Abstract
Bovine tuberculosis caused by Mycobacterium bovis remains a significant disease of farmed cattle in many countries despite ongoing tuberculosis eradication programs. Molecular typing methods such as restriction fragment length polymorphism (RFLP) analysis and spoligotyping have been used to identify related herd breakdowns in an attempt to identify more precisely the route of infection into cattle herds and to trace the transmission of bovine tuberculosis. A recent geographical survey of Irish M. bovis isolates demonstrated that a significant proportion of isolates (∼20%) exhibit a common strain type, limiting the value of current strain typing methods as an epidemiological tool. We have identified and cloned a region of the M. bovis genome, pUCD, which generates a clear, highly polymorphic banding pattern when used as an RFLP probe on AluI restriction-digested M. bovis genomic DNA and which effectively subdivides this common strain type. When used to type 60 Irish M. bovis isolates, pUCD exhibited greater discriminatory power than the commonly used mycobacterial RFLP probes IS6110, PGRS, and DR and detected an equivalent number of strain types to a combination of these three probes. pUCD also detected significantly more strain types than the spoligotyping technique, while maintaining a high level of concordance between epidemiologically related and unrelated herd breakdowns. The polymorphic element within pUCD remains to be fully characterized, however the potential for this probe to greatly decrease the workload necessary to genotype M. bovis by RFLP analysis is compelling.
Bovine tuberculosis (bovine TB) caused by Mycobacterium bovis remains a significant disease of farmed cattle in the Republic of Ireland, incurring substantial annual economic costs through surveillance and eradication programs (21, 29). Despite the implementation in this country of a comprehensive testing and eradication scheme operated by the Department of Agriculture and Food since 1954, which has dramatically reduced the incidence of the disease, bovine TB persists within the Irish cattle population (11). Various reasons have been proposed to explain this residual level of infection, including a relatively high level of cattle movement within the country (20) and reservoirs of infection maintained in other domestic, feral, and wild species (15, 21). Although cattle are the definitive host of this pathogen, M. bovis has an exceptionally broad host range and can be maintained in a variety of mammalian species to a greater or lesser extent (35). In Ireland and the United Kingdom, the badger (Meles meles) is known to represent a significant source of infection outside of the cattle population and is thought to be a contributory factor in the repeated herd breakdowns observed in certain areas, although the precise dynamics of TB transmission between badgers and cattle have never been firmly established (17, 20).
Epidemiological analyses of M. bovis infection have been hampered in the past by the difficulty associated with distinguishing between isolates originating from different sources and species. In recent years, molecular biological techniques have been employed to examine M. bovis bacilli at the DNA level and to identify genotypically distinct strain types that may be presumed to be epidemiologically unrelated. Such data may provide information pertinent to routine field investigations undertaken following herd breakdowns and provide a greater understanding of disease transmission within the national cattle herd and between cattle and nonbovine transmission vectors.
Several strategies for typing M. bovis isolates on the basis of DNA polymorphisms have arisen in recent years. Techniques commonly used internationally include restriction fragment length polymorphism (RFLP) analysis (4, 7, 9, 25, 30, 31, 37), spoligotyping (4, 7, 16, 27), pulsed-field gel electrophoresis (12), and PCR-based techniques such as “ampliprinting” (13, 22).
RFLP analysis has been demonstrated to be a robust and highly discriminatory typing procedure due to the availability of multiple DNA probes for the detection of polymorphic loci within the M. bovis genome and has been the method of choice, alongside spoligotyping, for typing M. bovis isolates from cattle and other hosts in Ireland (4, 5, 30, 31). The most commonly used genetic markers for this type of analysis include the mycobacterial insertion sequence IS6110 (33, 34), the highly repetitive polymorphic GC-rich repeat sequence (PGRS) (27), and the direct repeat (DR) region of the mycobacterial genome (14). IS6110 has been widely used as a tool for genotyping Mycobacterium tuberculosis (19, 36), in which it is usually present in up to 20 copies (24). This DNA element has since been adopted for use as an RFLP probe for typing M. bovis isolates also (8, 18). In M. bovis however, IS6110 is generally found in one to five copies, greatly limiting the ability of this element to discriminate between different strains of this organism (12, 30). The majority of Irish bovine-derived M. bovis isolates typed to date contain a single, monomorphic copy of IS6110 which is insufficiently sensitive for epidemiological purposes, necessitating supplementary typing with additional probes (4, 30, 31).
The PGRS-based RFLP probe appears to be the single most discriminatory of the probes currently available for M. bovis strain typing (4, 7, 9, 25) and can be present in up to 30 copies in members of the M. tuberculosis complex (23, 28). PGRS is present in multiple copies interspersed throughout the genome and exhibits a high level of polymorphism between unrelated isolates. However, the result of a PGRS DNA fingerprint is relatively complex, containing a great many bands that can be difficult to interpret and which can hinder computer-assisted band analysis, particularly when the final image is less than ideal.
