Abstract
We have previously shown that the control of cellular copper homeostasis by the copper-modulated transcription factor GRISEA has an important impact on the phenotype and lifespan of Podospora anserina. Here we demonstrate that copper depletion leads to the induction of an alternative respiratory pathway and to an increase in lifespan. This response compensates mitochondrial dysfunctions via the expression of PaAox, a nuclear gene coding for an alternative oxidase. It resembles the retrograde response in Saccharomyces cerevisiae. In P. anserina, this pathway appears to be induced by specific impairments of the copper-dependent cytochrome c oxidase. It is not induced as the result of a general decline of mitochondrial functions during senescence. We cloned and characterized PaAox. Transcript levels are decreased when cellular copper, superoxide, and hydrogen peroxide levels are raised. Copper also controls transcript levels of PaSod2, the gene encoding the mitochondrial manganese superoxide dismutase (PaSOD2). PaSod2 is a target of transcription factor GRISEA. During the senescence of wild-type strain s, the activity of PaSOD2 decreases, whereas the activity of the cytoplasmic copper/zinc superoxide dismutase (PaSOD1) increases. Collectively, the data explain the postponed senescence of mutant grisea as a defined consequence of copper depletion, ultimately leading to a reduction of oxidative stress. Moreover, they suggest that during senescence of the wild-type strain, copper is released from mitochondria. The involved mechanism is unknown. However, it is striking that the permeability of mitochondrial membranes in animal systems changes during apoptosis and that mitochondrial proteins with an important impact on this type of cellular death are released.
In biological systems, copper as a cofactor of different enzymes (e.g., cytochrome c oxidase [COX] and Cu/Zn superoxide dismutase [SOD1]) is an essential trace element. Apart from this essential role, elevated copper levels are cytotoxic. This effect is thought to be due to the copper-mediated generation of the highly toxic hydroxyl radical able to efficiently damage all biomolecules. The dual role of copper makes it crucial for biological systems to control cellular copper levels tightly. To date, the regulation of cellular copper homeostasis is best understood in Saccharomyces cerevisiae (for recent reviews, see references 25, 31, 59, and 60). In this yeast, control occurs at the transcriptional level via the two copper-sensing and copper-modulated transcription factors MAC1 and ACE1. MAC1 is active under low-copper conditions and induces the expression of four genes involved in high-affinity copper uptake. These are the genes encoding two high-affinity copper ion permeases, CTR1 and CTR3, and the metalloreductase FRE1 (10, 18, 26). The function of FRE7, the product of the fourth target gene of MAC1, is still unknown (34). Elevated cellular copper levels lead to a repression of MAC1. As a consequence, the expression of the mentioned target genes is reduced. In contrast to MAC1, ACE1 is active when cellular copper levels are high. Under these conditions, ACE1 induces the expression of genes encoding the copper-binding proteins CUP1 and CRS5, two yeast metallothioneins, and the cytoplasmic SOD1. These proteins act against the toxic effect of copper by binding this metal.
Several of the different components of the complex molecular system involved in the control of cellular copper homeostasis in S. cerevisiae have been identified in other organisms, including different yeasts, the filamentous fungus Podospora anserina, and plants and humans, thus stressing the importance of strict regulation of cellular copper levels (3, 8, 19, 22, 28, 62).
In P. anserina, GRISEA was identified as an ortholog of MAC1 (8, 41). A mutation in the Grisea gene leads to a different phenotype and a 60% increase in life span, further emphasizing the significance of copper homeostasis. Due to a specific mutation leading to a splice defect, Grisea is not expressed, resulting in a copper uptake defect. In mitochondria of the corresponding mutant, the reduction of copper levels was found to increase the stability of the mitochondrial DNA (mtDNA), a process significantly contributing to life span extension (7). mtDNA stabilization appears to be due to a reduced homologous recombination activity in mitochondria, indicating a copper dependence of the underlying mechanism (6).
In most obligate aerobes, energy transduction is strictly dependent on the availability of cellular copper since the COX complex, the terminal oxidase of the cyanide-sensitive respiratory chain, requires copper as a cofactor. However, higher plants, some protozoans, and some fungi can induce an alternative respiratory pathway. This pathway branches at the ubiquinone pool of the respiratory chain and is dependent on the expression of a gene coding for alternative terminal oxidase (AOX) (reviewed in references 14, 54 and 55). Instead of copper, the AOX utilizes iron as a cofactor (4, 53). The AOX is cyanide resistant but sensitive to salicylhydroxamic acid (SHAM).
In this study, we report data of investigations conducted to further elucidate the significance of cellular copper for senescence and lifespan control. In particular, we focused on the energy transduction pathways in mitochondria. We cloned and characterized a gene coding for the AOX of P. anserina and investigated the expression of this gene in the wild-type strain and in two mutants with an affected COX. In addition, we investigated the role of copper in the expression and activity of components of the defense system against oxidative stress and found that the cellular distribution of copper appears to change during senescence.
MATERIALS AND METHODS
Strains and media.
The wild-type strain s and the two mutant strains grisea and ex1 of P. anserina were used in this study (15, 43, 50). Cultures were grown on cornmeal agar at 27°C under light. In some of the experiments described below, cultures were subsequently grown in liquid complete medium (CM) for 2 to 3 days (6). To determine life span, cultures were grown in race tubes and examined until they stopped growing. Several supplements like bathocuproinedisulfonic acid (BCS; Sigma), paraquat, and H2O2, as well as different metals, were added to the autoclaved medium at a temperature of about 60°C. Metals and BCS were added after sterile filtration. To reduce Cu(II) to Cu(I), ascorbic acid was added in combination with the Cu(I) chelator BCS. Concentrations are indicated in the figure legends. Before preparation of RNA, different concentrations of paraquat (100, 250, and 500 μM) were added to the CM and cultures were incubated in this supplemented medium for different times (15 min, 30 min, 1 h, and 4 h). Accordingly, 0.01, 0.02, and 0.04% H2O2 was added 15 min, 30 min, 1 h, 4 h, 16 h, and 4 to 6 days before RNA isolation. These cultures were incubated at 27°C in the dark.
Isolation of mitochondria.
All isolation steps were performed at 4°C. Wet mycelium (30 to 40 g) was suspended in 200 ml of grinding buffer (350 mM mannitol, 30 mM morpholinepropanesulfonic acid [MOPS], 1 mM EDTA, and 0.2% bovine serum albumin [pH 7.6]). Immediately, 1.2 g of polyvinylpyrrolidone and 0.253 g of l-cysteine were added. Cell walls were broken in a Waring blender for 1 min. The cell debris was filtered through cheesecloth, and the filtrate was centrifuged at 6,500 rpm (SS34 rotor; Sorvall) for 2 min. Subsequently, the supernatant was centrifuged once again at 12,750 rpm for 5 min. The pellet was resuspended in 12 ml of wash medium (300 mM mannitol, 20 mM MOPS, 1 mM EDTA, and 0.2% bovine serum albumin [pH 7.2]) and homogenized using a glass homogenizer. Wash medium (28 ml) was added, and the solution was centrifuged at 6,500 rpm for 2 min. Twenty milliliters of the supernatant was layered onto 8 ml of 0.6 M sucrose in a centrifuge tube, and mitochondria were pelleted at 9,250 rpm for 20 min. Pellets were resuspended in 100 ml of suspension solution (250 mM sucrose, 30 mM MOPS [pH 7.2]). Mitochondria were subsequently used for COX activity measurements or for Western blot analysis.
Determination of COX activity.
