Abstract
RNA editing in kinetoplastid mitochondria inserts and deletes uridylates at multiple sites in pre-mRNAs as directed by guide RNAs. This occurs by a series of steps that are catalyzed by endoribonuclease, 3′-terminal uridylyl transferase, 3′-exouridylylase, and RNA ligase activities. A multiprotein complex that contains these activities and catalyzes deletion editing in vitro was enriched from Trypanosoma brucei mitochondria by sequential ion-exchange and gel filtration chromatography, followed by glycerol gradient sedimentation. The complex size is approximately 1,600 kDa, and the purified fraction contains 20 major polypeptides. A monoclonal antibody that was generated against the enriched complex reacts with an ∼49-kDa protein and specifically immunoprecipitates in vitro deletion RNA editing activity. The protein recognized by the antibody was identified by mass spectrometry, and the corresponding gene, designated TbMP52, was cloned. Recombinant TbMP52 reacts with the monoclonal antibody. Another novel protein, TbMP48, which is similar to TbMP52, and its gene were also identified in the enriched complex. These results suggest that TbMP52 and TbMP48 are components of the RNA editing complex.
Several mitochondrial RNAs are posttranscriptionally edited in kinetoplastid protozoa by the insertion and deletion of uridylates (U's) at multiple sites, to produce mature mRNAs. RNA editing creates initiation and termination codons and the likely functional open reading frames (ORFs). Indeed, translation of edited RNA has recently been directly demonstrated (11). The RNA editing appears to regulate mitochondrial respiration in different life cycle stages of Trypanosoma brucei. The insertion and deletion of U's is directed by small RNAs that are called guide RNAs (gRNAs). The editing occurs by a series of enzymatic steps. These steps include gRNA-directed cleavage of the pre-mRNA by endoribonuclease, U addition or removal at the 3′ end of the 5′ cleavage product by 3′-terminal uridylyl transferase (TUTase) or 3′-exouridylylase, respectively, and ligation of 5′ and 3′ cleavage products by RNA ligase (reviewed in references 6, 13, and 28).
RNA editing occurs in association with a ribonucleoprotein complex which sediments at 20S in glycerol gradients (4, 22). Fractionation and hence partial purification of the complex by glycerol gradient and liquid chromatographic techniques have been reported (4, 18, 22, 24). For the most part, these preparations were insufficient to identify specific proteins that are part of the editing complex. However, Rusché et al. (24) suggested that a complex of eight proteins could catalyze editing. They concluded that three of these proteins were adenylylatable and suggested that they represented the editing RNA ligase, although the role of these proteins has not yet been demonstrated. Indeed, little progress has been made on the definitive identification of proteins that are components of the editing complex. Three T. brucei mitochondrial proteins, gBP21 (15), DEAD box protein mHEL61p (19), and REAP1 (18), were identified as candidate components of the editing complex. In addition, two T. brucei mitochondrial poly(U) binding proteins, TBRGG1 (30) and RBP16 (10), were identified and suggested to have a role in RNA editing. Knockout of both gBP21 alleles (i.e., null mutations) had no effect on RNA editing in bloodstream-form T. brucei in vivo, indicating that gBP21 is not essential for editing (16). However, knockout of both mHEL61 alleles resulted in slow-growing insect procyclic forms. These cells are capable of in vitro editing but have a >70% reduction in edited mRNAs in vivo, which is restored upon reexpression of mHEL61p (19). These data suggest that mHEL61p may be a component of the editing complex, although not an essential one. Similar assays of the other candidate editing complex proteins have not yet been published.
The difficulty in identifying the protein components of the RNA editing complex reflects the apparent low cellular abundance of the complex, the low sensitivity of the in vitro editing assays, and the uncertainty that assays of endonuclease, exonuclease, TUTase, and RNA ligase are specific for activities associated with the intact complex. These factors, in addition to contamination from protein adsorption during fractionation, made protein identification by conventional microsequencing difficult. However, mass spectrometric analysis has been useful for identifying proteins that are present in small amounts and in mixtures of proteins (17). It was successfully used to identify components of multiprotein complexes, such as the U1 snRNP from the yeast Saccharomyces cerevisiae (21). Indeed, in organisms where the complete genome sequence is available, mass spectrometry can be used to identify the gene for virtually any protein that can be visualized by conventional staining methods. Few genomic sequence data were available for T. brucei until recently, as sequence data from the genome sequencing projects have been accumulating rapidly in the databases.
In this study, we report the biochemical fractionation of the RNA editing complex from T. brucei mitochondria. The fractionation was monitored using the in vitro deletion editing assay in an attempt to purify the complex that is capable of all steps of editing. The editing complex was isolated by sequential ion-exchange and gel filtration chromatography followed by sedimentation on a glycerol gradient. Two novel related proteins in the most purified fraction and their genes were identified using capillary liquid chromatography-tandem mass spectrometry (LC-MS/MS) and by comparison to the T. brucei genome sequence database. They were designated TbMP52 and TbMP48, based on the predicted mass of the preprocessed protein. One monoclonal antibody (MAb) from a panel that was generated against the isolated complex was specific for TbMP52 in Western analyses of native and recombinant protein. This MAb also immunoprecipitated the in vitro deletion editing activity. These data strongly suggest that TbMP52 and TbMP48 are components of the editing complex.
MATERIALS AND METHODS
Cell growth and isolation and fractionation of mitochondrial proteins.
T. brucei procyclic forms (strain IsTaR 1.7a) were grown to log phase in vitro as described previously (29). The mitochondrial vesicles were isolated (9) and stored at −70°C. Mitochondria from 5.1 × 1011 cells were lysed in 55 ml of buffer SP-A (10 mM Tris [pH 7.0], 10 mM MgCl2, 50 mM KCl, 1 mM dithiothreitol [DTT]) containing the protease inhibitors leupeptin (10 μg/ml), pepstatin (5 μg/ml), and Pefablock (1 μM). The lysis was carried out using 0.5% Triton X-100 for 15 min at 4°C with bidirectional mixing. The lysate was cleared by centrifugation at 17,500 × g for 30 min at 4°C. The cleared lysate was filtered through 0.2-μm-pore-size membranes and loaded onto a 10-ml SP Sepharose HR column (Pharmacia) at a 1-ml/min flow rate. All chromatographic steps were carried out using an automated fast protein liquid chromatography system (LKB-Pharmacia) at 4°C. The unbound proteins were washed away with 5 column volumes of buffer SP-A. The bound proteins were eluted with 80 ml of a linear salt gradient of 50 to 330 mM KCl, followed by an 80-ml linear gradient to 1 M KCl at a 2-ml/min flow rate. Fractions of 4 ml were collected and assayed for in vitro deletion RNA editing activity. All positive fractions (9 to 19) were pooled and further fractionated on a Q Sepharose column. Two HiTrap Q 1-ml columns (Pharmacia) were joined in series and equilibrated with buffer Q-A (10 mM Tris [pH 8.3], 10 mM MgCl2, 50 mM KCl, 1 mM DTT). The pooled fractions from the SP Sepharose column were diluted and pH adjusted to the same conditions as buffer Q-A. The sample was loaded onto a Q Sepharose column at a 1-ml/min flow rate, and unbound proteins were washed away with 5 column volumes of buffer Q-A. The elution was carried out with 16 ml of linear salt gradient to 330 mM KCl, followed by a 14-ml linear gradient to 1 M KCl, at a 0.5-ml/min flow rate. Fractions of 1 ml were collected and assayed for deletion RNA editing activity. The positive fractions (11 to 20) were pooled, and Triton X-100 was added to a final concentration of 0.1%. The sample was concentrated to 1/10 volume using Centricon-YM50 membrane (Amicon) at 3,000 × g. The proteins were size fractionated with a Superose 6 HR (10/30) column (Pharmacia). In each run a 250-μl sample was loaded onto the column at a 0.2-ml/min flow rate (the buffer was 10 mM Tris [pH 7.0], 10 mM MgCl2, 200 mM KCl, and 1 mM DTT). Fractions of 500 μl were collected and assayed for deletion RNA editing. The size of the complex in the peak editing fraction was estimated in comparison to globular protein size standards (gel filtration high-molecular-weight calibration kit; Pharmacia). The peak positive fractions (19 to 22) were pooled and concentrated as described above and sedimented on a 10-to-30% linear glycerol gradient. An 11-ml linear gradient was prepared in 10 mM Tris [pH 7.0]–10 mM MgCl2–100 mM KCl, and 500 μl of sample was layered on top of it. After centrifugation at 38,000 rpm for 5 h at 4°C (SW40 rotor; Beckman), 500-μl fractions were collected from the top and assayed for deletion RNA editing activity.
