Abstract
Five species of Bartonella have been reported to infect humans and cause a variety of diseases that can be difficult to diagnose. Four species of Bartonella have been reported to infect cats and dogs, and two of these species are considered to be zoonotic pathogens. Diagnosis of Bartonella infections is hampered by the slow, fastidious growth characteristics of Bartonella species. We report on the development of a single-step PCR-based assay for the detection and differentiation of medically relevant Bartonella species. PCR-mediated amplification of the 16S-23S rRNA intergenic region resulted in a product of a unique size for each Bartonella species, thereby allowing differentiation without the necessity of restriction fragment length polymorphism analysis or sequencing of the amplified product. The ability of the single-step PCR assay to differentiate between Bartonella species was determined with characterized isolates and blood samples from animals known to be infected with either Bartonella henselae, B. clarridgeiae, or B. vinsonii subsp. berkhoffii. The sensitivity of the single-step PCR assay relative to that of in vitro culture was determined with blood samples from B. henselae-infected cats. B. henselae target DNA was amplified from 100% of samples with greater than 50 CFU/ml and 80% of samples with 10 to 30 CFU/ml. The single-step assay described in the report expedites PCR-based detection and differentiation of medically relevant Bartonella species.
Bartonella species are gram-negative, fastidious, aerobic, short rod-shaped bacteria. The genus Bartonella includes 13 described species, 3 of which have been isolated from humans only (Bartonella bacilliformis, B. elizabethae, and B. quintana), 2 of which have been isolated from cats and dogs only (B. koehlerae and B. vinsonii subsp. berkhoffii, respectively), and 2 of which have been reported to infect both humans and cats (B. clarridgeiae and B. henselae). The remaining Bartonella species (B. doshiae, B. grahamii, B. peromysci, B. talpae, B. taylorii, and B. vinsonii [Baker strain]) have been isolated from the blood of rodents and moles (for reviews, see references 1 and 2). B. bacilliformis is the etiologic agent of Oroya fever and verruga peruana in South America. B. elizabethae was isolated from the blood of a patient with endocarditis. B. quintana is the etiologic agent of trench fever and has been shown to cause bacillary angiomatosis and endocarditis in immunocompromised individuals. B. henselae is most frequently associated with cat-scratch disease (CSD) and has also been associated with bacillary angiomatosis and endocarditis in immunocompromised individuals. B. clarridgeiae was originally isolated from the blood of a cat (7) and was subsequently associated with CSD in humans (19, 23). B. henselae and B. clarridgeiae are frequently isolated from domestic cats and are considered to be zoonotic pathogens. Cats can be persistently infected with B. henselae or B. clarridgeiae without obvious signs of illness (17, 30), although histopathologic lesions (11, 18) and a pathogenic strain of B. henselae which causes swelling at the site of inoculation, fever, and lethargy have been described (29). Recently, B. koehlerae, a proposed new species belonging to the genus Bartonella, has also been isolated from cats (9). Infection of domestic dogs with B. henselae and the association of CSD in humans with scratches from dogs have been described (16, 35). B. vinsonii subsp. berkhoffii has been reported to cause endocarditis in dogs (6).
Bartonella species are associated with a variety of human diseases. Unfortunately, diagnosis of diseases caused by these species is hampered by their slow, fastidious, growth characteristics. A variety of rapid detection assays have been developed, including PCR amplification of the 16S-23S rRNA intergenic region with genus- and species-specific primer sets (26), species-specific amplification of ftsZ gene sequences (15), repetitive-element PCR (32), restriction fragment length polymorphism (RFLP) analysis of PCR-amplified 16S rRNA genes (4, 8), RFLP analysis of the PCR-amplified 16S-23S rRNA intergenic region (24, 34), RFLP analysis of the PCR-amplified citrate synthase gene (28), and sequence analysis of the PCR-amplified citrate synthase gene (5, 13). However, each of these methods requires multiple PCR amplifications or additional sample-processing steps beyond the primary PCR amplification. Additionally, assay performance was demonstrated only with cultured bacteria for all but one (8) of the assays described above.
We report on the development of a single-step PCR-based assay for the differentiation of Bartonella species from one another. PCR is used to amplify a fragment of the 16S-23S rRNA intergenic region with primers complementary to sequences conserved in all medically relevant Bartonella species. The 16S-23S rRNA intergenic region was chosen because of its uniqueness compared to the sequences of other bacteria (33). Identification of conserved 16S-23S rRNA intergenic sequences for relevant Bartonella species required sequencing of the 16S-23S rRNA intergenic region from B. clarridgeiae and B. vinsonii subsp. berkhoffii. After amplification, PCR products are size fractionated by gel electrophoresis and Bartonella species are identified by the unique size of the amplified product without the necessity of further sample processing. The utility of this assay was tested by detection of Bartonella species directly from blood samples derived from bacteremic cats and dogs.
