Abstract
The transcriptional activity of nuclear receptors is mediated by coactivator proteins, including steroid receptor coactivator 1 (SRC1) and its homologues and the general coactivators CREB binding protein (CBP) and p300. SRC1 contains an activation domain (AD1) which functions via recruitment of CBP and and p300. In this study, we have used yeast two-hybrid and in vitro interaction-peptide inhibition experiments to map the AD1 domain of SRC1 to a 35-residue sequence potentially containing two α-helices. We also define a 72-amino-acid sequence in CBP necessary for SRC1 binding, designated the SRC1 interaction domain (SID). We show that in contrast to SRC1, direct binding of CBP to the estrogen receptor is weak, suggesting that SRC1 functions primarily as an adaptor to recruit CBP and p300. In support of this, we show that the ability of SRC1 to enhance ligand-dependent nuclear receptor activity in transiently transfected cells is dependent upon the integrity of the AD1 region. In contrast, the putative histone acetyltransferase domain, the Per-Arnt-Sim basic helix-loop-helix domain, the glutamine-rich domain, and AD2 can each be removed without loss of ligand-induced activity. Remarkably, a construct corresponding to residues 631 to 970, which contains only the LXXLL motifs and the AD1 region of SRC1, retained strong coactivator activity in our assays.
The nuclear receptors (NRs) are ligand-regulated transcription factors that mediate the effects of steroids, retinoids, and other lipophilic hormones on gene expression (32). In common with other transcriptional activators, NRs stimulate transcription by promoting the local modification of chromatin structure and recruitment of a preinitiation complex (59). This is achieved via two transcriptional activation functions (AF1 and AF2) which provide molecular surfaces for the recruitment of transcriptional coactivator proteins (17, 28, 36, 60).
The AF2 surfaces of the ligand binding domains (LBDs) of NRs appear to be the principal sites for coactivator recruitment. Far-Western experiments detected two major classes of proteins in nuclear extracts (with apparent molecular masses of 160 and 140 kDa) which bind to the LBD of the estrogen receptor (ER) in the presence of ligand (5, 14). At least three distinct p160 proteins have been identified, including steroid receptor coactivator 1 (SRC1) (39), transcription intermediary factor 2 (TIF2) (54) and its murine homologue GRIP1 (18), and p300–CBP cointegrator-associated protein (pCIP) (50), which is the mouse homologue of the human protein AIB1 (1), also known as ACTR (8), RAC3 (29), or TRAM1 (49). These proteins appear to be bona fide coactivators, as they enhance the activity of NRs in both in vitro and in vivo experimental systems. The p140 class appears to consist chiefly of the nuclear protein RIP140 (6). The function of RIP140 is unknown, although it has been shown to down-regulate NR-mediated transcription in transient-reporter assays, possibly via competition with p160s for the LBD (15, 27, 35, 51). Other AF2 binding proteins of different apparent molecular weights have also been identified by alternative approaches (13). The thyroid receptor-associated protein (TRAP) complex (12) and the very similar vitamin D receptor-interacting protein (DRIP) complex (44) have been shown to be important for the transcriptional activity of NRs in vitro. These contain mammalian homologues of the SRB and MED proteins and are related to the yeast Mediator complex, which is required for activated transcription (19). PGC-1 is a cold-inducible coactivator required for the function of peroxisome proliferator-activated receptor γ (PPARγ) in adaptive thermogenesis and is highly expressed in brown adipose tissue and skeletal muscle (41). Other AF2 binding proteins, such as the mouse SUG1 (56) and transcriptional intermediary factor 1 (TIF1) (25) may not have a direct role in transcriptional activation by this domain.
We and others have shown that interaction of the p140 and p160 proteins with the LBD are mediated by the LXXLL motif (16, 50). This sequence forms part of an amphipathic α-helix, which binds in a conserved hydrophobic cleft on the surface of liganded LBDs (37). The TRAP-DRIP complex has been shown to bind NRs via the TRAP220-DRIP205 component, which contains two LXXLL motifs (43, 63). Similarly, PGC-1 interaction with PPARγ is mediated by LXXLL motifs (52). CREB binding protein (CBP) and p300 have been reported to interact directly with retinoid receptors (7, 22) and PPARs (11). However, as shown here and in other studies (30, 34, 40, 41; D. M. Heery, S. Hoare, S. Hussain, M. G. Parker, and H. M. Sheppard, submitted for publication), this interaction is far weaker than the binding of p160s with NRs. Nonetheless, we have demonstrated that these weak interactions are mediated by LXXLL sequences close to the N and C termini of CBP and p300 (16; Heery et al., unpublished). In addition, the p300-CBP-associated factor (PCAF) has been reported to bind directly to NRs in a ligand-independent manner involving the DNA binding domain (DBD) (4).
CBP, p300, and PCAF have each been shown to possess histone acetyltransferase (HAT) activities (2, 38, 61). The isolated HAT domains of these proteins activate transcription when fused to a heterologous DBD, and this activity is dependent on the HAT function (33). Mutations that disrupt the HAT activity of p300 or CBP abrogate the ability of these coactivators to enhance transcription mediated by ER (24) or TR-RXR (30) on reconstituted chromatin templates in vitro. SRC1 and ACTR have also been reported to possess HAT activity (8, 47). In our hands, under conditions where CBP or PCAF HAT activities are readily observed, SRC1 HAT activity was below the limit of detection. Similarly, Voegel et al. (53) were unable to detect HAT activity associated with TIF2. In contrast to the HAT domains of CBP and PCAF, the sequence encoding the proposed HAT domain of SRC1 did not activate transcription when fused to a GAL4 DBD (21). To our knowledge, it has not been demonstrated that mutations in the SRC1 HAT region affect the ability of this protein to enhance NR-mediated transcription. In addition, experiments in which antibodies were used to block specific HAT activities in microinjected cell lines suggested different requirements for the HAT activities of CBP and PCAF, but not SRC1, for the transcriptional activities of RAR, STAT-1, and CREB on different promoters (23). Thus, the role of the SRC1 HAT activity in NR-mediated transcription remains to be clarified.
SRC1 and CBP are known to associate both in vitro and in vivo (22, 50, 62). The SRC1 sequences required for this interaction have been mapped to amino acids 896 to 1200 (23), 788 to 980 (21), and 900 to 990 (34) of SRC1. This region colocalizes with the potent transcriptional activation domain AD1, which has been shown to be CBP dependent (8, 21, 23, 50, 53). A number of studies have underlined the functional importance of p160-p300 interactions. It has been shown that deletion of amino acids 1018 to 1088 in ACTR, which include the CBP interaction domain AD1, negates the ability of ACTR to stimulate glucocorticoid receptor-mediated transcription in transiently transfected cells (8). Similarly, it has been shown that a deletion in the region encoding SRC1 AD1 (amino acids 900 to 950) abrogated the ability of SRC1 to enhance ligand-dependent transcription by the androgen receptor (AR) (3). However, the AR is somewhat atypical of NRs in that the majority of its transcriptional activity is associated with the N-terminal AF1 domain. A mutant form of CBP containing a deletion in the p160 binding region (amino acids 2098 to 2163) inhibited RAR-mediated transcription in microinjected cells (34). More recently, similar deletions in p300 were shown to abolish its ability to enhance NR-mediated transcription in in vitro transcription assays (24, 30). In another approach, microinjection of mammalian cells with affinity-purified antibodies against pCIP reduced NR-mediated transcription, which was only relieved by coinjection of vectors expressing both pCIP and CBP (50). CBP-p300 has been shown to acetylate ACTR at a lysine residue adjacent to an LXXLL motif, resulting in the dissociation of ACTR from the LBD (9). Thus, it has been suggested that CBP may both facilitate and attenuate NR-mediated transcription. In support of this, it has been shown that estradiol-induced histone hyperacetylation is transient in vivo, reaching a peak 1 to 3 h after hormone induction and being strongly down-regulated thereafter (9). Thus, there is substantial evidence indicating that the interaction of p160s with CBP-p300 is critical for NR-mediated transcription.