The third most commonly used RFLP probe for this kind of analysis is the DR region. Unique to members of the TB complex, the DR region consists of a series of identical 36-bp DR sequences interspersed with variable spacer sequences from 35 to 41 bp in length (14). This region has found application both as a target for RFLP typing and for the PCR amplification-based spoligotyping technique, both of which give a satisfactory level of strain discrimination (1, 7). The DR RFLP probe is targeted at the 36-bp DR sequences found in multiple copies within the DR region, whereas spoligotyping targets the spacer sequences in between. As both DR RFLP analysis and spoligotyping target the same genomic locus, albeit different areas within that same locus, the results of these two methods tend to be comparable in terms of strain discrimination and sensitivity (8). While having the advantages of being a considerably faster and less labor-intensive method to perform than RFLP analysis, spoligotyping alone does not usually provide sufficient discrimination between strains of M. bovis to be used as a sole typing method and is often combined with supplementary techniques such as RFLP typing with IS6110 and PGRS (4, 8, 26).
A recent geographical survey of Irish M. bovis isolates in which 452 M. bovis isolates were examined by RFLP analysis with IS6110, PGRS, and DR and also by spoligotyping (4) showed that 20% of isolates analyzed exhibited a common RFLP strain type, designated strain type A1 A1 A (IS6110, PGRS, and DR, respectively). This strain type, which cannot be further subdivided by existing RFLP methods, occurred in a variety of host species and was widely distributed across the country, appearing in geographically distant regions of the country where genetically isolated genotypes might be expected to arise. This strain type is similar to the most frequently occurring strain type identified from isolates in Northern Ireland (30). This may be due to an overall genetic homogeneity within the Irish M. bovis population and may represent a significant limitation on the usefulness of strain typing as an aid to the epidemiological investigation of herd breakdowns. As the collection and cultivation of isolates for typing is time-consuming and labor-intensive, there has been a need for supplementary methods of dividing this common type in order to maximize the amount of data that can be obtained from cultured material.
In this study, a new RFLP probe has been identified from an M. bovis genomic DNA library which is capable of splitting the most common M. bovis strain type observed in Irish isolates. Preliminary analysis indicates that this probe yields a clear, highly polymorphic banding pattern that has a discriminatory ability comparable to the current combination of IS6110, PGRS, and DR probes.
MATERIALS AND METHODS
The definitions suggested by Tenover et al. (32) have been adopted throughout this study. The term “isolate” refers to cultured bacteria grown from a single colony taken from a primary isolation plate, and an isolate is presumed to be derived from a single organism. Epidemiologically related isolates are isolates collected from a well-defined area as part of an epidemiological investigation that suggests that they may be derived from a common source. Similarly, “strain type” refers to an isolate or group of isolates that can be distinguished from other isolates by genotypic characteristics and may be considered a descriptive subdivision of a species.
Generation of an M. bovis genomic DNA library.
Total genomic DNA was extracted from cultured M. bovis bacilli as described previously (31). Genomic DNA was partially digested with the restriction enzyme MboI and fragments in the 5-to-6-kb size range were recovered by preparative gel electrophoresis (2). Purified genomic fragments were ligated into BamHI-digested plasmid pGEM4Z (Promega) and transformed into Escherichia coli DH10B cells (Gibco BRL). Transformants were plated onto 24- by 24-cm agar plates containing ampicillin at 100 μg/ml overlaid with a positively charged nylon membrane (Roche Molecular Biochemicals) and grown overnight at 37°C.
Primary library screening.
Approximately 3,000 recombinant E. coli colonies were screened for potentially repetitive mycobacterial genomic inserts. Colonies were lysed in situ, and plasmid DNA was covalently bound to nylon by UV cross-linkage. Filters were hybridized with total M. bovis genomic DNA labeled with digoxygenin (DIG; Roche) at a concentration of 30 ng per ml of hybridization buffer. Due to the highly repetitive nature of PGRS, steps were taken to minimize the changes of detecting recombinant clones containing PGRS by masking these sequences through the inclusion of unlabeled PGRS (in the form of plasmid pTBN12 [28]) at 100 μg/ml in both prehybridization and hybridization solutions. Prehybridization and hybridization solutions also contained unlabeled E. coli chromosomal DNA at 100 μg/ml to prevent cross-hybridization of M. bovis and E. coli genomic DNA. Prehybridization of membranes with unlabeled DNA was allowed to proceed for 4 h. Hybridization with labeled M. bovis genomic DNA was limited to 4 h to allow the preferential binding of repetitive elements over unique sequence, and signals were recorded by a brief flash exposure to X-ray film to detect the most strongly hybridizing colonies. All probes were labeled using the nonradioactive DIG system (Roche), and signal detection was carried out using anti-DIG alkaline phosphatase and the chemiluminescent substrate CDP-Star according to the manufacturer's instructions (Roche).
Secondary library screening.
Recombinant colonies that generated hybridization signals with M. bovis genomic DNA above background levels in duplicate screens were subcultured and subjected to a secondary screening process. Mycobacterial genomic inserts were excised by EcoRI and HindIII digestion and probed sequentially with M. bovis genomic DNA and the PGRS sequence contained within plasmid pTBN12. Inserts that gave strong hybridization signals with genomic DNA without binding PGRS were selected for further analysis. Clones that generated a hybridization signal with PGRS were discarded.
Probe generation.