In vitro deletion editing.
The in vitro deletion editing assay was carried out using 3′-labeled A6-U5 pre-mRNA substrate and gA6[14]Δ16G gRNA as described previously (26). The reaction was carried out for 2 h in a 30-μl final volume using 4 to 8 μl of test sample. Reaction products were run on 9% polyacrylamide–7 M urea gels and were detected with a Storm PhosphorImager screen (Molecular Dynamics). Quantification was performed with ImageQuaNT software.
MAb.
The RNA editing complex was fractionated in three batches as described above, from a total of 1.64 × 1012 cells. The peak editing fractions from glycerol gradients were pooled and concentrated using a Centricon-YM50 membrane and used as immunogens. MAbs were produced at Biologics Production Facility, Fred Hutchinson Cancer Research Center, Seattle, Wash. Supernatants from hybridomas were screened for production of antibody against the editing complex (sequentially fractionated by SP Sepharose, Q Sepharose, and Superose 6 columns) using the enzyme-linked immunosorbent assay (ELISA). All ELISA-positive samples were screened by Western blot analysis using SP and Q Sepharose-fractionated editing complexes as the antigens. The contents of 15 ELISA- and Western blot-positive hybridoma wells were subcloned by dilution. The supernatants were screened for MAb production by both ELISA and Western analyses.
Western blot analysis.
RNA editing complex that was partially purified by SP and Q Sepharose chromatography as described above was used as the antigen for Western analysis. One microgram of protein sample was separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a nitrocellulose filter. The filter was blocked overnight with 5% nonfat milk powder in PBST (10 mM phosphate buffer [pH 7.2], 150 mM NaCl, 0.05% Tween 20) at 4°C. It was washed three times with PBST and incubated with tissue culture supernatant diluted 1:100 in PBST. The incubation was carried out at room temperature for 2 h with gentle shaking. The filter was washed three times with PBST and incubated for 1 h with horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) (Bio-Rad) at a 1:2,000 dilution in PBST. The filter was washed four times with PBST and developed with an ECL kit (Amersham) according to the manufacturer's instructions. To assay the distribution of TbMP52 in the glycerol gradient, 5 μl of sample from each fraction was separated on an SDS-PAGE gel and examined by Western analysis.
Immunoprecipitation of editing complex.
Immunomagnetic beads (Dynabeads M-450; Dynal) coated with goat anti-mouse IgG were coupled to MAbs from tissue culture supernatants. A total of 4 × 107 beads were incubated with 1 ml of tissue culture supernatant and 1% bovine serum albumin at 4°C with bidirectional mixing for 1 h. The beads were washed three times with immunoprecipitation buffer (IP buffer; 10 mM Tris [pH 7.2], 10 mM MgCl2, 200 mM KCl, 0.1% Triton X-100). The mitochondria were lysed in IP buffer with 0.5% Triton X-100, cleared by centrifugation, and fractionated on a 10-to-30% glycerol gradient as described above. The antibody-bound beads were incubated with the mitochondrial 20S fraction (50 μg of proteins) in 1× IP buffer and 1% bovine serum albumin for 1 h at 4°C with bidirectional mixing. The beads were washed four times (each wash of 5 min duration with bidirectional mixing) with IP buffer and twice with buffer containing 25 mM HEPES (pH 7.9), 10 mM magnesium acetate [Mg(OAc)2], 50 mM KCl, 1 mM EDTA, and 0.1% Triton X-100 and directly assayed for deletion editing and adenylylatable proteins. Similarly, immunoprecipitation experiments were carried out with IP buffer containing 300 or 400 mM KCl, and beads were assayed for deletion editing and adenylylatable proteins. In addition, the mitochondrial 20S fraction was first adenylylated with [α-32P]ATP and immunoprecipitated as described above with IP buffer containing 400 mM KCl.
Adenylylation.
The presence of the adenylylatable proteins in the samples was determined as described earlier (25), with some modifications. Adenylylation reactions were carried out with 2.5 μCi of [α-32P]ATP in buffer containing 25 mM HEPES (pH 7.9), 10 mM Mg(OAc)2, 50 mM KCl, 0.5 mM DTT, and 10% dimethyl sulfoxide for 15 min at 28°C. The proteins were separated by SDS-PAGE, and radiolabeled proteins were detected by phosphorimaging.
Identification of proteins.
The proteins were separated on an SDS-PAGE gel (20 cm long) and visualized either by silver nitrate or Coomassie blue staining. The protein bands were excised from the gel, and in-gel tryptic digestion was carried out as described previously (27). The tryptic peptides were analyzed by microcapillary LC-MS with automated switching to MS/MS mode for peptide fragmentation and sequence analysis (7). The collision-induced dissociation (CID) spectra were compared with the OWL nonredundant protein sequence database and then with a trypanosome nucleotide database. The database included Trypanosoma genomic and expressed sequence tag sequences from the National Center for Biotechnology Information database and from The Institute for Genomic Research (http://www.tigr.org) and Sanger Center (http://www.sanger.ac.uk) genome sequencing projects. The search was carried out against all six ORFs for nucleotide sequences using the SEQUEST program, which matches theoretical and acquired tandem mass spectra (31). A protein match was determined from the number of identified peptides, their cross-correlation scores, and their predicted theoretical molecular weights compared to migration in the gel.
Cloning and expression of TbMP52 and TbMP48.
