Abstract
Coagulase-negative staphylococci, especially Staphylococcus epidermidis, are increasingly important nosocomial pathogens, particularly in critically ill neonates. A 3-year prospective surveillance of nosocomial infections in a neonatal intensive care unit (NICU) was performed by traditional epidemiologic methods as well as molecular typing of microorganisms. The aims of the study were (i) to quantify the impact of S. epidermidis on NICU-acquired infections, (ii) to establish if these infections are caused by endemic clones or by incidentally occurring bacterial strains of this ubiquitous species, (iii) to evaluate the use of different methods for the epidemiologic typing of the isolates, and (iv) to characterize the occurrence and the spread of staphylococci with decreased glycopeptide susceptibility. Results confirmed that S. epidermidis is one of the leading causes of NICU-acquired infections and that the reduced glycopeptide susceptibility, if investigated by appropriate detection methods such as population analysis, is more common than is currently realized. Typing of isolates, which can be performed effectively through molecular techniques such as pulsed-field gel electrophoresis but not through antibiograms, showed that many of these infections are due to clonal dissemination and, thus, are potentially preventable by strict adherence to recommended infection control practices and the implementation of programs aimed toward the reduction of the unnecessary use of antibiotics. These strategies are also likely to have a significant impact on the frequency of the reduced susceptibility of staphylococci to glycopeptides, since this phenomenon appears to be determined either by more resistant clones transmitted from patient to patient or, to a lesser extent, by strains that become more resistant as a result of antibiotic pressure.
Coagulase-negative staphylococci (CoNS) and, specifically, the dominant species Staphylococcus epidermidis have emerged in recent years as pathogens in a growing number of serious nosocomial infections in neonatal intensive care units (NICUs), particularly bloodstream infections (16, 19). Especially worrisome are the increased numbers of S. epidermidis isolates that show a low-level resistance to glycopeptide agents (14, 39, 40, 42), with an additional and probably greater concern which relates to the emergence of Staphylococcus aureus strains with reduced susceptibility to vancomycin (1, 6, 7, 21, 22, 41). These observations immediately prompted the rapid publication of guidelines for infection control management of infections caused by staphylococci intermediately resistant to vancomycin (5, 50).
Knowledge of the epidemiology of S. epidermidis infections is important for control of further spread of glycopeptide resistance among staphylococci. In this study molecular typing techniques as well as more traditional investigations conducted by infection control professionals were used to characterize the epidemiology of S. epidermidis in a NICU of a university hospital in Italy. The purposes of the study were (i) to quantify the impact of S. epidermidis on NICU-acquired infections, (ii) to establish if these infections are caused by endemic clones or by incidentally occurring bacterial strains of this ubiquitous species, (iii) to evaluate the use of different methods for the epidemiologic typing of the isolates, and (iv) to characterize the occurrence and the spread of staphylococci with decreased glycopeptide susceptibility.
MATERIALS AND METHODS
Setting.
The teaching hospital of the University “Federico II” of Naples is built on a 40-ha site and consists of 19 buildings, each of which contains one or more departments, connected by tunnels and passages and endowed with 1,470 beds. One building contains the pediatric department with three wards and one NICU; a connected building contains the obstetric department and the nursery. The NICU consists of three rooms with a maximal capacity of eight neonates per room. An additional smaller room is available for infants in need of isolation. Washing sinks are available in each room. Gloves, but no masks and gowns, are used routinely.
Surveillance procedures.
Nosocomial infection surveillance in the NICU was performed by a trained physician, who reviewed the following sources for evidence of infection: physician and nurse personnel in the unit, patient charts, and X rays and diagnostic bacteriology laboratory culture reports. Data collected on nosocomial infections included sites of infection, pathogens, time of acquisition from admission, birth weight, and major risk factors. Information about infections in the unit were recorded on a standardized form by the surveillance physician and were reviewed regularly with the attending pediatrician and the hospital epidemiologist. Surveillance of nosocomial infections after discharge was not performed. The infection surveillance reported here covers the period from January 1996 to December 1998.
Nosocomial infections were defined by standard Centers for Disease Control and Prevention definitions adapted to neonatal pathology (13). In particular, infants were considered to have a bloodstream infection if they had at least one positive blood culture and a compatible clinical presentation, provided that they received specific antibiotic therapy. Pneumonia was diagnosed, in the presence of a compatible clinical presentation, on chest X-ray evidence and the isolation of a microorganism from a tracheal or bronchial aspirate or a blood culture. The diagnosis of urinary tract infection required a urine culture with ≥105 colonies/ml and a compatible clinical presentation, with institution of specific antibiotic therapy. Meningitis was diagnosed if infants had a positive culture of cerebrospinal fluid and a compatible clinical presentation, provided that they received an appropriate antimicrobial therapy. The diagnosis of conjunctivitis required the isolation of a microorganism from a culture of a purulent exudate and the institution of appropriate antibiotic therapy. Infections resulting from passage through the birth canal or from transplacental transmission were excluded. In general, infections that occurred after 48 h of hospital stay were assumed to be acquired in the NICU, but each instance of infection was considered individually because of the late onset of some perinatally acquired infections.
All infection rates, including patient, patient-day, and device-associated infections, were calculated according to the formulas of the National Nosocomial Infections Surveillance (NNIS) high-risk nursery (HRN) component (15).
Microbiological methods.
All S. epidermidis strains isolated from clinical specimens during three “window” periods in 1996, 1997, and 1998 were collected, isolated in pure cultures, and stored at −80°C with glycerol for subsequent phenotypic and genotypic typing. The three window periods were of 5 months in 1996, 8 months in 1997, and 10 months in 1998.
Antimicrobial susceptibility testing.
