | ENSG00000229807/Xq13.2 | ↑ | 86 pituitary neuroendocrine tumors and 23 normal tissues | - | miR-424-5p↓, bFGF↑ | - | Xist expression was evaluated by RT-qPCR. XIST downregulation reduces migration, invasion, cell cycle progression, proliferation capacity, and increased apoptosis rate. | [12] |
| H19 | ENSG00000130600/11p15.5 | ↓ | 9 tumor and 9 normal tissues, athymic female nude mice | GH3, HEK293T | 4E-BP1 | - | H19 upregulation, evaluated by qRT-PCR reduces tumor progression and cell proliferation by inhibiting the mTORC1 normal function that mediates 4E-BP1 phosphorylation. | [14] |
| ↓ | 2 resistant and 3 sensitive prolactinoma tissues, female BALB/c athymic nude mice |
GH3 | miR-93a↓, ATG7↑ | - | By sequestering miR-93a, H19 increases ATG7 expression, influencing resistance to dopamine agonists. H19 expression was evaluated by qRT-PCR. | [15] | ||
| LINC00473 | ENSG00000112541/6q27 | ↑ | Invasive and non-invasive pituitary adenoma, each with 20 cases, athymic female nude mice | AtT-20, GT1-1 | miR-502-3p↓, KMT5A↑, cyclin D1↑, CDK2↑ | - | LINC00473 increases cell cycle progression, proliferation, and tumor growth. RNA-sequencing and qRT-PCR were used to assess LINC00473 expression. | [16] |
| SNHG7 | ENSG00000233016/9q34.3 | ↑ | 30 pituitary tumor and ANTs, nude mice | GH1, RC-4B/C, GH3, MMQ | miR-449a↓, Ki67↑ | - | SNHG7 upregulation, evaluated by RT-PCR, results in enhanced cell migration, invasion, tumor growth, and reduced apoptosis. | [17] |
| MEG8 | ENSG00000225746/14q32.2-q32.31 | ↑ | 20 bone-invasive and 20 non-invasive pituitry adenoma, male BALB/c nude mice | 293T, RAW264.7 | miR-454-3p↓, TNF-α↑ | - | RT-PCR confirmed MEG8 upregulation, which leads to TNF-α increase. The increased TNF-α enhances osteoclast activity which results in increased bone destruction. | [18] |
| MEG3 | ENSG00000214548/14q32.2 | ↓ | 34 tumors and ANTs | GH3, MMQ | miR-23b-3p↑, FOXO4↓ | - | RT-qPCR indicated MEG3 reduction in tumors. MEG3 diminishes cell proliferation, invasion, migration, EMT, and upregulates apoptosis rate. | [19] |
| ↓ | 30 tumors and 12 normal pituitary tissues, nude mice | PDFS | miR-376B-3P↓, HMGA2↑ | - | MEG3 downregulation was analyzed by qRT-PCR. Upregulation of MEG3 and MIR-376B-3P suppresses tumorigenesis and enhances apoptosis. | [20] | ||
| SNHG6 | ENSG00000245910/8q13.1; 8q13 | ↑ | Invasive and non-invasive pituitary tissues, each containing 30 cases. | HP75 | miR-944↓, RAB11A↑ | - | SNHG6 upregulation, measured by qRT-PCR, improves the proliferation, invasion, migration, viability, and EMT rate of tumor cells. | [21] |
| PCAT6 | ENSG00000228288/1q32.1 | ↑ | Tumors and ANTs from 20 invasive and 20 non-invasive cases, nude mice | RC-4B/C, GH3 | miR-139-3p↓, BRD4↑ | - | Located in the cytoplasm, PCAT6 increases cell proliferation, viability, invasion, cell cycle progression, migration but decreases apoptosis rate. RT-qPCR was used to evaluate PCAT6 expression. | [22] |
| lncRNA-m433s1 | -/6q32 | ↑ | Male and female SD rats | - | miR-433↓, Fshβ↑ | - | RT-qPCR confirmed lncRNA-m433s1 upregulation. As an intergenic lncRNA, this non-coding RNA is located in the cytoplasm and upregulates follicle-stimulating hormone. | [23] |
| UCA1 | ENSG00000214049/19p13.12 | ↑ | 30 pituitary tumors and 30 normal tissues | GH3, MMQ | prolactin↑, HK2↑, LDHA↑ | - | qRT-PCR evaluation showed UCA1 overexpression. UCA1 enhances glycolysis, prolactin secretion, and cell growth. | [24] |
| C5orf66-AS1 | ENSG00000249082/5q31.1 | ↓ | 11 patients and 4 normal cases | GT1-1 | SCGB3A1↓ | - | In this experiment RNA-sequencing, microarray analysis and qRT-PCR methods were used. C5orf66-AS1 upregulation leads to a marked reduction in cell viability and invasion. | [25] |
| PVT1 | ENSG00000249859/8q24.21 | ↑ | 86 pituitary adenoma and ANTs | GH3, HP75, HNPG | β-Catenin↑, c-Myc↑, Cyclin D1↑ | Wnt/β-catenin | qRT-PCR analysis indicated PVT1 upregulation. Cell proliferation, migration, and EMT are all improved by PVT1. | [26] |
| CCAT2 | ENSG00000280997/8q24.21 | ↑ | 74 adenoma and corresponding normal tissues | HP75 | PTTG1↑, SOX2↑, DLK1↑, MMP2↑, MMP13↑/E2F1 | - | CCAT2 boosts cell proliferation, invasion, and migration but impedes apoptosis. Its expression was determined by RT-PCR. | [27] |
| TUG1 | ENSG00000253352/22q12.2 | ↑ | 55 pituitary adenoma cases and 11 normal participants, BALB/c nude mice | HP75, GH3 | miR-187-3p↓, p65↑, IκB-α↑, TESC↑ | NF-κB | RT-qPCR assessed TUG1 expression. While TUG1 enhances the cell proliferation rate, invasion, migration, and EMT, the apoptosis rate is diminished. | [28] |
| AFAP1-AS1 | ENSG00000272620/4p16.1 | ↑ | 60 pituitary adenoma and ANTs | GH3, MMQ | PTEN↓, PI3K↑, p-AKT↑ | PTEN-PI3K-AKT | qRT-PCR showed a significant upregulation in AFAP1-AS1. Cell cycle progression and proliferation rates positively correlate with AFAP1-AS1 expression; in contrast to its negative correlation with the apoptosis rate. | [29] |
| IFNG-AS1 | ENSG00000255733/12q15 | ↑ | 20 pituitary adenoma and ANTs | HP75 | ESRP2↓ | - | IFNG-AS1 overexpression, evaluated by qRT-PCR, significantly increases cell proliferation, invasion, migration, and lowers the apoptosis rate. | [30] |
| SNHG1 | ENSG00000255717/11q12.3 | ↑ | 48 invasive and 10 non-invasive pituitary tumor tissues, nude mice | GH1, RC-4B/C | miR-302↓, miR-372↓, miR-373↓, miR-520↓, TGFBR2↑, RAB11A↑ | Wnt/β-catenin, TGF-β | Although SNHG1 overexpression—appraised by RT-PCR—decreases the apoptosis rate, proliferation, cell cycle progression, invasion, migration, and EMT rates are all improved. | [31] |
Figure 1.
