Abstract
In the last 15 years, Burkholderia cepacia has emerged as a significant pathogen in cystic fibrosis (CF) patients, mainly due to the severity of infection observed in a subset of patients and the fear of transmission of the organism to noncolonized patients. Although patients who deteriorate rapidly cannot be predicted by microbiological characteristics, three genetic markers have been described for strains that spread between patients. These are the cblA gene, encoding giant cable pili; a hybrid of two insertion sequences, IS1356 and IS402; and a 1.4-kb open reading frame known as the B. cepacia epidemic strain marker (BCESM). The latter two are of unknown function. An epidemic strain lineage was previously identified among CF patients in the United Kingdom that apparently had spread from North America and that was characterized by a specific random amplified polymorphic DNA (RAPD) pattern. We searched for the described genetic markers using specific PCR assays with 117 patient isolates of B. cepacia from 40 United Kingdom hospitals. Isolates were grouped according to genomovar and epidemic strain lineage RAPD pattern with a 10-base primer, P272. A total of 41 isolates from patients in 12 hospitals were classified as the epidemic strain, and 40 of these were distributed in genomovars IIIa (11 isolates), IIIb (1 isolate), and IIIc (28 isolates). All isolates of the epidemic strain were positive for the cblA gene and BCESM, but two lacked the insertion sequence hybrid. None of the 76 sporadic isolates contained cblA or the insertion sequence hybrid, but 11 of them were positive for BCESM. Nonepidemic isolates were distributed among genomovars I or IV (9), II (49), IIIa (11), IIIb (3), and IIIc (4). There were three clusters of cross-infection (one involving two patients and two involving three patients) with isolates of genomovar II. We conclude that in the United Kingdom, a single clonal lineage has spread between and within some hospitals providing care for CF patients. The presence of the cblA gene is the most specific marker for the epidemic strain. We recommend that all isolates of B. cepacia from CF patients should be screened by PCR to influence segregation and infection control strategies.
The acquisition of Burkholderia cepacia in the lungs of cystic fibrosis (CF) patients is viewed with great concern. The reason is that 20 to 30% of patients who become colonized with B. cepacia experience a rapidly fatal necrotizing pneumonia with septicemia (the “cepacia syndrome”) (3). Cross-infection between patients with the organism has been documented both in and out of the hospital (2). A single strain which had originally spread between clinics in Edinburgh and Manchester during the late 1980s was recognized as the predominant strain in many regional centers in the United Kingdom and was found in about one-third of all patients colonized with B. cepacia (9). This strain was representative of the original North American clone, designated ET 12 (4).
Since the transfer of B. cepacia to the genus Burkholderia, of which it is the type species (16), the phenotypic and genotypic definitions of B. cepacia have received increasing attention. It is now widely accepted that, rather than being a single species, B. cepacia is a complex of closely related species (11, 14, 15). According to the scheme of Vandamme et al. (14) that described five genomovars of B. cepacia, transmissible strains from CF patients fall mainly into genomovar III, although recent French experience suggests that three of their highly transmissible strains share characteristics with more than one genomovar (11).
Various markers have been associated with transmissible strains of B. cepacia. Giant fibers, known as cable pili, are found on the surfaces of some isolates of B. cepacia, and these fibers mediate adherence to respiratory mucins (10). The genetic relatedness of isolates expressing cable pili from outbreaks in CF patients suggested that this property was linked to the epidemic spread of strains (12). A hybrid of two insertion sequences (IS), IS402 and IS1356, was also found to be exclusive to epidemic strains from Ontario, Canada, and the United Kingdom (13); recently, a conserved 1.4-kb open reading frame was identified in strains of B. cepacia recovered from incidents of cross-infection between CF patients but was absent from sporadic strains (6). The open reading frame showed homology with a family of negative transcriptional regulatory genes and was termed the B. cepacia epidemic strain marker (BCESM). Transmissible strains also correlated with specific random amplified polymorphic DNA (RAPD) patterns (5). However, none of these factors has been positively associated with invasive strains, and patients who succumb to cepacia syndrome cannot yet be predicted.