MATERIALS AND METHODS
Bacterial strains.
B. bacilliformis ATCC 35685, B. clarridgeiae ATCC 51734 and ATCC 700095, B. elizabethae ATCC 49927, B. quintana ATCC 51694, B. vinsonii subsp. berkhoffii ATCC 51572, and Brucella canis ATCC 23365 were obtained from the American Type Culture Collection (Rockville, Md.). B. henselae isolates Houston-1 (ATCC 49882), Oklahoma (ATCC 49793), Marseilles, MO-2, SA-1, CA-4, Tiger-2, and Lassiter were kindly provided by Russell Regnery, Viral and Rickettsial Diseases Branch, Centers for Disease Control and Prevention, Atlanta, Ga.
Clinical samples.
Blood was obtained by aseptic procedures from the jugular veins of cats or dogs and placed in tubes that contained EDTA anticoagulant. Molecular characterization of the B. henselae, B. clarridgeiae, and B. vinsonii subsp. berkhoffii isolates from these naturally infected cats and dogs has been reported previously (17).
Determination of number of Bartonella species CFU per milliliter of blood.
Blood from cats experimentally infected with the Houston-1 strain of B. henselae was collected and was placed in tubes with EDTA as an anticoagulant. The samples were stored at −70°C until they were processed for culture. A total of 100 μl of blood was serially diluted 10-fold three times in brain heart infusion broth. A total of 100 μl of blood and 100 μl of each serial dilution were plated onto heart infusion agar plates containing 5% rabbit blood (catalog no. 4321-356; BBL). Agar plates were placed inside plastic bags and were then incubated at 32°C with 5% CO2 for 1 week. Cultures were read at 1 week; negative plates were incubated for 5 weeks before they were discarded as negative.
DNA extraction and PCR amplification of the 16S-23S rRNA intergenic region.
DNA for PCR amplification was prepared from pure cultures of each bacterial strain by using the QIAamp DNA Mini Kit (QIAGEN Inc., Valencia, Calif.) and from blood by using the QIAamp DNA Blood Mini Kit. PCR amplifications were performed in mixtures of 50 μl containing 10 mM Tris (pH 8.3), 50 mM KCl, 3.5 mM MgCl2, 200 μM each dATP, dCTP, and dGTP, 400 μM dUTP, 1 μM each primer, and 2.5 U of Amplitaq Gold DNA polymerase (PE Applied Biosystems, Foster City, Calif.). Amplification buffer was optimized with dUTP for use with uracil glycosylase to prevent PCR amplification product carryover. Optimum primer annealing temperatures were determined in a RoboCycler Gradient Temperature Cycler (Stratagene, La Jolla, Calif.). Amplifications were performed in a GeneAmp PCR System 9700 thermal cycler (PE Applied Biosystems) by a timed-release PCR protocol (14), as follows: 10 min of incubation at 20°C, followed by 2 min of denaturation at 95°C and then 45 cycles of 1 min of denaturation at 95°C, 1 min of annealing at 60°C, and 30 s of extension at 72°C. PCR amplification products were identified by ethidium bromide fluorescence after electrophoresis in 3% agarose gels.
DNA sequencing.
PCR amplification of the entire 16S-23S rRNA intergenic region was accomplished with primers described by Matar et al. (24) and Roux and Raoult (33, 34). PCR products amplified from the 16S-23S rRNA intergenic regions of B. clarridgeiae and B. vinsonii subsp. berkhoffii were sequenced with an ABI PRISM model 377 with XL upgrade DNA Sequencer (PE Applied Biosystems) after product labeling with the PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems) by the manufacturer's protocol. Sequence alignments and phylogenetic comparisons were done with the DNA analysis computer program DNAsis (Hitachi Software Engineering America Ltd., South San Francisco, Calif.).
Nucleotide sequence accession numbers.
The 16S-23S rRNA intergenic sequences for B. clarridgeiae and B. vinsonii subsp. berkhoffii have been submitted to GenBank under accession nos. AF167989 and AF167988, respectively.
RESULTS
Comparison of 16S-23S rRNA intergenic sequences of Bartonella species.