A schematic representation of the role of several non-coding RNAs in regulating the MAPK/ERK, PI3K/AKT, Wnt/β-Catenin, and BMP signaling pathways in pituitary gland tumors and meningiomas. The figure represents the potential crosstalk between various signaling cascades modulated via several ncRNAs in triggering the development of tumor cells. WNT-signaling is a crucial part of the crosstalk between key oncogenic cascades involved in pituitary gland tumors. Elements of the WNT cascades both could be regulated through diverse pathways, including MAPK/ERK, PI3K/AKT, and BMP, as well as transcriptional regulators containing p53 and MITF [32]. In addition, an accumulation of β-catenin in the cytoplasm could, in turn, lead to its translocation to the nucleus, where it could create a complex with TCF/LEF to trigger the transcription of RUNX2, Cyclin D1, and PIT1/2. Additionally, RUNX2 could modulate the transcription of various targets, including OCN, OSX, OPN, MMP13, and ALP [33]. Furthermore, BMP receptors could phosphorylate receptor-SMADs upon ligand binding. TCF12 and TWIST1 are basic helix-loop-helix transcription factors that could play an effective role in heterodimerization and suppressing transcription downstream of the BMP cascade [34]. According to the current report, lncRNA AFAP1-AS1 could enhance growth and inhibit apoptosis in pituitary adenomas through promoting PTEN expression and suppressing the expression levels of PI3K and AKT in tumor cells [29]. Moreover, another research has denoted that LINC00460 could elevate meningioma progression and metastasis through promoting the expression levels of MMP-2, MMP-9, and ZEB1 by sponging miR-539 and thereby acting as an oncogenic RNA in the meningioma malignancy [35]. Moreover, based on recent study, lncRNA SNHG1 via targeting miR-556-5p could elevate TCF12 expression, thereby promoting tumorigenesis of meningioma through the Wnt signaling cascade. In fact, TCF12 expression was positively modulated via SNHG1, and TCF12 could, in turn, enhance transcription of SNHG1 through binding with the promoter region of SNHG1 [36]. Green arrows indicate the upregulation of target genes modulated via ncRNAs (miRNAs and lncRNAs); red arrows depict inhibition regulated by them. All the information regarding the role of these ncRNAs in modulating pituitary gland tumors can be seen in Table 1, Table 2, Table 3, Table 4 and Table 5.
2.2. miRNAs
Dysregulation of miRNAs is also implicated in the pathogenesis of pituitary gland tumors. While almost all related miRNAs have been assessed in a single study, the expression of miR-34a and miR-145 has been evaluated in independent studies. Notably, the results of these studies are contradictory. Yang et al. have assessed the expression of miR-34a in rat pituitary tumor cells versus normal pituitary cells. The expression of this miRNA is lower in tumor cells compared with that in normal ones. Moreover, upregulation of miR-34a could suppress the proliferation of tumor cells and promote cell apoptosis via the regulation of expression of SOX7 [37]. On the other hand, another experiment in AIPmut+ cells as a genetic model of hereditary somatotropinoma has shown upregulation of miR-34a and miR-145 in AIPmut+ compared with AIPmut− somatotropinomas. Ectopic expression of AIPmut (p.R271W) in Aip−/− mouse embryonic fibroblasts led to enhancement of miR-34a and miR-145 levels, demonstrating a pivotal correlation between AIPmut and miRNAs signature. Upregulation of miR-34a enhances proliferation, colony-forming ability, and migration of rat pituitary cells and inhibits their apoptosis. miR-145 could also affect the proliferation and apoptosis of these cells to a lesser extent. Overexpression of miR-34a enhances intracellular levels of the mitogenic factor cAMP and reduces octreotide-mediated growth hormone inhibition and antiproliferative impacts. miR-34a has been found to target the Gnai2 gene, a gene that encodes a G protein subunit suppressing cAMP synthesis. Taken together, miR-34a has been identified as a downstream target of mutant AIP that confers a cellular phenotype reflecting the aggressive manifestations of AIPmut+ acromegaly [38].
Contrary to this report, the expression of miR-145-5p has been shown to be lower in bromocriptine-resistant prolactinoma clinical specimens and cell lines compared with that in sensitive samples and cells. TPT1 has also been identified as the direct target of miR-145-5p. Forced overexpression of miR-145-5p has increased the sensitivity of prolactinoma cells to bromocriptine through enhancing the expression of TPT1. Thus, miR-145-5p has been identified as a critical modulator of drug resistance in prolactinoma [39].
The expression of miR-378 has also been found to be decreased in pituitary adenoma tissues. This miRNA has been shown to downregulate the expression of RNF31. RNF31 silencing has remarkably inhibited the proliferation and migration of GH3 pituitary adenoma cells [40]. On the other hand, the expression of miR-543 has been found to be upregulated in pituitary adenoma tissues parallel with downregulation of Smad7. Smad7 has been verified as a target gene of miR-543. Overexpression of miR-543 has enhanced the proliferation, migration, and invasiveness of HP75 cells. This miRNA also reduces the apoptosis of these cells and decreases the expression of cleaved caspases 3 and 8 [41]. Table 2 shows the role of miRNAs in pituitary gland tumors. Figure 2 illustrates the role of various ncRNAs in pituitary gland tumors through regulating the TGF-β/SMAD signaling pathway.
Table 2.
miRNAs and pituitary gland tumors (ANTs: adjacent non-tumor samples).
| miRNA | Accession Number/Location | Pattern of Expression | Clinical Samples/Animal Model | Assessed Cell Lines | Targets/Regulators | Signaling Pathways | Description | Reference |
|---|---|---|---|---|---|---|---|---|
| miR-34a | ENSG00000284357/1p36.22 | ↓ | female Rattus norvegicus | GH4C1 | SOX7↑ | - | miR-34a upregulation decreases cell proliferation and increases apoptosis. miR-34a downregulation was evaluated by qRT-PCR. | [37] |
| ↑ | 42 cases: 32 somatotropinomas and 10 prolactinomas, AIP knockout mice | GH3, HEK293, GH4C1 | Gαi2↓, cAMP↑ | - | Somatotropinomas with AIP mutations lead to enhanced miR-34a expression, upregulated intracellular cAMP concentration, and reduced Gαi2, resulting in somatostatin resistance. Additionally, miR-34a limits the apoptosis rate. To assess miRNA expression, microarray analysis and qRT-PCR were used. | [38] | ||
| miR-338-3p | ENSG00000283604/17q25.3 | ↑ | 10 microadenoma within sella turcica and 13 invading cavernous sinus macroadenoma cases | GH3 | Pttg1↑, GH↑, prolactin↑ | - | miR-338-3p improves cells’ invasiveness, migration, and proliferation rate. miRNA and qRT-PCR were used. | [42] |
| miR-378 | ENSG00000199047/5q32 | ↓ | 25 tumors and ANTs | GH3 | RNF31↑ | - | RT-qPCR confirmed miR-378 downregulation. miR-378 reduces proliferation and migration rates. | [40] |
| miR-543 | ENSG00000212040/14q32.31 | ↑ | 71 invasive and 66 non-invasive tumor tissues | HP75 | Smad7↓ | Wnt/β-catenin | miR-543 overexpression, appraised by RT-qPCR, resulted in improved cell proliferation, invasion, and migration but reduced apoptosis. | [41] |
| miR-134 | ENSG00000207993/14q32.31 | ↓ | 29 patients affected by nonfunctioning pituitary neuroendocrine tumor | αT3-1 | VEGFA↑/SDF-1α↑ | - | Diminished levels of miR-134 were analyzed by qRT-PCR. SDF-1α decreases miR-134 and improves VEGFA to expand cell proliferation, viability, and cell cycle progression capacity. | [43] |
| miR-193a-3p | ENSG00000207614/17q11.2 | ↓ | 82 patients with pituitary adenoma: 42 nonfunctional, 32 prolactinomas, 5 growth hormone-secreting, and 2 follicle-stimulating hormone | - | - | - | qRT-PCR confirm miR-193a-3p downregulation. Its expression has a negative correlation with tumor size and recurrence rate. | [44] |
| miR-448 | ENSG00000199001/Xq23 | ↓ | Pituitary adenoma and ANTs | MMQ, HP75 | BCL2↑ | - | miR-448 downregulation was analyzed by qRT-PCR. Its overexpression restricts cell proliferation and migration and increases apoptosis. | [45] |