Current guidelines from the CF associations in both the United Kingdom and the United States advise patients colonized with B. cepacia to avoid contact in confined areas with other patients to reduce the risk of cross-infection. This has led to the segregation of colonized from noncolonized patients, with its attendant psychosocial impact. The differentiation of transmissible from sporadic strains may therefore have a role in infection control in this patient group by identifying patients least likely to transmit infection to others. We set out to determine the frequency of genes for the three markers (cable pili, IS hybrid, and BCESM) in a collection of B. cepacia strains from CF patients and examined their association with the classification of a strain as epidemic or sporadic by epidemiological origin, RAPD pattern, and genomovar.
MATERIALS AND METHODS
Bacterial isolates.
One hundred seventeen sputum isolates of B. cepacia from 114 CF and 3 non-CF patients in 40 hospitals in the United Kingdom were examined. Control strains included the index Edinburgh epidemic strain (CF 5610), which was a gift from J. R. W. Govan, University of Edinburgh Medical School, Edinburgh, United Kingdom, and reference strain NCTC 10661. Isolates were confirmed as B. cepacia by PCR with specific 16S rRNA primers as previously described (1). All isolates were tested in the API 20NE gallery (bioMérieux, Basingstoke, United Kingdom) and examined for Gram stain reaction; motility; hydrolysis of casein, gelatin, starch, tyrosine, Tween 20, and Tween 80; production of DNase, catalase, oxidase, nitrate reductase, and lecithinase; growth on MacConkey agar and B. cepacia selective agar (Mast, Bootle, United Kingdom); acid production from ammonium salt sugars; and growth at 42°C. Isolates were allocated to a genomovar on the basis of reactions in selected tests (14).
Detection of genes for transmissibility factors.
The oligonucleotide primers (Cruachem Ltd., Glasgow, United Kingdom) for the amplification of genes encoding transmissibility factors are shown in Table 1. Bacterial DNA was prepared by emulsifying five colonies of 48-h growth from nutrient agar into 100 μl of sterile tissue culture water (Sigma, Dorset, United Kingdom). The DNA was denatured in a Dri-block (Techne, Duxford, United Kingdom) at 100°C for 5 min, vortexed, and centrifuged at 13,000 × g for 5 min, and 3 μl of the supernatant was added to 12 μl of water. A water blank (15 μl) was prepared. PCR was carried out with a 25-μl volume for each epidemic marker primer pair. This volume contained 100 pmol of each primer, 50 pmol of MgCl2, 2.5 pmol of each deoxynucleotide triphosphate, 1.25 U of Taq DNA polymerase, and 2.5 μl of 10× PCR buffer (Life Technologies, Paisley, United Kingdom). Amplification was carried out with a GeneE thermal cycler (Techne). Products were separated in a 1.5% Nusieve agarose gel (Flowgen, Sittingbourne, United Kingdom) at 100 V for 1.5 h. Amplicon size was determined by comparison with a 123-bp ladder (Life Technologies).
TABLE 1.
Primers and conditions for PCR of transmissibility marker genes of B. cepacia
| Target | Primer (reference) | Sequence | PCR conditions | Amplicon size (bp) |
|---|---|---|---|---|
| cblA | CBL1 (10) | 5′ CCAAAGGACTAACCCA 3′ | 96°C for 5 min, 1 cycle | 664 |
| CBL2 | 5′ AGCCGATGTCCATCACA 3′ | 96°C for 15 s, 58°C for 30 s, 72°C for 90 s, 25 cycles; 70°C for 5 min, 1 cycle | ||
| IS hybrid | 402A (11) | 5′ CAACCGAGACTGAGGAGATG 3′ | 96°C for 5 min, 1 cycle | 592 |
| 1356B | 5′ TCCGGCGACACCTCGATGCC 3′ | 96°C for 15 s, 63°C for 30 s, 72°C for 90 s, 25 cycles; 70°C for 5 min, 1 cycle | ||
| Epidemic marker | BCESM1 (13) | 5′ CCACGGACGTGACTAACA 3′ | 96°C for 5 min, 1 cycle | 1,400 |
| BCESM2 | 5′ CGTCCATCCGAACACGAT 3′ | 96°C for 15 s, 63°C for 30 s, 72°C for 90 s, 25 cycles; 70°C for 5 min, 1 cycle | ||
| RAPD | P272 (12) | 5′ AGCGGGCCAA 3′ | 95°C for 5 min, 1 cycle; 95°C for 1 min, 36°C for 5 min, 72°C for 2 min, 45 cycles |
RAPD profiling.
