Abstract
Sheep (Ovis aries) and goats (Capra hircus) have, for more than a millennia, been a source of fibres for human use, be it for use in clothing and furnishings, for insulation, for decorative and ceremonial purposes, or for combinations thereof. While use of these natural fibres has in some respects been superseded by the use of synthetic and plant-based fibres, increased accounting for the carbon and water footprint of these fibres is creating a re-emergence of interest in fibres derived from sheep and goats. The keratin-associated proteins (KAPs) are structural components of wool and hair fibres, where they form a matrix that cross-links with the keratin intermediate filaments (KIFs), the other main structural component of the fibres. Since the first report of a complete KAP protein sequence in the late 1960s, considerable effort has been made to identify the KAP proteins and their genes in mammals, and to ascertain how these genes and proteins control fibre growth and characteristics. This effort is ongoing, with more and more being understood about the structure and function of the genes. This review consolidates that knowledge and suggests future directions for research to further our understanding.
Keywords: KAP, KRTAP, variation, polymorphism, wool, fibre, sheep, goat
1. Introduction
The keratin-associated proteins (KAPs) are structural components of the fibres that form the pelage of mammals. They are part of a matrix, which cross-links with the keratin (K) protein containing keratin intermediate filaments (KIFs), which are the other main structural components of the fibre. In the wool follicles of sheep, the KAPs are produced soon after the synthesis of keratins during the development of fibres in the follicle, and they are thought to cross-link with the KIFs by forming disulphide bonds with cysteine residues in the head and tail domains of the keratins [1]. The nature of this cross-linking is not well understood, and while co-immunoprecipitation studies have demonstrated an interaction between the head domain of human keratin K86 and KAP2-1 [2], and Western blot studies have demonstrated an interaction between the head domain of K85 and KAP8-1 [2], a complete understanding of which K and KAP cysteine thiol groups form disulphide bridges (be they inter- or intra-chain) is not well known, although the bulk of the most readily accessible cysteines in the KAPs are reported to be found close to either the N- or C-terminal domains in these proteins [3]. The KAPs are thought to play a critical role in regulating the physico-mechanical properties of the fibre.
With sheep and goats, the hair and wool fibres are produced by follicles that are located in the skin, but the value of these fibres varies considerably depending on their qualities, including their fineness (mean fibre diameter), their uniformity, their length, and their colour.
Since the first report of a complete KAP sequence in the late 1960s [4], considerable effort has been made to identify the KAP proteins and their genes in both sheep and goats. Research has also focused on ascertaining how these genes and proteins control and affect fibre qualities. Our understanding has progressed rapidly over the last two decades, especially since the sheep and goat genome sequences have become available. This has enabled more ovine and caprine KAP genes to be identified and characterized.
2. Keratin-Associated Proteins and the Genes That Encode Them
The KAPs are small (ca. 10–30 kDa) and possess either a high cysteine or a high glycine and tyrosine content [1,5]. These proteins were originally categorized into three broad groups based on their amino acid composition: the high sulphur (HS; ≤30 mol% cysteine), the ultra-high sulphur (UHS; >30 mol% cysteine) and the high glycine and tyrosine (HGT; 35–60 mol% glycine and tyrosine) KAPs [1]. Despite being glycine and tyrosine rich, the HGT-KAPs also contain an abundance of cysteine, with the apparent exception of KAP36-1, a recently identified protein from sheep that is deficient in cysteine [6].
The absence of cysteine in ovine KAP36-1 suggests the possibility of other forms of cross-linking or interaction for the HGT-KAPs. In this respect the presence of tyrosine in the KAPs may be of significance. Tyrosine is an aromatic amino acid and contains a benzene ring in its side-chain. The benzene ring may allow the tyrosine residues in the HGT-KAPs to interact with other tyrosine residues, or other aromatic amino acids in the KAPs and/or the KIFs, using a ‘ring-stacking’ mechanism. This cross-linking mechanism has been described for other aromatic amino acid-containing proteins [7]. In the HGT-KAPs, the tyrosine residues are usually surrounded by glycine residues. Having a small amino acid such as glycine in proximity to tyrosine would allow the tyrosine residues greater freedom to move their benzene rings into a preferred spatial orientation (i.e., conformational freedom), and thus enable the formation of amino acid to amino acid interactions. Tyrosine also possesses a reactive hydroxyl group at the end of its side-chain, which can act as a hydrogen donor, and thus potentially form hydrogen bonds with the centre of the benzene ring from another tyrosine, or another aromatic amino acid [8]. This would make the ring-stacking interaction even stronger, and may result in the wool fibre being further strengthened, while simultaneously giving some degree of pliability [9].
All of the KAPs are encoded by single exon (intron-less) genes called the KRTAPs and, accordingly, the KRTAPs are understandably small in size (with an open reading frame of usually less than 800 bp). They often share sequence similarities with each other, and can be assigned into families based on sequence similarity. Extensive bioinformatics analyses of the human genome have resulted in the identification of 88 functional KRTAPs, which is probably close to the complete catalogue of these genes in humans [10,11,12]. These KRTAPs have been assigned into 25 KAP families, numbered from KAP1 to KAP27, with the exclusion of KAP14 and KAP18. The KAP14 family name has been used for two murine HS-KAP genes, KRTAP14-1 (previously named mKAP13) [13] and KRTAP14-2 (previously called pmg1) [14], whereas the KAP18 family has been used for a different murine HGT-KAP gene [4]. Intact orthologues of these genes are not found in humans [15], suggesting that these KRTAPs may be mouse-specific.
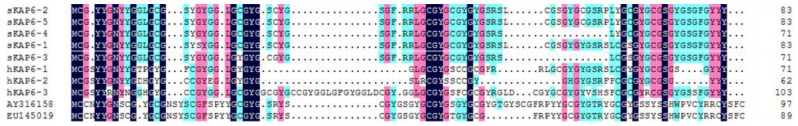
Numerous KRTAPs have been described in sheep and goats, with the identification of 30 ovine KRTAPs and 18 caprine KRTAPs to date. These KRTAPs represent 18 ovine KAP gene families (Table 1) and 12 caprine KAP families (Table 2). There are four other ovine KRTAP sequences reported that probably represent four other KRTAPs in sheep [16], but because these sequences are only described as partial coding sequences, their identity awaits further investigation. They are, accordingly, not included in the ‘identified’ KRTAPs described in Table 1. Two other goat KRTAP sequences, EU145019 [17] and AY316158 [18], are reported to be “alleles of caprine KRTAP6-2”, but a sequence analysis suggests that these two sequences are quite different to the KRTAP6-n sequences from sheep and humans (Figure 1). This suggests that these may not be caprine KRTAP6-n sequences, especially given other similarities between the sheep and goat genomes at the nucleotide sequence level, albeit not at the level of chromosomal arrangement. Accordingly, their true identity appears to await further investigation.
Table 1.
Summary of the ovine KRTAPs that have been identified.
| KRTAP | Number of Variants | Number of SNPs | Length Variation | GenBank Accession Numbers | References |
|---|---|---|---|---|---|
| KRTAP1-1 | 3 | 3 | ± 30-bp repeats | L33885-L33887 | [19] |
| KRTAP1-2 | 11 | 10 | No | HQ897973-HQ897982, KM105941-KM105942 | [20,21] |
| KRTAP1-3 | 9 | 17 | No | AY835589-AY835597 | [22] |
| KRTAP1-4 | 9 | 14 | No | GQ507741-GQ507749 | [23] |
| KRTAP2-1 | 4 | 9 | No | [24] | |
| KRTAP3-1 | Unknown | Unknown | Unknown | M21099 | [25] |
| KRTAP3-3 | Unknown | Unknown | Unknown | N21103 | [25] |
| KRTAP4-3 | Unknown | Unknown | Unknown | EU239778 | [26] |
| KRTAP5-1 | Unknown | Unknown | Unknown | X55294 | [27] |
| KRTAP5-4 | 5 | 6 | ±30-bp repeats | GU255997-GU256001 | [28] |
| KRTAP6-1 | 5 | 4 | ± 57-bp | GU319873, GU319875 | [29,30] |
| KRTAP6-2 | 6 | 5 | No | KT725827-KT725832 | [31] |
| KRTAP6-3 | 7 | 5 | ± 45-bp | KT725833-KT725837, GU319876 | [31,32,33] |
| KRTAP6-4 | 3 | 3 | No | KT725838-KT725840 | [31] |
| KRTAP6-5 | 6 | 5 | ± 18-bp | KT725841-KT725846 | [31] |
| KRTAP7-1 | 2 | 1 | No | JN091630, JN091631 | [34] |
| KRTAP8-1 | 5 | 4 | No | JN091632-JN091636 | [34] |
| KRTAP8-2 | 3 | 2 | No | KF220646-KF220647 | [35,36] |
| KRTAP11-1 | 6 | 5 | No | HQ595347-HQ595352 | [37] |
| KRTAP13-3 | 5 | 4 | No | JN377429-JN377433 | [38] |
| KRTAP15-1 | 4 | 4 | No | MH742372-MH742375 | [39] |
| KRTAP20-1 | 8 | 6 | ±12-bp | MH243552-MH243559 | [40] |
| KRTAP20-2 | 2 | 1 | No | MH071391, MH071392 | [41] |
| KRTAP21-1 | 3 | 2 | No | MF143980-MF143983 | [42] |
| KRTAP21-2 | 5 | 4 | No | MF143975-MF143979 | [43] |
| KRTAP22-1 | 3 | 2 | No | KX377616-KX377618 | [44] |
| KRTAP24-1 | 4 | 7 | No | JX112014-JX112017 | [45] |
| KRTAP26-1 | 4 | 7 | No | KX644903–KX644906 | [46] |
| KRTAP28-1 | 6 | 8 | ±2-bp repeats | MN053915-MN053920 | [47] |
| KRTAP36-1 | 3 | 4 | No | MK770620-MK770622 | [6] |
Table 2.