Both the PGRS and DR probes were obtained as commercially synthesized oligonucleotides based on the consensus sequence of the PGRS repetitive element (5′ CCG TTG CCG CCG TTG CCG CCG TTG CCG CCG) (10) and the DR 36-bp repeat (5′ GTC GTC AGA CCC AAA ACC CCG AGA GGG GAC GGA AAC) (14), respectively. A DIG label was incorporated into these probes during synthesis. IS6110 was amplified by PCR using the primer 5′ ACG TAC GAA TTC TGA ACC GCC CCG G 3′ on the basis of its terminal inverted repeat sequence (34). The 1.3-kb IS6110 region was labeled with DIG by random primed DIG labeling (Hi-Prime DIG labeling Kit; Roche) and used as a probe in its entirety. Plasmid pUCD was extracted and purified by standard methods, and the complete plasmid was labeled with DIG, as the pGEM4Z vector backbone of pUCD had previously been hybridized to M. bovis DNA and did not generate a hybridization pattern.
RFLP methods.
Mycobacterial DNA extraction was carried out using cetyltrimethylammonium bromide extraction protocol as described previously (31). Genomic DNA (1 μg) was digested overnight at 37°C with 10 U of the restriction enzyme AluI for strain typing with pUCD, PGRS, and DR and with 10 U of PvuII for strain typing with IS6110. Digested DNA was electrophoresed for 12 to 16 h in 25-cm 1% agarose gels at 2 V/cm in Tris-borate-EDTA (TBE) electrophoresis buffer, transferred to a positively charged nylon membrane (Roche) by overnight Southern capillary blotting, and covalently bound by UV cross-linkage. Internal molecular size ladders were not included; instead, a DIG-labeled molecular size ladder (Roche Molecular Size Ladder VII) was included in the outside lanes. Hybridization was carried out overnight in a 50% formamide buffer at 42°C.
M. bovis isolates.
Nylon membranes containing AluI-digested genomic DNA samples from M. bovis isolates originating from different geographical regions of the country and from different host species were available for analysis with pUCD following routine typing. Isolates were obtained during the period 1993 to 1998 as part of routine epidemiological investigations and were identified as M. bovis by standard biochemical tests as described previously (6). Isolates were subcultured on 50 ml of Middlebrook 7H9 medium with oleic acid, albumin, dextrose, citric acid enrichment (Difco, Detroit, Mich.) for 4 weeks at 37°C and were harvested after inactivation by heating at 75°C for 1 h.
A total of 81 isolates of M. bovis, which were divided into two sample sets, were used in this preliminary study. The first set was composed solely of type A1 A1 A strains and contained 21 isolates which had all previously been identified as RFLP strain type A1 A1 A by IS6110, PGRS, and DR RFLP analysis, respectively, and type A1 by spoligotyping. This sample contained both epidemiologically related and nonrelated isolates originating from cattle (n = 15), pigs (n = 3), deer (n = 2), and a badger (n = 1). Eleven of the bovine isolates from five different herds were classed as being epidemiologically linked. Data pertaining to these isolates are listed in Table 1.
