Abstract
The HOG (high-osmolarity glycerol) mitogen-activated protein kinase (MAPK) pathway regulates the osmotic stress response in the yeast Saccharomyces cerevisiae. Three type 2C Ser/Thr phosphatases (PTCs), Ptc1, Ptc2, and Ptc3, have been isolated as negative regulators of this pathway. Previously, multicopy expression of PTC1 and PTC3 was shown to suppress lethality of the sln1Δ strain due to hyperactivation of the HOG pathway. In this work, we show that PTC2 also suppresses sln1Δ lethality. Furthermore, the phosphatase activity of these PTCs was needed for suppression, as mutation of a conserved Asp residue, likely to coordinate a metal ion, inactivated PTCs. Further analysis of Ptc1 function in vivo showed that it inactivates the MAPK, Hog1, but not the MEK, Pbs2. In the wild type, Hog1 kinase activity increased transiently, ∼12-fold in response to osmotic stress, while overexpression of PTC1 limited activation to ∼3-fold. In contrast, overexpression of PTC1 did not inhibit phosphorylation of Hog1 Tyr in the phosphorylation lip, suggesting that Ptc1 does not act on Pbs2. Deletion of PTC1 also strongly affected Hog1, leading to high basal Hog1 activity and sustained Hog1 activity in response to osmotic stress, the latter being consistent with a role for Ptc1 in adaptation. In vitro, Ptc1 but not the metal binding site mutant, Ptc1D58N, inactivated Hog1 by dephosphorylating the phosphothreonine but not the phosphotyrosine residue in the phosphorylation lip. Consistent with its role as a negative regulator of Hog1, which accumulates in the nucleus upon activation, Ptc1 was found in both the nucleus and the cytoplasm. Thus, one function of Ptc1 is to inactivate Hog1.
Mitogen-activated protein kinase (MAPK) pathways comprise three sequentially acting kinases, MEKK (or Raf), MEK, and MAPK, known as the MAPK module 16, 22, 48. MEKKs can be activated by interaction with upstream components and phosphorylation on Ser, Thr, and Tyr residues. Activated MEKKs then activate MEK by phosphorylating Ser/Thr residues. Activated MEK activates MAPK by phosphorylating a Thr and a Tyr residue in the phosphorylation lip. Phosphorylation of both residues is required for full MAPK activation. There are three groups of MAPKs, classified by the signals that activate them and their phosphorylation lip sequence. One group, including vertebrate ERK1 and ERK2, contains the phosphorylation lip sequence TEY and is activated by growth factors, mitogens, and cytokines (22, 48). A second group, vertebrate c-Jun N-terminal kinase (JNK)–stress-activated protein kinase, contains the phosphorylation lip sequence TPY and is activated by UV light, osmotic stress, tumor necrosis factor, and interleukin-1 (22, 48). A third group, including Saccharomyces cerevisiae Hog1, Schizosaccharomyces pombe Spc1, and vertebrate p38, contains the phosphorylation lip sequence TGY and is activated by osmotic stress and other environmental stresses (16, 22, 48).
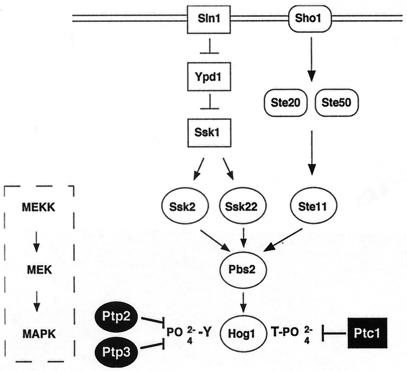
At least six MAPK cascades operate in S. cerevisiae to regulate physiologically distinct responses (16). The HOG pathway allows yeast to grow in high-osmolarity environments by inducing the expression of osmoprotectants. The upstream portion of this pathway has two branches (Fig. 1). One branch is the two-component signaling system comprising Sln1, Ypd1, and Ssk1 (25, 33, 37). Genetic and biochemical data support the following model for activation of this pathway. Sln1 is a plasma membrane-bound His/Asp kinase that is phosphorylated in the absence of stress. Osmotic stress causes its dephosphorylation, leading to dephosphorylation of Ypd1, a His kinase, and Ssk1, an Asp kinase (37). Unphosphorylated Ssk1 activates the MEKKs, Ssk2 and Ssk22, by binding to their N-terminal inhibitory domains (23). For Ssk2, this has been shown to result in autophosphorylation of a Thr residue and kinase activation (34). Activated Ssk2 and Ssk22 then activate the MEK, Pbs2, by phosphorylating a Ser and a Thr residue in the T loop. Activated Pbs2 activates the MAPK, Hog1, by phosphorylation of a Thr and Tyr residue in the phosphorylation lip (4, 42). A second branch of the HOG pathway is activated by the osmosensor Sho1 (23), which signals to Ste20, Ste50, Ste11, and then Pbs2 (32, 35, 36, 38, 50).
FIG. 1.
Protein phosphatase inactivation of the HOG pathway. The HOG pathway is regulated by two membrane-bound osmosensors, Sln1 and Sho1. Both Sln1 and Sho1 activate the MAPK cascade (boxed). The two-component regulators, Sln1, Ypd1, and Ssk1, negatively regulate the two MEKKs, Ssk2 and Ssk22, while Sho1, Ste20, and Ste50 positively regulate the MEKK, Ste11. The MEKKs activate a MEK, Pbs2, and the MAPK, Hog1. Two protein tyrosine phosphatases, Ptp2 and Ptp3, inactivate Hog1 by dephosphorylating the pY residue in the phosphorylation lip, while the type 2C Ser/Thr phosphatase, Ptc1, dephosphorylates the pT residue in the phosphorylation lip.
The identity of physiologically relevant protein phosphatases that inactivate MAPK pathways is less well established. Protein phosphatases can inactivate MEKK, MEK, and MAPK, since they require phosphorylation on Ser, Thr, and Tyr residues for activity (21, 22). Vertebrate MEKK and MEK are inactivated by the type 2A Ser/Thr phosphatase (PP2A) in vitro and may be inactivated by PP2A in vivo (1, 2, 17, 45). Stress-activated vertebrate MEKs, MKK6 and SEK1 (MKK4/JNKK1), are also inactivated by the type 2C Ser/Thr-specific phosphatase (PP2C) in vivo and in vitro (46). Since MAPKs require phosphorylation of a Thr and Tyr residue in the phosphorylation lip to be active, they can be inactivated by Ser/Thr phosphatases, protein tyrosine phosphatases (PTPs) specific for phosphotyrosine (pY), and dual-specificity phosphatases, capable of dephosphorylating both the phosphorylation lip phosphothreonine (pT) and pY residues (21, 22). Among the Ser/Thr phosphatases, PP2A has been shown to inactivate MAPK (1, 15, 45), and PP2C has been found to inactivate the stress-activated MAPK, p38 (46). In S. pombe, there are conflicting reports regarding the activity of PP2C on the stress-activated MAPK, Spc1/Sty1. Two PP2C phosphatases, Ptc1 and Ptc3, were shown to be important for inactivating Spc1/Sty1 activated by heat stress (31), while Ptc1 was shown to be unimportant for inactivating Spc1/Sty1 activated by osmotic stress (13).
In S. cerevisiae, MAPKs have been shown to be inactivated by a dual-specificity phosphatase, PTPs, and potentially PP2Cs. The dual-specificity phosphatase Msg5 inactivates Fus3 (11) but has not been shown to inactivate other MAPKs. The PTPs Ptp2 and Ptp3 inactivate Hog1, Fus3, and Mpk1 with different specificities (Fig. 1) (19, 27, 51, 52). The identity of Ser/Thr phosphatases that inactivate S. cerevisiae MAPKs has not been established. Two PP2Cs, Ptc1 and Ptc3, have been implicated as negative regulators of the HOG pathway by genetic means, but their substrates have not been identified. Initially, Maeda et al. identified PTC1 as a gene whose mutation produced a synthetic growth defect with deletion of PTP2 (24). We showed that this growth defect was suppressed by deletion of HOG1 (19), further suggesting that Ptc1 negatively regulates the HOG pathway. Previous studies also showed that overexpression of PTC1 or PTC3 suppressed lethality conferred by deletion of SLN1, which is known to hyperactivate the HOG pathway (25). In this work, we show that PTC2, encoding a type 2C Ser/Thr phosphatase closely related to Ptc3, also inactivates this pathway, and that Ptc1 inactivates the HOG pathway by dephosphorylating the MAPK, Hog1. Unlike vertebrate PP2C, Ptc1 does not have a strong effect on the MEK, Pbs2.
