Abstract
A set of universal oligonucleotide primers specific for the conserved regions of the eubacterial 16S rRNA gene was designed for use with the real-time PCR Applied Biosystems 7700 (TaqMan) system. During the development of this PCR, problems were noted with the use of this gene as an amplification target. Contamination of reagents with bacterial DNA was a major problem exacerbated by the highly sensitive nature of the real-time PCR chemistry. This was compounded by the use of a small amplicon of approximately 100 bases, as is necessary with TaqMan chemistry. In an attempt to overcome this problem, several methodologies were applied. Certain treatments were more effective than others in eliminating the contaminating DNA; however, to achieve this there was a decrease in sensitivity. With UV irradiation there was a 4-log reduction in PCR sensitivity, with 8-methoxypsoralen activity facilitated by UV there was between a 5- and a 7-log reduction, and with DNase alone and in combination with restriction digestion there was a 1.66-log reduction. Restriction endonuclease treatment singly and together did not reduce the level of contaminating DNA. Without the development of ultrapure Taq DNA polymerase, ultrapure reagents, and plasticware guaranteed to be free of DNA, the implementation of a PCR for detection of eubacterial 16S rRNA with the TaqMan system will continue to be problematical.
Recently, PCR assays for the improved nonculture diagnosis of meningococcal and other bacterial diseases have been developed as a result of the growing discrepancy between the number of clinically diagnosed cases of meningococcal infection about which the Office of National Statistics is notified and culture-confirmed cases identified by the Public Health Laboratory Service Meningococcal Reference Unit (14). Additionally, accurate disease surveillance is essential when polysaccharide-protein conjugate vaccines for meningococcal serogroup C (5, 26) and pneumococcal disease (23) are used and soon to be introduced into the national immunization schedules.
The advent of molecular techniques, notably, PCR, makes it possible to identify the presence of bacterial DNA in culture-negative samples from patients with suspected infection (1, 4). This approach allows nonculture confirmation of meningitis and septicemia, which leads to improved disease surveillance and which provides guidance on appropriate antibiotic usage and patient management.
Universal PCR can be used as a tool for the rapid detection of bacteria in normally sterile clinical samples and, as such, would be useful in differentiating bacterial from viral infections. This would confirm the necessity for antibiotic treatment and would influence patient management. Numerous workers (8, 16, 21, 24, 33) have used the 16S rRNA gene as a target for nonculture detection, and it has been the most widely used target for universal PCR amplification of DNAs from a broad range of organisms (17). The 16S rRNA gene is present in multiple copies in the genomes of all known human bacterial pathogens that belong to the eubacterial kingdom. Many bacterial species contain up to seven copies of the gene (3). A gene target that is present in multiple copies increases the possibility of detection of small numbers of pathogens over an assay that detects a single copy gene target. A large amount of 16S rRNA sequence data is available, and these data indicate the highly conserved nature of the gene across the eubacterial kingdom. In addition, there is sufficient variation within the 16S rRNA gene to provide species-specific discrimination of some of the major causative agents of meningitis and septicemia, namely, Neisseria meningitidis, Escherichia coli, Haemophilus influenzae, Streptococcus pneumoniae, and Listeria monocytogenes (24).
PCR is capable of 106- to 107-fold amplification of a single copy of template DNA (29), making minor contamination of the PCR mixture with exogenous DNA a problem. This problem is exaggerated by the use of a highly conserved multiple-copy amplification target. The implementation of a universal 16S rRNA PCR can be hindered by problems with contamination of reagents which may be derived from a bacterial source, such as Taq DNA polymerase and uracil-N-glycosylase (UNG). During enzyme production, nucleic acid, including ribosomal DNA sequences, are copurified. It has been well documented (2, 12, 25, 31) that Taq DNA polymerase enzyme may contain a source of contaminating DNA as a result of its manufacture and incomplete purification. The enzyme is commonly expressed as a recombinant protein in E. coli or is obtained as a native protein from Thermus aquaticus (19). Several investigators (6, 10, 13, 27, 32) have encountered and attempted to overcome this problem by a strategy of Taq DNA polymerase treatment by physical, chemical, and enzymatic means. However, these strategies used low-sensitivity detection by agarose gel electrophoresis with ethidium bromide staining and visualization with UV light and are not optimal for the detection of small numbers of the bacterial template DNA copies often found in culture-negative clinical samples.
Recently, a new PCR technology that combines enhanced specificity and enhanced sensitivity has been developed by Perkin-Elmer Applied Biosystems (PE-ABI; Foster City, Calif.). The ABI Prism 7700 Sequence Detector, known as the TaqMan system, uses a fluorogenic probe-based 5′ exonuclease technology in a closed-tube format with a resulting amplicon of approximately 100 bases. This system has been evaluated for its ability to detect meningococcal DNA (7a) and has been found to improve the laboratory confirmation of meningococcal disease (15). This study describes a number of methodologies used to overcome the problem of contaminating DNA when using the sensitive real-time TaqMan system.