Susceptibilities to common antibiotics were determined by disk diffusion methods (33, 34). The following antibiotics were tested: ampicillin (10 μg), methicillin (5 μg), chloramphenicol (30 μg), erythromycin (15 μg), tetracycline (30 μg), rifampin (5 μg), ciprofloxacin (5 μg), trimethoprim-sulfamethoxazole (1.25 + 27.75 μg), gentamicin (10 μg), netilmicin (10 μg), teicoplanin (30 μg), and vancomycin (30 μg). Isolates showing an intermediate level of susceptibility were classified as resistant. Susceptibility to glycopeptides was further investigated by population analysis profiles (PAPs) (39, 40, 45). Aerobically grown overnight cultures were plated at four dilutions (10−1, 10−2, 10−4, and 10−6) on plates that contained serial (twofold) dilutions of teicoplanin or vancomycin at concentrations of 0 and 0.8 to 100 μg/ml. Colonies were counted after incubation for 48 h at 37°C.
Preparation of chromosomal DNA for PFGE and restriction digestion.
The preparation of chromosomal DNA for pulsed-field gel electrophoresis (PFGE) and restriction digestion was performed as described by de Lencastre et al. (8). DNA restriction was done with SmaI according to the manufacturer's recommendations (New England Biolabs).
PFGE.
Gels for PFGE were run in a CHEF-DR II apparatus (Bio-Rad) by using the following conditions: run time, 23 h; temperature, 11.8°C; voltage, 200 V; initial forward time, 1 s, final forward time, 30 s.
DNA-DNA hybridization.
Chromosomal DNAs were digested with the restriction endonuclease ClaI, separated by conventional gel electrophoresis, and then transferred to nylon membranes with a vacuum blotting apparatus (VacuGene XL; Pharmacia), according to the manufacturer's instructions. Hybridization was done with two probes, one bearing the Tn554 transposon (11, 27) and the other bearing the mecA gene (8, 29, 30). Probe labeling and hybridization were done by using an enhanced chemiluminescence nonradioactive labeling kit (RPN3000; Amersham), according to the manufacturer's instructions.
RESULTS
Nosocomial infections.
During the 3-year period of study, 1,010 infants were admitted to the unit. Within 2 days after admission, 28 infants died or were transferred from the unit. Data for these infants were excluded from the study since nosocomial infections in this population would not have been identified by our surveillance methods. The remaining 982 patients, 556 males (56.6%) and 426 females (43.4%), were included in the study. Most babies (87.5%) were born in the obstetric department of the teaching hospital of University “Federico II” of Naples, and the remaining 12.5% were transported from other hospitals in the area. The 982 patients spent a total of 22,740 days in the NICU (average, 23.2 days; standard deviation, 24.1 days), with device utilization ratios of 0.48 and 0.37 for umbilical or central line and ventilator, respectively. The total mortality rate in the study population was 8.3% (82 of 982).
A total of 184 infections were detected among 142 patients, for an overall nosocomial infection rate of 18.7% or 8.1 per 1,000 patient-days (Table 1). The most common infections were bloodstream infections (47.8%), surface infections (mainly conjunctivitis) (25.5%), pneumonia (11.5%), and urinary tract infections (8.7%). Twelve cases of meningitis were observed (6.5%). The umbilical or central line-associated bloodstream infection rate was 8.0 per 1,000 device-days, ranging from 14.8 per 1,000 device-days in neonates with birth weights of 1,000 g or less to 3.5 per 1,000 device-days in neonates with birth weights of more than 2,500 g. The ventilator-associated pneumonia rate was 2.5 per 1,000 device-days, with the highest rate (3.8 per 1,000 device-days) observed among infants with birth weights of ≤1,000 g and the lowest rate (1.8 per 1,000 device-days) observed among infants with birth weights of 1,001 to 1,500 g. Both bloodstream and pneumonia rates decreased throughout the study period.
TABLE 1.
Incidence of nosocomial infections in the NICU of University “Federico II” during the period from 1996 to 1998
| Parameter | Incidence rate
|
|||
|---|---|---|---|---|
| 1996 | 1997 | 1998 | Total | |
| Overall nosocomial infection rate (per 100 patients at risk) | 25.4 | 15.2 | 15.1 | 18.7 |
| Overall nosocomial infection rate (per 1,000 patient-days) for infants with birth weight (g) of: | 10.7 | 6.6 | 6.7 | 8.1 |
| ≤1,000 | 16.2 | 13.6 | 15.1 | 14.4 |
| 1,001–1,500 | 11.6 | 6.0 | 6.5 | 8.4 |
| 1,501–2,500 | 12.4 | 2.9 | 3.4 | 7.1 |
| >2,500 | 4.3 | 6.2 | 6.4 | 5.7 |
| S. epidermidis nosocomial infection rate (per 1,000 patient-days) | 2.1 | 2.7 | 2.6 | 2.5 |
| S. aureus nosocomial infection rate (per 1,000 patient-days) | 3.2 | 2.8 | 0.5 | 2.2 |
| K. pneumoniae nosocomial infection rate (per 1,000 patient-days) | 3.3 | 0.3 | 0.1 | 1.3 |
| C. albicans nosocomial infection rate (per 1,000 patient-days) | 0.7 | 0.4 | 1.0 | 0.7 |
| Bloodstream infection rate (per 1,000 umbilical and central line-days) for infants with birth weight (g) of: | 8.7 | 7.8 | 7.0 | 8.0 |
| ≤1,000 | 9.6 | 14.7 | 19.0 | 14.8 |
| 1,001–1,500 | 12.6 | 6.0 | 6.5 | 8.7 |
| 1,501–2,500 | 9.5 | 5.8 | 4.3 | 7.2 |
| >2,500 | 2.1 | 4.5 | 3.7 | 3.5 |
| Pneumonia rate (per 1,000 ventilator days) for infants with birth weight (g) of: | 3.1 | 2.6 | 0.7 | 2.5 |
| ≤1,000 | 7.2 | 3.4 | 1.9 | 3.8 |
| 1,001–1,500 | 1.9 | 4.4 | 0.0 | 1.8 |
| 1,501–2,500 | 4.1 | 0.0 | 0.0 | 2.1 |
| >2,500 | 0.0 | 2.2 | 1.9 | 2.8 |
S. epidermidis was responsible for 56 of the 184 infections which occurred in the unit during the study period (30.4%). More precisely, S. epidermidis accounted for 35 bloodstream infections (39.8%), 14 surface infections (29.8%), and 7 cases of meningitis (58.3%). Other less frequently isolated pathogens were S. aureus (27.2% of all infections), Klebsiella pneumoniae (16.3%), and Candida albicans (9.2%). Interestingly, whereas the overall nosocomial infection rate during the study period decreased from 10.7 per 1,000 patient-days in 1996 to 6.7 per 1,000 patient-days in 1998, the S. epidermidis infection rate actually rose slightly from 2.1 per 1,000 patient-days in 1996 to 2.6 per 1,000 patient-days in 1998 (Table 1).
