Abstract
Following hepatic injury or stress, gluconeogenic and acute-phase response genes are rapidly upregulated to restore metabolic homeostasis and limit tissue damage. Regulation of the liver-restricted insulin-like growth factor binding protein 1 (IGFBP-1) gene is dramatically altered by changes in the metabolic state and hepatectomy, and thus it provided an appropriate reporter to assess the transcriptional milieu in the liver during repair and regeneration. The cytokine interleukin-6 (IL-6) is required for liver regeneration and repair, and it transcriptionally upregulates a vast array of genes during liver growth by unknown mechanisms. Evidence for a biologic role of IL-6 in IGFBP-1 upregulation was demonstrated by increased expression of hepatic IGFBP-1 in IL-6 transgenic and following injection of IL-6 into nonfasting animals and its reduced expression in IL-6−/− livers posthepatectomy. In both hepatic and nonhepatic cells, IL-6 -mediated IGFBP-1 promoter activation was via an intact hepatocyte nuclear factor 1 (HNF-1) site and was dependent on the presence of endogenous liver factor HNF-1 and induced factors STAT3 and AP-1 (c-Fos/c-Jun). IL-6 acted through the STAT3 pathway, as dominant negative STAT3 completely blocked IL-6-mediated stimulation of the IGFBP-1 promoter via the HNF-1 site. HNF-1/c-Fos and HNF-1/STAT3 protein complexes were detected in mouse livers and in hepatic and nonhepatic cell lines overexpressing STAT3/c-Fos/HNF-1. Similar regulation was demonstrated using glucose-6-phosphatase and α-fibrinogen promoters, indicating that HNF-1/IL-6/STAT3/AP-1-mediated transactivation of hepatic gene expression is a general phenomenon after liver injury. These results demonstrate that the two classes of transcription factors, growth induced (STAT3 and AP-1) and tissue specific (HNF-1), can interact as an adaptive response to liver injury to amplify expression of hepatic genes important for the homeostatic response during organ repair.
The liver, which plays an important role in maintaining metabolic and synthetic homeostasis, constitutes a conditional renewal system in which parenchymal cells normally in G0 may be induced to proliferate following toxic damage, hepatitis, and surgical resection that culminates in the rapid restoration of hepatic parenchyma (35). To maintain glucose balance following the acute loss of liver mass posthepatectomy, phosphoenolpyruvate carboxykinase (PEPCK), glucose-6-phosphatase (G6Pase), and other genes involved in gluconeogenesis are rapidly upregulated in the regenerating liver (14, 37, 54, 55). However, the molecular mechanism by which the liver maintains metabolic homeostasis despite the acute loss of two-thirds of hepatic tissue or after injury is not known.
Liver regeneration, a hyperplastic response, involves the proliferation of the mature functioning cells composing the intact organ (35, 54, 55). Of the known cytokines released after liver injury or hepatectomy, interleukin-6 (IL-6) has been shown to be required for normal liver regeneration and repair (8, 24). In IL6−/− mice, a highly significant reduction in hepatocyte DNA synthesis, increased liver necrosis, discrete G1-phase abnormalities including absence of STAT3 activation, reduction in AP-1 activation, and selective abnormalities in gene expression are observed posthepatectomy and after carbon tetrachloride injury, all of which are corrected by injection with IL-6.
Among those genes whose expression is abnormal in IL-6−/− livers after partial hepatectomy are those encoding proteins involved in cell cycle progression such as AP-1 factors, c-Myc, and cyclin D1. However, a number of other genes with less clear connection to cell growth show blunted induction in the absence of IL-6, including the insulin-like growth factor binding protein 1 (IGFBP-1) gene. The mechanism by which IL-6 activates such a vast array of genes is unknown. We chose to study IGFBP-1 because of its proposed role in both hepatic growth and metabolism.
IGFBP-1 is an immediate-early gene induced at the transcriptional level in the remnant liver following partial hepatectomy (26, 38, 50). It is distinct in that its plasma level is dynamically regulated by changes in the metabolic state and after hepatic injury. Of the known upregulators of IGFBP-1 transcription, only IL-6 and phorbol esters have been demonstrated to overcome the strong inhibition of IGFBP-1 expression by insulin, at least in vitro (27).
The IGFBP-1 promoter has been extensively studied. Traditional promoter and deletion analyses indicate that highly conserved sequences within a few hundred bases upstream of the transcription initiation site confer liver specific and hormonal regulation. DNase I hypersensitivity analyses identified clusters of liver-restricted nuclease sensitive sites in the promoter region, −100 to −300, −2300, −3100, and −5000, along with other weak sites (9). This tissue-specific pattern of expression may be regulated in part by members of hepatocyte nuclear factor (HNF-1) family of proteins, as the HNF-1 forms are responsible for the basal IGFBP-1 promoter activity in hepatoma cells via a conserved site just upstream of the RNA initiation site (1, 2, 46, 53). HNF-1α, a homeodomain protein, regulates the expression of a number of hepatic genes, and contains a variant homeodomain that binds to DNA either as a homodimer or as a heterodimer with HNF-1β (4, 60).
In this report, we propose a mechanism by which HNF-1α coordinates the interaction of STAT3/IL-6 and c-Fos, leading to synergistic transcriptional upregulation of promoters like the IGFBP-1, G6Pase, and α-fibrinogen promoters. We also provide evidence showing that the two classes of transcription factors, growth induced (STAT3 and AP-1) and tissue specific (HNF-1), can interact as an adaptive response to liver injury to amplify expression of hepatic genes important for the homeostatic response during organ repair.
MATERIALS AND METHODS
Partial hepatectomy and IL-6 injection.
Twelve- to 16-week-old male IL-6+/+ and IL-6−/− mice (45), generated on C57BL/6 backgrounds, were used. Seventy percent partial hepatectomy was performed as described elsewhere (8, 16). IL-6-treated mice were injected subcutaneously with recombinant, human IL-6 (rhIL-6; 1 mg/kg of body weight) as described elsewhere (8). Animals were sacrificed at indicated time points after partial hepatectomy or IL-6 injection.
Analysis of IGFBP-1 protein and mRNA expression.
Animals were sacrificed at the indicated times posthepatectomy or after IL-6 injection, and total liver RNA preparation, Northern blotting, and hybridization were performed as described elsewhere (38). Total RNA samples from the IL-6 transgenic mice were provided by Gennaro Ciliberto (33). For immunoblots, 5 μl of serum was electrophoresed on sodium dodecyl sulfate (SDS)–12% polyacrylamide gels, transferred to polyvinylidene difluoride membranes (Bio-Rad), and detected by enhanced chemiluminescence (Amersham). A 1:1,000 dilution of the IGFBP-1 antibody (Cocalico Biologicals, Inc., Reamstown, Pa) followed by a 1:10,000 dilution of goat anti-rabbit horseradish-conjugated secondary antibody (Zymed) was used for immunoblotting (26).