MATERIALS AND METHODS
Measures such as the provision of dedicated rooms and the use of dedicated equipment were taken to reduce the possibility of contamination at all times (18). Sterile water from two manufacturers (Sigma Chemicals, Poole, England; Phoenix Pharmaceuticals, Gloucester, England) was examined to assess the possible differences in the levels of contaminating DNA.
Primer and probe design.
The primers were designed with the Primer Express software package (PE-ABI, Warrington, England) to amplify a product of 87 bases from E. coli. The fluorescent probe was the reverse complement of the universal bacterial probe (RDR245), as described by Greisen et al. (7).
Bacterial strains and culture.
N. meningitidis serogroup B and C isolates referred to the Meningococcal Reference Unit (Manchester Public Health Laboratory, Withington Hospital, Manchester, United Kingdom) were used along with E. coli isolates from blood culture specimens sent to the routine clinical microbiology laboratory. The organisms were recovered from Microbank vials (Pro-Lab Diagnostics, Neston, Wirral, United Kingdom) that had been stored at −80°C and were cultured overnight on blood agar (Oxoid, Basingstoke, United Kingdom) at 37°C in 5% CO2.
DNA extraction. (i) E. coli.
Five colonies of pure bacterial cultures were emulsified in 500 μl of sterile injectable water (Phoenix Pharmaceuticals). The suspension was boiled for 10 min and was then rapidly cooled and held at −20°C for 1 min to denature the DNA. Following centrifugation at 12,000 × g for 5 min, the extracted DNA pellet was distributed into autoclaved sterile vials in 50-μl aliquots with presterilized pipette tips (Molecular BioProducts, San Diego, Calif.) and was stored at −80°C until required.
(ii) N. meningitidis.
In a microbiological class II safety cabinet, a sweep from a pure culture with a sterile cotton swab was emulsified in 2 ml of sterile injectable water. By using a spectrophotometer (Pharmacia, St. Albans, England) set at 650 nm, the meningococcal suspension was standardized to an optical density of 0.1 and was adjusted to a concentration of approximately 20,000 bacteria/ml, which represents 40 bacteria per 2 μl of inoculum. DNA denaturation and aliquot distribution were as described above for E. coli.
PCR components and amplification profile.
On the basis of a volume of 23 μl per reaction mixture, the master mixture was prepared from the TaqMan Core Reagent kit (PE-ABI) on the basis of the manufacturer's recommendations. Briefly, this comprises 200 nM each oligonucleotide primer (forward primer, 5′1320-CCATGAAGTCGGAATCGCTAG-13413′; reverse primer, 5′1431-ACTCCCATGGTGTGACGG1413-3′), 100 nM fluorescent labeled probe (6-FAM-5′-CGGTGAATACGTTCCCGGGCCTTGTAC-3′-TAMRA [where 6-FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine]), GeneAmp 10× PCR Buffer II, 6 mM MgCl2, 200 μM each deoxynucleoside triphosphate (dNTP; dATP, dCTP, dGTP, and dUTP) and 0.125 U of Low-DNA AmpliTaq polymerase LD (PE-ABI).
Master mixture preparation (UV only and 8-methoxypsoralen [8-MOP] plus UV).
The master mixture for the treatments involving UV light were prepared as outlined above by adding the UV absorbent (6), dNTPs, and oligonucleotide primers after UV exposure of the other components.
Controls.
Negative controls consisted of 23 μl of the master mixture with 2 μl of sterile injectable water (Phoenix Pharmaceuticals) as a template. An aliquot (2 μl) of extracted E. coli and N. meningitidis DNAs suspended in sterile injectable water was added to the master mixture in the same way to constitute the positive control.
Amplification of the DNA was performed on the ABI 7700 Sequence Detection system (TaqMan system). The parameters used were as follows: 50°C for 2 min and 95°C for 10 min, followed by 45 cycles at 95°C for 15 s and 60°C for 1 min.
Taq DNA polymerase source comparison.
Taq DNA polymerases from several sources were tested to determine whether different levels of contamination were present in Taq DNA polymerases from a number of manufacturers. Previously unopened vials of native Taq DNA polymerase (Life Technologies, Paisley, Scotland), Taq DNA Polymerase Batch 901AA (BioGene Limited, Kimbolton, England), AmpliTaq DNA Polymerase (PE-ABI), and Low-DNA AmpliTaq polymerase LD (PE-ABI) were compared.
Taq DNA polymerase treatment. (i) UV irradiation.
The modified master mixture was irradiated with the UV Linker (Oncor Appligene, Chester-le-Street, England) within a spectrum of 312 and 365 nm with concentrations of UV light between 1 and 12 J/cm2. Following irradiation and the addition of dNTPs, primers, and template, PCR was performed as described above.