The 16S-23S rRNA intergenic sequences for B. bacilliformis (GenBank accession no. L26364 [27]), B. elizabethae (GenBank accession no. L35103 [33]), B. henselae (GenBank accession no. L35101 [33]), B. quintana (GenBank accession no. L35100 [33]), B. vinsonii (Baker strain) (GenBank accession no. L35102 [33]), and B. abortus (GenBank accession no. X95889 [31]) were aligned by using the DNA analysis computer program DNAsis (Hitachi Software Engineering America Ltd.). Figure 1 illustrates the alignment of approximately 200 nucleotides in the 5′ region of the 16S-23S intergenic sequences. In this region, a nonconserved area is bordered by two areas with high degrees of homology. The sequences of individual Bartonella species differ in the nonconserved region primarily due to sequence insertions and/or deletions. The extent of variation suggested that PCR primers designed to amplify across the nonconserved region would generate amplified products of different sizes for each species of Bartonella. A PCR assay was designed to amplify the region shown in Fig. 1. Template DNA was obtained from B. bacilliformis, B. clarridgeiae, B. elizabethae, B. henselae, B. quintana, and B. vinsonii subsp. berkhoffii. B. koehlerae was not available for analysis at the time that this work was performed. Analysis of B. vinsonii (Baker strain) was not included because this Bartonella species has not been associated with disease in either humans or domestic animals. Template DNA was amplified using 5′-(C/T)CTTCGTTTCTCTTTCTTCA-3′ (B. henselae nucleotides [nt] 302 to 321) and 5′-GGATAAACCGGAAAACCTTC-3′ (B. henselae nt 448 to 429) as forward and reverse primers, respectively. The 16S-23S rRNA intergenic sequences predict that these primers should amplify products of 186 bp (B. bacilliformis), 216 bp (B. elizabethae), 147 bp (B. henselae), and 132 bp (B. quintana). A predicted product size could not be determined for B. clarridgeiae or B. vinsonii subsp. berkhoffii because the sequence of the 16S-23S rRNA intergenic region for these Bartonella species had not been reported. After amplification, the PCR products were electrophoresed on a 3% agarose gel, stained with ethidium bromide, and photographed. Products of the expected size were amplified from B. bacilliformis, B. elizabethae, B. henselae, and B. quintana template DNAs. The template DNA from B. vinsonii subsp. berkhoffii yielded a PCR product of approximately 235 bp rather than the 172-bp product predicted from the B. vinsonii (Baker strain) sequence. PCR amplification of the B. clarridgeiae template DNA yielded no product in reactions with these primers (data not shown).
FIG. 1.
Nucleotide sequence alignment of a portion of the 16S-23S rRNA intergenic region of B. bacilliformis (GenBank accession no. L26364), B. clarridgeiae (GenBank accession no. AF167989), B. elizabethae (GenBank accession no. L35103), B. henselae (GenBank accession no. L35101), B. quintana (GenBank accession no. L35100), B. vinsonii (Baker strain) (GenBank accession no. L35102), B. vinsonii subsp. berkhoffii (GenBank accession no. AF167988), and B. abortus (GenBank accession no. X95889). The corresponding GenBank nt numbers are indicated at the beginnings and ends of the sequences. The thin-line arrows designate the original PCR primer positions. The thick-line arrow designates the position of the revised reverse primer.
Sequencing of the 16S-23S rRNA intergenic region for B. clarridgeiae and B. vinsonii subsp. berkhoffii.
To investigate the failure to amplify a product from B. clarridgeiae and the discrepancy between the amplification product from B. vinsonii subsp. berkhoffii and the size predicted from the B. vinsonii (Baker strain) sequence, the 16S-23S rRNA intergenic regions from B. clarridgeiae and B. vinsonii subsp. berkhoffii were sequenced (GenBank accession nos. AF167989 and AF167988, respectively). Alignment of the B. vinsonii subsp. berkhoffii 16S-23S rRNA intergenic region revealed 63 bp inserted in the target region relative to the B. vinsonii (Baker strain) sequence (Fig. 1). Analysis of the B. clarridgeiae 16S-23S rRNA intergenic region sequence revealed that the 3′ nucleotide of the reverse PCR primer sequence is not conserved in B. clarridgeiae (Fig. 1). Annealing of the 3′ nucleotide is critical for extension by Taq polymerase, thus explaining the inability to amplify a PCR product from B. clarridgeiae template DNA.
PCR-based differentiation of Bartonella species.
