La is a conserved RNA-binding phosphoprotein that interacts with a large variety of ligands. The most ubiquitous function of La is association with newly synthesized RNA polymerase (Pol) III transcripts via their common UUU-OH 3′ termini and stabilization of these against exonucleolytic digestion. Accumulating evidence also indicates an activity for La in internal ribosome entry site-mediated translation in mammalian cells and in the metabolism of a subset of 3′-processed snRNA intermediates that end in uridylates but are synthesized by Pol II. The most highly conserved region of La resides in the N-terminal domain (NTD), and this appears to mediate high-affinity UUU-OH recognition. As critically reviewed here by comparison to a consensus core RNA recognition motif (RRM) structure, the NTD can be modeled into a pair of tandem RRMs. In addition to the conserved NTD, human La (hLa) protein contains a C-terminal domain (CTD) that harbors a third RRM and a potential Walker A motif that appears to recognize the 5′-ppp ends of nascent RNAs. The resulting bipartite mode of RNA binding can account for previously unexplained observations and may underlie a unifying principle of La function. While a role for hLa in transcription remains controversial, its presence in a Pol III holoenzyme suggests a role reminiscent of the CTD of Pol II, as an integrator of transcriptional and posttranscriptional activities that include 5′- and 3′-RNA metabolism. Evidence that the 5′-end-RNA recognition activity of hLa can be modulated by phosphorylation provides mechanistic insight into the signal transduction function of this protein.
La was first described as a human autoantigen more than 20 years ago and has since been identified in many eukaryotes from yeasts to humans (79, 143). La has been implicated in several cellular and viral RNA-associated processes, the most ubiquitous of which is the binding to and stabilization of newly synthesized RNA polymerase (Pol) III transcripts (6, 17, 41, 44, 58, 67, 88, 103, 143, 153, 154). These transcripts are the precursors to tRNAs, 5S rRNA, U6 snRNA, 7SL (SRP) RNA, 7SK snRNA, hY scRNAs, 4.5S I RNA, B1-Alu, Alu RNAs, and other RNAs (24, 43, 76, 81, 86, 89, 113–115). Binding occurs via the common UUU-OH 3′-terminal motif which results from transcription termination within the Pol III termination signal, oligo(dT) (58, 89, 113, 115, 136). As a result of UUU-OH binding and other activities, human La (hLa) also appears to function in transcription termination and reinitiation by Pol III (29, 42, 44–47, 87, 88), although this remains controversial (43, 82, 146, 154).
La proteins have been differentially expanded in the mass and domain complexity of their C-terminal domains (CTD) and utilize different pathways of nuclear import in different species. La protein of the yeast Saccharomyces cerevisiae is imported into the nucleus via a pathway that is different from the pathway used by the fission yeast, Schizosaccharomyces pombe, and higher eukaryotes (117). The type of nuclear localization signal (NLS) as well as the transport factors that mediate nuclear import of S. cerevisiae La is different from the NLS type and the import factors used in fission yeast and other eukaryotes (117). Thus, the NLS resides in the conserved N-terminal domain (NTD) of S. cerevisiae La but has been relegated to the CTD in all other eukaryotes examined, removed from the RNA-binding domain (117). It was suggested previously that this reorganization was associated with the acquisition of additional activities of La during evolution (117). In any case, it should be kept in mind that, while all identifiable La proteins share a highly conserved NTD, the trans-acting factors that recognize these proteins and their biological functions may differ significantly.
hLa is a 47-kDa phosphoprotein that is present at approximately 2 × 107 molecules per cell, mostly nuclear, although nucleolar (11, 36, 37, 48, 58) and cytoplasmic (54, 58, 93) localization has also been documented. The determinants in La responsible for nucleolar localization have not been elucidated (36, 37, 48, 58). Most La is extractable in a readily soluble form while a significant amount appears to be tightly associated with the nuclear matrix or other complexes (54). In addition to an NLS in the CTD, La also contains sequence elements that retain it in the nucleus (131). Accordingly, La can mediate transport of some RNAs into the nucleus and, for these and others, can retain them there, provided that they retain their 3′ UUU termini (15, 49, 132). Recently, Euplotes telomerase holoenzyme was found associated with a protein homolog of La. Since Euplotes telomerase RNA is a Pol III product that retains its UUU terminus as a small nuclear RNA, this La homolog may function as an ancillary factor to chaperone or retain telomerase RNA in the nucleus (reference 2 and references therein).
In addition to nascent Pol III transcripts, La also associates with other small RNAs. The first of this class to be described were the leader transcripts generated from vesicular stomatitis virus (78), although there are now reports of binding to the 5′ regions of the transcripts of many viruses (see below). Accumulating evidence indicates that La also associates with precursor intermediates of U1 to U5 snRNAs as well as U3 snoRNA, which are synthesized by Pol II (77, 85, 150). Apparently, the ends of these intermediates are exonucleolytically trimmed to expose 3′ uridylates prior to La binding (77, 85, 150). La was also found associated with an intermediate in a histone mRNA decay pathway whose 3′ end was processed at a position that would be expected to expose terminal uridylates (91). Thus, for many if not all of the above transcripts, binding appears to depend on La's best-known RNA-binding activity, recognition of 3′-terminal uridylates.
Many if not all of the above transcripts would presumably also share a triphosphate moiety on their initiating ends. Phosphatase treatment, which would remove the 5′ phosphates, has been shown to decrease La's affinity for UUU-OH-containing pre-tRNA in a functionally relevant manner (41). Direct binding as well as RNase P protection assays using a panel of mutant La proteins suggests that recognition of the 5′-ppp-RNA motif may be mediated by a Walker A motif (WAM) in the CTD of hLa (41). The modulation of this RNA recognition activity by phosphorylation and its apparent role in RNA expression in vivo (41, 67) will be reviewed below.
The hLa antigen.
Autoantigenic epitopes correspond to three major regions of the hLa protein, RRM-2 > RRM-1 > CTD (22, 25), and subpopulations of antibodies directed to these can vary during the course of autoimmune disease (23, 135). It should be noted that autoantigenic epitopes can differ from epitopes recognized during experimental immunization. La is targeted for proteolysis during infection by certain viruses and in the early phase of apoptosis, and recent evidence suggests that this may result in its autoantigenicity in some patients (19, 118). Accordingly, proteolytically exposed epitopes recognized during this phase of the immune response may be inaccessible in the native protein. Although some anti-La sera recognize more than one antigen, monospecific sera, e.g., in recognition of a single antigen by immunoblotting and/or other assays, are also available (e.g., see references 25 and 58).
The highly conserved NTD of La can be modeled into a tandem RRM domain.
After cloning of the cDNAs encoding La and other RNA-binding proteins, La was recognized as a founding member of a family of RNA-binding proteins that contain one or more versions of a domain known as the RNA recognition motif (RRM; also known as the RNA-binding domain) (22, 26, 112). A hallmark of the RRM is the presence of two short sequence motifs, RNP-2 and RNP-1 (1, 73, 119). Although attempts in multiple labs have not yet led to a crystal or nuclear magnetic resonance structure for La, the structures of other proteins have revealed the RRM as a globular domain with a β1-α1-β2-β3-α2-β4 topology (31, 38, 55, 61, 102, 122). In the RRM structure, the four β strands form a β sheet that contacts RNA on one surface while the other surface is packed by the two α helices, contributing to a hydrophobic core structure. The two RNP motifs occupy central locations on the β sheet, one each on the two internal β strands (RNP-2 on β1 and RNP-1 on β3). Some of the amino acids in the RNP motifs are oriented toward and contribute to the hydrophobic core, while others face outward as solvent-exposed residues involved in RNA recognition. Residues in the loops that connect the strands and helices have also been found to contact RNA (95, 144). RRM-containing proteins that harbor atypical RNP-1 and/or RNP-2 motifs are also known, although these can be more difficult to identify by sequence search programs, the majority of which presently rely on the RNP motifs (14, 73). For example, the polypyrimidine tract-binding protein contains four RRMs with little similarity to the prototypical RNPs (31, 73). Several proteins harbor multiple RRMs, each comprising a globular domain separated by a linker (31, 38, 55), the length of which can be a determinant of RNA binding cooperativity between the RRMs (123). In the solved structures of some of these, adjacent RRMs form a trough or binding cleft for their RNA ligands and therefore appear to form one functional RNA-binding unit from two RRMs (4, 38, 55).
The highest degree of identity among all known La homologs is concentrated in a region that corresponds to residues 20 to 92 of human La (121, 143, 153). Kenan's proposal 5 years ago that this region would adopt an RRM structure (72) is critically reviewed here with the aid of several new La sequences from diverse organisms, a program that predicts secondary structure from a set of aligned sequences, and, most importantly, comparison to a consensus RRM core structure that was previously derived from 70 RRM sequences. Thirteen sequences representing the La proteins of plants, trypanosomes, yeasts, insects, amphibians, and rodents and other mammals, including humans, allow us to identify the invariant and highly conserved residues in these sequences and examine the potential for RRM structure (Fig. 1). Although the hLa structure is represented by three RRMs in addition to other motifs, only the putative RRM-1 will be reviewed in detail here since this is the most highly conserved region and, as will be discussed below, appears to be involved in UUU-OH recognition. Alignments of RRM-2 and, for metazoans, RRM-3 have been reported previously (8, 14, 73, 83, 117, 121, 143, 155). In the alignment in Fig. 1, invariant residues are in black rectangles, with the amino acid (single-letter designation) in white. Conserved residues, i.e., side chains with similar chemical groups, are in gray rectangles.
FIG. 1.