The entire ORF representing TbMP52 was amplified from genomic DNA with primers Tb10-2261P (ACT GCA GAT GCA ACT CCA AAG G) and Tb10-3690M (AGA ATT CGC AGT AT CAT TCG CC) (the restriction sites are italicized). The amplified DNA was cloned into the pGEM-T easy vector (Promega), and the sequence was confirmed. The insert was released with PstI and EcoRI enzymatic digestion and cloned into the pRSET C vector (Invitrogen) cut with the same enzymes. Similarly, the TbMP48 ORF was PCR amplified with the primer pairs Tb07-1423P (TGG ATC CTG AAG ATG TTG CGT C) and Tb07-2679M (AGG TAC CAT TCG CTA AAG TCA GG) and cloned into the pRSET C vector at the BamHI-KpnI site. The plasmids were transformed into BL21 DE3-LysS cells, and recombinant proteins were expressed with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) induction. The Escherichia coli cells expressing recombinant proteins were separated by SDS-PAGE and either stained with Coomassie blue or transferred onto nitrocellulose paper. These were reacted with individual MAb supernatants (1:100 dilution) as described above. E. coli cells transformed with vector only were used as negative controls.
Subcellular localization of TbMP52 by immunofluorescence.
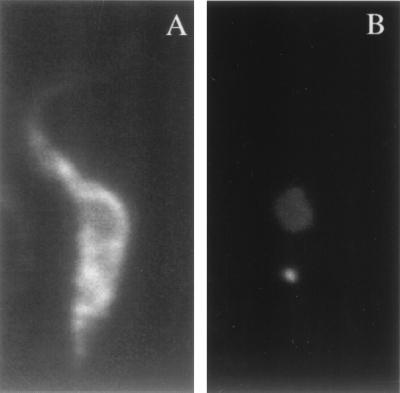
Procyclic-form trypanosomes were fixed onto microscope slides, and immunofluorescence reactions were carried out as previously described (1). Briefly, the cells were incubated with MAb P3C1-G2 (tissue culture supernatant diluted 1:10 in 1× phosphate-buffered saline [PBS]) for 1 h, washed, and incubated with fluorescein isothiocyanate-conjugated goat anti-mouse IgG (diluted 1:200 in PBS) for 1 h. The washed cells were treated with 4,6-diamidino-2-phenylindole (DAPI; 0.5 μg/μl in PBS) to stain DNA. Fluorescence was observed with a Nikon fluorescence microscope equipped with the appropriate filters.
Nucleotide sequence accession numbers.
The nucleotide and protein sequences have been submitted to GenBank and SWISS-PROT with accession numbers AY009110, AY009111, P82863, and P82864.
RESULTS
Enrichment of RNA editing complex.
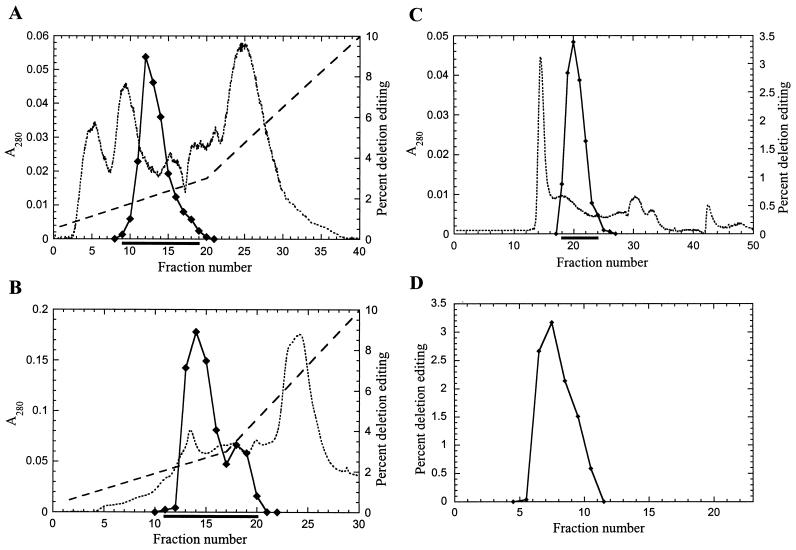
The RNA editing complex was enriched from T. brucei mitochondrial lysate by sequential fractionation by two ion-exchange columns, a gel filtration column, and glycerol gradient sedimentation (Fig. 1). The fractionation of the functional complex was monitored using the in vitro assay for deletion editing (26). The peak of in vitro editing activity from cleared mitochondrial lysate, which was prepared as described in Materials and Methods, eluted from the SP Sepharose (cation-exchange) column at about 200 mM KCl (Fig. 1A). Similarly, the peak of the editing activity from the pooled SP Sepharose fractions containing the editing activity eluted from the Q Sepharose (anion-exchange) column at about 225 mM KCl (Fig. 1B). There was no detectable editing activity in the fractions that did not bind to these columns. The editing activity from the pooled active fractions from the Q Sepharose column eluted from the Superose 6 (gel filtration) column with a peak which centered at an elution position corresponding to about 1,600 kDa compared to globular protein size standards (Fig. 1C and data not shown). The buffer used for size fractionation on the Superose 6 column contained 200 mM KCl in order to avoid nonspecific association of other proteins with the editing complex. We observed that the complex eluted from Superose 6 with the same apparent mass when a buffer containing up to 300 mM KCl was used. However, it eluted with a greater apparent mass (in void volume) with a buffer containing 50 mM KCl (data not shown), perhaps indicating its association with other proteins or complexes or self aggregation. The pooled peak of deletion editing activity from the Superose 6 column, as well as the bulk of the total protein (see Fig. 2), sedimented with a peak centered at ∼20S on a 10-to-30% glycerol gradient (Fig. 1D). The fractions containing the editing activity represent about 1/8,500 of the protein in the original cleared mitochondrial lysate. The fractions positive for RNA editing contained precleaved insertion editing (12), endonuclease (8), TUTase (4), 3′-exouridylylase, and RNA ligase activities, as well as adenylylatable proteins (25) (results not shown). Northern analysis revealed the presence of gRNAs and preedited mRNAs in the peak editing fraction isolated by sequential ion-exchange and gel filtration chromatography (results not shown).
FIG. 1.
Fractionation of RNA editing complex from T. brucei mitochondria. In vitro deletion RNA editing was used as the functional assay to monitor purification of the complex. (A) Cleared mitochondrial lysate prepared with 0.5% Triton X-100 was fractionated on an SP Sepharose column. (B and C) Fractions containing editing activity, as indicated by the dark lines below each panel, were sequentially fractionated on Q Sepharose (B) and Superose 6 (C) columns. (D) The complex was further purified by sedimentation on a 10-to-30% glycerol gradient, with fraction 1 being the top of the gradient. Diamonds, deletion editing; dotted line, absorbance at 280 nm; dashed line, KCl gradient profile (the KCl concentration [molar units] is the value on the righthand y axis divided by 10).
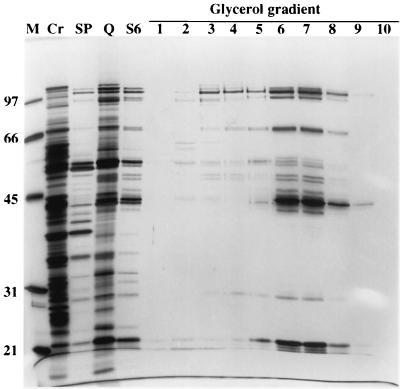
FIG. 2.