Molecular typing of S. epidermidis isolates.
Eighty-one S. epidermidis isolated during three window periods from 1996 to 1998 were available for molecular typing. The strains originated from blood (33.3%), vascular catheters (17.3%), eye (24.7%), endotracheal tubes or bronchial aspirates (18.5%), and liquor (6.2%). Half of these strains (50.6%) were judged to be involved with infection.
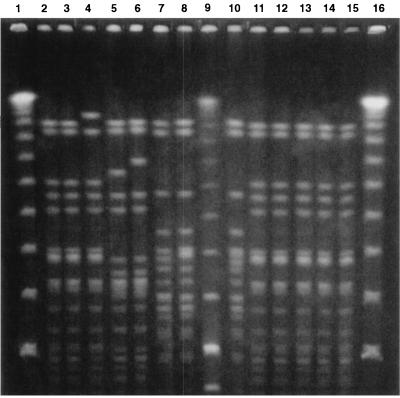
All these strains were typed by macrorestriction analysis of chromosomal DNA with SmaI and PFGE (Fig. 1). By assuming that a single base mutation in the chromosomal DNA could introduce maximally a three-fragment difference in the restriction pattern (44), strains with more than three fragment variations were assumed to represent strains with major patterns (assignment of capital letters), while strains with one to three fragment differences were considered to represent subtypes (capital letter with numerical subscripts). By these criteria, 25 major patterns (patterns A to Z except X) were identified. However, more than half of the isolates (45 of 81; 55.5%) were of four PFGE types (types G, C, E, and A). PFGE type G, which could be further classified into seven subtypes (subtypes G1 to G7), was detected for 22 strains (27.2%). Ten (12.3%) and seven (8.6%) strains were PFGE types C and E, respectively, and these types showed two and three subtypes, respectively. PFGE type A, with four subtypes, was found for six strains (7.4%). Two other PFGE types (types D and N), each of which had two subtypes, were found for four and three strains, respectively. The remaining PFGE patterns had no subtypes and, except for PFGE types H, K, O, P, V, and Y, were found only for single isolates.
FIG. 1.
SmaI PFGE patterns of S. epidermidis isolates in the NICU of University “Federico II” during the period from 1996 to 1998. Lanes 7, 8, and 10, subtypes of PFGE pattern G; lanes 2, 3, 4, and 11 to 15, strains of PFGE pattern C; lanes 5 and 6, subtypes of PFGE pattern E; lanes 1, 9, and 16, molecular weight markers (bacteriophage lambda ladder).
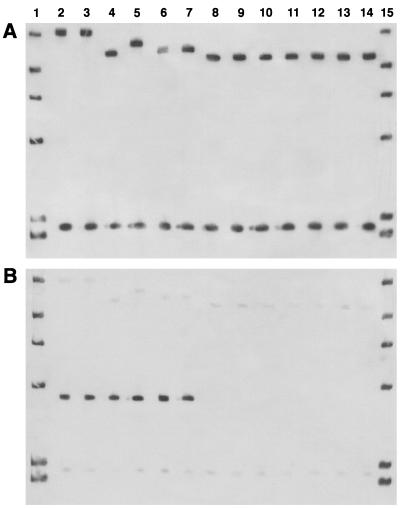
ClaI digests of the genomic DNAs of all S. epidermidis isolates were separated by conventional electrophoresis, transferred to a nylon membrane, and hybridized with the mecA probe (Fig. 2A). By considering differences in a single band as different mecA polymorphisms (21, 28), 14 ClaI-mecA types were identified. However, the majority of the isolates (55 of 81; 67.9%) were of four ClaI-mecA types (types I, II, VII, and VIII). Twenty-one of the 22 strains with PFGE pattern G and the 2 strains with PFGE pattern Y were found to be of ClaI-mecA type II. ClaI-mecA type VII was found only for the 10 strains with PFGE pattern C. All strains with PFGE pattern A were of ClaI-mecA type VIII, but this type was also associated with PFGE patterns V (two strains), P (two strains), and G (one strain). ClaI-mecA type I was found for six of the seven strains with PFGE pattern E and all strains with PFGE patterns D (four strains) and L (one strain). The other strain with PFGE pattern E was ClaI-mecA type XII. The remaining nine ClaI-mecA types were associated with strains with unique PFGE patterns, with the exception of type V (which was associated with PFGE patterns F and M) and type VI (which was found for strains with PFGE patterns O and Q). The mecA gene was absent from six strains already classified as methicillin susceptible by the disk diffusion test.
FIG. 2.
mecA polymorphs (A) and the associated Tn554 patterns (B) shown by S. epidermidis isolates in the NICU of University “Federico II” during the period from 1996 to 1998. (A) Lanes 2 and 3, mecA type VII; lanes 4, 6, and 7, mecA type I; lane 5, mecA type XII; lanes 8 to 14, mecA type II. (B) Lanes 2 to 7, Tn554 positive. Lanes 1 and 15, molecular weight markers (bacteriophage lambda ladder).
The same gels used for the analysis of the mecA gene were rehybridized with the Tn554 probe (Fig. 2B). The transposon was present in 21 of the 81 isolates (25.9%) in a unique insertion pattern (single insertion with loss of the ClaI restriction site, with only one hybridization band of approximately 4 kb). More precisely, the presence of Tn554 was documented in all isolates of ClaI-mecA types VII (10 strains), V (2 strains), XII (1 strains), and IV (1 strain). Moreover, the transposon was present in 7 of the 11 strains of ClaI-mecA type I, namely, those with PFGE patterns E (6 strains) and L (1 strain).