The p160 coactivators contain several other functional domains, including a basic helix-loop-helix Per-Arnt-Sim (PAS) domain (22, 62), a central NR interaction domain (NID) containing three LXXLL motifs (16, 50, 53), and a glutamine-rich sequence implicated in binding the ligand-independent AF1 domains of NRs (3, 31, 57). A second activation domain located close to the C termini of the p160s (AD2; amino acids 1240 to 1345 in SRC1) has recently been shown to bind CARM1, a protein with arginine methyltransferase activity that methylates histones in vitro (10). The goal of this study was to map the SRC1-CBP interaction interface in detail and to investigate the importance of the different functional domains of SRC1 for its coactivator function.
MATERIALS AND METHODS
Plasmids and strains.
The following plasmids used in transient-transfection experiments have been described previously: pSG5-SRC1e and p3ERE-TATA-LUC (21); pSG5-SRC1eΔAD1, pSG5-SRC1(1-1240), pSG5-SRC1(1-1100), and pSG5-SRC1(1-1100) (3); and pSG424 (46) and pGAL4-RXR (21). pGAL4-E16ΔLUC was a gift from M. Dickens. pGAL4-SRC 926-970 was created by cloning a PCR fragment into pSG424. The deletion construct pSG5-SRC1(631-970) was created by cloning a PCR fragment into a modified version of the cloning vector pSG5. PCR was also used to generate pSG5-SRC1(626-970) constructs containing mutations in the LXXLL motifs, using appropriate template DNA as described in Kalkhoven et al. (21). All PCR fragments were amplified with Elongase enzyme mix (Gibco BRL) and verified by sequence analysis.
For in vitro glutathione S-transferase (GST) pull-down assays, the control GST was a modified version of pGEX2TK empty vector (Pharmacia). The constructs GST-CBP (referred to here as GST–CBP-C), GST-AF2 (16), and GST-AD1 (21) have been described previously. pGEXm2TK-AF1MOR (referred to here as GST-AF1) was a gift from M. Parker. The plasmids pSG5-hSRC1e, pSG5-mCBP 1891-2441 (21), pCI-PCAF (a gift from Y. Nakatani), and pBSSK-HA-CBPFL (a gift from A. Bannister and T. Kouzarides) were used to produce 35S-labeled in vitro-translated proteins. Plasmids for use in yeast two-hybrid analysis were constructed as follows. PCR fragments flanked by appropriate restriction enzyme sites were cloned in frame with the LexA DBD or the VP16 acidic activation domain. The vectors encoding the domains were modified versions of pBTM116 (55) and pASV3 (26), respectively. All constructs were verified by sequencing, and expression of the fusion proteins in yeast was monitored by Western blotting using antibodies raised against VP16 or LexA (Santa Cruz Biotechnology). The Saccharomyces cerevisiae strain L40 (55) was used for yeast two-hybrid experiments.
GST pull-down assays.
Recombinant cDNAs in the pSG5 or pBS expression vectors were transcribed and translated in vitro in reticulocyte lysate (Promega) in the presence of [35S]methionine. GST fusion proteins were expressed in Escherichia coli and purified on glutathione-Sepharose beads (Pharmacia). Expression levels were verified by separating the proteins on sodium dodecyl sulfate (SDS)-polyacrylamide gels that were subjected to Coomassie blue staining. GST fusion proteins were incubated with 35S-radiolabeled protein as described previously (21). GST-AF1 or GST-AF2 experiments were performed in the presence of 10−6 M 17-β-estradiol (E2) or vehicle. The beads were washed three times, and bound proteins were separated on SDS–10% polyacrylamide gels, which were subsequently fixed, treated with Amplify (Amersham Life Sciences), and vacuum dried. Radiolabeled proteins were visualized by autoradiography. Peptide competition assays were performed as described previously using full-length CBP (16).
Immunoprecipitation assays.
Recombinant cDNAs in the pSG5 or pBS expression vectors were transcribed and translated in vitro in reticulocyte lysate (Promega) in the presence of [35S]methionine. The proteins were incubated with 5 μl of anti-hemagglutinin (HA) tag antibody (F-7; Santa Cruz Biotechnology) and 20 μl of protein A-protein G PLUS agarose beads (Santa Cruz Biotechnology) in 500 μl of NETN buffer (20 mM Tris [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40) for 3 h at 4°C. The beads were washed three times, and bound proteins were separated on SDS–10% polyacrylamide gels, which were subsequently fixed, treated with Amplify, and vacuum dried. Radiolabeled proteins were visualized by autoradiography.
Cell culture and transient transfections.
Cos-1 and HeLa cells were routinely maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Twenty-four hours prior to transfection, the cells were plated in six-well plates (Helena Biosciences) in phenol red-free Dulbecco's modified Eagle's medium supplemented with 5% dextran charcoal-stripped fetal calf serum. The cells were transfected by calcium phosphate (Clontech) coprecipitation according to the manufacturer's instructions. The transfected DNA included pJ7-lacZ control plasmid (500 ng/well), p3ERE-TATA-LUC (1 μg), or pGAL4-E1bΔ (500 ng) reporter plasmids with either pMT-MOR (100 ng) or GAL4-RXR (250 ng) NR expression plasmids and SRC1 constructs (500 ng) or empty vector. After 16 h, fresh medium containing either vehicle, 10−8 M estradiol (E2), or 10−7 M 9-cis retinoic acid was added. After a further 24 h, the cells were harvested; extracts were assayed for luciferase activity, using a Luciferase Assay System (Promega); and values were normalized relative to β-galactosidase activity, which was measured with a Galacto-Light chemiluminescent assay (Tropix).
RESULTS
The CBP interaction domain (AD1) of SRC1.
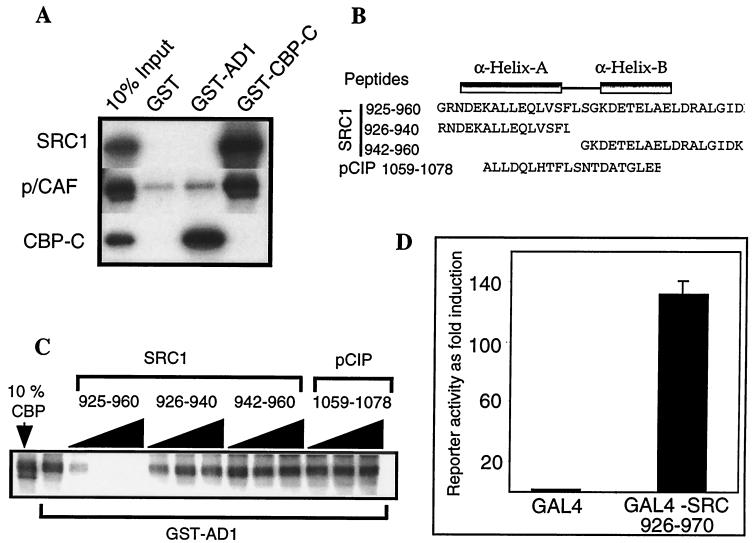
AD1 and the CBP interaction domain have been shown to be coincident in SRC1, ACTR, pCIP, and TIF2 (8, 21, 50, 53). AD1 has been mapped to amino acids 1041 to 1106 in TIF2 (53) and 1039 to 1088 in ACTR (8). At the outset of this study, the CBP interaction domain of SRC1 had been localized to amino acids 781 to 988 (21). Therefore, we used the yeast two-hybrid system in order to define the boundaries of this domain in SRC1 more precisely (Fig. 1A). A C-terminal fragment of CBP (CBP-C; amino acids 1891 to 2165), which is known to bind p160s (22, 62), was fused to the LexA DBD and tested for interaction with a series of SRC1 fragments fused to the VP16 activation domain (VP16 AD [Fig. 1A]). While VP16 AD alone did not interact with CBP-C, fusion of VP16 AD with amino acids 867 to 990 (data not shown) or 900 to 990 of SRC1 induced a strong interaction with the bait. This region of SRC1 spans a sequence predicted to fold into two α-helical structures (referred to as helices A and B) and is well conserved among the known p160 proteins (Fig. 1B). The minimum region required for this interaction was contained within amino acids 926 to 970. No binding was observed if either helix A (constructs 940-990 and 940-970) or helix B (constructs 900-940 and 911-940) was deleted. In addition, the presence of a complete helix A and a partial helix B (constructs 867-950 and 900-950) was not sufficient to maintain interaction with CBP.