SDS-PAGE profile of fractions from complex purification. A sample from each step of purification was separated by SDS-PAGE and stained with silver nitrate. Results for protein size standards (M), cleared mitochondrial lysate (Cr), and pooled editing activity-positive fractions from SP Sepharose (SP), Q Sepharose (Q), and Superose 6 (S6) columns are presented. Glycerol gradient fractions 1 to 10 (fraction 1 is at the top) are shown (fractions 11 to 23 are not shown since essentially no protein was detected in these fractions). The numbers on left indicate the sizes of molecular mass markers, in kilodaltons. The most purified editing activity-positive fraction from the glycerol gradient (7) shows 20 major polypeptide bands.
The protein profiles of the pooled active fractions from each column and from the glycerol gradient fractions were analyzed by electrophoresis in SDS-PAGE followed by silver staining (Fig. 2). The protein profiles from each pooled fraction from the columns were substantially different from each other and showed a progressively simpler pattern. This is consistent with the absorbance profiles from the columns, which indicate that most of the proteins that are not associated with the editing activity were eliminated. The profile of the pooled fractions from the Superose 6 column was very similar to that of the glycerol gradient fractions that showed the greatest editing activity. Examination of glycerol gradient fraction 7, which contains the greatest editing activity, reveals 20 major polypeptides with apparent masses of 20, 22, 29, 34, 42, 43, 44, 45, 47, 49, 50, 53, 55, 57, 69, 72, 90, 99, 106, and 114 kDa. While the 22-, 44-, and 90-kDa protein bands stained more intensely than the other proteins, this may or may not reflect the ratio of the proteins in the complex, since different proteins stain at different intensities with silver nitrate. In addition, editing was not detected in fractions 2 to 4, although many of these 20 proteins were present, and there was more editing in fraction 9 than in fraction 5, although it had less protein (Fig. 1D and 2). Perhaps there is a smaller proportion of complete (i.e., fully functional) complexes in the upper (lower sedimentation value) fractions than in fraction 9.
Immunoprecipitation of editing complex using MAbs.
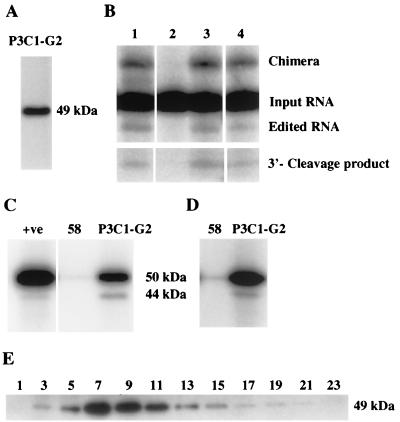
A panel of MAbs was generated using the enriched complex as the immunogen, to generate reagents that are specific to components of the editing complex and that can be used to identify and characterize these components. A total of 19 independent MAbs were isolated, and they were directed against seven different proteins, based on Western analyses. One MAb, P3C1-G2, reacted with an ∼49-kDa protein in Western analyses and immunoprecipitated in vitro deletion editing activity (Fig. 3A and B). The immunoprecipitations and washes were performed with the 20S fraction of glycerol gradient-fractionated mitochondrial lysate using 0.1% nonionic detergent and either 200, 300, or 400 mM KCl (to reduce nonspecific associations) as described in Materials and Methods. The editing activity did not immunoprecipitate with MAb 58 (Fig. 3B, lane 2), which is directed against the T. brucei mitochondrial pyruvate dehydrogenase E2 subunit (reference 1 and unpublished results) or with MAb P7D9-A12, which reacts with a protein in an unidentified 40S complex. Even material that was immunoprecipitated with 400 mM KCl was able to catalyze in vitro editing, although the activity was reduced by 30% compared to the activity of the material that was immunoprecipitated with 200 mM KCl (Fig. 3B, lane 4). The immunoprecipitated material also contained component editing activities (precleaved insertion editing, endonuclease, TUTase, 3′-exouridylylase, and RNA ligase activities [results not shown]). This demonstrates that MAb P3C1-G2 can immunoprecipitate the active editing complex.
FIG. 3.
Immunoanalysis of the editing complex using a MAb. (A) MAb P3C1-G2 raised against the purified complex reacts with an ∼49-kDa protein in Western analysis of a partially purified complex. (B) MAb P3C1-G2 specifically immunoprecipitates editing activity from the 20S mitochondrial fraction (see Materials and Methods for details) (lanes 3 and 4). Edited RNA, chimeras, and 3′ cleavage products and the input RNA from which they are derived are indicated. MAb 58 (1), the negative control, did not immunoprecipitate these activities (lane 2). Editing activity immunoprecipitated with 200 and 400 mM KCl (lanes 3 and 4, respectively). The positive deletion editing control using the mitochondrial 20S fraction is also shown (lane 1). (C) MAb P3C1-G2 immunoprecipitates both 50- and 44-kDa adenylylatable proteins. MAb 58, which was used as a negative control, essentially did not immunoprecipitate the adenylylation activity. The control (+ve) for these proteins using the mitochondrial 20S fraction is shown. (D) MAb P3C1-G2 also immunoprecipitated both the 50- and 44-kDa proteins from the mitochondrial 20S fraction in a buffer containing 400 mM KCl following their adenylylation. (E) Western analysis of glycerol gradient-fractionated cleared mitochondrial lysate using MAb P3C1-G2.
We also tested the ability of MAb P3C1-G2 to immunoprecipitate the 50- and 57-kDa adenylylatable proteins, which were reported to be RNA ligases with a possible role in RNA editing (25). These proteins were also reported to cofractionate with editing activities (18, 24). The material that was immunoprecipitated by MAb P3C1-G2 contained two adenylylatable proteins (Fig. 3C). In addition, MAb P3C1-G2 was able to immunoprecipitate two proteins that were adenylylated prior to the immunoprecipitation, even in buffer containing 400 mM KCl and 0.1% Triton X-100 (Fig. 3D). Essentially no adenylylatable proteins or preadenylylated proteins immunoprecipitated with MAb 58 (1), which was used as the negative control. Unlike the data in earlier reports, the major adenylylatable and preadenylylated proteins had apparent sizes of 44 and 50 kDa. Western analysis with MAb P3C1-G2 of fractions from a 10-to-30% glycerol gradient of mitochondria that were lysed in buffer with 200 mM KCl showed the bulk of the 49-kDa protein sediments at ∼20S, similar to the deletion editing activity (Fig. 1D and 3E).
Identification of TbMP52 and TbMP48 proteins.
The 49-kDa protein that reacts with MAb P3C1-G2 and its corresponding gene were identified by a combination of high-performance LC-MS/MS and analyses of T. brucei genome sequence data. Mass spectrometric analyses were performed on both the fraction containing the editing complex and on individual bands that were cut from SDS-PAGE gels (e.g., Fig. 2, lanes S6 and 7). LC-MS/MS is highly sensitive and can allow identification of proteins in mixtures (17). Protein was digested in gel with trypsin, and the resulting peptides were fractionated by on-line capillary liquid chromatography and eluted directly into the mass spectrometer, where they were analyzed. Peptide masses and amino acid sequences were determined by automated selection and fragmentation of specific ionized peptides. This entailed switching between MS and MS/MS modes and resulted in the CID MS/MS spectrum of each peptide. Individual CID spectra were compared to predicted spectra from sequence databases by using computer algorithms to identify the gene and hence the protein. This analysis identified multiple peptides that corresponded to multiple CID MS/MS spectra from the same protein.