The caprine KRTAPs that have been identified.
| KRTAP | Number of Variants | Number of SNPs | Length Variation | GenBank Accession Numbers | References |
|---|---|---|---|---|---|
| KRTAP1-1 * | 7 | 5 | No | [48] | |
| KRTAP1-2 | 6 | 5 | ±60-bp and 15-bp | [49] | |
| KRTAP1-3 * | Unknown | Unknown | Unknown | JQ772533 | [50] |
| KRTAP1-4 | 6 | 8 | No | N012101, JN012102, JN000317, JN000318, JQ436929, JQ627657 | [51] |
| KRTAP3-1 | Unknown | Unknown | Unknown | NM_001285774 | [52] |
| KRTAP7-1 | 2 | 1 | No | AY510121 | [53] |
| KRTAP8-1 | 4 | 2 | No | AY510122, EU595394, EU595395 | [48,54] |
| KRTAP8-2 | 3 | 2 | No | AY510123 | [55] |
| KRTAP9-2 | 3 | 1 | ±30-bp | AY510124, EU430080, | [56,57] |
| KRTAP11-1 | Unknown | Unknown | Unknown | JQ795995 | [58] |
| KRTAP13-3 | 18 | 17 | No | JX426138-JX426145 | [48,59] |
| KRTAP13-n | 2 | 1 | No | AY510115 | [60] |
| KRTAP15-1 | 6 | 8 | No | [61] | |
| KRTAP20-1 | 4 | 6 | No | MG742218- MG742221 | [62] |
| KRTAP20-2 | 3 | 4 | No | MF973462-MF973464 | [63] |
| KRTAP24-1 | 4 | 9 | No | MG996011-MG996014 | [64] |
| KRTAP27-1 | 3 | 2 | No | MN934937- MN934939 | [65] |
| KRTAP28-1 | 5 | 8 | ±2-bp repeats | [66] |
* The gene appears to have been identified, but is not well characterised.
Figure 1.
Comparison of the predicted amino acid sequences of two goat “KRTAP6-2” sequences with sheep and human KRTAP6 sequences. The goat “KRTAP6-2” sequences are indicated with the GenBank accession number AY316158 and EU145019. The sheep sequences are indicated with the prefix “s”, while the human sequences are marked with the prefix “h”. The GenBank accession numbers of these sequences are NM_001193399 (sKAP6-1), KT725832 (sKAP6-2), KT725837 (sKAP6-3), KT725840 (sKAP6-4), KT725845 (sKAP6-5), NM_181602 (hKAP6-1), NM_181604 (hKAP6-2), and NM_181605 (hKAP6-3). The numbers on the right of the sequences represent the length of the proteins. Amino acid positions with high levels of homology are coloured, with black indicating 100% homology, red indicating greater than or equal to 75%, and blue indicating greater than or equal to 50%.
Orthologues for all of the human KRTAPs can be found in sheep and goats, with the exception of KRTAP25-1. A recent search for sequences comparable to human KRTAP25-1 in the sheep genome assembly did not reveal any comparable sequences [47], but in the chromosome region, where the human KRTAP25-1 orthologue was expected to be found, there was an apparently unique KRTAP sequence, which has been named KRTAP28-1 [47].
In contrast, several KRTAPs that have not been described in humans are found in sheep and goats, including KRTAP8-2 [35,55], KRTAP6-4 [31], KRTAP6-5 [31], and KRTAP36-1 [6]. All of these ‘additional’ KRTAPs encode HGT-KAPs, which suggests that sheep possess more HGT-KRTAPs than humans.
Within families, the KRTAPs can exhibit a high degree of nucleotide sequence similarity, particularly in their coding regions, and some KRTAPs even have identical coding sequences. For example, KRTAP6-2 variants B, C, and D have coding sequences that are identical to KRTAP6-5 variants B and D (Figure 2). These genes and their variants can only be differentiated by variation in their 3′ and 5′ flanking sequences.
Figure 2.
Sequence comparison of variants of ovine KRTAP6-2 and KRTAP6-5. The nucleotides within the coding region are shown in upper-case text, while those in the flanking regions are shown in lower-case text. Dashes indicate nucleotide sequences identical to the top sequence, and dots have been introduced to improve the alignment. The start and stop codons are shown in bold.
The high sequence similarity in coding regions can make it difficult (and sometimes impossible) to assign KAP protein sequences or partial gene sequences into families. Equally, it is also difficult to determine whether different KAP protein sequences represent different family members, or just variant sequences of the same family member. This hampers the application of proteomic approaches to KAP research. It also highlights the critical importance of having extended and comprehensive gene sequences for the KRTAPs, along with an idea of their location on chromosomes, if one is to accurately identify and classify both the KAP and the KRTAP sequences.
3. Variation in the KRTAPs
Nucleotide sequence variation has been explored for many of the ovine and caprine KRTAPs. To date, all of the ovine and caprine KRTAPs that have been investigated are polymorphic, but the extent and nature of the polymorphism varies between the genes. Some KRTAPs exhibit a low level of polymorphism, such as ovine and caprine KRTAP7-1 [34,53] and ovine KRTAP20-2 [41]. Each of these genes has only two sequence variants. On the other hand, some KRTAPs possess a high level of polymorphism, such as ovine KRTAP1-2 [20,21], ovine KRTAP1-4 [23], and caprine KRTAP13-3 [48,59], for which nine or more sequence variants have been identified. The majority of KRTAPs exhibit a moderate level of polymorphism, with the number of sequence variants ranging from three to eight.
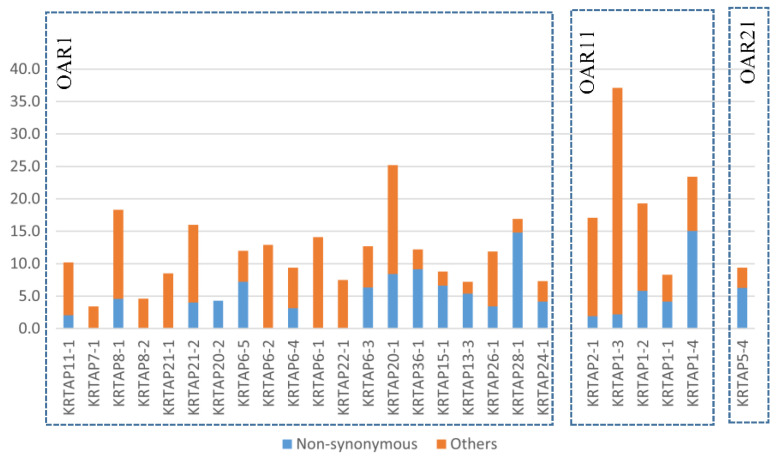
The polymorphism described in the KRTAPs includes single nucleotide polymorphisms (SNPs), and insertions and deletions (indels). With the exception of KRTAP6-1, for which all of the SNPs are found either upstream or downstream of the coding region [31,67], SNPs in all of the other KRTAPs mostly occur in the coding region. The SNP density, and the proportion of non-synonymous SNPs, varies considerably between the KRTAPs. Some, such as ovine KRTAP1-3, KRTAP1-4, and KRTAP20-1, have a density of over 20 SNPs per kb, while others, such as ovine KRTAP7-1, KRTAP8-2, and KRTAP20-2, have a density of less than five SNPs per kb (Figure 3). Overall, the SNP density in the majority of KRTAPs is higher than the average density of 4.9 SNP per kb that has been suggested to occur across the sheep genome [68], albeit that estimate is now quite dated. There does not appear to be any obvious pattern with respect to the chromosomal location of the polymorphism, and this suggests that the generation and accumulation of the SNPs in any given KRTAP may be, at least in part, independent of other KRTAPs.
Figure 3.
Density of SNPs in selected ovine KRTAPs. The density is expressed as the number of SNPs per kilobase. The order of KRTAPs on the x-axis represents their relative location on the chromosomes 1, 11, and 21. The SNPs are divided into non-synonymous SNPs (in blue) and others (including synonymous SNPs in the coding region and SNPs in the flanking regions).