FIG. 1.
The CBP interaction domain in SRC1 maps to amino acids 926 to 970. (A) The interaction between SRC1 and a C-terminal region of CBP was examined in a yeast two-hybrid system. S. cerevisiae L40 was cotransformed with LexA-CBP 1982-2163 and the plasmid pASV3 expressing the acidic activation domain (411 to 490) of VP16 (VP16 AD), or a series of SRC1 constructs were fused in frame with VP16 AD. The SRC1 sequences are represented schematically by grey rectangles, and the putative helices A and B are depicted by black boxes. Reporter activity in cell extracts is expressed in terms of units of β-galactosidase activity. The results from a representative experiment are shown, and similar results were obtained in triplicate experiments. Western blots using anti-LexA and anti-VP16 AD antibodies (Santa Cruz) confirmed that the levels of the bait protein did not vary significantly between different clones and that the levels of VP16 AD1 were similar in all transformants. (B) Sequence alignment of the CBP interaction domain in SRC1 with the corresponding regions in TIF2 and pCIP. The positions of the predicted α-helices and the CBP interaction domain as identified in panel A are indicated. Identical residues present in all three proteins are boxed.
To confirm the yeast two-hybrid data, the interaction between AD1 and CBP was tested in vitro using GST pull-down experiments. A GST-SRC1 fusion protein containing AD1 and spanning amino acids 781 to 988 of SRC1 strongly interacted with 35S-labeled CBP-C (amino acids 1891 to 2165), as shown previously (21) (Fig. 2A). A similar result was obtained by using full-length in vitro-translated CBP (data not shown). In the reciprocal experiment, GST–CBP-C (1891-2165) strongly interacted with 35S-labeled full-length SRC1e. GST–CBP-C also bound strongly to 35S-labeled full-length PCAF as expected (61), but negligible interaction was detected between GST-AD1 and PCAF (Fig. 2A). The control GST alone failed to bind any of the in vitro-translated proteins. In order to confirm the relative importance of the helix A and B sequences of AD1 to the SRC1-CBP interaction, competitive inhibition assays were performed using peptides which corresponded to either both helices (925 to 960), helix A alone (926 to 940), or helix B alone (942 to 960). An additional peptide corresponding to the homologous region of pCIP that partially spans helices A and B was also used (Fig. 2B). The peptide corresponding to helices A and B (925 to 960) effectively competed with CBP for binding to GST-AD1 (Fig. 2C). However, the addition of peptides spanning individual A or B helices, or partially spanning both helices, failed to inhibit the SRC1-CBP interaction, even at high concentrations of peptide. Thus, in agreement with the yeast two-hybrid data, our results suggest that the core CBP binding domain of SRC1 contains two putative α-helices and lies within the sequence 926 to 960. To test whether the core CBP binding domain of SRC1 retained transcriptional activity when tethered to DNA, a vector expressing GAL4 DBD fused to amino acids 926 to 970 of SRC1 was generated and tested in transient-transfection assays (Fig. 2D). Transcription in the presence of GAL4-SRC 926-970 was 130-fold higher than with GAL4 DBD alone, confirming that the core CBP binding domain of SRC1 colocalizes with AD1.
FIG. 2.
Two predicted α-helices in the CBP interaction domain of SRC1 are required in order to maintain interaction with CBP. (A) Glutathione-Sepharose-bound GST, GST-AD1 (781 to 988 of SRC1), and GST–CBP-C (1891 to 2165) were incubated with 35S-labeled full-length SRC1e, pCAF, or CBP-C. Bound proteins were eluted and analyzed by SDS-PAGE and autoradiography. One-tenth of the total labeled protein used in each binding reaction is shown for comparative purposes (10% input). (B) Sequences of synthetic peptides used in competition experiments are shown, with the positions of the predicted helices indicated. (C) Competition experiment showing the effect of increasing concentrations of the competitor peptides on the interaction of GST-AD1 with in vitro-translated 35S-labeled full-length SRC1. (D) Cos-1 cells were transiently transfected as described in Materials and Methods with a GAL4 reporter construct (pGAL4-E16ΔLUC) and 1 μg of vector expressing either GAL4 or the fusion protein GAL4-SRC 926-970. Luciferase activity was measured 48 h later, and the data were normalized to β-galactosidase activity. The activity of GAL4 alone was set at 1, and GAL4-SRC 926-970 activity is expressed relative to it. The values shown represent the average of triplicate samples, and the error bars indicate standard deviation.
The SID of CBP.