Gel mobility shift assays.
Electrophoretic mobility shift assays (EMSAs) were performed as described elsewhere (8). Binding reactions were performed using 10 μg of nuclear extracts from cultured HepG2 and HeLa cells. Where indicated, HepG2 and HeLa cells were treated with rhIL-6 (100 ng/ml) for 20 min prior to harvest. In addition, 1 ng of radiolabeled oligonucleotide and 2 μg of poly (dI-dC) (Pharmacia) as a nonspecific DNA competitor were also included in the binding reactions. The mixtures were incubated for 15 min at room temperature in binding buffer containing 10 mM HEPES (pH 7.9), 50 mM NaCl, 1 mM EDTA, and 10% glycerol. For competition assays, a 100-fold excess of unlabeled oligonucleotide was incubated with extracts for 15 min at room temperature prior to addition of the radiolabeled probe. For supershift experiments, 1 μl of antibody targeted to HNF-1α (sc-6547X), HNF-1β (sc-7411X), or USF1 (sc-229X) (all from Santa Cruz Biotechnology) was incubated with the extracts for 2 h at 4°C prior to addition of the radiolabeled probe. The reactions were analyzed on 5% nondenaturing gels in 0.5× Tris-borate-EDTA buffer, and the level of protein expression was assessed by densitometric scanning. Complementary oligonucleotide pairs for gel retardation experiments were obtained from Life Technologies Co. and annealed in a buffer containing 250 mM Tris-HCl (pH 7.6). The annealed products were purified by polyacrylamide gel electrophoresis (PAGE) and end labeled using T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP (Dupont NEN). Sequences for -70/-44 wild-type (WT) and M1, M2, M3, M4, M5, and CCGTT mutant oligonucleotides are shown in Fig. 3B. Oligonucleotide sequences for E2, c-Fos STAT3, C/EBPα/β, TTR-HNF-3 (−111/−85), and −3118/−3094 were described by Crissey et al. (9). The sequences for AP-1 and HNF4 are 5′-GATCCTTGAGTCACATCGATTGAGTCACG-3′ and 5′-GGAAAGGTCCAAAGGGCGCCTTG-3′, respectively.
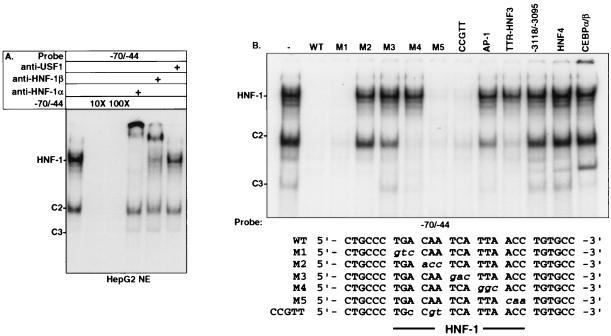
FIG. 3.
Characterization of proteins binding to region −70/−44 of the mouse IGFBP-1 promoter. (A) EMSA using HepG2 nuclear extracts (NE) showing competition and supershift by HNF-1α and -β antibodies. The indicated radiolabeled probe was incubated with HepG2 nuclear extracts in the absence or presence of anti-HNF-1α, anti-HNF-1β, or anti-USF1. For competition assays, a 10- or 100-fold molar excess of unlabeled oligonucleotide of the same sequence was used. Reaction products were fractionated on a 5% nondenaturing acrylamide gel. (B) Effects of selective mutations to HNF-1 site analyzed by EMSA using 100-fold molar excesses of the indicated competitors. Sequences for the wild-type and mutant oligonucleotides between −70 and −44 are shown; sequences for the other competitors are described in Materials and Methods.
Cell culture and transient transfections.
HepG2 hepatoma cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco/BRL), and HeLa cells were cultured in Iscove's medium. Both media were supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U of penicillin, and 50 U of streptomycin. Transient transfections were carried out in 24-well plates or 60-mm-diameter dishes, as indicated, using the calcium phosphate technique. Twenty-four hours after seeding, the cells were incubated with DNA precipitate containing the reporter and expression plasmids indicated in the figure legends. The cells were kept in media containing 0.2% FBS for 18 h, treated with 100 ng of rhIL-6 per ml for 4 h, and harvested with 1× Reporter Lysis Buffer (Promega Co.). Luciferase activities were measured using a Luminant luminometer (EG&G Wallac, Gaithersburg, Md.).
Plasmids.
The constructs pIBP-6.6, pIBP-0.23, pIBP-0.12, and pIBP-0.056 have been described elsewhere (9). The fragments for pIBP-0.07, pIBP-0.07 CCGTT, pIBP-0.07 M2, pIBP-0.07 M3, and pIBP-0.07 M4 were amplified by PCR using primer 5′-ATGCCAAGCTTGGCCGTGTG-3′ and primers 5′-CTGCCCTGACAATCATTAACCTGTGCC-3′, 5′-CTGCCCTGCCGTTCATTAACCTGTGCC-3′, 5′-CTGCCCTGAACCTCATTAACCTGTGCC-3′, 5′-CTGCCCTGACAAGACTTAACCTGTGCC-3′, and 5′-CTGCCCTGACAATCAGGCACCTGTGCC-3′, respectively. The fragments were digested with HindIII and cloned into pGL2 Basic between the HindIII and blunted SacI sites. The 800-bp mouse pGL-G6Pase promoter was obtained by PCR amplification using primers 5′-GATCCTCGAGCAGAGCCCGTGCAGTGAGTCCAAGC-3′ and 5′-GATCCGGTACCGTCGACGGTATCGATAAGCTTGAT-3′. The resulting fragment was cloned into pGL2 Basic between the KpnI and XhoI sites. The promoter for the human α-fibrinogen gene was obtained by cloning the fragment 5′-GATCTAGGACAAAGCCAATGATTAACCAAACCTCTTGCAGATTTA AATAGGATGGGAACTAGGAGTGGCGGCAATCCTTTCTTTCAGCTGG AGTGCTCA-3′ into pGL2 Basic, between the BglII and HindIII sites. Promoters PRL-1 pP1-Sma and HRS/SRp40 pGL0.1 were constructed as described previously (13, 43, 44). The fragment for pIBP-0.12 M3/HNF-1 was obtained by PCR amplification using primers 5′-ATGCCAAGCTTGGCCGTGTG-3′, 5′-CTGCCCTGACAAGACTTAACCTGTGCC-3′, and 5′-CTCACAAGCAAAACAAACTTA-3′. To make pIBP-1.7 M3/HNF-1, the region between −1718 and −115 of the IGFBP-1 promoter was isolated after digestion of pIBP-3.4 with SacI. The resulting fragment was inserted between the SacI sites of pIBP-0.12 M3/HNF-1. To make pIBP-3.4 M3/HNF-1 and pIBP-6.6 M3/HNF-1, the fragment between −1155 and +15 was first removed using HindIII and SacII. After digestion of pIBP-1.7 M3/HNF-1 with HindIII and SacII, the resulting fragment (− 1155/+15) containing the M3/HNF-1 mutation was cloned into pIBP-3.4 and pIBP-6.6, between the HindIII and SacII sites. All constructs were confirmed by sequencing.
