Abstract
Host factors that influence Congenital Zika Syndrome (CZS) outcome remain elusive. Interferons have been reported as the main antiviral factor in Zika and other flavivirus infections. Here, we accessed samples from 153 pregnant women (77 without and 76 with CZS) and 143 newborns (77 without and 66 with CZS) exposed to ZIKV conducted a case-control study to verify whether interferon alfa receptor 1 (IFNAR1) and interferon lambda 2 and 4 (IFNL2/4) single nucleotide polymorphisms (SNPs) contribute to CZS outcome, and characterized placenta gene expression profile at term. Newborns carrying CG/CC genotypes of rs2257167 in IFNAR1 presented higher risk of developing CZS (OR=3.41; IC=1.35-8.60; Pcorrected=0.032). No association between IFNL SNPs and CZS was observed. Placenta from CZS cases displayed lower levels of IFNL2 and ISG15 along with higher IFIT5. The rs2257167 CG/CC placentas also demonstrated high levels of IFIT5 and inflammation-related genes. We found CZS to be related with exacerbated type I IFN and insufficient type III IFN in placenta at term, forming an unbalanced response modulated by the IFNAR1 rs2257167 genotype. Despite of the low sample size se findings shed light on the host-pathogen interaction focusing on the genetically regulated type I/type III IFN axis that could lead to better management of Zika and other TORCH (Toxoplasma, Others, Rubella, Cytomegalovirus, Herpes) congenital infections.
Keywords: Congenital Zika Syndrome, rs2257167, placenta, type I interferon, type III interferon
Introduction
Zika virus (ZIKV) is a single stranded positive-sense RNA virus that belongs to the Flaviviridae family. Zika infection is mostly asymptomatic or associated with mild symptoms. After the outbreak in the Americas in 2015, the virus spread across 59 countries and more than 500.000 suspected cases were reported (1, 2). In a short while, there was a rise in cases of congenital abnormalities, including microcephaly, cerebral anomalies, congenital contractures, ocular alterations among other neurological abnormalities known as Congenital Zika Syndrome (CZS) (3–8). In a prospective cohort study, our group observed that 46% of the infants that were born to ZIKV-infected mothers bore abnormal clinical or brain imaging findings, including four infants with microcephaly, regardless of the trimester in pregnancy (7). Indeed, in a little while, a case-control study confirmed the association between the infection and CZS, and ZIKV epidemic was declared a public health emergence of international concern (3, 9). Nevertheless, not all infants that are born to ZIKV-infected mothers will develop CZS, and it is not clear what maternal and/or fetal factors contribute to infant adverse neurologic outcomes. One important risk factor for CSZ is infection within the first trimester of pregnancy, which poses almost twice as high a risk of severe outcomes such as CNS abnormalities when compared with third trimester infections (7). Furthermore, maternal nutritional and social factors, such as consumption of improper water and poor protein diet, have been related to CZS development (10, 11). These environmental factors do not completely explain CZS outcomes, and it has been reported that genetic background can influence these outcomes. Thus, studies testing other populations in Brazil identified maternal adenylate cyclase, and newborn collagen-encoding genes associated with CZS (12–14). Further, a previous study with a similar approach testing candidate SNPs demonstrated Toll-like receptor 3 (TLR3) rs3775291 and Tumor Necrosis Factor (TNF) rs1799964 associated with abnormal outcomes due to ZIKV infection during pregnancy (15).
During other congenital infections, namely TORCH (Toxoplasma, Others, Rubella, Cytomegalovirus, Herpes), which may cause congenital anomalies, placenta has been described as playing a crucial role in mother to fetus transmission (16). In zika, one of the hypotheses for the emergence of adverse neurological outcomes is that ZIKV can infect and cross trophoblast cell layers as cargo, ultimately reaching the fetal neurologic system and causing direct damage. On the other hand, ZIKV infection causes an innate immunological imbalance, excessive inflammation and vascular permeability dysfunction in the placenta, which may contribute to disrupting embryonic brain development (17–24).
Interferons (IFN) are key players of the innate immune response against viral infection, inducing hundreds of interferon-stimulated genes (ISGs) that act directly against virus components (25). Among these ISGs, ubiquitin-like protein ISG15, induced by type I IFNs, is one of the most strongly and rapidly induced, inhibiting viral replication and modulating host immunity (26–28). Another ISG, IFN-induced protein with tetratricopeptide repeats 5, IFIT5 (ISG58), activates IRF3/NF-κB pathway, which induces higher type I IFNs and proinflammatory mediators (29). It has been described that ZIKV disrupts type I IFN, harming phosphorylation of STAT1 and STAT2 (22, 30). In addition to a major role in antiviral defense, an exacerbated type I IFN response was demonstrated to be threatening for newborn development (31), indicating that a balanced production of type I IFNs could be effective in controlling infection and inflammation. Type III interferons (a.k.a. IFN-λ 1-4) present augmented expression during ZIKV infection in susceptible placental cells and higher levels of IFN-λ antagonize type I IFNs (21, 32–34). In this regard, administration of exogenous IFN-λ in mice led to signatures with balanced expression of ISGs (IFI44L, OASL, OAS1, and MX1) and inhibition of ZIKV replication, suggesting a therapeutic potential (35, 36).SNPs in the vicinity of IFNAR1 and IFNL1-4 loci have been associated with outcomes of viral infections, such as hepatitis B and C (37–39). Variants within IFNAR1 have been associated with an error of innate immunity related to severe viscerotropic adverse events following vaccination with another flavivirus: attenuated yellow fever virus (40). The IFNL4 rs12979860 CC genotype has been associated with persistent low levels of ISGs IFIT1, IFIT2, IFIT3, and OAS1 in postpartum normal pregnancy (41). In another flavivirus infection, hepatitis C, IFNL SNPs rs12979860, rs8099917, rs8109886, and rs368234815 are markers for good prognosis in chronic patients treated with IFNα and ribavirin (39, 42–44).
In this paper, we describe the association between the genetic background of newborns and mothers from ZIKV-infected pregnancy and CZS development, focusing on SNPs in IFNAR1, IFNL2 and IFNL4 loci, as well as the functional consequences of specific genotypes for the immunological imbalance in at term placentas from pregnant women exposed to ZIKV.
Materials and Methods
Human Subjects and Sample Collection
Our studies made use of the ongoing prospective clinical cohort study of ZIKV+ pregnant women and their infants at maternal and child hospital (IFF/Fiocruz) in Rio de Janeiro, Brazil (IRB/CAAE: 52675616.0.000.5269). In this cohort, pregnant women who were ZIKV+ received their prenatal care at IFF/Fiocruz. Since December 2015, a total of 301 mothers who were suspected of having been infected by ZIKV during gestation were referred to IFF — a major public reference hospital in Rio de Janeiro for congenital infections and congenital anomalies. Here, we utilized a subpopulation of 143 newborns and 153 mothers from the IFF cohort, including 3 pairs of bi-chorionic and bi-amniotic twins, with availability of samples, and confirmed ZIKV infection during pregnancy by ZIKV PCR of urine, blood, or placenta samples from mothers or newborns as inclusion criteriaFrom those cases, 84 placentas (74 from congenital ZIKV infections and 10 from uninfected patients) were accessed, processed and analyzed for gene expression. Samples from mothers were tested for HIV, evidence of past Dengue virus (DENV) infection (by DENV IgG and IgM), and Chikungunya virus (CHIKV) (blood PCR). Maternal demographic, medical/prenatal history and clinical findings were entered into case-report forms. All infants underwent routine clinical and extensive neurologic evaluation at the time of birth and were tested for CHIKV infection (blood PCR), syphilis and TORCH infections (toxoplasmosis, rubella, CMV, and herpes simplex virus as determined by standard testing). Infants were evaluated for the following adverse neurologic outcomes: (a) microcephaly (head-circumference z score of less than -2), (b) abnormal brain imaging by pre- or post-natal ultrasound (e.g., computed tomography and/or magnetic resonance imaging), and/or (c) abnormal clinical examination (including neurologic, ocular, and/or auditory with abnormalities confirmed by a multidisciplinary team of neonatologists, neurologists, infectious disease specialists, geneticists, ophthalmologists, and physical therapists). Our study included ZIKV+ pregnant adult women >18 years of age and their infants. Exclusion criteria included maternal HIV infection and pregnancies complicated by other congenital infections, known to cause infant neurologic damage (e.g., TORCH, CHIKV). Placental samples were collected at the time of delivery from the umbilical cord insertion region and stored in RNA later until RNA extraction. For DNA analysis, 5 mL of blood was collected from pregnant women at study enrollment and an oral swab was collected from newborns. Rational and workflow of sample analysis in Supplementary Figure 1 .