| miR-219a-2-3p | ENSG00000284185/9q34.11 | ↓ | - | AtT-20, GT1.1, MPC | MDM2↑, p53↓ | - | RT-qPCR was used to assess miR-219a-2-3p expression. miR-219a-2-3p improves apoptosis and inhibits cell proliferation. | [46] |
| miR-1299 | ENSG00000275377/9p11.2 | ↑ | 12 drug-resistant and 6 sensitive patients | MMQ | FOXO1↓, prolactin↑ | - | miR-1299 is upregulated in drug-resistant cases, which further inhibits FOXO1 expression. MicroRNA sequencing analysis and qRT-PCR were used in this experiment. | [47] |
| miR-410-3p | ENSG00000199092/14q32.31 | ↑ | 75 pituitary adenoma tissues: 34 gonadotroph, 30 somatotroph, 5 corticotroph, 3 plurihormonal, and 3 null cell tumors | RC-4B/C, AtT-20, GH3 | cyclin B1↑, p14↓, Wee1↓ | MAPK, PTEN/AKT, STAT3 | Invasive tumors have a higher miR-410-3p expression level, which leads to a higher proliferation rate, invasion, and cell cycle progression rates in RC-4B/C and AtT-20 cells. GH3 cells showed a precisely opposite result. qRT-PCR was used to assess miR-410-3p expression. | [48] |
| miR-137 | ENSG00000284202/1p21.3 | ↓ | 15 invasive and 15 non-invasive prolactinoma tissues, female F344 rats | MMQ, GH3 | MITF↑, WIF-1↓ | Wnt/β-catenin | miR-137 lowers cell proliferation, invasion, and β-catenin nuclear translocation rates. Tissue microarray and qRT-PCR were used in this project. | [49] |
| miR-205-5p | ENSG00000284485/1q32.2 | ↓ | - | GH3, MMQ, HEK293T | CBX1↑ | - | qRT-PCR confirmed miR-205-5p downregulation. miR-205-5p significantly reduces cell proliferation and migrations rates. | [50] |
| miR-93-5p | ENSG00000207757/7q22.1 | ↑ | 8 fibrous and 33 nonfibrous prolactinoma tissues | MMQ, HS27 | Smad7↓, TGF-β1↑ | TGF-β1/Smad3 | Small RNA sequencing and qRT-PCR methods were used in this experiment. miR-93-5p induces fibrosis in prolactinoma cases through regulating the TGF-β1/Smad3 pathway. | [51] |
| miR-370 | ENSG00000199005/14q32.31 | ↓ | 24 nonfunctional pituitary adenoma tissues | - | HMGA2↑/CXCL12↑ | - | miR-370 expression negatively correlates with higher tumor grades and Ki-67-positive cells. It also restricts cell proliferation rate and boosts cell apoptosis rate. miR-370 downregulation was indicated by RT-PCR. | [52] |
| miR-145-5p | ENSG00000276365/5q32 | ↓ | 11 normal pituitary tissues, 24 bromocriptine-sensitive and 8 resistant samples, female nude mice | MMQ | TPT1↑ | - | As confirmed by qRT-PCR, miR-145-5p is decreased in bromocriptine-sensitive tissues and highly reduced in bromocriptine-resistant tissues. On top of that, miR-145-5p upregulation reduces cell viability. | [39] |
| miR-16 | ENSG00000208006/13q14.2 | ↓ | 36 patients and 8 healthy controls | HP75 | p27↓, Bax↓, VEGFR2↓ | NF-κB | RT-qPCR confirmed miR-16 downregulation. Cell proliferation and apoptosis rates are, respectively, positively and negatively correlated with miR-16 overexpression. In addition, this microRNA inhibits angiogenesis. | [53] |
| miR-124 | ENSG00000284321/8p23.1 | ↓ | 68 invasive pituitary tissues: 7 growth hormone-secreting and 61 non-invasive pituitary adenomas | GH3 | PTTG1IP↑/Cav-1, EGR1, KLF5 | - | Caveolin-1 inhibits EGR1 translocation into the nucleus. Therefore, KLF5 does not interact with EGR1. Without the inhibitory effect of EGR1, KLF5 increases these miRNAs expression, which ultimately reduces PTTG1IP, FSCN1, and EZR levels—prohibiting cell invasion and migration. Microarray assay and qRT-PCR were used to assess miRNAs expression. | [54] |
| miR-145 | ENSG00000276365/5q32 | ↓ | FSCN1↑/Cav-1, EGR1, KLF5 | |||||
| miR-183 | ENSG00000207691/7q32.2 | ↓ | EZR↑/Cav-1, EGR1, KLF5 | |||||
| miR-148-3p | ENSG00000199085/7p15.2 | ↓ | 10 invasive and 10 non-invasive pituitary adenoma tissues | GH3, MMQ | ALCAM↑ | - | The qRT- PCR method evaluated these miRNAs’ expression. Proliferation and invasion rates are reduced by these miRNAs, but apoptosis is enhanced. | [55] |
| miR-152 | ENSG00000207947/17q21.32 | ↓ | ||||||
| miR-524-5p | ENSG00000283289/19q13.42 | ↓ | 20 adenoma and 8 normal tissues, BALB/c female nude mice | PDFS, HEK293FT | PBF↑ | - | Proliferation, invasion, clonogenicity, tumor growth, and migration are all inversely correlated with miR-524-5p expression. qRT- PCR was used to analyze miR-524-5p expression. | [56] |
| miR-153 | ENSG00000207647/2q35 | ↓ | - | MMQ | Skp↑ | - | miR-153 activates caspase-3 to increase apoptosis and decrease the proliferation rate. | [57] |
| miR-106b | ENSG00000208036/7q22.1 | ↑ | 32 invasive and 18 non-invasive adenoma tissues, 10 healthy control cases | HP75 | PTEN↓ | PI3K/AKT | Cell cycle progression, invasion, migration, and proliferation are markedly improved by miR-106b upregulation, which itself was analyzed by qRT-PCR. | [58] |
| miR-26a | ENSG00000199075/3p22.2 | ↑ | 12 normal, 31 invasive, and 39 non-invasive pituitary tissues | - | PLAG↓ | - | Through downregulating PLAG, miR-26a enhances tumor invasiveness. miR-26a expression was evaluated by qRT-PCR. | [59] |
| miR-133 | ENSG00000283927/18q11.2 | ↓ | 6 pituitary tumors and ANTs | HP75 | FOXC1↑ | - | miR-133 downregulation was assessed by RT-PCR. Cell migration, EMT, and invasion are inversely related to miR-133 expression. | [60] |
Figure 2.
A schematic diagram of the role of various ncRNAs in modulating the TGF-β/SMAD signaling pathway in pituitary gland tumors. According to this cascade, it could be triggered through the binding of active TGF-β with TβRII and forming the TβRI-TβRII heteromeric complex, resulting in phosphorylation of Smad2/3, oligomerization with Smad4, and consequent nuclear translocation to modulate the transcription of ECM genes. Furthermore, Smad7 could play a remarkable role as a negative modulator of the TGF-β cascade. In addition, TGF-β has a significant part in triggering the activation of downstream signaling pathways containing MAPK, modulated by the Ras-Raf-MEK-ERK cascade, and TAK1, regulated by the TAB1 pathway. This could also lead to mediating the activation of MKK4-JNK and MKK3-p38 cascades and upregulation of AP-1 and ATF-2, respectively, and the overexpression of NF- κB to modulate profibrotic responses [61]. Previous studies have authenticated that several ncRNAs could have a significant part in regulating the TGF-β/SMAD cascade in pituitary gland tumors. As an illustration, recent literature has detected that overexpression of lnc-SNHG1 could considerably elevate the expression level of TβRII through activating TβRII/SMAD3 in invasive pituitary tumor cells via sponging miR-302/372/373/520 [31]. Furthermore, other research has indicated that upregulation of miR-93-5p could downregulate the expression level of Smad7, thereby activating the TGF-β1/Smad3 signaling-mediated fibrosis of prolactinoma cells [51]. Green arrows indicate the upregulation of target genes modulated via ncRNAs (miRNAs and lncRNAs); red arrows depict inhibition regulated by them.
2.3. circRNAs
Du et al. have assessed circRNAs signature in growth hormone-secreting pituitary adenoma using a circRNA microarray. They have reported upregulation of more than 1900 circRNAs and downregulation of about 1600 circRNAs in this type of adenoma compared with that in normal control. Ten most overexpressed circRNAs have been shown to be mainly enriched in the mTOR and the Wnt signaling pathway. Upregulation of hsa_circ_0001368 has also been confirmed by qRT-PCR. This circRNA has been found to be specifically overexpressed in this type of adenoma in correlation with the invasive properties and serum levels of growth hormone. Hsa_circ_0001368 silencing suppresses proliferation, invasion, and secretion of growth hormone from primary cultured cells. Additionally, levels of hsa_circ_0001368 have been positively correlated with the pituitary-specific transcription factor Pit-1 [62]. CircOMA1 is an upregulated circRNA in nonfunctioning pituitary adenomas which sponges miR-145-5p, a miRNA that inhibits the growth of this type of tumor through targeting TPT1 [63]. Table 3 shows the role of circRNAs in pituitary adenomas.