MATERIALS AND METHODS
Strains, media, and genetic techniques.
Yeast strains (Table 1) were derived from BBY45 (MATa leu2-3,112 trp1-1 his3-Δ200 ura3-52 lys2-801) (3) except where noted. Isolation of PTC genes and examination of PTC mutants was performed using IMY101, a sln1Δ::HIS3 strain carrying a low-copy-number centromere (CEN)-based plasmid containing the wild-type SLN1 gene and the URA3 gene (19). Hog1 kinase assays were performed using IMY102, a hog1Δ::TRP1 strain carrying pHOG1-ha2, and IMY104, a hog1Δ::TRP1 ptc1Δ::LEU2 strain carrying pHOG1-ha2. The hog1Δ::TRP1 strain IMY100 (19) and the ptc1Δ::LEU2 strain ASY1 (19) were crossed, sporulated, and dissected to produce the ptc1Δ::LEU2 hog1Δ::TRP1 strain IMY103. PTC1 and HOG1 were overexpressed from the GAL1 promoter in IMY105, a hog1Δ strain produced by transformation of the galactose-inducible strain 334 (Table 1) (18) with the hog1::hisG allele (32). Hog1 kinase assays in strains overexpressing PTC1 were performed using IMY105, carrying pHOG1-ha2 and pKT-PTC1, a plasmid overexpressing PTC1 (see below). The control strain was IMY105 carrying pHOG1-ha2 and pEG(KT) (29). Glutathione S-transferase (GST)–Hog1 was isolated from JWY1, which is IMY105 carrying pKT-HOG1. Growth of strains expressing the hyperactive SSK2ΔN allele was assessed in the wild type, JD53 (Table 1) (10), and IMY107, a ptc1Δ::LEU2 strain produced by transforming JD53 with the tpd1::LEU2-3 allele (40).
TABLE 1.
Yeast strains used
| Strain | Genotype | Reference or source |
|---|---|---|
| Derived from BBY 45 | ||
| BBY45 | MATa trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 3 |
| IMY101 | MATa sln1Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 19 |
| IMY100 | MATa hog1Δ::TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 19 |
| ASY1 | MATα ptc1Δ::LEU2 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 19 |
| IMY103 | MATa hog1Δ::TRP1 ptc1Δ::LEU2 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY120 | MATa ptc2Δ::TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY121 | MATa ptc3Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY122 | MATa ptc1Δ::LEU2 ptc2Δ::TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY123 | MATa ptc1Δ::LEU2 ptc3Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY124 | MATa ptc2Δ::TRP1 ptc3Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY125 | MATa ptc1Δ::LEU2 ptc2Δ::TRP1 ptc3Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| ASY2 | MATa ptc1Δ::LEU2 ptp2Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 19 |
| ASY3 | MATa ptc1Δ::LEU2 ptp2Δ::HIS3 hog1Δ::TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 19 |
| IMY126 | MATa ptc2Δ::TRP1 ptp2Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY127 | MATa ptc3Δ::HIS3 ptp2Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY128 | MATα ptc2Δ::TRP1 ptc3Δ::HIS3 ptp2Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY129 | MATa ptc1Δ::LEU2 ptp3Δ::TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY71b | MATα ptp3Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY130 | MATa ptc2Δ::TRP1 ptp3Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY131 | MATa ptc3Δ::HIS3 ptp3Δ::TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY132 | MATa ptc2Δ::TRP1 ptc3Δ::HIS3 ptp3Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| Derived from 334 | ||
| 334 | MATα leu2-3,112 gal1 reg1-501 ura3-52 pep4-3 prb1-1122 | 18 |
| IMY105 | MATα hog1::hisG leu2-3,112 gal1 reg1-501 ura3-52 pep4-3 prb1-1122 | This work |
| Derived from JD53 | ||
| JD53 | MATα trp1-Δ63 ura3-52 his3-Δ200 leu2-3,112 lys2-801 GAL+ | 13a |
| IMY107 | MATα ptc1Δ::LEU2 trp1-Δ63 ura3-52 his3-Δ200 leu2-3,112 lys2-801 GAL+ | This work |
PTC3 isolation.
PTC3 was isolated as a negative regulator of the HOG pathway by selecting for plasmids from a yeast genomic library that suppressed lethality of the sln1Δ strain in the previously described 5-fluoro-orotic acid (5-FOA) assay (19). Strain IMY101, a sln1Δ strain carrying pSLN1-URA3, is necessarily Ura+ since it requires the SLN1 gene for viability. IMY101 is inviable on media containing the drug 5-FOA, since it is toxic to strains that are Ura+. IMY101 was transformed with a multicopy yeast genomic library based in the vector YEp13 (30) (American Type Culture Collection). Transformants capable of vigorous growth on 5-FOA were identified, and the plasmid DNA was isolated using standard methods. For further study, PTC3 was subcloned as a 4.6-kb BamHI fragment into the multicopy vector YEplac181 (14).
Plasmids.
PTC1, previously identified as negative regulator of this pathway, and PTC2, a gene closely related to PTC3, were isolated by PCR. A 1.5-kb fragment of PTC1, containing 304 bp of 5′ and 308 bp of 3′ flanking sequence, and a 2-kb fragment of PTC2, containing 275 bp of 5′ and 325 bp of 3′ flanking sequence, were subcloned into YEplac181 (14). Mutants Ptc1D58N, Ptc2D62N, and Ptc3D62N were produced by site-directed mutagenesis using the oligonucleotides containing the underlined mutated codons, 5′-GCGGTGTTTAATGGACATGCTGGG-3′, 5′-TTTATGGTATATTTAACGGTCATGGTG-3′, and 5′-TTTACGGTATATTCAATGGTCATGGTGGC-3′, respectively. PTC1 was overexpressed as a fusion to GST from the GAL1 promoter in the vector pEG(KT) (29).
To delete PTC2 and PTC3, the following plasmids were constructed. pPTC2Δ::TRP1 contains the ptc2Δ::TRP1 allele. To construct this plasmid, PCR was used to produce a BamHI site 725 bp upstream and a SmaI site 27 bp downstream of the start codon. The 752-bp BamHI-SmaI fragment containing the 5′ end of the PTC2 gene and a 780-bp EcoRV-SalI fragment containing the 3′ end were ligated into pBluescript II KS (Stratagene) to produce pPTC2Δ. This plasmid was digested with SmaI and EcoRV and ligated to an 850-bp EcoRI-BglII fragment of TRP1 (19) that was filled in with Klenow polymerase I. The resulting plasmid pPTC2Δ::TRP1, digested with BamHI and SalI, was transformed into the wild-type strain, BBY45 (3). pPTC3Δ::HIS3 contains the ptc3Δ::HIS3 allele. To construct this plasmid, PCR was used to introduce a BamHI site 275 bp upstream of the start codon and a KpnI site 215 bp downstream of the stop codon. The 5′ end of the PTC3 gene, contained in a 286-bp BamHI-SspI fragment, and the 3′ end of the gene, contained in a 210-bp SspI-KpnI fragment, were ligated into the vector pCRII (Invitrogen) to produce pPTC3Δ. This plasmid was digested with SspI and ligated to the HIS3 gene contained in a ∼1.8-kb BamHI fragment which was filled in with Klenow polymerase I (19). The resulting plasmid, pPTC3Δ::HIS3, was digested with BamHI and BsrI and transformed into BBY45 (3). That the ptc2Δ::TRP1 and ptc3Δ::HIS3 alleles integrated at the PTC2 and PTC3 loci, respectively, was confirmed by Southern analysis.
For expression in Escherichia coli, Ptc1 and Ptc1D58N were fused to six repeats of His (His6) at the amino terminus, using the vector pRSETa (Invitrogen). Using PCR-based methods, a BamHI site was engineered upstream of the start codon using the oligonucleotide 5′-GCGGATCCATGAGTAATCATTCTGAAATC-3′ (start codon underlined) and pairing it with the oligonucleotide 5′-CCTCCAGCCGACAACGGTGAAG-3′ downstream of the stop codon. The PCR product, digested at the engineered BamHI site and a genomic BamHI site, was cloned into the pRSETa vector.