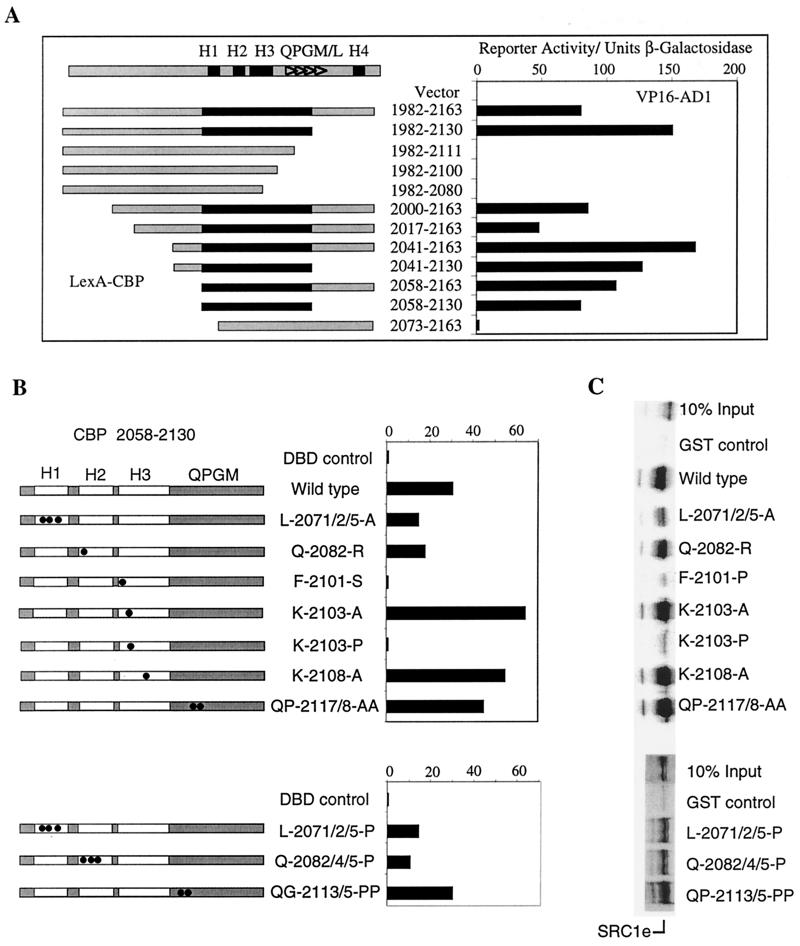
Previous studies have shown that the C-terminal sequence comprising amino acids 2058 to 2163 of CBP is required to bind SRC1 (22). We used the yeast two-hybrid system to confirm this and to analyze the region further. Fragments of CBP fused to the LexA DBD were used as bait and tested for interaction with the AD1 sequence of SRC1 (amino acids 926 to 970) fused to VP16 AD. VP16 AD1 did not interact with LexA DBD but displayed a strong interaction with amino acids 1982 to 2163 of CBP (Fig. 3A). Secondary-structure analysis predicts that this sequence has the potential to fold into four α-helical structures (referred to as H1 to H4). It also contains multiple repeats of the sequence QPGM/L between H3 and H4, although this repeated-motif region is not fully conserved in p300. As shown in Fig. 3A, the minimum region of CBP required for interaction with AD1 mapped to amino acids 2058 to 2130, suggesting that the H4 sequence is not required for the interaction. Deletions which removed all four QPGM/L motifs (CBP 1982 to 2111) or which truncated the H3 (CBP 1982 to 2100) or H3 and H2 (CBP 1982 to 2080) sequences resulted in a complete loss of interaction with SRC1. This suggests that the C-terminal boundary of the SRC1 interaction domain (SID) lies between amino acids 2111 and 2130. Deletion of H1 (construct 2073-2163) also led to a dramatic loss of binding between CBP and SRC1, suggesting that H1 to H3 and three repeats of the QPGM/L motif are necessary and sufficient to maintain an interaction with SRC1. Similar results were obtained in GST pull-down experiments, and in agreement with the yeast two-hybrid data, a strong interaction was observed between GST-CBP 2058-2130 and in vitro-translated full-length SRC1e (Fig. 3C).
FIG. 3.
The SID in CBP maps to amino acids 2058 to 2130. (A) Yeast two-hybrid interactions between VP16 AD1 (SRC1 900-970) and a series of LexA-CBP fusion proteins were assayed as described in the legend to Fig. 1. A schematic representation of the CBP sequence 1982 to 2163 is shown at the top, with the relative positions of four putative α-helices (H1 to H4 [black boxes]) and the QPGM/L repeat sequences (triangles) indicated. CBP sequences are represented schematically below by grey rectangles, whereas the black rectangle denotes the minimal SRC1 interaction domain mapped in these experiments. Reporter activity is expressed in terms of units of β-galactosidase activity, and the results of a representative experiment are shown. Similar results were obtained in triplicate experiments. Western blots confirmed that all LexA constructs were expressed at comparable levels and that VP16 AD1 levels were similar in all cell extracts. (B) Effect of mutations in the SID sequence on its interaction with SRC1 AD1. Yeast two-hybrid interactions between LexA-CBP 2058-2130 mutants and VP16 AD1 were assayed. The LexA-CBP constructs are shown schematically, and the boxed regions represent the relative positions of H1 to H3 and the QPGM/L region. Construct L-2071/2/5-A indicates alteration of leucines at positions 2071, 2072, and 2075 to alanine; similar nomenclature is used for the other constructs. The relative position of each mutation in relation to the predicted α-helices or the QPGM/L motifs is indicated with black circles. Western blots confirmed similar expression of LexA constructs. (C) GST pull-down experiments showing the effects of mutations in the SID on binding of SRC1e. The CBP 2058-2130 fragments identical to those shown in panel B were expressed as GST fusion proteins, and their abilities to bind in vitro-translated 35S-labeled full-length SRC1e were assayed as for Fig. 2A.
After the SID of CBP was mapped to amino acids 2058 to 2130, the relative importance of each of the three potential α-helices within this region was examined by testing the effects of mutations in the SID on its interaction with AD1 in yeast two-hybrid (Fig. 3B) and GST pull-down (Fig. 3C) assays. Mutations in H1 (LLL-2071, 2072, 2075-AAA and LLL-2071, 2072, 2075-PPP) resulted in a reproducible reduction (approximately 50%) in the level of reporter activity in the two-hybrid assay, and a similar reduction in binding was observed in vitro (Fig. 3B and C). Western blot analysis confirmed that wild-type and mutant proteins were expressed at comparable levels in the two-hybrid experiments (data not shown). The H1 region contains an LXXLL motif and mediates a weak interaction of the C terminus of CBP with NRs (Heery et al., submitted). Thus, in the context of the AD1-SID interaction, the conserved leucine residues do not appear to be of critical importance, and their conversion to alanines or prolines results in similar levels of disruption. Mutation of a Q-2082-R in H2 had some effect on the AD1-SID interaction (approximately 60% of wild type). A construct in which the glutamines in H2 at positions 2082, 2084, and 2085 were replaced with prolines (QQQ-2082, 2084, 2085-PPP) retained about 30% reporter activity. However, point mutations in H3, which could potentially disrupt α-helical structure (i.e., K-2101-P, K-2101-S, and K-2103-P), resulted in a complete loss of binding between AD1 and SID, indicating that the H3 region is critical to interaction. In contrast, the mutations K-2103-A and K-2108-A had no adverse effect but appeared to moderately enhance the interaction of SRC1 and CBP in both the yeast two-hybrid and pull-down assays. Mutation of the first QPGM/L repeat to PPPM or of the QP in the second repeat to AA did not affect binding. We conclude that each of the predicted helical regions contributes to the interaction between SRC1 and CBP, with residues in H3 being most critical. In contrast, mutations in the proximal QPGM/L sequences had little impact on the SID-AD1 interaction.
Direct interaction of CBP with ER is weak.
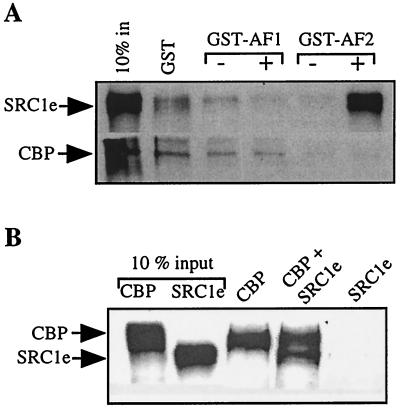
CBP has been reported to interact directly with nuclear receptors (7, 22). To verify whether direct binding of CBP to the ER is important in estrogen signaling, we compared the relative strengths of interaction of in vitro-translated full-length SRC1e and CBP proteins with the AF1 (GST-AF1) and AF2 (GST-AF2) domains of ER (Fig. 4A). There was no significant interaction between SRC1 and AF1. However, a strong ligand-dependent interaction was observed between the LBD of ER (GST-AF2) and SRC1. In contrast, interaction between CBP and either the AF1 or AF2 domain of ER was barely detectable in the presence or absence of ligand. These results are in agreement with yeast two-hybrid data, which indicate that reporter activities due to the interaction between fragments of SRC1 and the LBD of ER or other NRs are significantly stronger (approximately 100-fold) than those between fragments of CBP and ER (Heery et al., unpublished). Although interaction between ER and fragments of CBP, i.e., amino acids 1 to 101, was detected in GST pull-down assays, as reported by Kamei et al. (22), this binding was significantly less sensitive to ligand than ER-SRC1 interactions (data not shown). Immunoprecipitation assays were performed with the in vitro-translated full-length proteins which had been used in the GST pull-down assays described above (Fig. 4B). Anti-HA tag antibody specifically interacted with HA-tagged CBP and did not interact with SRC1 alone (Fig. 4B, compare lanes CBP and SRC1e). However, when incubated together, both CBP and SRC1 were immunoprecipitated, suggesting that although CBP interaction with ER is barely detectable, there is a strong interaction between CBP and SRC1 in vitro. Taken together, our results strongly suggest that SRC1 or other p160s are required to recruit CBP-p300 to ER-regulated promoters.