S. epidermidis isolates can be defined as clones on the basis of mecA::Tn554::PFGE profiles, similar to the criteria already used for methicillin-resistant S. aureus (MRSA) (8, 9, 36, 43, 48). Together the three genotypic methods used in the present study allowed us to define 28 clones among the 81 strains isolated during a 3-year period in the NICU of University “Federico II” of Naples (Table 2). Four predominant clones were identified: II::neg::G (21 strains), VII::pos::C (10 strains), I::pos::E (6 strains), and VIII::neg::A (6 strains), where neg is negativity for the presence of Tn554 and pos is positivity for the presence of Tn554. These clones were isolated during the entire study period from different clinical sources. For example, clone II::neg::G included strains isolated during 1996 (four strains), 1997 (eight strains), and 1998 (nine strains) from blood (six strains), eye (five strains), tubes or bronchial aspirates (six strains), and vascular catheters (four strains). Analogously, clone VII::pos::C comprised isolates from 1996 (one strain), 1997 (three strains), and 1998 (six strains) cultured from blood (five strains), endotracheal tubes or bronchial aspirates (two strains), vascular catheters (two strains), and an eye (one strain). Therefore, the predominant clones found in the NICU did not showed either evident temporal clusters or an association with particular clinical sources.
TABLE 2.
S. epidermidis clones identified in the NICU of University “Federico II” during the period from 1996 to 1998
| mecA polymorpha (n)b | Tn554 patterna (n) | PFGE patterna (n) | Clone | No. of strainsc | Resistance phenotypesa (n) | PAPd (n)
|
|
|---|---|---|---|---|---|---|---|
| Vancomycin | Teicoplanin | ||||||
| II (23) | Neg (23) | G (21) | II::neg::G | 21 (9) | 3a (1), 4d (1), 4e (1), 5a (2), 5d (1), 5e (1), 5m (2), 6b (7), 6d (3), 6h (1), 7a (1) | II (20), III (1) | II (21) |
| Y (2) | II::neg::Y | 2 (0) | 6g (2) | I (1), II (1) | I (1), II (1) | ||
| I (11) | Pos (7) | E (6) | I::pos::E | 6 (6) | 4a (1), 6a (1), 6f (2), 7a (1), 9a (1) | II (4), III (2) | III (6) |
| L (1) | I::pos::L | I (1) | 5d (1) | II (1) | II (1) | ||
| Neg (4) | D (4) | I::neg::D | 4 (4) | 6e (4) | I (1), II (3) | I (1), II (3) | |
| VIII (11) | Neg (11) | A (6) | VIII::neg::A | 6 (3) | 3b (1), 4d (1), 5a (1), 6b (1), 7a (1), 7b (1) | I (1), II (5) | I (1), II (5) |
| P (2) | VIII::neg::P | 2 (0) | 5f (2) | II (2) | II (2) | ||
| V (2) | VIII::neg::V | 2 (0) | 3a (2) | I (2) | I (2) | ||
| G (1) | VIII::neg::G | 1 (1) | 5d (1) | II (1) | II (1) | ||
| VII (10) | Pos (10) | C (10) | VII::pos::C | 10 (5) | 6a (2), 6d (2), 7a (2), 8b (3), 9a (1) | I (2), II (8) | I (5), II (5) |
| III (4) | Neg (4) | K (4) | III::neg::K | 4 (2) | 3c (1), 4b (1), 4c (2) | II (4) | II (4) |
| IX (4) | Neg (4) | H (4) | IX::neg::H | 4 (4) | 5a (3), 8a (1) | II (4) | II (4) |
| VI (3) | Neg (3) | O (2) | VI::neg::O | 2 (0) | 4a (2) | II (2) | II (2) |
| Q (1) | VI::neg::Q | 1 (1) | 5a (1) | II (1) | II (1) | ||
| V (2) | Pos (1) | F (1) | V::pos::F | 1 (1) | 5l (1) | II (1) | II (1) |
| Pos (1) | M (1) | V::pos::M | 1 (0) | 2a (1) | I (1) | I (1) | |
| X (2) | Neg (2) | N (2) | X::neg::N | 2 (0) | 5a (1), 5h (1) | I (2) | I (2) |
| IV (1) | Pos (1) | I (1) | IV::pos::I | 1 (0) | 5i (1) | II (1) | II (1) |
| XI (1) | Neg (1) | T (1) | XI::neg::T | 1 (0) | 5a (1) | II (1) | II (1) |
| XII (1) | Pos (1) | E (1) | XII::pos::E | 1 (1) | 5b (1) | II (1) | II (1) |
| XIII (1) | Neg (1) | Z (1) | XIII::neg::Z | 1 (0) | 5c (1) | II (1) | II (1) |
| XIV (1) | Neg (1) | J (1) | XIV::neg::J | 1 (0) | 6d (1) | II (1) | II (1) |
| Negative (6) | Neg (6) | N (1) | neg::neg::N | 1 (0) | 2c (1) | I (1) | I (1) |
| B (1) | neg::neg::B | 1 (1) | 2b (1) | I (1) | I (1) | ||
| R (1) | neg::neg::R | 1 (1) | 2d (1) | I (1) | I (1) | ||
| S (1) | neg::neg::S | 1 (1) | 2b (1) | II (1) | II (1) | ||
| U (1) | neg::neg::U | 1 (0) | 1a (1) | II (1) | II (1) | ||
| W (1) | neg::neg::W | 1 (0) | 3d (1) | II (1) | II (1) | ||
See text for definitions.
n, number of strains.
The numbers of strains considered to be causative of infection are indicated in parentheses.
See text for definitions.
Strains of S. epidermidis were judged to be involved with infection if they represented the only microorganisms isolated from a significant clinical specimen (blood, liquor, or purulent exudate) in the presence of clinical signs or symptoms of infection and the institution of a specific antibiotic therapy (mainly with teicoplanin or vancomycin) driven by the results of antibiograms. On the basis of these criteria, the majority of the clones found in this study included strains that caused both infection and colonization; the exception was clones of the ClaI-mecA type I polymorph, all of which were considered to be causative of infections. The latter observation suggests the hypothesis (which has yet to be confirmed) that some clones of S. epidermidis may be more pathogenic than others.