Hog1 yeast expression plasmids were produced as follows. For kinase assays, hemagglutinin (HA) epitope-tagged Hog1 (Hog1-HA) was expressed from its endogenous promoter using the plasmid pHOG1-ha2. The 5′ end of the HOG1 gene, a 1.5-kb ClaI-BstXI fragment, and the 3′ end of HOG1 fused to the HA epitope, a 290-bp BstXI-Hind III fragment from pHOG1-ha (19), were cloned into the multicopy vector pRS423 (8). GST-Hog1 was produced by cloning HOG1 into the vector pEG(KT) (29), containing GST under regulation of the GAL1 promoter. Using PCR, a BamHI site was introduced upstream of the start codon using the oligonucleotide 5′-GCGGATCCATGACCACTAACGAGGAATTC-3′ (start codon underlined), which was paired with an oligonucleotide containing a SacI site 102 bp downstream of the stop codon, 5′-CGCGAGCTCCCTCTATACAACTATATACG-3′. The 1.4-kb BamHI-SacI fragment was cloned into the vector pRSETa and then subcloned as a BamHI-HindIII fragment into pEG(KT). Both Hog1-HA2 and the GST-Hog1 fusion proteins were functional, since they suppressed the osmosensitivity of a hog1Δ strain. The kinase-inactive Hog1 mutant protein, HOG1-K52M, was derived from plasmid pHOG1-K52M-GFP (26), and the HA epitope was fused to its carboxy terminus in pRS423.
The hyperactive PBS2 allele, PBS2EE, bearing Glu residues that mimic phosphorylation at Ser514 and Thr518, was produced by PCR-based methods. Two PCR products that overlapped at the mutated Ser514 and Thr518 codons were combined in a third PCR to produce the full-length PBS2EE mutant. The 5′ fragment was produced by engineering a BamHI site upstream of the start codon using the oligonucleotide 5′-CGGGATCCGATGGAAGACAAGTTTGCT-3′ (start codon underlined) and pairing this with the mutagenic oligonucleotide 5′-CCAATATTTTCCCTCGCTAATTCTGCCACCAAATTACC-3′ (mutated codons underlined). The 3′ end of the PBS2 gene, containing the same mutated sites, was produced by pairing the mutagenic oligonucleotide 5′-GCAGAATTAGCGAAGGAAAATATTGGTTGTCAGTC-3′ with an oligonucleotide 330 bp downstream of the stop codon, 5′-GCTCTAGAGGAGTCGATGGCCGTAGC-3′. In the third PCR, gel-purified PCR products were combined and amplified with the outermost 5′ and 3′ primers above. The full-length PBS2EE PCR product was cloned into pRSETa for expression in E. coli.
To examine the subcellular localization of Ptc1, GFP was fused to the amino terminus of Ptc1 and expressed from the PTC1 promoter in multicopy and low-copy-number plasmids. GFP fusions to the carboxy terminus of Ptc1 did not fluoresce. To produce GFP-Ptc1, the PTC1 promoter, contained in an EcoRI-NotI fragment, was generated by PCR using the oligonucleotides 5′-TTGAGCTCGGCCTTCGCGAATTCTAACTGCAAAG-3′ and 5′-TTGCGGCCGGCTCATTATAATGATTTTTAAAAGATAAATGC-3′. The GFP fragment, contained in a NotI-BamHI fragment, was isolated by PCR using two oligonucleotides, 5′-TTGCGGCCGCAGTAAAGGAGAAGAAC-3′ and 5′-GCGGATCCACCACCACCACCTTTGTATAGTTCATCCATGCCATGTG-3′, and the template, pGFP (a gift from J. Hirsch, Columbia University). The PTC1 promoter, GFP, and the PTC1 open reading frame were cloned into pRS423 and the CEN-based vector pRS313 (8). Both 2μm and CEN GFP-Ptc1 fusion plasmids complemented the temperature sensitivity of the ptc1Δ strain at 37°C (40).
Immunoblot analysis.
Levels of singly (Hog1-pY) and dually (Hog1-pT,pY) phosphorylated Hog1 were examined by immunoblotting with anti-pY antibody (PY20; Santa Cruz Biotechnology) and antibody specific for dually phosphorylated p38 (phospho-p38 antibody; New England Biolabs), respectively. Cells from 4.5 ml of culture at 1 U (A600) were harvested by centrifugation, boiled in L1 buffer, and diluted in L2 buffer as described previously (27). Protein samples, 82 μg for the Hog1-pY blot and 113 μg for the phospho-p38 blot, were boiled in sample buffer, and Western analysis was performed (19). Hog1-HA and Hog1-K52M-HA were detected by immunoblotting with antibody 12CA5 (BAbCo), and GST and GST-Ptc1 were detected using anti-GST antibody (Pharmacia). For all immunoblots, alkaline phosphatase-conjugated secondary antibodies were used, and the blots were visualized with 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium (Promega).
Hog1 kinase assay.
Hog1 kinase activity in unstressed and osmotically stressed cells was examined as follows. Hog1-HA was isolated from strains grown to exponential phase, ∼1 U (A600), that were untreated or exposed to 0.4 M NaCl for various times. Cells were harvested by centrifugation and disrupted by glass bead lysis in buffer A (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 0.1% TX-100, and 1 mM dithiothreitol [DTT], containing protein phosphatase inhibitors (50 mM β-glycerophosphate and 1 mM sodium vanadate) and protease inhibitors (aprotinin, leupeptin, antipain, and chymostatin [each at 20 μg/ml] and 1 mM phenyl methylsulfonyl fluoride [PMSF]). The lysates were clarified by centrifugation, and Hog1-HA was immunoprecipitated by incubation with 2.5 μl of anti-HA antibody 12CA5 (1 mg/ml; BAbCo) for 1 h at 4°C and 12.5 μl of protein A-Sepharose for 0.5 h at 4°C. The immunoprecipitates were washed once with buffer A, twice with buffer B (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 1 mM DTT, 50 mM β-glycerophosphate, 1 mM sodium vanadate, 1 mM PMSF), and once with kinase assay buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 2 mM EGTA, 10 mM MgCl2, 50 mM β-glycerophosphate, 0.1 mM sodium vanadate, 1 mM DTT). To assay Hog1 kinase activity, Hog1-HA bound to protein A-Sepharose was incubated with [γ-32P]ATP (0.1 mM, 8,000 cpm/pmol) and myelin basic protein (MBP; 1 μM; Sigma) for 30 min at 30°C. Incorporation of 32P into MBP was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and quantified by PhosphorImager (Molecular Dynamics) analysis.
Ptc1 inactivation of Hog1 kinase in vitro.
GST-Hog1 was isolated from strain JWY1 grown to exponential phase in synthetic medium lacking uracil and containing 2% galactose. To activate Hog1, cells in exponential phase (∼1 U [A600]) were exposed to 0.4 M NaCl for 3 min at 30°C. Cells were isolated by centrifugation, and ∼1.5 × 108 cells were lysed by glass beading in 1 ml of buffer A. The lysate was clarified by centrifugation and mixed with 180 μl of a 1:1 slurry of glutathione-Sepharose (Pharmacia) for 1 h at 4°C. The resin was washed twice with 600 μl of buffer A, twice with 600 μl of buffer C (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 1 mM DTT, 50 mM β-glycerophosphate, 1 mM sodium vanadate, 1 mM PMSF), and twice with 600 μl of protein phosphatase buffer (50 mM Tris-HCl [pH 7], 0.1 mM EGTA, 5 mM MgCl2, 0.1% 2-mercaptoethanol). GST-Hog1 bound to resin was resuspended in protein phosphatase buffer and aliquoted into eight 15-μl fractions, each containing ∼170 ng of GST-Hog1. The bound GST-Hog1 was incubated with Ptc1 or Ptc1D58N (0 to 0.3 μg/μl) for 30 min at 37°C in phosphatase buffer. The reaction was terminated by addition of 300 μl of ice-cold buffer B, centrifugation, and washing as described below. Ptc1 inactivation assays were also done in the presence of Mn2+. For these assays, MgCl2 in buffers A, B, and C was replaced by 5 mM MnCl2 and phosphatase buffer contained 20 mM MnCl2. To assay Hog1 kinase activity, the resin was washed twice with kinase assay buffer and incubated with MBP and [γ-32P]ATP as described above. Samples were examined by SDS-PAGE and PhosphorImager analysis.