Mitochondria were diluted at a concentration of 30 μg/ml in assay buffer (30 mM MOPS, 20 mM KCl, 1 mM EDTA [pH 7.2]). To break the mitochondrial membranes, samples were sonicated three times (30% duty cycle) at 1-min intervals on ice using a Branson sonifier. Horse heart cytochrome c (Sigma), used as a substrate, was reduced by sodium dithionite and eluted by gel filtration on a Sephadex G-25 column. Measurements were performed in a spectrophotometer (Uvikon) at 550 nm in a 1-ml cuvette containing 10 μl of 20 μM reduced cytochrome c in 890 μl of assay buffer and 100 μl of mitochondria (containing about 3 μg of protein). As a control, the reaction was inhibited after 2 min by the addition of 10 μl of 350 mM KCN. COX activity was calculated in units per milligram by using an ɛ of 19.4 mM−1 cm−1.
Western blot analysis.
Mitochondria (5 to 15 μg of protein) were boiled for 2 min in loading buffer (0.1 Tris [pH 6.8], 6% sodium dodecyl sulfate (SDS), 6% glycerol, 0.6 M β-mercaptoethanol) and were separated on a 16% denaturing polyacrylamide gel using a Protean II electrophoresis unit (Bio-Rad). Subsequently, proteins were transferred to a nitrocellulose membrane by using an electroblotting device (Bio-Rad). Standard protocols were followed. Immunoblots were probed with an anti-AOX mouse monoclonal antibody (called AOA) that was generated against the AOX of Sauromatum guttatum (13). Additionally, blots were reprobed with anti-βATPase rabbit monoclonal antibodies (37) to confirm equal loading of mitochondrial proteins. Detection was performed by using the Western Light kit (Tropix) according to the protocol of the supplier.
Oxygen uptake measurements.
Mitochondrial respiration was measured with a Clark-type electrode (Rank Bros.) in oxygen uptake buffer containing 0.1 M potassium phosphate buffer (pH 6.0), 0.1% glucose, and 2 mM succinate. About 200 mg of wet mycelial pellets was washed in oxygen uptake buffer and transferred into the reaction vessel. The COX-dependent and alternative pathways were inhibited by the addition of 1 mM KCN and 4 mM SHAM, respectively. To calculate the amount of oxygen uptake, the dry weight of the mycelium used in each measurement was determined afterwards.
Cloning of PaAox and PaSod2.
Partially degenerated primers (AOX1, 5′-RCGMGAYAAYGGMTGGAT-3′; AOX2, 5′-TCCTCCTCRAGGTAMCCGAC-3′) were deduced from a conserved part of the Aox genes from Neurospora crassa, Aspergillus niger, and Magnaporthe grisea. These primers were used to amplify cDNA from the wild-type strain of P. anserina. The PCR was performed at an annealing temperature of 50°C. A single product of about 200 bp was obtained. This fragment was cloned and sequenced and used as a probe to screen a genomic DNA library and a cDNA library of the wild-type strain. The cDNA library was constructed starting from RNA of P. anserina cultures grown in medium that was depleted of copper by the addition of 33 μM BCS and 1 mM ascorbic acid. In the corresponding cDNA library, the probability to select for PaAox, the expression of which is induced under copper-depleted conditions, should be increased. This screen revealed a 1.4-kb cDNA clone and a 8.5-kb EcoRI genomic fragment. Sequence analysis confirmed that both clones contained the PaAox gene. Sequence alignments were performed using the BLASTN program (2).
A conserved part of the manganese superoxide gene (PaSod2) was cloned by PCR amplification using the two partly degenerated primers MnSOD1 (5′-AAGCACCAYCARACYCTAYGSA-3′) and MnSOD2 (5′-GTAGTASGCRTGYTCCCACAT-3′) deduced from the MnSOD genes of N. crassa, S. cerevisiae, and Penicillium chrysogenum. Amplification was performed using the cDNA of wild-type strain s at an annealing temperature of 48°C. The product of about 400 bp was cloned in the SmaI site of pUC18 and sequenced. The sequence showed 74% identity to the Sod-2 gene of N. crassa. This cloned fragment was used as a probe for Northern blot analysis and to screen a cDNA library. The Gpd gene of P. anserina (accession no. X62824) was amplified by using primers Gpd1 (5′-CAAACATGACTGTCAAGG-3′) and Gpd2 (5′-GGAACCTACGAATCAACTAG-3′) and served as an internal control.
Northern blot analysis.
Cultures were grown for 2 to 3 days on agar plates and 2 to 3 days in CM. Approximately 10 g of mycelium was harvested and ground in liquid nitrogen. The frozen mycelial powder was transferred to 30 ml of prewarmed (60°C) GTC buffer (5.5 M guanidine thiocyanate, 25 mM sodium citrate, 0.5% N-lauroylsarcosine, 0.2 M β-mercaptoethanol [pH 7.0]), mixed, and incubated for 10 min at 60°C. The sample was centrifuged (10 min, 10,000 rpm, room temperature, Sorvall SS34 rotor), and the supernatant was layered onto 3 ml of CsCl2 (5.7 M CsCl2, 0.1 M EDTA [pH 7.4], refraction index of 1.400) in an ultracentrifuge tube. RNA was centrifuged at 34,000 rpm (18 h, Sorvall TH-641 rotor) at 20°C overnight. The pellet was washed with 70% ethanol and dissolved in dimethyl pyrocarbonate (DMPC)-treated H2O. For Northern blot analysis, 10 to 20 μg of RNA was separated on a standard formaldehyde agarose gel and subsequently blotted onto a nylon membrane using a vacuum blotting device (Amersham Pharmacia Biotech). Hybridizations were performed in 6× SSC (1× SSC is 0.15 M NaCl plus 15 mM sodium citrate [pH 7.0]), 5× Denhardt's solution, 0.5% SDS, 50% formaldehyde, and 100-μg/ml denatured salmon sperm DNA. A radioactive probe was added at a concentration of 10 to 100 ng/ml. Hybridization was performed at 37°C overnight. Blots were washed at 37°C in 2× SSC–0.5% SDS twice for 10 min and subsequently at 50°C in 0.1× SSC–0.1% SDS twice for 15 min. Blots were exposed to X-ray films for 24 h to 7 days.
SOD activity assay.
Total proteins were isolated by grinding 5 g of wet mycelium in liquid nitrogen. The mycelial powder was dissolved in 20 ml of extraction buffer (1 mM EDTA, 20 mM HEPES [pH 7.5]) and was incubated on ice for 30 min. The sample was centrifuged (13,000 rpm, 10 min, Sorvall GSA rotor), and the supernatant was stirred for 30 min. During this time, ammonium sulfate salt was slowly added until a final concentration of 0.66 g per ml of supernatant was reached. The sample was stirred for an additional hour at 4°C and centrifuged (8,500 rpm, 50 min, Sorvall GSA rotor), and the protein pellet was resuspended in 0.5 to 1 ml of extraction buffer. The concentration of the protein sample was determined according to a modified protocol as described by Bradford using Roti-nanoquant (Roth). To detect SOD activity, proteins (50 to 75 μg) were separated on a 8.5% nondenaturing polyacrylamide gel and stained using nitroblue tetrazolium, riboflavin, and N,N,N′,N′-tetramethylethylenediamine as described previously (16).
Nucleotide sequence accession numbers.
The sequences of PaAox and PaSod2 have been submitted to GenBank and the EMBL Data Bank. The accession numbers are AJ290969 and AJ278985.
RESULTS
Life span of P. anserina is modulated by cellular copper levels.