Antimicrobial resistance of the isolates.
The antibiotic susceptibility data showed that large proportions of the isolates were resistant to ampicillin (100%), methicillin (92.6%), gentamicin (80.2%), erythromycin (65.4%), tetracycline (63.0%), and chloramphenicol (50.6%). Various but limited numbers of strains appeared to be resistant to trimethoprim-sulfamethoxazole (43.2%), netilmicin (23.5%), and ciprofloxacin (3.7%). All strains were susceptible to rifampin, teicoplanin, and vancomycin. In the attempt to use the antibiotic resistance profile as an epidemiologic marker, we defined resistance phenotypes through a number (representing the number of antibiotics to which the strain was resistant) with a letter subscript (indicating the particular combination of antibiotics to which the isolate was resistant). Altogether, 36 distinct resistance phenotypes were found, with a poor correlation with the clonal types defined by the molecular methods (Table 2). For example, the most frequent resistance phenotype (5a; which was found for 11.1% of the isolates and which was characterized by resistance to ampicillin, methicillin, gentamicin, erythromycin, and tetracycline) was present in strains of six distinct clonal types, as well as the major clonal type II::neg::G strains, with 11 different resistance phenotypes. It is interesting that isolates of the four main clones found in this study generally appeared to be resistant to more antibiotics than the other strains. Moreover, the vast majority of these isolates (94.6%) were fully resistant to both antibiotics used as empiric treatment in the NICU (ampicillin and gentamicin), whereas 63.1% of the strains of less frequently occurring clonal types were resistant to both antibiotics.
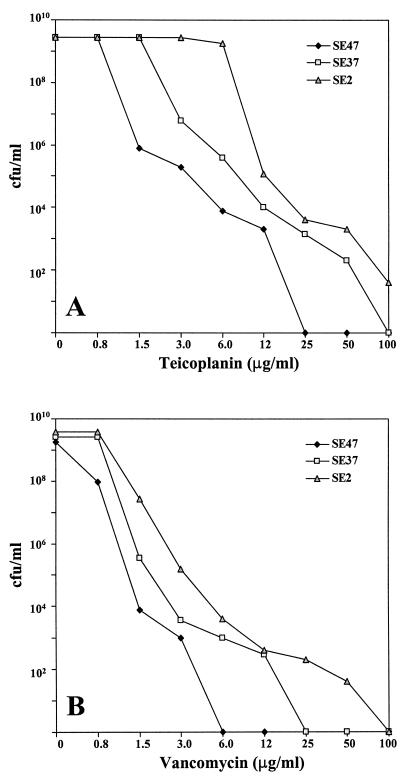
The glycopeptide susceptibilities of the isolates were investigated more extensively through the PAP approach, as described in Materials and Methods. Since the first experiments we realized that not all strains were equally susceptible to teicoplanin and vancomycin. Therefore, we defined three expression classes on the basis of PAPs (Fig. 3). Although the definition of these classes is arbitrary, class I through class III could be considered to represent three different increasing levels of glycopeptide resistance. In cultures of strains of class III, 100% of the cells grew in the presence of 3 to 6 μg of teicoplanin per ml or 0.8 to 1.5 μg of vancomycin per ml, with a small fraction able to grow in the presence of 100 μg of teicoplanin per ml or 50 to 100 μg of vancomycin per ml. By contrast, the great majority of cells in cultures of class I strains were inhibited by low concentrations of glycopeptides, with a teicoplanin MIC of 25 μg/ml and a vancomycin MIC of 6 μg/ml being found for only a small number of isolates. Class II strains exhibited PAPs that were intermediate between those of class I and III isolates. By these criteria, most of the S. epidermidis isolates were assigned to vancomycin class II (80.2%), with only three strains (3.7%) having a class III profile. The PAPs in tests with teicoplanin were similar, with 72.8 and 7.4% of strains displaying class II and III profiles, respectively. It is interesting that although some clones (such as clone I::pos::E) appeared to be more resistant than others (e.g., clone VII:pos::C), it was possible that strains of the same clonal type exhibited different PAPs (Table 2).
FIG. 3.
Phenotypic expression of teicoplanin (A) and vancomycin (B) resistance observed in S. epidermidis isolates in the NICU of University “Federico II” during the period from 1996 to 1998. Strains SE47 (class I), SE37 (class II), and SE2 (class III) are representatives of clones VII::pos::C, II::neg::G, and I::pos::E, respectively.
DISCUSSION
Reports on S. epidermidis infections in NICUs remained infrequent until the early 1980s. Hemming et al. (20) found no cases in 4 years (1970 to 1974) at the University of Utah Medical Center, confirming the results on the rates of S. epidermidis bacteremias obtained by McCracken and Shinefield (32) at New York Hospital and by Freedman et al. (12) at Yale-New Haven Hospital. More frequent reports on S. epidermidis sepsis began to appear in the early 1980s, with Goldmann et al. (17) finding that although gram-negative bacilli were still the predominant cause of nosocomial infections at Children's Hospital in Boston from 1974 to 1978, S. epidermidis was the principal gram-positive organism recovered during this period. Similar findings were reported by Hoogkamp-Korstanje et al. (23) at Wilhemina Children's Hospital in Utrecht, The Netherlands, in 1977, with 33% of all NICU-acquired infections caused by CoNS. Summary data from the Center for Diseases Control and Prevention's NNIS system show that CoNS accounted for more than 30% of all nosocomial infections in NICUs both for 1985 to 1986 (25) and for 1986 to 1994 (16).