Modeling of the proposed RRM-1 in the highly conserved, N-terminal region of La protein. Alignment of 13 La sequences (beginning with H. sapiens La and ending with A. thaliana La) available in the protein database was performed by ClustalW. Only the sequences that appeared to be full-length La proteins were included in the alignment, and of these, several have been characterized biochemically as authentic RNA-binding proteins (see text). The sequences aligned here correspond to residues 20 to 92 of hLa (numbered above only for H. sapiens La). Invariant residues are reverse shaded in black columns; conserved residues are in gray columns. The top line reflects the output of the Quadratic logistic secondary structure prediction with homologous sequences (QL w/hom) program (39); H, α helix; E, extended β strand; C, non-α-helix, non-β-strand. Below this is indicated the RRM topology β1-loop 1-α1-loop 2-β-loop 3-β3-loop 4-α2-loop 5-β4 that is used for aligning the RRM core hydrophobic residues according to the work of Birney et al. (14). Below the alignment set is the La consensus (“La cons”) line, in which invariant residues are shown in uppercase; Φ = Y or F; other conserved residues are represented according to the designations of Birney; U = uncharged (L, I, V, A, G, F, W, Y, C, or M); Z = U + S, T. Candidate residues that may be involved in uridylate recognition are underlined (see text). The positions of the putative RNP-2 (in β1) and RNP-1 (in β3) motifs of RRM-1 (La RNPs-2&1) are indicated by horizontal lines. The “La RRM CORE” is shown for comparison to the conserved “RRM CORE” structure according to reference 14. The “CORE con” line shows the RRM core with spacing according to reference 14. The sequences and accession numbers are as follows: H. sapiens (Hs), P05455; B. taurus (Bt), P10881; R. norvegicus (Rn), P38656; M. musculus (Mm), P32067; X. laevis La-B (Xb), P28049; X. laevis La-A (Xa), P28048; A. albopictus (Aa), Q26457; Caenorhabditis elegans (Ce), T30953; D. melanogaster (Dm), P40796; T. brucei (Tb), AAF34598.1; S. pombe (Sp), AAB82145.1; S. cerevisiae (Sc), P33399; A. thaliana (At), AAF09063.
The uppermost line of Fig. 1, labeled “QL w/hom,” reflects the output of a program that predicted the secondary structure elements of the set of 13 sequences in the alignment below it. In this output, H indicates α helix, E indicates extended β strand, and C indicates nonhelix and nonstrand. Below this line are indicated the proposed β strands, α helices, and connecting loops for La RRM-1 according to the RRM topology (14). This reveals general correspondence between the predicted structure of the α helices, β strands, and connecting loops and the consensus RRM topology. A noticeable lack of correspondence was found only in the β3 region in which no extended β strand was predicted for the set of sequences. However, it must be noted that examination of the individual La sequences by a sister program (QL without homologs; see Fig. 1 legend) revealed one or more predicted extended β strands for six of the La sequences, those from Homo sapiens, Bos taurus, Rettus norvegieus, Mus musculus, Aedes albopictus, and Trypanosoma brucei (i.e., one less than required for consensus), in the β3 region (data not shown). Therefore, it is highly possible that some if not all La sequences may adopt a β strand in this region and that the structure prediction programs, while helpful, are not reliable enough to accurately predict structural elements in all contexts. In the next paragraph, we compare the La sequences to a consensus RRM core structure.
Most relevant to the issue of whether the N-terminal La region can form an RRM is the part of Fig. 1 that compares the La sequences to a consensus of 70 RRMs. This consensus reflects the hydrophobic core structure of the RRM (14). The consensus RRM core is represented by the last two rows of Fig. 1, the upper of which (“RRM CORE”) shows only the conserved core residues while the lower (“CORE con”) shows the consensus with spaces (see the legend for designations). The “La RRM core” is also shown for convenience, also using the designations of Birney et al. (14).
The highest concentration of invariant residues in La proteins is from Q20 to L36. This includes residues within and surrounding the putative RNP-2 motif of the predicted β1 of the proposed RRM-1, in agreement with the fact that RNP motifs are generally regions of high conservation in RRMs (73). The putative RNP-2 motif (indicated by the horizontal bar in Fig. 1) contains two invariant aromatic residues, Y24 and F25, as well as a conserved aromatic residue, Y or F, at position 23, the latter of which appears to correspond to the solvent-exposed Y or F found in 73% of 179 RRMs (14). The conservation of these residues together with the conserved hydrophobic residue at position 21 would appear to constitute a reasonably good RNP-2 motif.
D33, F35, and L36 would reside in the proximal part of α1. As α1 is an important structural element of the RRM, a high degree of conservation in this region is consistent with the proposed RRM structure. Poly(A)-binding proteins contain several invariant amino acids in their α1 regions (38). Indeed, the invariant L36 of hLa corresponds to the L16 in 54 of the 70 RRM sequences (77%) in the alignment by Birney et al. (14). Another region of high conservation is a tetrapeptide, 45GWVP48, of which G45 and V47 are invariant, which would correspond to loop 2 of RRM-1. Glycines analogous to G45 were found at the homologous position in loops 2 of 58 of the 70 RRMs (83%) aligned by Birney et al., corresponding to the most highly conserved of all of the hydrophobic core residues (14) (Fig. 1). The invariant Val resides 2 amino acids (aa) downstream of G45 in hLa, also corresponding to a conserved hydrophobic core residue (14).
The invariant residue F55 would reside at the end of the β2 strand with the invariant residue R57 at the proximal end of loop 3 (31, 102). Since amino acids corresponding to these latter positions reside in a region involved in protein side chain contacts with sequence-specific bases in other RRM-RNA complexes and do not correspond to RRM core residues, their conservation in La suggests involvement in UUU-OH recognition (14). By similar reasoning, the invariant amino acids Q20, N29, F55, R57, and R91 do not appear to represent core hydrophobic residues and may be considered as candidates involved in specifying uridylate recognition.
Although no invariant residues were found in the predicted β3 of the putative RRM-1, all of the La proteins have either an Ile or a Val corresponding to aa 68 of hLa and a Val, Leu, Ile, or Ala corresponding to aa 69, which would appear to be compatible with the hydrophobic core of the RRM (Fig. 1) (14). It is also noteworthy that the mammalian La proteins all contain V at position 67, precisely matching the V in the RRM core, and that all of the vertebrate La proteins contain a phenylalanine (F65 in hLa) in the β3 region. Lack of invariant residues in the predicted β3 would suggest that any contacts with RNA might be through the main chain protein backbone (or via stacking) which is known to occur for β3 (31, 102). A71, which is invariant in all La proteins, as well as the all-hydrophobic amino acids at positions 72, 79, 80, 82, and 89, would also be expected to contribute to the core structure (Fig. 1) (14, 73). Curiously, the nearly invariant S75 of hLa (T in Arabidopsis thaliana) occupies a position predicted at the beginning of α2, precisely where a serine resides in RRM-1 of the Sex-lethal protein (55). In addition to conservation of hydrophobic core residues, the superimposition of the invariant residues in and around β1 and loop 1, as well as β2-loop 3 and β4, onto this core RRM structure suggests the potential for RNA recognition as indicated by comparison to documented RRM-RNA structures (see Fig. 1 in reference 31).
In summary, the analysis represented by Fig. 1 suggests that the most conserved region of the La protein (Q20 to S92; numbered according to H. sapiens La) can adopt an RRM fold (14). Three characteristics support this: (i) invariant and highly conserved hydrophobic residues that are positioned to provide a core RRM structure; (ii) predicted topology for five out of six of the α helices and β strands, as well as the connecting regions found in other RRMs; and (iii) invariant and highly conserved residues positioned in regions known to participate in RNA recognition in other RRMs.
A phylogenetic tree developed from a prior sequence alignment of nine La homologs revealed four major branches (117). Figure 2 shows a schematic alignment of four La proteins that have been characterized as bona fide UUU-OH-binding proteins (83, 136, 139, 153), each representing one of the four major branches of the La phylogenetic tree (117). In this schematic, the thin vertical bars connecting the four proteins represent the 14 invariant residues identified in our 13 sequences. As shown above, all of the invariant residues reside in the RRM-1 region. The region corresponding to aa 110 to 190 of hLa represents a canonical RRM, and its conservation in La proteins from diverse organisms has been noted elsewhere (73). This RRM, designated RRM-2, is connected to RRM-1 by a linker of 15 aa in all vertebrates and insects, and 12, 10, and 8 aa in A. thaliana, S. pombe, and T. brucei, respectively. Thus, RRM-1 and -2 are arranged in tandem in La proteins, with the first residing close to the N terminus (133). Despite the overall conservation of RRM-2 in the La sequences, which includes identifiable RNP-1 and RNP-2 motifs as well as other conserved residues (23, 79, 83, 121, 140, 143, 153), no invariant residues were found in this region. This degree of conservation is consistent with the idea that RRM-2 might contribute to overall affinity for RNA more so than uridylate recognition per se (see below).
FIG. 2.
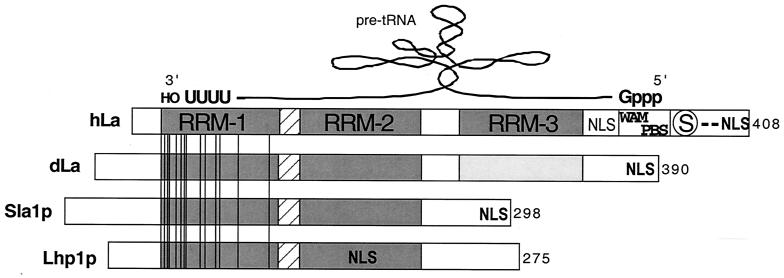
Schematic representation of hLa and its apparent mode of bipartite interaction with a model precursor tRNA and comparison to three other La proteins. Cartoon alignment of the La proteins of human (hLa), D. melanogaster (dLa), S. pombe (Sla1p), and S. cerevisiae (Lhp1p), representatives of four phylogenetic branches according to the work of Rosenblum et al. (117). Note that these represent four polypeptides that have been characterized as UUU-OH-binding proteins that associate with nascent Pol III transcripts in vivo (83, 136, 139, 153). Their lengths, in amino acids, are indicated at the right. RRM-1, -2, and -3 domains are shaded and labeled for hLa only. Invariant residues in the 13 sequences are shown as vertical lines; note that these are limited to RRM-1. The invariant residues (numbered according to hLa) are Q20, E22, Y24, F25, N29, D33, F35, L36, G45, V47, F55, R57, A71, and R91. The linker between RRM-1 and RRM-2 is hatched. A putative WAM and PBS and serine 366 (encircled S) are shown (see text). Positions of the NLS are indicated in boldface according to reference 117. A consensus NLS in hLa, as noted previously (131), is shown in lightface. A schematized pre-tRNA has been imposed above the hLa cartoon to illustrate its proposed orientation according to a model based on prior work (41, 67) as described in the text.
Features of UUU-OH recognition.
Although La will bind to almost any RNA (as will most RRM proteins), affinity increases as the number of terminal uridylates increases to a maximum of four (136). While as few as one or two terminal U's can contribute to La binding, three and four U's constitute progressively more stable interactions, demonstrating that the optimal number of U's for La binding matches the template requirement for termination by Pol III (136) and, accordingly, the number of terminal U's at the 3′ ends of nascent Pol III transcripts (47, 90, 115, 156).