Mouse HNF-1α deletion constructs.
The mouse pBJ5-HNF-1α expression plasmid was kindly provided by Gerald Crabtree. HNF-1α (amino acids [aa] 1 to 628) was generated by digesting pBJ5-HNF-1α with NotI and EcoRV, and the fragment was inserted between the NotI and EcoRV sites of pcDNA3.1/Myc-His(−) A MCS (Invitrogen). HNF-1α (aa 1 to 481) was made by digesting HNF-1α (aa 1 to 628) with PmlI and EcoRV followed by religation using T4 DNA ligase (New England Biolabs). To make HNF-1α (aa 1 to 295), pBJ5-HNF-1α was first digested with XmaI and then filled in using T4 DNA polymerase (New England Biolabs). After digestion of the fragment with HindIII, the HNF-1α (aa 1 to 295) fragment was cloned into pcDNA3.1/Myc-His(-) A MCS between the NotI and blunted HindIII sites. All of the HNF-1α constructs were confirmed by sequencing, and the expression levels were assessed by transfecting the constructs into HepG2 and HeLa cells followed by immunoblotting using anti-HNF-1α (sc-6547 and sc-6548; Santa Cruz) and anti-Myc (generated by the Cell Center Services at the University of Pennsylvania).
Coimmunoprecipitation analyses.
HepG2 hepatoma cells were cultured in DMEM (Gibco/BRL), and HeLa cells were cultured in Iscove's medium. Both media were supplemented with 10% FBS, 2 mM l-glutamine, 100 U of penicillin, and 50 U of streptomycin. Transient transfections using HNF-1, pRcCMV-STAT3-Flag, and pCMV-c-fos expression plasmids were carried out in 100-mm-diameter plates by the calcium phosphate technique. After overnight transfection, the cells were kept in DMEM with 0.2% FBS. The following day, the cells were treated with 100 ng of rhIL-6 per ml for 30 min. The cells were solubilized in TNTE (50 mM Tris [pH 7.4], 150 mM NaCl, 1mM EDTA, 0.5% Triton- X-100, 8% glycerol). Lysates were immunoprecipitated with antibodies to STAT3 (sc-482; Santa Cruz), c-Fos (sc-7202; Santa Cruz), and Flag (Sigma). The precipitates and total cell lysates were subjected to SDS-PAGE followed by immunoblotting with anti-Myc. For coimmunoprecipitation analyses using C57BL/6 mice, the animals were injected subcutaneously with rhIL-6 for 30 min as described previously (8). For the quiescent time point, the mice were sacrificed by cervical dislocation. Total liver lysates were prepared and immunoprecipitated with antibodies to C/EBP (sc-150), c-Jun (sc-1694), STAT3 (sc-482), c-Fos (sc-7202), c-Met (sc-162G), and HNF-1α (sc-6547) (all from Santa Cruz). Whole-cell lysates and precipitates were resolved by SDS-PAGE and assayed by immunoblotting using STAT3 antibody (sc-482; Santa Cruz) and HNF-1α antibody (sc-6547; Santa Cruz).
RESULTS
IL-6-mediated upregulation of hepatic IGFBP-1 mRNA and protein.
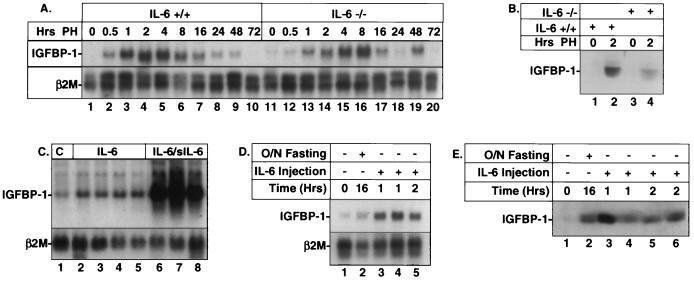
Previous studies showed that tumor necrosis factor alpha, IL-1β, or IL-6 could enhance IGFBP-1 secretion by the HepG2 human hepatoma cell line (49). However, it was not clear whether IL-6 is directly involved in regulating IGFBP-1 mRNA expression. Data from a hepatectomy time course study (Fig. 1A) indicated that the induction of IGFBP-1 mRNA is delayed (peak induction, 7̃0-fold in +/+ and 20-fold in −/− mice) in IL-6−/− livers, peaking at 8 h instead of at 2 h as seen in the IL-6+/+ livers (lanes 4 and 16). A fourfold decrease in the concentration of serum IGFBP-1 was also noted in IL-6−/− livers 2 h posthepatectomy (Fig. 1B). The difference in IGFBP-1 expression between the IL-6+/+ and IL-6−/− livers 8 h posthepatectomy (Fig. 1A, lanes 6 and 16) was due to loading difference as shown by β2-microglobulin.
FIG. 1.
Enhanced expression of IGFBP-1 in IL-6 transgenic mice, in IL-6+/+ livers posthepatectomy, and after IL-6 treatment. (A) Delayed expression of IGFBP-1 mRNA in IL6−/− livers posthepatectomy (PH). RNA was prepared from IL-6−/− and IL-6+/+ livers at the indicated times after hepatectomy. RNA (10 μg) was gel electrophoresed and probed with nick-translated rat IGFBP-1 cDNA probe. β2-Microglobulin (β2M) was used as a normalizing control. This Northern blot is representative of three. (B) Diminished serum IGFBP-1 protein level in IL-6−/− livers posthepatectomy. Five-microliter aliquots of serum were fractionated on an SDS–12% polyacrylamide gel, blotted, and incubated with the IGFBP-1 antibody. (C) Elevated expression of IGFBP-1 mRNA in IL-6 transgenic and IL-6/IL-6 soluble receptor (IL-6/sIL-6) double-transgenic mice. C, control animal. (D) Induction of hepatic IGFBP-1 mRNA after 16-h (overnight [O/N]) fast or 1 to 2 h after IL-6 injection. The animals were injected subcutaneously with rhIL-6 (1 mg/kg) for 1 or 2 h. (E) Induction of serum IGFBP-1 after overnight fasting or 1 to 2 h after IL-6 treatment.
