Abstract
DNA amplification of lngA, the structural gene of longus type IV pilus produced by human enterotoxigenic Escherichia coli (ETEC) was achieved by the use of specific oligonucleotide primers designed from the nucleotide sequence of lngA. A 630-bp fragment representing the entire lngA gene was amplified in eight prototype strains previously characterized as longus positive. Five ETEC strains producing colonization factor antigen III (CFA III) (also a type IV pilus) were also positive by PCR, confirming the DNA homology between CFA III and longus. None of the non-ETEC and non-E. coli enteropathogens studied showed the 0.63-kbp amplicon. The procedure thus detected only ETEC strains harboring type IV pili genes with or without other colonization factors. Except for five lngA PCR-positive, probe-positive strains, all lngA PCR-positive strains produced the pilin as demonstrated by immunoblotting. To test the amplification procedure in a clinical setting, a collection of 264 fresh clinical E. coli strains isolated from 88 Mexican children with diarrhea was screened by PCR. Among 82 ETEC isolates found, 30 (36.5%) were lngA PCR-positive. Twenty-seven percent of the children shed ETEC that possessed lngA. In parallel with DNA probes or PCR protocols to detect enterotoxin genes, the lngA PCR method may prove useful for detection of ETEC harboring type IV pilus genes in epidemiological studies.
Some bacterial enteropathogens produce surface appendages termed type IV pili that promote colonization of the intestinal tract (7, 13, 29, 30). For example, colonization of the gut mucosa by Vibrio cholerae requires expression of a toxin-coregulated pilus (TCP) (30). The bundle-forming pilus (BFP) elaborated by enteropathogenic Escherichia coli (EPEC) serotypes that produce the attaching and effacing lesion is believed to be responsible for the localized adherence observed in cultured epithelial cells and in intestinal biopsy specimens (7, 23). Enterotoxigenic E. coli (ETEC) produces a watery diarrhea similar to that caused by V. cholerae due to the elaboration of a cholera-like heat-labile toxin (LT) and/or a heat-stable toxin (ST) (23). Worldwide, ETEC is responsible for high rates of morbidity and mortality among children (1, 2, 14, 19, 24). This organism produces a spectrum of distinct surface-adhesive filaments termed colonization factors (CFs), all of which contribute to the recognition of different receptors in the intestinal mucosa, leading to efficient colonization (3, 4, 6, 23). In addition, human ETEC strains produce a type IV pilus termed longus which is encoded by large virulence plasmids also associated with the production of CF antigens (CFAs) and enterotoxins (9). Longus is composed of a 22-kDa subunit (LngA) which shares considerable N-terminal sequence similarities with the CFA III pilin subunit CofA of ETEC, TcpA, and BfpA (13, 17). Recently, the nucleotide sequence of the structural gene encoding longus (lngA) has been reported (15). lngA is 708 bp long and encodes a predicted protein of 236 amino acids which is processed by a prepilin peptidase, yielding a mature pilin of 206 residues (15). While lngA and cofA are closely related in terms of their amino acid and nucleotide sequence, less homology is observed with TcpA, and even less homology is observed with BfpA.
The variety of ETEC virulence factors explains why immunity against ETEC disease is dependent upon the acquisition of several infections during the first few years of life (2, 19, 20, 23). Studies in different parts of the world have shown that a considerable number of strains (30 to 50%) do not possess any known CFs, suggesting the presence of as yet undescribed fimbrial antigens (20–24). It is becoming apparent that a multifimbria-based vaccine would induce better protective immunity (6, 12, 20, 32). It is therefore important to identify the most common adhesive factors in areas of the world where ETEC represents a major health problem. Longus has been detected in strains producing the known CFs as well as in strains lacking these structures, suggesting that it is widely distributed among ETEC strains (10, 25). Thus, detection of longus DNA sequences in epidemiological studies would further assist in the identification of ETEC. Detection of CFs by either immunological or molecular assays employing DNA probes and DNA amplification have proven useful for identification of ETEC in diarrheal stools (5, 14, 19, 24, 31).