Alteration of the terminal 3′-OH group to a phosphate has been shown to decrease RNA binding by yeast, insect, amphibian, and human La proteins (136, 139, 153). The high degree of conservation of this unique feature suggests (i) that it is critical to function and (ii) that some of the invariant residues of La might contribute to a specific binding site for the terminal ribose 3′-OH, which cannot as easily accommodate a phosphate group. Therefore, although predictions indicate an RRM as a likely structure in the NTD, the actual structure would be expected to exhibit two features to accommodate the unique mode of RNA binding by La: (i) recognition of three to four U's specifically and (ii) sensitivity to the state of the 3′ group on the terminal ribose.
In the context of hLa, high-affinity RNA binding requires RRM-1 and RRM-2, as RRM-1 alone does not bind RNA (28, 44, 72). Deletion of the most N-terminal residues of RRM-1, as in the mutants hLa 22-408 and 26-408, decreases affinity for RNA (28, 44, 72), although this can be overcome by increasing the concentration of hLa in the binding reaction mixture (44). Since deletion of N-terminal amino acids from La leads to a dramatic loss of binding to UUU-OH-containing RNAs and since the UUU-OH motif comprises a high-affinity binding site for La, it would appear that the N-terminal amino acids in La mediate UUU-OH binding. This reasoning is consistent with experiments that showed that, even at very high concentrations, hLa lacking only the first 25 aa of RRM-1 binds to RNA but cannot efficiently protect nascent UUU-OH-containing RNAs against 3′-exonuclease digestion while the full-length protein can do so at lower concentrations (41, 44). The data suggest that, while RRM-1 does not provide high-affinity binding on its own, it contributes significantly to UUU-OH recognition, consistent with a contact surface of 4 to 6 nucleotides per RRM (38, 55, 122). RRM-2 would serve to increase affinity for RNA, although a role in uridylate recognition cannot yet be ruled out for RRM-2 (28, 44, 72). Thus, it would appear as if RRM-1 and RRM-2 of La work as a pair of RRMs to mediate high-affinity binding to nascent RNAs that contain UUU-OH.
It is interesting to note the crystal structure of Drosophila melanogaster Sex-lethal (Sxl) protein complexed with its target ligand, the UGUUUUUUUU sequence of tra precursor mRNA (55). In the cocrystal structure, the first of a tandem pair of Sxl RRMs recognizes the 3′ region of the target RNA, which is composed exclusively of U residues, while the second RRM recognizes the 5′ nucleotides of the target ligand. Among the contacts with the uracil bases are hydrogen bonds formed by Asn in RNP-2, as well as Gln, Asn, and other residues in Sxl RRM-1 (55). It is noteworthy to recall here that, in addition to the invariant aromatic residues within the RNP-2 motif of La RRM-1, RNP-2 is also flanked by two invariant amino acids, Gln20 and Asn29, residues capable of side chain hydrogen bonds to uracil-specific nucleotides (55). The effect of deleting Gln20, as in hLa 22-408 and 26-408, a profound loss of RNA binding and RNA 3′ protection, is consistent with a role for this invariant residue in UUU-OH recognition. The arrangement of Sxl, namely, that RRM-1 of a tandem RRM protein recognizes the 3′ U-rich end of its ligand and confers U-specific recognition while RRM-2 recognizes the 5′ end of the ligand, is similar to the UUU-OH binding arrangement proposed for hLa (Fig. 2) (72).
UUU-OH recognition and biological function.
As noted earlier, yeast La associates with certain Pol II-synthesized RNA intermediates via their terminal uridylates (77, 150). Although binding of La to a U1 snRNA precursor in HeLa cells was previously documented (85), it will be important to determine whether La binding to the Pol II snRNAs is more prevalent in yeast than in other eukaryotes. Since S. cerevisiae snRNA intermediates do not appear to cycle through the cytoplasm for maturation as they do in mammalian cells (see Discussion in reference 150) and since S. cerevisiae La uses a different nuclear import pathway from that of mammalian cells (see above), the nuclear retention-transport function of La and involvement in snRNA metabolism could have evolved differently in yeasts and higher eukaryotes (49).
While Pol III transcription is not a prerequisite for association with La, a transcription termination signal that encodes the UUU-OH motif ensures that all Pol III transcripts will be bound by La (136, 139, 153). Pol III termination-dependent positioning affords La the opportunity to serve as a primary determinant in the posttranscriptional processing and/or the ordered assembly of these transcripts into specific ribonucleoproteins (24, 27, 53, 56, 86, 137, 139). Identification of the yeast homolog of La set the stage for a most important phase of La research, genetic manipulation and elucidation of function in vivo (83, 153). With regard to Pol III transcripts, an essential tRNASer gene with a mutation that would disrupt base pairing in the anticodon stem of the tRNA was selected in a synthetic-lethal screen of La-deficient S. cerevisiae (154). In this case, the defect was at the level of pre-tRNA stability and processing. Another mutation isolated by the synthetic-lethal screen was in Lsm8p, a polypeptide component of a U6 snRNP (103). In this case, U6 precursors were unstable and inefficiently incorporated into U6 RNPs. Independent genetic analyses that are sensitive to alterations in tRNA expression revealed a role for La in stabilizing nascent pre-tRNAiMet (6, 17). These and another in vivo study indicate that La acts to stabilize various Pol III transcripts at the precursor stage only, maintaining them through a critical maturation process, after which time the mature species can accumulate and function in the absence of La (6, 17, 67, 103, 154). The necessity for this type of precursor-specific stabilizing activity may be explained by the possibility that, while some sequences may be optimal for function in the mature transcript, these same sequences may be suboptimal for stability in the context of a longer precursor transcript. Alternatively, precursor transcripts may contain sequences that target them for digestion that are masked by La. In any case, it appears that La serves as a molecular link between Pol III termination and posttranscriptional processing and as a chaperone that protects nascent transcripts during the time between synthesis and maturation (103). Stabilization by La could also provide a quality control function that would ensure that RNA-modifying enzymes or other factors (e.g., those that influence transcript stability) would have ample time to act on the precursor substrates (41).
Emergence of an accessory CTD in metazoan La proteins.
As mentioned in the introduction, an NLS was relegated from the N-terminal RNA-binding domain to the CTD, and the domain complexity of the CTD of La was expanded during evolution (Fig. 2). A third RRM, represented by aa 220 to 300 of hLa, was apparently acquired by the metazoan La proteins; this motif was identified as part of a general search of the protein database for atypical RRMs (14). The RRMs of this class were found to be enriched in metazoan proteins involved in RNA metabolism (14), consistent with the absence of RRM-3 in the yeast La proteins Sla1p and Lhp1p (Fig. 2). The RRM-3 is highly conserved in the vertebrate La proteins, while the three insect, one plant, and one trypanosome sequences exhibit significantly less similarity in this region. Functional involvement of RRM-3 in pre-tRNA processing has been demonstrated elsewhere for hLa (67).
The most significant variability among the vertebrate La proteins is in the region beyond RRM-3. Residues 316 to 332 of hLa conform perfectly to a consensus bipartite NLS, the sequence of which is well conserved only in the vertebrates. This is followed by a basic region (aa 328 to 363) in which 40% of the residues are positively charged, an acidic region (aa 366 to 390), and a terminal NLS (Fig. 2). The C-terminal NLS of hLa (aa 390 to 402) is functional, while the more fitting bipartite consensus NLS at aa 316 to 332 of hLa appears nonfunctional or masked in Xenopus laevis oocytes, although the effect of the consensus NLS at aa 316 to 332 was not examined in tissue culture cells (131).
Identification of a WAM, 333GRRFKGKN340, conforming to the consensus A/GXXXXGKX in hLa suggested the potential for ATP binding and helicase activity (140), although recent results suggest that this motif is not involved in helicase activity but is involved in 5′-ppp-RNA recognition (Fig. 2; also see below). A sequence tract that matches this consensus WAM is found in all vertebrate La proteins but not in those from other species. The rodent sequences contain a 16-aa expansion immediately upstream of the WAM. Farther downstream lies the major phosphorylation site of hLa, S366 (16, 42), which accentuates a transition from basic (aa 328 to 363) to acidic (aa 367 to 375) residues (42). The identity of the residues comprising and/or surrounding the S366 phosphorylation site in hLa exhibits significant differences in other species, although the residues are generally conserved (42). Sequences downstream of the acidic region of hLa are expanded in amphibian La (121). In summary, while the NTD of La is highly conserved, the CTD is not, being largely absent in yeasts and differentially expanded in metazoans.
A potential WAM in the CTD of hLa and its possible involvement in recognizing the 5′-ppp end of a nascent transcript.
The unexpected ability of hLa to protect various precursor tRNAs from 5′ processing led to a structure-function analysis that suggested that the WAM in the CTD could interact with the 5′-ppp end of a nascent transcript (41). This mode of interaction was suggested by an RNase P protection assay as well as direct binding studies that examined a collection of mutant La proteins and various forms of pre-tRNAs (41). Specifically, digestion of the RNA with phosphatase, which would remove the 5′-terminal phosphates, decreased interaction with La as monitored by direct binding and RNase P protection assays (41). 5′-ppp binding may also explain Stefano's observation (136) that even his best binding substrates (i.e., 5′-p-tRNA-UUUU-OH) formed complexes with La that were less stable than bona fide tRNA precursors. Thus, it was concluded that “the constructs used…lack some feature(s) recognized by the La protein on naturally occurring RNAs” (136). It must now be suspected that one outstanding feature was a 5′-ppp, which was missing on the mature 5′-p-tRNA-UUUU-OH ligand used in that study.
Interestingly, the BLOCKS program identifies aa 348 to 368 of hLa as a potential phosphate binding site (PBS), just downstream of the WAM, that is rich in conserved basic residues (59). The possibility that both of these motifs, the WAM and the putative PBS, contribute to 5′-ppp-RNA recognition in vivo is consistent with functional analyses in which hLa constructs lacking either of these regions, residues 328 to 344 or residues 345 to 363, cannot stabilize pre-tRNA against 5′ processing (67). We envisage that the WAM may recognize the initiating base, which is usually a purine in Pol III transcripts (58, 138, 157), while the PBS may recognize the 5′ phosphates.