FIG. 4.
Direct binding of CBP to the ER is weak. (A) GST pull-down assays were performed as described in the legend to Fig. 2A. Glutathione-Sepharose-bound GST, GST-AF1 (containing the ligand-independent activation function of ER), or GST-AF2 (containing the ligand-dependent activation function of ER) was incubated with 35S-labeled full-length SRC1e or CBP in the presence of 10−6 M E2 or vehicle as indicated. Bound proteins were eluted and analyzed by SDS-PAGE and autoradiography. Ten percent of the total labeled protein used in each binding reaction is shown for comparative purposes (10% in). (B) Immunoprecipitation assays were performed by incubating anti-HA tag antibody and protein A-protein G agarose beads with 35S-labeled full-length CBP, which contains an N-terminal HA tag, and/or SRC1e as indicated. Bound proteins were eluted and analyzed by SDS-PAGE and autoradiography. Ten percent of the total labeled protein used in each reaction is shown for comparative purposes (10% input).
SRC1 coactivator function requires AD1.
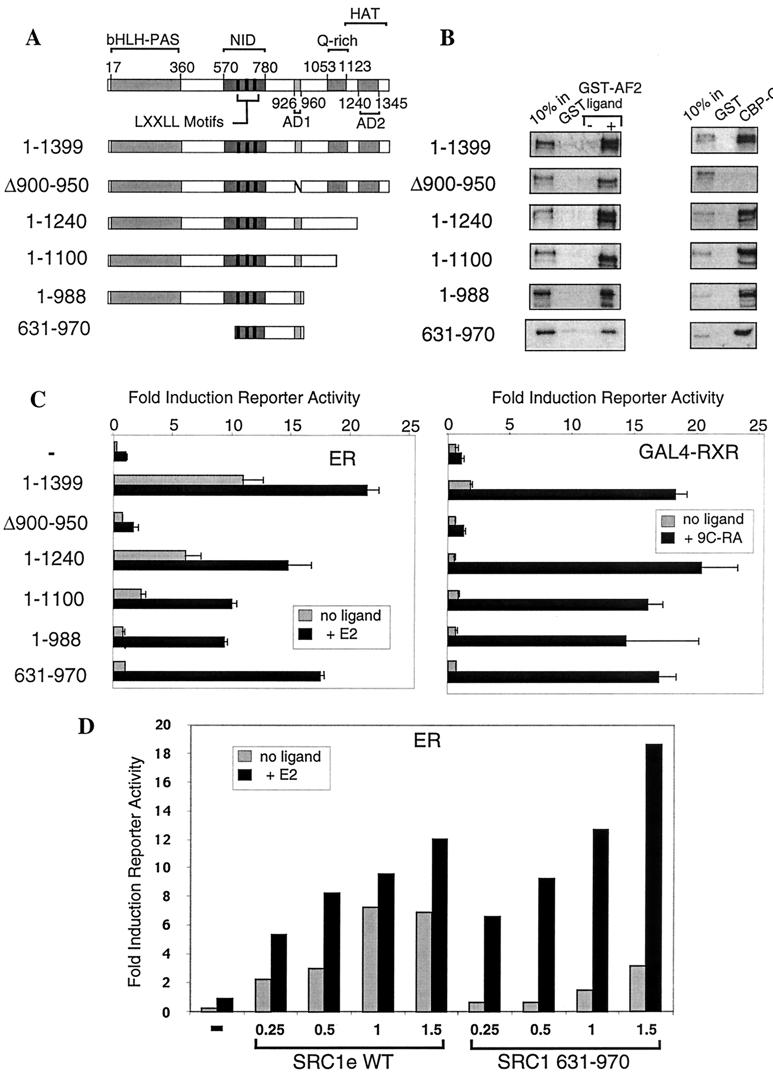
Having demonstrated that CBP interaction with ER is most likely indirect and mediated by SRC1, our next aim was to determine the importance of the different domains of SRC1 to its function as a coactivator of NR activity. To achieve this, a series of mutant SRC1e proteins was constructed in which various functional domains were fully or partially deleted (Fig. 5A). All constructs contained the NID and were capable of ligand-dependent binding to GST-AF2 (Fig. 5B). We also confirmed that each construct was capable of binding GST–CBP-C, with the exception of SRC1 Δ900-950, in which the AD1 sequence is deleted (Fig. 5B).
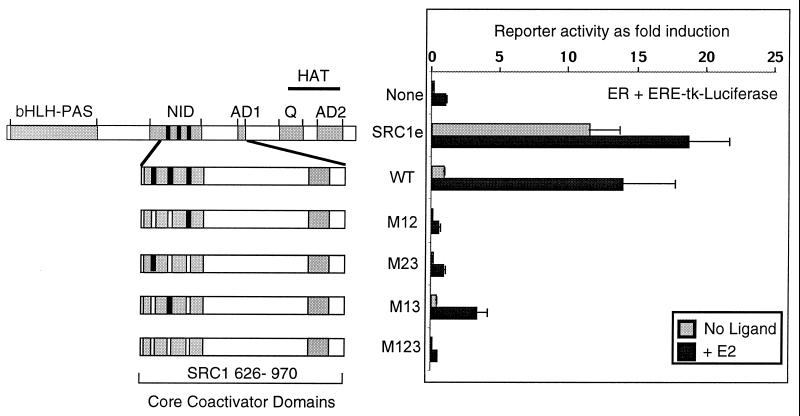
FIG. 5.
AD1 is necessary and, when fused to the NID, sufficient to maintain the coactivator potential of SRC1 in transient-transfection assays. (A) Schematic diagram of the functional domains (indicated by grey boxes) identified in SRC1 and the deletion constructs used in subsequent experiments: bHLH-PAS, sequence similarity with basic helix-loop-helix and Per-Arnt-Sim motifs; NID, with LXXLL motifs indicated by black bars; AD1 and AD2, two autonomous activation domains; Q-rich, a glutamine-rich sequence; and a putative HAT domain. (B) GST pull-down assays performed as described in the legend to Fig. 2A. Glutathione-Sepharose-bound GST, GST-AF2 (containing the ligand-dependent activation function of ER), and GST–CBP-C (1891 to 2165, containing the SID) were incubated with 35S-labeled SRC1, full length or with deletions as indicated. Ligand (10−6 M; E2) or vehicle was added to the binding reaction mixture as appropriate. (C) Cos-1 cells were transiently transfected as described in Materials and Methods with appropriate reporter constructs, a vector expressing either ER or GAL4-RXR, and SRC1e, full-length (1 to 1399) or with deletion mutants, as indicated. Luciferase activity was measured 24 h after the addition of appropriate ligand, and the data were normalized to β-galactosidase activity. The activity of each NR in the presence of ligand was set at 1 for each experiment, and other values are expressed relative to it. The values shown represent the average of triplicate samples, and the error bars indicate standard deviation. These results are representative of experiments performed at least three times. (D) Titration experiment to determine whether luciferase reporter activities induced by SRC1e wild type (WT) or SRC1 631-970 as shown in panel C are at saturating levels. Cos-1 cells were transiently transfected as in panel C using the estrogen-responsive ERE reporter, internal control reporter, ER expression vector, and increasing amounts (expressed in micrograms) of SRC1e wild type or SRC1 631-970 expression vector as indicated. A representative experiment is shown, and similar results were obtained in three replications.