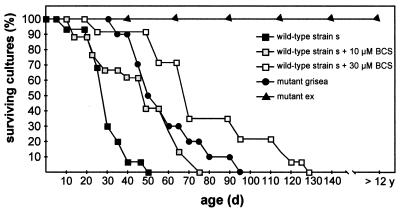
In previous work, the nuclear gene Grisea from P. anserina has been cloned and characterized (6–8, 41). A loss-of-function mutation in this gene has a pleiotropic effect on cultures: the pigmentation of the mycelium and of the ascospores is changed, the formation of female gametangia is affected, and life span is increased (43). In the corresponding mutant grisea, a high-affinity copper transporter gene is not expressed (unpublished data), consequently leading to decreased cellular copper levels. The phenotype of the mutant can be rescued to wild-type characteristics by growing cultures in medium containing high amounts of copper, most likely due to the uptake of copper via a low-affinity system (7, 33). These data clearly indicate an important effect of copper on the lifespan of P. anserina. In order to verify and further support this conclusion, we grew the wild-type strain on solid medium depleted of copper by the addition of different amounts of the copper chelator BCS. These conditions lower cellular copper levels. Since copper excess represses the activity of GRISEA, copper depletion results in a strong activation of this transcription factor (8). As may be seen from Fig. 1, the addition of BCS to the medium resulted in a clear extension of life span. Growth in a medium containing 30 μM BCS led to mean and maximum life spans that are greater than even the corresponding life span of mutant grisea. These results demonstrate that life span extension in long-lived mutant grisea is due to the defect in the control of the high-affinity copper uptake system and not due to the compromised regulation of unidentified target genes of GRISEA.
FIG. 1.
Copper depletion leads to an extension of life span. Fifteen P. anserina cultures of the wild-type strain derived from independent mononucleate ascospores were grown to senescence on cornmeal agar containing 10 μM BCS and 0.33 mM ascorbic acid or containing 30 μM BCS and 1 mM ascorbic acid or without supplements. Accordingly, 15 cultures of mutant grisea were grown on cornmeal agar. All cultures were grown in race tubes at 27°C in the light. A single culture of mutant ex1 has been growing in the laboratory on cornmeal agar plates for over 12 years without signs of senescence.
An alternative respiratory pathway is induced in long-lived mutant grisea.
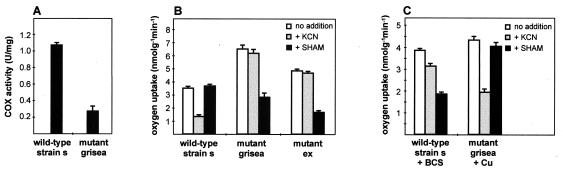
It was previously demonstrated that the long-lived mutant strain ex1 does not respire via the cyanide-sensitive, COX-dependent respiratory chain but via a cyanide-resistant, SHAM-sensitive pathway (17, 51). This switch results from the deletion of parts of the CoxI gene coding for the first subunit of the COX complex and seems to lead to eternal life (Fig. 1). Since copper is a cofactor of COX and cellular copper levels are reduced in mutant grisea, we analyzed the function of the mitochondrial respiratory chain in this mutant. First, we determined the activity of COX in mitochondria from middle-aged cultures of the wild-type strain and of mutant grisea. In mutant mitochondria, the COX activity was found to be reduced approximately fivefold in comparison to that from the wild-type strain (Fig. 2A). The subsequent analysis of oxygen uptake as a measure of mitochondrial respiration revealed that in mutant grisea, respiration is largely resistant to cyanide but sensitive to SHAM. This is similar to the situation in mutant ex1 but in sharp contrast to that in the wild-type strain (Fig. 2B). Growth of mutant grisea on medium supplemented with 100 μM CuSO4 led to a reversion of SHAM sensitivity to the characteristic cyanide sensitivity of the wild type. Vice versa, the wild-type strain grown in a medium depleted of copper by the addition of BCS becomes sensitive to SHAM and more resistant to KCN (Fig. 2C). Clearly, it appears that mitochondria of P. anserina affected at complex IV of the respiratory chain lead to an induction of the alternative respiratory pathway.
FIG. 2.
Copper depletion leads to reduced activity of COX and to the induction of the alternative oxidase. (A) Photometric determination of COX activity in the wild-type strain and mutant grisea. Three micrograms of mitochondrial protein was incubated with reduced cytochrome c as described in Materials and Methods. The protein preparation and measurements were repeated three times. (B) Oxygen uptake measurements of the wild-type strain, mutant grisea, and mutant ex1, without addition of respiratory inhibitors, with addition of 1 mM KCN, or with addition of 4 mM SHAM. (C) Oxygen uptake measurements of the wild-type strain grown on 30 μM BCS and 1 mM ascorbic acid and of mutant grisea grown on 100 μM CuSO4. Experiments described for panels B and C were carried out at least in triplicate with two or three independent isolates of each strain.
Cloning and transcriptional analysis of PaAox.
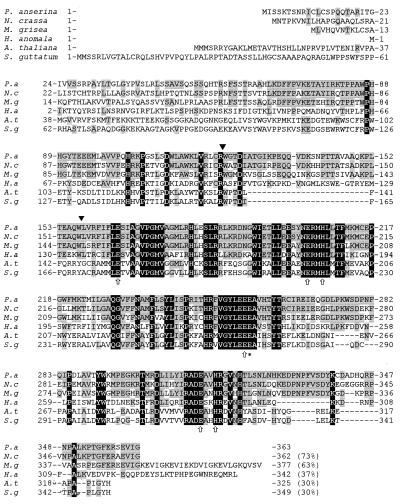
Since the alternative respiratory pathway is induced by cellular copper depletion and since the induction of this pathway appears to have a significant impact on longevity (12, 17, 51), we set out to clone and characterize the gene coding for the AOX of P. anserina. The gene, named PaAox, was cloned following a PCR cloning strategy. Degenerated primers were synthesized to conserved parts of the Aox gene from different organisms (see Materials and Methods). These primers were used to amplify the conserved part of PaAox from a cDNA library of P. anserina. The resulting amplification product containing a part of PaAox was used to isolate the full-length cDNA and the complete gene from a genomic library. The sequence of the corresponding insert fragments was found to contain an open reading frame encoding a peptide with a high degree of sequence identity to other AOX sequences, in particular to those of fungal and plant origin (Fig. 3). The open reading frame of the genomic fragment is interrupted by two introns of 72 and 64 bp. These introns are located in the first half of the gene (Fig. 3). In PaAOX the six amino acids proposed to be involved in iron binding (4, 53) and the glutamine essential for the catalytic activity of the alternative oxidase of S. guttatum (1) were found at the correct position. Hybridization of total DNA with the PaAox probe revealed that the gene is present as a single-copy genome of P. anserina (data not shown).
FIG. 3.
Multiple protein alignment of the deduced amino acid sequences of P. anserina PaAOX with N. crassa AOD-1 (29), M. grisea MgAOX (61), Hansenula anomala (49), Arabidopsis thaliana AOX1B (48), and S. guttatum AOX1 (46). The area with black background corresponds to residues completely conserved between all species, while the area with gray background displays homology of PaAOX with the AOX sequence of several but not all examples (indicated in percent at the end of the sequence). The amino acids indicated by arrows were proposed to form the binuclear iron center on the matrix side of the mitochondrial inner membrane, whereas Glu270 (∗) was found to be essential for the catalytic activity of the alternative oxidase from S. guttatum (1, 4, 53). These amino acids are also conserved in PaAOX. The positions of two introns in the nucleotide sequence of PaAox are indicated by black triangles.
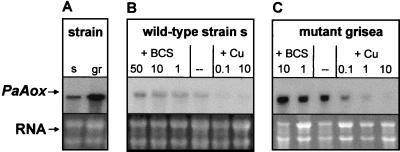
After cloning PaAox, we analyzed the expression of this gene at the transcriptional level. Transcript levels are higher in the copper-deficient mutant grisea than in the wild-type strain (Fig. 4A). The expression is clearly modulated by cellular copper. Supplementation of the growth medium with CuSO4 led to decreased transcript levels, whereas copper depletion resulted in an increase (Fig. 4B). Basically, the same response was observed in mutant grisea (Fig. 4C). These data indicate a copper-dependent expression of PaAox modulated at the transcriptional level. Since the copper-dependent regulation of PaAox is effective in mutant grisea lacking the functional copper-regulated transcription factor GRISEA, regulators other than GRISEA must be responsible for the observed regulation of this gene.