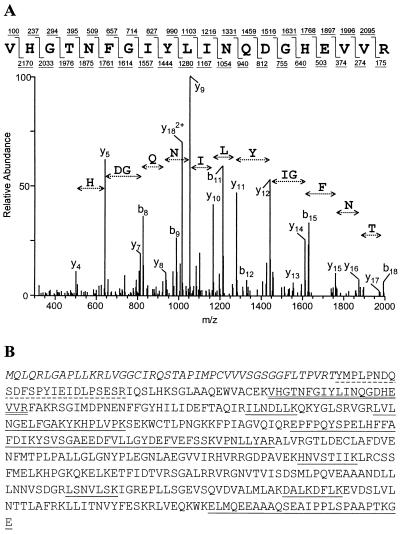
An example of a CID spectrum of a peptide that corresponds to a tryptic peptide with the amino acid sequence VHGTNFGIYLINQGDHEVVR from the 49-kDa protein is shown in Fig. 4A. The results of a database search that compared this CID spectrum with CID spectra predicted from Trypanosoma sequences are shown in Table 1. This search identified a candidate matching peptide sequence in a 1,410-nucleotide ORF in The Institute for Genomic Research T. brucei genome sequencing project database. The confidence level for this match (at positions 50,980 to 52,389; accession no. 7330318 and AC013484.9 [GenBank]; chromosome IX clone RPCI93-1L12) was quite high. Similar analyses identified 10 additional tryptic peptides with matches to this ORF (Fig. 4B). Thus, the probability that this gene corresponds to the 49-kDa protein is very high. The protein predicted from the ORF has a mass of 52 kDa, but its N-terminal sequence has an amphiphilic helix that is predicted by Gene Runner (Hastings Software, Inc.) and a mitochondrial targeting signal that is predicted by the PSORT II algorithm (http://psort.ims.u-tokyo.ac.jp/form2.html). In addition, the CID spectrum of a peptide from the 49-kDa protein identified the peptide (T)YMPLPNDQSDFSPYIEIDLPSESR from this ORF (Fig. 4B). The N-terminal amino acid Y in the peptide could be generated if in vivo processing removed the signal peptide, and the amino acid T, being nontryptic, may be part of the signal peptide. These data are consistent with localization of the 49-kDa protein within the mitochondrion after cleavage of the mitochondrial signal peptide. The protein is designated TbMP52, for “T. brucei mitochondrial protein,” and has a preprocessed molecular mass of 52 kDa; the gene is designated TbMP52. The first 44 amino acids in the N-terminal region may form a signal peptide, and this predicts a 48-kDa mature protein, which is consistent with its apparent migration size of 49 kDa. Immunofluorescence analysis with MAb P3C1-G2 showed staining of the mitochondrion, indicating that TbMP52 is localized in this organelle (Fig. 5).
FIG. 4.
Identification of TbMP52. (A) Sample tandem mass spectrum derived by CID of a peptide precursor ion, m/z 1135.0 (bottom), and the peptide sequence predicted by SEQUEST (top) showing b- and y-type ions (above and below the sequence, respectively). (B) Amino acid sequence of the complete ORF identified by 11 tryptic peptide matches with CID spectra. The identified peptides are underlined (at two different positions, two and three peptides were contiguous). The dashed underline indicates the probable N-terminal peptide in the mature protein that is nontryptic, and the double underline indicates the peptide with the highest correlation score (Table 1). The predicted mitochondrial targeting signal is italicized.
TABLE 1.
SEQUEST output file of the TbMP52 CID spectrum showing the correlation between peptides from the trypanosome database and the observed peptide CID spectrum
| (M+H)+ | Scorea | Database reference | Peptide sequencec |
|---|---|---|---|
| 2269.5 | 4.9026 | 7330318b | (K)VHGTNFGIYLINQGDHEVVR |
| 2268.7 | 2.7907 | 2K9.TV | (S)IRTRLWKVIDVGCRRPVE |
| 2267.7 | 2.6511 | 2312.TF | (Q)SSSYITGEINLLVIRKVVKM* |
| 2268.8 | 2.5782 | 4827345b | (T)KKLRERQINFKKGSLPVLL |
| 2269.8 | 2.5587 | 5119828b | (K)SRKHMAHRIVLLGVINITPL |
Cross-correlation score.
GenBank sequence identification number.
*, modified methionine.
FIG. 5.
Immunofluorescence with MAb P3C1-G2, which is specific for TbMP52. Procyclic T. brucei cells were stained with MAb (A) and DAPI (B), showing the nucleus and smaller kinetoplast.
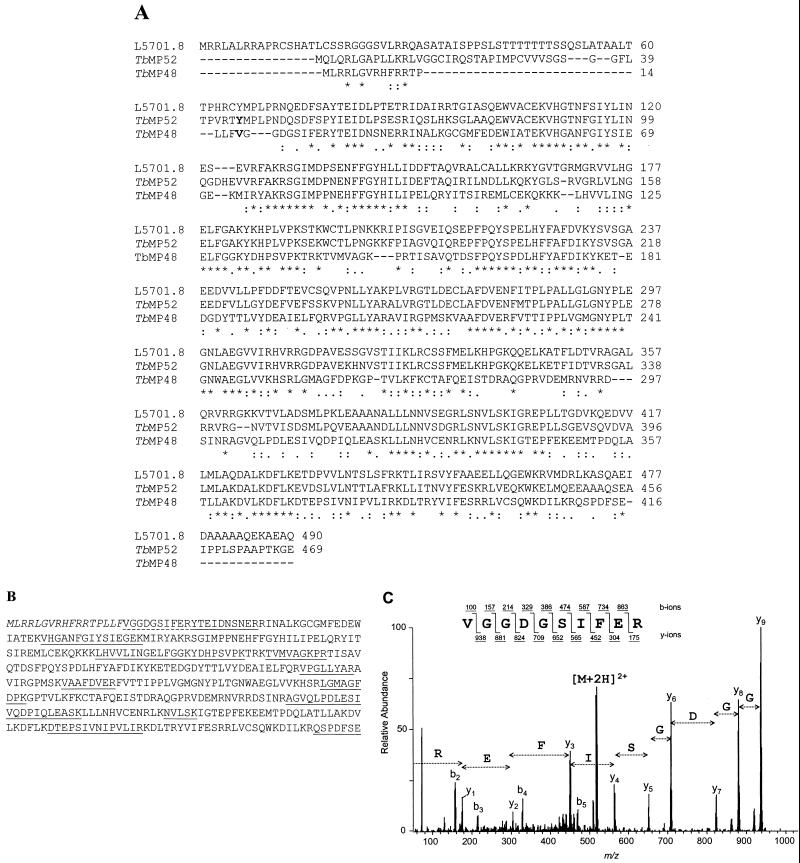
Another gene that is related to TbMP52 and its corresponding protein, which copurifies with the editing activity, were identified by a combination of mass spectrometry and database analyses (Fig. 6). A search of the Sanger Center T. brucei genome project database identified a 1,251-nucleotide ORF (positions 98742 to 97492 of contig TRYP1.0.7383 of chromosome I) that has significant homology to TbMP52 (Fig. 6A). This ORF encodes a 48-kDa protein, and similar to TbMP52, the N-terminal sequence of the 48-kDa protein predicts a mitochondrial targeting signal sequence. There is a deletion/insertion site near the N terminus of TbMP48 relative to TbMP52 that appears to be located within the signal peptide. LC-MS/MS analysis of an ∼44-kDa protein from the SDS-PAGE gel (Fig. 2, lane 7) identified 13 peptides that correspond to peptides predicted from this ORF (Fig. 6B). A peptide with the amino acid sequence (F)VGGDGSIFER was identified from near the N terminus of the predicted protein (Fig. 6B and C and Table 2). The amino acid (F) N-terminal to V, since it is not tryptic, may be part of the mitochondrial targeting signal. Thus, the first 17 amino acids, which also can fold into an amphiphilic helix (results not shown), are predicted to form the mitochondrial targeting signal. Hence, this second protein, designated TbMP48, also appears to be localized in the mitochondrion after removal of the signal peptide, resulting in a 45-kDa mature protein. The mature TbMP48 migrates as an ∼44-kDa band in SDS-PAGE gels.