Although it was thought that the WAM would be involved in the helicase-ATPase activity that had been noted elsewhere for La (7, 65, 149), it was recently documented that double-stranded nucleic acid binding by La does not require either the WAM or a putative double-stranded RNA (dsRNA)-binding motif, and La appears not to unwind dsRNA (70). ATP hydrolysis has never been reported for La, and attempts to demonstrate this using purified La protein have been unsuccessful (72). Thus, while WAMs in other proteins contribute to ATP binding, it is tempting to speculate that, in hLa, this motif instead has been recruited to recognize the 5′-pppG/A moieties found at the initiating ends of nascent Pol III transcripts (41, 67).
A two-domain structure, as previously proposed for hLa (23), would allow bipartite RNA binding; the conserved NTD can interact with the UUU-OH 3′ end of the RNA and the CTD can interact with the 5′-pppG/A end, both of which constitute unique determinants on all nascent Pol III transcripts. The predicted structures of many Pol III transcripts would place the 5′ and 3′ termini in close proximity, apparently well suited for bipartite interaction with La. According to the bipartite binding model, hLa would bind principally to the terminal regions of a pre-tRNA while leaving the body of the tRNA accessible to other factors, perhaps somewhat similar to what has been visualized elsewhere for EF-Tu and tRNA (41, 99, 100). That other proteins and/or modifying enzymes can recognize the RNA components of nascent RNA-La RNPs is indicated by modifications found on pre-tRNAs that coimmunoprecipitate with La (58).
A conserved role in precursor tRNA processing and maturation.
Nascent pre-tRNAs must undergo 5′-end processing by RNase P, 3′-end processing, splicing (if befitting), base modifications, 3′-CCA addition, aminoacylation, and nuclear export prior to function in the cytoplasm (52, 64, 148). Various aspects of tRNA maturation, including splicing, aminoacylation, and nuclear transport, have been reviewed recently (52, 64, 148). Many of the tRNA maturation-related activities may be interrelated (66, 120, 124, 125) and/or integrated with other aspects of gene expression; for example, accumulation of unprocessed pre-tRNAs can induce a nutritional stress response in yeast, and independently, the unfolded protein response is controlled by a key catalytic component of the tRNA splicing machinery (32, 111, 128–130). As the first protein that interacts with pre-tRNAs and serves to determine their metabolism, the La phosphoprotein appears to have been integrated into some of these pathways.
La's role in the expression pathways for tRNAs has been examined in recent studies (6, 17, 41, 67, 143, 154). Removal of the pre-tRNA 5′ leader is performed by RNase P, and in many cases, this appears to be the first step in the tRNA maturation pathway (5, 148). 3′-end formation can occur either by an endonuclease or by one or more exonucleases (21, 80, 97, 110, 154). The robust exonucleolytic digestion of the 3′ ends of nascent Pol III transcripts that occurs in La-depleted extracts and in La− cells is prevented by La (41, 44, 82, 88, 154; also see reference 110). In the case of pre-tRNAs, protection from 3′-exonucleolytic digestion might be sufficient to promote default processing by the pre-tRNA 3′ endonuclease, although recruitment of the endonuclease by La is also possible (154).
As mentioned above, the CTD of hLa unexpectedly interfered with 5′-end maturation of pre-tRNA (41), surprising because 5′ cleavage is a necessary step in tRNA maturation. The finding that S366 phosphorylation could reverse the inhibitory effect of hLa and render the pre-tRNA susceptible to 5′ processing by RNase P reconciled this paradox and suggested a mechanism by which tRNA maturation might be regulated (41). To examine whether this activity might be operational in vivo, a genetic system was developed using alleles of a nonessential tRNASerUGA opal suppressor that suppress ade6-704 and the accumulation of red pigment in fission yeast (67). This allowed examination of the expression pathway of a nonessential tRNA reporter gene by a colorimetric assay in vivo. Results obtained with this system confirmed the observations of Wolin and colleagues that hLa could substitute for fission yeast La in its ability to protect nascent pre-tRNAs from 3′-exonucleolytic digestion (143). However, hLa constructs bearing nonphosphorylatable residues at position 366 were found to block pre-tRNA processing at the 5′-end maturation step and were unable to support tRNA maturation and suppression (67, 143). Biochemical analyses confirmed that hLa is faithfully phosphorylated on S366 in fission yeast and allowed the conclusion that this modification promotes tRNASerUGA maturation (67).
Since RNase P components, their pre-tRNA substrates, and La have each been localized to the nucleolus as well as the nucleoplasm, a challenge is to determine whether phosphorylation-mediated control of La function in pre-tRNA 5′-end maturation is compartment specific (13, 69, 71, 148).
The hLa phosphoprotein: signal transduction through the CTD.
Approximately 80% of hLa isolated from HeLa cells is phosphorylated on serine 366 (42). Phosphorylation can also be detected at other sites, although only a very small fraction (probably <5%) of hLa is modified at these sites, and the significance is unknown (16). The phosphate on La exhibits a relatively short half-life in HeLa cells, indicating multiple cycles of phosphorylation and dephosphorylation (107). S366 of hLa constitutes a casein kinase II (CKII) target site that conforms to the consensus S/TXXD/E, although whether CKII phosphorylates this site in vivo has yet to be demonstrated (42, 67). A consensus CKII target site can also be identified in the homologous regions of the La proteins of other species, although amino acid identity is not highly conserved (42; R. Maraia, unpublished observations). While yeast La proteins are phosphorylated, the site of modification has not been reported (143). Although the phosphatase responsible for dephosphorylating hLa has not been identified, a PP2A-like activity is suspected (118).
The only activities of La that have been reported to be regulated by phosphorylation are those related to the synthesis and processing of Pol III-transcribed RNAs (41, 42, 67), as nuclear import, dsRNA binding, and inhibition of PKR activity have been shown to be unaffected by phosphorylation (16, 70). As mentioned above, La is dephosphorylated during the early stages of induced apoptosis (16, 70).
The basic region from aa 328 to 363 of hLa includes the WAM and putative PBS and appears to recognize the 5′-ppp end of a nascent RNA, while phosphorylation of S366 negatively modulates this activity (41, 42). In the model depicted in Fig. 3, the phosphate group on S366 not only would alter the conformation of the CTD but might also compete for the PBS. A more detailed mechanism can also be envisaged. In the absence of a 5′-ppp-RNA, phosphoserine 366 and nearby acidic side chains might mimic 5′-ppp and interact with the PBS. This putative interaction could be stabilized by hydrophobic interactions contributed by conserved aromatic residues and could be intramolecular or intermolecular as the amino acids required for homodimerization overlap the basic region (33). The effect of phosphorylation on the dimerization of La has not been reported.
FIG. 3.
A model of phosphorylation-regulated, bipartite precursor tRNA binding by the hLa protein. The model shown here depicts bipartite interactions with a nascent pre-tRNA according to previous data (41, 67) and the model in Fig. 2. The tandem RRM-1 and -2 of the NTD mediate high-affinity binding to UUU-OH-containing RNAs. The CTD of hLa contains a basic region and an acidic region, shown as +++ and −−, respectively; residues 328 to 344 conform to a WAM, and residues 348 to 368 represent a putative PBS (see Fig. 2 and text). Together, the WAM and PBS can recognize the 5′-pppG/A motif that comprises the 5′ ends of nascent Pol III transcripts (41, 67). Serine 366, which resides in a region that demarcates a transition from basic to acidic residues, is shown as S in unphosphorylated La and as P in the phosphoserine form.
Extending the potential biological implications of 5′-ppp binding is the finding that 5′-end recognition can be modulated by phosphorylation and is consistent with the proposed role for La as a quality control factor (41, 67). The CTD could afford La the ability to retain any nascent Pol III transcript (or its 5′-end region) in a stable form until it receives a signal, i.e., in the form of S366 phosphorylation, to release it (41).
Evidence for and against a role for hLa in transcription by Pol III.
HeLa cell extracts immunodepleted of La lost 99% of their Pol III transcription activity, and the few transcripts synthesized were foreshortened on their 3′ ends, suggesting that La is required for termination and high-efficiency transcription (47). Foreshortening probably reflects exonuclease activity in the absence of La. Repletion with biochemically purified La restored transcript length but not transcription efficiency (46, 47). Experiments using transcription complexes isolated on immobilized DNA suggest that La stimulates Pol III recycling by increasing transcript release and the rate of reinitiation (87, 88). The use of a set of B1-Alu gene mutants that differ only in the sequence context of their terminator provided additional insight into the transcription activity of hLa (45). For some of these, the inability to engage La in a manner that protects the nascent transcript from 3′-exonucleolotic digestion appears also to render the transcription complex a poor substrate for reinitiation by Pol III (45). This line of study supports the idea that Pol III termination and reinitiation are linked through La.
A short basic region that includes the WAM is required for the Pol III reinitiation activity of hLa (44). Fractionation of native hLa into a phosphoserine 366-containing form and a nonphosphorylated form revealed that only the nonphosphorylated form of La stimulates Pol III recycling in vitro (42). The latter study also provided an explanation for the low transcription activity of biochemically purified La, as the fractionation scheme employed by Gottlieb and Steitz yields mostly phosphorylated La which is inactive for transcription (42, 46, 47). These structure-function analyses of hLa suggest a model in which the WAM and PBS act as positive determinants of 5′-pppG/A recognition and transcription factor activity. The downstream acidic region would act as a negative influence on the activity of the WAM and PBS, accentuated and enhanced by phosphorylation of S366 (Fig. 3).
Publications from three laboratories indicate a role for La in transcription while others contest this activity (29, 42, 44–47, 82, 87, 88, 154). In HeLa cells, La accumulates to ∼2 × 107 molecules/cell while Pol III appears to accumulate to only ∼104 molecules/cell (109). Incomplete depletion might leave a surplus of La relative to Pol III and may also reflect La's association with a Pol III holoenzyme (82, 145, 146). Thus, while recent results raise significant doubt as to a role for La in Pol III transcription, the above concerns illuminate the need to address the differences noted in the different transcription systems (82, 145, 146), especially considering the intriguing correlations reviewed in the next paragraph.