Bacterial DNA for RAPD profiling was prepared by GES extraction (8), and 50 ng was added to 15 μl of sterile tissue culture water for PCR. The PCR reagents were as for the epidemic markers, except that 25 pmol of primer P272 (Table 1) was used. PCR was carried out with an Omnigene thermal cycler (Hybaid, Ashford, United Kingdom). Products were separated as described above, and profiles were compared with that of CF 5610.
RESULTS
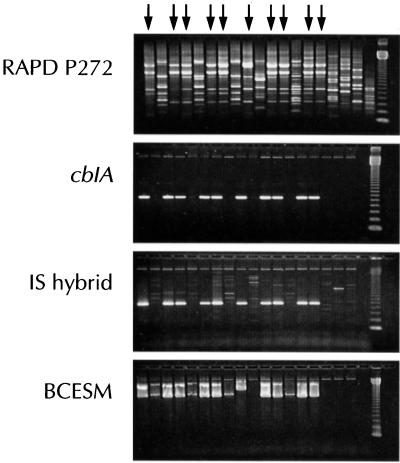
The control strain CF 5610 was positive for all three epidemic markers by PCR, in contrast to NCTC 10661, which was negative for each amplicon. Table 2 summarizes the results for 117 clinical isolates identified as B. cepacia from 40 hospitals in the United Kingdom. Of these isolates, 41 were identified as the same clone as strain CF 5610 by RAPD patterns and were isolated from patients in 12 hospitals. Each of these isolates, except 1, was classified in genomovar III—28 in IIIc (alkaline reaction in oxidation-fermentation medium and negative reaction in ammonium salt sugars), 11 in IIIa, and 1 in IIIb; the other isolate had a biochemical profile indistinguishable from that described for B. gladioli (14). All 41 isolates were positive for cblA and BCESM, and 39 were positive for the IS hybrid. Figure 1 illustrates the PCR assay results for the three markers. The cblA amplicon gave a clear strong band in all positive tests. Faint product bands were visible in the lanes of some nonepidemic strains in the IS hybrid PCR, although these were clearly distinguished from the specific product by molecular size. Two distinct bands were obtained with BCESM primers on most occasions, the lower-molecular-weight band corresponding to the predicted product size.
TABLE 2.
Genomovar classification and reaction in PCR for transmissibility factors of epidemic and nonepidemic B. cepacia strains
| Strains and marker | No. of strains with the following genomovar:
|
||||||
|---|---|---|---|---|---|---|---|
| I or IV | II | IIIa | IIIb | IIIc | B. gladioli | Total | |
| Epidemic, tested by RAPD | 0 | 0 | 11 | 1 | 28 | 1 | 41 |
| cblA | 11 | 1 | 28 | 1 | |||
| IS hybrid | 11 | 1 | 26 | 1 | |||
| BCESM | 11 | 1 | 28 | 1 | |||
| Nonepidemic, tested by RAPD | 9 | 49 | 11 | 3 | 4 | 0 | 76 |
| cblA | 0 | 0 | 0 | 0 | 0 | ||
| IS hybrid | 0 | 0 | 0 | 0 | 0 | ||
| BCESM | 2 | 6 | 2 | 1 | 0 | ||
| Control | |||||||
| CF 5610 | 1 | ||||||
| NCTC 10661 | 1 | ||||||
FIG. 1.
RAPD profile and presence of amplicons for the cblA gene, IS hybrid, and BCESM in PCR assays. Arrows denote lanes with DNA from epidemic strains. The first lane shows the epidemic control strain CF 5610. The last two lanes are a water control and a 123-bp ladder, respectively. Contamination of water with DNA was a common finding with primer P272.
Of the 76 isolates with a RAPD profile distinct from that of CF 5610, none was positive for cblA or the IS hybrid, but 11 produced a product with the BCESM primers. More than half of these isolates were genomovar II (B. multivorans); 11 were genomovar IIIa, 3 were IIIb, 4 were IIIc, and 9 were I or IV. The 11 nonepidemic BCESM-positive isolates were randomly distributed among hospitals and genomovars, and all appeared to be unique.