To further verify IL-6's biologic role, we examined the expression of IGFBP-1 mRNA in IL-6-overexpressing transgenic mice (33). As shown in Fig. 1C, the expression of IGFBP-1 mRNA in livers overexpressing IL-6 was elevated compared to normal livers (lanes 2 to 5). This elevation was even more pronounced in the livers of transgenic mice overexpressing both IL-6 and IL-6 soluble receptor (lanes 6 to 8). The direct effect of IL-6 in enhancing the induction of IGFBP-1 mRNA was further demonstrated in experiments in which IL-6 was injected into non fasting mice. As shown in Fig. 1D, the increase in hepatic IGFBP-1 mRNA expression after 1 to 2 h of IL-6 injection was comparable to if not greater than that for the overnight fasted liver. IL-6-induced changes in IGFBP-1 serum protein levels were also increased, though not to as great an extent as mRNA expression (Fig. 1E).
Localization of an IL-6-regulated sequence within the IGFBP-1 promoter to the HNF-1 DNA binding element.
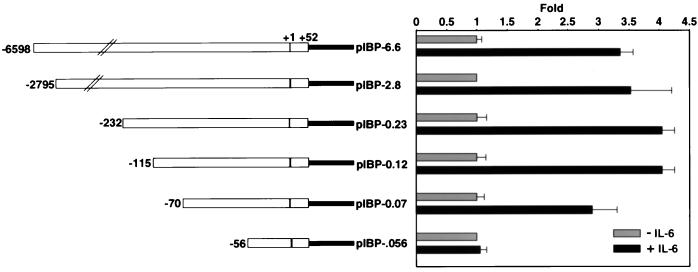
Previous studies identified a number of hypersensitive sites around the IGFBP-1 gene that showed variable hypersensitivity correlating with the proliferative state of the liver (9). We found that the same hypersensitive sites at −5000, −3100, and −100 to −300 that showed increased hypersensitivity posthepatectomy in normal mice showed a relative decrease in hypersensitivity in hepatectomized IL-6−/− livers (data not shown). To begin to identify the potential regulatory regions within the IGFBP-1 promoter that respond to IL-6, sequential 5′-deletion constructs of the mouse IGFBP-1 promoter were made and tested in HepG2 cells. HepG2 cells were used for these studies because STAT3 DNA binding activity is rapidly induced in HepG2 cells treated with IL-6, indicating that the IL-6 signaling pathway is intact in these cells (see Fig. 5B, lanes 3 and 4). As shown in Fig. 2, IL-6 stimulation of HepG2 cells transfected with various constructs led to approximately fourfold induction relative to the nonstimulated IGFBP-1 promoter constructs. The IL-6 response was substantially decreased when the fragment between −70 and −56 was deleted. These results indicated that the region between −70 and −56 contains sequences responsible for the majority of the IL-6 responsiveness in this cell line.
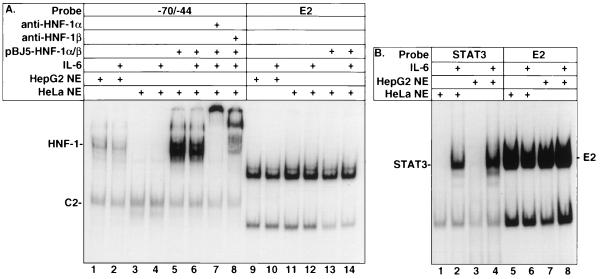
FIG. 5.
HeLa cells respond to IL-6 and do not contain HNF-1. (A) EMSA using HepG2 and HeLa (NE) with and without 20 min of IL-6 treatment (100 ng/ml) as well as HeLa nuclear extract containing overexpressed HNF-1α and -β in the presence and absence of IL-6 treatment. Anti-HNF-1α and anti-HNF-1β were used in the supershift analyses to verify the overexpressed HNF-1α and -β. (B) HeLa cells contain an intact IL-6 pathway. The indicated radiolabeled probes were incubated with HepG2 and HeLa nuclear extracts (10 μg). Reaction products were fractionated using a 5% nondenaturing acrylamide gel. The E2 probe was used as a normalizing control for loading.
FIG. 2.
Functional analyses of the mouse IGFBP-1 promoter in HepG2 cells and deletion mapping of the mouse IGFBP-1 promoter. Left, schematic diagrams of the various mouse IGFBP-1 deletion constructs; right, graphical representation of relative luciferase activity after normalization to β-galactosidase activity. To determine enzyme activity, 0.5 μg of the indicated reporter and 1 μg of pRSV-β-galactosidase were transfected in HepG2 cells by the calcium phosphate precipitation method using the 60-mm-diameter dishes. The cells were treated with rhIL-6 (100 ng/ml) for 4 h. The luciferase activity was expressed as fold induction relative to the basal activity of the reporter construct in the absence of IL-6 treatment. Six independent determinants were made for each construct by performing duplicates in three separate experiments. The values were plotted as averages ± standard deviations.
The region between −64 and −50 in the mouse IGFBP-1 promoter or −74 and −60 in the human promoter contains a highly conserved HNF-1 binding site. Studies have shown that HNF-1 binds to this element and transactivates the human IGFBP-1 promoter in HepG2 cells. To assess the binding of nuclear proteins to the HNF-1 core-like sequence within the mouse IGFBP-1 promoter, EMSAs coupled with supershift analyses were performed using the duplex oligonucleotide containing the HNF-1 core-like sequence extending from −70 to −44. As shown in Fig. 3A, analyses using anti-HNF-1 α and anti-HNF-1β clearly demonstrated the ability of HNF-1 to interact with the HNF-1 binding site. The EMSAs also identified other specific complexes that interacted with this site, C2 and C3. Since IL-6 is an activator of AP-1 and STAT3 expression in regenerating liver (8), additional double-stranded oligonucleotides spanning positions −70 to +52, −70 to −4, −70 to −24, −115 to −86, and −86 to −53 were made and tested by EMSA, competition, and supershift analyses (data not shown). However, no STAT3 and AP-1 DNA binding site were found within or near the HNF-1 binding site (data not shown).