The ratio of non-synonymous SNPs to synonymous SNPs does not have any obvious pattern, but the KRTAPs that are located close together on the chromosomes appear to have a similar ratio. For example, ovine KRTAP36-1, KRTAP15-1, and KRTAP13-3 are located close together on ovine chromosome 1, and they all have a high proportion of non-synonymous SNPs. The same is true for ovine KRTAP28-1 and KRTAP24-1 (Figure 3). A low proportion of non-synonymous SNPs is observed for ovine KRTAP6-2, KRTAP6-4, KRTAP6-1, and KRTAP22-1, which are clustered in proximity to each other on ovine chromosome 1, and also for ovine KRTAP1-2 and KRTAP1-3 on chromosome 11. Further investigation of variation in other KRTAPs as they are found and characterized may provide more information about this effect. It also is notable that all of the non-synonymous SNPs revealed to date in the ovine KRTAPs are missense, with the exception of a single nonsense SNP in ovine KRTAP20-2 [41].
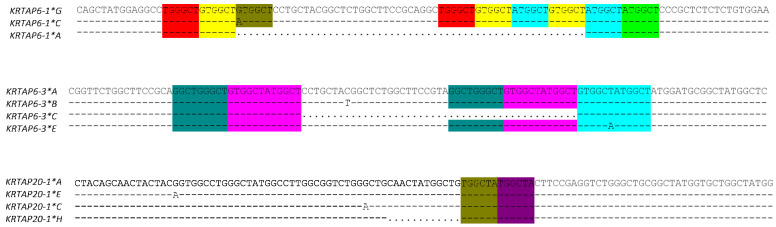
Beside the presence of SNPs, the KRTAPs also contain indels. For sheep, this has been described for numerous KRTAPs, including KRTAP1-1, KRTAP5-4, KRTAP6-1, KRTAP6-5, KRTAP20-1, and KRTAP28-1 [19,28,29,31,40,47], and KRTAP9-2 and KRTAP28-1 in goats [56,66]. For ovine KRTAP1-1, KRTAP5-4, and KRTAP6-5, and for caprine KRTAP9-2, the indels occur within tandem repeat regions of the coding sequence, and they lead to variation in the number of tandem repeats that are present. In ovine and caprine KRTAP28-1, the indels are located within dinucleotide repeats (microsatellites), while the indels in ovine KRTAP6-1, KRTAP6-3, and KRTAP20-1 are not located in the tandem repeat region, but instead occur in regions that are flanked by sequence repeats (Figure 4). All of the indels identified in the KRTAPs are in multiples of three nucleotides in size, and hence they preserve the reading frame. The exception is the dinucleotide repeats found in ovine and caprine KRTAP28-1.
Figure 4.
The indels identified in ovine KRTAP6-1, KRTAP6-3, and KRTAP20-1. The sequence repeats flanking the indels are shaded in different colours. In the region that is shown, variants B, D, E, and F of ovine KRTAP6-1 have a sequence identical to ovine KRTAP6-1 variant A; variants D and F of ovine KRTAP6-3 have a sequence identical to the ovine KRTAP6-3 variant A; and variants B, D, F, and G of ovine KRTAP20-1 have a sequence identical to ovine KRTAP20-1 variant A; hence, only one variant from the identical sequences is shown.
4. Mechanisms for the Generation of KRTAP Variation
In the KRTAPs, transition SNPs predominate, and they account for over 70% of all SNPs. Among these transition SNPs, the G/C to A/T transition (52.4% of occurrences) happens at nearly three times the frequency of the A/T to G/C transition (18.1%). This effect is still pronounced when the ratio of A/T (approximately 46%) to G/C (approximately 54%) base pairs (1:1.19) in ovine KRTAPs is taken into account. Such a transitional bias should create a pressure towards ovine KRTAPs having a higher AT content, but some other pressure must operate in the other direction to maintain GC ratio.
For over twenty years, there has been a strong belief that biased gene conversion (BGC) is important for shaping the GC content in the genomes of mammals and other eukaryotes [69]. The BGC theory is based on DNA repair processes inside heteroduplexes, the double-stranded DNA segments that form during meiosis at crossover and non-crossover recombination sites. The theory has it that one DNA strand of a heteroduplex has a maternal origin, while the complementary strand is paternal. The heterozygous sites that occur in heteroduplexes create mismatches, and these mismatches are non-randomly resolved in favour of G/C over A/T nucleotides, which leads to an increase of GC content in the sequence [69,70]. The polymorphic nature of the KRTAPs could potentially increase the chance of forming heteroduplexes, and hence elevate BGC. The effect of BGC, if strong enough, can overcome purifying selection and lead to an increased ratio of non-synonymous SNPs to synonymous SNPs [70], as is found for some of KRTAPs.
Indels are in abundance in eukaryotic genomes, and in humans they are the second most abundant form of genetic variation, after SNPs [71]. Mechanisms have been proposed to explain the generation of indels, including replication slippage (also known as slipped-strand mispairing) [72], unequal crossing-over (also known as non-reciprocal recombination) [73], and transposition (also known as translocation) [74]. It is thought that replication slippage is the principal mechanism responsible for the majority of small indels in the human genome [75].
Replication slippage is a mutation process that occurs during DNA replication and also during the DNA synthesis step of DNA repair processes. It requires DNA polymerase pausing to occur within a short direct repeat. The paused polymerase dissociates from the DNA, and then the terminal portion of the newly synthesized strand separates from the template and anneals to another direct repeat, after which replication resumes [76]. A slippage event normally occurs when a sequence of repetitive nucleotides (e.g., tandem repeats) are found at the site of replication, and strand misalignment at repeated sequences leads to genetic rearrangements, resulting in the insertion or deletion of nucleotides [77]. Slipped-strand mispairing can also occur with non-continuous repeat sequences, and results in longer insertions or deletions of intervening sequences flanked by the repeats [78].
Many KRTAPs are characterized by having an abundance of nucleotide repeats [4,16,19,28,31]. The KRTAPs are also GC-rich, with an average GC content of over 54% in all of the ovine KRTAPs identified to date. It has been reported that a high GC content results in reduced DNA polymerase processivity and increased DNA polymerase slippage, and consequently it can lead to elevated rates of mutation, including the creation of indels and nucleotide substitutions [79].
There is an association between the occurrence of indels and nucleotide substitutions, and this association appears to be universal to all prokaryotic and eukaryotic genomes examined so far [80,81,82]. McDonald et al. [82] propose that it is not the indels per se, but instead the sequence in which the indels occur that causes the accumulation of nucleotide substitutions. Repeat sequences can promote an increased probability of replication fork arrest and are prone to causing the stalling of high-fidelity DNA polymerase. This can lead to the recruitment of error-prone (low-fidelity) DNA polymerases to replicate the surrounding sequence with a higher-than-average error rate [82].
Sequence analyses of two of the most variable KRTAPs in sheep (KRTAP1-2 and KRTAP1-4), reveal that short segments of DNA exchange may have occurred and contributed to the generation of the different nucleotide sequences (Figure 5). This suggests gene conversion or non-reciprocal genetic exchange is one of the mechanisms for creating sequence diversity in some KRTAPs. Unique sequence motifs have been postulated to promote genetic recombination, including the crossover hotspot instigator (Chi) sequence (5′-GCTGGTGG-3′) or its complementary sequence, which are abundant in the genomes of bacteriophage and Escherichia coli [83]. Chi and Chi-like sequences, or their complementary sequences, have been reported in KRTAPs [19,61,64,65]. It is proposed that Chi-like sequences, with minor nucleotide variations to the consensus Escherichia coli Chi sequence, may have partial ‘hotspot’ activity [84]. The presence of Chi and Chi-like, or their complementary sequences in KRTAPs suggests recombination may play a role in creating sequence diversity.
Figure 5.
Sequence comparisons revealing potential DNA exchanges between variants of ovine KRTAP1-2 (A) and KRTAP1-4 (B). Nucleotides identical to the top sequences are presented as dashes, and the numbering of the nucleotide positions follows the Human Genome Variation Society (HGVS) nomenclature (http://varnomen.hgvs.org/; accessed on 21 June 2021).