FIG. 4.
Transcription of the PaAox gene is dependent on the cellular copper concentration. (A) Transcript level of PaAox for the wild-type strain (s) compared to that for mutant grisea (gr). (B and C) Northern blot analysis of PaAox in wild-type strain s (B) and mutant grisea (C) at low or high copper concentrations in the medium. The copper concentration was reduced by the addition of 10 μM BCS and 0.33 mM ascorbic acid or 30 μM BCS and 1 mM ascorbic acid or 50 μM BCS and 1.7 mM ascorbic acid. Copper levels were increased by the addition of 0.1, 1.0, or 10 μM CuSO4 to the medium. In the lower part of the figure, the ethidium bromide-stained RNA is shown as loading control.
The alternative pathway is not induced during aging of P. anserina cultures.
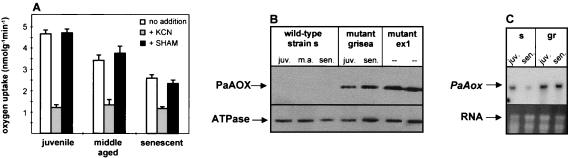
As mentioned above, dysfunctional mitochondria affected at the COX complex of the respiratory chain of P. anserina lead to the induction of PaAox. Dysfunctional mitochondria are not restricted to particular mutants of P. anserina but regularly arise during aging of these cultures. Functional impairment, at least in part, is the consequence of extensive rearrangements of the mtDNA. These rearrangements result in deletions of large regions of DNA encoding different proteins of the respiratory chain (9, 27). It thus was reasonable to surmise that a switch from the COX-dependent to AOX-dependent respiration may occur regularly during aging of wild-type cultures. We investigated this possibility by the analysis of oxygen uptake in cultures of different ages. Clearly, the general performance of mitochondria was found to decline during aging. However, the response to the specific inhibitors of the two terminal oxidases did not change significantly (Fig. 5A). These results were confirmed by Western blot analysis using a monoclonal antibody against the AOX of S. guttatum (13). In both long-lived mutants, a single band of about 39 kDa reacted with the plant antibody. Interestingly, PaAOX levels were highest in the immortal mutant strain ex1 (Fig. 5B). In mutant grisea, the amount of the transcript and of the protein did not change during aging (Fig. 5B and C). In the wild-type strain PaAox, transcripts were detected and were found to decline during aging. The protein was only detectable in very low amounts. At the protein level, the difference between the wild-type strain and mutant grisea appears to be more pronounced than at the transcript level, suggesting differences in the posttranscriptional regulation of PaAox in the two analyzed strains. Taken together, the data from the Western blot analysis and those from the transcript analysis of the wild-type strain were consistent with the data from respiration measurements showing that an induction of the alternative respiratory pathway does not occur during senescence.
FIG. 5.
PaAOX-dependent respiration is not induced in senescent cultures of the wild-type strain. (A) Oxygen uptake measurements of juvenile (10 days), middle-aged (15 days), and senescent cultures of the wild-type strain. Respiration was measured without addition of respiratory inhibitors, after the addition of 1 mM KCN, or after the addition of 4 mM SHAM. (B) Western blot analysis of PaAOX proteins of juvenile (juv.), middle-aged (m.a.), and senescent (sen.) cultures of the wild-type strain and of mutant grisea. In addition, mitochondrial protein preparations of mutant ex1 were analyzed. PaAOX was detected with AOA monoclonal antibodies directed against the AOX of S. guttatum strain Schott and visualized by enhanced chemiluminescence. An antibody directed against the β subunit of the ATPase complex was used as a loading control. (C) Northern blot analysis of PaAox transcription in juvenile and senescent cultures of the wild-type strain and of mutant grisea. The ethidium bromide-stained RNA is shown as a loading control.
Oxidative stress reduces PaAox transcript levels.
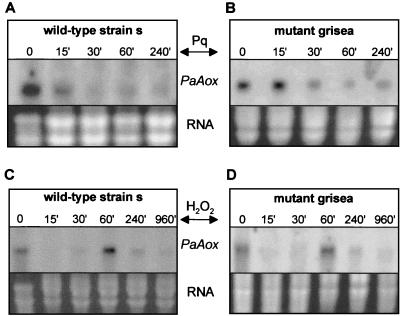
In higher plants, the activity of the AOX was demonstrated to increase by reactive oxygen species (ROS) (36, 57). In the fungus M. grisea, transcription of the MgAox gene was found to increase after addition of H2O2 (61). To test whether this is also the case in P. anserina, we incubated cultures of wild-type strain s and of mutant grisea for different times in medium containing either H2O2 or paraquat. The latter compound was added as a superoxide generator. Northern blot analysis revealed that a long-term induction of PaAox expression is not observed under these conditions (Fig. 6). The reduction in transcript levels can be observed after a few minutes of incubation in paraquat-containing medium. The effect remains stable for at least 4 h. The addition of higher levels of paraquat (250 and 500 μM) reduced PaAox transcript levels even more (not shown). Growing cultures in H2O2 also led to an overall decrease in transcript levels, although after an hour of incubation, transcript levels recovered to the initial level. Later, levels decreased again. The addition of higher levels of H2O2 (0.02 and 0.04%) also reduced transcript levels. The same results were obtained in the wild type and in mutant grisea. In both strains, it appears that either the transcription of PaAox is repressed or the stability of the transcript is affected by the two tested additives. Thus, increased PaAox transcript levels were not detected in P. anserina under oxidative stress conditions as in M. grisea (61).
FIG. 6.
Transcription of PaAox is reduced by the addition of paraquat and hydrogen peroxide. (A and B) Northern blot analysis of PaAox transcripts in the wild-type strain and in mutant grisea. Paraquat (Pq, 100 μM) was added to the medium 15, 30, 60, and 240 min before RNA preparation. (C and D) H2O2 (0.01%) was added to the medium 15, 30, 60, 240, and 960 min before RNA preparation. These cultures were grown in the dark.
Expression of SODs in P. anserina.
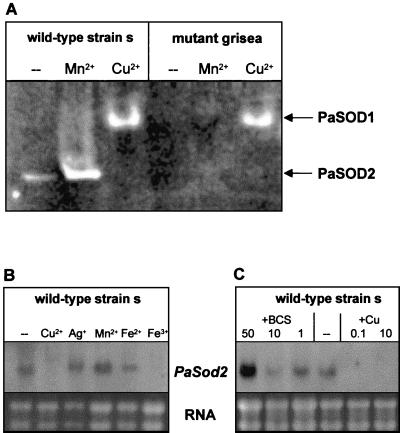
The extension of life span in mutants grisea and ex1 correlates with the expression of PaAox. Since it is known from plants and from P. anserina that the alternative pathway results in reduced formation of ROS (12, 35, 44, 58), we surmised that in mitochondria of long-lived mutant grisea, oxidative stress is reduced. In order to verify this idea, mitochondrial ROS production was determined indirectly by investigating the activity of the manganese SOD (PaSOD2), a mitochondrial scavenger of superoxide. In the wild-type strain, one SOD band was detected on nondenaturing polyacrylamide gels. The intensity of this band increased after addition of large amounts of manganese to the growth medium (Fig. 7A). Since it is known from other systems (23) that manganese is involved in the expression of Sod2, we surmised that the corresponding protein band represents PaSOD2. Interestingly, the corresponding band does not appear in mutant grisea, regardless of the conditions under which this strain was cultivated. In addition to this manganese-inducible SOD, another SOD band migrating much more slowly was identified in both wild-type and mutant grisea when strains were cultivated in medium supplemented with 100 μM CuSO4. Since the activity of SOD1 (Cu/Zn SOD) is dependent on the availability of copper, we surmised that this band represents PaSOD1. Interestingly, the activity of PaSOD2 is reduced in cultures of the wild-type strain grown in medium containing 100 μM CuSO4.