There were three small clusters of cross-infection in three hospitals with genomovar II isolates and one hospital with a nonepidemic IIIa isolate. Two clusters involved three patients, and the other involved two patients. All of these isolates were BCESM negative, and none had the epidemic strain RAPD profile.
DISCUSSION
To minimize the risk of cross-infection, CF patients colonized with B. cepacia usually are treated on separate wards and attend different outpatient clinics than their noncolonized counterparts. They are also advised to avoid social contact with noncolonized patients. This is an emotive issue, as contact with other patients both in and out of the hospital is considered to be important for their general well-being. This fear is due mainly to the unpredictable course following acquisition of B. cepacia and the lack of knowledge of bacterial and host factors that influence clinical outcome. Evidence suggests that where segregation is practiced in hospitals, patients tend to acquire unique strains, whereas endemic strains predominate in hospitals with no segregation policy (7). Govan et al. showed that the main cause of an increase in B. cepacia colonization among CF patients in Edinburgh and Manchester in the late 1980s was an epidemic strain that had spread between centers (2). They also demonstrated that in one instance where a patient was infected with the epidemic strain and a second strain of B. cepacia, only the epidemic strain was passed on to another patient. Subsequently, the Edinburgh strain of B. cepacia was identified as being responsible for the colonization of approximately 30% of CF patients in 10 of 16 regional centers in the United Kingdom (9). It may therefore be of value for infection control purposes to identify patients colonized by strains with the potential to spread and to differentiate such patients from those harboring sporadic strains. Segregation of patients would thus be based on the properties of the strains rather than simply the presence or absence of the organism.
The description of putative transmissibility factors in CF-associated B. cepacia by North American workers prompted our investigation of the specificity of the factors among isolates from patients in the United Kingdom. We have shown here that the cblA gene is specific for and that the IS hybrid is indicative of strains of the epidemic lineage. These findings support the original findings that cblA and the hybrid of IS1356 inserted within IS402 were associated specifically with the North American epidemic strain lineage which subsequently emerged in Britain (12, 13). All but 2 of the 41 epidemic strains examined here contained the IS hybrid, a frequency almost identical to that reported previously by Tyler et al. for the North American clone (13). The IS hybrid may be linked to the apparent increase in transmissibility of the epidemic strain or may simply be a marker for this lineage. Mahenthiralingam et al. (5) identified a conserved region in RAPD profiles of seven strains of B. cepacia that had been associated with outbreaks of infection within groups of patients, and only one of these strains was cblA positive. They suggested that the presence of BCESM was related to the ability of a strain to spread between multiple patients. We identified BCESM in all isolates of the United Kingdom epidemic strain but also in 11 strains in all four genomovars of B. cepacia. These strains appeared to be unique, and there was no evidence of spread either within or between centers. We did, however, identify small clusters of cross-infection with strains that were BCESM negative.
The finding that all isolates, except one, of the epidemic strain were genomovar III was consistent with previous reports (14). However, Segonds et al. (11), using restriction fragment length polymorphism (RFLP) analysis of amplified 16S ribosomal DNA, found that three of five highly transmissible clones of B. cepacia in French CF centers belonged to a genomovar I- or III-related RFLP group. They also observed that one of the clones associated with fatal septicemia had a B. multivorans (genomovar II) RFLP pattern. It is noteworthy that more than half of the epidemic isolates described here, however, were genomovar IIIc, which is characterized by a loss of the ability to utilize all saccharide substrates. This feature could contribute to poor identification of asaccharolytic isolates and underlines the need for the confirmation of all biochemically atypical isolates by PCR with B. cepacia-specific primers (1, 15).