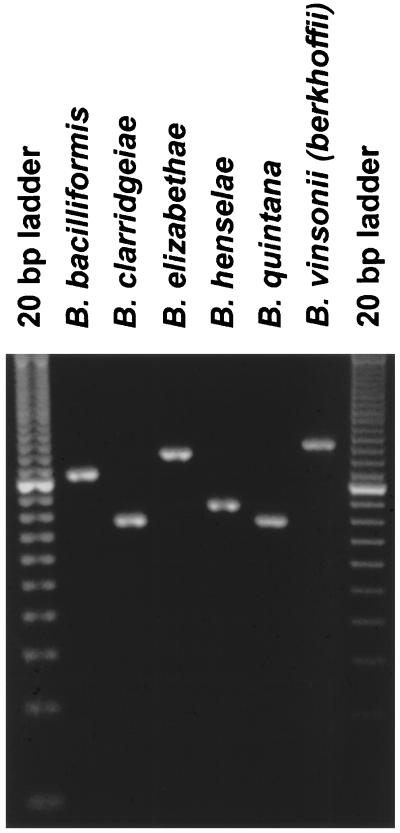
To detect and differentiate medically relevant Bartonella species, a new reverse primer complementary to the 16S-23S rRNA intergenic region sequences shared by all of the Bartonella species was selected for PCR amplification (Fig. 1). Amplification of template DNA with 5′-(C/T)CTTCGTTTCTCTTTCTTCA-3′ (B. henselae nt 302 to 321) and 5′-AACCAACTGAGCTACAAGCC-3′ (B. henselae nt 473 to 454) as forward and reverse primers, respectively, resulted in amplified products corresponding to those of the predicted size, namely, 211 bp (B. bacilliformis), 154 bp (B. clarridgeiae), 241 bp (B. elizabethae), 172 bp (B. henselae), 157 bp (B. quintana), and 260 bp (B. vinsonii subsp. berkhoffii) (Fig. 2). PCR products from amplification of B. clarridgeiae (154 bp) and B. quintana (157 bp) DNAs are not readily differentiated by 3% agarose gel electrophoresis (Fig. 2) but can be differentiated by 4.5% agarose or 10% polyacrylamide gel electrophoresis (data not shown). Amplification of template DNAs derived from the CA-4, MO-2, SA-1, Houston, Lassiter, Marseilles, Oklahoma, and Tiger-2 isolates of B. henselae yielded amplification products of the same size, demonstrating conservation of this target region among different isolates of B. henselae (data not shown). Amplification of template DNAs derived from Clostridium perfringens, Enterobacter cloacae, Escherichia coli, Ehrlichia canis, Ehrlichia chaffeensis, Ehrlichia equi, Ehrlichia ewingii, Ehrlichia risticii, Fusobacterium necrophorum, Klebsiella pneumoniae, Salmonella choleraesuis, and Staphylococcus intermedius did not result in product amplification (data not shown). Amplification of template DNA derived from Brucella canis, a member of the alpha-2 subdivision of the class Proteobacteria that is closely related to the genus Bartonella (10), yielded a product of 188 bp which was easily differentiated from the B. henselae (172-bp) and the B. bacilliformis (211-bp) products (data not shown). Amplification with forward and reverse primers that comprise sequences from adjacent regions, namely, 5′-CTCTTTCTTCAGATGATGATCC-3′ (B. henselae nt 311 to 332) and 5′-AACCAACTGAGCTACAAGCCCT-3′ (B. henselae nt 473 to 452) (Fig. 1), respectively, resulted in amplified products of approximately 202 bp (B. bacilliformis), 145 bp (B. clarridgeiae), 232 bp (B. elizabethae), 163 bp (B. henselae), 148 bp (B. quintana), and 251 bp (B. vinsonii subsp. berkhoffii) but failed to amplify a product from Brucella canis template DNA (data not shown).
FIG. 2.
PCR-based identification of Bartonella species. An ethidium bromide-stained agarose gel (3%) demonstrating amplified products from template DNAs derived from Bartonella species is shown. B. bacilliformis, B. clarridgeiae, B. elizabethae, B. henselae, B. quintana, and B. vinsonii subsp. berkhoffii yielded expected products of 211, 154, 241, 172, 157, and 260 bp, respectively. The first and last lanes each contain a 20-bp ladder.
Phylogenetic analysis of Bartonella species.