(ii) 8-MOP and UV irradiation.
8-MOP (Sigma Chemicals) dissolved in dimethyl sulfoxide (DMSO; BDH Chemicals Ltd., Poole, England) was added to the modified master mixture to give a working concentration of 25 μg/ml and a final concentration of 1% DMSO in the PCR assay. The same concentration of DMSO was included in the control mixture. The mixture was incubated for 1 h in the dark at room temperature and was then irradiated with the UV Linker (Oncor Appligene) at 365 nm for between 1 and 5 min. Following treatment and the addition of template, PCR was performed with the TaqMan system.
DNase I enzyme.
DNase I enzymes were obtained from two different manufacturers, Promega Corporation (Southampton, England) and Life Technologies. In both cases the oligonucleotide primers, probe, and dNTPs were added after treatment of the Low-DNA AmpliTaq DNA polymerase LD and MgCl2 in sterile injectable water and PCR buffer. Amplification was performed with the TaqMan system.
The DNase from Promega was diluted in sterile water to which 0.125 U of Low-DNA AmpliTaq DNA polymerase LD per reaction mixture had been added to give concentrations of 100, 30, 25, 20, 10, and 5 U per liter. The solution was incubated at 37°C for 10 min and was then denatured at 95°C for 5 min. The Low-DNA AmpliTaq DNA polymerase LD was buffered by the addition of 1% bovine serum albumin and 10 mM MgCl2, as recommended by the manufacturer. The DNase from Life Technologies was diluted in 1× DNase buffer (Life Technologies) and sterile water in the range 100 to 10 U per liter. The Low-DNA AmpliTaq DNA polymerase LD was added to give a final concentration of 0.125 U per PCR mixture. The solution was incubated at room temperature for 15 min, followed by the addition of 0.2 μl of EDTA per reaction mixture and heat denaturation at 70°C for 10 min.
Restriction endonuclease digestion.
Five restriction endonuclease enzymes, AvaI, HaeIII, HinfI, Sau3AI, and SmaI (Pharmacia Biotech, St. Albans, England), were selected for use in the pretreatment of the PCR master mixture on the basis of the restriction sites identified in the 16S rRNA sequence within the primer binding sites by use of the Genetics Computer Group MAP software program (Program Manual for the Wisconsin Package, version 8, Aug. 1994, Genetics Computer Group, Madison, Wis.). The ability of each enzyme to digest a false-positive product was demonstrated by incubating 1 μl of each enzyme with 15 μl of product at 37°C overnight in the presence of One-phor-all buffer (supplied with the enzymes by the manufacturer). Following digestion, restriction enzymes were heat inactivated according to the manufacturer's recommendations. (Program Manual for the Wisconsin Package, version 8, Aug. 1994, Genetics Computer Group, Madison, Wis.). Restriction digests (10 μl) mixed with ethidium bromide were analyzed by 2% agarose (Oncor Appligene) gel electrophoresis for 30 min. The DNA intercalated with ethidium bromide was visualized by using a UV fluorescence transilluminator (Genetic Research Instruments, Braintree, United Kingdom). The restriction enzymes were used to treat the Low-DNA AmpliTaq polymerase LD, both with and without the other master mixture reagents, singly or in combination in the presence of One-phor-all buffer. To minimize damage to the Taq DNA polymerase caused by heat, the enzymes were denatured at temperatures below 85°C. After treatment, PCR was performed as described above.
Restriction endonuclease digestion and DNase I enzyme concentration.
The restriction endonuclease digestion was performed as described above for the treatment of Low-DNA AmpliTaq DNA polymerase LD in sterile water. DNase I was added to the digestion mixture at a concentration of 16.6 U per liter, and the mixture was incubated at room temperature for 15 min, followed by denaturation at 70°C for 10 min to minimize damage to the Taq DNA polymerase caused by heat. The remaining master mixture reagents were added, and DNA amplification was performed with the TaqMan system.
The GenBank accession number for the E. coli sequence is X80733.
RESULTS
The threshold/cycle (CT) value is a measure of the dye fluorescence generated by cleavage of probe against a fixed baseline threshold. A comparison of the sterile water from different sources showed no variation in the false-positive CT value. No false-positive signal was detected when the primers and probe diluted in sterile water were amplified in the reaction plate.
Comparison of Taq DNA polymerases from different sources.
Taq DNA polymerases from all sources gave false-positive results for the no-template (water) controls, with similar CT values.
UV irradiation.
The CT values for the positive and negative controls increased with intensity (as joules per square centimeter) of UV exposure (Table 1). A UV dose of 4 J/cm2 necessary to eliminate endogenous DNA resulted in a 4-log-unit loss in the amount of DNA detected; this is equivalent to an increase of 12 PCR cycles in the CT value for the positive control.
TABLE 1.