The ability of the SRC1 deletion mutants to function as coactivators of NR activity was investigated in transiently transfected Cos-1 cells expressing full-length ER or a GAL4-RXR with appropriate reporter genes (Fig. 5C). Full-length SRC1e potentiated ER activity both in the presence and absence of ligand (Fig. 5C, left), as previously shown (21). A C-terminal deletion resulting in loss of AD2 and part of the HAT domain (construct 1-1240) retained strong coactivator function. Similarly, the construct SRC1 1-1100, in which AD2, the HAT domain, and part of the Q-rich domain are deleted, strongly enhanced the ligand-dependent activity of full-length ER in these assays. However, a significantly reduced enhancement of the ligand-independent activity of ER was observed with this construct. Further truncation deleting the entire Q-rich domain (SRC1 1-988) resulted in almost complete loss of ligand-independent enhancement of ER activity. This indicates that the Q-rich domain of SRC1, located between amino acids 1053 and 1123, mediates the SRC1 enhancement of ER ligand-independent activity, consistent with previous observations that this is the case for steroid receptors (3, 31, 57). In contrast, enhancement of the ligand-dependent ER activity was largely unaffected. Remarkably, the construct SRC1 631-970, in which both the N- and C-terminal regions of SRC1 are deleted, enhanced ligand-dependent ER activity to almost the same level as that of full-length SRC1e (Fig. 5C and D). The only known domains in this construct are the NID and AD1. This result suggests that the principal role of SRC1 in these experiments is to recruit CBP-p300, or other AD1 binding proteins, to the NR dimers. A deletion that truncates AD1 (SRC1 Δ900-950) resulted in a complete abrogation of both ligand-dependent and ligand-independent activities of ER. In addition, we found that this protein behaves as a dominant negative in a dose-dependent manner, which reduces ER activity to levels below that seen in the absence of added SRC1 (data not shown). The SRC1 mutants gave similar results on other estrogen-responsive reporters and in transiently transfected HeLa cells (data not shown).
We also examined the ability of the SRC1 mutant proteins to enhance the activity of GAL4-RXR, which contains the LBD of human RXRα fused to the GAL4 DBD. Results similar to those in the ER experiments were obtained in that all constructs containing a functional AD1, including SRC1 631-970, were as potent as full-length receptor in increasing GAL4-RXR activity by 15- to 20-fold, whereas SRC1 Δ900-950 showed no ability to enhance GAL4-RXR activity (Fig. 5C, right). Due to the absence of an AF1 function in this construct, ligand-independent activation of GAL4-RXR mediated via the Q-rich region was not observed.
To establish that the reporter activities shown in Fig. 5C were not at saturation levels for any of the constructs shown, we performed a series of titration experiments by measuring reporter activity over a range of amounts of transfected DNA (200 to 1,500 ng per well) for each construct. This confirmed that full-length wild-type SRC1e (1-1399) and SRC1 631-970 enhanced ER-mediated reporter activities to similar levels over a range of expression levels (Fig. 5D). Similarly, the SRC1 1-1100 construct stimulated reporter activity to levels similar to those with SRC1e, whereas SRC1 Δ900-950 failed to stimulate reporter activity above the endogenous level at any amount of transfected DNA tested (data not shown). These results confirm our conclusion that a minimal construct containing only the NID and AD1 functions as a potent coactivator in vivo.
It was shown previously that the binding of SRC1 to ER requires at least two functional LXXLL motifs and that motif 2 is critical for optimal binding (21). As shown in Fig. 6, the ability of the minimal coactivator SRC1 631-970 to potentiate ER activity was also dependent on functional LXXLL motifs in the NID. Constructs carrying LXXAA mutations in motifs 1 and 2 (M12), 2 and 3 (M23), or all three motifs (M123) failed to enhance ER activity (Fig. 6). While the binding of these constructs to GST–CBP-C was unaffected, they failed to bind GST-AF2 in the presence of ligand in GST pull-down assays (data not shown). The mutant containing a functional motif 2 (M13) enhanced ER activity two- to threefold in transfection experiments (Fig. 6) and displayed weak binding to GST-AF2 in vitro (data not shown). In contrast, a full-length SRC1e mutant in which only motif 2 is functional retained significant coactivator function in similar experiments (21). This suggests that sequences outside the core NID may contribute to stabilizing contacts with the NR.
FIG. 6.
Mutation of LXXLL motifs in the context of the SRC1 minimal coactivator abrogates activity. A schematic diagram is shown representing full-length SRC1 (see the legend to Fig. 5A for details of the functional domains indicated by grey boxes) and the deletion constructs spanning the core coactivator domains and containing mutations in which the leucine doublet in two or more LXXLL motifs was mutated to alanines. Cos-1 cells were transfected as described in Materials and Methods with appropriate reporter constructs, vector expressing ER, and the SRC1 constructs as indicated. Luciferase activity was measured 24 h after the addition of 10−8 M E2, and the data were normalized to β-galactosidase activity. The activity of ER in the presence of ligand was set at 1, and the other values are expressed relative to it. The values shown represent the average of triplicate samples, and the error bars indicate standard deviation. The results shown are representative of experiments performed at least three times.
Taken together, our results clearly demonstrate that the coactivator function of SRC1 is dependent on its ability to bind to NRs via LXXLL motifs in the NID and to recruit CBP-p300 or other factors via the AD1 sequence. In contrast, the PAS helix-loop-helix domain and sequences encoding the AD2, the HAT domain, and the Q-rich region can be deleted without affecting the ability of SRC1 to enhance AF2 activity in these assays. In summary, we have defined a minimal p160 coactivator protein that functions by recruitment of CBP to the LBDs of NRs.
DISCUSSION
In this study we have used yeast two-hybrid, GST pull-down, and peptide competition experiments to precisely map the sequences which contribute to the interaction interface of SRC1 and CBP-p300. We have localized the core CBP binding domain (AD1) of SRC1 to the sequence 926 to 960 and have shown that amino acids 926 to 970 function as an autonomous activation domain (Fig. 1 and 2). This sequence potentially contains two α-helices (helix A and helix B) and corresponds to the predicted helices present in the analogous region of TIF2, referred to as H1 and H2 (53). Our data are in agreement with the results of Chen et al. (8), who localized the CBP binding sequence of ACTR to residues 1039 to 1088, corresponding to residues 910 to 959 of SRC1. Two leucine-rich sequences (LXD4 and LXD5) have been proposed to be important for the interaction of SRC1 with CBP (34). LXD5 corresponds to the helix A sequence defined here, and the minimal CBP binding sequence defined by this group (SRC1 900-970) also contains the sequence corresponding to helix B. However, our finding that SRC1 926-970 is sufficient to bind CBP in the two-hybrid system indicates that the sequence corresponding to LXD4 is not essential for the SRC1-CBP interaction. Similarly, deletion of the region corresponding to LXD4 in TIF2 (construct TIF2.19) did not affect the transcriptional activity or CBP binding property of the TIF2 AD1 region (53). We also demonstrated that the 926-to-970 sequence behaves as a potent transcription activation domain in mammalian cells, thus confirming the hypothesis that AD1 and the CBP binding domain are coincident. Based on these observations and the data presented here, we conclude that the core CBP binding sequence resides within a 35-residue sequence of SRC1. Nonetheless, it is possible that sequences outside of the core domain may help to stabilize these interactions.