5. The Chromosomal Clustering of KRTAPs and Evolution
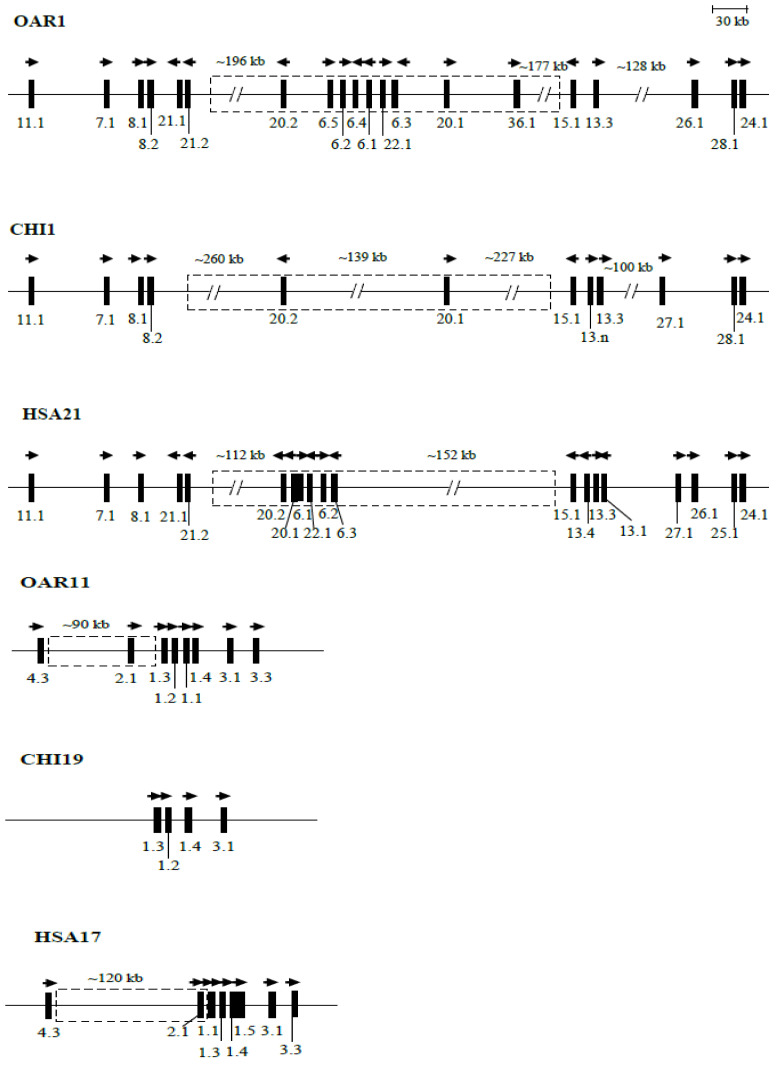
The KRTAPs are clustered and located in several chromosomal regions. In humans, where a full complement of KRTAPs has possibly been identified, there are five clusters of genes located on three chromosomes [10]. Based on the number of KAP genes in each cluster, ranging from the largest to the smallest, these are: cluster 1, containing 15 KAP gene families (KAP6 to KAP8, KAP11, KAP13, KAP15, and KAP19 to KAP27) located on chromosome 21q22.1; cluster 2, containing seven KAP gene families (KAP1 to KAP4, KAP9, KAP16, and KAP17) located on chromosome 17q21.2; cluster 3, containing two KAP gene families (KAP10 and KAP12) located on chromosome 21q22.3; cluster 4, containing six KAP5 genes (KRTAP5-1 to KRTAP5-6) located on chromosome 11p15.5; and cluster 5, which contains the other six KAP5 genes (KRTAP5-7 to KRTAP5-12) located on chromosome 11q13.4 [10,11].
In sheep and goats, all of the KRTAPs identified to date are from cluster 1 and cluster 2, except ovine KRTAP5-4, a cluster 4 gene on ovine chromosome 21. In these two species, the cluster 1 KRTAPs are located on chromosome 1, but the cluster 2 KRTAPs are located on chromosome 11 in sheep, and on chromosome 19 in goats (Figure 6).
Figure 6.
Chromosomal locations of the KRTAPs identified in sheep and goats together with their human orthologues. A vertical bar represents each KRTAP and the arrowheads above the bars indicate the direction of transcription. The numbers below the bars indicate the name of the KRTAPs (i.e., 11.1 represents KRTAP11-1). The distances between the KRTAPs are only approximate. The dashed-line boxes represent the chromosome regions that appear to be markedly different between sheep/goats and humans. Note that human KRTAP1-4, KRTAP1-3, KRTAP1-1, and KRTAP1-5 are the orthologues of ovine KRTAP1-1, KRTAP1-2, KRTAP1-3, and KRTAP1-4, respectively.
Within clusters, the KRTAPs are unevenly distributed on the chromosome [10,16]. Despite having different transcriptional directions for the KRTAPs found in each cluster, the genes that are near to each other tend to have the same direction of transcription (Figure 6 and [5]). This may suggest that some genes are under a shared form of transcriptional control.
The identification of numerous KRTAPs from cluster 1 and cluster 2 in sheep and goats enables a comparison of these two ruminant KRTAP clusters and the matching human cluster. From this, it can be observed that the overall configuration of KRTAPs is similar across these species, but that sheep and goats are more similar to each other (as might be expected), and have some differences to humans. In cluster 1, a major difference is observed in the region between KRTAP21-2 and KRTAP15-1. In humans, this region is estimated to be 306 kb in size, and it contains four KAP gene families (two KRTAP20s, three KRTAP6s, one KRTAP22, and seven KRTAP19s; [15]). Genes of the KAP19 family have not been identified in sheep and goats, but for the other KRTAPs that have been identified, their locations are quite different to those described in humans (Figure 6). This region is estimated to be 560 kb in length in sheep and contains two additional KRTAP6s and one new KAP gene named KRTAP36-1 [6,31].
An obvious difference in cluster 2 is located between KRTAP4-3 and KRTAP1-3 (Figure 6). This region is approximately 120 kb in length in humans, but it is estimated to be 90 kb in size in sheep. At a finer level in humans, KRTAP2-1 is approximately 5 kb away from human KRTAP1-1, which corresponds to ovine KRTAP1-3, but the distance between KRTAP2-1 and KRTAP1-3 is approximately 28 kb in sheep. This region contains numerous human KRTAP4s and KRTAP2s [85], but only one KRTAP4 and one KRTAP2 have been identified in this region in sheep. The identification of more ovine and caprine KRTAPs in this region would assist in providing a better understanding of the similarities and differences between the regions in the different species.
The clustering of genes that produce proteins that are involved in key metabolic pathways has been accepted for many eukaryotes, but the evolutionary causes or benefits of clustering remain controversial [86]. One hypothesis put forward to explain the clustering of genes involved in metabolic pathways is that they have ‘arrived’ in genomes as a group, following horizontal gene transfers from bacteria [87].
Given the intron-less character of the KRTAPs, the possibility of individual genes or groups of KRTAPs having prokaryotic origins cannot be excluded. Research in humans suggests that the cluster-3 KRTAPs are located within introns of the thrombospondin-type laminin G domain and EAR repeat-containing protein gene (TSPEAR) on chromosome 21 [88]. This rather strange location could suggest that this KRTAP cluster has been inserted into this position, and then subsequently, gene duplication and divergence may have enlarged the cluster.
Gene duplication can arise via several mechanisms, with the major mechanisms including unequal crossing-over, retroposition and chromosomal duplication events (large-scale duplications) [89]. Large-scale duplications, as a consequence of polyploidy, are reported to occur frequently in plants, but are much less frequent in animals [89]. However, duplications of large genomic segments (segmental duplications) are abundant in animals such as primates [90] and rodents [91]. Unequal crossing-over, along with gene conversion, is believed to be the main driver for the generation of gene duplications, but the possibility of retroposition should not be ignored.
Retroposition is an RNA-mediated process that occurs when a message RNA is retro-transcribed to complementary DNA (cDNA), and the resulting cDNA is inserted back into the genome. Retrogenes are therefore expected to lack introns and regulatory sequences (which sits well with the nature of the KRTAPs), but instead contain poly A tracts and flanking short direct repeats [89]. Further bioinformatics analyses of the KRTAPs and their flanking sequences may shed more light on the evolution of the KRTAP clusters.