Results of this study confirm that the impact of S. epidermidis on NICU-acquired infection is substantial. Both the umbilical or central line-associated bloodstream infection rate and the ventilator-associated pneumonia rate were within the NNIS system benchmarks for high-risk nurseries of hospitals that are part of the NNIS system, as were the device utilization ratios for all birth weight categories (4). S. epidermidis accounted for 30.4% of all NICU-acquired infections, confirming the high frequency of CoNS infections reported by most recent studies. Although the overall nosocomial infection rate decreased during the study period, the S. epidermidis infection rate rose slightly. It should be noted, however, that the decrease in the overall infection rate was observed mainly in neonates weighing more than 1,000 g, whereas CoNS infections are known to occur predominately in patients with very low birth weights (19).
When studying the epidemiology of CoNS infections, it is important to determine whether these infections are caused by several different strains or by one or a few epidemic clones, since measures to prevent cross-infections are likely to have a major impact only on the latter isolates. While some investigators found evidence of an endogenous origin of CoNS in NICUs (35, 46), this study demonstrates that a significant proportion of S. epidermidis infections may be attributable to transmission among patients and that certain strains can become endemic over long periods in this setting. These results extend the findings of previous studies that have suggested the importance of cross-infection with CoNS in the NICU (3, 24, 28, 46, 47, 49). The success of the predominant clones in this and other studies may be related to yet uncharacterized factors that provide the organisms with advantages in colonization or in their ability to infect patients. One of these factors may be antibiotic resistance (3, 47), since the four main clones found in this study were more resistant than the sporadic strains, particularly to antibiotics used as empiric treatment in the NICU.
Many typing techniques have recently been applied to studies of the epidemiology of CoNS. The study described here confirms that the comparative analysis of drug resistance patterns is unsatisfactory (24, 28), since some monoclonal strains differed by their drug resistance patterns, and some polyclonal strains had the same resistance phenotype. With respect to genotyping, we used three molecular typing tools that have already demonstrated good discriminatory power in the analysis of MRSA (8, 9, 36, 43, 48). Our results indicate that the discriminatory power of the mecA probe may be higher than that reported for MRSA isolates. Only a relatively few mecA polymorphs have been identified among several hundred MRSA strains collected from a wide variety of geographic sources (27). This finding is in sharp contrast to the observation described in this study: the S. epidermidis isolates collected at a single NICU showed a wide range of variation in the vicinity of the mecA gene, with 15 distinct patterns among 81 isolates (including the mecA-negative strains). The use of the Tn554 probe was of limited value, since transposon-like elements were present only in a minority of the isolates without heterogeneity. By contrast, PFGE showed high resolution, detecting at least 25 different patterns and identifying the four main clones found in this study with better precision than the use of the mecA probe alone. The simultaneous use of the three techniques allow us only a modest increase in sensitivity, differentiating 28 clones among the 81 isolates.
The recent reports from the United States and Japan of clinical isolates of S. aureus with reduced susceptibility to vancomycin (1, 4, 6, 7, 21, 22) have renewed the interest in glycopeptide resistance in CoNS. Isolation of S. epidermidis strains with reduced susceptibility or frank resistance to glycopeptides has been reported from both Europe and the United States in the last several years (2, 14, 18, 26, 31, 37, 39, 40). Together with those observations, the results of this study suggest that intermediate glycopeptide resistance in S. epidermidis may be more prevalent than is currently realized. All S. epidermidis isolates, when analyzed by the method of population analysis, exhibited a heterogeneous phenotype with respect to glycopeptide susceptibility, with the vast majority of isolate cultures containing cells capable of growing in the presence of 12 μg of vancomycin per ml or 50 μg of teicoplanin per ml. A small percentage of S. epidermidis isolates were assigned to what we defined as class III, since their cultures contained a small fraction of cells capable of forming colonies on agar containing elevated concentrations of vancomycin (50 to 100 μg/ml) and teicoplanin (100 μg/ml). These more substantially resistant bacteria would not have been detected by the routine methods used in the clinical microbiology laboratory because of their low frequency.
The basis of glycopeptide resistance in S. epidermidis is unknown, but it appears to differ substantially from that observed in enterococci. Prior vancomycin or teicoplanin therapy was common in many reports of S. epidermidis clinical isolates with reduced resistance to glycopeptides, and stepwise in vitro exposure of strains to increasing concentrations of glycopeptides easily generates resistance. The addition of the drug to cultures caused the appearance of cellular aggregates, the inhibition of autolysis, and the removal of the antibiotic from the growth medium, features that resemble those of a recently described vancomycin-resistant laboratory mutant of S. aureus (38, 39). The antibiotic pressure exerted by glycopeptide agents on the infants included in this investigation was substantial, since, on average, neonates spent 4.9% of their hospital stay under therapy with vancomycin or teicoplanin. Likewise, in vitro experiments aimed at the selection of isolates with frank resistance by selection with vancomycin or teicoplanin were successful (data not shown). While this study did not investigate the mechanisms of glycopeptide resistance of S. epidermidis, the results of the molecular typing of the isolates allow us to identify some determinants that contribute to the frequency of CoNS with reduced susceptibility to glycopeptides in the NICU setting. In particular, two findings need to be emphasized (see Results): some clones appeared to be more resistant than others, and isolates of the same clonal type had different glycopeptide PAPs. In combination, these observations suggest that the prevalence of reduced susceptibility to glycopeptides in staphylococci may be due to the cross-transmission of intermediate resistant clones as well as, to a lesser extent, to the selective pressure exerted in vivo by vancomycin or teicoplanin use.
In conclusion, this study confirmed that S. epidermidis is one of the leading causes of NICU-acquired infections and that the reduced glycopeptide susceptibility, if investigated by appropriate detection methods, may be more common than is currently estimated. Typing of isolates, which can be performed effectively through molecular techniques such as PFGE but not through antibiograms, showed that many of these infections are due to clonal dissemination and, thus, are potentially preventable by strict adherence to recommended infection control practices and the implementation of programs aimed at the reduction of unnecessary use of antibiotics. These strategies are also likely to have a significant impact on the frequency of staphylococci with reduced glycopeptide susceptibility, since this phenomenon appears to be determined either by more resistant clones transmitted from patient to patient or by strains that change from susceptible to resistant as a result of antibiotic pressure.