Table 3.
circRNAs and pituitary gland tumors.
| circRNA | Pattern of Expression | Clinical Samples/Animal Model | Assessed Cell Lines | Targets/Regulators | Signaling Pathways | Description | Reference |
|---|---|---|---|---|---|---|---|
| hsa_circ_0001368 | ↑ | Growth hormone-secreting pituitary adenoma: 19, nonfunctioning pituitary adenoma: 20, prolactin-secreting adenoma: 18, ACTH-secreting adenoma: 12 | - | Pit-1↑ | mTOR, Wnt | Proliferation, invasion, and growth hormone-secretion levels are positively related to this circRNA expression. circRNA microarray, RNA-seq, and qRT-PCR were used in this project. | [62] |
| circOMA1 | ↑ | 50 nonfunctioning adenomas and 15 normal tissues, BALB/c nude mice | PDFS, HEK293T | miR-145-5p↓, TPT1↑, Mcl-1↑, Bcl-xL↑, Bax↓ | - | The qRT-PCR method was used to appraise circOMA1 expression. Tumor invasion and cell proliferation are enhanced by circOMA1, whereas the apoptosis rate is decreased. | [63] |
2.4. Prognostic/Diagnostic Value of Non-Coding RNAs in Pituitary Gland Tumors
The prognostic/diagnostic value of non-coding RNAs has been assessed in pituitary gland tumors (Table 4). Downregulation of SNHG7 [17], CCAT2 [27], and IFNG-AS1 [30] lncRNAs has been shown to increase the survival of patients with pituitary adenomas. miR-193a-3p upregulation results in a lower relapse-free survival rate [44]. On the other hand, upregulation of miR-137 expression is related to a higher recurrence-free survival rate [49].
Table 4.
Prognostic/diagnostic value of non-coding RNAs in pituitary gland tumors (OS: overall survival, ANTs: adjacent non-tumor samples).
| Non-Coding RNA | Accession Number/Location | Clinical Cases | AUC | Kaplan–Meier Analysis | Univariate/Multivariate Cox Regression | Reference |
|---|---|---|---|---|---|---|
| SNHG7 | ENSG00000233016/9q34.3 | 30 pituitary tumors with high and low expression | - | A lower SNHG7 expression results in a higher OS rate. | - | [17] |
| CCAT2 | ENSG00000280997/8q24.21 | 74 adenomas and corresponding normal tissues | - | Higher CCAT2 expression leads to a lower OS rate. | - | [27] |
| IFNG-AS1 | ENSG00000255733/12q15 | 20 tumors and ANTs | - | Higher IFNG-AS1 expression is an indicator of a lower survival rate. | - | [30] |
| miR-193a-3p | ENSG00000207614/17q11.2 | High: 29 Low: 53 |
- | miR-193a-3p upregulation leads to a lower relapse-free survival rate. | - | [44] |
| miR-137 | ENSG00000284202/1p21.3 | High: 16 Low: 14 |
- | Upregulated miR-137 expression is related to a higher recurrence-free survival rate. | - | [49] |
| miR-16 | ENSG00000208006/13q14.2 | 36 patients were divided into high- and low-expression groups | - | miR-16 overexpression marks longer OS and disease-free survival rates. | - | [40] |
| miR-26a | ENSG00000199075/3p22.2 | 12 normal, 31 invasive and 39 non-invasive pituitary tissues | 0.818 | Downregulated miR-26a represents a shorter survival rate. | Tumor invasiveness, miR-26a, and PLAG1 expression could be used as survival risk factors. | [59] |
3. Meningioma
Meningiomas are another group of brain tumors that are mostly encapsulated lesions. These benign lesions are typically associated with few types of genetic aberrations, yet their intracranial location can result in serious and possibly fatal consequences [64]. The expression of a number of lncRNAs and miRNAs has been dysregulated in meningioma.
3.1. LncRNAs
SNHG1 is an upregulated lncRNA in meningioma cell lines. SNHG1 silencing blocks cell growth and induces their apoptosis. SNHG1 could function as a sponge for miR-556-5p and enhance the expression of TCF12. In fact, the SNHG1/miR-556-5p/TCF12 axis could promote the proliferation of meningioma cells and suppress their apoptosis by enhancing the activity of Wnt signaling. Moreover, TCF12 has been shown to increase the expression of SNHG1 via binding with its promoter [36]. LINC00702 is another upregulated lncRNA in meningioma which regulates proliferation and migration of these cells via the miR-4652-3p/ZEB1 axis [65]. In addition, LINC00460 has been found to increase cell invasion and proliferation and decrease apoptosis rate through sponging miR-539 [35]. On the other hand, MEG3 is a possible tumor suppressor lncRNA in meningioma, which modulates invasive properties of these cells through the miR-29c/AKAP12 axis [66]. Table 5 shows the role of lncRNAs in meningiomas.
Table 5.
LncRNAs and meningioma (ANTs: adjacent non-tumor samples).
| lncRNA | Accession Number/Location | Pattern of Expression | Clinical Samples/Animal Model | Assessed Cell Lines | Targets/Regulators | Signaling Pathways | Description | Reference |
|---|---|---|---|---|---|---|---|---|
| SNHG1 | ENSG00000255717/11q12.3 | ↑ | - | CH157-MN, HBL-52, BEN-MEN-1, IOMM-Lee |
miR-556-5p↓, TCF12↑/TCF12 | Wnt/β-catenin | SNHG1 overexpression, indicated by qRT-PCR, has a marked impact on elevating cell proliferation and growth rates and inhibiting apoptosis. | [36] |
| LINC00702 | ENSG00000233117/10p15.1 | ↑ | 88 malignant meningioma and ANTs | OMM-Lee, KT21, CH157-MN, HBL-52, Ben-Men-1 | miR-4652-3p↓, ZEB1↑ | Wnt/β-catenin | This lncRNA overexpression, as formerly showed by qRT-PCR, has a significant correlation with a lower OS rate. Cell migration and proliferation are positively associated with its expression. | [65] |
| LINC00460 | ENSG00000233532/13q33.2 | ↑ | 33 meningioma and 10 normal meninges tissues | IOMM-Lee, CH157-MN, Ben-Men-1 | miR-539↓, MMP-2↑, MMP-9↑, ZEB1↑ | - | LINC00460 escalates cell invasion and proliferation and lowers the apoptosis rate. qRT-PCR was used to evaluate this lncRNA’s expression. | [35] |
| MEG3 | ENSG00000214548/14q32.2 | ↓ | 5 healthy meninges and 32 meningioma tissues | (IOMM-Lee, CH157-MN | miR-29c↑, AKAP12↓ | - | qRT-PCR showed MEG3 downregulation in tumor tissues. Cell cycle progression, proliferation, migration, and invasion are negatively correlated with MEG3 expression. | [66] |
3.2. miRNAs
Negroni et al. have reported that the expression of the miR-497~195 cluster in meningioma is reduced with increasing tumor grade. Notably, Cyclin D1 upregulation has been correlated with a decrease in the levels of the miR-497~195 cluster. In fact, the effect of GATA binding protein 4 in enhancing the viability of meningioma cells is exerted through the regulation of expression of this miRNA cluster, which finally results in enhancement of Cyclin D1 level. Finally, serum exosome levels of miR-497 are lower in patients with high-grade meningioma compared to those with benign lesions [67].
Expression of miRNAs and mRNAs in benign and malignant meningiomas has also been assessed by RNA sequencing and miRNA microarray. This study has led to identifying upregulation of fatty acid synthase (FASN) in malignant lesions compared with benign ones. This gene has been shown to be targeted by miR-195. Overexpression of miR-195 could inhibit proliferation, migration, and invasiveness of malignant meningioma cells. Taken together, miR-195 has been verified as a tumor-suppressive miRNA in malignant meningioma by targeting FASN. NUP210, SPIRE2, SLC7A1, and DMTN are also among competing endogenous RNAs that modulate the expression of FASN through sponging miR-195 [68].