Effect of UV dose on CT value for 16S rRNA PCR with positive (N. meningitidis) and negative (template) controls
| Control |
CT value for the following UV exposure (J/cm2):
|
||||
|---|---|---|---|---|---|
| None | 1 | 2 | 3 | 4 | |
| Positive | 23.71 | 23.16 | 24.07 | 29.10 | 35.45 |
| Negative | 27.21 | 30.66 | 32.26 | 39.12 | >45.0a |
A value of >45.0 is deemed to be negative.
8-MOP and UV irradiation.
At concentrations greater than or equal to 50 μg/ml, 8-MOP was found to inhibit the PCR. A concentration of 25 μg/ml was not inhibitory and so was selected for use in further studies. The final DMSO concentration in the PCR mixture was fixed at 1%, as DMSO concentrations higher than 5% have been shown to inhibit the PCR (21). UV exposure times were optimized in the range of 1 to 5 min. The CT value for the negative control which included 8-MOP and no UV activation was 23.05, which increased to >45.0 after 5 min of exposure. Similarly, the CT value for the positive controls increased from 19.23 to 35.57 with E. coli and 21.77 to 43.79 with N. meningitidis with increasing UV exposure times (Table 2). This equates to a reduction in PCR sensitivity of 5 log for E. coli and 7 log for N. meningitidis controls.
TABLE 2.
Effects of 8-MOP dissolved in DMSO and UV irradiation time on CT value for 16S rRNA PCR-positive (E. coli and N. meningitidis) and -negative (template) controls
| Control |
CT value for the following UV exposure time (min):
|
|||||
|---|---|---|---|---|---|---|
| None | 1 | 2 | 3 | 4 | 5 | |
| Positive, N. meningitidis | 21.77 | 24.90 | 27.76 | 27.02 | 29.75 | 43.79 |
| Positive, E. coli | 19.23 | 19.77 | 20.39 | 21.67 | 25.30 | 35.57 |
| Negative | 23.05 | 26.64 | 25.31 | 30.15 | 35.24 | >45.0a |
A CT value of >45.0 is deemed to be negative.
DNase I.
Concentrations of DNase I from Promega of greater than 30 U per liter inhibited the PCR. The concentration necessary to eliminate a false-positive signal, 5 U per liter, resulted in a loss of sensitivity equivalent to that of 3 PCR cycles, or 1 log unit. The product yield (ΔRn) value was reduced from 1.4 to 0.35. The DNase I from Life Technologies at a concentration of 100 U per liter eliminated the false-positive signal, with a loss of sensitivity equivalent to that of 6 PCR cycles, or 2 log units (Table 3).
TABLE 3.
DNase I concentration optimization
| DNase I source and control |
CT value (ΔRn value) with DNase I at the following concn (U/liter):
|
||||||
|---|---|---|---|---|---|---|---|
| 100 | 30 | 25 | 20 | 10 | 5 | No enzyme | |
| Promega | |||||||
| Positive | >45.0 | >45.0 | 23.65 | 24.04 | 27.71 | 26.88 (0.35) | 23.5 (1.4) |
| Negative | >45.0a | >45.0 | 31.80 | 28.81 | 31.79 | >45.0 | 28.93 |
| Life Technologies | |||||||
| Positive | 29.40 | 25.27 | 24.49 | 24.42 (1.4) | 23.31 (3.0) | ||
| Negative | >45.0 | 31.67 | 29.74 | 31.19 | 30.07 | ||
A CT value of >45.0 is deemed to be negative.
Restriction endonuclease digestion.
Restriction endonuclease digestion of false-positive product resulted in two bands by UV visualization. Restriction endonuclease treatment either reduced the CT value or had very little effect on the CT value for the false-positive signal (Table 4).
TABLE 4.
Effects of restriction endonucleases singly and in combination on CT value for positive 16S rRNA (E. coli and N. meningitidis) and negative (sterile water) template controls
| Control |
CT value with the following restriction enzyme (enzyme combinationa):
|
|||||
|---|---|---|---|---|---|---|
| No enzyme | AvaI | HaeIII | HinfI | Sau3AI | SmaI | |
| Positive, E. coli | 17.26 | 17.26 | 16.77 | 17.10 | 16.84 | 17.12 |
| Positive, N. meningitidis | 24.08 | 23.70 | 23.77 | 23.32 | 23.49 | 23.65 |
| Negative | 28.61 | 29.20 | 28.53 (28.54) | 28.81 (26.39) | 27.80 (28.65) | 27.80 (30.01) |
Combinations included AvaI and the enzyme for which data are provided in the column.
Restriction endonuclease digestion and DNase I enzyme concentration.