Purification of Ptc1, Ptc1D58N, and Pbs2EE from E. coli.
His6-Ptc1, His6-Ptc1D58N, and His6-Pbs2EE were expressed from the vector pRSETa in E. coli BL21(DE3)pLysS. The strains were grown in 2×YT medium at 37°C to early log phase, (∼0.5 U [A600]), cooled to 23°C, and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside until the culture reached 1.0 unit (A600). To isolate His6-tagged proteins, cells were lysed in sonication buffer (20 mM Tris-HCl [pH 8], 100 mM NaCl) and centrifuged at 17,000 × g for 15 min to clarify the supernatant, and 25 ml of lysate was incubated with 1.25 ml of Co2+-immobilized metal affinity resin (Talon; Clontech) for 1 h at 4°C. The resin was transferred to a column and washed with 20 mM Tris-HCl (pH 8), 100 mM NaCl, and 10 mM imidazole, and the His6-tagged proteins were eluted with the same buffer containing 75 mM imidazole. Fractions containing His6-tagged proteins were identified by immunoblotting using anti-His6 antibody (BAbCo), and the amount of His6-tagged protein was quantified using the Pierce bicinchoninic acid protein assay.
In vitro phosphorylation and dephosphorylation of Hog1.
GST-Hog1 was phosphorylated with Pbs2EE and [γ-32P]ATP in vitro. GST-Hog1 was isolated from ∼1.5 × 108 JWY1 cells grown to exponential phase in synthetic medium lacking uracil and containing 2% galactose. Cells were isolated by centrifugation and lysed in 600 μl of buffer A. The lysate was clarified by centrifugation and mixed with 180 μl of a 1:1 slurry of glutathione-Sepharose (Pharmacia) for 1 h at 4°C. The beads were washed twice with 600 μl of buffer A, twice with 600 μl of buffer C, and twice with 100 μl of kinase assay buffer. GST-Hog1 bound to resin was resuspended in kinase assay buffer and incubated with His6-Pbs2EE (0.8 μg/μl) and [γ-32P]ATP (0.1 mM, 80,000 cpm/pmol) for 90 min at 30°C. To dephosphorylate Hog1, bead-bound GST-Hog1 was washed twice with 100 μl of phosphatase assay buffer and incubated with Ptc1 (0 or 1.2 μg/μl) for 30 min at 37°C. GST-Hog1 was isolated for phosphoamino acid analysis by SDS-PAGE and transferred to a polyvinylidene difluoride membrane.
Phosphoamino acid analysis.
32P-labeled GST-Hog1, untreated or incubated with Ptc1, was examined by phosphoamino acid analysis (20). The polyvinylidene difluoride membrane with bound GST-Hog1 was rinsed in methanol and then water and hydrolyzed in 200 μl of 5.7 N HCl at 110°C for 1 h. The filter was removed, and the sample was lyophilized and resuspended in 10 μl of pH 1.9 buffer (2.5% formic acid, 7.8% glacial acetic acid) containing 2.5 nmol of each of the phosphoamino acid standards, pT, pY, and phosphoserine (Sigma). The sample was applied to a cellulose thin-layer chromatography plate and electrophoresed in the first dimension in pH 1.9 buffer and in the second dimension in pH 3.5 buffer (0.5% pyridine, 5.0% glacial acetic acid). The plate was treated with ninhydrin to visualize phosphoamino acid standards, and PhosphorImager analysis was used to examine the radiolabeled amino acids.
RESULTS
Ptc1 inactivates the HOG pathway.
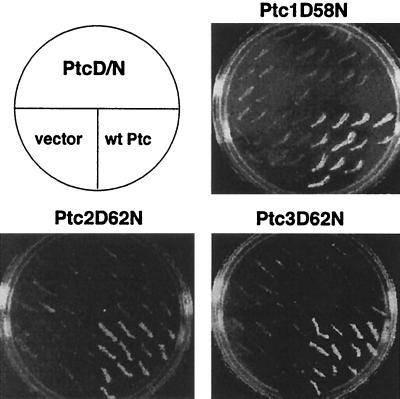
Yeast strains that constitutively activate the HOG pathway exhibit growth defects due to hyperactivation of the MAPK module comprising the MEKKs, Ssk2 and Ssk22, the MEK, Pbs2, and the MAPK, Hog1 (Fig. 1). A strain that lacks the receptor histidine kinase response regulator, SLN1, hyperactivates the downstream MAPK cascade and is lethal (25, 33). Overexpression of protein phosphatases restores viability by inactivating the MAPK cascade. For example, overexpression of PTP2 or PTP3, encoding protein tyrosine phosphatases that dephosphorylate Hog1, suppresses lethality of the sln1Δ strain by inactivating Hog1 (19, 51). Three genes, PTC1, PTC2, and PTC3, suppressed sln1Δ lethality from multicopy (Fig. 2) but not low-copy-number CEN-based plasmid (data not shown).
FIG. 2.
PTC1, PTC2, and PTC3 suppress growth defects due to Hog1 hyperactivation. Deletion of SLN1 hyperactivates the downstream MAPK cascade and is lethal. Expression of PTC1, PTC2, and PTC3 from multicopy plasmids suppressed lethality of the sln1Δ strain, while overexpression of the metal binding site mutations ptc1D58N, ptc2D62N, and ptc3D62N did not. The ability of PTCs and their mutant counterparts to suppress sln1Δ lethality was examined using the 5-FOA assay. IMY101, the sln1Δ strain bearing pSLN1(URA3), a CEN-based plasmid expressing the SLN1 and URA3 genes, was transformed with multicopy plasmids bearing wild-type (wt) PTC genes (pPTC1, pPTC2, and pPTC3), mutant PTCs (pPTC1D58N, pPTC2D62N, and pPTC3D62N), or empty vector (YEplac181). The transformants were patched on synthetic medium lacking leucine and containing 5-FOA and then examined for growth after 3 days at 30°C.
PTC1, PTC2, and PTC3 encode proteins belonging to the PP2C class of highly conserved protein phosphatases found in all eukaryotes. Ptc1, Ptc2, and Ptc3 show a high degree of similarity to each other and to the vertebrate enzymes. For example, Ptc1 is 33% identical and 44% similar to the human PP2Cα catalytic domain. Ptc2 and Ptc3 are 75% identical to each other, likely being related by a genome duplication event (49), and differ from Ptc1 in having C-terminal noncatalytic domains of ∼170 amino acids. One other yeast gene, PTC4 (YBR125c), encodes a protein most closely resembling Ptc1, Ptc2 and Ptc3, being 29% identical in the catalytic domain. Ptc4 has been shown to have Ser/Thr phosphatase activity (7); however, its expression from a multicopy plasmid did not suppress lethality of the sln1Δ strain (data not shown). Thus, Ptc1, Ptc2, and Ptc3 likely possess characteristics in addition to PP2C activity that enable them to inactivate the HOG pathway.
We next tested whether the protein phosphatase activity of PTCs is required to inactivate the HOG pathway. PP2Cs have been characterized as monomeric enzymes that require Mn2+ or Mg2+ for activity and are insensitive to okadaic acid. The crystal structure of human PP2Cα reveals two Mn2+ ions coordinated by five acidic residues, one Glu and four Asp, and the carbonyl oxygen of a Gly residue (9). The yeast PTCs align well with the primary structure of the human enzyme and show conservation of the metal-coordinating residues. Mutation of one of the acidic residues in each of the three PTCs was sufficient to inactivate them in vivo. The mutants Ptc1D58N, Ptc2D62N, and Ptc3D62N could not suppress lethality of the sln1Δ strain (Fig. 2), indicating that their protein phosphatase activity is required for HOG pathway inactivation.