TABLE 1.
Breakdown of isolates by RFLP analysis and spoligotyping resultsa
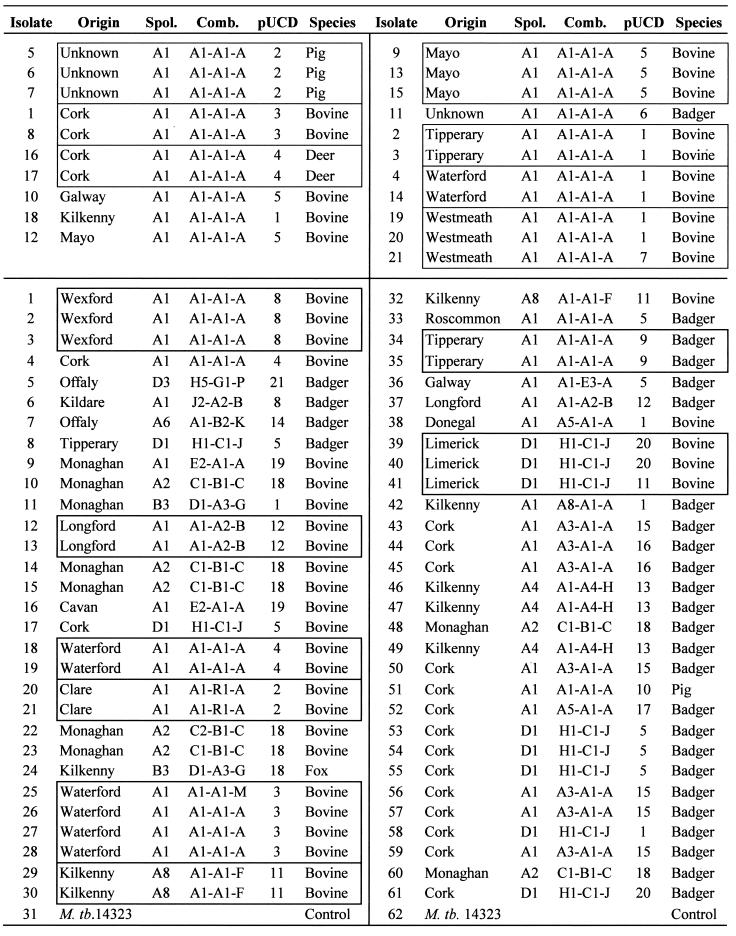
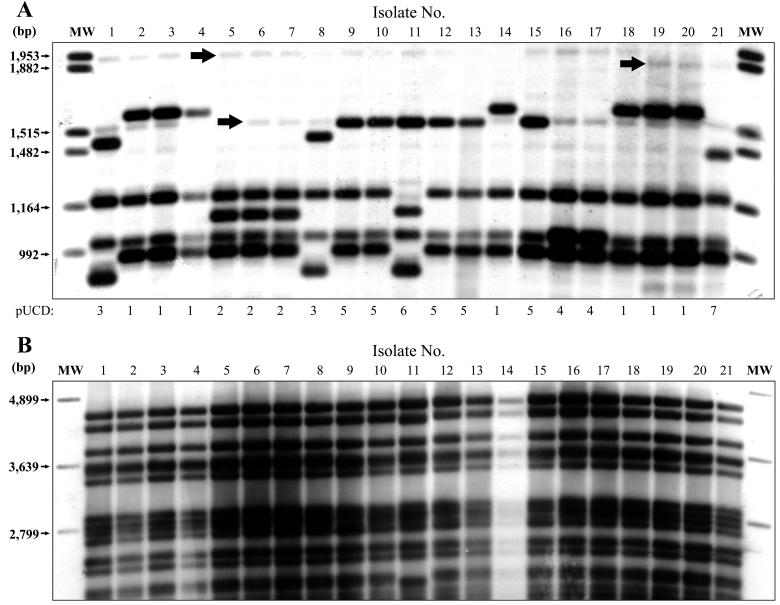
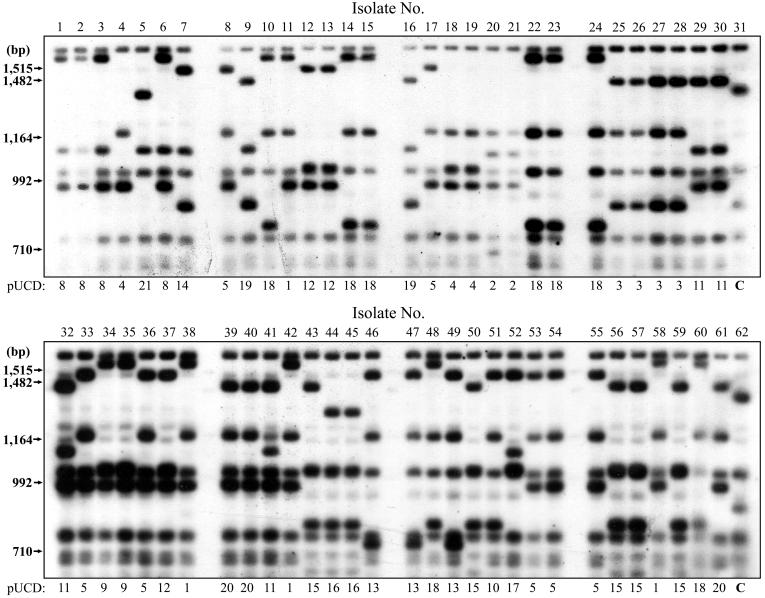
The top part of the table shows the breakdown for 21 isolates with the predominant strain type A1 A1 A. Isolate numbers in the top part of the table refer to the order in which they appear in Fig. 1, which illustrates the pUCD strain types obtained. The bottom part of the table shows strain types identified for a further 60 M. bovis isolates by RFLP analysis with IS6110, PGRS, DR, and pUCD and by spoligotyping. pUCD strain types for these isolates are illustrated in Fig. 2. Isolates originating from the same property which may be presumed to be epidemiologically related are shown boxed. Origin, geographic region of the country from which isolates were originally collected; Spol, spoligotype identified for each isolate; Comb., strain types identified by an IS6110-PGRS-DR probe combination. M. tb., M. tuberculosis.
A second, larger sample set contained a further 60 isolates, both epidemiologically related and nonrelated, from different geographic regions and was made up of isolates originating from cattle (n = 30), badgers (n = 28), a pig (n = 1), and a fox (n = 1). This set contained an additional 13 isolates of the A1 A1 A strain type. Eighteen of the bovine isolates from seven different properties were classed as epidemiologically linked, along with two badger isolates which were collected from the same property. The origin of these isolates is listed in Table 1.
Nomenclature.
The RFLP naming system adopted by Costello et al. (4) was used throughout this study, where each strain type is assigned an alphanumeric designation. Spoligotype patterns were identified by a letter and a number, and pUCD strain types were identified numerically.
Characterization of plasmid clone pUCD.
The M. bovis genomic insert contained within plasmid pUCD was partially characterized by subcloning and sequencing. The insert DNA was digested with AvaI to produce fragments of suitable size for subcloning. AvaI fragments were blunted by incubation with Taq DNA polymerase in the presence of deoxynucleoside triphosphates for 15 min at 70°C, ligated into a T vector (pGEM T-Easy; Promega), and transformed into E. coli DH10B cells (Gibco BRL). Recombinants were subcultured, and the plasmid DNA was extracted and purified for sequencing. DNA sequencing was carried out using an automated DNA sequencer (model 370A; Applied Biosystems).
RESULTS
Recovery of repetitive DNA elements from a genomic library.
A genomic DNA plasmid library prepared from M. bovis was screened for repetitive sequence elements by selecting recombinant clones that generated a strong hybridization signal with total M. bovis genomic DNA after a brief hybridization. Twenty recombinant E. coli colonies that exhibited hybridization signals above background levels were selected in this manner, 14 of which were examined for potential value as RFLP probes.