FIG. 7.
PaSOD2 is active in the wild-type strain and is dependent upon cellular copper. (A) SOD activity assay. Proteins were prepared from the wild-type strain and from mutant grisea after cultivation of these strains in CM without supplements (−) or in CM containing 100 μM MnCl2 (Mn2+) or 100 μM CuSO4 (Cu2+), run on a native polyacrylamide gel, and stained for SOD activity. (B) Northern blot analysis of PaSod2 transcription. Different metals (100 μM CuSO4 [Cu2+], 100 μM AgCl [Ag+], 100 μM MnCl2 [Mn2+], 100 μM FeSO4 [Fe2+], 100 μM FeCl3 [Fe3+]) were added to the medium. −, control. (C) Northern blot analysis of PaSod2 transcription at different copper concentrations. To reduce the copper concentration, cultures were grown in medium containing 1 μM BCS and 33 μM ascorbic acid or 10 μM BCS and 0.33 mM ascorbic acid or 50 μM BCS and 1.7 mM ascorbic acid. Copper levels were increased by the addition of 0.1 or 10 μM CuSO4 to the medium. −, no addition of CuSO4 (control).
To verify the nature of the different SODs identified in native protein gels more specifically, we cloned a part of PaSod2 and used this sequence as a probe for Northern analysis. Cloning was achieved following a PCR strategy. First, we amplified a specific cDNA of the corresponding PaSod2 gene using partly degenerated primers. Subcloning and sequencing confirmed that a conserved part of the gene was amplified. The amino acid sequence derived from the cloned cDNA shares high homology with other fungal SOD2 proteins (e.g., N. crassa). Subsequently, we isolated a cDNA clone from a P. anserina cDNA library and determined the nucleotide sequence. The sequence verified that PaSod2 was isolated. Northern blot analysis in which RNA of wild-type cultures cultivated in standard medium demonstrated expression of the gene in standard medium. In accordance with the predictions from the SOD activity analysis, supplementation of the medium with Mn2+ leads to increased transcript levels. The addition of Ag+ and Fe2+ ions had no significant effect on transcription, whereas Cu2+ and Fe3+ led to decreased transcript levels (Fig. 7B). The effect of copper was more thoroughly investigated by analyzing RNA isolated from cultures grown in medium containing different amounts of copper and BCS (Fig. 7C). These experiments clearly demonstrated that PaSod2 transcript levels are highest under copper-depleted conditions.
PaSod2 is not expressed in long-lived mutant grisea.
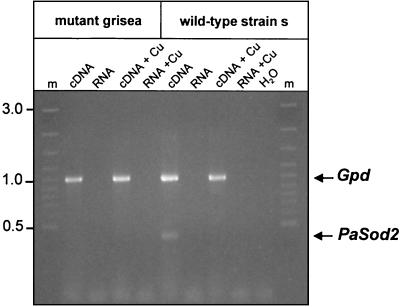
The transcript analysis data indicate that the SOD activity of the faster-migrating protein in the activity gels that appears to be induced by manganese corresponds to PaSOD2. Interestingly, although PaSod2 transcription was found to be increased by the addition of manganese or under copper-deficient conditions, the activity of this protein was not detected in the copper uptake mutant grisea. The addition of 100 μM Mn2+ did not restore this defect. We surmised that PaSod2 is a target gene of transcription factor GRISEA. To verify this assumption, Northern analysis of PaSod2 transcripts was performed, confirming that this gene is not expressed in mutant grisea (not shown). The same results were obtained with a more sensitive reverse transcription-PCR analysis, which was performed using RNA preparations of the wild-type strain and of mutant grisea. Transcripts were amplified using a set of specific PaSod2 primers. As an internal control, a pair of specific primers for the glyceraldehyde dehydrogenase gene (Gpd) of P. anserina was included in the PCR (Fig. 8). A PCR product was obtained only in cDNA preparations of the wild-type strain grown in standard medium but not in medium containing additional copper. No transcript was detected in the corresponding cDNA preparation of mutant grisea, supporting the idea that PaSod2 is a target gene of transcription factor GRISEA.
FIG. 8.
PaSod2 is a putative target gene of transcription factor GRISEA. RNA was isolated from wild-type and grisea cultures grown with 100 μM CuSO4 or without supplements. RNA was treated with DNase I to reduce DNA contamination and subsequently reverse transcribed by using an oligo(dT) primer. Reverse-transcribed RNA samples (cDNA) were used as a template. The PaSod2 transcripts were amplified by using primers MnSOD1 and MnSOD2 (see Materials and Methods). The Gpd gene was amplified in the same reaction and served as an internal control (positive control). DNase I-treated RNA (RNA) was included to exclude amplification of DNA (negative control). Lane m, 100-bp size standard. Numbers on the left indicate fragment sizes in kilobase pairs.
The activity of PaSOD2 decreases during aging of wild-type cultures of P. anserina.
Mitochondrial oxidative stress is thought to be a main contributor to aging processes in different biological systems and is thought to generally increase during aging (for a review see reference 38). In P. anserina, the increase in ROS during aging is thought to be the result of two processes and consequences: first, protein damage of the respiratory chain by ROS leaking from the electron transfer chain during energy transduction; and second, the inability of senescent cultures to replace affected respiratory chains. This inability is due to extensive rearrangements of the mtDNA occurring during aging, which lead to the deletion of genes coding for different components of the respiratory chain (39). Consequently, the performance of the respiratory chain becomes reduced during senescence, and the generation of ROS increases. Since PaSOD2 is part of the mitochondrial defense system directed against oxidative stress, we investigated whether levels of PaSOD2 increase during aging of the wild-type strain. SOD activity was determined in protein extracts of juvenile and senescent cultures grown in standard medium. In addition, extracts of juvenile cultures grown in a medium supplemented with 100 μM paraquat and 100 μM Mn2+ were investigated. Whereas manganese clearly induced PaSOD2 activity, paraquat did not lead to such an induction. Moreover, in the wild-type strain, PaSOD2 levels were significantly reduced in senescent cultures (Fig. 9), whereas PaSOD1 levels increased. These data clearly demonstrated that the oxidative stress defense capacity of mitochondria is reduced in senescent cultures. The expression of PaSod2 is most probably reduced due to the previously observed increase in cytoplasmic copper in senescent cultures, which also explains the increased activity of PaSOD1 in these cultures. We have shown that the transcript levels of PaMt1, coding for a Cu-metallothionein, greatly increase during senescence (5). Therefore, the observed decrease is not just a general degenerative effect observed later in the lifespan.
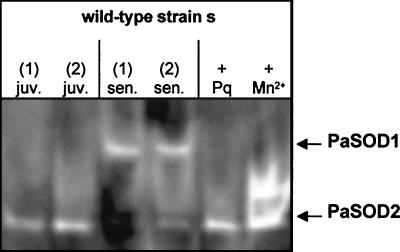
FIG. 9.
The activity of PaSOD2 is reduced in senescent cultures of the wild-type strain. Proteins of two independent juvenile (juv.) and two senescent (sen.) cultures were isolated, as well as proteins from a juvenile strain grown for 4 h on 100 μM paraquat (+ Pq) or for 4 days on 100 μM MnCl2 (+ Mn2+). These proteins were subjected to native polyacrylamide gel electrophoresis. SOD activity was determined by staining of the gel.