In the present study, a PCR procedure based on the DNA sequence of lngA has been applied to identify type IV pilin genes among human ETEC strains, isolated from different regions of the world, that do or do not express any of the known CFs. We evaluated and demonstrated the usefulness of the procedure in a collection of prototype ETEC strains and in a subset of fresh clinical isolates obtained from children with diarrhea. The lngA-based PCR procedure is specific for the identification of ETEC harboring type IV pili and is a relatively rapid and simple method.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Human ETEC and non-ETEC diarrheagenic E. coli strains employed in this study are listed in Table 1. These included a well-characterized collection of ETEC strains isolated from diarrheal patients (adults and children) in different countries, including Bangladesh and Chile. E9034A (O8:H9) is a prototype ETEC strain which harbors CS3 and longus genes in a large 90-kbp virulence plasmid (pE9034A) and was used as a positive control in our experiments (9). As negative controls we used ETEC strains E9034P (a plasmidless derivative of E9034A which does not produce longus), and longus probe-negative M145-C2, H10407A, D19C1, D226C1, and C117C2. Non-ETEC strains used to test for specificity were E. coli K-12 DH5α, EPEC E2348/69 and B171, enteroinvasive E. coli (EIEC) E11, enteroaggregative E. coli (EAEC) C17 and 236 (isolated from diarrheal cases in Mexico and Brazil, respectively), and reference enterohemorrhagic E. coli (EHEC) 86-24 and EDL933. Non-E. coli strains (Citrobacter rhodentium DBS13, Klebsiella pneumoniae KPF28, Proteus mirabilis MA424, Shigella flexneri 2457T, Salmonella typhi CVD908, and V. cholerae O395) obtained from the Center for Vaccine Development were also included in the study to further test the specificity of the PCR. All strains had been maintained at −70°C in Luria broth containing 15% glycerol. The cultures were grown on Luria agar at 37°C for DNA amplification and on Trypticase soy agar supplemented with 5% defibrinated sheep blood (TSAB) for expression of longus (15).
TABLE 1.
Expression of longus pilin and amplification of lngA in ETEC and non-ETEC strains
| Category and strain | Serotype | Pilus(i) | Toxin type(s) | Reactiona
|
Reference or sourceb | ||
|---|---|---|---|---|---|---|---|
| IngA probe | IngA PCR | Immunoblot | |||||
| Prototype strains | |||||||
| E9034A | O8:H9 | CS3 | LT, ST | + | + | + | 9 |
| B2C | O6:H16 | CS1, CS3 | LT, ST | + | + | + | 9 |
| M447-C4 | NDc | CS1, CS3 | ND | + | + | + | 9 |
| M633C1 | O20:H− | CFA I | LT, ST | + | + | + | 9 |
| B7A | O148:H28 | CS6 | LT, ST | + | + | + | 9 |
| M415-C1 | OX:H− | CFA I | ST | + | + | + | 9 |
| M452-C1 | O20:H− | CFA I | LT, ST | + | + | + | 9 |
| M111C5 | ND | CS4, CS6 | ND | + | + | − | 9 |
| Negative controls | |||||||
| M145-C2 | O128:HS4 | CS3 | LT, ST | − | − | − | 9 |
| H10407A | O78:H11 | CFA I | LT, ST | − | − | − | 9 |
| E9034P | O:H9 | ND | − | − | − | 9 | |
| CFA III-producing ETEC strains | |||||||
| E34420A | O25:H− | CFA III, CS6 | LT | + | + | + | 15 |
| Z128-6 | O?:H− | CFA III | LT, ST | + | + | + | 15 |
| Z84-1 | O25:H− | CFA III | ND | + | + | + | 15 |
| D117-5 | Rough:H33 | CFA III | ND | + | + | + | 15 |
| MP215-1 | O78:H12 | CFA III | ST | + | + | + | 15 |
| Isolates from Bangladesh and Chile | |||||||
| D56C1 | ND | ND | LT | + | + | − | 24 |
| D19C1 | ND | ND | LT | + | + | − | 24 |
| D89C2 | ND | ND | LT | − | − | − | 24 |
| D226C1 | ND | ND | ST | − | − | − | 24 |
| C117C2 | ND | ND | ST | − | − | − | 24 |
| BD1 | ND | CFA I | ST | ND | + | + | 24 |
| BD2 | ND | CFA I | ST | ND | + | + | 24 |
| BD3 | ND | CS1, CS3 | LT, ST | ND | + | + | 24 |
| BD4 | ND | CFA I | ST | ND | + | + | 24 |
| BD5 | ND | − | ST | ND | + | + | 24 |