FIG. 6.
(A) Alignment of predicted amino acid sequences of TbMP52 and related proteins TbMP48 and L5701.8. Potential N-terminal amino acids are in bold. The TbMP48 gene sequence is from contig TRYP1.0.7383 of chromosome I (Sanger Center T. brucei database) and the L5701.8 ORF is from L. major chromosome 1 (20). The alignment indicates amino acids that are conserved (∗), semiconserved (:), and partially conserved (.) among all these proteins. (B) Predicted amino acid sequence of TbMP48 showing the 12 tryptic peptides (two of which were contiguous) that were identified by mass spectrometric analysis (underlined). The first N-terminal peptide (dashed underline) is nontryptic, and the 17 amino acids at the N terminus (italicized) are a predicted mitochondrial targeting signal. (C) CID spectrum of the likely N-terminal peptide of TbMP48.
TABLE 2.
SEQUEST output file for the CID spectrum of TbMP52 N-terminal peptide
| (M+H)+ | Scorea | Database reference | Peptide sequence |
|---|---|---|---|
| 1037.1 | 3.9923 | TRYP1.0.7383 | (F)VGGDGSIFER |
| 1037.2 | 2.9684 | 6A9.TR | (V)VGGIGTTFER |
| 1037.2 | 2.8651 | 1395223b | (A)RFFEAGNVP |
| 1037.1 | 2.7472 | 18L22.TF | (R)VDDSGKMER |
| 1037.1 | 2.7390 | trypEf4.p1p | (S)VDDAYM*IGH |
Cross-correlation score.
GenBank sequence identification number.
Additional BLAST searching of the National Center for Biotechnology Information database revealed that the Leishmania major gene L5701.8 (accession no. AAC24666 [20]) has significant homology to TbMP52 at both the nucleic acid and predicted amino acid sequence levels, while the relationship of TbMP48 to TbMP52 is evident only at the predicted amino acid sequence level. L5701.8 has 66% identity and 78% similarity to TbMP52 at the amino acid level, while TbMP48 has 41% identity and 60% similarity to TbMP52. Thus, L5701.8 appears to be an ortholog of TbMP52, while TbMP48 is a paralog of TbMP52. Database searches for homology, motif searches, a ProfileScan of the PROSITE database, and a search of the BLOCKS database revealed no homologs of these proteins, nor any functional motifs. Thus, these proteins appear to be novel.
Association of TbMP52 and TbMP48 with the RNA editing complex.
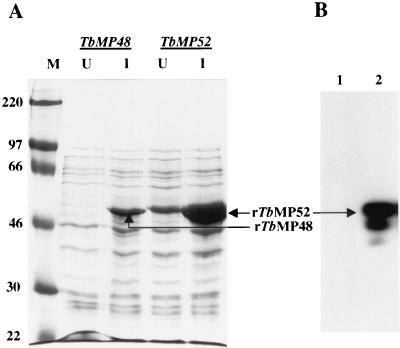
The association of the 45- and 48-kDa proteins with the editing complex was assessed by immunoprecipitation, since the cofractionation of these proteins with the editing activity suggests that they may both be associated with the editing complex. Recombinant TbMP52 (rTbMP52) and rTbMP48 proteins were expressed in E. coli with an N-terminal six-His tag. Abundant expression of these proteins was evident as bands in SDS-PAGE gels of IPTG-induced E. coli that stained intensely with Coomassie blue (Fig. 7A). None of the MAbs that we isolated after immunization with the purified editing complex reacted with rTbMP48. However, Western analysis of the recombinant proteins revealed that MAb P3C1-G2 specifically reacts with rTbMP52 (Fig. 7B). This further confirms that the TbMP52 gene encodes the protein identified by mass spectrometry analysis of the ∼49-kDa protein present in the purified editing complex. This observation, along with the finding that MAb P3C1-G2 specifically immunoprecipitates RNA editing activity from the 20S mitochondrial fraction in high KCl concentrations (Fig. 3B), shows that TbMP52 is tightly associated with the RNA editing complex. Peptides corresponding to both TbMP52 and TbMP48 were identified in the immunoprecipitated editing complex by mass spectrometry analysis (results not shown).
FIG. 7.
Expression of rTbMP48 and rTbMP52 with an N-terminal six-His tag. (A) Coomassie blue-stained gel of total E. coli lysates separated on SDS-PAGE showing protein size standards (M; sizes [in kilodaltons] are on the left), uninduced cells (U), and cells 3 h after induction with 1 mM IPTG (I). (B) Western analysis showing the reaction of MAb P3C1-G2 with rTbMP52 (lane 2). E. coli cells expressing rTbMP48 (lane 1) were used as a negative control.
DISCUSSION
This study reports enrichment of the RNA editing complex from T. brucei mitochondria and the identification of two genes that encode 45- and 48-kDa (mature) proteins that are tightly associated with the editing complex. The complex was fractionated by a combination of cation-exchange, anion-exchange, and gel filtration chromatography followed by glycerol gradient sedimentation using the in vitro deletion editing assay to monitor purification. The enriched complex contains all of the catalytic activities that are associated with editing. These include the gRNA-directed endoribonuclease, 3′-TUTase, 3′-exouridylylase, and RNA ligase (and adenylylation) activities that are predicted by models of the general mechanism of editing (2, 14, 26). A MAb that was prepared by using the enriched complex is specific for both native and recombinant 48-kDa protein and also immunoprecipitates the in vitro deletion editing activity as well as the associated catalytic activities.