The DNase I enzyme concentration was optimized at 16.6 U of DNase I per liter and was used in combination with the AvaI restriction endonuclease (Table 5). This combined treatment eliminated the false-positive signal, with no loss of PCR sensitivity, as demonstrated by the CT values for the positive control with and without treatment, which were almost identical. The ΔRn value for the positive control increased with increasing DNase I concentration. Lower concentrations of DNase I are ineffective at reducing a false-positive CT value. Stronger dilutions of DNase I caused a minimum loss of PCR sensitivity equivalent to that of 5 cycles, or 1.66 log units.
TABLE 5.
Optimization of DNase I enzyme concentration with AvaI restriction enzyme
| Control |
CT value (ΔRn value) with the following DNase I concn (U/liter):
|
||||||
|---|---|---|---|---|---|---|---|
| 30 | 25 | 20 | 16.6 | 10 | 6.6 | No enzyme | |
| Positive, E. coli | 24.90 (0.4) | 23.20 (0.8) | 21.60 (1.4) | 16.44 (2.0) | 16.44 | 17.4 | 16.35 (3.0) |
| Negative | >45.0a | >45.0 | >45.0 | >45.0 | 30.1 | 29.3 | 28.4 |
A CT value of >45.0 is deemed to be negative.
DISCUSSION
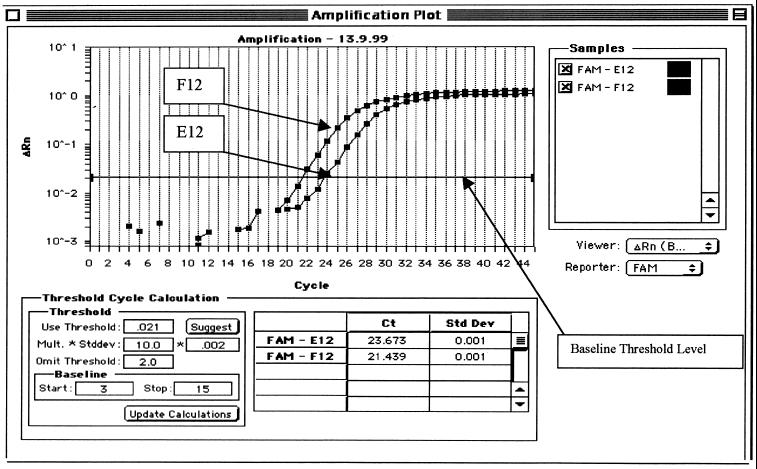
The PCR assay with the TaqMan system uses a fluorogenic probe labeled at the 5′ end with a reporter dye (6-FAM) and at the 3′ end with the quencher dye (TAMRA). When the sequence-specific probe is cleaved by Taq DNA polymerase 5′ nuclease activity, the reporter dye is separated from the quencher dye, generating a fluorescent, sequence-specific signal. The TaqMan system monitors the level of fluorescence at every cycle. In this way, the CT value can be determined and the real-time progress of the PCR can be monitored. The endpoint measurement of the amount of accumulated PCR product is referred to as the ΔRn value (Fig. 1) (9, 11, 20). Figure 1 also demonstrates a typical amplification plot in a negative control reaction (E12) without any Low-DNA AmpliTaq polymerase LD treatment.
FIG. 1.
Typical 16S rRNA PCR logarithmic amplification plot for N. meningitidis (F12) and no-template (E12) control fluorescent signal (no treatment).
A number of previously described methodologies for the elimination of endogenous DNA were evaluated. The eubacterial TaqMan system primers were found to amplify rRNA sequences when no exogenous DNA had been added to the negative (no-template) control. DNA sequencing of the contaminating product with the PE-ABI 310 Genetic Analyzer revealed more than one sequence; therefore, it was not possible to identify one contaminating organism. By amplification of the primers and probe diluted in sterile water, nonspecific interactions with the plasticware were eliminated as a source of a false-positive signal. The PCR product was of the predicted size for bacterial 16S rRNA, as determined by agarose gel electrophoresis. The water from Sigma is filtered through a 0.2-μm-pore-size filter, which will remove most microorganisms. Filtering will not, however, remove the nucleic acids from the lysed organisms. Thus, the water source cannot be completely discounted as a source of contamination. In addition, water contamination could occur postfiltration. Taking this into consideration, it is very difficult to determine the source of the contaminating DNA. Similarly, plasticware and reagents may become contaminated at the time of manufacture.
The similar CT values obtained by the Taq DNA polymerase source comparison are at odds with the findings of Böttger (2), who identified quantitative differences in Taq polymerases from different manufacturers. Low-DNA AmpliTaq polymerase LD was selected for further studies on the basis of the following: (i) the quality control procedure during manufacture limits the number of copies of 16S ribosomal DNA to 10 or fewer per 2.5 U of enzyme, (ii) this enzyme gave the highest ΔRn value with the N. meningitidis and E. coli controls, and (iii) Greisen et al. (7) and Meier et al. (21) recommend the use of Low-DNA AmpliTaq polymerase LD.