| BD6 | ND | CS1 | LT, ST | ND | + | + | 24 |
| BD7 | ND | CFA I | ST | ND | + | + | 24 |
| BD8 | ND | CFA I | ST | ND | + | + | 24 |
| BD9 | ND | CS1, CS3 | LT, ST | ND | + | + | 24 |
| BD10 | ND | CS2, CS3 | ST | ND | + | + | 24 |
| BD11 | ND | CFA I | ST | ND | + | − | 24 |
| 11381a | ND | − | ST | + | + | + | 19 |
| 10001a | O6:NM | CS1, CS3 | ST | + | + | + | 19 |
| 10154a | ND | − | ST | + | + | + | 19 |
| 2998a | ND | − | ST | + | + | + | 19 |
| E379a | ND | − | ND | + | + | − | 19 |
| Non-ETEC E. coli strains (n)d | |||||||
| EPEC (2) | BFP | − | − | − | 7 | ||
| EIEC (1) | − | − | − | 5 | |||
| EHEC (2) | O157:H7 | − | − | − | CVD | ||
| EAEC (2) | − | − | − | CVD | |||
| K-12 DH5α | − | − | − | CVD | |||
| Enteric pathogens | |||||||
| C. rhodentium DBS13 | − | − | − | CVD | |||
| K. pneumoniae KPF-28 | − | − | − | ADM | |||
| V. cholerae O395 | TCP | − | − | − | CVD | ||
| P. mirabilis MA424 | MR/P | − | − | − | CVD | ||
| S. flexneri 2457T | − | − | − | CVD | |||
| S. typhi CVD908 | − | − | − | CVD | |||
+, positive; −, negative.
CVD, Center for Vaccine Development, University of Maryland; ADM, Arlette Darfeuille-Michaud, Facultés de Pharmacie et Médecine, Clermont-Ferrand, France.
ND, not determined.
Number of isolates tested.
Clinical isolates.
Stool samples from 88 children (from 3 to 36 months of age) with acute diarrhea who attended the Hospital del Niño Poblano in the City of Puebla, Mexico, were processed for isolation of gram negative enteric pathogens as previously described (14, 19). This hospital provides free medical care to low-income families in Puebla. Clinical features such as type of diarrhea, vomiting, and fever (>38°C) were recorded. To evaluate the use of lngA PCR in a clinical laboratory, three lactose-fermenting E. coli colonies cultured on MacConkey agar were picked from each stool culture and kept in Luria broth with glycerol at −70°C until tested for the presence of lngA.
Amplification of lngA.
A 0.63-kbp fragment comprising the entire lngA gene was amplified using oligonucleotide primers JG1 (5′-CGGAATTCATGAGCCTGCTGGAAGTTATCA-3′) and JG2 (5′-CGGAATTCCGGCTACCTAAAGTAATTGAGT-3′), derived from the lngA DNA sequence recently obtained from pJAG1 (14). pJAG1 contains a 7-kbp BamHI fragment isolated from pE9034A, which encodes lngA and other accessory genes involved in the biogenesis of the pilus (15). The amplification reaction was performed in a 50-μl volume and contained 500 mM KCl, 100 mM Tris-HCl (pH 8.3), 1 mM MgCl2, a 0.25 mM concentration of each deoxynucleoside triphosphate, and 0.5 U of Taq polymerase (Boehringer Mannheim). A suitable amount of bacteria was picked from a colony on a Luria agar plate and suspended in the PCR mixture. The PCR consisted of 10 min of heating at 95°C; 35 cycles of 1-min denaturation at 95°C, 3-min annealing at 50°C, and 3-min primer extension at 72°C; and a final extension of 10 min at 72°C (5). The amplified DNA fragments were resolved by 1% agarose gel electrophoresis and visualized under UV transillumination after staining with ethidium bromide (26).
SDS-PAGE and immunoblottings of whole cell extracts.
To test for longus production, all the PCR lngA-positive prototype strains, including those producing CFA III, were grown on TSAB at 37°C. The bacteria were resuspended to the same concentration in 50 mM phosphate-buffered saline, pH 7.4, and boiled for 5 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. These whole-cell extracts were subjected to electrophoresis in 16% polyacrylamide gels (18). Separated proteins were transferred onto nitrocellulose membranes and analyzed by Western blotting using rabbit anti-longus antibodies and the appropriate secondary antibodies (9). The blots were developed with a mixture of nitroblue tetrazolium and 5-bromo-chloro-indolyl-phosphate (Sigma).