Genetic Studies: SNP Selection and Linkage Disequilibrium Analysis
Selection of candidate SNPs for the case-control association study was performed by integrating different tools: Principal Component Analysis (PCA), ANNOVAR (45), allele frequencies, literature and HAPLOVIEW (46). First, all SNPs located in the IFNL (chr19:39,733,272-39,736,609-GRCh37/hg19) and IFNAR1 regions (chr21:34,696,734-34,732,168- GRCh37/hg19) were recovered from African (ENS, GWD, LWK, MSL, and YRI) and European (CEU, FIN, GBR, IBS, and TSI) populations from phase 3 of the 1000 Genomes Project (47). Then, Principal Component Analysis (PCA) was performed using EIGENSOFT4.2 (46). The use of this strategy in the selection of functional SNPs assumes that, since the analyzed variability is of a functional genome region (meaning: a gene), the clusters generated by PCA would be mainly influenced by functionality. Thus, SNPs with high weight for principal component 1 (PC1) could be potential candidates for having a functional role. SNPs were thus sorted by decreasing values of “SNP weight” for PC1, and functional annotation of all SNPs was performed using ANNOVAR (45), with ref Gene hg19 (11 Dez 2015). According to the functional category identified by ANNOVAR, “SNP weight” for PC1 (with SNP weight values within the highest 30, called top SNPs), minimal allele frequencies (MAF) in African and European populations (> 0.1) and associations with infectious diseases already reported in the literature, SNPs present in the IFNAR1 and IFNL region were selected for genotyping and haplotype construction. Haplotype inferences using selected SNPs, haplotype frequencies and linkage disequilibrium (LD) analysis for all studied populations were performed using HAPLOVIEW (46). To select SNPs, PCA was used to retrieve those located either in the IFNAR1 or IFNL regions, which are found among the African and European populations from the 1000 Genomes Project ( Supplementary Figure 2A ). Within IFNL, we selected four representative SNPs (rs12979860, rs4803222, rs8109886, rs8099917, and rs368234815). In the IFNAR1 region, SNPs rs2843710, rs2257167, rs17875834, rs2834202 were selected to construct the haplotypes in parental populations. Allele frequencies, annotation, and reference of the selected SNPs are described in Supplementary Table 1 . Linkage disequilibrium (LD) analysis and haplotype arrangements indicated ancestry-specific patterns for these two genomic regions. ( Supplementary Figure 2B ). The IFNAR1 arranged haplotypes suggest rs2843710, rs2257167 are tags to discriminate Europeans and Africans, while IFNL rs12979860 and rs8109886 SNPs also present very different frequencies among the major Brazilian parental populations ( Supplementary Table 2 ).
Genomic DNA Extraction and SNP Genotyping Analysis
DNA extraction was performed from saliva swabs or whole blood cells collected from each individual newborn (n=143) and mothers (n=153), respectively, using the salting out method. Following extraction, DNA was quantified with a Nanodrop ND 1000 spectrophotometer (Nanodrop Technologies). After PCA, LD and haplotype analysis, the following tag polymorphisms were genotyped due to their representativeness within the corresponding genomic regions: IFNL2-IFNL4: rs8099917 (C11710096_10) located 8.9 kb upstream of the IFNL4 (T > G) start codon and rs8109886 (C11710100_10) located 3.3 kb upstream of the IFNL4 (A > C) start codon; IFNL4: rs12979860 (C7820464_10) in intron 4 (C > T), rs4803222 (C7820457_10) in the 5’ UTR (C > G), and rs368234815 an indel in exon 1 (TT>ΔG) as described by Prokunina-Olsson et al, 2013 (44); IFNAR1: rs2257167 (C:16076297_10) located within exon 4 of IFNAR1 (G>C) and rs2843710 (C:26796048_10) located within the promoter region of IFNAR1 (C > G). All SNPs were genotyped using the allelic discrimination method for real-time TaqMan assays (Applied Biosystems) using Viia7 Real-time PCR System. Approximately 30 ng of DNA was used in the genotyping reaction. Statistical analyses were performed using “snpassoc”, “genetics” and “haplo.stats” packages in software R version 2.11.1, as previously described (48). Briefly, genotype frequencies were tested for HWE using a Chi-square test. The genotypic, allelic, and carrier frequencies were calculated and compared in cases and controls by conditional logistic regression adjusted for ancestry and trimester of infection. Next, we compared the frequencies between CZS and no CZS, separately. Linkage disequilibrium values for SNPs studied in IFNAR1 and IFNL were estimated by r2 and haplotype frequencies were compared between cases and controls by logistic regression, also adjusted for ancestry and trimester of infection. For mother-child SNP interaction, we used Estimation of Maternal, Imprinting and Interaction Effects using multinomial modeling analysis using a multinomial model to test the existence (and estimate) of maternal genotype relative risk parameters that may increase (or decrease) the possibility that a child is affected, as described previously (49).
Ancestry Analysis
Since the Brazilian population is highly admixed and ethnic classification is not uniformly defined, ancestry data is necessary to adjust the logistic regression and eliminate bias in genetic associations (50). Thus, DNA samples were genotyped for 46 Ancestry Informative Markers (AIM)-Indels in a multiplex PCR system followed by capillary electrophoresis in an ABI 3500 Genetic Analyzer (Thermo Fisher), as described previously (51, 52). Allele calls were obtained by GeneMapper v.4.1 and results for individual and global ancestry estimates were performed by using the HGDP-CEPH diversity panel as a reference (European, African and Native-American; K=3) in STRUCTURE v2.3. In the logistic regression performed in R, covariates AFR+EUR were used to control for population stratification along with trimester of infection.
ZIKV PCR Detection
RT-qPCR was performed using the 2x QuantiTect Probe RT-PCR kit (Qiagen, Valencia, CA, USA) with the same primers and cycle times as previously described (53). All the assays were carried out in triplicate and fluorescence curves that crossed the threshold within or below 38 cycles were considered positive.
Gene Expression Profile Analysis
Analysis of gene expression in placental tissue, specifically from the region of umbilical cord insertion, from pregnant mothers (control, with or without CZS samples) was performed using Fluidigm (Biomark platform) assays. Detailed data available under request. Our experimental design followed a previously described workflow (54).
Real-Time RT-PCR Expression Analysis
From routines created in R for parsing raw foreground and background intensities, we carried out background correction and exploratory data analysis: fluorescence accumulation and melting curve graphs of Rn for each reaction with each gene. For relative quantification of expression, the fluorescence accumulation data of each sample were used for fitting four parameter sigmoid curves using the qPCRlibrary from R statistical package version 3.4.1 (48). For each amplification, the cycle of quantification was determined as the maximum of the second derivative of the fit sigmoid curve and the efficiency, as the ratio between the fluorescence of the cycle of quantification and the fluorescence of the cycle that immediately preceded that. For each gene, efficiency was estimated by the mean of all the efficiencies for each amplification reaction for that gene. Endogenous controls used for normalizing between different amplified samples were selected by the geNorm method. Normalization factors were estimated for each sample using the geometric average of the selected normalized genes (55).
Statistical Analysis of Gene Expression
Pairwise comparisons of log-transformed (base 2) normalized expression means between/among groups of interest were performed by contrasts/differences (fold-changes) obtained after both bi- and multivariate linear models adjusted by ordinary least square regressions. Whenever the variable of interest had more than two levels, p-values were corrected by the Tukey Honest Significant Difference post-Hoc method (56). After gene-per-gene pairwise comparisons, we conducted a Type I error adjustment for multiple comparisons by the Holm-Bonferroni method (57). Different sets of confounding variables were selected by clinical experts and included in the multivariate models to adjust the fit effects for different variables of interest for all genes. To obtain the marginal means expected values for the variable of interest, we kept the confounding variables in the multivariate models in their mean values or equal proportions in all models. For the analysis, two-tailed levels of significance ≤ 0.01, 0.05, and 0.1 were considered as “highly significant”, “significant”, and “suggestive”, respectively.
Results
Newborn IFNAR1 rs2257167 Are Associated With CZS Outcome
The DNA samples from whole blood admixed ancestry population of 143 newborns and 153 mothers from ZIKV-infected pregnancies, with development of CZS (cases) or otherwise (controls) ( Supplementary Tables 3, 4 ), were genotyped for SNPs encompassing IFNAR1 (rs2257167, rs2843710, rs2834202, and rs17875834), IFNL4 genes (rs12979860, rs368234815, and rs4803222), and within IFNL2 and IFNL4 genes (rs8099917 and rs8109886). The frequency of each SNP was verified in cases and controls and CZS outcome was evaluated. Genotype frequencies were found to be in HWE for all SNPs tested. Data were adjusted by genetic ancestry and the trimester of pregnancy in which ZIKV infection occurred (when symptoms of ZIKV infection were detected). CZS risk was observed for CG/CC carriers of SNP rs2257167 following FDR correction, OR = 3.42; CI = 1.35-8.6, Pcorrected = 0.032 ( Table 1 ). No significant differences were observed in the frequencies between cases and controls in any other SNP tested. Other analyses did not show any significant results including: IFNAR1 newborn haplotypes, IFNAR1 mother genotypes and haplotypes, IFNL genotypes or haplotypes, as well as the Estimation of Maternal, Imprinting and Interaction Effects using multinomial modeling analysis to investigate if risk-associated newborn genotypes could possibly present an additive contribution of maternal same genotype ( Supplementary Tables 5–7 ).