DISCUSSION
In this study, we investigated the impact of cellular copper on the longevity of the filamentous ascomycete P. anserina. We first demonstrated that a reduction of the copper concentration in the growth medium led to an increased life span for the wild-type strain. This is not in concordance with a direct role of the active copper-modulated transcription factor GRISEA in bringing life to an end, as it may be concluded from the demonstration that Grisea is not expressed in the long-lived loss-of-function mutant grisea. The data presented in this work underscore the significant role of cellular copper, which has also been suggested by previous mutant rescue experiments (8, 33).
A retrograde response in P. anserina.
Since there is a huge body of experimental data demonstrating a crucial role for mitochondria in senescence in the two filamentous ascomycetes P. anserina and N. crassa (for review see references 21, 39, and 40), and because copper is a cofactor of the mitochondrial COX complex, we investigated the performance of mitochondria in wild-type and nuclear long-lived mutant grisea. As expected, the copper uptake mutant showed a clear reduction in COX activity. Energy transduction proceeds mainly via the cyanide-resistant alternative oxidase, PaAOX. The cloning and characterization of the corresponding gene revealed a strong conservation of the protein with all of the previously demonstrated crucial amino acid residues. However, in sharp contrast to plants, in which the expression of the AOX becomes induced by copper (42), in P. anserina, higher copper levels lead to a reduction of PaAox transcripts.
The expression of PaAox under low-copper conditions appears not to be directly related to cellular copper depletion, e.g., as the result of the activation of a copper-repressed transcription factor, but is rather due to the fact that reduced copper levels give rise to dysfunctional mitochondria. This conclusion can be drawn from the demonstration that the same response is induced in the two COX-deficient, long-lived strains ex1 and cox5::BLE (12, 50). In these two strains, the uptake of copper is not affected. It thus appears that in P. anserina, like in yeast, dysfunctional mitochondria signal to the nucleus and induce the expression of nuclear genes to compensate for the defect in mitochondria. In yeast, such a mechanism was previously named the retrograde response (30). It is controlled by the three regulatory proteins RTG1, RTG2, and RTG3 (32, 47, 52). Importantly, it was recently shown that the induction of the retrograde response postpones senescence, leading to an increased replicative life span for the corresponding yeast strains (24). Although we currently do not know whether homologs of the regulator proteins involved in the retrograde response in yeast play a role in P. anserina, the situation in the long-lived mutants of P. anserina mentioned above clearly suggests that a retrograde response with an impact on life span can be induced in P. anserina. Moreover, it is intriguing that this response appears to be specifically induced in mutants with a compromised COX function. This was clearly demonstrated not only by oxygen uptake measurements but also, for the first time, by Western blot analysis. Regardless of whether complex IV is impaired by deletions of mitochondrial or nuclear genes coding for subunits of this respiratory chain complex or by the depletion of copper as a crucial cofactor of COX, the outcome seems to be the same: the signaling of dysfunctional mitochondria to the nucleus and the induction of PaAox.
Since the mtDNA of P. anserina encoding different subunits of complex I (NADH dehydrogenase) and of complex V (ATP synthetase) of the inner mitochondrial membrane is rearranged during senescence, resulting in dysfunctional mitochondria, it was reasonable to expect that a retrograde response is also induced during senescence. We were surprised to see that this is not the case. However, if one looks more closely at the effect of an induction of PaAox, the obtained results can be explained by the fact that the expression of PaAox can rescue mitochondria only if complex I is functional. At this point it needs to be emphasized that P. anserina is a strict aerobe depending on respiration. Since the alternative oxidase is located upstream of complexes III and IV, generation of a proton gradient, a prerequisite for ATP synthesis at complex V, is dependent on a functional complex I. It thus makes sense that the retrograde response is induced only if an impaired function can be rescued by the induction of specific nuclear genes. In different long-lived strains of P. anserina, this is clearly the case (12, 50). However, mitochondrial dysfunction as the result of age-dependent reorganization of the mtDNA leads to impairments of the whole respiratory chain, including complex I. Thus, an induction of the retrograde response in senescent cultures cannot be expected.
Age-related increase of copper stress.
What is the molecular basis of the observed failure to induce the alternative pathway in senescent cultures of P. anserina, cultures which are clearly affected in their mitochondrial functions? The investigations addressing the age-related capacity of the enzymatic defense system against copper and oxidative stress, which were performed in a previous study as well as in this one, provided clear evidence that cytoplasmic copper stress increases during aging of wild-type cultures of P. anserina. First, we demonstrated that the expression of a gene, PaMt1, encoding a Cu-metallothionein strongly increases during senescence of wild-type cultures. In the copper uptake mutant grisea, this was not the case (5). Second, transcript levels of PaSod2, a target gene of transcription factor GRISEA, decrease (this study). Third, the copper-dependent activity of PaSOD1 increases during aging of wild-type cultures (this study). Fourth, the expression of PaCtr3, which encodes a high-affinity copper transporter, is copper regulated and repressed during senescence (unpublished data). Consequently, since the expression of the gene coding for the alternative oxidase was also found to be repressed by elevated copper levels, the reduced PaAox transcript levels and the almost undetectable protein levels in senescent cultures can easily be explained by increased levels of cytoplasmic copper. In addition, the reduced activity of PaSOD2, the impairments of the respiratory chain, and the increased copper concentration observed during aging result in increased levels of ROS, contributing to the repression of PaAox.
But what are the reasons for the suggested cytoplasmic copper increase? In this respect the role of mitochondria in apoptosis in animal systems is of special interest. It is clear that mitochondria, as the result of different cellular stresses, change their membrane permeability characteristics. It has been shown in animal systems that cytochrome c and the Smac/DIABLO protein are released from mitochondria and give rise to apoptosis (11, 20, 45, 56). We suggest here that during the senescence of P. anserina, copper normally bound to COX is released from mitochondria and induces the different molecular pathways mentioned above. This idea is supported by reduced cyanide-sensitive respiration in senescent P. anserina cultures. Moreover, it is intriguing that PaMt1 transcript levels in senescent cultures of the long-lived mutant grisea are not increased in comparison to those in juvenile cultures. Only in middle-aged cultures is a moderate increase observed. These data seem to suggest that copper from a limited number of functional COX complexes is released rather early during the fungal life span. In later stages, there is no mitochondrial copper reservoir left that can lead to the marked effect on PaMt1, PaCtr3, and PaSod2 transcript levels observed in the wild-type strain (5).
Life span extension in the copper uptake mutant grisea.
Previous investigations and the data from this study clearly demonstrate the important impact of cellular copper levels on longevity. From these data it is clear that impairments of the molecular machinery regulating cellular copper levels have specific consequences. In long-lived mutant grisea, the primary cause of such an impairment is a loss-of-function mutation in the gene coding for the copper-modulated transcription factor GRISEA. As a consequence, a gene coding for a high-affinity copper transporter is not expressed, and import of copper is restricted to a low-affinity uptake system. Under these conditions, complex IV of the respiratory chain is affected. Only a small number of functional COX-dependent respiratory chains are assembled. Most mitochondria are dysfunctional and signal to the nucleus to induce PaAox. Energy transduction via the alternative pathway leads to a reduced generation of mitochondrial ROS. Therefore, a block in the expression of PaSod2 (a target gene of GRISEA), as the result of the absence of this transcription factor in mutant grisea, is not a major problem. In addition, in contrast to that of the wild-type strain, the mtDNA of mutant grisea remains stable during senescence and is available to replace damaged mtDNA-encoded components of the respiratory chain. Finally, the inactivity of PaSOD1 might explain why the increase in life span is not as pronounced as in mutant ex1 and most cox5::BLE strains.