Detection of CFs.
CFs were detected by monoclonal antibodies and dot blot enzyme-linked immunosorbent assay (ELISA) as previously described (27, 28).
DNA probing.
The production of ST and LT enterotoxins in the Bangladeshi strains was analyzed by ELISA (19), and in the Mexican isolates the presence of the enterotoxin genes was probed by DNA hybridization using nonradioactively labeled DNA probes as previously described (5, 8). Briefly, for detection of the sequences encoding LT and ST, bacterial colonies were transferred onto nylon membranes and lysed with proteinase K (1 mg/ml; Sigma) at 50°C for 1 h. The DNA was immobilized by UV treatment for 2 min, and the DNA strands were separated by 0.5 NaOH treatment of the membranes and then neutralized with 5 mM Tris (pH 7.2) and 1.5 mM NaCl. The membranes were hybridized with digoxigenin-labeled ST and LT probes as previously described (5).
RESULTS
lngA DNA amplification and specificity.
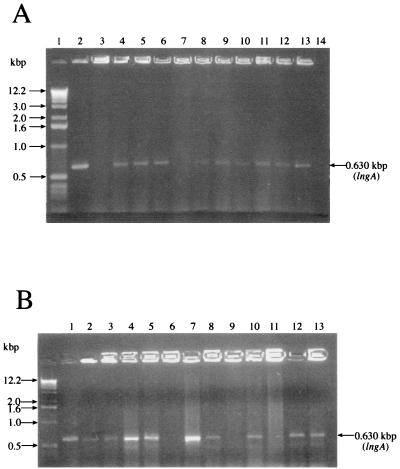
In an earlier study, prototype ETEC strains expressing longus were shown to hybridize with an oligonucleotide probe derived from the N-terminal region of the longus pilin (9) (Table 1). These strains were used in this study to evaluate the PCR procedure involving amplification of lngA. The lngA DNA sequence obtained from E9034A showed that this gene is contained in a plasmid-derived 0.708-kbp fragment and encodes a protein of 236 residues (15). Oligonucleotide primers derived from the 5′ and 3′ ends of the DNA encoding the mature pilus peptide (210 residues long) amplified a 630-bp fragment in lngA-containing ETEC (Fig. 1A; Table 1). No amplification products were seen in ETEC negative controls E9034P (the E9034A-derived plasmidless strain that does not produce longus), M145-C2, or H10407A. ETEC strains previously designated as CFA III positive were shown to also amplify the 0.63-kbp amplicon. These results are not surprising since both longus and CFA III share significant DNA homologies, and in fact, antigenic cross-reactivities between these two have been shown before (15, 29). The expected PCR product was also obtained in ETEC strains previously isolated from diarrheal cases in Bangladesh and Chile (Fig. 1B; Table 1). From the latter collection, three isolates (D89C2, D226C1, and C117C2) known to lack lngA did not show the PCR product.
FIG. 1.
Amplification of lngA in ETEC strains. (A) Ethidium bromide-stained agarose gel of lngA PCR-amplified products from prototype ETEC strains using the primers JG1 and JG2. Lanes: 1, 1-kbp ladder; 2, E9034A; 3, E9034P (negative control); 4, B2C; 5, M633C1; 6, M447-C4; 7, H10407A (negative control); 8, B7A; 9, M415C1, 10, M111C5; 11, M452C1; 12, E34420A; 13, Z128-6; 14, V. cholerae O395 (negative control). (B) Amplification of lngA in clinical isolates from various countries. Lane 1, D117-5; 2, D56C1; 3, 11381a; 4, BD1; 5, BD2; 6, D226C1(−); 7, BD3; 8, BD4; 9, C117C2(−); 10, BD5; 11, D89C2 (−); 12, 10001a; 13, 10154a. (−) denotes lack of amplification product.
A collection of several enteric pathogens such as S. typhi, S. flexneri, P. mirabilis, K. pneumoniae, and C. rhodentium, non-ETEC diarrheagenic E. coli (EIEC, EHEC, and EAEC), and K-12 DH5α were used to further evaluate the specificity of the lngA-based PCR assay. None of these organisms amplified a lngA PCR product, indicating the lack of lngA-related sequences in these organisms (Table 1). Furthermore, other bacteria producing type IV pili, namely, EPEC expressing BFP and V. cholerae O395 expressing TCP, were negative in the PCR procedure, indicating that there is no DNA homology between these type IV pili (Table 1).