Table 1.
Association study with newborn IFNAR1 and IFNL SNPs and CZS abnormalities.
| No CZS findings | CZS | Adjusted by trimester of ZIKV exposure and ancestry | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | OR | lower | upper | p-value | FDR p-value | |
| rs2257167 | |||||||||
| G/G | 56 | 73.7 | 38 | 58.5 | 1 | 0.023 | 0.068 | ||
| C/G | 18 | 23.7 | 24 | 36.9 | 3.13 | 1.19 | 8.21 | ||
| C/C | 2 | 2.6 | 3 | 4.6 | 5.72 | 0.77 | 42.47 | ||
| G/G | 56 | 73.7 | 38 | 58.5 | 1 | 0.007 | 0.032 | ||
| C/G-C/C | 20 | 26.3 | 27 | 41.5 | 3.41 | 1.35 | 8.6 | ||
| G/G-C/G | 74 | 97.4 | 62 | 95.4 | 1 | 0.171 | 0.308 | ||
| C/C | 2 | 2.6 | 3 | 4.6 | 3.83 | 0.55 | 26.97 | ||
| G/G-C/C | 58 | 76.3 | 41 | 63.1 | 1 | 0.032 | 0.072 | ||
| C/G | 18 | 23.7 | 24 | 36.9 | 2.71 | 1.07 | 6.92 | ||
| Additive | 76 | 53.9 | 65 | 46.1 | 2.76 | 1.29 | 5.91 | 0.006 | 0.032 |
| rs2843710 | |||||||||
| G/G | 30 | 40 | 24 | 36.4 | 1 | 0.210 | 0.629 | ||
| C/G | 36 | 48 | 27 | 40.9 | 1.15 | 0.47 | 2.82 | ||
| C/C | 9 | 12 | 15 | 22.7 | 2.65 | 0.85 | 8.31 | ||
| C/C | 30 | 40 | 24 | 36.4 | 1 | 0.340 | 0.765 | ||
| C/G-G/G | 45 | 60 | 42 | 63.6 | 1.49 | 0.65 | 3.41 | ||
| C/C-C/G | 66 | 88 | 51 | 77.3 | 1 | 0.082 | 0.525 | ||
| G/G | 9 | 12 | 15 | 22.7 | 2.46 | 0.87 | 6.92 | ||
| C/C-G/G | 39 | 52 | 39 | 59.1 | 1 | 0.641 | 1.000 | ||
| C/G | 36 | 48 | 27 | 40.9 | 0.83 | 0.37 | 1.85 | ||
| Additive | 75 | 53.2 | 66 | 46.8 | 1.55 | 0.89 | 2.69 | 0.117 | 0.525 |
| rs2834202 | |||||||||
| A/A | 44 | 58.7 | 40 | 62.5 | 1 | 0.398 | 1 | ||
| A/G | 29 | 38.7 | 20 | 31.2 | 0.87 | 0.37 | 2.04 | ||
| G/G | 2 | 2.7 | 4 | 6.2 | 3.26 | 0.48 | 21.89 | ||
| A/A | 44 | 58.7 | 40 | 62.5 | 1 | 0.923 | 1 | ||
| A/G-G/G | 31 | 41.3 | 24 | 37.5 | 1.04 | 0.46 | 2.33 | ||
| A/A-A/G | 73 | 97.3 | 60 | 93.8 | 1 | 0.186 | 1 | ||
| G/G | 2 | 2.7 | 4 | 6.2 | 3.41 | 0.52 | 22.42 | ||
| A/A-G/G | 46 | 61.3 | 44 | 68.8 | 1 | 0.605 | 1 | ||
| A/G | 29 | 38.7 | 20 | 31.2 | 0.8 | 0.35 | 1.85 | ||
| Additive | 75 | 54 | 64 | 46 | 1.21 | 0.62 | 2.36 | 0.567 | 1 |
| rs17875834 | |||||||||
| C/C | 72 | 93.5 | 56 | 88.9 | 1 | 0.162 | 0.729 | ||
| C/T | 5 | 6.5 | 7 | 11.1 | 2.76 | 0.63 | 12.06 | ||
| Additive | 77 | 55 | 63 | 45 | 2.76 | 0.63 | 12.06 | 0.162 | 0.729 |
| rs12979860 | |||||||||
| C/C | 31 | 40.3 | 22 | 33.3 | 1 | 0.608 | 1 | ||
| C/T | 34 | 44.2 | 28 | 42.4 | 1.3 | 0.54 | 3.15 | ||
| T/T | 12 | 15.6 | 16 | 24.2 | 1.78 | 0.56 | 5.68 | ||
| C/C | 31 | 40.3 | 22 | 33.3 | 1 | 0.406 | 1 | ||
| C/T-T/T | 46 | 59.7 | 44 | 66.7 | 1.42 | 0.62 | 3.25 | ||
| C/C-C/T | 65 | 84.4 | 50 | 75.8 | 1 | 0.419 | 1 | ||
| T/T | 12 | 15.6 | 16 | 24.2 | 1.54 | 0.54 | 4.38 | ||
| C/C-T/T | 43 | 55.8 | 38 | 57.6 | 1 | 0.858 | 1 | ||
| C/T | 34 | 44.2 | 28 | 42.4 | 1.08 | 0.48 | 2.39 | ||
| Additive | 77 | 53.8 | 66 | 46.2 | 1.33 | 0.76 | 2.33 | 0.319 | 1 |
| rs8099917 | |||||||||
| T/T | 58 | 75.3 | 43 | 65.2 | 1 | 0.309 | 0.695 | ||
| G/T | 17 | 22.1 | 18 | 27.3 | 2.09 | 0.78 | 5.61 | ||
| G/G | 2 | 2.6 | 5 | 7.6 | 1.66 | 0.27 | 10.11 | ||
| T/T | 58 | 75.3 | 43 | 65.2 | 1 | 0.130 | 0.542 | ||
| G/T-G/G | 19 | 24.7 | 23 | 34.8 | 2 | 0.81 | 4.95 | ||
| T/T-G/T | 75 | 97.4 | 61 | 92.4 | 1 | 0.706 | 1 | ||
| G/G | 2 | 2.6 | 5 | 7.6 | 1.41 | 0.23 | 8.44 | ||
| T/T-G/G | 60 | 77.9 | 48 | 72.7 | 1 | 0.154 | 0.542 | ||
| G/T | 17 | 22.1 | 18 | 27.3 | 2.02 | 0.76 | 5.41 | ||
| Additive | 77 | 53.8 | 66 | 46.2 | 1.63 | 0.78 | 3.37 | 0.181 | 0.542 |
| rs8109886 | |||||||||
| A/A | 24 | 31.2 | 22 | 33.3 | 1 | 0.795 | 1 | ||
| C/A | 36 | 46.8 | 31 | 47 | 0.86 | 0.34 | 2.16 | ||
| C/C | 17 | 22.1 | 13 | 19.7 | 0.68 | 0.22 | 2.11 | ||
| A/A | 24 | 31.2 | 22 | 33.3 | 1 | 0.622 | 1 | ||
| C/A-C/C | 53 | 68.8 | 44 | 66.7 | 0.8 | 0.34 | 1.91 | ||
| A/A-C/A | 60 | 77.9 | 53 | 80.3 | 1 | 0.548 | 1 | ||
| C/C | 17 | 22.1 | 13 | 19.7 | 0.74 | 0.28 | 1.98 | ||
| A/A-C/C | 41 | 53.2 | 35 | 53 | 1 | 0.971 | 1 | ||
| C/A | 36 | 46.8 | 31 | 47 | 1.01 | 0.46 | 2.23 | ||
| Additive | 77 | 53.8 | 66 | 46.2 | 0.83 | 0.47 | 1.45 | 0.505 | 1 |
| rs4803222 | |||||||||
| G/G | 33 | 42.9 | 33 | 50.8 | 1 | 0.909 | 1 | ||
| C/G | 39 | 50.6 | 26 | 40 | 0.84 | 0.36 | 1.92 | ||
| C/C | 5 | 6.5 | 6 | 9.2 | 0.84 | 0.2 | 3.56 | ||
| G/G | 33 | 42.9 | 33 | 50.8 | 1 | 0.662 | 1 | ||
| C/G-C/C | 44 | 57.1 | 32 | 49.2 | 0.84 | 0.38 | 1.86 | ||
| G/G-C/G | 72 | 93.5 | 59 | 90.8 | 1 | 0.904 | 1 | ||
| C/C | 5 | 6.5 | 6 | 9.2 | 0.92 | 0.23 | 3.67 | ||
| G/G-C/C | 38 | 49.4 | 39 | 60 | 1 | 0.713 | 1 | ||
| C/G | 39 | 50.6 | 26 | 40 | 0.86 | 0.39 | 1.91 | ||
| Additive | 77 | 54.2 | 65 | 45.8 | 0.88 | 0.48 | 1.64 | 0.694 | 1 |
| rs368234815 | |||||||||
| TT/TT | 34 | 44.2 | 24 | 37.5 | 1 | 0.872 | 1 | ||
| TT/ΔG | 28 | 36.4 | 27 | 42.2 | 1.22 | 0.5 | 2.95 | ||
| ΔG/ΔG | 15 | 19.5 | 13 | 20.3 | 1.27 | 0.43 | 3.72 | ||
| TT/TT | 34 | 44.2 | 24 | 37.5 | 1 | 0.604 | 1 | ||
| TT/ΔG-ΔG/ΔG | 43 | 55.8 | 40 | 62.5 | 1.24 | 0.55 | 2.76 | ||
| TT/TT-TT/ΔG | 62 | 80.5 | 51 | 79.7 | 1 | 0.775 | 1 | ||
| ΔG/ΔG | 15 | 19.5 | 13 | 20.3 | 1.16 | 0.43 | 3.1 | ||
| TT/TT-ΔG/ΔG | 49 | 63.6 | 37 | 57.8 | 1 | 0.771 | 1 | ||
| TT/ΔG | 28 | 36.4 | 27 | 42.2 | 1.13 | 0.5 | 2.54 | ||
| Additive | 77 | 54.6 | 64 | 45.4 | 1.14 | 0.68 | 1.92 | 0.624 | 1 |
DNA samples from 143 newborns with congenital infection by ZIKV, with or without CZS, were genotyped for IFNAR1 SNPs rs2257167, rs2843710, rs2834202, and rs17875834; and IFNL SNPs rs8109886, rs12979860 rs8099917, rs4803222, and rs368234815. Data were adjusted considering the trimester of the first symptoms or asymptomatic ZIKV infections and the percentage of the African and European ancestry of each individual. The total number of genotyped samples for each SNP may vary due to genotype miscalling. Major allele was used as baseline. Odds ratio (OR) with 95% confidence interval (CI) and p-values. We conducted type I error adjustment of multiple comparisons by the False Discovery Rate (FDR) method.