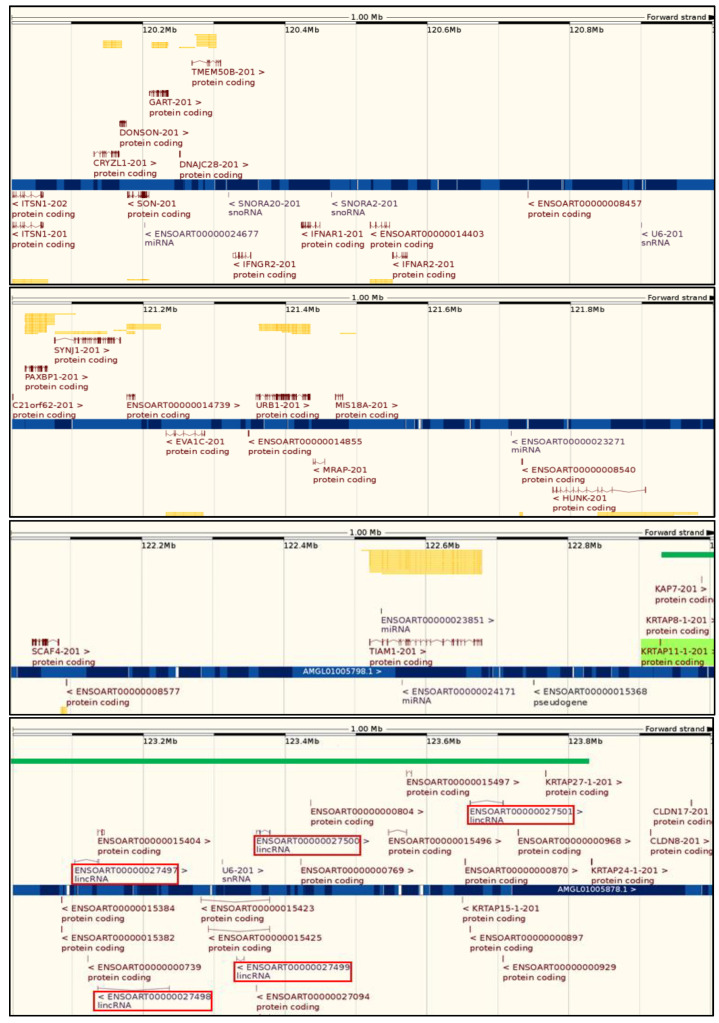
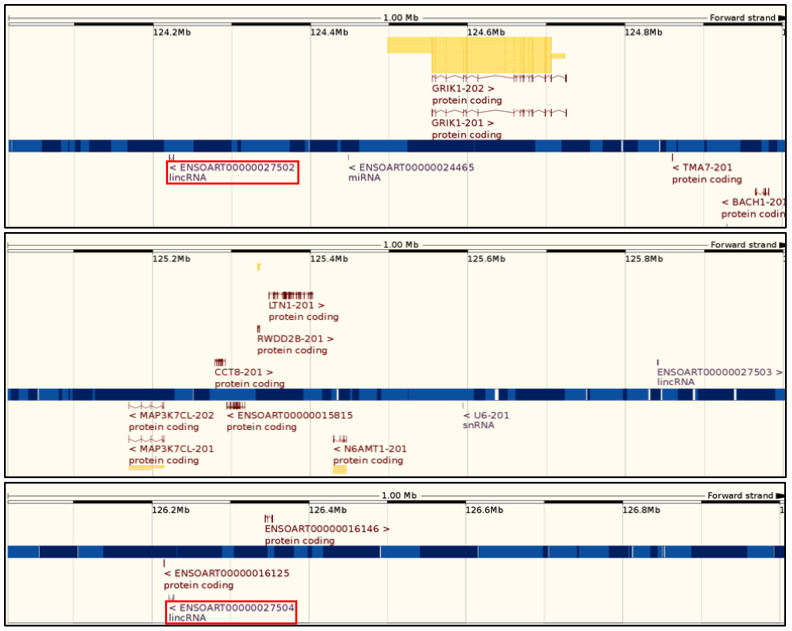
A preliminary sequence analysis of the sheep chromosome 1 region that contains KRTAPs reveals five long intergenic non-coding RNA (lincRNA) genes within the cluster region (spanning approximately 0.9 Mb). However, there is no lincRNA gene found in the approximately 2.9 Mb upstream region, and only two lincRNA genes are found in the approximately 3.2 Mb downstream region (Figure 7). The exact functions of lincRNAs are not well known, but it is proposed that they broadly serve to fine-tune the expression of neighbouring genes with tissue specificity, and with a diversity of mechanisms [92]. Analysis of human lincRNAs reveals one feature, a high prevalence of transposable elements (TEs) [93], or repetitive mobile genetic sequences that are capable of duplicating genes or gene fragments [94]. Whether these lincRNAs play a role in the evolution of KRTAPs awaits further investigation.
Figure 7.
Sequence analysis of the sheep chromosome 1 region reveals the presence of more long intergenic non-coding RNA (lincRNA) genes within the KRTAP cluster region than in its flanking regions. The KRTAP cluster, spanning from KRTAP11-1 to KRTAP24-1, is indicated by the green bar. The lincRNA genes, identified based on the sheep assembly sequence Oar_rambouillet_v1.0 (GCA_002742125.1) using Ensembl (http://asia.ensembl.org/; accessed on 21 June 2021), are marked in red boxes.
6. The Effect of KRTAP Variation
The proteins encoded by KRTAPs serve as a matrix embedding the KIFs. They form one of the main components of the wool/hair fibre, and thus it is thought that variation in KRTAPs may affect fibre properties, possibly in three ways.
Firstly, non-synonymous SNPs and insertions/deletions in the coding region will alter the protein sequence. This may affect the structure and/or properties of the protein, and consequently its interaction with KIFs and/or other KAP proteins, which may then affect fibre properties. As an example, a nonsense mutation in ovine KRTAP20-2 has been shown to be associated with variation in the curvature of wool fibres [41], and a 57-bp insertion/deletion in the coding region of ovine KRTAP6-1 is associated with variation in the fibre diameter traits [29]. The mean fibre diameter (MFD) of wool is a key determinant of value, with finer wools of a mean diameter of 19 microns or less being considerably more valuable than strong wools of a mean diameter over 36 microns.
Secondly, synonymous SNPs and SNPs in the upstream and downstream of the coding region may affect gene expression and consequently alter the amount of that protein in the wool/hair fibres. Despite synonymous SNPs not causing amino acid changes in the protein, research has shown that synonymous SNPs can affect the stability and structure of mRNA, and also the folding of protein [95]. In felting lustre mutant wool follicles, Li et al. [96] reported that the expression of KRTAP2-12 and KRTAP4-2 was un-regulated, whereas the expression of KRTAP6-1, KRTAP7, and KRTAP8 was down-regulated. In wool from sheep on a restricted diet, KAP13-1 and KAP6-n protein levels were increased, and this was found to be associated with a decrease in the fibre diameter [97].
Lastly, variation in KRTAPs may also affect the post-translational modification of the protein. A recent proteomic study revealed a differential abundance of some phosphorylated KAPs and keratins, when comparing the crimped and straight wool of Tan sheep [96], and provided evidence that phosphorylation of KAPs and keratins can occur. Bioinformatics analyses of ovine KRTAP11-1 and KRTAP13-3 reveals some non-synonymous SNPs that would alter putative phosphorylation sites in proteins derived from these genes [37,38], and the phosphorylation of the KAPs may alter the structural conformation and interactions with KIFs and/or other KAPs, and consequently affect the properties of the wool fibres [98].
7. Concluding Remarks and Future Research Directions
To date, 30 KRTAPs from 18 different families have been identified in sheep and 18 KRTAPs from 12 families have been reported in goats. Most of these genes are present in humans, but some are absent. This suggests that sheep and goats may possess more KRTAPs than humans. The ovine and caprine KRTAPs are unevenly clustered on chromosomes and translated in alternating directions. The configuration of the KRTAPs in the sheep and goat genomes are similar to the configuration reported in humans, but differences occur too. All of the sheep and goat KRTAPs are polymorphic, but the extent and nature of polymorphism varies between the genes.
Our current understanding of KRTAPs is based primarily on their chromosomal location and sequence, but little is known about how the genes have evolved and the mechanisms that underlie the generation of variation in the genes. Further investigation into their sequences, especially of their flanking regions, may shed more light on their evolutionary origin and how natural selection may have created or enhanced their diversity. It also must not be forgotten that for many hundreds of years humans have been selecting and breeding sheep and goats for fibres, meat, and milk traits; hence, there may evolutionary dead-ends or rare sequences that will be hard, if not impossible, to place in that evolutionary history.
Ongoing investigations are also needed in other important areas. First is the ongoing need to identify and characterize new KRTAPs that are likely present in the sheep and goat genomes. This should, in time, lead to the definition of a full catalogue of KRTAPs in these species, and the complete annotation of the genes in the reference genomes. This will doubtlessly be enhanced by the use of high-throughput and rapid genome-sequencing techniques, whereby hundreds or thousands of sheep can be rapidly sequenced, subjected to bioinformatics analysis, and publicly recorded and indexed.
Second is the pressing need to better understand the temporal and spatial nature of KRTAP expression. Questions about how, when, and why the genes are expressed will need to be addressed, especially if the fibres from sheep and goats are to be improved and better fitted to purpose. Given the potential number of KRTAPs that exist, the number of variants for each KRTAP, and the diploid nature of the sheep and goat genomes, this will be an immense challenge. If all of the KRTAPs are expressed, then there will, by definition, be a large diversity of KAP proteins, and more importantly different permutations of those proteins and the KIFs in the matrix of the fibres. When fully understood, this may enable a greater fibre uniformity to be selected for breeding sheep and goats, and enable us to address one of the bigger constraints on fibre use: its natural variability. This will not be a small task, because while quantitative analytical techniques can be used to investigate whether variation in KRTAPs affects gene expression or post-translational modifications of individual KRTAPs/KAPs, it is still difficult to unravel the effect of individual KRTAPs because of the potentially large numbers of KAP and keratin proteins in fibres. Given the adage, backed by evidence, that ‘there is more variation within any given fleece than between fleeces in any given flock’, the size of this task should not be underestimated. Current studies describing associations between variation in individual KRTAPs and variation in fibre traits is a start, but much more research and new multiplex analytical approaches will be needed.