The sensitivity of the 5′-ppp-RNA-binding activity to a panel of mutations in hLa was indistinguishable from the sensitivity of the transcription initiation factor activity of La to the same collection of mutations (41, 44). This correlation was strengthened by the finding that both activities are negatively modulated by phosphorylation of hLa S366 and strengthened the idea that they are mechanistically linked, i.e., that the transcription initiation factor activity of hLa depends on recognition of the initiating 5′-pppG/A nucleotide of the nascent transcript (42, 44). The presence of La in a human Pol III holoenzyme independently suggests involvement in RNA production (145). These observations provide a compelling reason to continue to consider the transcription factor activity of hLa seriously. We envisage that, just after transcription initiation, as the transcript emerges from the polymerase, La that is associated with the Pol III holoenzyme would capture the 5′-pppG/A end of the elongating RNA. As the transcript nears completion and the 3′ terminus is formed, hLa would exchange its contact with the holoenzyme for high-affinity contact with UUU-OH, dissociating from the enzyme complex with the nascent RNA securely attached. Unphosphorylated La would remain attached to both ends of the RNA (41, 67). Phosphorylation would initiate the dissociation of the nascent RNA and La. Accordingly, a phosphatase would then prepare phospho-La for involvement in the next round of RNA synthesis (42).
The ability of hLa to recognize both the 5′ end and 3′ end of a nascent Pol III transcript, the presence in a Pol III holoenzyme, and modulation by phosphorylation is reminiscent of the role of the CTD of Pol II in mRNA expression. The Pol II CTD recruits factors that modify both the 5′ and 3′ ends (5′ capping and 3′ cleavage and polyadenylation) of nascent Pol II transcripts, and it contributes to transcription efficiency by a mechanism that is regulated by phosphorylation and dephosphorylation (75, 98). A major challenge for the future is to develop a genetic or other in vivo system that can also be used for in vitro biochemistry with which to examine the transcription factor activity of hLa in more detail.
Interaction with the internal ribosome entry sites (IRES) and other elements in viral and cellular mRNAs.
As mentioned above and previously reviewed, La appears to function in a pathway of translation initiation that is used by viral and cellular mRNAs that contain IRES elements (12, 62). Although first described for poliovirus mRNA (105), the list of IRES-containing mRNAs has been expanded to cellular mRNAs that encode cell cycle-dependent transcripts as well as other mRNAs (31a, 63, 84, 96, 101, 110a, 142, 152). Of the cellular mRNAs, some have recently been found to interact with La (12, 62). Viral mRNAs that have been reported to interact with La are not limited to those containing IRES and include mRNAs from poliovirus, hepatitis C virus, rubella virus, vesicular stomatitis virus, encephalomyocarditis virus, Sindbis virus, and human immunodeficiency virus (28, 40, 57, 68, 74, 78, 104, 108, 134).
La has also been reported to interact with the U-rich 5′ regions of ribosomal protein mRNAs in vitro and to stimulate their translation in vivo in a Xenopus tissue culture system (34, 106). Because many of the reported interactions between La and mRNAs occur near the 5′ end of the RNAs, it seems noteworthy that 5′-pppG/A recognition [or m7G(5′)ppp(5′)-cap recognition] may be involved, since the region required for this activity includes the WAM and because La was found in close proximity (i.e., could be cross-linked) to a 5′-cap structure on a U1-like snRNA (41, 49). Consistent with this, residues within the region of aa 293 to 348 of hLa, which leads to dimerization, contain the WAM and are required for La's activity in IRES-mediated translation (33).
Fruit fly larvae carrying a null allele of La die as an apparent result of gastrointestinal defects that are associated with the loss of Ultrabithorax (Ubx) mRNA (9), suggesting that Drosophila La may influence Ubx mRNA metabolism, perhaps involving the IRES of Ubx (152).
Alternative isoforms of La mRNA, in addition to the major isoform that encodes nuclear La, have been identified in lymphocytes (18, 141). Some isoforms are tissue specific and, when expressed in recombinant form, give rise to La proteins that differentially localize to the nucleus or cytoplasm (50, 51, 60). Intriguingly, these La mRNA isoforms contain functional IRES, a finding that led to the proposal that synthesis of La may occur under conditions when cap-dependent translation is compromised, such as inflammation, apoptosis, or certain viral infections (18).
Alternative modes of RNA binding by La?
As discussed above, a highly conserved feature of La RNA binding is recognition of a terminal 3′-OH group (136, 139, 153). High-affinity interaction has been reported for a small RNA probe (which ended in two U's) that was used to study the poliovirus IRES element (Kd, ∼4 nM) (93), comparable to the affinity for UUU-OH-containing RNAs (28, 44). Yet, it is not clear that La would recognize native internal IRES elements via the UUU-OH mode of binding since these elements would not be expected to contain a 3′-OH group. A similar conundrum exists for TAR RNA (28). In an attempt to understand the apparent mechanism of recognition for TAR RNA, Chang et al. examined binding to various RNAs (28). Although more than a sixfold decrease in binding affinity was revealed by deletion of terminal uridylates, the analysis also suggested a mode of binding distinct from terminal uridylate recognition (28). Yet, the most N terminal of La residues, thought to be involved in UUU-OH recognition, contributed significantly as a primary determinant of binding in that study, regardless of which RNA was examined (28). In other studies, the RNA probes that contained terminal uridylates bound best to, or competed best for, La (3, 57, 92, 93) (note that EcoRI restriction leaves a 3′ end that would encode two terminal U's in the runoff transcript). In general, it appears that if the only difference between two RNA probes is the presence of two or more terminal U's, this feature is a significant determinant of La binding, although it is not clear that such probes would best represent IRES elements. It should also be noted that, in the cases where the RNA probe ends with uridylates, some as yet unknown feature of internal sequences can further contribute to La binding, suggesting the existence of an alternative mode of binding in these cases (28, 57, 72). Further documentation and characterization of these features and the mechanism by which they contribute to the binding of viral and cellular mRNAs by La are outstanding issues.
La is targeted by viruses.
Some viruses engage La either by binding their small RNAs or by modifying La by proteolysis, presumably to facilitate the execution of their own genetic programs. Adenovirus and Epstein-Barr virus encode small RNAs (VAI and EBER RNAs, respectively) that are synthesized at high levels by Pol III and are complexed with La (116). VAI RNA serves to inactivate the dsRNA-activated protein kinase (PKR) that allows viral mRNAs to be translated by the cap-dependent mechanism (147). Although EBER RNAs can also bind PKR, its role in the Epstein-Barr virus life cycle is less clear (155). Unlike most Pol III transcripts, which are only transiently associated with La, these RNAs remain associated with La as they accumulate to high levels, although the consequences of this are unknown. It has been proposed elsewhere that La can regulate PKR by virtue of an RNA-unwinding or helicase activity that was presumed to be dependent on ATP hydrolysis and the presence of the WAM (65, 149). However, as noted above, recent studies indicate that the WAM does not mediate this activity (70). Also, a region that resembles dsRNA-binding proteins that was thought to be responsible for dsRNA binding by La has subsequently been shown to be dispensable for dsRNA binding (70).
Finally, we note that poliovirus protease 3C cleaves hLa at residue 358 (127). This protease also cleaves and inactivates other factors involved in small RNA expression and causes apoptosis (10, 30, 35, 94, 126, 151). This cleavage would leave the dimerization and RNA-binding domains, including the 5′-ppp-interaction motif, intact (131). Thus, poliovirus causes La to accumulate in the cytoplasm, wherein the truncated protein would presumably be engaged in the IRES-dependent translation of poliovirus mRNA (see above). Induced apoptosis leads to a caspase-dependent cleavage of hLa at residue 371 or 374 (118). The role of La cleavage in apoptosis is unknown (19, 20, 118), although a function in the regulated expression of the X-linked inhibitor of apoptosis mRNA is suggested by recent work (62).
Conclusions.
hLa antigen is a ubiquitous, abundant nuclear phosphoprotein that is found associated with a large variety of RNA ligands. La facilitates the expression of RNA Pol III-synthesized RNAs by stabilizing the precursor forms of these transcripts and can integrate activities that control the 5′- and 3′-end metabolism of these RNAs. A significant amount has been learned about the structural basis of RNA recognition by La and the relationship of this to activity. Although the atomic structure of La remains to be determined, its most conserved region, the NTD, can be readily modeled into a tandem pair of RRMs with features consistent with UUU-OH recognition. While the NTD recognizes the UUU-OH 3′-terminal motif that results from transcription termination by Pol III, the CTD of hLa recognizes the 5′-ppp-RNA motif that results from transcription initiation by Pol III. La is also found associated with certain precursor forms of snRNAs and other small RNAs in vivo, which are synthesized by Pol II, although in these cases it appears that posttranscriptional processing exposes 3′-terminal uridylates prior to La binding. For nascent Pol III transcripts, the 5′-ppp and UUU-OH-3′ motifs are often found in close proximity in the structured RNA, affording high-affinity and specific, bipartite binding by La.
Although La is not essential in yeast, its presence in all eukaryotes and apparent essentiality in D. melanogaster suggest that it provides a function that has been repeatedly retained and perhaps refined during the radiation of eukaryotes. It is becoming increasingly clear that La is not simply a passive chaperone for Pol III transcripts but indeed serves a significant function in RNA expression. While all La proteins function by UUU-OH binding, it is not clear if the La proteins of yeast can mediate 5′-ppp-RNA recognition or if this was added to the repertoire of La activity during the evolution of higher eukaryotes. The 5′-end-RNA recognition activity of hLa can be modulated by phosphorylation of S366, and this can control tRNA maturation and perhaps other activities that might be mediated by this mode of RNA recognition.
Another activity of La is the apparent involvement in the expression of a subset of mRNAs, both viral and cellular, that rely on the IRES-mediated mechanism of translation, although the details of RNA recognition in these cases are not yet clear. Certain viruses engage La by employing its RNA Pol III-related activities, while others appear to relocalize it to the cytoplasm, apparently to direct IRES-dependent translation of the virus mRNA. A similar proteolysis-mediated localization mechanism may be used during apoptosis.
To date, only the Pol III-related functions of human La have been reported to be regulated by phosphorylation of S366, although rapid dephosphorylation occurs during the early phase of apoptosis. While an atomic structure for the conserved NTD will be highly informative, visualization of the structure of hLa's CTD will also be important.
ACKNOWLEDGMENTS
We thank D. Kenan and J. Keene for numerous discussions and encouragement, Y. Huang and an anonymous reviewer for comments on the manuscript, T. Cech and S. Aigner for communicating results prior to publication, and Cheryl Varron for assistance with preparation of the manuscript.
R.V.A.I. was supported by an NIH Visiting Fellow Award.