To delineate the recognition sequence for HNF-1, C2, and C3 complexes, further EMSAs using spanning mutant oligonucleotides were performed. As shown in Fig. 3B, M1, M5, and CCGTT mutant duplex oligonucleotides partially blocked HNF-1 binding, whereas M2, M3, and M4 failed to block the binding of HNF-1. Molar excess of unlabeled M1, M4, M5, and CCGTT mutant oligonucleotides, but not M2 and M3, blocked the binding of the C2 complex. The binding of C3 was impeded by M1, M2, M4, and M5 mutant oligonucleotides but not by M3. The binding of C2 was partially competed by an HNF-3 oligonucleotide but not by an HNF-4, AP-1, or C/EBP consensus oligonucleotide.
Cooperative induction of the IGFBP-1 promoter via the HNF-1 site by STAT3/IL-6 and c-Fos/c-Jun overexpression in hepatic cells.
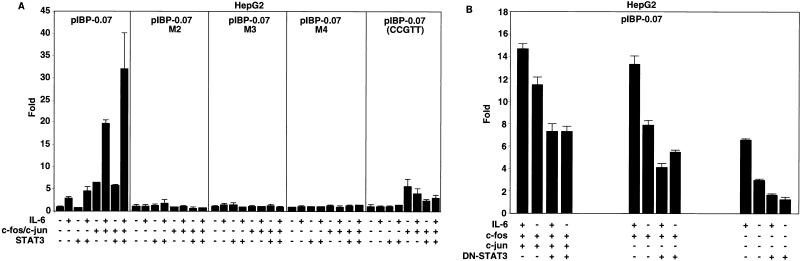
Transfection analyses were performed to ascertain whether definitive loss of HNF-1 binding and loss or impaired binding of other complexes correlated with the loss of IL-6 induction noted in HepG2 cells. IL-6 signals via STAT pathways (56, 57). Both STAT3 and IGFBP-1 are strongly induced in liver regeneration, and STAT3 activation during liver regeneration is strictly modulated by IL-6 (8). Thus, we overexpressed STAT3 in HepG2 cells as a means to determine whether such overexpression coupled with IL-6 stimulation could further enhance the IL-6 responsiveness noted in the −70/+52 WT construct. As shown in Fig. 4A, cotransfecting the STAT3 expression plasmid coupled with IL-6 treatment further enhanced the IL-6 responsiveness. IL-6/STAT3 responsiveness was reduced with the CCGTT construct and abolished with the mutated HNF-1 site constructs (M2, M3, and M4), indicating that IL-6 responsiveness was dependent on an intact HNF-1 element.
FIG. 4.
Regulation of the mouse IGFBP-1 promoter activity by STAT3, c-Fos/c-Jun, and IL-6. (A) An intact HNF-1 binding site coupled with STAT3/c-Fos/c-Jun overexpression in the presence of IL-6 is required for maximal stimulation of the IGFBP-1 promoter in HepG2 cells. (B) Suppression of IL-6-mediated stimulation of the IGFBP-1 promoter via the HNF-1 site by DN-STAT3. HepG2 cells were transfected with the indicated wild-type and mutant pIBP-0.07 constructs (36 ng) in a 24-well plate. For panel A, 36 ng of pCMV-STAT3 with or without 36 ng of pCMV-c-jun and pCMV-c-fos was used. Luciferase activity was expressed as fold induction relative to the basal activity of the reporter construct in the absence of IL-6 treatment and in the absence of the STAT3 or c-Fos/c-Jun expression plasmid. For panel B, 36 ng of pCMV-c-fos with or without 36 ng of pCMV-c-jun or 71 ng of DN-STAT3, as indicated, was used. Luciferase activity was expressed as fold induction relative to the activity of the reporter construct in the presence of DN-STAT3 but in the absence of IL-6 treatment. As indicated, the transfected cells were treated with rhIL-6 (100 ng/ml) for 4 h. Nine independent determinants were made for each construct by performing triplicates in three separate experiments. The values were plotted as averages ± standard deviations.
Because IL-6 is an activator of AP-1 expression in regenerating liver (8) and AP-1 interacts with STAT3 to enhance transcription (64), we sought to determine whether overexpression of c-Jun/c-Fos coupled with IL-6 stimulation could further enhance the transcriptional activity of the −70/+52 WT construct. As shown (Fig. 4A), overexpression of c-Jun/c-Fos enhanced the transcriptional activity of the −70/+52 WT construct approximately five fold, and an additional fourfold induction was observed after IL-6 treatment. Approximately 33-fold induction of the IGFBP-1 promoter was achieved by overexpressing c-Fos/c-Jun and STAT3 in combination with IL-6 stimulation, implying synergistic enhancement. Most of this induction was eliminated by mutations within the HNF-1 site, further suggesting that cooperative interactions between STAT3, c-Fos, and c-Jun at the HNF-1 site could be occurring.
The STAT3 mutant Y705F (dominant negative [DN-STAT3]) has previously been shown to act in a dominant negative manner to inhibit endogenous STAT3 phosphorylation and activation by IL-6 in HepG2 cells by competing for the binding to the phosphotyrosine(s) on gp130 (23). We overexpressed DN-STAT3 in HepG2 cells to determine if the STAT3 pathway is required for IL-6-mediated upregulation of IGFBP-1 expression. Overexpression of DN-STAT3 inhibited IGFBP-1 promoter activation by IL-6 (Fig. 4B) and in fact reduced the basal activity. A reduction but not elimination of c-Jun/c-Fos stimulation by DN-STAT3 was noted. In the majority of cell types, the levels of expression of fos, jun, and related genes are relatively low (10, 39). In HepG2 cells transfected with DN-STAT3, a low amount of phosphorylated STAT3 could still be detected (23). Thus, the lack of complete repression of the IGFBP-1 promoter by DN-STAT3 in the presence of overexpressed c-Jun/c-Fos could be attributed to the presence of residual amount of phosphorylated STAT3 which could still interact with c-Jun/c-Fos. On the other hand, the specific mitogen-activated protein kinase inhibitor PD98059 failed to block IL-6-mediated stimulation (data not shown). These results strongly suggested that the major mechanism by which IL-6 stimulates the region −70/+52 of the IGFBP-1 promoter is via STAT3.
Requirement of HNF-1 in the induction of IGFBP-1 promoter activity by IL-6/STAT3 and c-Jun/c-Fos in nonhepatic cells.