REFERENCES
- 1.Ariza J, Pujol M, Cabo J, Pena C, Fernandez N, Linares J, Ayats J, Gudiol F. Vancomycin in surgical infections due to methicillin-resistant Staphylococcus aureus with heterogeneous resistance to vancomycin. Lancet. 1999;353:1587–1588. doi: 10.1016/s0140-6736(99)01017-x. [DOI] [PubMed] [Google Scholar]
- 2.Bannerman T L, Wadiak D L, Kloos W E. Susceptibility of Staphylococcus species and subspecies to teicoplanin. Antimicrob Agents Chemother. 1990;34:901–903. doi: 10.1128/aac.35.9.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnie J P, Naderi-Nasab M, Loudon K W, Matthews R C. An epidemiological study of blood cultures isolates of coagulase-negative staphylococci demonstrating hospital acquired infection. J Clin Microbiol. 1997;35:1746–1750. doi: 10.1128/jcm.35.7.1746-1750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC NNIS System. National Nosocomial Infections Surveillance (NNIS) report, data summary from October 1986 to April 1998, issued June 1998. Am J Infect Control. 1998;26:522–533. doi: 10.1016/s0196-6553(98)70026-4. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Interim guidelines for prevention and control of staphylococcal infection associated with reduced susceptibility to vancomycin. Morbid Mortal Weekly Rep. 1997;46:626–635. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Staphylococcus aureus with reduced susceptibility to vancomycin—United States. Morbid Mortal Weekly Rep. 1997;46:765–766. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Update: Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morbid Mortal Weekly Rep. 1997;46:813–815. [PubMed] [Google Scholar]
- 8.de Lencastre H, Couto I, Santos I, Melo-Cristino J, Torres-Pereira A, Tomasz A. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur J Clin Microbiol Infect Dis. 1994;13:64–73. doi: 10.1007/BF02026129. [DOI] [PubMed] [Google Scholar]
- 9.Dominguez M A, de Lencastre H, Linares J, Tomasz A. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J Clin Microbiol. 1994;32:2081–2087. doi: 10.1128/jcm.32.9.2081-2087.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domingez M A, Linares J, Pulido A, Perez J L, de Lencastre H. Molecular tracking of coagulase-negative staphylococci from catheter-related infections. Microb Drug Resist. 1996;2:423–429. doi: 10.1089/mdr.1996.2.423. [DOI] [PubMed] [Google Scholar]
- 11.Figueiredo A M S, Ha E, Kreiswirth B, de Lencastre H, Noel G J, Senterfit L, Tomasz A. In vivo stability of heterogeneous expression classes in clinical isolates of methicillin-resistant staphylococci. J Infect Dis. 1991;164:883–887. doi: 10.1093/infdis/164.5.883. [DOI] [PubMed] [Google Scholar]
- 12.Freedman R M, Ingram D L, Gross L. A half century of neonatal sepsis at Yale. Am J Dis Child. 1981;135:140–144. doi: 10.1001/archpedi.1981.02130260032010. [DOI] [PubMed] [Google Scholar]
- 13.Garner J S, Jarvis W R, Emori T G, Horan T C, Hughers J M. CDC definitions for nosocomial infections. Am J Infect Control. 1988;16:128–140. doi: 10.1016/0196-6553(88)90053-3. [DOI] [PubMed] [Google Scholar]
- 14.Garrett D O, Jochimsen E, Murfitt K, Hill B, McAllister S, Nelson P, Spera R V, Sall R K, Tenover F C, Johnston J, Zimmer B, Jarvis W R. The emergence of decreased susceptibility to vancomycin in Staphylococcus epidermidis. Infect Control Hosp Epidemiol. 1999;20:167–170. doi: 10.1086/501605. [DOI] [PubMed] [Google Scholar]
- 15.Gaynes R P, Horan T C. Surveillance of nosocomial infections. In: Mayhall C G, editor. Hospital epidemiology and infection control. Baltimore, Md: The Williams & Wilkins; 1996. pp. 1017–1031. [Google Scholar]
- 16.Gaynes R P, Edwards J R, Jarvis W R, Culver D H, Tolson J S, Martone and the National Nosocomial Infections Surveillance System W J. Nosocomial infections among neonates in high-risk nursery in the United States. Pediatrics. 1996;98:357–361. [PubMed] [Google Scholar]
- 17.Goldmann D A, Durbin W A, Freeman J. Nosocomial infections in a neonatal intensive care unit. J Infect Dis. 1981;144:449–458. doi: 10.1093/infdis/144.5.449. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein F W, Coutrot A, Sieffer A, Acar J F. Percentages and distribution of teicoplanin- and vancomycin-resistant strains among coagulase-negative staphylococci. Antimicrob Agents Chemother. 1990;34:899–900. doi: 10.1128/aac.34.5.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall S L. Coagulase-negative staphylococcal infections in neonates. Pediatr Infect Dis J. 1991;10:50–67. doi: 10.1097/00006454-199101000-00012. [DOI] [PubMed] [Google Scholar]
- 20.Hemming V G, Overall J C, Britt M R. Nosocomial infections in a newborn intensive-care unit. N Engl J Med. 1976;294:1310–1316. doi: 10.1056/NEJM197606102942403. [DOI] [PubMed] [Google Scholar]
- 21.Hiramatsu K, Hanaky H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 22.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1999;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 23.Hoogkamp-Korstanje J A A, Cats B, Senders R C, van Ertbruggen I. Analysis of bacterial infections in a neonatal intensive care unit. J Hosp Infect. 1982;3:275–284. doi: 10.1016/0195-6701(82)90046-9. [DOI] [PubMed] [Google Scholar]
- 24.Huebner J, Pier G B, Maslow J N, Muller E, Shiro H, Parent M, Kropec A, Arbeit R D, Goldmann D A. Endemic nosocomial transmission of Staphylococcus epidermidis bacteremia isolates in a neonatal intensive care unit over 10 years. J Infect Dis. 1994;169:526–531. doi: 10.1093/infdis/169.3.526. [DOI] [PubMed] [Google Scholar]
- 25.Jarvis W R. Epidemiology of nosocomial infections in pediatric patients. Pediatr Infect Dis J. 1987;7:344–351. doi: 10.1097/00006454-198704000-00003. [DOI] [PubMed] [Google Scholar]