IFNAR1 rs2257167 CG/CC Genotypes as CZS Risk Factors in ZIKV Infection During Second and Third Trimester of Pregnancy
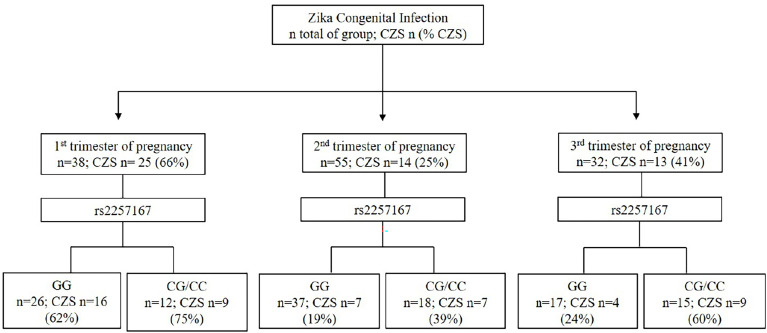
Following previous clinical studies (7), in our cohort, the determination of the trimester of pregnancy in which ZIKV infection occurs was a strong predicting factor for CZS outcome, along with IFNAR1 genotype ( Figure 1 ). Taking into account only newborns with information of rs2257167 genotype and trimester of infection (n=125), results show that ZIKV infection during first trimester of pregnancy culminates in with 66% of CZS cases, in contrast with second (25%) or third (41%) trimesters. Interestingly, data showed that newborns with rs2257167 CG/CC genotypes presented twice the frequency compared with newborns with the GG genotype in second (19%-GG vs 39% -CG/CC) and third (24%-GG vs 60%-CG/CC) trimesters.
Figure 1.
Event-based flowchart of CZS occurrence. The trimester of pregnancy in which first Zika symptoms occur and newborn genotypes of rs2257167 were used as independent variables to determine the association with CZS outcome. Total number of newborns with full information of both trimesters of infection and rs2257167 genotypes (n=125) was used to calculate the absolute number of newborns per group (n) and CZS percentage. Concerning infections in the first trimester of pregnancy, we found that irrespective of rs2257167 genotypes, 66% of the cases developed CZS, in contrast with second and third trimesters. Considering rs2257167, newborn CG/CC genotype seems to be associated with CZS risk in the second and third trimesters.
Congenital ZIKV Infection Leads to an Immunological Imbalance in Placenta
To functionally verify how ZIKV could influence severe congenital outcomes across associated IFNAR1 genotypes, we performed gene expression analyses from the placental tissues, most of which were fetal, obtained at time of delivery from 10 uninfected pregnant woman and 74 congenital ZIKV cases ( Supplementary Table 8 ). First, all ZIKV RT-PCR positive samples at term showed higher gene expression of most genes analyzed. ZIKV congenital infections occurring in the third trimester of pregnancy resulted in highest gene expression levels. Finally, mothers exhibiting ≥ 40 years of age (y/o) expressed lower levels of inflammatory genes( Supplementary Table 9 ). Because of these intrinsic differential expression profiles, ZIKV RT-qPCR positive placenta, trimester of exposure to ZIKV, and mothers’ age (≥ 40 y/o) were considered as variables in gene expression analysis of congenital ZIKV cases.
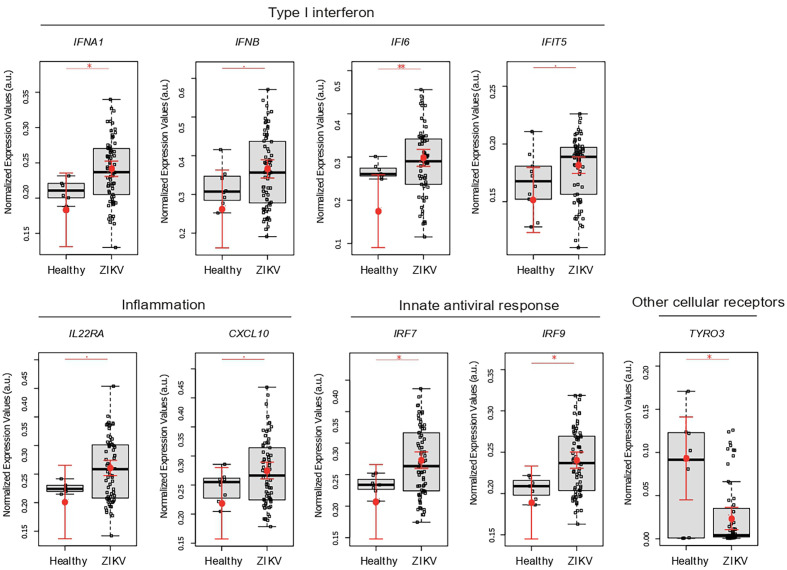
By comparing placental gene expression from congenital ZIKV infections vs uninfected pregnant women, in spite of small sample number in this group, results showed that ZIKV leads to a typical inflammatory response in the placenta. This response includes higher expression of: anti-viral type I IFN genes (IFIT5, IFNA1, and IFNB), type II interferon (IFI16), cytokine signaling (IL22RA and IP10), and interferon regulatory factors (IRF7 and IRF9); together with decreased expression of TYRO3 ( Figure 2 ).
Figure 2.
ZIKV infection leads to an immunological imbalance in placenta. Placental gene expression profile in healthy (N = 10) versus ZIKV-infected women (N = 74). Each dot corresponds to one placenta analyzed. The number of dots varies according to gene analyzed due to failed amplifications. Median and standard deviation of gene expression values are normalized by the housekeeping genes selected by the geNorm and NormFinder as well as 18S ribosomal RNA and RLP13 ribosomal protein L13 (grey boxes). Values are adjusted by mothers’ age (below or equal to/above 40 years of age) and trimester of infection (the trimester of pregnancy in which the first Zika symptoms occur or asymptomatic ZIKV infections). P-values ** ≤ 0.01, * ≤ 0.05 and ≤ 0.1.