In summary, the data presented in this study underscore the role of copper in the control of life span. This becomes clear from the analysis of both the wild-type strain and a copper uptake mutant. A molecular mechanism able to sense mitochondrial dysfunction and to adapt the system to altered situations, the so-called retrograde response, was demonstrated in P. anserina. The induction of the retrograde response appears to have a significant impact on life span, indicating that metabolism plays an important role. An alternative oxidase was found to be part of this pathway. The expression of the gene encoding PaAOX is controlled in a way different from that found in higher plants. Copper was found to play a significant role in the expression of PaAox and of PaSod2. Finally, and very surprisingly, different lines of evidence suggest the age-related release of copper from mitochondria. Such a process is probably the result of changes in the permeability of mitochondrial membranes that are demonstrated to occur during apoptosis in animal systems. It will be of particular interest to investigate these similarities between senescence in P. anserina and apoptosis in more detail.
ACKNOWLEDGMENTS
We thank Bernd Ludwig and Ute Pfitzner (Frankfurt, Germany) for introducing cytochrome oxidase measurements and for providing anti-βATPase antibodies.
This work was supported by a grant of the Deutsche Forschungsgemeinschaft (Bonn, Germany) to H.D.O.
REFERENCES
- 1.Albury M S, Affourtit C, Moore A L. A highly conserved glutamate residue (Glu-270) is essential for plant alternative oxidase activity. J Biol Chem. 1998;273:30301–30305. doi: 10.1074/jbc.273.46.30301. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amarvadi R, Glerum D M, Tzagoloff A. Isolation of a cDNA encoding the human homolog of COX17, a yeast gene essential for mitochondrial copper recruitment. Hum Genet. 1997;99:329–333. doi: 10.1007/s004390050367. [DOI] [PubMed] [Google Scholar]
- 4.Andersson M E, Nordlund P. A revised model of the active site of alternative oxidase. FEBS Lett. 1999;449:17–22. doi: 10.1016/s0014-5793(99)00376-2. [DOI] [PubMed] [Google Scholar]
- 5.Averbeck, N. B., C. Borghouts, A. Hamann, V. Specke, and H. D. Osiewacz. Molecular control of cellular copper homeostasis in filamentous fungi: increased expression of a metallothionein gene during aging of Podospora anserina. Mol. Gen. Genet., in press. [DOI] [PubMed]
- 6.Borghouts C, Kerschner S, Osiewacz H D. Copper-dependence of mitochondrial DNA rearrangements in Podospora anserina. Curr Genet. 2000;37:268–275. doi: 10.1007/s002940050528. [DOI] [PubMed] [Google Scholar]
- 7.Borghouts C, Kimpel E, Osiewacz H D. Mitochondrial DNA rearrangements of Podospora anserina are under the control of the nuclear gene Grisea. Proc Natl Acad Sci USA. 1997;94:10768–10773. doi: 10.1073/pnas.94.20.10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borghouts C, Osiewacz H D. GRISEA, a copper-modulated transcription factor from Podospora anserina involved in senescence and morphogenesis, is an ortholog of MAC1 in Saccharomyces cerevisiae. Mol Gen Genet. 1998;260:492–502. doi: 10.1007/s004380050922. [DOI] [PubMed] [Google Scholar]
- 9.Cummings D J, McNally K L, Domenico J M, Matsuura E T. The complete DNA sequence of the mitochondrial genome of Podospora anserina. Curr Genet. 1990;17:375–402. doi: 10.1007/BF00334517. [DOI] [PubMed] [Google Scholar]
- 10.Dancis A, Haile D, Yuan D S, Klausner R D. The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J Biol Chem. 1994;269:25660–25667. [PubMed] [Google Scholar]
- 11.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 12.Dufour E, Boulay J, Rincheval V, Sainsard-Chanet A. A casual link between respiration and senescence in Podospora anserina. Proc Natl Acad Sci USA. 2000;97:4138–4143. doi: 10.1073/pnas.070501997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elthon T E, Nickels R L, McIntosh L. Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 1989;89:1311–1317. doi: 10.1104/pp.89.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elthon T E, Stewart C R. A chemiosmotic model for plant mitochondria. BioScience. 1983;33:687–692. [Google Scholar]
- 15.Esser K. Podospora anserina. In: King R C, editor. Handbook of genetics. New York, N.Y: Plenum Press; 1974. pp. 531–551. [Google Scholar]
- 16.Flohe L, Ötting F. Superoxide dismutase assays. Methods Enzymol. 1984;105:93–104. doi: 10.1016/s0076-6879(84)05013-8. [DOI] [PubMed] [Google Scholar]
- 17.Frese D, Stahl U. Oxidative stress and ageing in the fungus Podospora anserina. Mech Ageing Dev. 1992;65:277–288. doi: 10.1016/0047-6374(92)90041-b. [DOI] [PubMed] [Google Scholar]
- 18.Georgatsou E, Mavrogiannis L A, Fragiadakis G S, Alexandraki D. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J Biol Chem. 1997;272:13786–13792. doi: 10.1074/jbc.272.21.13786. [DOI] [PubMed] [Google Scholar]
- 19.Glerum D M, Shtanko A, Tzagoloff A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J Biol Chem. 1996;271:14504–14509. doi: 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
- 20.Green D R, Reed J C. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths A J. Fungal senescence. Annu Rev Genet. 1992;26:351–372. doi: 10.1146/annurev.ge.26.120192.002031. [DOI] [PubMed] [Google Scholar]
- 22.Kampfenkel K, Kushnir S, Babiychuk E, Inzé D, Van Montagu M. Molecular characterization of a putative Arabidopsis thaliana copper transporter and its yeast homologue. J Biol Chem. 1995;270:28479–28486. doi: 10.1074/jbc.270.47.28479. [DOI] [PubMed] [Google Scholar]
- 23.Kim Y C, Miller C D, Anderson A J. Transcriptional regulation by iron of genes encoding iron- and manganese-superoxide dismutases from Pseudomonas putida. Gene. 1999;239:129–135. doi: 10.1016/s0378-1119(99)00369-8. [DOI] [PubMed] [Google Scholar]
- 24.Kirchman P A, Kim S, Lai C Y, Jazwinski S M. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight S A B, Koch K A, Thiele D J. Yeast metallothionein gene regulation. In: Silver S, Walden W, editors. Metal ions in gene regulation. New York, N.Y: Chapman & Hall; 1997. pp. 316–341. [Google Scholar]
- 26.Knight S A B, Labbe S, Kwon L F, Kosman D J, Thiele D J. A widespread transposable element masks expression of a yeast copper transport gene. Genes Dev. 1996;10:1917–1929. doi: 10.1101/gad.10.15.1917. [DOI] [PubMed] [Google Scholar]
- 27.Kück U, Stahl U, Esser K. Plasmid-like DNA is part of mitochondrial DNA in Podospora anserina. Curr Genet. 1981;3:151–156. doi: 10.1007/BF00365719. [DOI] [PubMed] [Google Scholar]
- 28.Labbe S, Pena M M, Fernandes A R, Thiele D J. A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. J Biol Chem. 1999;274:36252–36260. doi: 10.1074/jbc.274.51.36252. [DOI] [PubMed] [Google Scholar]