The efficacy of each of the recovered clones as a potentially useful RFLP probe was gauged on the basis of its ability to subdivide a group of 21 AluI-digested genomic DNA samples from epidemiologically related and unrelated M. bovis isolates previously identified as RFLP strain type A1 A1 A. The spoligotyping technique was also unable to distinguish between these isolates, identifying them all as type A1. Thirteen of the clones produced a large number of bands on hybridization with M. bovis genomic DNA, indicating that they were most likely derived from sequence elements that are repetitive in nature, but did not demonstrate notable levels of polymorphism between A1 A1 A isolates, save for minor pattern differences evident at one or two band positions (results not shown). No further analysis was carried out on these clones.
In contrast, one clone exhibited a relatively simple banding pattern with a considerable level of banding polymorphism between isolates. This clone, referred to here as pUCD, successfully distinguished between A1 A1 A isolates and was selected for further study on the basis of this ability.
Differentiation of the A1 A1 A strain type.
Plasmid pUCD subdivided 21 isolates of the prevalent strain type A1 A1 A into seven different banding patterns, as illustrated in Fig. 1A. These isolates were confirmed as being type A1 A1 A by typing with IS6110, PGRS, and DR, and the banding patterns generated by PGRS are reproduced in Fig. 1B for comparison. Known epidemiological links between the isolates were conserved among the strain types produced by pUCD, as demonstrated in Table 1. It can be seen that A1 A1 A isolates of different geographical origin have been successfully split with pUCD but remain grouped with IS6110, PGRS, and DR and also by spoligotyping, whereas isolates that originated from a single property remain identical.
FIG. 1.
(A) Twenty-one M. bovis isolates of the predominant strain type A1 A1 A subdivided by pUCD into seven strain types. pUCD strain types are shown underneath the panel. Arrows indicate some of the background banding evident with this probe that would not be included in strain type identification. Data relating to these isolates are listed in Table 1. MW, molecular weight. (B) Banding pattern produced when the same isolates are probed with PGRS. Molecular sizes are shown to the left. Images were acquired using a Microtek flatbed scanner and labeled using Adobe Photoshop version 4.0.
Performance comparison of pUCD with IS6110, PGRS, and DR RFLP analysis and spoligotyping.
Sixty epidemiologically related and nonrelated M. bovis isolates from across the country were typed by spoligotyping and RFLP analysis using IS6110, PGRS, DR, and pUCD (Fig. 2). Spoligotyping of these isolates resulted in 8 strain types. RFLP analysis using IS6110, PGRS, and DR identified 10, 11, and 10 strain types, respectively, and in combination split the group into 18 different strain types. pUCD typing alone resulted in 19 strain types. This group included an additional 13 isolates of the A1 A1 A strain type, which were subdivided into six strain types with pUCD.
FIG. 2.
Patterns generated with 60 M. bovis isolates by RFLP analysis using pUCD. The strain type identification is listed underneath each panel. Lanes 31 and 62 contain M. tuberculosis control strain 14323. Details relating to these isolates are listed in Table 1. Molecular sizes are shown to the left. Images were acquired using a Microtek flatbed scanner and labeled using Adobe Photoshop version 4.0.
A combination of the results obtained with pUCD and DR increased the total number of strain types detected to 26. A combination of all of the four probes discussed here yielded a total of 28 strain types for the 60 isolates; however, this does not represent a great improvement over the pUCD-DR combination and would demand substantially more time to perform.
It can be seen from Table 1 that, for the most part, isolates that originated from the same property and that so might be presumed to be of a single clonal origin are not subdivided by any of the probes. Isolates from different areas which might be presumed to be genetically unrelated have been distinguished by pUCD but remain grouped by RFLP analysis using IS6110, PGRS, and DR and by spoligotyping.
Characterization of pUCD.
The genomic insert contained within plasmid pUCD was subcloned and partially sequenced in order to characterize the DNA element(s) responsible for generating the polymorphism observed between M. bovis isolates. The sequence data obtained from three subclones were compared with the M. tuberculosis genome database maintained at the Sanger Centre (http://www.sanger.ac.uk/Projects/M_tuberculosis) using the BLAST search algorithm. Each subclone was aligned within a 4-kb region of the M. tuberculosis genome (bases 2163227 to 2166681) with >90% identity, indicating that a homologous region of the M. bovis genome is likely represented within pUCD. Subsequent bioinformatics analysis and comparison of the sequence data against the M. tuberculosis complete genome database at GenBank identified segment 85/162 of the M. tuberculosis H37Rv complete genome (3) as an analogous region (accession no. Z97193, bases 40021 to 43561). This region was identified as part of the PPE gene family, found widely dispersed throughout the mycobacterial genome (3). This sequence includes a series of five near-perfect 69-bp repeat units with the following sequence: TCGGTCCGATTGTGGTGCCGGATAT TACT T TCCTGGTAT TCCGTTGAGCCTGAACGCGCTGGGTGGTG. This region also contained three copies of a second repeat unit with the following sequence: TTAATCCGTTCGGTTTGAATATTCCGCTGAGCGGTGCTACCAACGCTGTCACGATCCCTGGTTTCGCGA. A subsequent BLAST search of this region against the M. tuberculosis database produced significant alignments with sequences also identified as PPE gene partial coding sequences (accession no. AF082288, AF082289, and AF086633) which contained similar repeat units.