Since overexpressing PTC1, PTC2, and PTC3 inactivated the HOG pathway, we tested whether their deletion would exacerbate growth defects due to hyperactivation of the HOG pathway. Deletion of PTC genes alone or in combination did not produce growth defects that were dependent on the HOG pathway, as indicated in Table 2. For example, the ptc1Δ mutant had a modest growth defect at 30°C and was temperature sensitive at 37°C as reported previously (40). However, the temperature sensitivity of this strain was not due to Hog1 hyperactivation, as the ptc1Δ hog1Δ strain also exhibited a temperature-sensitive defect. The ptc2Δ, ptc3Δ, and ptc2Δ ptc3Δ mutants grew similarly to the wild type, as previously reported (7). The ptc1Δ ptc2Δ ptc3Δ triple mutant had a growth defect more severe than that of the ptc1Δ strain, but this was not suppressed by deletion of HOG1 (Table 2).
TABLE 2.
Growth of ptcΔ and ptcΔ ptpΔ mutant strainsa
| Mutant strain | Growth at:
|
|
|---|---|---|
| 30°C | 37°C | |
| Wild type | +++ | +++ |
| ptc1Δ | ++ | − |
| ptc1Δ hog1Δ | ++ | − |
| ptc2Δ | +++ | +++ |
| ptc3Δ | +++ | +++ |
| ptc1Δ ptc2Δ | ++ | − |
| ptc1Δ ptc3Δ | ++ | − |
| ptc2Δ ptc3Δ | +++ | +++ |
| ptc1Δ ptc2Δ ptc3Δ | + | − |
| ptc1Δ ptc2Δ ptc3Δ hog1Δ | + | − |
| ptc1Δ ptp2Δ | +/− | − |
| ptc1Δ ptp2Δ hog1Δ | ++ | − |
| ptc1Δ ptp3Δ | ++ | − |
| ptc2Δ ptp2Δ | +++ | +/− |
| ptc3Δ ptp2Δ | +++ | +/− |
| ptc2Δ ptc3Δ ptp2Δ | +++ | +/− |
| ptc2Δ ptp3Δ | +++ | +++ |
| ptc3Δ ptp3Δ | +++ | +++ |
| ptc2Δ ptc3Δ ptp3Δ | +++ | +++ |
ptcΔ strains were compared to the wild-type parent on standard rich medium (YPD).
Growth defects due to Hog1 hyperactivation can be observed when PTC1 is deleted together with PTP2, encoding the nucleus-localized PTP that dephosphorylates Hog1-pY (19) (Table 2). However, deletion of PTC1 and PTP3, encoding the weaker, cytoplasm-localized PTP, was not lethal (19). We previously showed that the severe growth defect of the ptc1Δ ptp2Δ strain was likely due to Hog1 hyperactivation, as deletion of HOG1 suppressed this defect (19). To test whether deletion of PTC2 and/or PTC3 would produce synthetic growth defects in combination with ptp2Δ or ptp3Δ, the strains listed in Table 2 were produced. Strains lacking PTC2 and/or PTC3 did not show growth defects in combination with ptp2Δ or ptp3Δ at 30°C (Table 2). However, the ptc2Δ ptp2Δ, ptc3Δ ptp2Δ, and ptc2Δ ptc3Δ ptp2Δ strains showed stronger growth defects at 37°C. We have not yet identified the cause of these defects. Since the ptc1Δ ptp2Δ mutant exhibited a severe growth defect at 30°C dependent on HOG1, we first examined the role of Ptc1 in the HOG pathway.
Ptc1 can function downstream of the MEKK Ssk2.
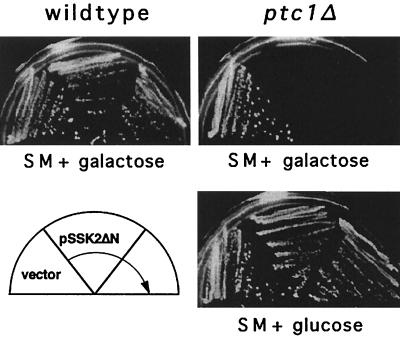
Since overexpressing PTC1 suppressed lethality of the sln1Δ strain, Ptc1 could conceivably inactivate any component downstream of Sln1 that requires Ser or Thr phosphorylation for activity. The MEKKs, Ssk2 and Ssk22, the MEK, Pbs2, and the MAPK, Hog1, are all potential candidates for Ptc1 inactivation. In an attempt to delineate the point of action of Ptc1 in this pathway, we examined the effect of overexpressing or deleting PTC1 in a strain expressing a hyperactivated MEKK allele, SSK2ΔN, that lacks the amino-terminal domain inhibitory for kinase activity. Overexpression of this allele was previously shown by Maeda et al. (23) to be lethal due to hyperactivation of Pbs2 and Hog1. In our strain background, however, overexpressing SSK2ΔN from the GAL1 promoter produced only a slight growth defect (Fig. 3). Overexpression of PTC1 in this strain did not improve growth (data not shown). Such an improvement may be difficult to detect since SSK2ΔN conferred only a mild growth defect. In contrast, deletion of PTC1 in the SSK2ΔN overexpressor was lethal (Fig. 3). Therefore, Ptc1 may inactivate the HOG pathway by acting downstream of MEKK, although this experiment does not rule out the possibility that Ptc1 acts on MEKKs or upstream of them.
FIG. 3.
Ptc1 acts downstream of the MEKK, Ssk2. Deletion of PTC1 exacerbated growth defects due to overexpression of SSK2ΔN, consistent with its role as a negative regulator. The wild type and a ptc1Δ strain were transformed with pSSK2ΔN, a multicopy plasmid expressing the hyperactive MEKK allele, SSK2ΔN, under regulation of the GAL1 promoter, or the empty vector pYES2. Growth on galactose conferred only a slight defect in the wild type but was lethal in a ptc1Δ strain. Growth of strains on selective medium (SM) containing galactose was assessed after 3 days at 30°C.
Ptc1 inactivates Hog1 but has little effect on Pbs2 in vivo.
We next used another assay to examine the point of action of Ptc1 in this pathway. If Ptc1 inactivates upstream kinases that activate Hog1, such as Pbs2, then overexpression of PTC1 should inhibit activation of Hog1. To assess Hog1 activation, we examined the level of Tyr176 phosphorylation in the phosphorylation lip. We chose this assay, since it should be unaffected by potential Ptc1-dependent dephosphorylation of Hog1-pT. Overexpression of PTC1 did not reduce the level of Hog1-pY induced by osmotic stress (Fig. 4A), suggesting that Ptc1 does not inactivate components upstream of Hog1. Since Hog1-pY could be due to autophosphorylation, we also performed this experiment with the catalytically inactive mutant Hog1-K52M (26). Osmotic stress also induced phosphorylation of Hog1-K52M in the PTC1 overexpressor (data not shown), suggesting that Ptc1 does not strongly affect Pbs2.
FIG. 4.
Ptc1 inhibits the activity of Hog1 but not Pbs2 in vivo. (A) The effect of PTC1 overexpression on Pbs2 activity was examined by monitoring the level of Hog1-pY, using anti-PY antibody. Hog1-pY was examined in the wild-type strain [334 carrying empty vector pEG(KT)] and in a strain overexpressing PTC1 (334 carrying pKT-PTC1). Levels of GST and GST-Ptc1 were examined using anti-GST antibody. (B) Hog1 kinase activity was examined in the wild type, IMY105 carrying pHOG1-ha and empty vector pEG(KT), and in a strain overexpressing PTC1, IMY105 carrying pHOG1-ha and pKT-PTC1. Hog1-HA was isolated from yeast that were untreated or exposed to 0.4 M sodium chloride for the indicated times. Immunoprecipitated Hog1-HA was incubated with MBP and [γ-32P]ATP, and the radiolabel incorporated into MBP was quantified by PhosphorImager analysis. The level of Hog1-HA in lysates was assessed by immunoblotting with anti-HA antibody.