- 29.Li Q, Ritzel R G, McLean L L, McIntosh L, Ko T, Bertrand H, Nargang F E. Cloning and analysis of the alternative oxidase gene of Neurospora crassa. Genetics. 1996;142:129–140. doi: 10.1093/genetics/142.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao X, Butow R A. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- 31.Liu X D, Thiele D J. Yeast metallothionein gene expression in response to metals and oxidative stress. Methods. 1997;11:289–299. doi: 10.1006/meth.1996.0423. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Butow R A. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marbach K, Fernandez-Larrea J, Stahl U. Reversion of a long-living, undifferentiated mutant of Podospora anserina by copper. Curr Genet. 1994;26:184–186. doi: 10.1007/BF00313809. [DOI] [PubMed] [Google Scholar]
- 34.Martins L J, Jensen L T, Simon J R, Keller G L, Winge D R, Simons J R. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J Biol Chem. 1998;273:23716–23721. doi: 10.1074/jbc.273.37.23716. [DOI] [PubMed] [Google Scholar]
- 35.Maxwell D P, Wang Y, McIntosh L. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA. 1999;96:8271–8276. doi: 10.1073/pnas.96.14.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minagawa N, Koga S, Nakano M, Sakajo S, Yoshimoto A. Possible involvement of superoxide anion in the induction of cyanide-resistant respiration in Hansenula anomala. FEBS Lett. 1992;302:217–219. doi: 10.1016/0014-5793(92)80444-l. [DOI] [PubMed] [Google Scholar]
- 37.Nelson N, Schatz G. Energy-dependent processing of cytoplasmically made precursors to mitochondrial proteins. Proc Natl Acad Sci USA. 1979;76:4365–4369. doi: 10.1073/pnas.76.9.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osiewacz H D. Genetic regulation of aging. J Mol Med. 1997;75:715–727. doi: 10.1007/s001090050158. [DOI] [PubMed] [Google Scholar]
- 39.Osiewacz H D, Borghouts C. Mitochondrial oxidative stress and aging in the filamentous fungus Podospora anserina. Ann N Y Acad Sci. 2000;908:31–39. doi: 10.1111/j.1749-6632.2000.tb06633.x. [DOI] [PubMed] [Google Scholar]
- 40.Osiewacz H D, Kimpel E. Mitochondrial-nuclear interactions and lifespan control in fungi. Exp Gerontol. 1999;34:901–909. doi: 10.1016/s0531-5565(99)00063-7. [DOI] [PubMed] [Google Scholar]
- 41.Osiewacz H D, Nuber U. GRISEA, a putative copper-activated transcription factor from Podospora anserina involved in differentiation and senescence. Mol Gen Genet. 1996;252:115–124. doi: 10.1007/BF02173211. [DOI] [PubMed] [Google Scholar]
- 42.Padua M, Aubert S, Casimiro A, Bligny R. Arrest of mitochondrial biogenesis in copper-treated sycamore cells. FEBS Lett. 1996;398:248–252. doi: 10.1016/s0014-5793(96)01260-4. [DOI] [PubMed] [Google Scholar]
- 43.Prillinger H, Esser K. The phenoloxidases of the ascomycete Podospora anserina. XIII. Action and interaction of genes controlling the formation of laccase. Mol Gen Genet. 1977;156:333–345. doi: 10.1007/BF00267190. [DOI] [PubMed] [Google Scholar]
- 44.Purvis A C. Role of the alternative oxidase in limiting superoxide production by plant mitochondria. Physiol Plant. 1997;100:165–170. [Google Scholar]
- 45.Reed J C. Cytochrome c: can't live with it—can't live without it. Cell. 1997;91:559–562. doi: 10.1016/s0092-8674(00)80442-0. [DOI] [PubMed] [Google Scholar]
- 46.Rhoads D M, McIntosh L. The salicylic acid-inducible alternative oxidase gene aox1 and genes encoding pathogenesis-related proteins share regions of sequence similarity in their promoters. Plant Mol Biol. 1993;21:615–624. doi: 10.1007/BF00014545. [DOI] [PubMed] [Google Scholar]
- 47.Rothermel B A, Thornton J L, Butow R A. Rtg3p, a basic helix-loop-helix/leucine zipper protein that functions in mitochondrial-induced changes in gene expression, contains independent activation domains. J Biol Chem. 1997;272:19801–19807. doi: 10.1074/jbc.272.32.19801. [DOI] [PubMed] [Google Scholar]
- 48.Saisho D, Nambara E, Naito S, Tsutsumi N, Hirai A, Nakazono M. Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol Biol. 1997;35:585–596. doi: 10.1023/a:1005818507743. [DOI] [PubMed] [Google Scholar]
- 49.Sakajo S, Minagawa N, Komiyama T, Yoshimoto A. Molecular cloning of cDNA for antimycin A-inducible mRNA and its role in cyanide-resistant respiration in Hansenula anomala. Biochim Biophys Acta. 1991;1090:102–108. doi: 10.1016/0167-4781(91)90043-l. [DOI] [PubMed] [Google Scholar]
- 50.Schulte E, Kück U, Esser K. Multipartite structure of mitochondrial DNA in a fungal longlife mutant. Plasmid. 1989;21:79–84. doi: 10.1016/0147-619x(89)90089-9. [DOI] [PubMed] [Google Scholar]
- 51.Schulte E, Kück U, Esser K. Extrachromosomal mutants from Podospora anserina: permanent vegetative growth in spite of multiple recombination events in the mitochondrial genome. Mol Gen Genet. 1988;211:342–349. [Google Scholar]
- 52.Sekito T, Thornton J, Butow R A. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors rtg1p and rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siedow J N, Umbach A L, Moore A L. The active site of the cyanide-resistant oxidase from plant mitochondria contains a binuclear iron center. FEBS Lett. 1995;362:10–14. doi: 10.1016/0014-5793(95)00196-g. [DOI] [PubMed] [Google Scholar]
- 54.Sluse F E, Jarmuszkiewicz W. Alternative oxidase in the branched mitochondrial respiratory network: an overview on structure, function, regulation, and role. Braz J Med Biol Res. 1998;31:733–747. doi: 10.1590/s0100-879x1998000600003. [DOI] [PubMed] [Google Scholar]
- 55.Vanlerberghe G C, McIntosh L. Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:703–734. doi: 10.1146/annurev.arplant.48.1.703. [DOI] [PubMed] [Google Scholar]
- 56.Verhagen A M, Ekert P G, Pakusch M, Silke J, Connolly L M, Reid G E, Moritz R L, Simpson R J, Vaux D L. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 57.Wagner A M. A role for active oxygen species as second messengers in the induction of alternative oxidase gene expression in Petunia hybrida cells. FEBS Lett. 1995;368:339–342. doi: 10.1016/0014-5793(95)00688-6. [DOI] [PubMed] [Google Scholar]
- 58.Wagner A M, Moore A L. Structure and function of the plant alternative oxidase: its putative role in the oxygen defence mechanism. Biosci Rep. 1997;17:319–333. doi: 10.1023/a:1027388729586. [DOI] [PubMed] [Google Scholar]
- 59.Winge D R, Jensen L T, Srinivasan C. Metal-ion regulation of gene expression in yeast. Curr Opin Chem Biol. 1998;2:216–221. doi: 10.1016/s1367-5931(98)80063-x. [DOI] [PubMed] [Google Scholar]
- 60.Winge D R, Sewell A K, Yu W, Thorvaldsen J L, Farrell R. Metal ion stress in yeast. In: Silver S, Walden W, editors. Metal ions in gene regulation. New York, N.Y: Chapman & Hall; 1997. pp. 279–315. [Google Scholar]
- 61.Yukioka H, Inagaki S, Tanaka R, Katoh K, Miki N, Mizutani A, Masuko M. Transcriptional activation of the alternative oxidase gene of the fungus Magnaporthe grisea by a respiratory-inhibiting fungicide and hydrogen peroxide. Biochim Biophys Acta. 1998;1442:161–169. doi: 10.1016/s0167-4781(98)00159-6. [DOI] [PubMed] [Google Scholar]
- 62.Zhou B, Gitschier J. hCTR1: a human gene for copper uptake identified by complementation in yeast. Genetics. 1997;94:7481–7486. doi: 10.1073/pnas.94.14.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]