In conclusion, certain isolates of B. cepacia have a propensity for epidemic spread in the CF community; (i) these isolates are members of the same genomovar, (ii) they share a characteristic DNA fingerprint, and (iii) they possess a unique genetic marker that could have a profound affect on patient care. Patients colonized with transmissible strains may have to be segregated from patients colonized with sporadic strains, who themselves may not need to be segregated. In the United Kingdom, we have found only one clonal lineage of B. cepacia that has spread within and between hospitals. Almost all isolates of this strain possess genetic markers associated with transmissibility, the most specific of which is the presence of the cblA gene. We consider it necessary, therefore, for all isolates of B. cepacia recovered from CF patients to be tested for confirmation of species identity and transmissibility factors by PCR. The question remains as to whether patients colonized with epidemic strains are at greater or lesser risk of cepacia syndrome than those colonized with nonepidemic strains.
REFERENCES
- 1.Clode F E, Kaufmann M E, Malnick H, Pitt T L. Evaluation of three oligonucleotide primer sets in PCR for the identification of Burkholderia cepacia and their differentiation from Burkholderia gladioli. J Clin Pathol. 1999;52:173–176. doi: 10.1136/jcp.52.3.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Govan J R W, Brown P H, Maddison J, Doherty C J, Nelson J W, Dodd M, Greening A P, Webb A K. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet. 1993;342:15–19. doi: 10.1016/0140-6736(93)91881-l. [DOI] [PubMed] [Google Scholar]
- 3.Govan J R W, Nelson J W. Microbiology of cystic fibrosis lung infections: themes and issues. J R Soc Med. 1993;86(Suppl. 20):11–18. [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson W M, Tyler S D, Rozee K R. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J Clin Microbiol. 1994;31:924–930. doi: 10.1128/jcm.32.4.924-930.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahenthiralingam E, Campbell M E, Henry D A, Speert D P. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J Clin Microbiol. 1996;34:2914–2920. doi: 10.1128/jcm.34.12.2914-2920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahenthiralingam E, Simpson D A, Speert D P. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul M, Pegler M, Benn R. Molecular epidemiology of Burkholderia cepacia in two Australian cystic fibrosis centres. J Hosp Infect. 1998;38:19–26. doi: 10.1016/s0195-6701(98)90171-2. [DOI] [PubMed] [Google Scholar]
- 8.Pitcher D, Saunders N, Owen R. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 9.Pitt T L, Kaufmann M E, Patel P S, Benge L C A, Gaskin S, Livermore D M. Type characterisation and antibiotic susceptibility of Burkholderia (Pseudomonas) cepacia isolates from patients with cystic fibrosis in the United Kingdom and the Republic of Ireland. J Med Microbiol. 1996;44:203–210. doi: 10.1099/00222615-44-3-203. [DOI] [PubMed] [Google Scholar]
- 10.Sajjan U S, Sun L, Goldstein R, Forstner J F. Cable (Cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J Bacteriol. 1995;177:1030–1038. doi: 10.1128/jb.177.4.1030-1038.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Segonds C, Heulin T, Marty N, Chabanon G. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J Clin Microbiol. 1999;37:2201–2208. doi: 10.1128/jcm.37.7.2201-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, Jiang R-Z, Steinbach S, Holmes A, Campanelli C, Forstner J, Sajjan U, Tan Y, Riley M, Goldstein R. The emergence of a highly transmissible lineage of cbl+Pseudomonas (Burkholderia) cepacia causing CF centre epidemics in North America and Britain. Nat Med. 1995;1:661–666. doi: 10.1038/nm0795-661. [DOI] [PubMed] [Google Scholar]
- 13.Tyler S D, Rozee K R, Johnson W M. Identification of IS1356, a new insertion sequence, and its association with IS402 in epidemic strains of Burkholderia cepacia infecting cystic fibrosis patients. J Clin Microbiol. 1996;34:1610–1616. doi: 10.1128/jcm.34.7.1610-1616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandamme P, Holmes B, VanCanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 15.van Pelt C, Verduin C M, Goessens W H F, Vos M C, Tummler B, Segonds C, Reubsaet F, Verbrugh H, van Belkum A. Identification of Burkholderia spp. in the clinical microbiology laboratory: comparison of conventional and molecular methods. J Clin Microbiol. 1999;37:2158–2164. doi: 10.1128/jcm.37.7.2158-2164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yabuuchi E, Kosaka Y, Oyaizu H, Kano I, Hotta H, Hashimoto Y, Ekazi T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]