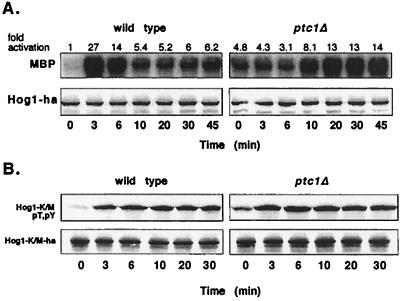
To test whether Ptc1 inactivates Hog1, we examined Hog1 kinase activity, which requires phosphorylation of both Thr174 and Tyr176 for full activation (42). Hog1 was isolated from a wild-type strain and a strain overexpressing PTC1, and its kinase activity was examined before and after exposure to osmotic stress. To assess Hog1 kinase activity, Hog1-HA was immunoprecipitated from yeast lysates and incubated with the MAPK substrate MBP and [γ-32P]ATP. In the wild type, Hog1 kinase activity was low before osmotic stress but increased ∼12-fold after 5 min of osmotic stress (Fig. 4B). This was followed by a rapid decrease in kinase activity, reaching nearly basal activity by ∼30 min. In the strain overexpressing PTC1, the level of Hog1 kinase activity prior to osmotic stress was nearly the same as for the wild type, but treatment with osmotic stress stimulated Hog1 kinase activity only threefold (Fig. 4B).
Since overexpression of PTC1 inhibited activation of Hog1, we examined whether a strain lacking PTC1 would express a higher level of Hog1 activity. To this end, we used a different strain background (BBY48 rather than 334); thus, Hog1 kinase activity was reassessed in the wild type and compared to the isogenic ptc1Δ strain. Before osmotic stress, Hog1 kinase activity was 4.8-fold higher in the ptc1Δ strain than in the wild type (Fig. 5A), suggesting that Ptc1 acts as a negative regulator to maintain a low basal level of Hog1 kinase activity. This was due to increased Hog1 activity and not to an increase in Hog1 protein, as judged by immunoblotting (Fig. 5A). In both wild type and mutant, osmotic stress induced Hog1 kinase activity. However, in response to continuous exposure to osmotic stress, Hog1 activity decreased rapidly in the wild type, while the ptc1Δ strain displayed a strong defect in Hog1 inactivation (Fig. 5A). In the wild type, Hog1 activity began decreasing after 3 min of osmotic stress, but in the ptc1Δ strain, Hog 1 activity did not decrease below maximal levels after 45 min of osmotic stress. Further exposure, for up to 1 h of osmotic stress, did not appreciably lower Hog1 kinase activity (data not shown). These results suggest that Ptc1 inactivates Hog1 during adaptation.
FIG. 5.
A strain lacking PTC1 shows defects in Hog1 activation and inactivation. (A) Hog1 kinase activity and the level of Hog1 protein were examined in the wild-type strain IMY102 and in ptc1Δ strain IMY103. Hog1-HA was isolated from yeast cells that were untreated or exposed to 0.4 M sodium chloride for the indicated times, and Hog1 kinase activity was assessed as described in the legend to Fig. 4. The level of Hog1-HA was assessed using anti-HA antibody. (B) Dual phosphorylation of kinase-inactive Hog1-K52M is rapid in both wild-type ptc1Δ strains. PTC1 hog1Δ and ptc1Δ hog1Δ strains expressing Hog1-K52M-HA were exposed to 0.4 M NaCl for the indicated times. Activation of Hog1-K52M-HA was monitored using antibody specific for dually phosphorylated Hog1, and its relative level of expression was assessed using anti-HA antibody.
Significant differences between the wild-type and ptc1Δ strains were also observed in the kinetics of Hog1 activation. The ptc1Δ strain showed an obvious defect in rapidly activating Hog1; kinase activity reached a high level only after 20 min of osmotic stress (Fig. 5A). In contrast, the wild type showed maximal Hog1 kinase activity after 3 min of osmotic stress. The ptc1Δ strain grew more slowly than the wild type in liquid culture (doubling times of 117 and 95 min, respectively) which might explain differences in Hog1 activation. To test this possibility, the doubling time of the wild type was increased to 165 min by incubation at 23°C rather than 30°C. Under these conditions, the kinetics of Hog1 activation were not appreciably altered from those of the wild type grown at 30°C (data not shown).
Another possible explanation for the delayed activation of Hog1 in the ptc1Δ strain is that the high basal Hog1 activity inhibits Hog1 activation. To test this idea, the kinase inactive mutant Hog1-K52M (26) was substituted for wild-type Hog1 in both wild-type and ptc1Δ strains. Hog1-K52M phosphorylation was measured using anti-phospho-p38 antibody, which is specific for dually phosphorylated Hog1 (32, 39). In contrast to wild-type Hog1, whose dual phosphorylation (data not shown) and kinase activation (Fig. 5A) were markedly delayed in the ptc1Δ strain, Hog1-K52M was rapidly phosphorylated in the ptc1Δ mutant (Fig. 5B). In the PTC1 strain, Hog1 and Hog1-K52M were both rapidly phosphorylated. These results indicate that Hog1 kinase activity is necessary for the delayed activation of Hog1 in the ptc1Δ strain.
Ptc1 inactivates Hog1 in vitro.
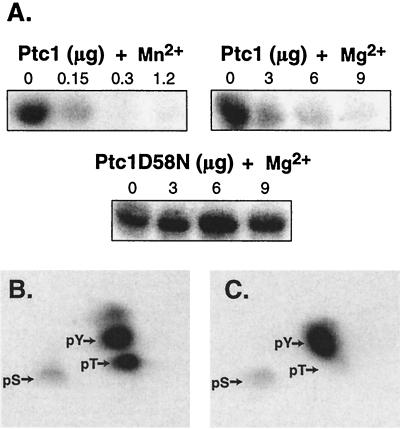
Since Ptc1 inactivates Hog1 in vivo, we asked whether Ptc1 could dephosphorylate Hog1 in vitro. Activated GST-Hog1 was isolated from yeast and treated with Ptc1 or the metal binding site mutant Ptc1D58N, purified from E. coli. GST-Hog1 kinase activity was then examined by incubation with MBP and [γ-32P]ATP. Treatment of GST-Hog1 with Ptc1 inactivated Hog1, while treatment with Ptc1D58N did not (Fig. 6A). We also examined the divalent cation requirement of Ptc1 for catalytic activity. Previously, a partially purified preparation of Ptc1 expressed in E. coli was shown to have greater activity with Mn2+ than with Mg2+, using phosphorylated casein as a substrate (40, 41). Using GST-Hog1 as a substrate, we found that Ptc1 had at least 10-fold-greater activity with Mn2+ than with Mg2+ (Fig. 6A).
FIG. 6.
Ptc1 inactivates Hog1 kinase activity in vitro by dephosphorylating pT. (A) Ptc1 inactivated Hog1 kinase activity in vitro. GST-Hog1 was isolated from osmotically stressed yeast strain JWY1 and incubated in the absence or presence of wild-type Ptc1 or mutant Ptc1D58N for 30 min at 30°C. Ptc1 or PtcD58N in the amounts specified (0 to 9 μg) was incubated in the presence of 5 mM Mg2+ or 20 mM Mn2+. The bead-bound GST-Hog1 was washed extensively and incubated with MBP and [γ-32P]ATP. Radiolabel incorporated into MBP was examined by PhosphorImager analysis. (B) Hog1 was phosphorylated at Thr and Tyr by the hyperactive MEK mutant, Pbs2EE, in vitro. GST-Hog1 was isolated from untreated yeast using glutathione-Sepharose. The bead-bound GST-Hog1 was phosphorylated by incubation with Pbs2EE purified from E. coli and [γ-32P]ATP. Phosphoamino acid analysis was performed to examine the level of pT and pY. Arrows indicate the position of phosphoamino acid standards as revealed by ninhydrin staining. pS, phosphoserine. (C) Ptc1 specifically dephosphorylates Hog1-pThr in vitro. 32P-phosphorylated GST-Hog1 was treated with Ptc1 and examined as above.
Since activation of Hog1 requires phosphorylation of both Thr174 and Tyr176 in the phosphorylation lip (42), we tested whether Ptc1 inactivates Hog1 by dephosphorylating either or both of these residues. Based on sequence similarity to PP2Cs, we expected that Ptc1 would specifically dephosphorylate the pT residue in the phosphorylation lip of Hog1. To test this, Hog1 was isolated from yeast grown under standard conditions and phosphorylated in vitro with radiolabeled ATP, using the hyperactive Pbs2 mutant Pbs2EE. Hog1 treated in this manner was phosphorylated predominantly at Thr and Tyr, with trace phosphorylation at Ser (Fig. 6B). Phosphorylation occurred exclusively at the phosphorylation lip Thr174 and Tyr176, since a parallel experiment performed with the mutant Hog1-T174A,Y176F showed no phosphorylation (data not shown). When 32P-labeled wild-type Hog1 was treated with Ptc1, it was dephosphorylated at pT but not at pY (Fig. 6C).