REFERENCES
- 1.Adam S A, Nakagawa T, Swanson M S, Woodruff T K, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aigner S, Lingner J, Goodrich K J, Grosshans C A, Shevchenko A, Mann M, Cech T R. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 2000;19:6230–6239. doi: 10.1093/emboj/19.22.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allain F H, Gilbert D E, Bouvet P, Feigon J. Solution structure of the two N-terminal RNA-binding domains of nucleolin and NMR study of the interaction with its RNA target. J Mol Biol. 2000;303:227–241. doi: 10.1006/jmbi.2000.4118. [DOI] [PubMed] [Google Scholar]
- 5.Altman S, Kirsebom L. Ribonuclease P. In: Gesteland R F, Chech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 351–380. [Google Scholar]
- 6.Anderson J, Phan L, Cuesta R, Carlson B A, Pak M, Asano K, Bjork G R, Tamame M, Hinnebusch A G. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachmann M, Pfeifer K, Schröder H-C, Müller W E G. Characterization of the autoantigen La as a nucleic acid-dependent ATPase/dATPase with melting properties. Cell. 1990;60:85–93. doi: 10.1016/0092-8674(90)90718-t. [DOI] [PubMed] [Google Scholar]
- 8.Bai C, Li Z, Tolias P P. Developmental characterization of a DrosophilaRNA-binding protein homologous to the human systemic lupus erythematosus-associated La/SS-B autoantigen. Mol Cell Biol. 1994;14:5123–5129. doi: 10.1128/mcb.14.8.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai C, Tolias P P. Genetic analysis of a La homolog in Drosophila melanogaster. Nucleic Acids Res. 2000;28:1078–1084. doi: 10.1093/nar/28.5.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barco A, Feduchi E, Carrasco L. Poliovirus protease 3C(pro) kills cells by apoptosis. Virology. 2000;266:352–360. doi: 10.1006/viro.1999.0043. [DOI] [PubMed] [Google Scholar]
- 11.Baserga S J, Steitz J A. The diverse world of small ribonucleoproteins. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 359–381. [Google Scholar]
- 12.Belsham G J, Sonenberg N, Svitkin Y V. The role of the La autoantigen in internal initiation. Curr Top Microbiol Immunol. 1995;203:85–98. doi: 10.1007/978-3-642-79663-0_4. [DOI] [PubMed] [Google Scholar]
- 13.Bertrand E, Houser-Scott F, Kendall A, Singer R H, Engelke D R. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birney E, Kumar S, Krainer A R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boelens W C, Palacios I, Mattaj I W. Nuclear retention of RNA as a mechanism for localization. RNA. 1995;1:273–283. [PMC free article] [PubMed] [Google Scholar]
- 16.Broekhuis C H, Neubauer G, van der Heijden A, Mann M, Proud C G, van Venrooij W J, Pruijn G J. Detailed analysis of the phosphorylation of the human La (SS-B) autoantigen. (De)phosphorylation does not affect its subcellular distribution. Biochemistry. 2000;39:3023–3033. doi: 10.1021/bi992308c. [DOI] [PubMed] [Google Scholar]
- 17.Calvo O, Cuesta R, Anderson J, Gutierrez N, Garcia-Barrio M T, Hinnebusch A G, Tamame M. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4167–4181. doi: 10.1128/mcb.19.6.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter M S, Sarnow P. Distinct mRNAs that encode La autoantigen are differentially expressed and contain internal ribosome entry sites. J Biol Chem. 2000;275:28301–28307. doi: 10.1074/jbc.M004657200. [DOI] [PubMed] [Google Scholar]
- 19.Casciola-Rosen L, Andrade F, Ulanet D, Wong W, Rosen A. Cleavage by granzyme B is strongly predictive of autoantigen status: implications for initiation of autoimmunity. J Exp Med. 1999;190:815–826. doi: 10.1084/jem.190.6.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casiano C A, Tan E M. Antinuclear autoantibodies: probes for defining proteolytic events associated with apoptosis. Mol Biol Rep. 1996;23:211–216. doi: 10.1007/BF00351171. [DOI] [PubMed] [Google Scholar]
- 21.Castano J G, Tobian J A, Zasloff M. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3′ terminus of human pre-tRNAimet(3′ pre-tRNase) J Biol Chem. 1985;260:9002–9008. [PubMed] [Google Scholar]
- 22.Chambers J C, Keene J D. Isolation and analysis of cDNA clones expressing human lupus La antigen. Proc Natl Acad Sci USA. 1985;82:2115–2119. doi: 10.1073/pnas.82.7.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chambers J C, Kenan D, Martin B J, Keene J D. Genomic structure and amino acid sequence domains of human La autoantigen. J Biol Chem. 1988;263:18043–18051. [PubMed] [Google Scholar]
- 24.Chambers J C, Kurilla M G, Keene J D. Association between the 7 S RNA and the lupus La protein varies among cell types. J Biol Chem. 1983;258:11438–11441. [PubMed] [Google Scholar]
- 25.Chan E K L, Francoeur A M, Tan Z M. Epitopes, structural domains and asymmetry of amino acid residues in SS-B/La nuclear protein. J Immunol. 1986;136:3744–3749. [PubMed] [Google Scholar]
- 26.Chan E K L, Sullivan K F, Tan E M. Ribonucleoprotein SS-B/La belongs to a protein family with consensus sequences for RNA-binding. Nucleic Acids Res. 1989;17:2233–2244. doi: 10.1093/nar/17.6.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang D Y, Hsu K, Maraia R J. Monomeric and dimeric Alu RNAs induced by adenovirus assemble into SRP9/14-containing RNPs in HeLa cells. Nucleic Acids Res. 1996;24:4165–4170. doi: 10.1093/nar/24.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang Y-N, Kenan D J, Keene J D, Gatignol A, Jeang K-T. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J Virol. 1994;68:7008–7020. doi: 10.1128/jvi.68.11.7008-7020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu W M, Ballard R E, Schmid C W. Palindromic sequences preceding the terminator increase polymerase III template activity. Nucleic Acids Res. 1997;25:2077–2082. doi: 10.1093/nar/25.11.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark M E, Hammerle T, Wimmer E, Dasgupta A. Poliovirus proteinase 3C converts an active form of transcription factor IIIC to an inactive form: a mechanism for inhibition of host cell polymerase III transcription by poliovirus. EMBO J. 1991;10:2941–2947. doi: 10.1002/j.1460-2075.1991.tb07844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conte M R, Grune T, Ghuman J, Kelly G, Ladas A, Matthews S, Curry S. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 2000;19:3132–3141. doi: 10.1093/emboj/19.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 32.Cox J S, Chapman R E, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craig A W, Svitkin Y V, Lee H S, Belsham G J, Sonenberg N. The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol Cell Biol. 1997;17:163–169. doi: 10.1128/mcb.17.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crosio C, Boyl P P, Loreni F, Pierandrei-Amaldi P, Amaldi F. La protein has a positive effect on the translation of TOP mRNAs in vivo. Nucleic Acids Res. 2000;28:2927–2934. doi: 10.1093/nar/28.15.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das S, Dasgupta A. Identification of the cleavage site and determinants required for poliovirus 3CPro-catalyzed cleavage of human TATA-binding transcription factor TBP. J Virol. 1993;67:3326–3331. doi: 10.1128/jvi.67.6.3326-3331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng J S, Takasaki Y, Tan E M. Nonhistone nuclear antigens reactive with autoantibodies. Immunofluorescence studies on distribution in synchronized cells. J Cell Biol. 1981;91:654–660. doi: 10.1083/jcb.91.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng J S, Tan E M. Effect of actinomycin D on the expression of nuclear antigen SS-B/La. J Investig Dermatol. 1985;84:225–228. doi: 10.1111/1523-1747.ep12265236. [DOI] [PubMed] [Google Scholar]
- 38.Deo R C, Bonanno J B, Sonenberg N, Burley S K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 39.Di Francesco V, Garnier J, Munson P J. Improving protein secondary structure prediction with aligned homologous sequences. Protein Sci. 1996;5:106–113. doi: 10.1002/pro.5560050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duncan R C, Nakhasi H L. La autoantigen binding to a 5′ cis-element of rubella virus RNA correlates with element function in vivo. Gene. 1997;201:137–149. doi: 10.1016/s0378-1119(97)00438-1. [DOI] [PubMed] [Google Scholar]
- 41.Fan H, Goodier J L, Chamberlain J, Engelke D R, Maraia R J. 5′ processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol Cell Biol. 1998;18:3201–3211. doi: 10.1128/mcb.18.6.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan H, Sakulich A L, Goodier J L, Zhang X, Qin J, Maraia R J. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–715. doi: 10.1016/s0092-8674(00)81913-3. [DOI] [PubMed] [Google Scholar]
- 43.Francoeur A M, Mathews M B. Interaction between VA RNA and the lupus antigen La: formation of a ribonucleoprotein particle in vitro. Proc Natl Acad Sci USA. 1982;79:6772–6776. doi: 10.1073/pnas.79.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodier J L, Fan H, Maraia R J. A carboxy-terminal basic region controls RNA polymerase III transcription factor activity of human La protein. Mol Cell Biol. 1997;17:5823–5832. doi: 10.1128/mcb.17.10.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodier J L, Maraia R J. Terminator-specific recycling of a B1-Alu transcription complex by RNA polymerase III is mediated by the RNA terminus-binding protein La. J Biol Chem. 1998;273:26110–26116. doi: 10.1074/jbc.273.40.26110. [DOI] [PubMed] [Google Scholar]
- 46.Gottlieb E, Steitz J A. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 1989;8:851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gottlieb E, Steitz J A. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J. 1989;8:841–850. doi: 10.1002/j.1460-2075.1989.tb03445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graus F, Cordon-Cardo C, Bonfa E, Elkon K B. Immunohistochemical localization of La nuclear antigen in brain. Selective concentration of the La protein in neuronal nucleoli. J Neuroimmunol. 1985;9:307–319. doi: 10.1016/s0165-5728(85)80028-x. [DOI] [PubMed] [Google Scholar]
- 49.Grimm C, Lund E, Dahlberg J E. In vivo selection of RNAs that localize in the nucleus. EMBO J. 1997;16:793–806. doi: 10.1093/emboj/16.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grolz D, Bachmann M. An altered intracellular distribution of the autoantigen La/SS-B when translated from a La mRNA isoform. Exp Cell Res. 1997;234:329–335. doi: 10.1006/excr.1997.3608. [DOI] [PubMed] [Google Scholar]