Author Contributions
Conceptualization: H.Z., J.W., Y.L. and J.G.H.H.; formal analysis: H.Z., J.W. and J.T., investigation, H.Z., H.G. and S.L., Writing—Original draft preparation: H.Z., H.G. and J.G.H.H.; Writing—Review and editing: H.Z., Y.L. and J.G.H.H.; funding acquisition: S.L., J.W., J.T. and J.G.H.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32060140), the Distinguished Young Scholars Fund of Gansu Province (21JR7RA857) and Basic Research Creative Groups of Gansu Province (18JR3RA190). This publication is partially financed by the Lincoln University Open Access Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Powell B.C., Rogers G.E. Formation and Structure of Human Hair. Birkhäuser; Basel, Switzerland: 1997. The Role of Keratin Proteins and Their Genes in the Growth, Structure and Properties of Hair; pp. 59–148. [DOI] [PubMed] [Google Scholar]
- 2.Fujikawa H., Fujimoto A., Farooq M., Ito M., Shimomura Y. Characterisation of the human hair keratin-associated protein 2 (KRTAP2) gene family. J. Invest. Dermatol. 2012;132:1806–1813. doi: 10.1038/jid.2012.73. [DOI] [PubMed] [Google Scholar]
- 3.Plowman J.E., Miller R.E., Thomas A., Grosvenor A.J., Harland D.P., Deb-Choudhury S. A detailed mapping of the readily accessible disulphide bonds in the cortex of wool fibres. Proteins. 2021;89:708–720. doi: 10.1002/prot.26053. [DOI] [PubMed] [Google Scholar]
- 4.Haylett T., Swart L. Studies on the high-sulfur proteins of reduced merino wool part III: The amino-acid sequence of protein SCMKB-IIIB2. Text. Res. J. 1968;39:917–929. doi: 10.1177/004051756903901004. [DOI] [Google Scholar]
- 5.Rogers M.A., Langbein L., Praetzel-Wunder S., Winter H., Schweizer J. Human hair keratin-associated proteins (KAPs) Int. Rev. Cytol. 2006;251:209–263. doi: 10.1016/S0074-7696(06)51006-X. [DOI] [PubMed] [Google Scholar]
- 6.Gong H., Zhou H., Wang J., Li S., Luo Y., Hickford J.G. Characterisation of an ovine keratin associated protein (KAP) gene, which would produce a protein rich in glycine and tyrosine, but lacking in cysteine. Genes. 2019;10:848. doi: 10.3390/genes10110848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGaughey G.B., Gagné M., Rappé A.K. π-stacking interactions alive and well in proteins. J. Biol. Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 8.Levitt M., Perutz M.F. Aromatic rings act as hydrogen bond acceptors. J. Mol. Biol. 1988;201:751–754. doi: 10.1016/0022-2836(88)90471-8. [DOI] [PubMed] [Google Scholar]
- 9.Fraser R.B., Parry D.A. Filamentous structure of hard β-keratins in the epidermal appendages of birds and reptiles. Subcell. Biochem. 2017;82:231–252. doi: 10.1007/978-3-319-49674-0_8. [DOI] [PubMed] [Google Scholar]
- 10.Rogers M.A., Schweizer J. Human KAP genes, only the half of it? Extensive size polymorphisms in hair keratin-associated protein genes. J. Invest. Dermatol. 2005;124:8–9. doi: 10.1111/j.0022-202X.2005.23728.x. [DOI] [PubMed] [Google Scholar]
- 11.Rogers M.A., Winter H., Langbein L., Wollschläger A., Praetzel-Wunder S., Jave-Suarez L.F., Schweizer J. Characterization of human KAP24.1, a cuticular hair keratin-associated protein with unusual amino-acid composition and repeat structure. J. Invest. Dermatol. 2007;127:1197–1204. doi: 10.1038/sj.jid.5700702. [DOI] [PubMed] [Google Scholar]
- 12.Rogers M.A., Langbein L., Praetzel Wunder S., Giehl K. Characterization and expression analysis of the hair keratin associated protein KAP26.1. Br. J. Dermatol. 2008;159:725–729. doi: 10.1111/j.1365-2133.2008.08743.x. [DOI] [PubMed] [Google Scholar]
- 13.Aoki N., Ito K., Ito M. Hair follicle has a novel anagen-specific protein, mKAP13. J. Invest. Dermatol. 1998;111:804–809. doi: 10.1046/j.1523-1747.1998.00387.x. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn F., Lassing C., Range A., Mueller M., Hunziker T., Ziemiecki A., Andres A.C. Pmg-1 and pmg-2 constitute a novel family of KAP genes differentially expressed during skin and mammary gland development. Mech. Dev. 1999;86:193–196. doi: 10.1016/S0925-4773(99)00115-X. [DOI] [PubMed] [Google Scholar]
- 15.Rogers M.A., Langbein L., Winter H., Ehmann C., Praetzel S., Schweizer J. Characterization of a first domain of human high glycine-tyrosine and high sulfur keratin-associated protein (KAP) genes on chromosome 21q22.1. J. Biol. Chem. 2002;277:48993–49002. doi: 10.1074/jbc.M206422200. [DOI] [PubMed] [Google Scholar]
- 16.Gong H., Zhou H., Forrest R.H., Li S., Wang J., Dyer J.M., Luo Y., Hickford J.G. Wool keratin-associated protein genes in sheep—a review. Genes. 2016;7:24. doi: 10.3390/genes7060024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao M., Wang X., Chen H., Lan X.Y., Guo Y.K., Li J.Y., Wei T.B., Jing Y.J., Liu S.Q., Zhang M.H., et al. The PCR-SSCP and DNA sequencing methods detecting a large deletion mutation at KAP6.2 locus in the cashmere goat. Small Rumin. Res. 2008;75:243–246. doi: 10.1016/j.smallrumres.2007.10.007. [DOI] [Google Scholar]
- 18.Yin J., Hu T.M., Li J.Q., Zhang C.l., Guo Z.Z., Zhou H.M. Construction of a skin cDNA library of cashmere goat and cloning of KAP6-2 full-length cDNA. Zool. Res. 2004;25:166–171. [Google Scholar]
- 19.Rogers G.R., Hickford J.G.H., Bickerstaffe R. Polymorphism in two genes for B2 high sulfur proteins of wool. Anim. Genet. 1994;25:407–415. doi: 10.1111/j.1365-2052.1994.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 20.Gong H., Zhou H., Yu Z., Dyer J., Plowman J.E., Hickford J. Identification of the ovine keratin-associated protein KAP1-2 gene (KRTAP1-2) Exp. Dermatol. 2011;20:815–819. doi: 10.1111/j.1600-0625.2011.01333.x. [DOI] [PubMed] [Google Scholar]
- 21.Gong H., Zhou H., Hodge S., Dyer J.M., Hickford J.G. Association of wool traits with variation in the ovine KAP1-2 gene in Merino cross lambs. Small Rumin. Res. 2015;124:24–29. doi: 10.1016/j.smallrumres.2015.01.009. [DOI] [Google Scholar]
- 22.Itenge-Mweza T.O., Forrest R.H., McKenzie G.W., Hogan A., Abbott J., Amoafo O., Hickford J.G. Polymorphism of the KAP1.1, KAP1.3 and K33 genes in Merino sheep. Mol. Cell. Probes. 2007;21:338–342. doi: 10.1016/j.mcp.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Gong H., Zhou H., Hickford J.G. Polymorphism of the ovine keratin-associated protein 1-4 gene (KRTAP1-4) Mol. Biol. Rep. 2010;37:3377–3380. doi: 10.1007/s11033-009-9925-4. [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Zhou H., Hickford J.G., Luo Y., Gong H., Hu J., Liu X., Li S., Song Y., Ke N. Identification of the ovine keratin-associated protein 2-1 gene and its sequence variation in four Chinese sheep breeds. Genes. 2020;11:604. doi: 10.3390/genes11060604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frenkel M.J., Powell B.C., Ward K.A., Sleigh M.J., Rogers G.E. The keratin BIIIB gene family: Isolation of cDNA clones and structure of a gene and a related pseudogene. Genomics. 1989;4:182–191. doi: 10.1016/0888-7543(89)90298-X. [DOI] [PubMed] [Google Scholar]
- 26.Yu Z., Gordon S.W., Nixon A.J., Bawden C.S., Rogers M.A., Wildermoth J.E., Maqbool N.J., Pearson A.J. Expression patterns of keratin intermediate filament and keratin associated protein genes in wool follicles. Differentiation. 2009;77:307–316. doi: 10.1016/j.diff.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 27.MacKinnon P., Powell B., Rogers G. Structure and expression of genes for a class of cysteine-rich proteins of the cuticle layers of differentiating wool and hair follicles. J. Cell Biol. 1990;111:2587–2600. doi: 10.1083/jcb.111.6.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong H., Zhou H., Plowman J.E., Dyer J.M., Hickford J.G. Analysis of variation in the ovine ultra-high sulphur keratin-associated protein KAP5-4 gene using PCR-SSCP technique. Electrophoresis. 2010;31:3545–3547. doi: 10.1002/elps.201000301. [DOI] [PubMed] [Google Scholar]
- 29.Zhou H., Gong H., Li S., Luo Y., Hickford J.G.H. A 57-bp deletion in the ovine KAP6-1 gene affects wool fibre diameter. J. Anim. Breed Genet. 2015;132:301–307. doi: 10.1111/jbg.12138. [DOI] [PubMed] [Google Scholar]