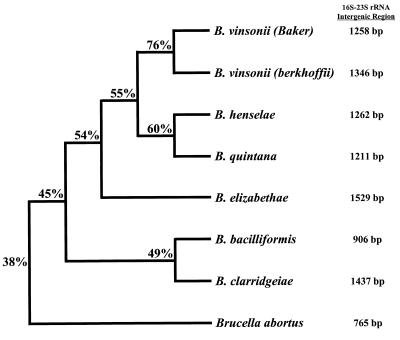
The entire 16S-23S rRNA intergenic sequence of B. clarridgeiae and B. vinsonii subsp. berkhoffii were compared with the reported sequences of B. bacilliformis, B. elizabethae, B. henselae, B. quintana, and B. vinsonii (Baker strain) (Fig. 3). The 16S-23S rRNA intergenic regions of Bartonella species vary from 906 bp in length (B. bacilliformis) to 1,529 bp (B. elizabethae) (33) and have from 45 to 76% homology. As expected, B. vinsonii subsp. berkhoffii is most closely related to B. vinsonii (Baker strain). On the basis of 16S-23S rRNA intergenic region sequences, B. clarridgeiae is most closely related to B. bacilliformis (Fig. 3). For comparison, Fig. 3 illustrates the relatedness of B. abortus to Bartonella species. The 16S-23S rRNA intergenic region of Brucella species are 765 bp in length, with approximately 99% homology among the six species (10, 31).
FIG. 3.
Phylogenetic comparison of 16S-23S rRNA intergenic region sequences for Bartonella species and B. abortus. Calculated matching percentages are indicated at each branch point of the dendrogram. The lengths of the horizontal and vertical lines are not significant. The lengths of the 16S-23S rRNA intergenic region sequences (in base pairs) are located at the right.
PCR detection of Bartonella species in clinical samples.
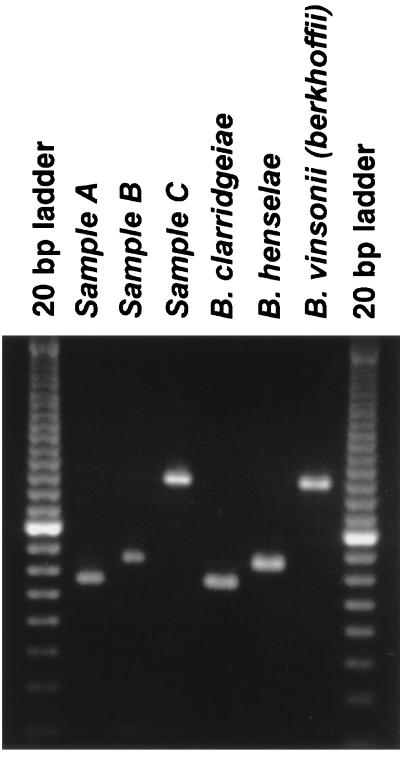
To evaluate the utility of the PCR assay for detection of Bartonella species in clinical samples, DNA was prepared from the blood of animals known to be infected with either B. henselae, B. clarridgeiae, or B. vinsonii subsp. berkhoffii. Briefly, DNA was extracted from 200 μl of blood by using the QIAamp Blood Kit (QIAGEN Inc., Santa Clarita, Calif.) and was eluted in a final volume of 200 μl according to the manufacturer's protocol. Samples (5 μl of template DNA) were amplified with the primers described above. After amplification, the PCR products were electrophoresed on a 3% agarose gel, stained with ethidium bromide, and photographed. As illustrated in Fig. 4, the single-step PCR assay is capable of detecting and differentiating infections with B. henselae, B. clarridgeiae, and B. vinsonii subsp. berkhoffii in clinical samples derived from naturally infected animals.
FIG. 4.
PCR-based identification of Bartonella species from animals known to be infected with either B. clarridgeiae (sample A), B. henselae (sample B), or B. vinsonii subsp. berkhoffii (sample C). DNA was extracted from 200 μl of blood and was eluted in a final volume of 200 μl, and then 5 μl of template DNA was used in each PCR amplification. After amplification, the PCR products were electrophoresed on a 3% agarose gel and stained with ethidium bromide. Amplified control template DNA derived from isolated B. clarridgeiae, B. henselae, and B. vinsonii subsp. berkhoffii strains yielded expected products of 154, 172, and 260 bp, respectively. The first and last lanes each contain a 20-bp ladder.
Sensitivity of PCR versus blood culture for detection of B. henselae.
To determine the sensitivity of the single-step PCR assay relative to that of blood culture, we purified template DNA from 200 μl of blood containing 10 to 100 CFU (per ml) of B. henselae derived from experimentally infected cats. DNA was eluted in 200 μl of buffer, and 5 μl was used as the template in the single-step PCR assay as described above. The single-step PCR assay detected B. henselae in 100% (12 of 12) of blood samples with 50 to 100 CFU/ml, 85% (6 of 7) of blood samples with 30 CFU/ml, and 75% (6 of 8) of blood samples with 10 to 20 CFU/ml.