Subcellular localization of Ptc1.
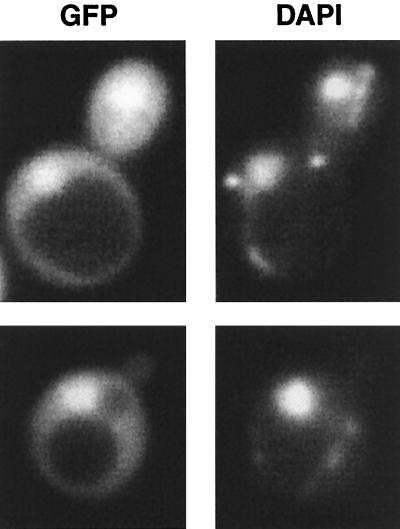
Since Hog1 accumulates in the nucleus upon osmotic stress (12, 26, 39), we predicted that at least a fraction of Ptc1 would be present in the nucleus. To address this question, GFP was fused to Ptc1 and expressed from either low-copy-number or multicopy plasmids in a ptc1Δ strain. In most cells, GFP-Ptc1 was found evenly distributed between the cytoplasm and the nucleus; in some cells, it was concentrated in the nucleus (Fig. 7). Osmotic stress did not alter the localization of Ptc1 (data not shown). Thus, the localization of Ptc1 is consistent with its ability to regulate the basal activity of Hog1, as well as dephosphorylating activated Hog1 found concentrated in the nucleus.
FIG. 7.
Ptc1 is both cytoplasmic and nuclear. The subcellular localization of a GFP-Ptc1 fusion protein expressed from a multicopy plasmid was examined in a ptc1Δ strain. Cells in exponential growth phase were visualized using a Zeiss fluorescence microscope with a charge-coupled device camera. 4′,6-Diamidino-2-phenylindole (DAPI) was used to visualize the nucleus.
DISCUSSION
In this study, we showed that Ptc1 inactivates the Hog1 MAPK in vivo and in vitro, establishing the importance of a PP2C in regulation of MAPK signaling in S. cerevisiae. The phosphatase activity of Ptc1 and of two closely related phosphatases, Ptc2 and Ptc3, was required for pathway inactivation since metal binding site mutants could not suppress the severe growth defect of the sln1Δ strain due to Hog1 hyperactivation. Ptc1 inhibited the HOG pathway by inactivating Hog1 but not Pbs2. Overexpression of PTC1 did not inhibit Tyr phosphorylation of Hog1, suggesting it does not act on Pbs2 (Fig. 4A). Similarly, overexpression of S. pombe Ptc1 and Ptc3 did not inhibit Tyr phosphorylation of Spc1/Sty1 (13, 31). Thus yeast PP2C appears to differ from mammalian PP2Cα, which inhibits the stress-activated MEKs, MKK6 and SEK1 (46). Further study of S. cerevisiae PTC1 showed that its overexpression inhibited osmotic stress-induced Hog1 kinase activation, while its deletion led to a defect in adaptation, elevated basal Hog1 kinase activity, and an inability to rapidly activate Hog1. The biochemical basis for Ptc1 effects on Hog1 activity was specific dephosphorylation of the pT but not the pY residue in the phosphorylation lip.
Examination of Hog1 kinase activity before and during treatment with osmotic stress showed that Ptc1 plays a role in adaptation. In the wild type, Hog1 reached a peak of activity at 3 min and declined rapidly thereafter, whereas in the ptc1Δ strain, Hog1 kinase activity did not decrease significantly for up to 1 h. In this respect, the role of Ptc1 is similar to that of the PTPs that dephosphorylate Hog1-pY. Previously, we and others showed that strains lacking the PTPs poorly dephosphorylated Hog1-pY during continuous exposure to osmotic stress (19, 51, 53). PTP-mediated adaptation may require their upregulation, since osmotic stress induces PTP2 and PTP3 transcripts in a Hog1-dependent manner (19). Unlike PTP transcripts, however, the PTC1 transcript did not increase in response to osmotic stress (data not shown).
A second role for Ptc1 is to maintain a low basal level of Hog1 activity. In the absence of osmotic stress, Hog1 kinase activity was higher in the ptc1Δ strain than in the wild type. We believe that increased Hog1 activity in the ptc1Δ strain is due to increased phosphorylation of Thr174, and not Thr174 and Tyr176, as Ptc1 is specific for the pT residue (Fig. 6C). That Thr174-phosphorylated Hog1 is active is supported by in vivo studies, where Hog1-Y176F, but not Hog1-T174A, allows growth on high-osmolarity media (42; S. S. Spencer and I. M. Ota, unpublished data).
Unexpectedly, maintaining a low basal Hog1 kinase activity was critical for its rapid signal-induced activation. Deletion of PTC1 led to a significant delay in Hog1 activation (Fig. 5A). One factor necessary for this delay was Hog1 kinase activity. Substitution of the wild-type Hog1 with the kinase-inactive Hog1-K52M led to a similar rapid activation in the ptc1Δ and wild-type PTC1 strains (Fig. 5B). One possible explanation for this delay could be Hog1 kinase-dependent activation of negative regulators. For example, PTP2 and PTP3 transcripts are upregulated by osmotic stress in a Hog1-dependent manner, and their constitutive overexpression in the ptc1Δ strain could inhibit Hog1 activation (19). Other mechanisms independent of gene expression could also come into play. For example, the level of negative regulators can be increased through their stabilization. Phosphorylation of the dual-specificity phosphatase MKP-1 by its substrate, ERK, was shown to increase MKP-1 activity by inhibiting its proteolysis (5).
An alternative explanation for delayed Hog1 activation in the ptc1Δ strain is that Ptc1 also acts as a positive regulator in the pathway. For example, MEK retrophosphorylated by MAPK at specific Thr residues is inhibitory for MEK activity (6). A Ser/Thr phosphatase that specifically dephosphorylates these sites could activate MEK. Whether Ptc1 could be such a phosphatase remains to be answered. A precedent for a Ser/Thr phosphatase acting as both a positive and a negative regulator has been observed in the Drosophila MAPK pathway regulating photoreceptor development. PP2A acts as a negative regulator downstream of Ras and a positive regulator downstream of Raf (47). Similarly, in Caenorhabditis elegans, PP2A acts as a positive regulator downstream of Ras (44). Further examination of these phosphatases will be required to determine the biochemical basis for their positive regulatory effect on MAPK pathways.
Genetic and biochemical evidence now show that Ptc1 and Ptp2 act together to regulate Hog1 activity in vivo. The synthetic growth defect of the ptc1Δ ptp2Δ strain due to Hog1 hyperactivation (19, 24) can be explained by increased phosphorylation of Hog1 at the phosphorylation lip residues. Increased phosphorylation of Thr174 in the ptc1Δ strain, or Tyr176 in the ptp2Δ strain, is not sufficient to produce a growth defect, but hyperphosphorylation of both residues in the ptc1Δ ptp2Δ strain is nearly lethal. The function of Ptc1 and PTPs separately regulating Hog1 activity via dephosphorylating pT or pY is not known. The S. pombe stress-activated MAPK, Spc1, is also regulated by PTPs and PP2Cs (28, 31, 43). Interestingly, Spc1-pY dephosphorylation is delayed compared to pT during adaptation to heat stress (31), but the biological function of this regulation is not known. Continued examination of the role of PTCs and how they act together with PTPs to regulate the HOG pathway should provide further insights into the regulation of MAPK pathways by protein phosphatases.
ACKNOWLEDGMENTS
This work was supported by grant RPG-98-353-01-TBE from the American Cancer Society. Dipesh Amin was supported by the Hughes Initiative.
We thank Tom Arnold, Tim Jacoby, and Anita Seto for construction of plasmids and Anne Burkholder, Chris Mattison, Christian Young, and Doris Heidysch for assistance with Hog1 phosphorylation assays. We also thank members of the lab and Natalie Ahn for critical reading of the manuscript.