DISCUSSION
Bovine TB remains a significant economic burden in Ireland despite 40 years of intensive control measures aimed at ensuring its complete eradication from the cattle population. Various authorities have concluded that, in countries where there is transmission of infection from spillover hosts into cattle, complete eradication is not a feasible goal and control measures must be applied indefinitely (11, 21, 29). However, the pathogen has been successfully eliminated in other European countries through similar practices, and the goals of the Department of Agriculture's eradication program remain unchanged. A more complete understanding of the epidemiology of M. bovis and its various routes of transmission into cattle will complement ongoing eradication strategies. The application of molecular typing technology is currently the most effective means of investigating this.
In a recent proposal for a standardized approach to DNA fingerprinting of M. bovis from an unknown population, Cousins et al. (8) recommended that isolates should be typed in the first instance with IS6110 in order to define the population, followed by either spoligotyping or RFLP analysis with PGRS, DR, or a combination of both when greatest sensitivity is required. The study recommended that, when isolates contain three or more copies of the IS6110 element, IS6110 RFLP typing should be sufficient for epidemiological purposes. In countries where the majority of bovine M. bovis isolates generally contain a single copy of IS6110, such as Ireland (4), Argentina (25), and Australia (7), genotyping with the IS6110 probe alone does not generate sufficient polymorphism to be of value to epidemiological studies, and secondary probes such as PGRS and DR are necessary to obtain suitable levels of discrimination. Whereas IS6110 generates maximal polymorphism when used to probe PvuII-digested genomic DNA, the restriction enzyme of choice for PGRS and DR analysis is AluI. The need for a second restriction enzyme when working with PGRS and DR necessitates the preparation of a second set of samples. In addition, samples to be analyzed with PGRS and DR should ideally be prepared separately, as each probe requires different electrophoresis conditions for optimum separation of the resulting bands. With PGRS the bands of interest are relatively large (2 to 5 kb), whereas DR detects much smaller bands (300 to 1,100 bp). This makes RFLP analysis a labor-intensive technique in situations where strain discrimination of the highest resolution is required, such as in areas that present a genetically homogeneous M. bovis population.
By screening a genomic library for potentially repetitive sequences, we have identified a region of the M. bovis chromosome which, used as an RFLP probe, may prove to be a useful adjunct to current genotyping protocols. This region, when hybridized to AluI-digested genomic DNA, generates a clear, highly polymorphic banding pattern which effectively split the common A1 A1 A strain type observed within Irish M. bovis isolates. A total of 34 A1 A1 A isolates were examined in this study, and these were subdivided into 10 strain types by pUCD. In addition, when used to strain type 60 isolates, pUCD detected a greater number of strain types than did IS6110, PGRS, DR, or spoligotyping and one more strain type than IS6110, PGRS, and DR in combination. As genomic DNA to be analyzed with both pUCD and DR is prepared using the restriction enzyme AluI, there is the option of using both of these probes consecutively on the same blot, as the gel running conditions normally applied for DR typing are also suitable for pUCD in terms of band spacing and definition. A pUCD-DR combination increased the total number of strain types detected within this study to 26. The restriction enzyme AluI was chosen for preparing template DNA, as membranes containing AluI-digested isolates were readily available from previous routine typing studies. pUCD has not as yet been assessed with different restriction enzymes which could potentially increase the discriminatory power of the probe, although such a change would preclude the subsequent use of the DR probe on the same membrane.
Discriminatory power alone is only one measure of the usefulness of an RFLP probe for epidemiological purposes and must be backed up by concordance with known data concerning specific disease outbreaks. The results observed for the set of isolates used in this study conformed well with known epidemiological information relating to the samples. A proportion of herd breakdowns will generally exhibit multiple strain types, particularly when several probes are used in the analysis, which may reflect infections with more than one clonal orgin. An example of this can be seen in Table 1, where samples 39, 40, and 41 represent three isolates originating from a single herd breakdown from County Limerick, Ireland. Isolates 39 and 40 were classed as pUCD strain type 20, whereas isolate 41 was identified as strain type 11, which suggests an additional route of infection into this herd. This occurrence was reiterated in samples 25, 26, 27, and 28, which originated from a single property in County Waterford, Ireland. (Table 1). In this case the DR probe identified a different strain type for sample 25.
The potential value of pUCD is illustrated in cases where M. bovis isolates cultured from neighboring herds or herd breakdowns in which an epidemiological link is not suspected are identical when genotyped with existing probes but may be distinguished by pUCD. An example of such a situation is contained within the sample of M. bovis isolates used in this study where isolates 19, 20, and 21 (Table 1; Fig. 1A) were obtained from herds in the same local region (County Westmeath, Ireland). Isolates 19 and 20 originated from the same herd, and isolate 21 originated from a neighboring property in the same local area. All three isolates were identical by RFLP analysis using IS6110, PGRS, and DR and also by spoligotyping, whereas RFLP analysis using pUCD displayed a different banding pattern for isolate 21.
The presence of a region homologous to the M. bovis-derived pUCD clone within the M. tuberculosis genome database suggests that this probe may also be of use for strain typing other mycobacterial species. M. tuberculosis 14323 was used as a control strain within this study and produced a banding pattern with pUCD, as would be expected. The polymorphic banding pattern produced by pUCD is suggestive of one or more repeated sequences approximately 50 to 100 bp long. The presence of ∼69-bp repeat units within the DNA sequences identified from the M. tuberculosis genome database seems to support this argument. In addition to the polymorphic bands of interest, the pattern displayed by pUCD includes a proportion of bands which do not contribute to strain differentiation. These bands are presumably derived from additional sequence elements contained within the cloned region, and it is hoped that further characterization of pUCD will allow the elimination of this background banding, simplifying the resulting pattern further.