The CBP sequence responsible for SRC1 binding has previously been localized to amino acids 2058 to 2163 (22). Secondary-structure analysis programs predict that this sequence has the potential to fold into four α-helices (34), and there are also four repeats of the sequence QPGM/L (Fig. 3). Using yeast two-hybrid and GST pull-down experiments, we have further refined this SID to residues 2058 to 2130, indicating that helix 4 and part of the QPGM/L sequence are not essential for the SRC1-CBP interaction (Fig. 3). The corresponding region of p300 (2025 to 2141) also interacts with AD1 in yeast two-hybrid experiments or with full-length SRC1 in GST pull-down experiments (data not shown). In a previous study, the construct GST-CBP 2058-2133 failed to bind SRC1 AD1 in a GST pull-down experiment (34). However, our results indicate that CBP 2058-2130 is necessary and sufficient to bind both the AD1 and full-length SRC1 proteins in two different assay systems.
Mutational analysis of the SID revealed that residues in the H3 sequence are critical for the SRC1-CBP interaction. The mutations F-2101-S, F-2101-P, and K-2103-P, which potentially disrupt the putative α-helical conformation of this sequence, resulted in a complete loss of SID-AD1 interaction in the two-hybrid experiments and the SID-SRC1e interaction in vitro (Fig. 3B and C). In contrast, the mutations K-2103-A and K-2108-A did not negatively affect SID-AD1 binding in either assay; thus, there is a discrepancy with the study of McInerney et al. (34), who reported that a K-2108-A mutation reduced interaction between CBP and AD1 in GST pull-down assays. In fact, we noted a moderate and reproducible increase in the interactions of K-2103-A and K-2108-A mutants with AD1 in both assay systems (Fig. 3B and C), despite the fact that Western blots showed that the expression level of these proteins was approximately twofold lower than that of the wild type in the yeast two-hybrid experiments (data not shown). Mutation of the conserved LXXLL motif in helix 1 to AXXAA or PXXPP reduced the interaction to about 50% of wild type in both assay systems. Mutations in the glutamine-rich sequence of helix 2 reduced the interaction by approximately two-thirds. However, mutations in QPGM/L motifs had little effect on SRC1-CBP interaction. Thus, while our conclusion that the relative importance of the putative helices to CBP-SRC1 interaction appears to be H3>H2>H1, where H3 is the most critical, is in agreement with the results of McInerney et al. (34), we clearly demonstrate that the H4 region is not required to maintain a fully functional interface for SRC1 binding.
In contrast to SRC1 and other p160 proteins, the ligand-dependent interaction of full-length CBP with either the N-terminal domain or the LBD of ER (Fig. 4A) or other NRs (data not shown) is very weak. In addition, fragments derived from the N terminus of CBP, which have been reported to bind NRs, displayed very weak ligand-dependent interactions with NRs in both yeast two-hybrid and GST pull-down experiments (data not shown). We have shown previously that LXXLL motifs at the N termini of CBP and p300 produce 50- to 100-fold-lower reporter activity via interaction with the LBD of ER in yeast two-hybrid experiments than the motifs present in SRC1 (16). In contrast, we observed a strong interaction between full-length CBP and SRC1 (Fig. 4B) and between fragments of these proteins in vitro (Fig. 1A, Fig. 2, and Fig. 3). Several groups have reported that deletion of the N-terminal NR binding region did not disrupt the ability of CBP-p300 to stimulate NR activity (24, 34, 58), whereas deletion of the SRC1 binding region markedly abrogates p300 coactivator potential (24, 30). In addition, a recent report suggests that there is minimal direct interaction between liganded TR-RXR and p300 and that p300 is recruited to TR-RXR via its interaction with SRC1 family members (30). Finally, we note that proteins with molecular weights corresponding to those of CBP and p300 were not readily detected in the original far-Western experiments using the LBD of ER (5, 14). Thus, while CBP-p300 HAT activity is necessary for ER function, our in vitro results indicate that it is the recruitment of CBP-p300 by SRC1, rather than direct interaction between the ER and CBP-p300, which is essential.
Using a series of deletion mutants in transiently transfected cells, we have shown that truncated SRC1 proteins that retain the ability to bind to NRs and CBP-p300 in vitro function as efficient coactivators of ER- and GAL4-RXR-mediated transcription. Thus, the conserved PAS helix-loop-helix domain, the CARM1 binding AD2 domain, the Q-rich region, and the HAT domain can all be deleted without affecting the ability of SRC1 to enhance ligand-dependent ER activity in these assays. While these conserved regions undoubtedly have functions in vivo, they appear to be dispensable in the transient-reporter expression assay system. This may reflect qualitative and quantitative differences in hormone-induced expression of genomic NR target genes in vivo and plasmidic reporter genes in transiently transfected cell lines. Nonetheless, there is evidence indicating that plasmid DNA is rapidly chromatinized in transfected cells (42, 45, 48). Histone deacetylase inhibitors, such as TSA, induce basal activity of transfected reporter genes, indicating that the promoter region of the reporter gene has nucleosome structure (20). In addition, the HAT domain of CBP-p300 has been shown to be required for its NR coactivator function in transient-transfection experiments or in microinjected cells, which again suggests that reporter genes are chromatinized (23, 30). However, the possibility that it is acetylation of nonhistone targets which is required to stimulate reporter activity in these assays cannot be ruled out.
Our data indicate that the Q-rich domain appears to mediate the ligand-independent activity of SRC1. This was observed only with full-length ER and not with the GAL4-RXR construct, which contains only the LBD of RXR, indicating that the Q-rich region may interact with the N-terminal activation domain AF1 present in NRs, as reported recently in other studies (3, 31, 57). However, no significant interaction was observed between SRC1 and the AF1 domain of ER in GST pull-down assays, perhaps indicating that if this interaction occurs it is weak and/or stabilized in the context of the full-length protein (Fig. 4). We noted that the ligand-independent ER activity mediated by the Q-rich domain was observed only when AD1 was intact (Fig. 5C). This indicates a requirement for the interaction of SRC1 with CBP-p300, or other AD1 binding proteins, in its ligand-independent activity.
The NIDs of SRC1 and other p160s contain three LXXLL motifs (16, 50). The crystal structure of the liganded LBD homodimer of PPAR γ complexed with an SRC1 polypeptide containing motifs 1 and 2 strongly supports the hypothesis that two LXXLL motifs are required to make efficient contacts with NR homodimers and that the stoichiometry of the complex is one p160 protein per NR dimer (37). In a previous study, it was shown that a full-length SRC1 mutant containing a single functional LXXLL motif (motif 2) retained significant ability to bind the ER and stimulate its activity (21). Similar mutants containing only motif 1 or motif 3 were significantly impaired in ER binding and coactivator functions. Our results using the minimal coactivator revealed that in contrast to full-length SRC1e mutants, SRC1 630-970 mutants containing a single functional motif are dramatically impaired in these functions (Fig. 5C). This implies that additional sequences outside the NID may stabilize contacts between SRC1 and NRs. The Q-rich region–AF1 interaction appears to be a good candidate for such an auxiliary binding domain.
In summary, our data have defined the boundaries of the minimal sequences required for the interaction of SRC1 and CBP and demonstrated the importance of CBP recruitment to NRs via SRC1. We have shown in principle that this interaction can be disrupted using short peptides, at least in vitro. Polypeptides encompassing AD1 and the SID have been purified to near homogeneity, and we have determined that they can associate in native polyacrylamide gels (data not shown). Crystallization trials are under way to enable us to probe the structure of this interface at the atomic level. Given the involvement of p160 and p300 proteins in cancer, e.g., MOZ-CBP and MOZ-TIF2 fusions in acute myeloid leukemias and the overexpression of AIB1 in breast cancer, it will be important to further understand the molecular structure of the p160-p300 interaction interface.