- 30.Tao J., Zhou H., Gong H., Yang Z., Ma Q., Cheng L., Ding W., Li Y., Hickford J.G. Variation in the KAP6-1 gene in Chinese Tan sheep and associations with variation in wool traits. Small Rumin. Res. 2017;154:129–132. doi: 10.1016/j.smallrumres.2017.08.001. [DOI] [Google Scholar]
- 31.Zhou H., Gong H., Wang J., Dyer J.M., Luo Y., Hickford J.G.H. Identification of four new gene members of the KAP6 gene family in sheep. Sci. Rep. 2016;6:24074. doi: 10.1038/srep24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong H., Zhou H., Hickford J.G.H. Diversity of the glycine/tyrosine-rich keratin-associated protein 6 gene (KAP6) family in sheep. Mol. Biol. Rep. 2011;38:31–35. doi: 10.1007/s11033-010-0074-6. [DOI] [PubMed] [Google Scholar]
- 33.Li S., Zhou H., Gong H., Zhao F., Wang J., Luo Y., Hickford J.G. Variation in the ovine KAP6-3 gene (KRTAP6-3) is associated with variation in mean fibre diameter-associated wool traits. Genes. 2017;8:204. doi: 10.3390/genes8080204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong H., Zhou H., Plowman J.E., Dyer J.M., Hickford J.G.H. Search for variation in the ovine KAP7-1 and KAP8-1 genes using polymerase chain reaction–single-stranded conformational polymorphism screening. DNA Cell Biol. 2012;31:367–370. doi: 10.1089/dna.2011.1346. [DOI] [PubMed] [Google Scholar]
- 35.Gong H., Zhou H., Dyer J.M., Hickford J.G. The sheep KAP8-2 gene, a new KAP8 family member that is absent in humans. SpringerPlus. 2014;3:1–5. doi: 10.1186/2193-1801-3-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullah F., Jamal S., Zhou H., Hickford J.G.H. Variation in ovine KRTAP8-2 and its association with wool characteristics in Pakistani sheep. Small Rumin. Res. 2021 (Under review) [Google Scholar]
- 37.Gong H., Zhou H., Dyer J.M., Hickford J.G. Identification of the ovine KAP11-1 gene (KRTAP11-1) and genetic variation in its coding sequence. Mol. Biol. Rep. 2011;38:5429–5433. doi: 10.1007/s11033-011-0697-2. [DOI] [PubMed] [Google Scholar]
- 38.Gong H., Zhou H., Dyer J.M., Plowman J.E., Hickford J.G.H. Identification of the keratin-associated protein 13-3 (KAP13-3) gene in sheep. Open J. Genet. 2011;1:60–64. doi: 10.4236/ojgen.2011.13011. [DOI] [Google Scholar]
- 39.Li W., Gong H., Zhou H., Wang J., Liu X., Li S., Luo Y., Hickford J.G.H. Variation in the ovine keratin-associated protein 15-1 gene affects wool yield. J. Agric. Sci. 2018;156:922–928. doi: 10.1017/S0021859618000953. [DOI] [Google Scholar]
- 40.Gong H., Zhou H., Bai L., Li W., Li S., Wang J., Luo Y., Hickford J.G.H. Associations between variation in the ovine high glycine-tyrosine keratin-associated protein gene KRTAP20-1 and wool traits. J. Anim. Sci. 2019;97:587–595. doi: 10.1093/jas/sky465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai L., Gong H., Zhou H., Tao J., Hickford J.G.H. A nucleotide substitution in the ovine KAP20-2 gene leads to a premature stop codon that affects wool fibre curvature. Anim. Genet. 2018;49:357–358. doi: 10.1111/age.12668. [DOI] [PubMed] [Google Scholar]
- 42.Li S., Zhou H., Gong H., Zhao F., Wang J., Liu X., Hu J., Luo Y., Hickford J.G.H. Identification of the ovine keratin-associated protein 21-1 gene and its association with variation in wool traits. Animals. 2019;9:450. doi: 10.3390/ani9070450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S., Zhou H., Gong H., Zhao F., Wang J., Liu X., Hu J., Luo Y., Hickford J.G.H. The mean staple length of wool fibre is associated with variation in the ovine keratin-associated protein 21-2 gene. Genes. 2020;11:148. doi: 10.3390/genes11020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S., Zhou H., Gong H., Zhao F., Wang J., Liu X., Luo Y., Hickford J.G.H. Identification of the ovine keratin-associated protein 22-1 (KAP22-1) gene and its effect on wool traits. Genes. 2017;8:27. doi: 10.3390/genes8010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou H., Gong H., Yan W., Luo Y., Hickford J.G.H. Identification and sequence analysis of the keratin-associated protein 24-1 (KAP24-1) gene homologue in sheep. Gene. 2012;511:62–65. doi: 10.1016/j.gene.2012.08.049. [DOI] [PubMed] [Google Scholar]
- 46.Li S., Zhou H., Gong H., Zhao F., Hu J., Luo Y., Hickford J.G.H. Identification of the ovine keratin-associated protein 26-1 gene and its association with variation in wool traits. Genes. 2017;8:225. doi: 10.3390/genes8090225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bai L., Wang J., Zhou H., Gong H., Tao J., Hickford J.G.H. Identification of ovine KRTAP28-1 and its association with wool fibre diameter. Animals. 2019;9:142. doi: 10.3390/ani9040142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrews M., Visser C., van Marle-Köster E. Identification of novel variants for KAP1.1, KAP8.1 and KAP13.3 in South African goats. Small Rumin. Res. 2017;149:176–180. doi: 10.1016/j.smallrumres.2017.02.014. [DOI] [Google Scholar]
- 49.Zhao M., Zhou H., Luo Y., Wang J., Hu J., Liu X., Li S., Zhang K., Zhen H., Hickford J.G.H. Variation in a newly identified caprine KRTAP gene is associated with raw cashmere fiber weight in Longdong cashmere goats. Genes. 2021;12:625. doi: 10.3390/genes12050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah R., Ganai T., Shanaz S., Ayaz A., Khan N. Allelic polymorphism of KAP1.3 gene in goats. Indian J. Small Rumin. 2017;23:257–260. doi: 10.5958/0973-9718.2017.00062.9. [DOI] [Google Scholar]
- 51.Shah R., Ganai T., Sheikh F., Shanaz S., Shabir M., Khan H. Characterization and polymorphism of keratin associated protein 1.4 gene in goats. Gene. 2013;518:431–442. doi: 10.1016/j.gene.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 52.Parris D., Swart L.S. Studies on the high-sulphur proteins of reduced mohair. The isolation and amino acid sequence of protein scmkb-m1.2. Biochem. J. 1975;145:459–467. doi: 10.1042/bj1450459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ayaz A., Singh N., Ganai N.A. Comparative sequence analysis of keratin associated protein (KAP7.1) gene in two indigenous Pashmina goat breeds of India. Int. J. Curr. Microbiol. App. Sci. 2017;6:3314–3318. [Google Scholar]
- 54.Liu H., Yue C.W., Zhang W., Zhu X., Yang G., Jia Z. Association of the KAP8.1 gene polymorphisms with fibre traits in inner mongolian cashmere goats. Asian-Australas. J. Anim. Sci. 2011;24:1341–1347. doi: 10.5713/ajas.2011.11120. [DOI] [Google Scholar]
- 55.Liu H., Li N., Jia C., Zhu X., Jia Z. Effect of the polymorphisms of keratin associated protein 8.2 gene on fibre traits in Inner Mongolia cashmere goats. Asian-Australas. J. Anim. Sci. 2007;20:821–826. doi: 10.5713/ajas.2007.821. [DOI] [Google Scholar]
- 56.Yu H., Wang X., Chen H., Wang M., Zhao M., Lan X.Y., Lei C.Z., Wang K.Y., Lai X.S., Wang X.L. The polymorphism of a novel 30 bp-deletion mutation at KAP9.2 locus in the cashmere goat. Small Rumin. Res. 2008;80:111–115. doi: 10.1016/j.smallrumres.2008.09.010. [DOI] [Google Scholar]
- 57.Wang X., Zhao Z., Xu H., Qu L., Zhao H., Li T., Zhang Z. Variation and expression of KAP9.2 gene affecting cashmere trait in goats. Mol. Biol. Rep. 2012;39:10525–10529. doi: 10.1007/s11033-012-1937-9. [DOI] [PubMed] [Google Scholar]
- 58.Jin M., Cao Q., Wang R., Piao J., Zhao F., Piao J. Molecular characterization and expression pattern of a novel Keratin-associated protein 11.1 gene in the Liaoning cashmere goat (Capra hircus) Asian-Australas. J. Anim. Sci. 2017;30:328–337. doi: 10.5713/ajas.16.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li M., Liu X., Wang J., Li S., Luo Y. Molecular characterization of caprine KRTAP13-3 in Liaoning cashmere goat in China. J. Appl. Anim. Res. 2014;42:140–144. doi: 10.1080/09712119.2013.822813. [DOI] [Google Scholar]
- 60.Fang Y., Liu W., Zhang F., Shao Y., Yu S. The polymorphism of a novel mutation of KAP13. 1 gene and its associations with cashmere traits on Xinjiang local goat breed in China. Asian J. Anim. Vet. Adv. 2010;5:34–42. doi: 10.3923/ajava.2010.34.42. [DOI] [Google Scholar]
- 61.Zhao M., Zhou H., Hickford J.G., Gong H., Wang J., Hu J., Liu X., Li S., Hao Z., Luo Y. Variation in the caprine keratin-associated protein 15-1 (KAP15-1) gene affects cashmere fibre diameter. Arch. Anim. Breed. 2019;62:125–133. doi: 10.5194/aab-62-125-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J., Hao Z., Zhou H., Luo Y., Hu J., Liu X., Li S., Hickford J.G.H. A keratin-associated protein (KAP) gene that is associated with variation in cashmere goat fleece weight. Small Rumin. Res. 2018;167:104–109. doi: 10.1016/j.smallrumres.2018.08.014. [DOI] [Google Scholar]