Decreased IFNL2 and Augmented Type I IFN in Placenta Aa Term Is Associated With Newborn CZS Abnormalities
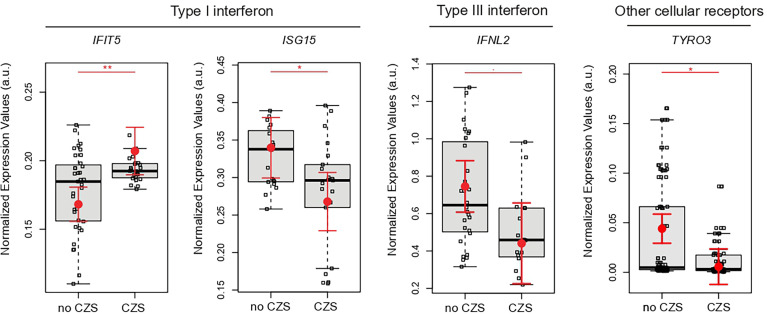
Next, we tested whether gene expression signatures of placentas from ZIKV-infected women could be associated with the presence or absence of CZS. These analyses illustrated IFNL2, ISG15, and TYRO3 significant decreases in newborns with CZS. On the other hand, IFIT5 increased significantly in newborns in the CZS group ( Figure 3 ).
Figure 3.
Placental gene expression associated with CZS. Detailed graphs of differentially expressed genes in placenta without CZS (No CZS; N = 45) or with CZS (CZS; N = 29). Each dot corresponds to one placenta analyzed. The number of dots varies according to gene analyzed due to failed amplifications. Median and standard deviation of gene expression values are normalized by housekeeping genes selected by geNorm and NormFinder as well as 18S ribosomal RNA and RLP13 ribosomal protein L13 (grey boxes). Values are adjusted by mothers’ age (below or equal to/above 40 years of age) and infection trimester (trimester of pregnancy in which the first Zika symptoms occur or asymptomatic ZIKV infections). P-values ** ≤ 0.01, * ≤ 0.05, and ≤0.1.
Genotypes rs2257167 CG/CC Are Associated With Increased Placental Type I IFN and Inflammatory Response
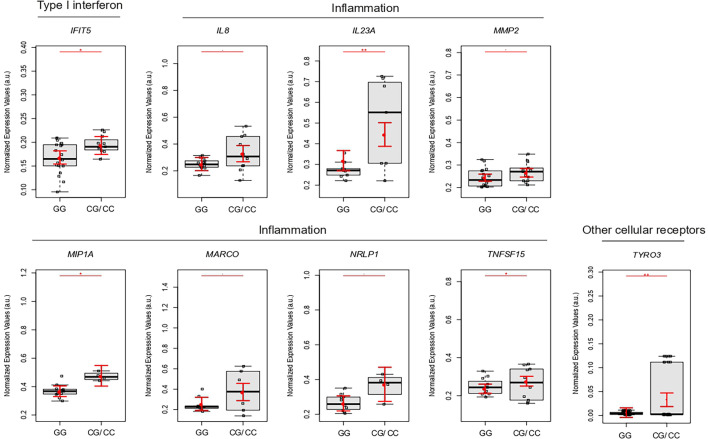
We clustered 39 newborns and 45 mothers according to GG or CG/CC genotypes of rs2257167 to assess how IFNAR1 newborn background influences the placental gene expression profile. Placentas from rs2257167 CG/GG newborns showed significantly increased expression of IFIT5 and genes related with the inflammatory response (IL8, IL23A, MMP9, MIP1A, MARCO, NRLP1, and TNFSF15) ( Figure 4 ). We could highlight also that placentas from GC/CC non-CZS babies have higher levels of most of the genes statistically different when compared to i) GG non-CZS babies or ii) GC/GG CZS newborns. One important marker is MIP1A that is significantly augmented in GC/CC compared with GG newborns with CZS ( Supplementary Figure 3 )
Figure 4.
Placental gene expression is modulated by newborn rs2257167 genotypes in ZIKV-infected pregnancy. Detailed graphs of differentially expressed genes in placenta from rs2257167 GG (N = 20) and CG/CC (N = 13) newborns. Each dot corresponds to one placenta analyzed. The number of dots varies according to gene analyzed due to failed amplifications. Median and standard deviation of gene expression values are normalized by housekeeping genes selected by geNorm and NormFinder as well as 18S ribosomal RNA and RLP13 ribosomal protein L13 (grey boxes). Values are adjusted by mothers’ age (below or equal to/above 40 years of age) and infection trimester (trimester of pregnancy in which first Zika symptoms or asymptomatic ZIKV infections occur) P-values**≤ 0.01, * ≤ 0.05, and ≤ 0.1.
Discussion
Despite the high risk, ZIKV infection during pregnancy is necessary, albeit not enough, to induce CZS. We hypothesized whether host genetic background, especially SNPs in IFNAR1 and IFNL, contributes to CSZ development and conducted a case-control study with CZS cases and healthy ZIKV+ mothers. Specifically, our results demonstrated that newborns who carried the CG/CC genotypes of SNP rs2257167 (IFNAR1) had a 3.4 higher risk of developing CZS, resulting from ZIKV infection during pregnancy, compared to those with the GG genotype. In chronic hepatitis B infected patients, the presence of the C allele was associated with higher plasma levels of the aspartate and alanine amino-transferase hepatic enzymes (58). Another study among chronic HBV-infected patients suggested that patients carrying the rs2257167 CC (leu/leu) genotype presented higher expression levels of IFNAR1 in PBMCs when compared with patients carrying the GG genotype (val/val) (59). Moreover, crystal structure studies showed that Val141 is distal to the ligand binding surface, hence Val141 (G)> Leu141(C) may influence the IFNAR1 downstream signaling (59). Here we observed that individuals Leu141(C) carriers presented a high IFIT5 and inflammatory placenta profile.
Our cohort also highlights ZIKV infection in the first trimester of pregnancy as a critical CZS risk-associated factor, and we do not observe an association with other previous arbovirus infections and CZS development in the cohorts from same hospitals (7). The immunomodulatory role of rs2257167, and how this SNP influences CZS frequency, was observed especially when ZIKV infections occur in the second and third trimesters of pregnancy. This data also shows the importance of IFNAR1 rs2257167 genetic background regulating placental gene expression culminating in CZS development. Studies using mice models and ex vivo placental cultures demonstrated that regions and maturity of placentas will provide different responses against ZIKV (36, 60). Generally, fetal-derived tissues developed from midgestational placenta are more restrictive to ZIKV replication (60). In fact, in vitro cultures show that ZIKV possesses high tropism for trophoblasts from the first trimester of pregnancy (61). Altogether, these data corroborate our findings, considering the hypothesis that ZIKV faces an IFN immunological barrier in midgestational or older placentas, while rs2257167 CG/CC carriers with higher type I and lower type III IFNs would unbalance type I/type III IFNs towards a pronounced and exacerbated type I IFN production leading to CZS susceptibility.
Notably, at term placenta from the CZS cases is associated with an increased expression of IFIT5, which is an important enhancer of type I IFN and a proinflammatory response (29). In parallel, placenta from CZS cases showed a decrease in ISG15 mRNA, which was already identified as being protective from CZS ocular manifestations (62). Another role of ISG15 is to modulate IFN responses since IFNλ4 blocks type I IFN response using the ISG15 and USP18 ubiquitin system (32). Further, although in vitro studies strongly suggest that TYRO3 is the main entry receptor for ZIKV (63, 64), ZIKV-infected placentas showed a decreased expression of TYRO3, corroborating recent findings in mice indicating that in complex organisms these receptors do not appear to be required for ZIKV infection (65). Another possibility is ZIKV downregulate entry receptors, as occurs in another viral infections (66, 67). Interestingly, here the diminished expression of TYRO3 in CZS cases highlight its role as a signaling receptor associated with inhibition of type I IFN and general innate immune responses, as well described in other viral infections (68). This profile of augmented type I IFN associated with severity is corroborated in a ZIKV-infected mice model (31) and other TORCH infections (69, 70). However, it is noteworthy that studies in mice models demonstrated that the lack of a type I IFN response also lead to CSZ (31, 71) indicating that only optimal levels of type I IFN could possibly confer a healthy pregnancy upon Zika and probably other congenital infections.
Indeed, functionally validating our association study, a high type I IFN expression phenotypic pattern in rs2257167 CG/CC individuals was observed suggesting they cannot efficiently regulate exacerbated type I IFN, which might be one of the factors leading to CZS. Besides, the pro-inflammatory profile rs2257167 CG/CC raised placental production of inflammatory mediators upon ZIKV may contribute to an environment more susceptible to infection, as observed by Rabelo and colleagues (24).