- 51.Grolz D, Laubinger J, Wilmer F, Troster H, Bachmann M. Transfection analysis of expression of mRNA isoforms encoding the nuclear autoantigen La/SS-B. J Biol Chem. 1997;272:12076–12082. doi: 10.1074/jbc.272.18.12076. [DOI] [PubMed] [Google Scholar]
- 52.Grosshans H, Simos G, Hurt E. Review: transport of tRNA out of the nucleus—direct channeling to the ribosome? J Struct Biol. 2000;129:288–294. doi: 10.1006/jsbi.2000.4226. [DOI] [PubMed] [Google Scholar]
- 53.Guddat U, Bakken A H, Pieler T. Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in Xenopus oocytes. Cell. 1990;60:619–628. doi: 10.1016/0092-8674(90)90665-2. [DOI] [PubMed] [Google Scholar]
- 54.Habets W J, denBrok J H, Boerbooms A M, van de Putte L B, van Venrooij W J. Characterization of the SS-B (La) antigen in adenovirus-infected and uninfected HeLa cells. EMBO J. 1983;2:1625–1631. doi: 10.1002/j.1460-2075.1983.tb01636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 56.Hashimoto C, Steitz J A. Sequential association of nucleolar 7-2 RNA with two different autoantigens. J Biol Chem. 1983;258:1379–1382. [PubMed] [Google Scholar]
- 57.Heise T, Guidotti L G, Chisari F V. La autoantigen specifically recognizes a predicted stem-loop in hepatitis B virus RNA. J Virol. 1999;73:5767–5776. doi: 10.1128/jvi.73.7.5767-5776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hendrick J P, Wolin S L, Rinke J, Lerner M R, Steitz J A. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol. 1981;1:1138–1149. doi: 10.1128/mcb.1.12.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henikoff S, Henikoff J G. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991;19:6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hilker M, Troster H, Grolz D, Hake U, Bachmann M. The autoantigen La/SS-B: analysis of the expression of alternatively spliced La mRNA isoforms. Cell Tissue Res. 1996;284:383–389. doi: 10.1007/s004410050599. [DOI] [PubMed] [Google Scholar]
- 61.Hoffman D W, Query C C, Golden B L, White S W, Keene J D. RNA-binding domain of the A protein component of the U1 small nuclear ribonucleoprotein analyzed by NMR spectroscopy is structurally similar to ribosomal proteins. Proc Natl Acad Sci USA. 1991;88:2495–2499. doi: 10.1073/pnas.88.6.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holcik M, Korneluk R G. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol Cell Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk R G. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 64.Hopper A K. Nuclear functions charge ahead. Science. 1998;282:2003–2004. doi: 10.1126/science.282.5396.2003. [DOI] [PubMed] [Google Scholar]
- 65.Huhn P, Pruijn G J, van Venrooij W J, Bachmann M. Characterization of the autoantigen La (SS-B) as a dsRNA unwinding enzyme. Nucleic Acids Res. 1997;25:410–416. doi: 10.1093/nar/25.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hurt D J, Wang S S, Lin Y H, Hopper A K. Cloning and characterization of LOS1, a Saccharomyces cerevisiaegene that affects tRNA splicing. Mol Cell Biol. 1987;7:1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Intine R V A, Sakulich A L, Koduru S B, Huang Y, Pierstorrf E, Goodier J L, Phan L, Maraia R J. Transfer RNA maturation is controlled by phosphorylation of the human La antigen on serine 366. Mol Cell. 2000;6:339–348. doi: 10.1016/s1097-2765(00)00034-4. [DOI] [PubMed] [Google Scholar]
- 68.Isoyama T, Kamoshita N, Yasui K, Iwai A, Shiroki K, Toyoda H, Yamada A, Takasaki Y, Nomoto A. Lower concentration of La protein required for internal ribosome entry on hepatitis C virus RNA than on poliovirus RNA. J Gen Virol. 1999;80:2319–2327. doi: 10.1099/0022-1317-80-9-2319. [DOI] [PubMed] [Google Scholar]
- 69.Jacobson M R, Cao L G, Taneja K, Singer R H, Wang Y L, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- 70.James M C, Jeffrey I W, Pruijn G J, Thijssen J P, Clemens M J. Translational control by the La antigen. Structure requirements for rescue of the double-stranded RNA-mediated inhibition of protein synthesis. Eur J Biochem. 1999;266:151–162. doi: 10.1046/j.1432-1327.1999.00839.x. [DOI] [PubMed] [Google Scholar]
- 71.Jarrous N, Wolenski J S, Wesolowski D, Lee C, Altman S. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J Cell Biol. 1999;146:559–572. doi: 10.1083/jcb.146.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kenan D J. RNA recognition by the human La protein and its relevance to transcription, translation and viral infectivity. Ph.D. thesis. Durham, N.C: Duke University; 1995. [Google Scholar]
- 73.Kenan D J, Query C C, Keene J D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 74.Kim Y K, Jang S K. La protein is required for efficient translation driven by encephalomyocarditis virus internal ribosomal entry site. J Gen Virol. 1999;80:3159–3166. doi: 10.1099/0022-1317-80-12-3159. [DOI] [PubMed] [Google Scholar]
- 75.Kobor M S, Archambault J, Lester W, Holstege F C, Gileadi O, Jansma D B, Jennings E G, Kouyoumdjian F, Davidson A R, Young R A, Greenblatt J. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 76.Kole R, Fresco L D, Keene J D, Cohen P L, Eisenberg R A, Andrews P G. Alu RNA-protein complexes formed in vitro react with a novel lupus autoantibody. J Biol Chem. 1985;260:11781–11786. [PubMed] [Google Scholar]
- 77.Kufel J, Allmang C, Chanfreau G, Petfalski E, Lafontaine D L, Tollervey D. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol Cell Biol. 2000;20:5415–5424. doi: 10.1128/mcb.20.15.5415-5424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurilla M G, Keene J D. The leader RNA of vesicular stomatitis virus is bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1983;34:837–845. doi: 10.1016/0092-8674(83)90541-x. [DOI] [PubMed] [Google Scholar]
- 79.Lerner M R, Boyle J A, Hardin J A, Steitz J A. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science. 1981;211:400–402. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- 80.Levinger L, Vasisht V, Greene V, Bourne R, Birk A, Kolla S. Sequence and structure requirements for Drosophila tRNA 5′- and 3′-end processing. J Biol Chem. 1995;270:18903–18909. doi: 10.1074/jbc.270.32.18903. [DOI] [PubMed] [Google Scholar]
- 81.Li W Y, Reddy R, Henning D, Epstein P, Busch H. Nucleotide sequence of 7 S RNA. Homology to Alu DNA and La 4.5 S RNA. J Biol Chem. 1982;257:5136–5142. [PubMed] [Google Scholar]
- 82.Lin-Marq N, Clarkson S G. Efficient synthesis, termination and release of RNA polymerase III transcripts in Xenopusextracts depleted of La protein. EMBO J. 1998;17:2033–2041. doi: 10.1093/emboj/17.7.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin-Marq N, Clarkson S G. A yeast RNA binding protein that resembles the human autoantigen La. J Mol Biol. 1995;245:81–85. doi: 10.1006/jmbi.1994.0008. [DOI] [PubMed] [Google Scholar]
- 84.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 85.Madore S J, Wieben E D, Pederson T. Eukaryotic small ribonucleoproteins. Anti-La human autoantibodies react with U1 RNA-protein complexes. J Biol Chem. 1984;259:1929–1933. [PubMed] [Google Scholar]
- 86.Maraia R, Zasloff M, Plotz P, Adeniyi-Jones S. Pathway of B1-Alu expression in microinjected oocytes: Xenopus laevisproteins associated with nuclear precursor and processed cytoplasmic RNAs. Mol Cell Biol. 1988;8:4433–4440. doi: 10.1128/mcb.8.10.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maraia R J. Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc Natl Acad Sci USA. 1996;93:3383–3387. doi: 10.1073/pnas.93.8.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maraia R J, Kenan D J, Keene J D. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol. 1994;14:2147–2158. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mathews M B, Francoeur A M. La antigen recognizes and binds to the 3′-oligouridylate tail of a small RNA. Mol Cell Biol. 1984;4:1134–1140. doi: 10.1128/mcb.4.6.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mazabraud A, Scherly D, Rungger D, Clarkson S G. Structure and transcription termination of a lysine tRNA gene from Xenopus laevis. J Mol Biol. 1987;195:835–845. doi: 10.1016/0022-2836(87)90488-8. [DOI] [PubMed] [Google Scholar]
- 91.McLaren R S, Caruccio N, Ross J. Human La protein: a stabilizer of histone mRNA. Mol Cell Biol. 1997;17:3028–3036. doi: 10.1128/mcb.17.6.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meerovitch K, Pelletier J, Sonenberg N. A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989;67:3798–3807. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- 93.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mittal V, Ma B, Hernandez N. SNAP(c): a core promoter factor with a built-in DNA-binding damper that is deactivated by the Oct-1 POU domain. Genes Dev. 1999;13:1807–1821. doi: 10.1101/gad.13.14.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nagai K, Oubridge C, Jessen T H, Li J, Evans P R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 96.Nanbru C, Lafon I, Audigier S, Gensac M C, Vagner S, Huez G, Prats A C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 97.Nashimoto M, Tamura M, Kaspar R L. Selection of cleavage site by mammalian tRNA 3′ processing endoribonuclease. J Mol Biol. 1999;287:727–740. doi: 10.1006/jmbi.1999.2639. [DOI] [PubMed] [Google Scholar]
- 98.Neugebauer K M, Roth M B. Transcription units as RNA processing units. Genes Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- 99.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark B F, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 100.Nyborg J, Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Clark B F. Structure of the ternary complex of EF-Tu: macromolecular mimicry in translation. Trends Biochem Sci. 1996;21:81–82. [PubMed] [Google Scholar]
- 101.Oh S K, Scott M P, Sarnow P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 102.Oubridge C, Ito N, Evans P R, Teo C H, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 103.Pannone B, Xue D, Wolin S L. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pardigon N, Strauss J H. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J Virol. 1996;70:1173–1181. doi: 10.1128/jvi.70.2.1173-1181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 106.Pellizzoni L, Cardinali B, Lin-Marq N, Mercanti D, Pierandrei-Amaldi P. A Xenopus laevis homologue of the La autoantigen binds the pyrimidine tract of the 5′ UTR of ribosomal protein mRNAs in vitro: implication of a protein factor in complex formation. J Mol Biol. 1996;259:904–915. doi: 10.1006/jmbi.1996.0368. [DOI] [PubMed] [Google Scholar]