DISCUSSION
Infections with Bartonella species have been associated with similar clinical signs. For example, B. elizabethae, B. henselae, and B. quintana have been isolated from people with endocarditis, CSD in humans can be caused by either B. henselae or B. clarridgeiae, and both B. henselae and B. quintana have been associated with bacillary angiomatosis in immunosuppressed individuals. Diagnosis of infections caused by Bartonella species by bacterial culture is difficult due to the fastidious, slow growth characteristics of Bartonella species. Serology is frequently used for the diagnosis of infections caused by Bartonella species. However, problems with both sensitivity (3, 21, 22) and specificity (20, 25) have been reported for serology-based diagnostic assays. Differences in in vitro susceptibilities to antimicrobial agents between various Bartonella species have been reported (12); however, in vitro activity is not necessarily predictive of treatment efficacy in people infected with Bartonella species (2). Identification of Bartonella species is important to clarify the epidemiology and clinical presentation of Bartonella species infections in humans and animals.
To expedite the diagnosis of Bartonella infections and identification of the Bartonella species causing infections, a number of diagnostic assays including PCR with species-specific primers (15, 26), RFLP analysis of PCR products (4, 8, 24, 28, 34), and sequencing of PCR products (5, 13) have been proposed for the differentiation of Bartonella species. While these methods detect and differentiate Bartonella species, all require either multiple PCR assays or additional sample-handling steps to obtain results. The present study was undertaken to develop a single-step PCR-based assay capable of detecting and differentiating Bartonella species. The 16S-23S rRNA intergenic region proved to be an appropriate target because it contains regions of sufficient sequence divergence to differentiate between Bartonella species as well as regions of adequate homology to enable the use of a single PCR primer pair (Fig. 1). A limitation of the single-step PCR assay described here is the inability to differentiate subspecies within different Bartonella species.
Phylogenetic analysis of Bartonella species has been based on the sequences of the 16S rRNA gene (5, 9); however, due to the highly conserved nature of this gene it is difficult to clearly distinguish relatedness. The sequence of the less conserved gltA gene has also been used for phylogenetic analysis of Bartonella species (5). On the basis of the gltA gene sequences, B. bacilliformis appears to be most closely related to B. henselae (5). Phylogenetic analysis based on sequences of the 16S-23S rRNA intergenic region suggests that B. bacilliformis is most closely related to B. clarridgeiae (Fig. 3). On the basis of both the gltA gene and the 16S-23S rRNA intergenic region sequences, B. henselae appears to be most closely related to B. quintana (5) (Fig. 3).
The single-step PCR assay described here can be used to directly screen samples from humans or animals, e.g., blood or tissue, for the presence of Bartonella species (Fig. 4). Using blood samples from B. henselae-infected cats, we compared the sensitivity of the single-step PCR assay to that of the culture assay. The theoretical sensitivity limit of the single-step PCR assay with 5 μl of template DNA, equivalent to 5 μl of blood, is 200 targets/ml. The observed sensitivity with 5 μl of template DNA was 100% for samples with 50 to 100 CFU/ml and 75% for samples with only 10 CFU/ml, or approximately 0.05 CFU. The ability to amplify B. henselae target DNA from template containing less than 1 CFU suggests amplification of target DNA derived from nonviable bacteria or, alternatively, inefficiency of the in vitro culture technique.
Although PCR-based assays for the detection and differentiation of Bartonella species have been described previously, a distinct advantage of the single-step PCR assay described here is that post-PCR sample handling is limited to gel electrophoresis. This technique dramatically decreases the cost and time required to obtain results as well as decreases the likelihood of sample contamination due to PCR product carryover. In conclusion, the single-step PCR assay described here provides a simple and rapid means of identifying pathogenic Bartonella species in humans and companion animals.