In another study, Katar et al. have shown significant enhancement in miR-21 levels with increasing grade of meningiomas, whereas there was a remarkable decrease in miR-107 levels with the increasing grade. Expressions of miR-137 and miR-29b have not been different in different histopathologic grades [69]. Table 6 shows the role of miRNAs in meningioma.
Table 6.
miRNAs and meningioma.
| miRNA | Accession Number/Location | Pattern of Expression | Clinical Samples/Animal Model | Assessed Cell Lines | Targets/Regulators | Description | Reference |
|---|---|---|---|---|---|---|---|
| miR-497~195 | ENSG00000267532/17p13.1 | ↓ | 80 meningioma and 25 primary meningioma cases | KT21-MG1-Luc5D, Ben-Men-1 | Cyclin D1↑/GATA-4↑ | GATA-4 upregulation restricts miR-497~195 cluster expression and increases cell viability. RT-PCR was used to assess miR-497 expression. | [67] |
| miR-195 | ENSG00000284112/17p13.1 | ↓ | 3 paired malignant and benign meningioma cases | IOMM-Lee | FASN↑/NUP210, SPIRE2, SLC7A1, DMTN | Migration, invasion, and proliferation rates of tumor cells are axiomatically elevated by miR-195 downregulation. RNA-sequencing, miRNA microarray, and qRT-PCR methods were used in this experiment. | [68] |
| miR-21 | ENSG00000284190/17q23.1 | ↑ | 50 patients affected with meningioma | - | - | miR-21 and miR-107 have positive and negative correlations with tumor grade, respectively. Their expression was evaluated by miRNA detection kit. | [69] |
| miR-107 | ENSG00000198997/10q23.31 | ↓ | |||||
| miR-34a-3p | ENSG00000284357/1p36.22 | ↓ | 35 meningioma cases | Ben-Men-1, HEK293T | SMAD4↑, FRAT1↑, BCL2↑ | miR-34a-3p downregulation was assessed by RT-PCR. While the proliferation rate is diminished after miR-34a-3p overexpression, the apoptosis rate is improved. | [70] |
| miR-29c-3p | ENSG00000284214/1q32.2 | ↑ | 58 meningioma tumor tissues | MEN-117, MEN-141 | PTX3↓ | Microarray analysis and RT-PCR were used in this experiment. Cell viability is improved by miR-29c-3p upregulation. In contrast, apoptosis is lowered by miR-29c-3p. | [71] |
| let-7d | ENSG00000199133/9q22.32 | ↓ | 17 meningioma samples | IOMM-Lee, CH-157MN | AEG-1↑ | qRT-PCR evaluated let-7d expression. Proliferation, invasion, and viability are effectively inhibited by let-7d, whereas the apoptosis rate is elevated. | [72] |
Finally, a number of studies have shown associations between genetic polymorphisms of lncRNAs or miRNAs and risk of meningioma (Table 7). For instance, the rs619586 A > G polymorphism of MALAT1 has been shown to affect expression levels of MALAT1 and COL5A1, resulting in lower invasiveness of meningioma [73]. Expression of MALAT1 has been shown to be reduced in a stepwise manner with enhancing levels of miR-145 in tumor/serum specimens having AA, AG, and GG genotypes of rs619586, respectively. In addition, levels of COL5A1 have been reduced in a similar stepwise manner in relation to the rs619586 genotypes. Thus, the rs619586A > G of the MALAT1 gene can decrease the expression of this lncRNA, influencing the impact of miR-145 on COL5A1. Consistently, meningioma cells harboring the G genotype of the rs619586 had higher levels of COL5A1 [73]. Moreover, certain haplotype blocks of miR-146a, miR-149, miR-196a2 and miR-499 have been shown to be associated with risk of meningioma [74].
Table 7.
Polymorphisms of non-coding RNAs in meningioma.
| lncRNA | Accession Number/Location | Clinical Samples | Assessed Cell Lines | Polymorphism | Description | References |
|---|---|---|---|---|---|---|
| MALAT1 | ENSG00000251562/11q13.1 | 427 invasive and 402 non-invasive meningioma cases | KNS-89, SNB-19 | A > G (rs619586) |
AA genotype increases invasive meningioma risk. | [73] |
| miR-146a | ENSG00000283733/5q33.3 | 69 meningioma and 183 healthy controls | - | C > G (rs2910164) |
These three haplotypes significantly increase the chances of meningioma: (1) miR-146a-miR-149-miR-196a2 -miR-499: G-T-C-G (2) miR-146a-miR-196a2 -miR-499: G-C-G (3) miR-149-miR-196a2 -miR-499: C-C-G |
[74] |
| miR-149 | ENSG00000207611/2q37.3 | T > C (rs4846049) |
||||
| miR-196a2 | ENSG00000207924/12q13.13 | T > C (rs11614913) |
||||
| miR-499 | ENSG00000207635/20q11.22 | A > G (rs3746444) |
4. Discussion
The contribution of lncRNAs, miRNAs, and circRNAs has been assessed in pituitary adenomas. lncRNAs that participate in the pathogenesis of pituitary gland tumors mainly serve as sponges for miRNAs. CLRN1-AS1/miR-217, XIST/miR-424-5p, H19/miR-93a, LINC00473/miR-502-3p, SNHG7/miR-449a, MEG8/miR-454-3p, MEG3/miR-23b-3p, MEG3/miR-376B-3P, SNHG6/miR-944, PCAT6/miR-139-3p, lncRNA-m433s1/miR-433, TUG1/miR-187-3p, SNHG1/miR-187-3p, SNHG1/miR-302, SNHG1/miR-372, SNHG1/miR-373, and SNHG1/miR-520 are identified lncRNA/miRNA pairs that are involved in this process.
The contribution of lncRNAs in meningioma is less studied. However, similar to pituitary adenomas, lncRNAs mainly serve as sponges for miRNAs. SNHG1/miR-556-5p, LINC00702/miR-4652-3p, LINC00460/miR-539, and MEG3/miR-29c are identified lncRNA/miRNA pairs in this type of lesion. Functional effects of miRNAs sponging are diverse, ranging from the enhancement of invasiveness of tumors to induction of epithelial-mesenchymal transition in these cells.
Exosomal levels of miRNAs in serum samples of patients have been associated with the presence of brain malignancies in some cases. However, the diagnostic and prognostic applications of these exosomes have not been assessed comprehensively. In other types of cancer, exosomal levels of non-coding RNAs represent an applicable source of biomarkers for prediction of course of cancer as well as response to therapies [75].
Expression of non-coding RNAs might also affect response to therapeutic regimens [76]. For instance, H19 increases ATG7 expression and influences resistance to dopamine agonists [15].
Theoretically, the expression of non-coding RNAs can be used for molecular classification of meningiomas. However, this approach has not been implemented yet. A previous study has used the DNA methylation signature for this purpose. This approach has captured clinically more homogenous groups. Moreover, it has been proved to be superior to WHO classification in prediction of tumor recurrence and prognosis [77].
Genetic variants within non-coding regions have the potential to affect the function of these transcripts or the regulatory impacts of other transcripts on non-coding ones. A number of these variants have been demonstrated to be associated with the risk of meningioma, yet their impacts on the risk of pituitary tumors have not been revealed.
Since aberrant expressions of lncRNAs/miRNAs can affect the response of tumor cells to therapeutic modalities, it is possible that targeted therapies for modulation of expression of lncRNAs/miRNAs not only reduce the invasiveness of these tumors but also increase their response to conventional therapies.
5. Conclusions
Cumulatively, non-coding RNAs represent an emerging class of transcripts with putative effects on the pathogenesis of pituitary adenomas as well as meningiomas. Future studies are needed to find possible specific markers for each of these tumors to help in the identification of tumors in a less invasive manner.
Abbreviations
Long non-coding RNAs (lncRNAs), microRNAs (miRNAs), circular RNAs (circRNAs), clarin 1 antisense RNA 1 (CLRN1-AS1), dickkopf WNT signaling pathway inhibitor 1 (DKK1), adjacent non-tumor samples (ANTs).