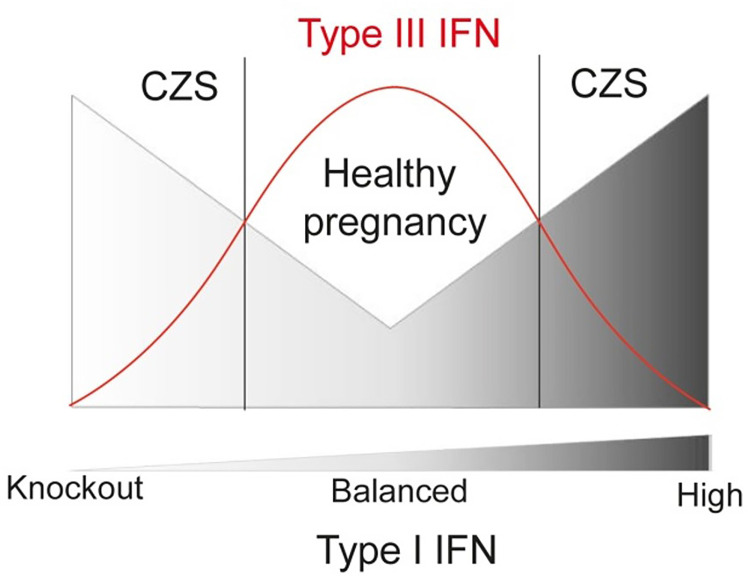
Here we showed the augmented compartmentalized expression of type III interferons (IFNL2) during ZIKV infection. The type III IFN present a remarkable role in limiting the inflammation process with a strong antiviral activity at local level, presenting therapeutic potential (35). In this regard, exacerbated levels of type III IFNs are associated with lung barrier damage induced by SARS-CoV-2, impairing lung epithelial cell proliferation during recovery (72, 73). Altogether, the placental barrier seems to require the balance between type I and III IFN, damage and immunosuppression, to confer a healthy pregnancy upon Zika and probably other congenital infections. Hence, we can hypothesize that well-adjusted production of type I and type III IFN levels in second and third trimester of pregnancy could lead to a proper protective response to ZIKV infection in the placenta, which could prevent CZS severe outcomes ( Figure 5 ).
Figure 5.
Schematic representation of functional relation between Type I and III IFN in congenital ZIKV infections regarding CZS, showing that lower risk of developing CZS is related to higher levels of IFNL and balanced levels of Type-I IFN.
Despite the sample size, the present association study is the bigger genetic study sampling effort of confirmed Zika cases during pregnancy (12–15). In Brazil, from April 2015 to November 2020 there are 3,590 cases of CZS with ZIKV infection confirmed by molecular diagnosis, making it difficult in obtaining large cohorts (74). Hence, genetic association of newborn IFNAR1 rs225167 with CZS should be independently replicated in other populations. The genetic findings indicate indeed that C-allele has a prominent role in CZS risk, since different genetic models showed association in the same direction (OR-values) irrespective of the p-value. As a strategy to validate our association study, here we focused on at term placenta gene expression study as a functional validation of association study that showed characterization of placenta response to ZIKV. It is important, here we demonstrate that newborns CC/CG rs2257167 indeed present a CZS-like placenta profile. Although, the present work encourages future in vitro investigation of IFNs unbalance and proinflammatory responses in placenta infected by ZIKV.
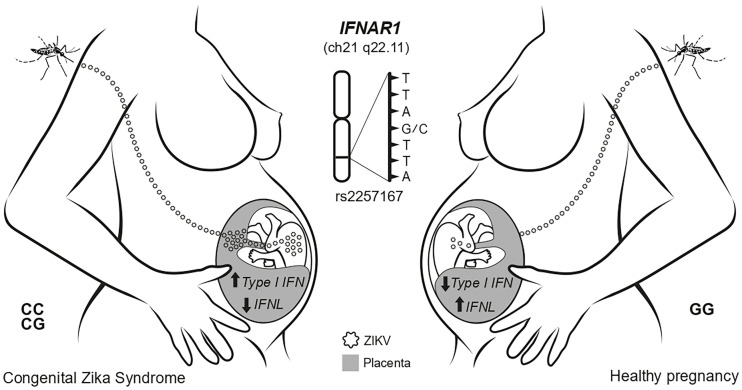
In summary, our study showed that intensity of immune responses during ZIKV infections in humans can be regulated by IFNAR1 rs2257167 genotypes. During pregnancy, genetic regulatory pathways control placental tissue-specific type I and type III IFN expression during ZIKV congenital infection influencing fetal neurological damage ( Figure 6 ). Understanding of this novel pathway may help in the development of a custom pharmacological intervention to normalize its levels, which would likely affect and disrupt CZS development.
Figure 6.
Schematic model showing the main findings of this study. The ZIKV infection during pregnancy faces placenta as an immunological barrier. In newborns with IFNAR1 rs2257167 CG/CC genotype the high levels of type I IFN and low type III IFN in placenta, culminating with CZS.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Zenodo, 10.5281/zenodo.5567327
Ethics Statement
The studies involving human participants were reviewed and approved by the Instituto Nacional de Saúde da Mulher, da Criança e do Adolescente Fernandes Figueira, Fiocruz (IRB/CAAE: 52675616.0.000.5269). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
Conceptualization: MOM, ZV, PN, and TA. Data curation: TA, DC, EP, LCG, LG, MLM, and ZV. Formal analysis: TA, FK, AS, CC, MR-A, TC, OB, FM, LF, and EC. Investigation: AS. Funding acquisition and resources: MM and MOM. Supervision: PN, ZV and MOM. Writing – original draft: TA. Writing – review and editing: DC, PN, ZV, and MOM. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Instituto Oswaldo Cruz (Rio de Janeiro, Brazil) and by the Instituto de Tecnologia em Imunobiológicos (Rio de Janeiro, Brazil).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Ms. Natália Pedra Gonçalves and Dr. Erica Louro da Fonseca from Vice Diretoria de Qualidade, Biomanguinhos, Fiocruz, for their help with ancestry analysis. We would also like to thank the team of Laboratório de Tecnologia Imunológica, Vice Diretoria de Desenvolvimento de Biomanguinhos, Fiocruz, including Ms. Camilla Bayma Fernandes, Ms. Jane da Silva, and Mr. Alessandro Fonseca de Souza for their technical and management support. This work was supported by Biomanguinhos and Instituto Oswaldo Cruz, Fiocruz, Brazil.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.764746/full#supplementary-material
References
- 1. Relich RF, Loeffelholz M. Zika Virus. Clin Lab Med (2017) 37:253–67. doi: 10.1016/j.cll.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 2. Petersen LR, Jamieson DJ, Powers AM, Honein MA. Zika Virus. N Engl J Med (2016) 374:1552–63. doi: 10.1056/NEJMra1602113 [DOI] [PubMed] [Google Scholar]
- 3. de Araújo TVB, Ximenes RA de A, Miranda-Filho D de B, Souza WV, Montarroyos UR, de Melo APL, et al. Association Between Microcephaly, Zika Virus Infection, and Other Risk Factors in Brazil: Final Report of a Case-Control Study. Lancet Infect Dis (2018) 18:328–36. doi: 10.1016/S1473-3099(17)30727-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adhikari EH, Nelson DB, Johnson KA, Jacobs S, Rogers VL, Roberts SW, et al. Infant Outcomes Among Women With Zika Virus Infection During Pregnancy: Results of a Large Prenatal Zika Screening Program. Am J Obstet Gynecol (2017) 216:292.e1–8. doi: 10.1016/j.ajog.2017.01.018 [DOI] [PubMed] [Google Scholar]
- 5. Wen Z, Song H, Ming G-L. How Does Zika Virus Cause Microcephaly? Genes Dev (2017) 31:849–61. doi: 10.1101/gad.298216.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jürgens I, Rey A. Ocular Findings in Patients With Microcephaly can Suggest Presumed Congenital Zika Virus Infection. Acta Ophthalmol (Copenh) (2018) 96:423–4. doi: 10.1111/aos.13548 [DOI] [PubMed] [Google Scholar]
- 7. Brasil P, Pereira JP, Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, et al. Zika Virus Infection in Pregnant Women in Rio De Janeiro. N Engl J Med (2016) 375:2321–34. doi: 10.1056/NEJMoa1602412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sousa AQ, Cavalcante DIM, Franco LM, Araújo FMC, Sousa ET, Valença-Junior JT, et al. Postmortem Findings for 7 Neonates With Congenital Zika Virus Infection. Emerg Infect Dis (2017) 23:1164–7. doi: 10.3201/eid2307.162019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. World Health Organization . Weekly Epidemiological Report. Geneva: World Health Organization; (2017). [Google Scholar]
- 10. Barbeito-Andrés J, Pezzuto P, Higa LM, Dias AA, Vasconcelos JM, Santos TMP, et al. Congenital Zika Syndrome Is Associated With Maternal Protein Malnutrition. Sci Adv (2020) 6:eaaw6284. doi: 10.1126/sciadv.aaw6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pedrosa C da SG, Souza LRQ, Gomes TA, de Lima CVF, Ledur PF, Karmirian K, et al. The Cyanobacterial Saxitoxin Exacerbates Neural Cell Death and Brain Malformations Induced by Zika Virus. PloS Negl Trop Dis (2020) 14:e0008060. doi: 10.1371/journal.pntd.0008060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caires-Júnior LC, Goulart E, Melo US, Araujo BHS, Alvizi L, Soares-Schanoski A, et al. Discordant Congenital Zika Syndrome Twins Show Differential In Vitro Viral Susceptibility of Neural Progenitor Cells. Nat Commun (2018) 9:475. doi: 10.1038/s41467-017-02790-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rossi ÁD, Faucz FR, Melo A, Pezzuto P, de Azevedo GS, Schamber-Reis BLF, et al. Variations in Maternal Adenylate Cyclase Genes Are Associated With Congenital Zika Syndrome in a Cohort From Northeast, Brazil. J Intern Med (2019) 285:215–22. doi: 10.1111/joim.12829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aguiar RS, Pohl F, Morais GL, Nogueira FCS, Carvalho JB, Guida L, et al. Molecular Alterations in the Extracellular Matrix in the Brains of Newborns With Congenital Zika Syndrome. Sci Signal (2020) 13:eaay6736. doi: 10.1126/scisignal.aay6736 [DOI] [PubMed] [Google Scholar]