Correlation between PCR positivity and longus expression.
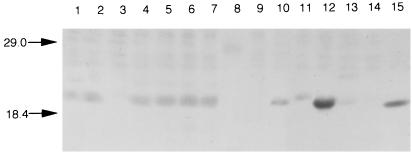
To correlate the presence of lngA by PCR with longus expression, whole-cell extracts of all PCR lngA-positive strains were subjected to immunoblotting using anti-longus antibodies. Figure 2 depicts the reactivity of anti-longus serum with some PCR-positive and PCR-negative ETEC isolates. Only those strains that showed the 0.63-kbp PCR product produced the 22-kDa pilin subunit (Fig. 2). Except for five PCR-positive isolates (M111C5, D19C1, D56C1, BD11, and E379a) that tested negative by immunoblotting, the lngA-based PCR procedure was in complete agreement with expression of longus. Failure to demonstrate longus in these isolates could be attributed to the employment of inadequate environmental growth conditions or the lack of a genetic element involved in longus regulation and synthesis. Although no differences in size was noted in the PCR products, differences in molecular mass were apparent between the pilin subunits in some of the isolates tested, suggesting heterogeneity among longus pili produced by various ETEC. Amino acid changes within the pilus protein might account for the differences in mobility in SDS-PAGE gels. CFA III-producing ETEC that were PCR positive for lngA also reacted with longus antiserum (Fig. 2; Table 1).
FIG. 2.
Analysis of whole-cell extracts of lngA-positive and-negative ETEC strains by immunoblotting. Lanes: 1, BD1; 2, BD2; 3, C117C2 (−); 4, BD3; 5, BD4; 6, BD5; 7, BD6; 8, D226C1 (−); 9, M145-C2 (−); 10, 11381a; 11, 10001a (±); 12, 10154a; 13, 2998a (±); 14 H10407A (−); 15, BD6. Molecular mass markers (in kilodaltons) are indicated by arrows. (−) denotes lack of reactivity, and (±) indicates a weak reaction.
Presence of lngA in fresh clinical isolates.
Among the 264 E. coli isolates obtained from the 88 Mexican children with diarrhea, 82 (31%) harbored enterotoxin genes. Among these 82 ETEC strains, 30 (36.5%) of them contained lngA, as determined by PCR. These 30 isolates were found in 24 (27%) of the 88 children studied. Taking into account that the children were perhaps not all infected with ETEC, but were perhaps infected with other bacterial, parasitic, or viral etiological agents, the frequency of ETEC (in particular lngA-positive ETEC) among these isolates is relevant. These 30 PCR lngA-positive E. coli strains possessed either the ST and/or LT genes, confirming the toxigenic nature of the strains. Eighteen of them contained the LT gene only, 11 hybridized with the ST and LT probes, and only 1 isolate contained the ST gene (data not shown). This is not by any means an epidemiological study of the burden of ETEC infection in the community. It is clear that the lngA-based PCR alone will not detect all ETEC strains in a particular study. Therefore, it will be necessary to include detection of ST and LT genes by other assays. Nevertheless, the procedure was useful for detecting ETEC harboring type IV pilin genes.
DISCUSSION
In areas of the world where diarrheal infections are endemic, detection of virulence markers by means of molecular probes, DNA amplification, and specific antibodies has been widely used to identify bacterial pathogens in epidemiological studies (1, 5, 10, 11, 14, 19, 24, 31). Several investigators have utilized ELISA, dot blotting, DNA hybridization, and PCR techniques to identify EPEC (8, 11, 16) and ETEC strains producing enterotoxins and the known CFs (1, 5, 10, 19, 21, 24, 31). In this study, we have developed a PCR procedure based on the identification of the nucleotide sequence encoding lngA, a type IV pilus gene of ETEC (15). The primers amplified a 0.63-kbp amplicon which corresponds to the DNA sequence encoding the mature pilus protein (∼22 kDa). First, the procedure was tested against a collection of prototype ETEC strains known to harbor lngA. All these strains amplified the expected PCR product. None of the longus-negative ETEC strains employed as negative controls showed the lngA product. A subset of well-characterized ETEC strains isolated from diarrheal patients in Bangladesh and Chile were also included in the study, and some of them were shown by PCR to contain lngA and, furthermore, to produce longus. Except for five isolates (M111C5, D19C1, D56C1, BD11, and E379a) which were positive by PCR and negative in immunoblots, the lngA-based PCR procedure was in complete agreement with the expression of longus. Failure to demonstrate longus in these isolates could be attributed to the employment of inadequate environmental growth conditions or the lack of a genetic element involved in longus regulation and synthesis.