Author Contributions
M.T. and S.G.-F. supervised the study, wrote the draft, and edited the submission. G.S., A.A. and B.M.H. performed the data collection, designed the tables and figures. All of the authors contributed equally and are fully aware of the submission. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare they have no conflicts of interests.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Kaiser J. DNA sequencing. A plan to capture human diversity in 1000 genomes. Sci. N. Y. 2008;319:395. doi: 10.1126/science.319.5862.395. [DOI] [PubMed] [Google Scholar]
- 3.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grillone K., Riillo C., Scionti F., Rocca R., Tradigo G., Guzzi P.H., Alcaro S., Di Martino M.T., Tagliaferri P., Tassone P. Non-coding RNAs in cancer: Platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020;39:1–19. doi: 10.1186/s13046-020-01622-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Y., Fullwood M.J. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genom. Proteom. Bioinform. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y., Tai W., Lu N., Li T., Liu Y., Wu W., Li Z., Pu L., Zhao X., Zhang T. lncRNA ZFAS1 promotes lung fibroblast-to-myofibroblast transition and ferroptosis via functioning as a ceRNA through miR-150-5p/SLC38A1 axis. Aging Albany N. Y. 2020;12:9085. doi: 10.18632/aging.103176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asa S.L. Tumors of the Pituitary Gland. Amer Registry of Pathology; London, UK: 1998. [Google Scholar]
- 9.Asa S.L., Ezzat S. The pathogenesis of pituitary tumors. Annu. Rev. Pathol. Mech. Dis. 2009;4:97–126. doi: 10.1146/annurev.pathol.4.110807.092259. [DOI] [PubMed] [Google Scholar]
- 10.Ezzat S., Asa S.L., Couldwell W.T., Barr C.E., Dodge W.E., Vance M.L., McCutcheon I.E. The prevalence of pituitary adenomas: A systematic review. Cancer. 2004;101:613–619. doi: 10.1002/cncr.20412. [DOI] [PubMed] [Google Scholar]
- 11.D’Angelo D., Mussnich P., Sepe R., Raia M., Del Vecchio L., Cappabianca P., Pellecchia S., Petrosino S., Saggio S., Solari D. RPSAP52 lncRNA is overexpressed in pituitary tumors and promotes cell proliferation by acting as miRNA sponge for HMGA proteins. J. Mol. Med. 2019;97:1019–1032. doi: 10.1007/s00109-019-01789-7. [DOI] [PubMed] [Google Scholar]
- 12.Zhou K., Li S., Du G., Fan Y., Wu P., Sun H., Zhang T. LncRNA XIST depletion prevents cancer progression in invasive pituitary neuroendocrine tumor by inhibiting bFGF via upregulation of microRNA-424-5p. OncoTargets Ther. 2019;12:7095. doi: 10.2147/OTT.S208329. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Wang C., Tan C., Wen Y., Zhang D., Li G., Chang L., Su J., Wang X. FOXP1-induced lncRNA CLRN1-AS1 acts as a tumor suppressor in pituitary prolactinoma by repressing the autophagy via inactivating Wnt/β-catenin signaling pathway. Cell Death Dis. 2019;10:499. doi: 10.1038/s41419-019-1694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z.R., Yan L., Liu Y.T., Cao L., Guo Y.H., Zhang Y., Yao H., Cai L., Shang H.B., Rui W.W. Inhibition of mTORC1 by lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary tumours. Nat. Commun. 2018;9:1–14. doi: 10.1038/s41467-018-06853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Z., Zheng Y., Xie W., Li Q., Zhang Y., Ren B., Cai L., Cheng Y., Tang H., Su Z. The long noncoding RNA-H19/miRNA-93a/ATG7 axis regulates the sensitivity of pituitary adenomas to dopamine agonists. Mol. Cell. Endocrinol. 2020;518:111033. doi: 10.1016/j.mce.2020.111033. [DOI] [PubMed] [Google Scholar]
- 16.Li J., Qian Y., Zhang C., Wang W., Qiao Y., Song H., Li L., Guo J., Lu D., Deng X. LncRNA LINC00473 is involved in the progression of invasive pituitary adenoma by upregulating KMT5A via ceRNA-mediated miR-502-3p evasion. Cell Death Dis. 2021;12:1–12. doi: 10.1038/s41419-021-03861-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue X., Dong C., Ye Z., Zhu L., Zhang X., Wang X., Mo F., Li Z., Pan B. LncRNA SNHG7 sponges miR-449a to promote pituitary adenomas progression. Metab. Brain Dis. 2021;36:123–132. doi: 10.1007/s11011-020-00611-5. [DOI] [PubMed] [Google Scholar]
- 18.Zhu H.-B., Li B., Guo J., Miao Y.-Z., Shen Y.-T., Zhang Y.-Z., Zhao P., Li C.-Z. LncRNA MEG8 promotes TNF-α expression by sponging miR-454-3p in bone-invasive pituitary adenomas. Aging Albany N. Y. 2021;13:14342. doi: 10.18632/aging.203048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Li X., Wang Z. lncRNA MEG3 inhibits pituitary tumor development by participating in cell proliferation, apoptosis and EMT processes. Oncol. Rep. 2021;45:1–11. doi: 10.3892/or.2021.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu D., Xiao Z., Wang Z., Hu B., Duan C., Zhu Z., Gao N., Zhu Y., Wang H. MEG3/MIR-376B-3P/HMGA2 axis is involved in pituitary tumor invasiveness. J. Neurosurg. 2020;134:499–511. doi: 10.3171/2019.10.JNS191959. [DOI] [PubMed] [Google Scholar]
- 21.Mao D., Jie Y., Lv Y. LncRNA SNHG6 Induces Epithelial–Mesenchymal Transition of Pituitary Adenoma Via Suppressing MiR-944. Cancer Biother. Radiopharm. 2020 doi: 10.1089/cbr.2020.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao P., Cheng J., Li B., Nie D., Wang H., Li C., Gui S., Zhang Y. LncRNA PCAT6 regulates the progression of pituitary adenomas by regulating the miR-139-3p/BRD4 axis. Cancer Cell Int. 2021;21:1–21. doi: 10.1186/s12935-020-01698-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Zhang S.Y., Huang S.H., Gao S.X., Wang Y.B., Jin P., Lu F.J. Upregulation of lncRNA RMRP promotes the activation of cardiac fibroblasts by regulating miR-613. Mol. Med. Rep. 2019;20:3849–3857. doi: 10.3892/mmr.2019.10634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu G., Wang L., Li Y. Inhibition of lncRNA-UCA1 suppresses pituitary cancer cell growth and prolactin (PRL) secretion via attenuating glycolysis pathway. Vitr. Cell. Dev. Biol. Anim. 2020;56:642–649. doi: 10.1007/s11626-020-00494-x. [DOI] [PubMed] [Google Scholar]
- 25.Yu G., Li C., Xie W., Wang Z., Gao H., Cao L., Hao L., Zhang Y. Long non-coding RNA C5orf66-AS1 is downregulated in pituitary null cell adenomas and is associated with their invasiveness. Oncol. Rep. 2017;38:1140–1148. doi: 10.3892/or.2017.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., Tan Y., Wang H., Xu M., Xu L. Long non-coding RNA plasmacytoma variant translocation 1 (PVT1) enhances proliferation, migration, and epithelial-mesenchymal transition (EMT) of pituitary adenoma cells by activating β-catenin, c-Myc, and cyclin D1 expression. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019;25:7652. doi: 10.12659/MSM.917110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Fu D., Zhang Y., Cui H. Long noncoding RNA CCAT2 is activated by E2F1 and exerts oncogenic properties by interacting with PTTG1 in pituitary adenomas. Am. J. Cancer Res. 2018;8:245. [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang R., Yang F., Fan H., Wang H., Wang Q., Yang J., Song T. Long non-coding RNA TUG1/microRNA-187-3p/TESC axis modulates progression of pituitary adenoma via regulating the NF-κB signaling pathway. Cell Death Dis. 2021;12:1–12. doi: 10.1038/s41419-021-03812-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang H., Hou B., Ye Z., Ling C., Guo Y. Knockdown of long non-coding RNA AFAP1-AS1 inhibits growth and promotes apoptosis in pituitary adenomas. Int. J. Clin. Exp. Pathol. 2018;11:1238. [PMC free article] [PubMed] [Google Scholar]