- 26.Krcmery V, Jr, Trupl J, Drgona L, Lacka J, Kukuckova E, Oravcova E. Nosocomial bacteremia due to vancomycin-resistant Staphylococcus epidermidis in four patients with cancer, neutropenia, and previous treatment with vancomycin. Eur J Clin Microbiol Infect Dis. 1996;15:259–261. doi: 10.1007/BF01591369. [DOI] [PubMed] [Google Scholar]
- 27.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 28.Lyytikainen O, Saxen H, Ryhanen R, Vaara M, Vuopio-Varkila J. Persistence of a multiresistent clone of Staphylococcus epidermidis in a neonatal intensive-care unit for a four-year period. Clin Infect Dis. 1995;20:24–29. doi: 10.1093/clinids/20.1.24. [DOI] [PubMed] [Google Scholar]
- 29.Matthews P, Tomasz A. Insertional inactivation of the mec gene in a transposon mutant of a methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1990;34:1777–1779. doi: 10.1128/aac.34.9.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews P R, Reed K C, Stewart P R. The cloning of chromosomal DNA associated with methicillin and other resistances in Staphylococcus aureus. J Gen Microbiol. 1987;133:1919–1929. doi: 10.1099/00221287-133-7-1919. [DOI] [PubMed] [Google Scholar]
- 31.Maugein J, Pellegrin J L, Brossard G, Fourche J, Leng B, Reiffers J. In vitro activities of vancomycin and teicoplanin against coagulase-negative staphylococci isolated from neutropenic patients. Antimicrob Agents Chemother. 1991;35:1919–1922. doi: 10.1128/aac.34.5.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCracken G H, Jr, Shinefield H R. Changes in the patterns of neonatal septicemia and meningitis. Am J Dis Child. 1996;112:33–39. doi: 10.1001/archpedi.1966.02090100069006. [DOI] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility test. Approved standard M2-A5. 5th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Sixth informational supplement: M100-S6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1995. [Google Scholar]
- 35.Nesin M, Projan S J, Kreiswirth B, Bolt J, Novick R P. Molecular epidemiology of Staphylococcus epidermidis blood isolates from neonatal intensive care unit patients. J Hosp Infect. 1995;31:111–121. doi: 10.1016/0195-6701(95)90166-3. [DOI] [PubMed] [Google Scholar]
- 36.Santos Sanches I, Ramirez M, Troni H, Abecassis M, Padua M, Tomasz A, De Lencastre H. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J Clin Microbiol. 1995;33:1243–1246. doi: 10.1128/jcm.33.5.1243-1246.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanyal D, Johnson A P, George R C, Edwards R, Greenwood D. In-vitro characteristics of glycopeptide resistant strains of Staphylococcus epidermidis isolated from patients on CAPD. J Antimicrob Chemother. 1993;32:267–278. doi: 10.1093/jac/32.2.267. [DOI] [PubMed] [Google Scholar]
- 38.Sieradzki K, Tomasz A. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J Bacteriol. 1997;179:2557–2566. doi: 10.1128/jb.179.8.2557-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sieradzki K, Villari P, Tomasz A. Decreased susceptibilities to teicoplanin and vancomycin among coagulase-negative methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1998;42:100–107. doi: 10.1128/aac.42.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sieradzki K, Roberts R B, Serur D, Hargrave J, Tomasz A. Heterogeneously vancomycin-resistant Staphylococcus epidermidis strains causing recurring peritonitis in a dialysis patient during vancomycin therapy. J Clin Microbiol. 1999;37:39–44. doi: 10.1128/jcm.37.1.39-44.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sieradzki K, Roberts R B, Haber S W, Tomasz A. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N Engl J Med. 1999;340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 42.Strausbaugh L J. Vancomycin-intermediate Staphylococcus epidermidis: curio or omen? Infect Control Hosp Epidemiol. 1999;20:163–165. doi: 10.1086/501604. [DOI] [PubMed] [Google Scholar]
- 43.Teixeira L A, Resende C A, Ormonde L R, Rosenbaum R, Figueiredo A M S, de Lencastre H, Tomasz A. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J Clin Microbiol. 1995;33:2400–2404. doi: 10.1128/jcm.33.9.2400-2404.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomasz A, Nachman S, Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1991;35:124–129. doi: 10.1128/aac.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valvano M A, Harstein A I, Morthland V H, Dragoon M E, Potter S A, Reynolds J W, Crosa J H. Plasmid DNA analysis of Staphylococcus epidermidis isolated from blood and colonization cultures in very low birth weight neonates. Pediatr Infect Dis J. 1988;7:116–120. doi: 10.1097/00006454-198802000-00010. [DOI] [PubMed] [Google Scholar]
- 47.Vermont C L, Hartwing N G, Fleer A, de Man P, Verbrugh H, van den Anker J, de Groot R, van Belkum A. Persistence of clones of coagulase-negative staphylococci among premature neonates in neonatal intensive care units: two-center study of bacterial genotyping and patient risk factors. J Clin Microbiol. 1998;36:2485–2490. doi: 10.1128/jcm.36.9.2485-2490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villari P, Farullo C, Torre I, Nani E. Molecular characterization of methicillin-resistant Staphylococcus aureus (MRSA) in a university hospital in Italy. Eur J Epidemiol. 1998;14:807–816. doi: 10.1023/a:1007506824091. [DOI] [PubMed] [Google Scholar]
- 49.Villari P, Iacuzio L, Torre I, Scarcella A. Molecular epidemiology as an effective tool in the surveillance of infections in the neonatal intensive care unit. J Infect. 1998;37:274–281. doi: 10.1016/s0163-4453(98)92107-7. [DOI] [PubMed] [Google Scholar]
- 50.Wenzel R P, Edmond M B. Vancomycin-resistant Staphylococcus aureus: infection control considerations. Clin Infect Dis. 1998;27:245–251. doi: 10.1086/514646. [DOI] [PubMed] [Google Scholar]