Another possible source of contaminating DNA in the TaqMan PCR master mixture is UNG, an enzyme expressed in an E. coli host. Elimination of UNG from the PCR mixture reduced the false-positive ΔRn value and the contaminating CT value by 1 PCR cycle, which indicates a reduction in the number of copies of contaminating DNA. As a result, UNG was eliminated from the PCR master mixture for the Low-DNA AmpliTaq DNA polymerase LD enzyme treatments.
The use of UV irradiation has been identified as a method for the potential reduction or elimination of contaminating DNA from PCR when endogenous DNA is a problem (6, 21, 30, 31). To find the most effective and reproducible method of UV treatment of Taq DNA polymerase, the enzyme was irradiated with a range of UV doses measured either as concentration (in joules per square centimeter) or for a given period (in minutes). The CT value for the positive and negative (no-template) controls increased in proportion to the dose of UV irradiation (Table 1). This is indicative of a decrease in PCR efficiency most likely due to damage of the Taq DNA polymerase enzyme by irradiation (22). Large molecules such as Taq DNA polymerase are more likely to be highly UV sensitive (30). With the dose of UV irradiation necessary to eliminate the false-positive signal by use of TaqMan chemistry, it is difficult to discern whether UV irradiation is degrading the contaminating DNA or whether a decrease in the false-positive signal is due to reduced Taq DNA polymerase activity. Any advantage of improved sensitivity resulting from the amplification of a multicopy gene target is negated by reduced sensitivity as a result of pretreatment to eliminate exogenous DNA from the PCR reagents.
Psoralens are known to intercalate into double-stranded nucleic acids and to form a covalent interstrand cross-link after photoactivation with light at between 320 and 400 nm (13, 21). The use of 8-MOP activated by UV irradiation without interfering with Taq DNA polymerase was suggested by Jinno et al. (13) and has since been used to successfully remove contaminating DNA from 16S rRNA PCR mixtures (12, 21). In this study it was not possible to eliminate endogenous contaminating DNA from the PCR mixture without seriously compromising the efficiency of the PCR. This was probably due to the damage to Taq DNA polymerase caused by UV irradiation. The price of eliminating contaminating DNA was a reduction in PCR sensitivity. An exposure time of 4 min in combination with a limitation of the number of PCR cycles to 30, as used by Hughes et al. (12), would obviate the false-positive signal (Table 2) (negative control CT value, 35.24). However, the loss of sensitivity, as can be seen when the highly sensitive TaqMan chemistry is used, could not be overlooked. As with UV irradiation treatment of Taq DNA polymerase, any advantage gained by targeting a multicopy gene would be seriously reduced. Maximization of the number of PCR cycles when amplification is continuously monitored is particularly important when attempting to detect small numbers of bacteria in a clinical specimen.
DNase I is capable of degrading single- and double-stranded DNAs, producing 3′ hydroxyl oligonucleotides. One unit of enzyme completely degrades 1 μg of DNA in 10 min at 37°C. By treating Low-DNA AmpliTaq DNA polymerase LD in this way, any contaminating DNA should be eliminated. DNase I treatment reduced the sensitivity of the PCR, with a reduced ΔRn value being indicative of limited primer or template availability for amplification. This may possibly be due to residual DNase I activity during the PCR procedure. Supporting evidence is provided in the form of negative results for both positive and negative controls at DNase I concentrations of greater than 25 U per liter; i.e., the enzyme is too concentrated. For complete denaturation, DNase I must be heated at 95°C for 50 min (10). Doing so would compromise the Taq DNA polymerase amplification efficiency, as the enzyme has a half-life of 40 min (10). A decrease in PCR sensitivity could be due to exposure of Taq DNA polymerase to 95°C for 5 min. For DNase I treatment to be optimally effective, therefore, it is necessary to obtain the most durable Taq DNA polymerase available. The DNase I from Life Technologies was selected on the basis of the fact that it has a denaturation temperature of 70°C, thus limiting Taq DNA polymerase damage by avoiding exposure to temperatures above 90°C.
A reduction in the CT value as a result of restriction endonuclease digestion indicates an increased template concentration, as an initial inoculum, in the PCR mixture. This suggests that additional contaminating DNA was introduced into the PCR mixture by the restriction endonuclease enzymes (13), and a further reduction in CT values for the false-positive control confirms this impression when the enzymes are used in combination as opposed to singly (Table 4). This may partially be due to the bacterial origin of the restriction enzymes; for example, AvaI is from Anabaena variabilis (28). The ineffectiveness of this set of restriction endonucleases in reducing the false-positive signal suggests that the contaminant may not be E. coli DNA (12). Rather, it indicates that the contaminating DNA may be introduced from an environmental source or from the buffer or chromatography columns during purification of the Taq DNA polymerase (25).