- 30.Lu G., Duan J., Zhou D. Long-noncoding RNA IFNG-AS1 exerts oncogenic properties by interacting with epithelial splicing regulatory protein 2 (ESRP2) in pituitary adenomas. Pathol. Res. Pract. 2018;214:2054–2061. doi: 10.1016/j.prp.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Wang H., Wang G., Gao Y., Zhao C., Li X., Zhang F., Jiang C., Wu B. Lnc-SNHG1 activates the TGFBR2/SMAD3 and RAB11A/Wnt/β-catenin pathway by sponging MiR-302/372/373/520 in invasive pituitary tumors. Cell. Physiol. Biochem. 2018;48:1291–1303. doi: 10.1159/000492089. [DOI] [PubMed] [Google Scholar]
- 32.Gajos-Michniewicz A., Czyz M. WNT signaling in melanoma. Int. J. Mol. Sci. 2020;21:4852. doi: 10.3390/ijms21144852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinhold S., Blankesteijn W.M., Foulquier S. The Interplay of WNT and PPARγ Signaling in Vascular Calcification. Cells. 2020;9:2658. doi: 10.3390/cells9122658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timberlake A.T., Wu R., Nelson-Williams C., Furey C.G., Hildebrand K.I., Elton S.W., Wood J.S., Persing J.A., Lifton R.P. Co-occurrence of frameshift mutations in SMAD6 and TCF12 in a child with complex craniosynostosis. Hum. Genome Var. 2018;5:1–5. doi: 10.1038/s41439-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing H., Wang S., Li Q., Ma Y., Sun P. Long noncoding RNA LINC00460 targets miR-539/MMP-9 to promote meningioma progression and metastasis. Biomed. Pharmacother. 2018;105:677–682. doi: 10.1016/j.biopha.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Yu R., Li Q., Li Y., Xuan T., Cao S., Zheng J. SNHG1/miR-556-5p/TCF12 feedback loop enhances the tumorigenesis of meningioma through Wnt signaling pathway. J. Cell. Biochem. 2020;121:1880–1889. doi: 10.1002/jcb.29423. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z., Zhang T., Wang Q., Gao H. Overexpression of microRNA-34a attenuates proliferation and induces apoptosis in pituitary adenoma cells via SOX7. Mol. Ther. Oncolytics. 2018;10:40–47. doi: 10.1016/j.omto.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogner E.M., Daly A.F., Gulde S., Karhu A., Irmler M., Beckers J., Mohr H., Beckers A., Pellegata N.S. miR-34a is upregulated in AIP-mutated somatotropinomas and promotes octreotide resistance. Int. J. Cancer. 2020;147:3523–3538. doi: 10.1002/ijc.33268. [DOI] [PubMed] [Google Scholar]
- 39.Jian M., Du Q., Zhu D., Mao Z., Wang X., Feng Y., Xiao Z., Wang H., Zhu Y. Tumor suppressor miR-145-5p sensitizes prolactinoma to bromocriptine by downregulating TPT1. J. Endocrinol. Investig. 2019;42:639–652. doi: 10.1007/s40618-018-0963-4. [DOI] [PubMed] [Google Scholar]
- 40.Qiu P., Xu T.J., Lu X.D., Yang W., Zhang Y.B., Xu G.M. MicroRNA-378 regulates cell proliferation and migration by repressing RNF31 in pituitary adenoma. Oncol. Lett. 2018;15:789–794. doi: 10.3892/ol.2017.7431. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Shen D.-W., Li Y.-L., Hou Y.-J., Xu Z.-D., Li Y.-Z., Chang J.-Y. MicroRNA-543 promotes cell invasion and impedes apoptosis in pituitary adenoma via activating the Wnt/β-catenin pathway by negative regulation of Smad7. Biosci. Biotechnol. Biochem. 2019;83:1035–1044. doi: 10.1080/09168451.2019.1591260. [DOI] [PubMed] [Google Scholar]
- 42.Lee Y.J., Cho J.M., Moon J.H., Ku C.R., Kim J., Kim S.H., Lee E.J. Increased miR-338-3p expression correlates with invasiveness of GH-producing pituitary adenomas. Endocrine. 2017;58:184–189. doi: 10.1007/s12020-017-1390-6. [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Fang Y., Zhou Y., Guo X., Xu K., Li C., Zhang J., Hong Y. SDF-1α/MicroRNA-134 axis regulates nonfunctioning pituitary neuroendocrine tumor growth via targeting VEGFA. Front. Endocrinol. 2020;11 doi: 10.3389/fendo.2020.566761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su W., Wang J., Ye M., Chen W., Liao C. Expression and clinical significance of miR-193a-3p in invasive pituitary adenomas. Eur. Rev. Med. Pharmacol. Sci. 2020;24:7673–7680. doi: 10.26355/eurrev_202007_22268. [DOI] [PubMed] [Google Scholar]
- 45.He C., Yang J., Ding J., Li S., Wu H., Zhou F., Teng L., Yang J. MiR-448 targets BLC2 and inhibits the growth of pituitary adenoma cells. Biochem. Cell Biol. 2020;98:511–517. doi: 10.1139/bcb-2019-0336. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., Zhao J., Zhang C., Wang P., Huang C., Peng H. MiR-219a-2-3p suppresses cell proliferation and promotes apoptosis by targeting MDM2/p53 in pituitary adenomas cells. Biosci. Biotechnol. Biochem. 2020;84:911–918. doi: 10.1080/09168451.2020.1715780. [DOI] [PubMed] [Google Scholar]
- 47.Xiao Z., Wang Z., Hu B., Mao Z., Zhu D., Feng Y., Zhu Y. MiR-1299 promotes the synthesis and secretion of prolactin by inhibiting FOXO1 expression in drug-resistant prolactinomas. Biochem. Biophys. Res. Commun. 2019;520:79–85. doi: 10.1016/j.bbrc.2019.09.070. [DOI] [PubMed] [Google Scholar]
- 48.Grzywa T.M., Klicka K., Rak B., Mehlich D., Garbicz F., Zieliński G., Maksymowicz M., Sajjad E., Włodarski P.K. Lineage-dependent role of miR-410-3p as oncomiR in gonadotroph and corticotroph pituitary adenomas or tumor suppressor miR in somatotroph adenomas via MAPK, PTEN/AKT, and STAT3 signaling pathways. Endocrine. 2019;65:646–655. doi: 10.1007/s12020-019-01960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lei C., Jing G., Jichao W., Xiaohui L., Fang Q., Hua G., Yazhou M., Zhang Y. MiR-137’s tumor suppression on prolactinomas by targeting MITF and modulating Wnt signaling pathway. J. Clin. Endocrinol. Metab. 2019;104:6391–6402. doi: 10.1210/jc.2018-02544. [DOI] [PubMed] [Google Scholar]
- 50.Hu A., Zhang Y., Zhao X., Li J., Ying Y. CBX1 is a direct target of miR-205-5p and contributes to the progression of pituitary tumor. Die Pharm. Int. J. Pharm. Sci. 2019;74:154–156. doi: 10.1691/ph.2019.8908. [DOI] [PubMed] [Google Scholar]
- 51.Gao Z.F., Ji X.L., Gu J., Wang X.Y., Ding L., Zhang H. microRNA-107 protects against inflammation and endoplasmic reticulum stress of vascular endothelial cells via KRT1-dependent Notch signaling pathway in a mouse model of coronary atherosclerosis. J. Cell. Physiol. 2019;234:12029–12041. doi: 10.1002/jcp.27864. [DOI] [PubMed] [Google Scholar]
- 52.Cai F., Dai C., Chen S., Wu Q., Liu X., Hong Y., Wang Z., Li L., Yan W., Wang R. CXCL12-regulated miR-370–3p functions as a tumor suppressor gene by targeting HMGA2 in nonfunctional pituitary adenomas. Mol. Cell. Endocrinol. 2019;488:25–35. doi: 10.1016/j.mce.2019.02.020. [DOI] [PubMed] [Google Scholar]