Studies have shown that HepG2 cells express IGFBP-1 (28) and HNF-1 (12, 15, 53), while HeLa cells do not (46). As shown in Fig. 5A, HeLa cells do not express HNF-1 (lanes 3 and 4). However, overexpression of HNF-1α/β expression plasmids resulted in HNF-1 DNA binding activity (lanes 5 and 6), as confirmed by supershift analyses (lanes 7 and 8). HeLa cells also contain complexes migrating at a position similar to C2. Like HepG2 cells, HeLa cells contain an intact IL-6 pathway, as demonstrated by induction of STAT3 DNA binding after IL-6 treatment (Fig. 5B, lanes 1 and 2).
We carried out transfection experiments using HeLa cells to distinguish the effects of HNF-1, c-Fos/c-Jun, and STAT3/IL-6 in transactivating the IGFBP-1 promoter. AP-1, STAT3, or IL-6 alone or in combination, in the absence of HNF-1, had a minimal effect on IGFBP-1 promoter activity (Table 1). Conversely, overexpression of HNF-1α/β, STAT3/HNF-1α/β, and c-Fos/c-Jun/HNF-1α/β coupled with IL-6 stimulation increased promoter activity approximately 4.1-, 6.5-, and 8.2-fold, respectively (Table 1). However, maximal upregulation of the IGFBP-1 promoter was achieved by overexpressing STAT3/IL-6, c-Fos/c-Jun, and HNF-1α. These results suggested that STAT3, c-Fos/c-Jun, HNF-1α, and IL-6 could act in synergy to transactivate the IGFBP-1 promoter. The data also showed that in the presence of overexpressed HNF-1α, HNF-1β is not needed to transactivate the IGFBP-1 promoter.
TABLE 1.
Maximal activation of the IGFBP-1 promoter in nonhepatic cells after HNF-1 and IL-6/STAT3/AP-1 cotransfectiona
| Cotransfection
|
Luciferase activity
|
||||
|---|---|---|---|---|---|
| IL-6/STAT3 | c-Fos/c-Jun | HNF-1α | HNF-1β | Fold (avg) | SD |
| − | − | − | − | 1.0 | 0.16 |
| + | − | − | − | 1.1 | 0.08 |
| − | + | − | − | 2.5 | 0.03 |
| + | + | − | − | 3.3 | 0.41 |
| − | − | + | + | 4.1 | 0.56 |
| + | − | + | + | 6.5 | 0.57 |
| − | + | + | + | 8.2 | 0.69 |
| + | + | + | + | 23.5 | 2.35 |
| + | + | + | − | 26.0 | 1.86 |
HeLa cells were transfected with the pIBP-0.07 construct (29 ng) in 24-well plates. For cotransfection experiments, 29 ng of pCMV-STAT3, 29 ng of pCMV-c-jun, 29 ng of pCMV-c-fos, 29 ng of pBJ5-HNF-1α, or 29 ng of pBJ5-HNF-1β was (+) or was not (−) used. The transfected cells were treated with rhIL-6 (100 ng/ml) for 4 h. Luciferase activity was expressed as fold induction relative to the basal activity of the reporter construct in the absence of IL-6 treatment and in the absence of any indicated expression plasmids. Nine independent determinants were made for each construct by performing triplicates in three separate experiments.
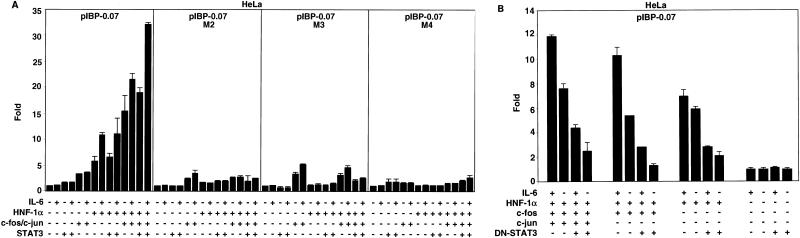
Since the synergistic effect was detected only by overexpressing HNF-1α in HeLa cells, we wanted to further discern whether the IL-6/STAT3/AP-1-mediated transcriptional induction noted in HepG2 cells was dependent on the presence of both HNF-1 protein and an intact HNF-1 site. Overexpression of HNF-1α, c-Fos/c-Jun, and STAT3 followed by IL-6 treatment was sufficient to upregulate the IGFBP-1 promoter approximately 33-fold, and mutations within the HNF-1 binding site (M2, M3, and M4) blocked the majority of the stimulation (Fig. 6A). These observations confirmed the requirement of HNF-1α for the effect mediated by STAT3 and c-Jun/c-Fos and showed that the effect is dependent on both an intact HNF-1 site and HNF-1α protein.
FIG. 6.
Activation of the IGFBP-1 promoter in HeLa cells with HNF-1α and IL-6/STAT3/AP-1 cotransfection. (A) An intact HNF-1 binding site coupled with HNF-1α/STAT3/c-Fos/c-Jun overexpression in the presence of IL-6 is required for maximal stimulation of the IGFBP-1 promoter in HeLa cells. (B) The ability of the exogenous HNF-1 with or without c-Fos or c-Fos/c-Jun to activate the IGFBP-1 promoter in HeLa cells is blocked by DN-STAT3. HeLa cells were transfected with the indicated pIBP-0.07 constructs (29 ng) in 24-well plates. For cotransfection experiments, 29 ng of pCMV-STAT3, 29 ng of pCMV-c-jun, 29 ng of pCMV-c-fos, 29 ng of pcDNA3.1-HNF-1α (aa 1 to 481), or 71 ng of DN-STAT3, as indicated, was used. The transfected cells were treated with rhIL-6 (100 ng/ml) for 4 h. Fold induction relative to the basal activity of the reporter construct in the absence of IL-6 treatment and in the absence of any indicated expression plasmids; fold induction relative to the activity of the reporter construct in the presence of DN-STAT3 but in the absence of IL-6 treatment. Nine independent determinants were made for each construct by performing triplicates in three separate experiments. The values were plotted as averages ± standard deviations.
DN-STAT3 was overexpressed in HeLa cells to determine if the STAT3-cytokine-mediated pathway was required to augment the expression of IGFBP-1 promoter as in HepG2 cells. As shown in Fig. 6B, the ability of exogenous HNF-1 to activate the IGFBP-1 promoter in HeLa cells was blocked by DN-STAT3. Unlike the case for HepG2 cells, transactivation of the IGFBP-1 promoter by HNF-1α/AP-1 and HNF-1α/c-Fos both in the presence and in the absence of IL-6 treatment was dramatically reduced by DN-STAT3 (Fig. 6B).