The fractionation reported here differs from previously reported purification of the editing complex (18, 24). This study used in vitro editing to monitor purification rather than following purification of adenylylatable proteins. It also enriched the complex sequentially with SP Sepharose (cation-exchange), Q Sepharose (anion-exchange), and Superose 6 (gel filtration) columns followed by sedimentation in a glycerol gradient. The purification achieved could not be calculated based on specific activity since the in vitro editing assay is not linear (our unpublished results). However, purification from cleared mitochondrial lysate is estimated to be ∼8,500-fold based on protein recovery and substantially more based on total cellular protein. The in vitro editing assays requires that all steps in editing occur. The intent of the purification approach used in this study was to select for the fully functional complex and to avoid activities that may resemble but not be part of the editing complex.
The complex that is fully functional in editing has an apparent mass of 1,600 kDa, based on gel filtration chromatography, and it sediments at ∼20S. Several of the editing-associated activities, such as endonuclease, TUTase, and RNA ligase activities, elute in a second peak with an apparent mass of 500 kDa (Panigrahi et al., unpublished results). Madison-Antenucci et al. (18) reported two complexes with masses of ∼700 and ∼450 kDa. The smaller masses may reflect the differences in isolation protocol used in the study. In addition, the isolated ∼1,600-kDa complex is capable of in vitro editing in the present study, while this was not shown for the complex isolated by Madison-Antenucci et al. (18). The complex isolated here contained gRNAs and preedited mRNAs, similar to that described by Madison-Antenucci et al. (18) but unlike the complex reported by Rusché et al. (24).
The composition of the fully functional editing complex is not known. The most purified fraction reported here contains 20 major proteins (Fig. 2) that together total 1,140 kDa, but at least three of these proteins may be contaminants, and editing complex proteins may not all be present in a 1:1 stoichiometry (Panigrahi et al., unpublished). The most purified fraction reported by Rusché et al. (24) contained eight major proteins, and those authors suggested that three of the proteins may represent RNA ligase based on their ability to be adenylylated. The most purified fraction isolated by Madison-Antenucci et al. (18) contained 13 major proteins. Since abundance of proteins in a purified fraction is not definitive evidence that a protein is a component of the editing complex, determination of which are components of the complex will await independent evidence. It seems unlikely that the fully functional editing complex contains only eight polypeptides, as suggested by Rusché et al. (24). Those authors suggested that three of the proteins may represent RNA ligase and that editing requires proteins that perform the endoribonuclease, exouridylylase, and TUTase functions. It is also likely that some proteins are involved in RNA-RNA positioning, annealing, and unwinding functions. Indeed, the complex isolated here contains gRNAs and preedited mRNAs, indicating the presence of RNA binding proteins. A helicase activity appears to be associated with RNA editing based on cofractionation (4) and experiments showing that mitochondrial helicase null mutants have reduced editing (19). Thus, helicase may be a component of the editing complex, although not absolutely essential for editing. The pre-mRNA and gRNA must be bound by the complex, perhaps by the anchor duplex, and the 5′ cleavage fragment must be retained after cleavage by the endoribonuclease. It also seems likely that proteins are needed to maintain the structure of the editing complex in order to position the catalytic sites of the editing enzymes and relocate the pre-mRNA and gRNA as each site is edited.
Several proteins have been suggested to be components of the editing complex based on approaches other than purification of the complex. Several proteins which specifically cross-link to gRNA upon UV irradiation have been described. Read et al. (23) demonstrated cross-linking of gRNA to 25- and 90-kDa proteins upon incubation with mitochondrial extract. The 90-kDa protein was shown to be specific for oligo(U), as is the case for the 16-kDa RBP16 (10) and the 75-kDa TBRGG1 (30). Allen et al. (1) identified a 55-kDa gRNA binding protein in an enriched RNA editing fraction. Wang et al. (personal communication) identified 50- and 70-kDa proteins in a partially purified (sequential SP Sepharose- and Q Sepharose-fractionated) editing complex fraction that specifically bind gRNA. The roles of these proteins in RNA editing have not been demonstrated. Indeed, while MAbs against gBP21, the 25-kDa protein that specifically binds gRNA, immunoprecipitate editing activity, this immunoprecipitation is prevented by micrococcal nuclease treatment (1), and gBP21 null mutants perform editing normally (16). Thus, while it appears that gBP21 is an RNA binding protein, its role in editing is uncertain. It does not appear to be an essential component of the editing complex and thus may have an accessory role, such as bringing gRNA to the complex. Alternatively, it may have precipitated editing activity simply based on its affinity for RNA. The 110-kDa glutamate dehydrogenase specifically cross-links to RNA in Leishmania tarentolae (3), but the knockout of the gene for this protein in T. brucei had no effect on editing (5). The roles of the other RNA binding proteins have not been tested. REAP1 was identified using MAbs made to an ∼40S fraction from glycerol gradient fractionation of a mitochondrial lysate. This antibody recognizes proteins in the ∼20S and ∼40S fractions, where in vitro editing is detected, and upon incubation with the mitochondrial fraction, the antibody inhibits in vitro insertion editing, suggesting that this protein may be a functional component of the complex (18). At this time, the role of REAP1 in RNA editing has not been determined.
The two proteins and corresponding genes that we identified here using a combination of mass spectrometry and immunoprecipitation and Western analyses are candidate components of the editing complex. A MAb that was raised against the purified native complex is specific for a 48-kDa protein that is primarily localized in the 20S fraction in a glycerol gradient of total cleared mitochondrial lysate (Fig. 3E), where editing primarily sediments. The MAb also immunoprecipitates in vitro deletion editing activity from the 20S fraction in addition to the precleaved insertion editing, endonuclease, TUTase, 3′-exouridylylase, and RNA ligase activities that are associated with editing. Furthermore, it immunoprecipitates the adenylylation activity and adenylylated proteins (Fig. 3C and D), which have been suggested to be the editing-associated RNA ligases. The immunoprecipitation was specific, since it occurred even in 400 mM KCl with 0.1% nonionic detergent and did not occur with antibodies that do not react with proteins in the fraction which contains the most in vitro editing activity. The 45-kDa protein is related to the 48-kDa protein and also cofractionates with the in vitro deletion editing activity. Both the 45- and 48-kDa proteins were identified in the immunoprecipitated sample by mass spectrometry (Panigrahi et al., unpublished). The genes for each of these proteins predict a mitochondrial targeting signal sequence, and TbMP52 was localized in mitochondria using the MAb. Thus, taken together the evidence strongly suggests that both proteins are associated with the editing complex. TbMP52 is an ortholog of the L. major gene L5701.8 (20), which has no known function. Database searching found no other homologs or functional motifs in the databases, and thus we are not able to assign a function to these proteins. Further biochemical and genetic studies on the roles of these proteins in RNA editing are in progress. Preliminary results suggest that TbMP52 has RNA ligase activity (Schnaufer et al., unpublished results).
Overall no protein has been shown to be essential for RNA editing. Of the candidates identified to date, a functional association with RNA editing has been demonstrated only for RNA helicase, mHEL61p (19). While several other candidates have been identified by a variety of approaches, functional analyses such as knockout and genetic modification in vivo have yet to show a definitive role in RNA editing or have yet to be done. However, the list of candidates has increased and the criteria for a possible role in editing have improved, suggesting that functions in editing will be demonstrated for several proteins in the near future. For example, what are the specific functions of the individual protein components of the complex, what is the structure of the editing complex and does it have subunits, and what is the biogenesis of the editing complex? Such studies are likely to make progress toward answering these and other questions about the editing complex.