- 107.Pfeifle J, Anderer F A, Franke M. Multiple phosphorylation of human SS-B/La autoantigen and its effect on poly(U) and autoantibody binding. Biochim Biophys Acta. 1987;928:217–226. doi: 10.1016/0167-4889(87)90124-8. [DOI] [PubMed] [Google Scholar]
- 108.Pogue G P, Hofmann J, Duncan R, Best J M, Etherington J, Sontheimer R D, Nakhasi H L. Autoantigens interact with cis-acting elements of rubella virus RNA. J Virol. 1996;70:6269–6277. doi: 10.1128/jvi.70.9.6269-6277.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pombo A, Jackson D A, Hollinshead M, Wang Z, Roeder R G, Cook P R. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Preiser P, Vasisht V, Birk A, Levinger L. Poly(U)-binding protein inhibits Drosophila pre-5 S RNA 3′-exonuclease digestion. J Biol Chem. 1993;268:11553–11557. [PubMed] [Google Scholar]
- 110a.Pyronnet, Pradayrol S L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 111.Qiu H, Hu C, Anderson J, Björk G, Sarkar S, Hopper A, Hinnebusch A G. Defects in tRNA processing and nuclear export induce GCN4 translation independently of phosphorylation of the subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2000;20:2505–2516. doi: 10.1128/mcb.20.7.2505-2516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Query C C, Bentley R C, Keene J D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- 113.Reddy R, Henning D, Tan E, Busch H. Identification of a La protein binding site in a RNA polymerase III transcript (4.5 I RNA) J Biol Chem. 1983;258:8352–8356. [PubMed] [Google Scholar]
- 114.Rinke J, Steitz J A. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 1985;13:2617–2629. doi: 10.1093/nar/13.7.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rinke J, Steitz J A. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1982;29:149–159. doi: 10.1016/0092-8674(82)90099-x. [DOI] [PubMed] [Google Scholar]
- 116.Rosa M D, Gottlieb E, Lerner M R, Steitz J A. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol. 1981;1:785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rosenblum J S, Pemberton L F, Bonifaci N, Blobel G. Nuclear import and the evolution of a multifunctional RNA-binding protein. J Cell Biol. 1998;143:887–899. doi: 10.1083/jcb.143.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rutjes S A, Utz P J, der Heijden A, Broekhuis C, van Venrooij W J, Pruijn G J. The La (SS-B) autoantigen, a key protein in RNA biogenesis, is dephosphorylated and cleaved early during apoptosis. Cell Death Differ. 1999;6:976–986. doi: 10.1038/sj.cdd.4400571. [DOI] [PubMed] [Google Scholar]
- 119.Sachs A B, Bond M W, Kornberg R D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 120.Sarkar S, Azad A K, Hopper A K. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:14366–14371. doi: 10.1073/pnas.96.25.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Scherly D, Stutz F, Lin-Marq N, Clarkson S G. La proteins from Xenopus laeviscDNA cloning and developmental expression. J Mol Biol. 1993;231:196–204. doi: 10.1006/jmbi.1993.1275. [DOI] [PubMed] [Google Scholar]
- 122.Shamoo Y, Abdul-Manan N, Patten A M, Crawford J K, Pellegrini M C, Williams K R. Both RNA-binding domains in heterogenous nuclear ribonucleoprotein A1 contribute toward single-stranded-RNA binding. Biochemistry. 1994;33:8272–8281. doi: 10.1021/bi00193a014. [DOI] [PubMed] [Google Scholar]
- 123.Shamoo Y, Abdul-Manan N, Williams K R. Multiple RNA binding domains (RBDs) just don't add up. Nucleic Acids Res. 1995;23:725–728. doi: 10.1093/nar/23.5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shen W C, Selvakumar D, Stanford D R, Hopper A K. The Saccharomyces cerevisiae LOS1 gene involved in pre-tRNA splicing encodes a nuclear protein that behaves as a component of the nuclear matrix. J Biol Chem. 1993;268:19436–19444. [PubMed] [Google Scholar]
- 125.Shen W C, Stanford D R, Hopper A K. Los1p, involved in yeast pre-tRNA splicing, positively regulates members of the SOL gene family. Genetics. 1996;143:699–712. doi: 10.1093/genetics/143.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen Y, Igo M, Yalamanchili P, Berk A J, Dasgupta A. DNA binding domain and subunit interactions of transcription factor IIIC revealed by dissection with poliovirus 3C protease. Mol Cell Biol. 1996;16:4163–4171. doi: 10.1128/mcb.16.8.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shiroki K, Isoyama T, Kuge S, Ishii T, Ohmi S, Hata S, Suzuki K, Takasaki Y, Nomoto A. Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J Virol. 1999;73:2193–2200. doi: 10.1128/jvi.73.3.2193-2200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sidrauski C, Chapman R, Walter P. The unfolded protein response: an intracellular signalling pathway with many surprising features. Trends Cell Biol. 1998;8:245–249. doi: 10.1016/s0962-8924(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 129.Sidrauski C, Cox J S, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 130.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 131.Simons F H, Broers F J, Van Venrooij W J, Pruijn G J. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp Cell Res. 1996;224:224–236. doi: 10.1006/excr.1996.0132. [DOI] [PubMed] [Google Scholar]
- 132.Simons F H, Rutjes S A, van Venrooij W J, Pruijn G J. Interactions with Ro60 and La differentially affect nuclear export of hY1 RNA. RNA. 1996;2:264–273. [PMC free article] [PubMed] [Google Scholar]
- 133.Sobel S G, Wolin S L. Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes. Mol Biol Cell. 1999;10:3849–3862. doi: 10.1091/mbc.10.11.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Spangberg K, Goobar-Larsson L, Wahren-Herlenius M, Schwartz S. The La protein from human liver cells interacts specifically with the U-rich region in the hepatitis C virus 3′ untranslated region. J Hum Virol. 1999;2:296–307. [PubMed] [Google Scholar]
- 135.St. Clair E W, Burch J A, Jr, Ward M M, Keene J D, Pisetsky D S. Temporal correlation of antibody responses to different epitopes of the human La autoantigen. J Clin Investig. 1990;85:515–521. doi: 10.1172/JCI114467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stefano J E. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 137.Steitz J A, Berg C, Hendrick J P, La Branche-Chabot H, Metspalu A, Rinke J, Yario T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J Cell Biol. 1988;106:545–556. doi: 10.1083/jcb.106.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Steitz J A, Wolin S L, Rinke J, Pettersson I, Mount S M, Lerner E A, Hinterberger M, Gottlieb E. Small ribonucleoproteins from eukaryotes: structures and roles in RNA biogenesis. Cold Spring Harbor Symp Quant Biol. 1983;47:893–900. doi: 10.1101/sqb.1983.047.01.103. [DOI] [PubMed] [Google Scholar]
- 139.Terns M P, Lund E, Dahlberg J E. 3′-end-dependent formation of U6 small nuclear ribonucleoprotein particles in Xenopus laevisoocyte nuclei. Mol Cell Biol. 1992;12:3032–3040. doi: 10.1128/mcb.12.7.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Topfer F, Gordon T, McCluskey J. Characterization of the mouse autoantigen La (SS-B) J Immunol. 1993;150:3091–3100. [PubMed] [Google Scholar]
- 141.Troster H, Metzger T E, Semsei I, Schwemmle M, Winterpacht A, Zabel B, Bachmann M. One gene, two transcripts: isolation of an alternative transcript encoding for the autoantigen La/SS-B from a cDNA library of a patient with primary Sjogren's syndrome. J Exp Med. 1994;180:2059–2067. doi: 10.1084/jem.180.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vagner S, Gensac M C, Maret A, Bayard F, Amalric F, Prats H, Prats A C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Van Horn D J, Yoo C J, Xue D, Shi H, Wolin S L. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA. 1997;3:1434–1443. [PMC free article] [PubMed] [Google Scholar]
- 144.Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- 145.Wang Z, Luo T, Roeder R G. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Weser S, Bachmann M, Seifart K H, Meißner W. Transcription efficiency of human polymerase III genes in vitro does not depend on the RNP-forming autoantigen La. Nucleic Acids Res. 2000;28:3935–3942. doi: 10.1093/nar/28.20.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wold W S, Hermiston T W, Tollefson A E. Adenovirus proteins that subvert host defenses. Trends Microbiol. 1994;2:437–443. doi: 10.1016/0966-842x(94)90801-x. [DOI] [PubMed] [Google Scholar]
- 148.Wolin S L, Matera A G. The trials and travels of tRNA. Genes Dev. 1999;13:1–10. doi: 10.1101/gad.13.1.1. [DOI] [PubMed] [Google Scholar]
- 149.Xiao Q, Sharp T V, Jeffrey I W, James M C, Pruijn G J, van Venrooij W J, Clemens M J. The La antigen inhibits the activation of the interferon-inducible protein kinase PKR by sequestering and unwinding double-stranded RNA. Nucleic Acids Res. 1994;22:2512–2518. doi: 10.1093/nar/22.13.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xue D, Rubinson D A, Pannone B K, Yoo C J, Wolin S L. snRNP assembly in yeast involves the La protein. EMBO J. 2000;19:1650–1660. doi: 10.1093/emboj/19.7.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yalamanchili P, Weidman K, Dasgupta A. Cleavage of transcriptional activator Oct-1 by poliovirus encoded protease 3Cpro. Virology. 1997;239:176–185. doi: 10.1006/viro.1997.8862. [DOI] [PubMed] [Google Scholar]
- 152.Ye X, Fong P, Iizuka N, Choate D, Cavener D R. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol Cell Biol. 1997;17:1714–1721. doi: 10.1128/mcb.17.3.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yoo C J, Wolin S L. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol. 1994;14:5412–5424. doi: 10.1128/mcb.14.8.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yoo C J, Wolin S L. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 155.Yu Y-T, Scharl E C, Smith C M, Steitz J A. The growing world of small nuclear ribonucleoproteins. In: Gesteland R F, Chech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 487–524. [Google Scholar]
- 156.Zasloff M, Santos T, Romeo P, Rosenberg M. Transcription and precursor processing of normal and mutant human tRNAimetgenes in a homologous cell-free system. J Biol Chem. 1982;257:7857–7863. [PubMed] [Google Scholar]
- 157.Zecherle G N, Whelen S, Hall B D. Purines are required at the 5′ ends of newly initiated RNAs for optimal RNA polymerase III gene expression. Mol Cell Biol. 1996;16:5801–5810. doi: 10.1128/mcb.16.10.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]