Transcriptional upregulation of the full-length IGFBP-1 promoter by IL-6/STAT3/AP-1 depends on both an intact HNF-1 site and HNF-1α protein.
A number of highly conserved functional DNA binding sites, such as the glucocorticoid response element, cyclic AMP response element, and USF1 element (9, 27), have been identified in the first 300 bp of the IGFBP-1 promoter. Additional functional tissue-specific sites that interacted with C/EBP, HNF3, and AP-1 transcription factors have also been identified in the −3100 region (9). Although we showed that the integrity of the HNF-1 cis element and the HNF-1α transcription factor play an important role in directing the basal IGFBP-1 promoter activity as well as in mediating the IL-6/STAT3/AP-1 response in the context of pIBP-0.07, we wanted to assess whether the same effect would be seen using the full-length IGFBP-1 construct (pIBP-6.6).
To verify whether the HNF-1 DNA binding site is important for IL-6 and AP-1 activation of the IGFBP-1 promoter in the context of the whole promoter, HepG2 cells were transfected with various wild-type and mutant constructs along with IL-6 or AP-1 (Table 2). Mutations within the HNF-1 binding site blocked the IL-6 and AP-1 (c-Fos/c-Jun) enhancement, irrespective of the length of the reporter constructs. In HeLa cells, transcriptional upregulation of pIBP-0.07, pIBP-0.12, and pIBP-3.4, and pIBP-6.6 was dependent on an intact HNF-1 cis element and overexpressed HNF-1α (data not shown), as mutations within the HNF-1 binding site blocked the transcriptional enhancement noted in the presence of exogenous HNF-1α. These finding reiterated the significance of the HNF-1 element and HNF-1α transcription factor, and they identified the HNF-1 binding site as the main mediating element for the IL-6/STAT3/AP-1 response.
TABLE 2.
Maximal activation of the full-length IGFBP-1 promoter by IL-6/STAT3/AP-1 depends on HNF-1α and an intact HNF-1 binding site in HepG2 cellsa
| Promoter | Cotransfection
|
Luciferase activity
|
||
|---|---|---|---|---|
| c-Fos/c-Jun | IL-6 | Fold (avg) | SD | |
| pIBP-0.07 | − | − | 1.0 | 0.11 |
| − | + | 2.7 | 0.08 | |
| + | − | 5.1 | 0.67 | |
| pIBP-0.07 M3/HNF-1 | − | − | 0.8 | 0.08 |
| − | + | 1.0 | 0.10 | |
| + | − | 0.5 | 0.14 | |
| pIBP-0.12 | − | − | 1.0 | 0.05 |
| − | + | 4.1 | 0.30 | |
| + | − | 9.2 | 0.62 | |
| pIBP-0.12 M3/HNF-1 | − | − | 0.8 | 0.07 |
| − | + | 1.3 | 0.04 | |
| + | − | 1.0 | 0.24 | |
| pIBP-3.4 | − | − | 1.0 | 0.12 |
| − | + | 4.5 | 1.38 | |
| + | − | 4.8 | 0.25 | |
| pIBP-3.4 M3/HNF-1 | − | − | 0.5 | 0.11 |
| − | + | 0.8 | 0.27 | |
| + | − | 0.9 | 0.35 | |
| pIBP-6.6 | − | − | 1.0 | 0.10 |
| − | + | 5.3 | 0.62 | |
| + | − | 9.3 | 1.56 | |
| pIBP-6.6 M3/HNF-1 | − | − | 0.5 | 0.06 |
| − | + | 0.5 | 0.06 | |
| + | − | 0.9 | 0.14 | |
HepG2 cells were transfected with the indicated reporter constructs (25 ng) in 24-well plates. For cotransfection experiments, 25 ng of pCMV-c-fos and 25 ng of pCMV-c-jun, as indicated, were (c-Fos/c-Jun) (+) or were not (−) used. The cells were kept in medium containing 0.2% FBS for 18 h and treated (+) or not treated with rhIL-6 (100 ng/ml) for 4h. Luciferase activity was expressed as fold induction relative to the basal activity of the wild-type reporter construct in the absence of IL-6 treatment and in the absence of any indicated expression plasmids. Nine independent determinants were made for each construct by performing triplicates in three separate experiments.
Detection of protein-protein interactions between STAT3/HNF-1 and c-Fos/HNF-1 in liver and transfected hepatic and nonhepatic cells.
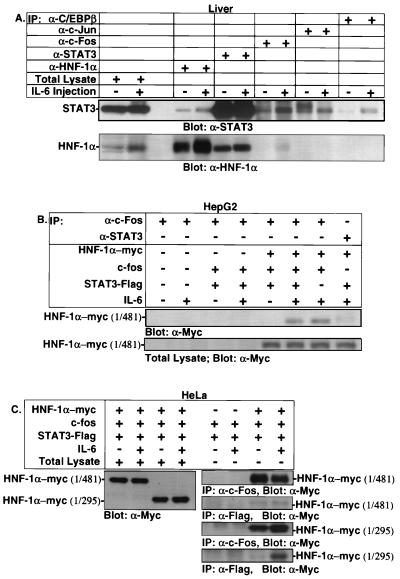
Based on the transfection analyses using both HepG2 and HeLa cells, it is conceivable that HNF-1α, c-Fos/c-Jun, and STAT3 physically interact to upregulate the expression of IGFBP-1. We used coimmunoprecipitation assays of whole-cell extracts to assess possible in vivo interactions of STAT3 and c-Fos/c-Jun with HNF-1 in wild-type livers. Mice were injected with rhIL-6 for 30 min, and whole-cell lysates were immunoprecipitated with appropriate antibodies. As shown in Fig. 7A (top), STAT3 interacted with HNF-1α, c-Fos, c-Jun, and C/EBPβ in the presence and absence of IL-6. Enhanced interactions between STAT3 and c-Fos and between STAT3 and C/EBPβ were noted after IL-6 injection. No interaction between STAT3 and a negative control, c-Met, was detected (data not shown). Immunoblotting using the HNF-1α antibody also revealed interactions between HNF-1α and STAT3 and between HNF-1α and c-Fos (Fig. 7A, bottom). In this experiment, the expression of HNF-1 protein in the total lysate appears higher after IL-6 injection. However, multiple Western and EMSA analyses using different liver whole-cell and nuclear extracts showed that the expression of HNF-1 is not enhanced after IL-6 injection (data not shown).
FIG. 7.