REFERENCES
- 1.Alessi D R, Gomez N, Moorhead G, Lewis T, Keyse S M, Cohen P. Inactivation of p42 MAP kinase by protein phosphatase 2A and a protein tyrosine phosphatase, but not CL100 in various cell lines. Curr Biol. 1995;5:283–295. doi: 10.1016/s0960-9822(95)00059-5. [DOI] [PubMed] [Google Scholar]
- 2.Anderson N G, Maller J L, Tonks N K, Sturgill T W. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature. 1990;343:651–653. doi: 10.1038/343651a0. [DOI] [PubMed] [Google Scholar]
- 3.Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brewster J L, Devaloir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 5.Brondello J M, Pouyssegur J, McKenzie F R. Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science. 1999;286:2514–2517. doi: 10.1126/science.286.5449.2514. [DOI] [PubMed] [Google Scholar]
- 6.Brunet A, Pages G, Pouyssegur J. Growth factor-stimulated MAP kinase induces rapid retrophosphorylation and inhibition of MAP kinase kinase (MEK1) FEBS Lett. 1994;346:299–303. doi: 10.1016/0014-5793(94)00475-7. [DOI] [PubMed] [Google Scholar]
- 7.Cheng A, Ross K E, Kaldis P, Solomon M J. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 1999;13:2946–2957. doi: 10.1101/gad.13.22.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 9.Das A K, Helps N R, Cohen P T W, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 Å resolution. EMBO J. 1996;15:6797–6809. [PMC free article] [PubMed] [Google Scholar]
- 10.Dohmen R J, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- 11.Doi K, Gartner A, Ammerer G, Errede B, Shinkawa H, Sugimoto K, Matsumoto K. MSG5, a novel protein phosphatase, promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 1994;13:61–70. doi: 10.1002/j.1460-2075.1994.tb06235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrigno P, Posas F, Koepp D, Saito H, Silver P A. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaits F, Shiozaki K, Russell P. Protein phosphatase 2C acts independently of stress-activated kinase cascade to regulate the stress response in fission yeast. J Biol Chem. 1997;272:17873–17879. doi: 10.1074/jbc.272.28.17873. [DOI] [PubMed] [Google Scholar]
- 13a.Ghislain M, Dohmen R J, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proleolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- 14.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 15.Gomez N, Cohen P. Dissection of the protein kinase cascade by which nerve growth factor activates MAP kinases. Nature. 1991;353:170–173. doi: 10.1038/353170a0. [DOI] [PubMed] [Google Scholar]
- 16.Gustin M C, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haystead T A J, Weiel J E, Litchfield D W, Tsukitani Y, Fisher E J, Krebs E G. Okadaic acid mimics the action of insulin in stimulating protein kinase activity in isolated adipocytes. J Biol Chem. 1990;265:16571–16580. [PubMed] [Google Scholar]
- 18.Hovland P, Flick J, Johnston M, Sclafani R A. Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- 19.Jacoby T, Flanagan H, Faykin A, Seto A G, Mattison C, Ota I. Two protein tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J Biol Chem. 1997;272:17749–17755. doi: 10.1074/jbc.272.28.17749. [DOI] [PubMed] [Google Scholar]
- 20.Kamps M P, Sefton B M. Acid and base hydrolysis of phosphoproteins bound to immobilon facilitates analysis of phosphoamino acids in gel-fractionated proteins. Anal Biochem. 1989;176:22–27. doi: 10.1016/0003-2697(89)90266-2. [DOI] [PubMed] [Google Scholar]
- 21.Keyse S M. Protein phosphatases and the regulation of mitogen-activated protein kinase signaling. Curr Opin Cell Biol. 2000;12:186–192. doi: 10.1016/s0955-0674(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 22.Lewis T S, Shapiro P S, Ahn N G. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 23.Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 24.Maeda T, Tsai A Y M, Saito H. Mutations in a protein tyrosine phosphatase gene (PTP2) and a protein serine/threonine phosphatase gene (PTC1) cause a synthetic growth defect in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5408–5417. doi: 10.1128/mcb.13.9.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 26.Mattison C P, Ota I M. Two protein tyrosine phosphatases, Ptp2 and Ptp3, modulate the subcellular localization of the Hog1 MAP kinase in yeast. Genes Dev. 2000;14:1229–1235. [PMC free article] [PubMed] [Google Scholar]
- 27.Mattison C P, Spencer S S, Kresge K A, Lee J, Ota I M. Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol Cell Biol. 1999;19:7651–7660. doi: 10.1128/mcb.19.11.7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millar J B A, Buck V, Wilkinson M G. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell D A, Marshall T K, Deschenes R J. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 30.Nasmyth K, Reed S. Isolation of genes by complementation in yeast: molecular cloning of a cell cycle gene. Proc Natl Acad Sci USA. 1980;77:2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen A N, Shiozaki K. Heat-shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes Dev. 1999;13:1653–1663. doi: 10.1101/gad.13.13.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Rourke S M, Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ota I M, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 34.Posas F, Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2 MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 36.Posas F, Witten E A, Saito H. Requirement of Ste50 for osmostress-induced activation of the Ste11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol Cell Biol. 1998;18:5788–5796. doi: 10.1128/mcb.18.10.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Posas F, Wurgler-Murphy S M, Maeda T, Witten E, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 38.Ramezani R M, Buhring J G, Hollenberg C P. Ste50 is involved in regulating filamentous growth in the yeast Saccharomyces cerevisiae and associates with Ste11p. Mol Gen Genet. 1998;259:29–38. doi: 10.1007/s004380050785. [DOI] [PubMed] [Google Scholar]
- 39.Reiser V, Ruis H, Ammerer G. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol Cell Biol. 1999;10:1147–1161. doi: 10.1091/mbc.10.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson M K, Van Zyl W H, Phizicky E M, Broach J R. TPD1 of Saccharomyces cerevisiae encodes a protein phosphatase 2C-like activity implicated in tRNA splicing and cell separation. Mol Cell Biol. 1994;14:3632–3645. doi: 10.1128/mcb.14.6.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson M K, Phizicky E M. Purification and assay of the Ptc1/Tpd1 protein phosphatase 2C from the yeast Saccharomyces cerevisiae. Methods Mol Biol. 1998;93:235–242. doi: 10.1385/0-89603-468-2:235. [DOI] [PubMed] [Google Scholar]
- 42.Schuller C, Brewster J L, Alexander M R, Gustin M C, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiozaki K, Russell P. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- 44.Sieburth D S, Sundaram M, Howard R M, Han M. A PP2A regulatory subunit positively regulates Ras-mediated signaling during Caenorhabditis elegans vulval induction. Genes Dev. 1999;13:2562–2569. doi: 10.1101/gad.13.19.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sontag E, Federov S, Kamibayashi C, Robbins D, Cobb M, Mumby M. The interaction of SV40 small tumour antigen with protein phosphatase 2A stimulates the map kinase pathway and induces cell proliferation. Cell. 1994;75:887–897. doi: 10.1016/0092-8674(93)90533-v. [DOI] [PubMed] [Google Scholar]
- 46.Takekawa M, Maeda T, Saito H. Protein phosphatase 2Cα inhibits the human stress-responsive p38 and JNK MAPK pathways. EMBO J. 1998;16:4744–4752. doi: 10.1093/emboj/17.16.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wassarman D A, Solomon N M, Chang H C, Karim F D, Therrien M, Rubin G M. Protein phosphatase 2A positively and negatively regulates Ras1-mediated photoreceptor development in Drosophila. Genes Dev. 1996;10:272–278. doi: 10.1101/gad.10.3.272. [DOI] [PubMed] [Google Scholar]
- 48.Widmann C, Gibson S, Jarpe M B, Johnson G L. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 49.Wolfe K H, Shields D C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 50.Wu C, Whiteway M, Thomas D Y, Leberer E. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J Biol Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- 51.Wurgler-Murphy S M, Maeda T, Witten E A, Saito H. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhan X-L, Deschenes R J, Guan K-L. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG in Saccharomyces cerevisiae. Genes Dev. 1997;11:1690–1702. doi: 10.1101/gad.11.13.1690. [DOI] [PubMed] [Google Scholar]
- 53.Zhan X L, Guan K L. A specific protein-protein interaction accounts for the in vivo substrate selectivity of Ptp3 towards the Fus3 MAP kinase. Genes Dev. 1999;13:2811–2827. doi: 10.1101/gad.13.21.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]