The results reported here suggest that the RFLP probe pUCD provides discriminatory power equivalent to that of a IS6110-PGRS-DR probe combination in a single hybridization step, with a concomitant reduction in the time taken to obtain a final result. The discriminatory power of this probe along with its ability to subdivide the A1 A1 A strain type suggests that it will prove to be a useful epidemiological tool in the study of bovine TB transmission in cases where current probes reveal a high proportion of a common strain type.
ACKNOWLEDGMENTS
We gratefully acknowledge the assistance of Michael Sheridan and Ian O'Boyle of ERAD for their contribution to the organization of this study. We acknowledge the contribution of Frances Quigley, Anthoney Gogarty, and John McGuirk of the Central Veterinary Research Laboratory, Abbotstown. We thank Ciaran O'Brien for photographic reproduction.
This project was funded by the Department of Agriculture and Food.
REFERENCES
- 1.Aranaz A, Liébana E, Mateos A, Dominguez L, Vidal D, Domingo M, Gonzáles O, Rodridues-Ferri E F, Bunshoten A, van Embden J, Cousins D. Spoligotyping of Mycobacterium bovis strains from cattle and other animals: a tool for epidemiology of tuberculosis. J Clin Microbiol. 1996;34:2734–2740. doi: 10.1128/jcm.34.11.2734-2740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. I. New York, N.Y: Wiley, Inc.; 1994. p. 6.4.6. [Google Scholar]
- 3.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares S, Squares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 4.Costello E, O'Grady D, Flynn O, O'Brien R, Rogers M, Quigley F, Egan J, Griffin J. Study of restriction fragment length polymorphism analysis and spoligotyping for epidemiological investigation of Mycobacterium bovis infection. J Clin Microbiol. 1999;37:3217–3222. doi: 10.1128/jcm.37.10.3217-3222.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costello E, O'Grady D, O'Brien R, Flynn O, Rogers M, Quigley F, Egan J, Griffin J M. Strain typing of Mycobacterium bovis isolates. Ir Vet J. 1998;51:133. [Google Scholar]
- 6.Costello E, Quigley F, Flynn O, Gogarty A, McGuirk J, Murphy A, Dolan L. Laboratory examination of suspect tuberculous lesions detected on abattoir post mortem examination of cattle from non-reactor herds. Ir Vet J. 1998;51:248–250. [Google Scholar]
- 7.Cousins D, Williams S, Liébana E, Aranaz A, Bunschoten A, van Embden J, Ellis T. Evaluation of four DNA typing techniques in epidemiological investigations of bovine tuberculosis. J Clin Microbiol. 1998;36:168–178. doi: 10.1128/jcm.36.1.168-178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cousins D V, Skuce R A, Kazwala R R, van Embden J D A. Towards a standardized approach to DNA fingerprinting of Mycobacterium bovis. Int J Tuberc Lung Dis. 1998;2:471–478. [PubMed] [Google Scholar]
- 9.Cousins D V, Williams S N, Ross B C, Ellis T M. Use of a repetitive element isolated from Mycobacterium tuberculosis in hybridisation studies with Mycobacterium bovis: a new tool for epidemiological studies of bovine tuberculosis. Vet Microbiol. 1993;37:1–17. doi: 10.1016/0378-1135(93)90178-a. [DOI] [PubMed] [Google Scholar]
- 10.Doran T J, Hodgson A L M, Davies J K, Radford A J. Characterisiation of a highly repeated DNA sequence from Mycobacterium bovis. FEMS Microbiol Lett. 1993;111:147–152. doi: 10.1111/j.1574-6968.1993.tb06377.x. [DOI] [PubMed] [Google Scholar]
- 11.Downey L. Bovine TB programme. What are the realistic expectations? Dublin, Ireland: Eradication of Animal Disease Board; 1991. [Google Scholar]
- 12.Feizabadi M M, Robertson I D, Cousins D V, Hampson D J. Genomic analysis of Mycobacterium bovis and other members of the Mycobacterium tuberculosis complex by isoenzyme analysis and pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:1136–1142. doi: 10.1128/jcm.34.5.1136-1142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glennon M, Jëger B, Dowdall D, Maher M, Dawson M, Quigley F, Costello E, Smith T. PCR-based fingerprinting of Mycobacterium bovis isolates. Vet Microbiol. 1997;54:235–245. doi: 10.1016/s0378-1135(96)01280-1. [DOI] [PubMed] [Google Scholar]
- 14.Hermans P W M, van Soolingen D, Bik E M, de Haas P E W, Dale J W, van Embden J D A. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes M S, Neill S D, Rogers M S. Vaccination of the badger (Meles meles) against Mycobacterium bovis. Vet Microbiol. 1996;51:363–379. doi: 10.1016/0378-1135(96)00051-x. [DOI] [PubMed] [Google Scholar]
- 16.Kamerbeek K, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Sjoukje S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krebs J R. Bovine tuberculosis in cattle and badgers. MAFF report. London, United Kingdom: Her Majesty's Stationery Office; 1997. [Google Scholar]