Furthermore, a collection of non-ETEC E. coli, including a K-12 strain, defined diarrheagenic E. coli strains such as EIEC, EHEC, and EAEC, and several other bacterial enteropathogens tested negative by the PCR procedure. Since longus shares similarities with the type IV pilus family (9, 13), we included V. cholerae and EPEC to further determine the specificity of the PCR. No PCR products were obtained in these type IV pilus-producing organisms, confirming a lack of DNA homology between longus, Bfp, and Tcp pilin genes (7, 9, 15). However, ETEC strains bearing CFA III (also a type IV pilus) were also detected by the PCR procedure, confirming the reported DNA homologies between these pili (15, 29). It is possible that some of these isolates previously identified as CFA III positive may be in fact longus-producing ETEC, specially those belonging to serogroups other than O25 (21). In a previous study, Gómez et al. showed that longus and CFA III are immunologically cross-reactive (15).
In order to validate the use of the lngA-based PCR in clinical epidemiological studies, we performed PCR in a collection of 264 E. coli strains isolated from children with diarrhea in the City of Puebla, Mexico. Among the 264 E. coli strains tested, 82 (31%) were identified as ETEC strains, of which 30 (36.5%) possessed lngA. These 30 strains were found in almost one-third of the children studied, suggesting that ETEC is an important pathogen in children in this community. No data are available in terms of the burden of ETEC disease for this part of Mexico, and we cannot estimate the importance of ETEC infections based on the PCR data because the collection of ETEC strains was relatively small. Nevertheless, the data are important considering that the frequency of CFA I and CFA II among ETEC strains in several countries may vary between 25 and 40% (1, 6, 10, 18, 19, 23, 31). The scope of this paper was to set up the basis for future epidemiological studies of the burden of ETEC in childhood diarrhea through detection of virulence genes by DNA amplification. Using an oligonucleotide longus probe, the frequency of longus among ETEC isolated from different regions of the world varied between 19 and 38% (10). In a different study in Bangladesh, using monoclonal antibodies Qadri et al. detected longus in 61 of 667 (8.5%) ETEC isolates obtained from diarrheal stools from children and adults (25). Of the isolates, 50 were positive for other CFs (61% for CFA II and 21% for CFA I), while 11 were negative for any of the other eight CFs tested for. Thus, as for other CFAs the frequency of longus among ETEC strains varies depending on the geographic region of the world studied.
The lngA PCR-based detection of ETEC strains has several advantages over DNA probing and hybridization techniques. It may take 2 to 3 days to obtain results using DNA probes, while PCR results are obtained in a few hours in the same day. Moreover, unlike the DNA probe method, no preparation of sample DNA is required in the PCR assay, as bacteria can be directly used in the amplification assay. In fact, pools of lactose-positive E. coli colonies from individual patients may be employed.
In summary, the lngA-based PCR procedure described is useful and specific in detecting ETEC harboring type IV pilus (longus and CFA III) genes, and it may assist in the identification of these ETEC strains in epidemiological studies in most settings. It is obvious that the use of the lngA PCR procedure alone as a screening test for ETEC in stool will not detect longus-negative ETEC. Thus, this procedure should be used in parallel with other assays to detect the most important virulence factors of ETEC, namely LT and ST enterotoxins, as well as the most common CFAs. Frankel et al. (5) described the use of PCR to simultaneously amplify three virulence genes in diarrheal stools. Thus, it may be possible to include other ETEC virulence genes in the lngA PCR protocol to detect ETEC directly in stools.
ACKNOWLEDGMENTS
This work was supported by Conacyt (México) (grant 3485P-M9607 to Jorge A. Girón), and by Sida-SAREC (grant 1998-05440) to Firdausi Qadri). The ICDDR,B Centre for Health and Population Research is supported by countries and agencies that share its concerns for the health problems of developing countries. Zita Gutiérrez thanks Conacyt for her scholar stipend.
We thank Alejandro Ruiz and Dvorak Condado for helpful assistance; Oscar Gómez for helpful criticism; and Yolande Bertin, David Schauer, and Harry Mobley for non-E. coli enteropathogens.