REFERENCES
- 1.Bass J W, Vincent J M, Person D A. The expanding spectrum of Bartonella infections. I. Bartonellosis and trench fever. Pediatr Infect Dis J. 1997;16:2–10. doi: 10.1097/00006454-199701000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Bass J W, Vincent J M, Person D A. The expanding spectrum of Bartonella infections. II. Cat-scratch disease. Pediatr Infect Dis J. 1997;16:163–179. doi: 10.1097/00006454-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Bergmans A M C, Peeters M F, Schellekens J F P, Vos M C, Sabbe L J M, Ossewaarde J M, Verbakel H, Hooft H J, Schouls L M. Pitfalls and fallacies of cat scratch disease serology: evaluation of Bartonella henselae-based indirect fluorescence assay and enzyme-linked immunoassay. J Clin Microbiol. 1997;35:1931–1937. doi: 10.1128/jcm.35.8.1931-1937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birtles R J. Differentiation of Bartonella species using restriction endonuclease analysis of PCR-amplified 16S rRNA genes. FEMS Microbiol Lett. 1995;129:261–266. doi: 10.1111/j.1574-6968.1995.tb07590.x. [DOI] [PubMed] [Google Scholar]
- 5.Birtles R J, Raoult D. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int J Syst Bacteriol. 1996;46:891–897. doi: 10.1099/00207713-46-4-891. [DOI] [PubMed] [Google Scholar]
- 6.Breitschwerdt E B, Kordick D L, Malarkey D E, Keene B, Hadfield T L, Wilson D. Endocarditis in a dog due to infection with a novel Bartonella subspecies. J Clin Microbiol. 1995;33:154–160. doi: 10.1128/jcm.33.1.154-160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarridge J E, Raich T J, Pirwani D, Simon B, Tsai L, Rodriguez-Barradas M C, Regnery R, Zollo A, Jones D C, Rambo C. Strategy to detect and identify Bartonella species in routine clinical laboratory yields Bartonella henselae from human immunodeficiency virus-positive patient and unique Bartonella strain from his cat. J Clin Microbiol. 1995;33:2107–2113. doi: 10.1128/jcm.33.8.2107-2113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dauga C, Miras I, Girmont P A. Identification of Bartonella henselae and B. quintana 16S rDNA sequences by branch-, genus-, and species-specific amplification. J Med Microbiol. 1996;45:192–199. doi: 10.1099/00222615-45-3-192. [DOI] [PubMed] [Google Scholar]
- 9.Droz S, Chi B, Horn E, Steigerwalt A G, Whitney A M, Brenner D J. Bartonella koehlerae sp. nov., isolated from cats. J Clin Microbiol. 1999;37:1117–1122. doi: 10.1128/jcm.37.4.1117-1122.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox K F, Fox A, Nagpal M, Steinberg P, Heroux K. Identification of Brucella by ribosomal-spacer-region PCR and differentiation of Brucella canis from other Brucella spp. pathogenic for humans by carbohydrate profiles. J Clin Microbiol. 1998;36:3217–3222. doi: 10.1128/jcm.36.11.3217-3222.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guptill L, Slater L, Wu C C, Lin T L, Glickman L T, Welch D F, HogenEsch H. Experimental infection of young specific pathogen-free cats with Bartonella henselae. J Infect Dis. 1997;176:206–216. doi: 10.1086/514026. [DOI] [PubMed] [Google Scholar]
- 12.Ives T J, Manzewitsch P, Regnery R L, Butts J D, Kebede M. In vitro susceptibilities of Bartonella henselae, B. quintana, B. elizabethae, Rickettsia rickettsii, R. conorii, R. akari, and R. prowazekii to macrolike antibiotics as determined by immunofluorescent-antibody analysis of infected vero cell monolayers. Antimicrob Agents Chemother. 1997;41:578–582. doi: 10.1128/aac.41.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joblet C, Roux V, Drancourt M, Gouvernet J, Raoult D. Identification of Bartonella (Rochalimaea) species among fastidious gram-negative bacteria on the basis of the partial sequence of the citrate-synthase gene. J Clin Microbiol. 1995;33:1879–1883. doi: 10.1128/jcm.33.7.1879-1883.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kebelmann-Betzing C, Seeger K, Dragon S, Schmitt G, Moricke A, Schild T A, Henze G, Beyermann B. Advantages of a new Taq DNA polymerase in multiplex PCR and time-release PCR. BioTechniques. 1998;24:154–158. doi: 10.2144/98241pf01. [DOI] [PubMed] [Google Scholar]
- 15.Kelly T M, Padmalayam I, Baumstark B R. Use of the cell division protein FtsZ as a means of differentiating among Bartonella species. Clin Diagn Lab Immunol. 1998;5:766–772. doi: 10.1128/cdli.5.6.766-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keret D, Giladi M, Kletter Y, Wientroub S. Cat-scratch disease osteomyelitis from a dog scratch. Br J Bone Joint Surg. 1998;80:766–767. [PubMed] [Google Scholar]