- 18.Liébana E, Aranaz A, Dominguez L, Mateos A, González-Llamazares O, Rodriguez-Ferri E F, Domingo M, Vidal D, Cousins D. The insertion sequence IS6110 is a useful tool for DNA fingerprinting Mycobacterium bovis isolates from cattle and goats in Spain. Vet Microbiol. 1997;54:223–233. doi: 10.1016/s0378-1135(96)01282-5. [DOI] [PubMed] [Google Scholar]
- 19.Mazurek G H, Cave M D, Eisenach K D, Wallace R J, Jr, Bates J H, Crawford J T. Chromosomal DNA fingerprint patterns produced with IS6110 as strain specific markers for epidemiologic study of tuberculosis. J Clin Microbiol. 1991;29:2030–2033. doi: 10.1128/jcm.29.9.2030-2033.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Connor R, O'Malley E. Badgers and bovine tuberculosis in Ireland. Dublin, Ireland: Eradication of Animal Disease Board; 1989. [Google Scholar]
- 21.O'Reilly L M, Daborn C J. The epidemiology of Mycobacterium bovis in animals and man: a review. Tuberc Lung Dis. 1995;76(Suppl. 1):1–46. doi: 10.1016/0962-8479(95)90591-x. [DOI] [PubMed] [Google Scholar]
- 22.Plikaytis B B, Crawford J T, Woodley C L, Butler W R, Eisenach K D, Cave M D, Shinnick T M. Rapid, amplification-based fingerprinting of Mycobacterium tuberculosis. J Gen Microbiol. 1993;139:1537–1542. doi: 10.1099/00221287-139-7-1537. [DOI] [PubMed] [Google Scholar]
- 23.Poulet S, Cole S T. Characterization of the highly abundant polymorphic GC-rich repetitive sequence (PGRS) present in Mycobacterium tuberculosis. Arch Microbiol. 1995;163:87–95. doi: 10.1007/BF00381781. [DOI] [PubMed] [Google Scholar]
- 24.Poulet S, Cole S T. Repeated DNA sequences in mycobacteria. Arch Microbiol. 1995;165:79–86. doi: 10.1007/BF00381780. [DOI] [PubMed] [Google Scholar]
- 25.Romano M I, Alito A, Fisanotti J C, Bigi F, Kantor I, Cicuta M E, Cataldi A. Comparison of different genetic markers for molecular epidemiology of bovine tuberculosis. Vet Microbiol. 1996;50:59–71. doi: 10.1016/0378-1135(95)00197-2. [DOI] [PubMed] [Google Scholar]
- 26.Roring S, Brittain D, Bunschoten A E, Hughes M S, Skuce R A, van Embden J D A, Neill S D. Spacer oligotyping of Mycobacterium bovis isolates compared to typing by restriction fragment length polymorphism using PGRS, DR and IS6110 probes. Vet Microbiol. 1998;61:111–120. doi: 10.1016/s0378-1135(98)00178-3. [DOI] [PubMed] [Google Scholar]
- 27.Roring S, Hughes M S, Beck L-A, Skuce R A, Neill S D. Rapid diagnosis and strain differentiation of Mycobacterium bovis in radiometric culture by spoligotyping. Vet Microbiol. 1998;61:71–80. doi: 10.1016/s0378-1135(98)00167-9. [DOI] [PubMed] [Google Scholar]
- 28.Ross B C, Raois K, Jackson K, Dwyer B. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiological tool. J Clin Microbiol. 1992;30:942–946. doi: 10.1128/jcm.30.4.942-946.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehy S J, Christiansen K H. Cost/Benefit analysis of Irish bovine tuberculosis eradication schemes. Dublin, Ireland: Eradication of Animal Disease Board; 1991. [Google Scholar]
- 30.Skuce R A, Brittain D, Hughes M S, Neill S D. Differentiation of Mycobacterium bovis isolates from animals by DNA typing. J Clin Microbiol. 1996;34:2469–2474. doi: 10.1128/jcm.34.10.2469-2474.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skuce R A, Brittain D, Hughes M S, Beck L-A, Neill S D. Genomic fingerprinting of Mycobacterium bovis from cattle by restriction fragment length polymorphism analysis. J Clin Microbiol. 1994;32:2387–2392. doi: 10.1128/jcm.32.10.2387-2392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thierry D, Brisson-Nöel A, Vincent-Lévy-Frébault V, Nguyen S, Guesdon J, Gicquel B. Characterization of a Mycobacterium tuberculosis insertion sequence IS6110, and its application in diagnosis. J Clin Microbiol. 1990;28:2668–2673. doi: 10.1128/jcm.28.12.2668-2673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thierry D, Cave M D, Eisenach K D, Crawford J T, Bates J H, Gicquel B, Guesdon J L. IS6110, an IS-like element of the Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorns C J, Morris J A. The immune spectrum of Mycobacterium bovis infections in some mammalian species: a review. Commonwealth Bur Anim Health Vet Bull. 1983;53:543–550. [Google Scholar]
- 36.Van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Soolingen D, de Haas P E W, Haagsma J, Eger T, Hermans P W M, Ritacco V, Alito A, van Embden J D A. Use of various genetic markers in differentiation of Mycobacterium bovis strains from animals and humans and for studying epidemiology of bovine tuberculosis. J Clin Microbiol. 1994;32:2425–2433. doi: 10.1128/jcm.32.10.2425-2433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]