REFERENCES
- 1.Abu-Elyazeed R, Wierzba T F, Mourad A S, Peruski L F, Kay B A, Rao M, Churrilla A M, Bourgeois A L, Mortagy A K, Kamal S M, Savarino S J, Campbell J R, Murphy J R, Naficy A, Clemens J D. Epidemiology of enterotoxigenic Escherichia coli diarrhea in a pediatric cohort in a periurban area of lower Egypt. J Infect Dis. 1999;179:382–389. doi: 10.1086/314593. [DOI] [PubMed] [Google Scholar]
- 2.Black R E, Merson M H, Rowe B, Taylor P R, Alim A R M A, Gross R J, Sack D A. Enterotoxigenic Escherichia coli diarrhoea: acquired immunity and transmission in an endemic area. Bull W H O. 1981;59:263–267. [PMC free article] [PubMed] [Google Scholar]
- 3.Evans D G, Evans D J, Tjoa W. Hemagglutination of human group A erythrocytes by enterotoxigenic Escherichia coli isolated from adults with diarrhea: correlation with colonization factor. Infect Immun. 1977;18:330–337. doi: 10.1128/iai.18.2.330-337.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans D G, Evans D J, Jr, Tjoa W S, Dupont H L. Detection and characterization of a colonization factor of enterotoxigenic Escherichia coli isolated from adults with diarrhea. Infect Immun. 1978;19:727–736. doi: 10.1128/iai.19.2.727-736.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frankel G, Girón J A, Schoolnik G K. Multi-gene amplification: simultaneous detection of three virulence genes in diarrheic stool. Mol Microbiol. 1989;3:1729–1734. doi: 10.1111/j.1365-2958.1989.tb00158.x. [DOI] [PubMed] [Google Scholar]
- 6.Gaastra W, Svennerholm A M. Colonization factors of human enterotoxigenic Escherichia coli (ETEC) Trends Microbiol. 1996;4:446–452. doi: 10.1016/0966-842x(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 7.Girón J A, Ho A S Y, Schoolnik G K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 8.Girón J A, Donnenberg M S, Martin W C, Jarvis K G, Kaper J B. Distribution of the bundle-forming pilus structural gene (bfpA) among enteropathogenic Escherichia coli (EPEC) J Infect Dis. 1993;168:1037–1041. doi: 10.1093/infdis/168.4.1037. [DOI] [PubMed] [Google Scholar]
- 9.Girón J A, Levine M M, Kaper J B. Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol Microbiol. 1994;12:71–82. doi: 10.1111/j.1365-2958.1994.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 10.Girón J A, Viboud G I, Sperandio V, Gómez-Duarte O G, Maneval D, Albert M J, Levine M M, Kaper J B. Prevalence and association of Longus pilus structural gene (lngA) with colonization factor antigens, ST/LT enterotoxin types, and serotypes of enteroroxigenic Escherichia coli. Infect Immun. 1995;63:4195–4198. doi: 10.1128/iai.63.10.4195-4198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girón J A, Qadri F, Azim T, Jarvis K G, Kaper J B, Albert M J. Monoclonal antibodies specific for the bundle-forming pilus of enteropathogenic Escherichia coli. Infect Immun. 1995;63:4949–4952. doi: 10.1128/iai.63.12.4949-4952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girón J A, Xu J G, González-Bonilla C, Hone D, Kaper J B, Levine M M. Simultaneous expression of CFA/I and CS3 colonization factor antigens of enterotoxigenic Escherichia coli by aroC, aroD Salmonella typhi vaccine strain CVD908. Vaccine. 1995;13:939–946. doi: 10.1016/0264-410x(95)00003-j. [DOI] [PubMed] [Google Scholar]
- 13.Girón J A, Gómez-Duarte O G, Jarvis K G, Kaper J B. Longus pilus of enterotoxigenic Escherichia coli and its relatedness to other type-4 pili: a minireview. Gene. 1997;192:39–43. doi: 10.1016/s0378-1119(97)00039-5. [DOI] [PubMed] [Google Scholar]
- 14.Gomes T A T, Rassi V, MacDonald K L, Ramos S R T S, Trabulsi L R, Vieira M A M, Guth B E C, Candeias J A N, Ivey C, Toledo M R F, Blake P A. Enteropathogens associated with acute diarrheal disease in urban infants in São Paulo, Brazil. J Infect Dis. 1991;164:331–337. doi: 10.1093/infdis/164.2.331. [DOI] [PubMed] [Google Scholar]