- 17.Kordick D L, Breitschwerdt E B. Persistent infection of pets within a household with three Bartonella species. Emerg Infect Dis. 1998;4:325–328. doi: 10.3201/eid0402.980225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kordick D L, Brown T T, Shin K, Breitschwerdt E B. Clinical and pathologic evaluation of chronic Bartonella henselae or Bartonella clarridgeiae infection in cats. J Clin Microbiol. 1999;37:1536–1547. doi: 10.1128/jcm.37.5.1536-1547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kordick D L, Hilyard E J, Hadfield T L, Wilson K H, Steigerwalt A G, Brenner D J, Breitschwerdt E B. Bartonella clarridgeiae, a newly recognized zoonotic pathogen causing inoculation papules, fever, and lymphadenopathy (cat scratch disease) J Clin Microbiol. 1997;35:1813–1818. doi: 10.1128/jcm.35.7.1813-1818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.La Scola B, Raoult D. Serological cross-reactions between Bartonella quintana, Bartonella henselae, and Coxiella burnetii. J Clin Microbiol. 1996;34:2270–2274. doi: 10.1128/jcm.34.9.2270-2274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Scola B, Raoult D. Culture of Bartonella quintana and Bartonella henselae from human samples: a 5-year experience (1993 to 1998) J Clin Microbiol. 1999;37:1899–1905. doi: 10.1128/jcm.37.6.1899-1905.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mainardi J L, Figliolini C, Goldstein F W, Blanche P, Baret- Rigoulet M, Galezowski N, Fournier P E, Raoult D. Cat scratch disease due to Bartonella henselae serotype Marseille (Swiss cat) in a seronegative patient. J Clin Microbiol. 1998;36:2800. doi: 10.1128/jcm.36.9.2800-2800.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margileth A M, Baehren D F. Chest-wall abscess due to cat-scratch disease (CSD) in an adult with antibodies to Bartonella clarridgeiae: case report and review of the thoracopulmonary manifestations of CSD. Clin Infect Dis. 1998;27:353–357. doi: 10.1086/514671. [DOI] [PubMed] [Google Scholar]
- 24.Matar G M, Swaminathan B, Hunter S B, Slater L N, Welch D F. Polymerase chain reaction-based restriction fragment length polymorphism analysis of a fragment of the ribosomal operon from Rochalimaea species for subtyping. J Clin Microbiol. 1993;31:1730–1734. doi: 10.1128/jcm.31.7.1730-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurin M, Eb F, Etienne J, Raoult D. Serological cross-reactions between Bartonella and Chlamydia species: implications for diagnosis. J Clin Microbiol. 1997;35:2283–2287. doi: 10.1128/jcm.35.9.2283-2287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minnick M F, Barbian K D. Identification of Bartonella using PCR; genus- and species-specific primer sets. J Microbiol Methods. 1997;31:51–57. [Google Scholar]
- 27.Minnick M F, Strange J C, Williams K. Characterization of the 16S-23S rRNA intergenic spacer of Bartonella bacilliformis. Gene. 1994;143:149–150. doi: 10.1016/0378-1119(94)90622-x. [DOI] [PubMed] [Google Scholar]
- 28.Norman A F, Regnery R, Jameson P, Greene C, Krause D C. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–1803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Reilly K L, Bauer R W, Freeland R L, Foil L D, Hughes K J, Rohde K R, Roy A F, Stout R W, Triche P C. Acute clinical disease in cats following infection with a pathogenic strain of Bartonella henselae (LSU16) Infect Immun. 1999;67:3066–3072. doi: 10.1128/iai.67.6.3066-3072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Regnery R L, Rooney J A, Johnson A M, Nesby S L, Manzewitsch P, Beaver K, Olson J G. Experimentally induced Bartonella henselae infections followed by challenge exposure and antimicrobial therapy in cats. Am J Vet Res. 1996;57:1714–1719. [PubMed] [Google Scholar]
- 31.Rijpens N P, Jannes G, Van Asbroeck M, Rossau R, Herman L M. Direct detection of Brucella spp. in raw milk by PCR and reverse hybridization with 16S-23S rRNA spacer probes. Appl Environ Microbiol. 1996;62:1683–1688. doi: 10.1128/aem.62.5.1683-1688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Barradas M C, Hamill R J, Houston E D, Georghiou P R, Clarridge J E, Regnery R L, Koehler J E. Genomic fingerprinting of Bartonella species by repetitive element PCR for distinguishing species and isolates. J Clin Microbiol. 1995;33:1089–1093. doi: 10.1128/jcm.33.5.1089-1093.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roux V, Raoult D. The 16S-23S rRNA intergenic spacer region of Bartonella (Rochalimaea) species is longer than usually described in other bacteria. Gene. 1995;156:107–111. doi: 10.1016/0378-1119(94)00919-j. [DOI] [PubMed] [Google Scholar]
- 34.Roux V, Raoult D. Inter- and intraspecies identification of Bartonella (Rochalimaea) species. J Clin Microbiol. 1995;33:1573–1579. doi: 10.1128/jcm.33.6.1573-1579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukahara M, Tsuneoka H, Iino H, Ohno K, Murano I. Bartonella henselae infection from a dog. Lancet. 1998;352:1682. doi: 10.1016/s0140-6736(05)61455-9. [DOI] [PubMed] [Google Scholar]