The effective elimination of false-positive amplification by restriction endonuclease digestion was demonstrated previously (32). The amplification target, however, was not the highly conserved 16S rRNA gene, and the detection system was agarose gel electrophoresis, not the more sensitive TaqMan chemistry. Until the organism that is the source of the contaminating DNA can be correctly identified, enabling the selection of appropriate enzymes, restriction endonuclease digestion as a means of decontaminating the 16S rRNA PCR will be relatively ineffective.
A double-treatment strategy for elimination of the contaminating DNA associated with Taq DNA polymerase enzyme was examined. This involved treatment with restriction enzymes followed by DNase I treatment, as it was thought that this might be more effective than either individual treatment. AvaI was selected as it was the restriction enzyme most effective in reducing the CT value for the false-positive control. As such, E. coli 16S rRNA would be specifically targeted, with DNase I enzyme treatment degrading any double- or single-stranded DNA. There were problems associated with this combined decontamination strategy in that the results were not easily reproducible. It was believed that 16.6 U of DNase I per liter was approaching the upper limit of enzyme activity, with additional problems being due to primer or template limitation as a result of residual DNase I activity.
In conclusion, to give consistent results, it was not possible to eliminate contaminating DNA from the PCR for detection of eubacterial 16S rRNA without a significant decrease in sensitivity. The loss in sensitivity obviates any advantage gained in amplifying a multicopy target, which, in this study, was the 16S rRNA gene.
Taq DNA polymerase has a high affinity for DNA (25); therefore, a certain amount of contaminating bacterial DNA may always remain protected from physical, chemical, or enzymatic treatment.
The most effective treatment was that with the DNase I enzyme, although it was not possible to fully denature the enzyme without compromising Taq DNA polymerase activity. It may be possible to improve on these results by using a more thermostable Taq DNA polymerase such as Deep VentR (exo) (New England Biolabs, Hitchin, England), which is from a Pyrococcus sp. (isolate GB-D) and which has a half-life of 23 h at 95°C and a half-life of 8 h at 100°C.
Reducing the number of PCR cycles would produce the false-positive signal outside the detection limits by use of real-time TaqMan chemistry. This approach, however, would not provide the improved nonculture means of detection needed for enhanced disease surveillance.
The small amplicon size coupled with the highly conserved nature of the 16S rRNA gene compounds any problem resulting from the lack of sequence variation between pathogenic bacterial sequences and contaminating bacterial sequences.
Without the development of ultrapure Taq DNA polymerase, ultrapure reagents, and plasticware guaranteed to be free of DNA, the implementation of a PCR for detection of eubacterial 16S rRNA by sensitive technologies, such as the TaqMan system, will continue to be problematical.
Thus, single-copy, species-specific PCR assays which may be used in multiplex formats are likely to prove to be the assays of choice for real-time PCR.
ACKNOWLEDGMENTS
This study was funded by the Meningitis Research Foundation, Bristol, United Kingdom.
Many thanks to Francesca Sadler for DNA sequencing reactions.
REFERENCES
- 1.Borrow R, Claus H, Guiver M, Smart L, Jones D M, Kaczmarski E B, Frosch M, Fox A J. Non-culture diagnosis and serogroup determination of meningococcal B and C infection by a siayltransferase (siaD) PCR ELISA. Epidemiol Infect. 1997;118:111–117. doi: 10.1017/s0950268896007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Böttger E C. Frequent contamination of Taq DNA polymerase with DNA. Clin Chem. 1990;36:1258–1259. [PubMed] [Google Scholar]
- 3.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Bacteriol. 1981;62:293–300. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 4.Davison E, Borrow R, Guiver M, Kaczmarski E B, Fox A J. The adaptation of the IS1106 PCR to a PCR ELISA format for the diagnosis of meningococcal infection. Serodiagn Immunother Infect Dis. 1996;8:51–56. [Google Scholar]
- 5.Fairley C K, Begg N, Borrow R, Fox A J, Jones D M, Cartwright K A V. Reactogenicity and immunogenicity of conjugate meningococcal serogroup A and C vaccine in UK infants. J Infect Dis. 1996;174:1360–1363. doi: 10.1093/infdis/174.6.1360. [DOI] [PubMed] [Google Scholar]
- 6.Frothingham R, Blitchington R B, Lee D H, Greene R C, Wilson K H. UV absorption complicates PCR decontamination. BioTechniques. 1992;13:208–210. [PubMed] [Google Scholar]
- 7.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Guiver, M., R. Borrow, J. Marsh, S. G. Gray, E. B. Kaczmarski, and A. J. Fox. Evaluation of the Applied Biosystems Automated TaqMan PCR System for the detection of meningococcal DNA. FEMS Immunol. Med. Microbiol., in press. [DOI] [PubMed]