- 15.Gómez-Duarte O G, Ruiz-Tagle A, Gómez D C, Viboud G I, Jarvis K G, Kaper J B, Girón J A. Identification of lngA, the structural gene of longus type IV pilus of enterotoxigenic Escherichia coli. Microbiology. 1999;145:1809–1816. doi: 10.1099/13500872-145-7-1809. [DOI] [PubMed] [Google Scholar]
- 16.Gunzburg S T, Tornieporth N G, Riley L W. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J Clin Microbiol. 1995;33:1375–1377. doi: 10.1128/jcm.33.5.1375-1377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda T, Arita M, Miwatani T. Characterization of new hydrophobic pili of human enterotoxigenic Escherichia coli: a possible new colonization factor. Infect Immun. 1984;43:959–965. doi: 10.1128/iai.43.3.959-965.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Levine M M, Ferreccio C, Prado V, Cayazzo M, Abrego O, Martínez J, Maggie L, Baldini M M, Martin W C, Maneval D R, Kay B, Guers L, Lior H, Wasserman S S, Nataro J P. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level periurban community in Santiago, Chile. Am J Epidemiol. 1993;138:849–869. doi: 10.1093/oxfordjournals.aje.a116788. [DOI] [PubMed] [Google Scholar]
- 20.Levine M M, Girón J A, Noriega F. Fimbrial vaccines. In: Klem P, editor. Fimbriae: adhesion, biogenics, genetics and vaccines. Boca Raton Fla: CRC Press; 1995. pp. 255–270. [Google Scholar]
- 21.McConnell M M, Rowe B. Prevalence of the putative colonization factors CFA/III and PCFO159:H4 in enterotoxigenic Escherichia coli. J Infect Dis. 1989;159:582–586. doi: 10.1093/infdis/159.3.582. [DOI] [PubMed] [Google Scholar]
- 22.McConnell M M. Newly characterized putative colonization factors of human enterotoxigenic Escherichia coli. In: Wädstrom T, editor. Molecular pathogenesis of gastrointestinal infections. New York, N.Y: Plenum Press; 1991. pp. 79–85. [Google Scholar]
- 23.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;5:109–114. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qadri F, Das S K, Faruque A S G, Fuchs G J, Albert M J, Sack R B, Svennerholm A-M. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J Clin Microbiol. 2000;38:27–31. doi: 10.1128/jcm.38.1.27-31.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qadri, F., J. A. Girón, J. Xicohténcatl-Cortes, Y. A. Begum, M. Asaduzzaman, E. Negrete, and J. M. Albert. Human antibody response to Longus type IV pilus and study of its prevalence among enterotoxigenic Escherichia coli in Bangladesh using monoclonal antibodies. J. Infect. Dis., in press. [DOI] [PubMed]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 27.Svennerholm A-M, Wiklund G. Rapid GM1-enzyme-linked immunosorbent assay with visual reading for identification of Escherichia coli heat-labile enterotoxin. J Clin Microbiol. 1983;17:596–600. doi: 10.1128/jcm.17.4.596-600.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svennerholm A-M, Wikstrom M, Lindblad M, Holmgren J. Monoclonal antibodies against Escherichia coli heat-stable toxin (STa) and their use in a diagnostic ST ganglioside GM1-enzyme-linked immunosorbent assay. J Clin Microbiol. 1986;24:585–590. doi: 10.1128/jcm.24.4.585-590.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taniguchi T, Fujino Y, Yamamoto K, Miwatani T, Honda T. Sequencing of the gene encoding the major pilin of pilus colonization factor antigen III (CFA/III) of human enterotoxigenic Escherichia coli and evidence that CFA/III is related to type IV pili. Infect Immun. 1995;63:724–728. doi: 10.1128/iai.63.2.724-728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viboud G I, Binsztein N, Svennerholm A M. Characterization of monoclonal antibodies against putative colonization factors of enterotoxigenic Escherichia coli and their use in an epidemiological study. J Clin Microbiol. 1993;31:558–564. doi: 10.1128/jcm.31.3.558-564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.World Health Organization. New frontiers in the development of vaccines against enterotoxigenic (ETEC) and enterohaemorrhagic (EHEC) Escherichia coli infections. Weekly Epidemiol Rec. 1999;13:98–100. [Google Scholar]