- 15. Santos CNO, Ribeiro DR, Cardoso Alves J, Cazzaniga RA, Magalhães LS, de Souza MSF, et al. Association Between Zika Virus Microcephaly in Newborns With the Rs3775291 Variant in Toll-Like Receptor 3 and Rs1799964 Variant at Tumor Necrosis Factor-α Gene. J Infect Dis (2019) 220:1797–801. doi: 10.1093/infdis/jiz392 [DOI] [PubMed] [Google Scholar]
- 16. Coyne CB, Lazear HM. Zika Virus - Reigniting the TORCH. Nat Rev Microbiol (2016) 14:707–15. doi: 10.1038/nrmicro.2016.125 [DOI] [PubMed] [Google Scholar]
- 17. de Noronha L, Zanluca C, Azevedo MLV, Luz KG, dos Santos CND. Zika Virus Damages the Human Placental Barrier and Presents Marked Fetal Neurotropism. Mem Inst Oswaldo Cruz (2016) 111:287–93. doi: 10.1590/0074-02760160085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hirsch AJ, Roberts VHJ, Grigsby PL, Haese N, Schabel MC, Wang X, et al. Zika Virus Infection in Pregnant Rhesus Macaques Causes Placental Dysfunction and Immunopathology. Nat Commun (2018) 9:263–78. doi: 10.1038/s41467-017-02499-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, et al. Zika Virus Targets Different Primary Human Placental Cells, Suggesting Two Routes for Vertical Transmission. Cell Host Microbe (2016) 20:155–66. doi: 10.1016/j.chom.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quicke KM, Bowen JR, Johnson EL, McDonald CE, Ma H, O’Neal JT, et al. Zika Virus Infects Human Placental Macrophages. Cell Host Microbe (2016) 20:83–90. doi: 10.1016/j.chom.2016.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques ETDA, et al. Type III Interferons Produced by Human Placental Trophoblasts Confer Protection Against Zika Virus Infection. Cell Host Microbe (2016) 19:705–12. doi: 10.1016/j.chom.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, et al. Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe (2016) 19:882–90. doi: 10.1016/j.chom.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Delorme-Axford E, Donker RB, Mouillet J-F, Chu T, Bayer A, Ouyang Y, et al. Human Placental Trophoblasts Confer Viral Resistance to Recipient Cells. Proc Natl Acad Sci (2013) 110:12048–53. doi: 10.1073/pnas.1304718110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rabelo K, de Souza LJ, Salomão NG, Machado LN, Pereira PG, Portari EA, et al. Zika Induces Human Placental Damage and Inflammation. Front Immunol (2020) 11:2146. doi: 10.3389/fimmu.2020.02146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sadler AJ, Williams BRG. Interferon-Inducible Antiviral Effectors. Nat Rev Immunol (2008) 8:559–68. doi: 10.1038/nri2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loeb KR, Haas AL. The Interferon-Inducible 15-Kda Ubiquitin Homolog Conjugates to Intracellular Proteins. J Biol Chem (1992) 267:7806–13. doi: 10.1016/S0021-9258(18)42585-9 [DOI] [PubMed] [Google Scholar]
- 27. Der SD, Zhou A, Williams BRG, Silverman RH. Identification of Genes Differentially Regulated by Interferon, or Using Oligonucleotide Arrays. Proc Natl Acad Sci (1998) 95:15623–8. doi: 10.1073/pnas.95.26.15623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perng Y-C, Lenschow DJ. ISG15 in Antiviral Immunity and Beyond. Nat Rev Microbiol (2018) 16:423–39. doi: 10.1038/s41579-018-0020-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang B, Liu X, Chen W, Chen L. IFIT5 Potentiates Anti-Viral Response Through Enhancing Innate Immune Signaling Pathways. Acta Biochim Biophys Sin (2013) 45:867–74. doi: 10.1093/abbs/gmt088 [DOI] [PubMed] [Google Scholar]
- 30. Bowen JR, Quicke KM, Maddur MS, O’Neal JT, McDonald CE, Fedorova NB, et al. Zika Virus Antagonizes Type I Interferon Responses During Infection of Human Dendritic Cells. PloS Pathog (2017) 13:e1006164. doi: 10.1371/journal.ppat.1006164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yockey LJ, Jurado KA, Arora N, Millet A, Rakib T, Milano KM, et al. Type I Interferons Instigate Fetal Demise After Zika Virus Infection. Sci Immunol (2018) 3:eaao1680. doi: 10.1126/sciimmunol.aao1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sung PS, Hong S-H, Chung J-H, Kim S, Park S-H, Kim HM, et al. IFN-Λ4 Potently Blocks IFN-α Signalling by ISG15 and USP18 in Hepatitis C Virus Infection. Sci Rep (2017) 7:3821. doi: 10.1038/s41598-017-04186-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bordi L, Lalle E, Lapa D, Caglioti C, Quartu S, Capobianchi MR, et al. Type III Interferon (IFN-Lambda) Antagonizes the Antiviral Activity of Interferon-Alpha In Vitro . J Biol Regul Homeost Agents (2013) 27:1001–9. [PubMed] [Google Scholar]
- 34. Bordi L, Lalle E, Caglioti C, Travaglini D, Lapa D, Marsella P, et al. Antagonistic Antiviral Activity Between IFN-Lambda and IFN-Alpha Against Lethal Crimean-Congo Hemorrhagic Fever Virus In Vitro . PloS One (2015) 10:e0116816. doi: 10.1371/journal.pone.0116816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Caine EA, Scheaffer SM, Arora N, Zaitsev K, Artyomov MN, Coyne CB, et al. Interferon Lambda Protects the Female Reproductive Tract Against Zika Virus Infection. Nat Commun (2019) 10:280. doi: 10.1038/s41467-018-07993-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jagger BW, Miner JJ, Cao B, Arora N, Smith AM, Kovacs A, et al. Gestational Stage and IFN-Λ Signaling Regulate ZIKV Infection In Utero . Cell Host Microbe (2017) 22:366–376.e3. doi: 10.1016/j.chom.2017.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Welzel TM, Morgan TR, Bonkovsky HL, Naishadham D, Pfeiffer RM, Wright EC, et al. Variants in Interferon-Alpha Pathway Genes and Response to Pegylated Interferon-Alpha2a Plus Ribavirin for Treatment of Chronic Hepatitis C Virus Infection in the Hepatitis C Antiviral Long-Term Treatment Against Cirrhosis Trial. Hepatology (2009) 49:1847–58. doi: 10.1002/hep.22877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He X-X, Chang Y, Jiang H-J, Tang F, Meng F-Y, Xie Q-H, et al. Persistent Effect of IFNAR-1 Genetic Polymorphism on the Long-Term Pathogenesis of Chronic HBV Infection. Viral Immunol (2010) 23:251–7. doi: 10.1089/vim.2009.0102 [DOI] [PubMed] [Google Scholar]
- 39. Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is Associated With Response to Chronic Hepatitis C Interferon-Alpha and Ribavirin Therapy. Nat Genet (2009) 41:1100–4. doi: 10.1038/ng.447 [DOI] [PubMed] [Google Scholar]
- 40. Hernandez N, Bucciol G, Moens L, Le Pen J, Shahrooei M, Goudouris E, et al. Inherited IFNAR1 Deficiency in Otherwise Healthy Patients With Adverse Reaction to Measles and Yellow Fever Live Vaccines. J Exp Med (2019) 216:2057–70. doi: 10.1084/jem.20182295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Price AA, Tedesco D, Prasad MR, Workowski KA, Walker CM, Suthar MS, et al. Prolonged Activation of Innate Antiviral Gene Signature After Childbirth is Determined by IFNL3 Genotype. Proc Natl Acad Sci USA (2016) 113:10678–83. doi: 10.1073/pnas.1602319113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y, et al. Hepatic ISG Expression Is Associated With Genetic Variation in Interleukin 28B and the Outcome of IFN Therapy for Chronic Hepatitis C. Gastroenterology (2010) 139:499–509. doi: 10.1053/j.gastro.2010.04.049 [DOI] [PubMed] [Google Scholar]