- 63.Wang J., Che L., Hickford J.G., Zhou H., Hao Z., Luo Y., Hu J., Liu X., Li S. Identification of the caprine keratin-associated protein 20-2 (KAP20-2) gene and its effect on cashmere traits. Genes. 2017;8:328. doi: 10.3390/genes8110328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J., Zhou H., Luo Y., Zhao M., Gong H., Hao Z., Hu J., Hickford J.G.H. Variation in the caprine KAP24-1 gene affects cashmere fibre diameter. Animals. 2019;9:15. doi: 10.3390/ani9010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao M., Zhou H., Luo Y., Wang J., Hu J., Liu X., Li S., Hao Z., Jin X., Song Y. Variation in the caprine keratin-associated protein 27-1 gene is associated with cashmere fiber diameter. Genes. 2020;11:934. doi: 10.3390/genes11080934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J., Zhou H., Hickford J.G., Zhao M., Gong H., Hao Z., Shen J., Hu J., Liu X., Li S. Identification of caprine KRTAP28-1 and its effect on cashmere fiber diameter. Genes. 2020;11:121. doi: 10.3390/genes11020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W., Gong H., Zhou H., Wang J., Li S., Liu X., Luo Y., Hickford J.G. Variation in KRTAP6-1 affects wool fibre diameter in New Zealand Romney ewes. Arch. Anim. Breed. 2019;62:509–515. doi: 10.5194/aab-62-509-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kijas J.W., Townley D., Dalrymple B.P., Heaton M.P., Maddox J.F., McGrath A., Wilson P., Ingersoll R.G., McCulloch R., McWilliam S. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE. 2009;4:e4668. doi: 10.1371/journal.pone.0004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duret L., Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Ann. Rev. Genom. Hum. Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 70.Galtier N., Duret L., Glémin S., Ranwez V. GC-biased gene conversion promotes the fixation of deleterious amino acid changes in primates. Trends Genet. 2009;25:1–5. doi: 10.1016/j.tig.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 71.1000 Genomes Project Consortium. Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levinson G., Gutman G.A. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 73.Szostak J.W., Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980;284:426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- 74.Huang C.R.L., Burns K.H., Boeke J.D. Active transposition in genomes. Ann. Rev. Genet. 2012;46:651–675. doi: 10.1146/annurev-genet-110711-155616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montgomery S.B., Goode D.L., Kvikstad E., Albers C.A., Zhang Z.D., Mu X.J., Ananda G., Howie B., Karczewski K.J., Smith K.S. The origin, evolution, and functional impact of short insertion–deletion variants identified in 179 human genomes. Genome Res. 2013;23:749–761. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Viguera E., Canceill D., Ehrlich S.D. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20:2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lovett S.T., Drapkin P.T., Sutera V., Gluckman-Peskind T.J. A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics. 1993;135:631–642. doi: 10.1093/genetics/135.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ripley L.S. Model for the participation of quasi-palindromic DNA sequences in frameshift mutation. Proc. Natl. Acad. Sci. USA. 1982;79:4128–4132. doi: 10.1073/pnas.79.13.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kiktev D.A., Sheng Z., Lobachev K.S., Petes T.D. GC content elevates mutation and recombination rates in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2018;115:E7109–E7118. doi: 10.1073/pnas.1807334115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tian D., Wang Q., Zhang P., Araki H., Yang S., Kreitman M., Nagylaki T., Hudson R., Bergelson J., Chen J.Q. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature. 2008;455:105–108. doi: 10.1038/nature07175. [DOI] [PubMed] [Google Scholar]
- 81.Chen J.Q., Wu Y., Yang H., Bergelson J., Kreitman M., Tian D. Variation in the ratio of nucleotide substitution and indel rates across genomes in mammals and bacteria. Mol. Biol. Evol. 2009;26:1523–1531. doi: 10.1093/molbev/msp063. [DOI] [PubMed] [Google Scholar]
- 82.McDonald M.J., Wang W.C., Huang H.D., Leu J.Y. Clusters of nucleotide substitutions and insertion/deletion mutations are associated with repeat sequences. PLoS Biol. 2011;9:e1000622. doi: 10.1371/journal.pbio.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith G.R., Kunes S.M., Schultz D.W., Taylor A., Triman K.L. Structure of chi hotspots of generalized recombination. Cell. 1981;24:429–436. doi: 10.1016/0092-8674(81)90333-0. [DOI] [PubMed] [Google Scholar]
- 84.Cheng K.C., Smith G.R. Recombinational hotspot activity of Chi-like sequences. J. Mol. Biol. 1984;180:371–377. doi: 10.1016/S0022-2836(84)80009-1. [DOI] [PubMed] [Google Scholar]
- 85.Rogers M.A., Langbein L., Winter H., Ehmann C., Praetzel S., Korn B., Schweizer J. Characterization of a cluster of human high/ultrahigh sulfur keratin-associated protein genes embedded in the type I keratin gene domain on chromosome 17q12-21. J. Biol. Chem. 2001;276:19440–19451. doi: 10.1074/jbc.M100657200. [DOI] [PubMed] [Google Scholar]
- 86.Nützmann H.W., Scazzocchio C., Osbourn A. Metabolic gene clusters in eukaryotes. Ann. Rev. Genet. 2018;52:159–183. doi: 10.1146/annurev-genet-120417-031237. [DOI] [PubMed] [Google Scholar]
- 87.Husnik F., McCutcheon J.P. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 2018;16:67–79. doi: 10.1038/nrmicro.2017.137. [DOI] [PubMed] [Google Scholar]
- 88.Shibuya K., Obayashi I., Asakawa S., Minoshima S., Kudoh J., Shimizu N. A cluster of 21 keratin-associated protein genes within introns of another gene on human chromosome 21q22.3. Genomics. 2004;83:679–693. doi: 10.1016/j.ygeno.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 89.Zhang J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 90.Marques-Bonet T., Girirajan S., Eichler E.E. The origins and impact of primate segmental duplications. Trends Genet. 2009;25:443–454. doi: 10.1016/j.tig.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bailey J.A., Church D.M., Ventura M., Rocchi M., Eichler E.E. Analysis of segmental duplications and genome assembly in the mouse. Genome Res. 2004;14:789–801. doi: 10.1101/gr.2238404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ransohoff J.D., Wei Y., Khavari P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kelley D., Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13:1–14. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cerbin S., Jiang N. Duplication of host genes by transposable elements. Cur. Opin. Genet. Dev. 2018;49:63–69. doi: 10.1016/j.gde.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 95.Hunt R., Sauna Z.E., Ambudkar S.V., Gottesman M.M., Kimchi-Sarfaty C. Silent (synonymous) SNPs: Should we care about them? Methods Mol. Biol. 2009;578:23–39. doi: 10.1007/978-1-60327-411-1_2. [DOI] [PubMed] [Google Scholar]
- 96.Li S.W., Ouyang H.S., Rogers G.E., Bawden C.S. Characterization of the structural and molecular defects in fibres and follicles of the merino felting lustre mutant. Exp. Dermatol. 2009;18:134–142. doi: 10.1111/j.1600-0625.2008.00774.x. [DOI] [PubMed] [Google Scholar]
- 97.Almeida A.M., Plowman J.E., Harland D.P., Thomas A., Kilminster T., Scanlon T., Milton J., Greeff J., Oldham C., Clerens S. Influence of feed restriction on the wool proteome: A combined iTRAQ and fiber structural study. J. Proteom. 2014;103:170–177. doi: 10.1016/j.jprot.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 98.He D., Chen L., Luo F., Zhou H., Wang J., Zhang Q., Lu T., Wu S., Hickford J.G.H., Tao J. Differentially phosphorylated proteins in the crimped and straight wool of Chinese Tan sheep. J. Proteom. 2021;235:104115. doi: 10.1016/j.jprot.2021.104115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.