ACKNOWLEDGMENTS
We thank Susan Hoare, Alison Davis, Anil Pancholi, and Jacquie Greenwood for technical assistance and members of the PNACL laboratory at Leicester University for automated sequencing. We are grateful to M. Parker, E. Kalkhoven, P. Chambon, Y. Nakatani, R. Goodman, A. Bannister, T. Kouzarides, and M. Dickens for generous gifts of materials. We also thank Peter Moody for useful discussions.
This work was supported by the Wellcome Trust.
REFERENCES
- 1.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X-Y, Sauter G, Kallioniemi O-P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 2.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 3.Bevan C L, Hoare S, Classens F, Heery D M, Parker M G. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol. 1999;19:8383–8392. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco J C, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavailles V, Dauvois S, Danielian P S, Parker M G. Interaction of proteins with transcriptionally active estrogen-receptors. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavailles V, Dauvois S, Lhorset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen-receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/p300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Lin R J, Schlitz R L, Chakravati D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen D, Ma H, Koh S S, Huang S-M, Schurter B T, Aswad D W, Stallcup M R. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 11.Dowell P, Ishmael J E, Avram D, Peterson V J, Nevrivy D J, Leid M T I. p300 functions as a coactivator for the peroxisome proliferator-activated receptor alpha. J Biol Chem. 1997;272:33435–33443. doi: 10.1074/jbc.272.52.33435. [DOI] [PubMed] [Google Scholar]
- 12.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 14.Halachmi S, Marden E, Martin G, Mackay H, Abbondanza C, Brown C. Estrogen receptor-associated proteins—possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 15.Harnish D C, Evans M J, Scicchitano M S, Bhat R A, Karathanasis S K. Estrogen regulation of the apolipoprotein AI gene promoter through transcription cofactor sharing. J Biol Chem. 1998;273:9270–9278. doi: 10.1074/jbc.273.15.9270. [DOI] [PubMed] [Google Scholar]
- 16.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 17.Heery D M, Parker M G. Ligand-induced transcription by nuclear receptors. Retinoids. 1997;13:26–30. [Google Scholar]
- 18.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito M, Yuan C-X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z-Y, Zhang X, Qin J, Roeder R G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 20.Jenster G, Spencer T E, Burcin M M, Tsai S Y, Tsai M-J, O'Malley B W. Steroid receptor induction of gene transcription: a two-step model. Proc Natl Acad Sci USA. 1997;94:7879–7884. doi: 10.1073/pnas.94.15.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and Ap-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 23.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 24.Kraus W L, Manning E T, Kadonaga J T. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Douarin B, Zechel C, Garnier J M, Lutz Y, Tora L, Pierrat B, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of ligand-dependent activation function (AF2) of nuclear receptors, is fused to B-Raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Douarin B, Pierrat B, vom Baur E, Chambon P, Losson R. A new version of the two-hybrid assay for detection of protein-protein interactions. Nucleic Acids Res. 1995;23:876–878. doi: 10.1093/nar/23.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee C H, Chinpaisal C, Wei L N. Cloning and characterization of mouse RIP140, a corepressor for nuclear orphan receptor TR2. Mol Cell Biol. 1998;18:6745–6755. doi: 10.1128/mcb.18.11.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemon B D, Freedman L P. Nuclear receptor cofactors as chromatin remodelers. Curr Opin Cell Biol. 1999;9:499–504. doi: 10.1016/s0959-437x(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor associated protein that is related to SRC-1 and TIF-2. Proc Natl Acad Sci USA. 1997;94:3895–3903. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, O'Malley B, Wong J. P300 requires its histone acetyltransferase activity and SRC-1 interaction domain to facilitate thyroid hormone receptor activation in chromatin. Mol Cell Biol. 2000;20:2031–2042. doi: 10.1128/mcb.20.6.2031-2042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma H, Hong H, Huang S-M, Irvine R A, Webb P, Kushner P J, Coetzee G A, Stallcup M R. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol Cell Biol. 1999;19:6164–6173. doi: 10.1128/mcb.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Balbas M M, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T-M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyata K S, McCaw S E, Meetens L M, Patel H V, Rachubinski R J P. Receptor-interacting protein 140 interacts with and inhibits transcription by peroxisome proliferator-activated receptor alpha and liver-X-receptor alpha. Mol Cell Endocrinol. 1998;25:69–76. doi: 10.1016/s0303-7207(98)00196-8. [DOI] [PubMed] [Google Scholar]
- 36.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 37.Nolte R T, Wisely G B, Westin S, Cobbs J E, Lambert M H, Kurokawa R, Rosenfeld M G, Wilson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 38.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;94:35–44. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 39.Onate S A, Tsai S T, Tsai M-J, O'Malley B W. Sequence and characterisation of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 40.Perissi V, Dasen J S, Kurokawa R, Wang Z Y, Korzus E, Rose D W, Glass C K, Rosenfeld M G. Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc Natl Acad Sci USA. 1999;96:3652–3657. doi: 10.1073/pnas.96.7.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puigserver P, Wu Z D, Park C W, Graves R, Wright M, Spiegelman B M A. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 42.Pullner A, Mautner J, Albert T, Eick D. Nucleosomal structure of active and inactive c-myc genes. J Biol Chem. 1996;271:31452–31457. doi: 10.1074/jbc.271.49.31452. [DOI] [PubMed] [Google Scholar]
- 43.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 44.Rachez C, Suldan Z, Ward J, Chang C P B, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. A novel protein complex that interacts with the vitamin D-3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeves R, Gorman C M, Howard B. Minichromosome assembly of non-integrated plasmid DNA transfected into mammalian cells. Nucleic Acids Res. 1985;13:3599–3615. doi: 10.1093/nar/13.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spencer T E, Jenster G, Burchin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M-J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 48.Stanfield-Oakley S A, Griffith J D. Nucleosomal arrangement of HIV-1 DNA: maps generated from an integrated genome and an EBV-based episomal model. J Mol Biol. 1996;256:503–516. doi: 10.1006/jmbi.1996.0104. [DOI] [PubMed] [Google Scholar]
- 49.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits properties distinct from steroid receptor co-activator 1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 50.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C, Rosenfield M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 51.Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson J A. A regulatory role for RIP140 in nuclear receptor activation. Mol Endocrinol. 1998;12:864–881. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- 52.Vega R B, Huss J M, Kelly D P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voegel J J, Heine M J S, Tini M, Vivat V, Chambon P, Gronemeyer H. The co-activator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function of AF-2 of nuclear receptors. EMBO J. 1996;15:101–108. [PMC free article] [PubMed] [Google Scholar]
- 55.Vojtek B A, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 56.vom Baur E V, Zechel C, Heery D, Heine M J S, Garnier J M, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- 57.Webb, P., P. Nguyen, J. Shinsako, C. Anderson, W. Feng, M. P. Nguyen, D. Chen, S.-M. Huang, S. Subramanian, E. McKinerney, B. S. Katzenellenbogen, M. R. Stallcup, and P. J. Kushner. 1998. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. 12:1605–1618. [DOI] [PubMed]
- 58.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 59.Wolffe A P, Wong J, Pruss D. Activators and repressors: making use of chromatin to regulate transcription. Genes Cells. 1997;2:291–302. doi: 10.1046/j.1365-2443.1997.1260323.x. [DOI] [PubMed] [Google Scholar]
- 60.Xu L, Glass C K, Rosenfeld M G. Co-activator and co-repressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 61.Yang X-J, Ogryzko V V, Nishikawa J I, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 62.Yao T-P, Gregory K, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;95:5527–5532. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan C X, Ito M, Fondell J D, Fu Z Y, Roeder R G. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;14:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]