- 43. Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, et al. IL28B Genotype Is Associated With Differential Expression of Intrahepatic Interferon-Stimulated Genes in Patients With Chronic Hepatitis C. Hepatology (2010) 52:1888–96. doi: 10.1002/hep.23912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, et al. A Variant Upstream of IFNL3 (IL28B) Creating a New Interferon Gene IFNL4 Is Associated With Impaired Clearance of Hepatitis C Virus. Nat Genet (2013) 45:164–71. doi: 10.1038/ng.2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang K, Li M, Hakonarson H. ANNOVAR: Functional Annotation of Genetic Variants From High-Throughput Sequencing Data. Nucleic Acids Res (2010) 38:e164–4. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics (2005) 21:263–5. doi: 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 47. The 1000 Genomes Project Consortium. Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, et al. An Integrated Map of Structural Variation in 2,504 Human Genomes. Nature (2015) 526:75–81. doi: 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. R Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Core Team; (2017). [Google Scholar]
- 49. Howey R, Cordell HJ. PREMIM and EMIM: Tools for Estimation of Maternal, Imprinting and Interaction Effects Using Multinomial Modelling. BMC Bioinf (2012) 13:149. doi: 10.1186/1471-2105-13-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barnholtz-Sloan JS, McEvoy B, Shriver MD, Rebbeck TR. Ancestry Estimation and Correction for Population Stratification in Molecular Epidemiologic Association Studies. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol (2008) 17:471–7. doi: 10.1158/1055-9965.EPI-07-0491 [DOI] [PubMed] [Google Scholar]
- 51. Manta FSN, Pereira R, Caiafa A, Silva DA, Gusmão L, Carvalho EF. Analysis of Genetic Ancestry in the Admixed Brazilian Population From Rio De Janeiro Using 46 Autosomal Ancestry-Informative Indel Markers. Ann Hum Biol (2013) 40:94–8. doi: 10.3109/03014460.2012.742138 [DOI] [PubMed] [Google Scholar]
- 52. Pereira R, Phillips C, Pinto N, Santos C, dos Santos SEB, Amorim A, et al. Straightforward Inference of Ancestry and Admixture Proportions Through Ancestry-Informative Insertion Deletion Multiplexing. PloS One (2012) 7:e29684. doi: 10.1371/journal.pone.0029684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, et al. Genetic and Serologic Properties of Zika Virus Associated With an Epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis (2008) 14:1232–9. doi: 10.3201/eid1408.080287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guerreiro LTA, Robottom-Ferreira AB, Ribeiro-Alves M, Toledo-Pinto TG, Rosa Brito T, Rosa PS, et al. Gene Expression Profiling Specifies Chemokine, Mitochondrial and Lipid Metabolism Signatures in Leprosy. PloS One (2013) 8:e64748. doi: 10.1371/journal.pone.0064748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol (2002) 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. WS Cleveland JT . The Collected Works of John W. Tukey. London: Taylor & Francis; (1984). [Google Scholar]
- 57. Holm . A Simple Sequentially Rejective Multiple Test Procedure, Vol. 6. (1979). pp. 65–70. [Google Scholar]
- 58. Song LH, Xuan NT, Toan NL, Binh VQ, Boldt AB, Kremsner PG, et al. Association of Two Variants of the Interferon-Alpha Receptor-1 Gene With the Presentation of Hepatitis B Virus Infection. Eur Cytokine Netw (2008) 19:204–10. doi: 10.1684/ecn.2008.0137 [DOI] [PubMed] [Google Scholar]
- 59. Zhou J, Smith DK, Lu L, Poon VKM, Ng F, Chen D-Q, et al. A non-Synonymous Single Nucleotide Polymorphism in IFNAR1 Affects Susceptibility to Chronic Hepatitis B Virus Infection. J Viral Hepat (2009) 16:45–52. doi: 10.1111/j.1365-2893.2008.01040.x [DOI] [PubMed] [Google Scholar]
- 60. Weisblum Y, Oiknine-Djian E, Vorontsov OM, Haimov-Kochman R, Zakay-Rones Z, Meir K, et al. Zika Virus Infects Early- and Midgestation Human Maternal Decidual Tissues, Inducing Distinct Innate Tissue Responses in the Maternal-Fetal Interface. J Virol (2017) 91:e01905–16. doi: 10.1128/JVI.01905-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. El Costa H, Gouilly J, Mansuy J-M, Chen Q, Levy C, Cartron G, et al. ZIKA Virus Reveals Broad Tissue and Cell Tropism During the First Trimester of Pregnancy. Sci Rep (2016) 6:35296. doi: 10.1038/srep35296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Singh PK, Guest J-M, Kanwar M, Boss J, Gao N, Juzych MS, et al. Zika Virus Infects Cells Lining the Blood-Retinal Barrier and Causes Chorioretinal Atrophy in Mouse Eyes. JCI Insight (2017) 2:e92340. doi: 10.1172/jci.insight.92340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, et al. Biology of Zika Virus Infection in Human Skin Cells. J Virol (2015) 89:8880–96. doi: 10.1128/JVI.00354-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bagasra O, Addanki KC, Goodwin GR, Hughes BW, Pandey P, McLean E. Cellular Targets and Receptor of Sexual Transmission of Zika Virus. Appl Immunohistochem Mol Morphol AIMM (2017) 25:679–86. doi: 10.1097/PAI.0000000000000580 [DOI] [PubMed] [Google Scholar]
- 65. Hastings AK, Yockey LJ, Jagger BW, Hwang J, Uraki R, Gaitsch HF, et al. TAM Receptors Are Not Required for Zika Virus Infection in Mice. Cell Rep (2017) 19:558–68. doi: 10.1016/j.celrep.2017.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schneider-Schaulies J, Schnorr JJ, Brinckmann U, Dunster LM, Baczko K, Liebert UG, et al. Receptor Usage and Differential Downregulation of CD46 by Measles Virus Wild-Type and Vaccine Strains. Proc Natl Acad Sci (1995) 92:3943–7. doi: 10.1073/pnas.92.9.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tanaka M, Ueno T, Nakahara T, Sasaki K, Ishimoto A, Sakai H. Downregulation of CD4 Is Required for Maintenance of Viral Infectivity of HIV-1. Virology (2003) 311:316–25. doi: 10.1016/S0042-6822(03)00126-0 [DOI] [PubMed] [Google Scholar]
- 68. Rothlin CV, Ghosh S, Zuniga EI, Oldstone MBA, Lemke G. TAM Receptors Are Pleiotropic Inhibitors of the Innate Immune Response. Cell (2007) 131:1124–36. doi: 10.1016/j.cell.2007.10.034 [DOI] [PubMed] [Google Scholar]
- 69. Scott GM, Chow SSW, Craig ME, Pang CNI, Hall B, Wilkins MR, et al. Cytomegalovirus Infection During Pregnancy With Maternofetal Transmission Induces a Proinflammatory Cytokine Bias in Placenta and Amniotic Fluid. J Infect Dis (2012) 205:1305–10. doi: 10.1093/infdis/jis186 [DOI] [PubMed] [Google Scholar]
- 70. Racicot K, Aldo P, El-Guindy A, Kwon J-Y, Romero R, Mor G. Cutting Edge: Fetal/Placental Type I IFN can Affect Maternal Survival and Fetal Viral Load During Viral Infection. J Immunol Baltim Md 1950 (2017) 198:3029–32. doi: 10.4049/jimmunol.1601824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Miner JJ, Cao B, Govero J, Smith AM, Fernandez E, Cabrera OH, et al. Zika Virus Infection During Pregnancy in Mice Causes Placental Damage and Fetal Demise. Cell (2016) 165:1081–91. doi: 10.1016/j.cell.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Broggi A, Ghosh S, Sposito B, Spreafico R, Balzarini F, Lo Cascio A, et al. Type III Interferons Disrupt the Lung Epithelial Barrier Upon Viral Recognition. Science (2020) 369:706–12. doi: 10.1126/science.abc3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Grajales-Reyes GE, Colonna M. Interferon Responses in Viral Pneumonias. Science (2020) 369:626–7. doi: 10.1126/science.abd2208 [DOI] [PubMed] [Google Scholar]
- 74. Secretaria de vigilância em saúde . Situação Epidemiológica Da Síndrome Congênita Associada À Infecção Pelo Vírus Zika Em 2020, Até a SE 45. Available at: https://www.gov.br/saude/pt-br/assuntos/media/pdf/2020/dezembro/11/boletim_epidemiologico_svs_47.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Zenodo, 10.5281/zenodo.5567327