- 53.Lu B., Liu G.-L., Yu F., Li W.-J., Xiang X.-X., Xiao H.-Z. MicroRNA-16/VEGFR2/p38/NF-κB signaling pathway regulates cell growth of human pituitary neoplasms. Oncol. Rep. 2018;39:1235–1244. doi: 10.3892/or.2018.6227. [DOI] [PubMed] [Google Scholar]
- 54.Yang W., Xu T., Qiu P., Xu G. Caveolin-1 promotes pituitary adenoma cells migration and invasion by regulating the interaction between EGR1 and KLF5. Exp. Cell Res. 2018;367:7–14. doi: 10.1016/j.yexcr.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 55.He W., Huang L., Li M., Yang Y., Chen Z., Shen X. MiR-148b, MiR-152/ALCAM axis regulates the proliferation and invasion of pituitary adenomas cells. Cell. Physiol. Biochem. 2017;44:792–803. doi: 10.1159/000485342. [DOI] [PubMed] [Google Scholar]
- 56.Zhen W., Qiu D., Zhiyong C., Xin W., Mengyao J., Dimin Z., Chonghui H., Haijun W., Yonghong Z. MicroRNA-524-5p functions as a tumor suppressor in a human pituitary tumor-derived cell line. Horm. Metab. Res. 2017;49:550–557. doi: 10.1055/s-0043-106437. [DOI] [PubMed] [Google Scholar]
- 57.Zhao Z., Li M., Shi P., Zhang P. MiRNA153 induces pituitary tumor MMQ cell line apoptosis through down-regulating Skp protein expression. Eur. Rev. Med. Pharmacol. Sci. 2017;21:1270–1275. [PubMed] [Google Scholar]
- 58.Zheng Z., Zhang Y., Zhang Z., Yang Y., Song T. Effect of miR-106b on invasiveness of pituitary adenoma via PTEN-PI3K/AKT. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2017;23:1277. doi: 10.12659/MSM.900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu C., Li J., Sun F., Cui J., Fang H., Sui G. Expression and clinical significance of miR-26a and pleomorphic adenoma gene 1 (PLAG1) in invasive pituitary adenoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016;22:5101. doi: 10.12659/MSM.898908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang D., Zhang H., Zhang B., Yuan Z., Yu Z., Yang T., Zhang S., Liu Y., Jia X. miR-133 inhibits pituitary tumor cell migration and invasion via down-regulating FOXC1 expression. Genet. Mol. Res. 2016;15:1–10. doi: 10.4238/gmr.15017453. [DOI] [PubMed] [Google Scholar]
- 61.Sureshbabu A., Muhsin S.A., Choi M.E. TGF-β signaling in the kidney: Profibrotic and protective effects. Am. J. Physiol. Ren. Physiol. 2016;310:F596–F606. doi: 10.1152/ajprenal.00365.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du Q., Zhang W., Feng Q., Hao B., Cheng C., Cheng Y., Li Y., Fan X., Chen Z. Comprehensive circular RNA profiling reveals that hsa_circ_0001368 is involved in growth hormone-secreting pituitary adenoma development. Brain Res. Bull. 2020;161:65–77. doi: 10.1016/j.brainresbull.2020.04.018. [DOI] [PubMed] [Google Scholar]
- 63.Du Q., Hu B., Feng Y., Wang Z., Wang X., Zhu D., Zhu Y., Jiang X., Wang H. circOMA1-mediated miR-145-5p suppresses tumor growth of nonfunctioning pituitary adenomas by targeting TPT1. J. Clin. Endocrinol. Metab. 2019;104:2419–2434. doi: 10.1210/jc.2018-01851. [DOI] [PubMed] [Google Scholar]
- 64.Wiemels J., Wrensch M., Claus E.B. Epidemiology and etiology of meningioma. J. Neurooncol. 2010;99:307–314. doi: 10.1007/s11060-010-0386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li T., Ren J., Ma J., Wu J., Zhang R., Yuan H., Han X. LINC00702/miR-4652-3p/ZEB1 axis promotes the progression of malignant meningioma through activating Wnt/β-catenin pathway. Biomed. Pharmacother. 2019;113:108718. doi: 10.1016/j.biopha.2019.108718. [DOI] [PubMed] [Google Scholar]
- 66.Ding C., Yi X., Xu J., Huang Z., Bu X., Wang D., Ge H., Zhang G., Gu J., Kang D. Long Non-Coding RNA MEG3 Modifies Cell-Cycle, Migration, Invasion, and Proliferation Through AKAP12 by Sponging miR-29c in Meningioma Cells. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.537763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Negroni C., Hilton D.A., Ercolano E., Adams C.L., Kurian K.M., Baiz D., Hanemann C.O. GATA-4, a potential novel therapeutic target for high-grade meningioma, regulates miR-497, a potential novel circulating biomarker for high-grade meningioma. EBioMedicine. 2020;59:102941. doi: 10.1016/j.ebiom.2020.102941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song L.-R., Li D., Weng J.-C., Li C.-B., Wang L., Wu Z., Zhang J.-T. MicroRNA-195 functions as a tumor suppressor by directly targeting fatty acid synthase in malignant meningioma. World Neurosurg. 2020;136:e355–e364. doi: 10.1016/j.wneu.2019.12.182. [DOI] [PubMed] [Google Scholar]
- 69.Katar S., Baran O., Evran S., Cevik S., Akkaya E., Baran G., Antar V., Hanimoglu H., Kaynar M.Y. Expression of miRNA-21, miRNA-107, miRNA-137 and miRNA-29b in meningioma. Clin. Neurol. Neurosurg. 2017;156:66–70. doi: 10.1016/j.clineuro.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 70.Werner T.V., Hart M., Nickels R., Kim Y.-J., Menger M.D., Bohle R.M., Keller A., Ludwig N., Meese E. MiR-34a-3p alters proliferation and apoptosis of meningioma cells in vitro and is directly targeting SMAD4, FRAT1 and BCL2. Aging Albany N. Y. 2017;9:932. doi: 10.18632/aging.101201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dalan A.B., Gulluoglu S., Tuysuz E.C., Kuskucu A., Yaltirik C.K., Ozturk O., Ture U., Bayrak O.F. Simultaneous analysis of miRNA-mRNA in human meningiomas by integrating transcriptome: A relationship between PTX3 and miR-29c. BMC Cancer. 2017;17:1–9. doi: 10.1186/s12885-017-3198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H., Zhao J. let-7d suppresses proliferation and invasion and promotes apoptosis of meningioma by targeting AEG-1. Onco Targets Ther. 2017;10:4895. doi: 10.2147/OTT.S141008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng J., Pang C.h., Du W., Wang L., Sun L.g., Xing Z.y. An allele of rs619586 polymorphism in MALAT1 alters the invasiveness of meningioma via modulating the expression of collagen type V alpha (COL5A1) J. Cell. Mol. Med. 2020;24:10223–10232. doi: 10.1111/jcmm.15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lim J., Kim J.O., Park H.S., Han I.B., Kwack K., Kim N.K., Cho K. Associations of miR-146aC> G, miR-149C> T, miR-196a2C> T and miR-499A> G polymorphisms with brain tumors. Oncol. Rep. 2018;40:1813–1823. doi: 10.3892/or.2018.6557. [DOI] [PubMed] [Google Scholar]
- 75.Da M., Jiang H., Xie Y., Jin W., Han S. The biological roles of exosomal long non-coding RNAs in cancers. OncoTargets Ther. 2021;14:271. doi: 10.2147/OTT.S281175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghafouri-Fard S., Agabalazadeh A., Abak A., Shoorei H., Hassanzadeh Taheri M.M., Taheri M., Sharifi G. Role of Long Non-Coding RNAs in Conferring Resistance in Tumors of the Nervous System. Front. Oncol. 2021;11:2054. doi: 10.3389/fonc.2021.670917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sahm F., Schrimpf D., Stichel D., Jones D.T.W., Hielscher T., Schefzyk S., Okonechnikov K., Koelsche C., Reuss D.E., Capper D., et al. DNA methylation-based classification and grading system for meningioma: A multicentre, retrospective analysis. Lancet Oncol. 2017;18:682–694. doi: 10.1016/S1470-2045(17)30155-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author upon reasonable request.