ACKNOWLEDGMENTS
We thank Barbara Morach, Brian Panicucci, and RoseMary Reed for technical help, Bingbing Wang for sharing unpublished results, Elizabeth Wayner for MAb production, and Peter Myler for helpful suggestions. We thank Najib M. El-Sayed for providing sequence information prior to publication.
Sequencing of the T. brucei genome was accomplished as part of the Trypanosoma Genome Network with support from The Wellcome Trust and NIAID. R.I. was supported by NIH postdoctoral fellowship AI10312. This work was supported in part by NIH grants RR1823 and AI141109 to R.A. and AI14102 and GM42188 to K.S.
REFERENCES
- 1.Allen T E, Heidmann S, Reed R, Myler P J, Göringer H U, Stuart K D. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol Cell Biol. 1998;18:6014–6022. doi: 10.1128/mcb.18.10.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 3.Bringaud F, Stripecke R, Frech G C, Freedland S, Turck C, Byrne E M, Simpson L. Mitochondrial glutamate dehydrogenase from Leishmania tarentolaeis a guide RNA-binding protein. Mol Cell Biol. 1997;17:3915–3923. doi: 10.1128/mcb.17.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corell R A, Read L K, Riley G R, Nellissery J K, Allen T E, Kable M L, Wachal M D, Seiwert S D, Myler P J, Stuart K D. Complexes from Trypanosoma bruceithat exhibit deletion editing and other editing-associated properties. Mol Cell Biol. 1996;16:1410–1418. doi: 10.1128/mcb.16.4.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estévez A M, Kierszenbaum F, Wirtz E, Bringaud F, Grunstein J, Simpson L. Knockout of the glutamate dehydrogenase gene in bloodstream Trypanosoma bruceiin culture has no effect on editing of mitochondrial mRNAs. Mol Biochem Parasitol. 1999;100:5–17. doi: 10.1016/s0166-6851(99)00024-9. [DOI] [PubMed] [Google Scholar]
- 6.Estévez A M, Simpson L. Uridine insertion/deletion RNA editing in trypanosome mitochondria—a review. Gene. 1999;240:247–260. doi: 10.1016/s0378-1119(99)00437-0. [DOI] [PubMed] [Google Scholar]
- 7.Gygi S P, Rochon Y, Fanza B R, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris M, Decker C, Sollner-Webb B, Hajduk S. Specific cleavage of pre-edited mRNAs in trypanosome mitochondrial extracts. Mol Cell Biol. 1992;12:2591–2598. doi: 10.1128/mcb.12.6.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris M E, Moore D R, Hajduk S L. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J Biol Chem. 1990;265:11368–11376. [PubMed] [Google Scholar]
- 10.Hayman M L, Read L K. Trypanosoma bruceiRBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J Biol Chem. 1999;274:12067–12074. doi: 10.1074/jbc.274.17.12067. [DOI] [PubMed] [Google Scholar]
- 11.Horváth A, Berry E A, Maslov D A. Translation of the edited mRNA for cytochrome b in trypanosome mitochondria. Science. 2000;287:1639–1640. doi: 10.1126/science.287.5458.1639. [DOI] [PubMed] [Google Scholar]
- 12.Igo R P, Jr, Palazzo S S, Burgess M L K, Panigrahi A K, Stuart K. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol Cell Biol. 2000;20:8447–8457. doi: 10.1128/mcb.20.22.8447-8457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kable M L, Heidmann S, Stuart K. RNA editing: getting U into RNA. Trends Biochem Sci. 1997;22:162–166. doi: 10.1016/s0968-0004(97)01041-4. [DOI] [PubMed] [Google Scholar]
- 14.Kable M L, Seiwert S D, Heidmann S, Stuart K. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- 15.Köller J, Müller U, Schmid B, Missel A, Kruft V, Stuart K, Göringer H U. Trypanosoma bruceigBP21: an arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J Biol Chem. 1997;272:3749–3757. doi: 10.1074/jbc.272.6.3749. [DOI] [PubMed] [Google Scholar]
- 16.Lambert L, Müller U F, Souza A E, Göringer H U. The involvement of gRNA-binding protein gBP21 in RNA editing—an in vitro and in vivoanalysis. Nucleic Acids Res. 1999;27:1429–1436. doi: 10.1093/nar/27.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Link A J, Eng J, Schieltz D M, Carmack E, Mize G J, Morris D R, Garvik B M, Yates J R. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 18.Madison-Antenucci S, Sabatini R S, Pollard V W, Hajduk S L. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J. 1998;17:6368–6376. doi: 10.1093/emboj/17.21.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Missel A, Souza A E, Norskau G, Göringer H U. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma bruceiaffects edited mRNAs. Mol Cell Biol. 1997;17:4895–4903. doi: 10.1128/mcb.17.9.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myler P J, Audleman L, deVos T, Hixson G, Kiser P, Lemley C, Magness C, Rickel E, Sisk E, Sunkin S, Swartzell S, Westlake T, Bastien P, Fu G, Ivens A, Stuart K. Leishmania majorFriedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc Natl Acad Sci USA. 1999;96:2902–2906. doi: 10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neubauer G, Gottschalk A, Fabrizio P, Seraphin B, Luhrmann R, Mann M. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc Natl Acad Sci USA. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollard V W, Harris M E, Hajduk S L. Native mRNA editing complexes from Trypanosoma bruceimitochondria. EMBO J. 1992;11:4429–4438. doi: 10.1002/j.1460-2075.1992.tb05543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Read L K, Göringer H U, Stuart K. Assembly of mitochondrial ribonucleoprotein complexes involves specific guide RNA (gRNA)-binding proteins and gRNA domains but does not require preedited mRNA. Mol Cell Biol. 1994;14:2629–2639. doi: 10.1128/mcb.14.4.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rusché L N, Cruz-Reyes J, Piller K J, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma bruceimitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabatini R, Hajduk S L. RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma bruceimRNA editing. J Biol Chem. 1995;270:7233–7240. doi: 10.1074/jbc.270.13.7233. [DOI] [PubMed] [Google Scholar]
- 26.Seiwert S D, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitrosuggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 27.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 28.Stuart K, Allen T E, Heidmann S, Seiwert S D. RNA editing in kinetoplastid protozoa. Microbiol Mol Biol Rev. 1997;61:105–120. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stuart K, Gobright E, Jenni L, Milhausen M, Thomashow L, Agabian N. The IsTaR 1 serodeme of Trypanosoma brucei: development of a new serodeme. J Parasitol. 1984;70:747–754. [PubMed] [Google Scholar]
- 30.Vanhamme L, Perez-Morga D, Marchal C, Speijer D, Lambert L, Geuskens M, Alexandre S, Ismaïli N, Göringer U, Benne R, Pays E. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitroRNA editing activity. J Biol Chem. 1998;273:21825–21833. doi: 10.1074/jbc.273.34.21825. [DOI] [PubMed] [Google Scholar]
- 31.Yates J R, Eng J K, McCormack A L. Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]