- 8.Hendolin P H, Markkanen A, Ylikoski J, Wahlfors J J. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J Clin Microbiol. 1997;35:2854–2858. doi: 10.1128/jcm.35.11.2854-2858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR: real-time monitoring of DNA amplification reactions. Bio/Technology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 10.Hilali F, Saulnier P, Chachaty E, Andremont A. Decontamination of polymerase chain reaction reagents for detection of low concentration of 16S rRNA genes. Mol Biotechnol. 1997;7:207–216. doi: 10.1007/BF02740812. [DOI] [PubMed] [Google Scholar]
- 11.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes M S, Beck L A, Skuce R A. Identification and elimination of DNA sequences in Taq DNA polymerase. J Clin Microbiol. 1994;32:2007–2008. doi: 10.1128/jcm.32.8.2007-2008.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinno Y, Yoshiura K, Niikawa N. Use of psoralen as extinguisher of contaminating DNA in PCR. Nucleic Acids Res. 1990;18:6739. doi: 10.1093/nar/18.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaczmarski E B. Meningococcal infections in England and Wales. Commun Dis Rep Rev. 1995;7:R55–R59. [PubMed] [Google Scholar]
- 15.Kaczmarski E B, Ragunathan P L, Marsh J, Gray S J, Guiver M. Creating a national service for the diagnosis of meningococcal disease by polymerase chain reaction. Commun Dis Public Health. 1998;1:54–56. [PubMed] [Google Scholar]
- 16.Klausegger A. Gram type-specific broad-range PCR amplification for rapid detection of 62 pathogenic bacteria. J Clin Microbiol. 1999;37:464–466. doi: 10.1128/jcm.37.2.464-466.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krzanowski J J. Broad-range polymerase chain reaction for detection and identification of bacteria. J Fla Med Assoc. 1994;81:835–837. [PubMed] [Google Scholar]
- 18.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1984;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 19.Lawyer F C, Stoffel S, Saiki R K, Myambo K, Drummond R, Gelfand D H. Isolation, characterisation, and expression in E. coli of the DNA polymerase gene from the extreme thermophile, Thermus aquaticus. J Biol Chem. 1989;264:6427–6437. [PubMed] [Google Scholar]
- 20.Livak K J, Flood S J, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 21.Meier A, Persing D, Finken M, Böttger E C. Elimination of contaminating DNA within polymerase chain reaction reagents: implication for a general approach to detection of uncultured pathogens. J Clin Microbiol. 1993;31:646–652. doi: 10.1128/jcm.31.3.646-652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ou C-Y. Use of UV irradiation to reduce false positivity in polymerase chain reaction. BioTechniques. 1991;10:442–444. [PubMed] [Google Scholar]
- 23.Poland G A. The burden of pneumococcal disease: the role of conjugate vaccines. Vaccine. 1999;17:1605–1611. doi: 10.1016/s0264-410x(98)00435-6. [DOI] [PubMed] [Google Scholar]
- 24.Rådström P, Bäckman A, Qian N, Kragsbjerg P, Påhlson C, Olcén P. Detection of bacterial DNA in cerebrospinal fluid by an assay for simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and streptococci using a seminested PCR strategy. J Clin Microbiol. 1994;32:2738–2744. doi: 10.1128/jcm.32.11.2738-2744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rand K H, Houck H. Taq polymerase contains bacterial DNA of unknown origin. Mol Cell Probes. 1990;4:445–450. doi: 10.1016/0890-8508(90)90003-i. [DOI] [PubMed] [Google Scholar]
- 26.Richmond P, Miller E, Borrow R, Clark S, Sadler F, Fox A J, Begg N T, Morris R, Cartwright K A V. Meningococcal serogroup C conjugate vaccine is immunogenic in infancy and primes for memory. J Infect Dis. 1999;179:1569–1572. doi: 10.1086/314753. [DOI] [PubMed] [Google Scholar]
- 27.Rochelle P A, Weightman A J, Fry J C. DNase I treatment of Taq DNA polymerase for complete PCR decontamination. BioTechniques. 1992;13:520. [PubMed] [Google Scholar]
- 28.Roizes G, Nardeaux P C, Monier R. A new specific endonuclease from Anabaena variabilis. FEMS Lett. 1979;104:39–44. doi: 10.1016/0014-5793(79)81081-9. [DOI] [PubMed] [Google Scholar]
- 29.Saiki R K. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 30.Sarkar G, Sommer S S. Removal of DNA contamination in polymerase chain reaction. Methods Enzymol. 1993;218:381–389. doi: 10.1016/0076-6879(93)18030-g. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt T M, Pace B, Pace N. Detection of DNA contamination in Taq DNA polymerase. BioTechniques. 1991;11:176–177. [PubMed] [Google Scholar]
- 32.Sharma J K, Gopalkrishna V, Das B C. A simple method for elimination of unspecific amplifications in polymerase chain reaction. Nucleic Acids Res. 1992;20:6117–6118. doi: 10.1093/nar/20.22.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson K H, Blitchington R B, Greene R C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]