Presence of STAT3/HNF-1α/c-Fos complex in livers, HeLa cells, and HepG2 cells. (A) Coimmunoprecipitation analyses using C57BL/6 mice. For the quiescent time point, the mice were sacrificed by cervical dislocation. For IL-6-treated mice, the animals were injected subcutaneously with rhIL-6 for 30 min. Whole-cell lysates were immunoprecipitated (IP) with the indicated antibodies and analyzed by immunoblotting using STAT3 antibody (top) and HNF-1α antibody (bottom). HepG2 and HeLa cells were transiently transfected with 2 μg of the indicated HNF-1α, 2 μg of pRcCMV-STAT3-Flag, and 2 μg of pCMV-c-fos expression plasmid in 100-mm-diameter plates using the calcium phosphate technique. The cells were kept in 0.2% FBS for 18 h and then incubated with rhIL-6 (100 ng/ml) for 30 min. Whole-cell lysates were immunoprecipitated with the indicated antibodies and analyzed by immunoblotting using the Myc antibody.
We performed coimmunoprecipitation assays using Myc-tagged HNF-1α and Flag-tagged STAT3 in HepG2 and HeLa whole-cell extracts to further verify the interaction between STAT3, HNF-1α, and c-Fos. As shown in Fig. 7B, HNF-1α formed a much more abundant protein complex with c-Fos when HepG2 cells were cotransfected with c-Fos and treated with IL-6 for 30 min. The c-Fos–HNF-1α interactions were independent of IL-6 stimulation in HeLa whole-cell extracts (Fig. 7C). The immunoprecipitates of Flag antibody (STAT3) contained more HNF-1α (aa 1 to 295) than HNF-1α (aa 1 to 481) when HeLa cells were treated with IL-6 (Fig. 7C), albeit the expression levels of HNF-1α (aa 1 to 295) and HNF-1α (aa 1 to 481) were comparable. An enhanced interaction between HNF-1α (aa 1 to 295) and STAT3-Flag was observed after IL-6 stimulation, even though similar expression of HNF-1α (aa 1 to 295) was detected in total lysates prepared from both untreated and IL-6-treated whole-cell HeLa extracts. In similar experiments using both HepG2 and HeLa cells and the full-length HNF-1α (aa 1 to 628) construct, however, protein expression was very low compared to levels for HNF-1α constructs encompassing aa 1 to 295 and aa 1 to 481 (data not shown).
Transcriptional upregulation of human α-fibrinogen and mouse G6Pase promoters by IL-6/HNF-1α/STAT3/c-Fos.
To investigate whether transcriptional enhancement of liver-specific promoters by IL-6/HNF-1α/STAT3/c-Fos is a general phenomenon and not limited to IGFBP-1, we performed transfection analyses using both G6Pase and α-fibrinogen promoters. The gene for G6Pase, the key enzyme in glucose homeostasis, is expressed in a tissue-specific manner in the liver and kidney (42) and is induced during liver regeneration following partial hepatectomy (14). Even though HNF-1 has been shown to transactivate the G6Pase promoter and mediate its glucocorticoid and cyclic AMP responses (30, 31, 52), it is not known whether IL-6/STAT3 is involved in regulating its expression. Fibrinogen, a hepatically derived class II acute-phase protein, is the product of three separate genes (Aα, Bβ, and γ), which are transcriptionally upregulated by IL-6 (32, 36) and glucocorticoids. Studies have shown that expression of the human and rat α and β chains (6, 11, 17, 18, 19) is mediated by HNF-1. Unlike the fibrinogen γ promoter, which contains known STAT3 binding sites (65), STAT3 does not bind to the identified IL-6-responsive element in the α-fibrinogen promoter (32). Like the case for the IGFBP-1 promoter, transcriptional upregulation of the α-fibrinogen promoter is dependent on an intact HNF-1 binding site, as mutations within the HNF-1 cis element blocked the transcriptional enhancement supported by sequences upstream of this region (17).
As for the IGFBP-1 promoter, similar synergistic transcriptional enhancements of the G6Pase and α-fibrinogen promoters were observed in the presence of HNF-1α coupled with IL-6/STAT3 and c-Fos overexpression in HeLa cells (Table 3). To show that HNF-1α is not a general transcription activator, promoters like PRL-1 pP1-Sma (a nuclear protein tyrosine phosphatase) (44, 57) and AP-1, both of which lack any known HNF-1 binding sites, were included in the analyses. As shown (Table 3), overexpression of the exogenous HNF-1α failed to enhance the expression of both PRL-1 pP1-Sma and AP-1.
TABLE 3.
Activation of α-fibrinogen and G6Pase promoters in HeLa cells with HNF-1α and IL-6/STAT3/c-Fos cotransfectiona
| Promoter | Cotransfection
|
Luciferase activity
|
||||
|---|---|---|---|---|---|---|
| HNF-1α | DN-STAT3 | STAT3/IL-6 | c-Fos | Fold (avg) | SD | |
| G6Pase | − | − | − | − | 1.0 | 0.3 |
| − | + | − | − | 1.0 | 0.3 | |
| − | − | − | + | 1.6 | 0.1 | |
| − | − | + | − | 1.8 | 0.0 | |
| − | − | + | + | 3.1 | 0.3 | |
| + | − | − | − | 4.7 | 0.8 | |
| + | + | − | − | 2.1 | 0.3 | |
| + | − | − | + | 8.1 | 2.1 | |
| + | + | − | + | 2.4 | 0.4 | |
| + | − | + | − | 10.2 | 0.4 | |
| + | − | + | + | 11.3 | 1.3 | |
| αFibrinogen | − | − | − | − | 1.0 | 0.2 |
| − | + | − | − | 1.0 | 0.0 | |
| − | − | − | + | 1.5 | 0.0 | |
| − | − | + | − | 1.0 | 0.4 | |
| − | − | + | + | 1.5 | 0.4 | |
| + | − | − | − | 14.1 | 0.2 | |
| + | + | − | − | 3.1 | 0.6 | |
| + | − | − | + | 19.4 | 0.1 | |
| + | + | − | + | 2.4 | 0.4 | |
| + | − | + | − | 30.6 | 1.1 | |
| + | − | + | + | 69.5 | 5.5 | |
| AP-1 | − | − | 1.0 | 0.1 | ||
| − | + | 0.2 | 0.0 | |||
| + | − | 1.1 | 0.0 | |||
| + | + | 0.4 | 0.0 | |||
| PRL-1 pP1-Sma | − | − | 1.0 | 0.1 | ||
| − | + | 1.4 | 0.2 | |||
| + | − | 1.1 | 0.2 | |||
| + | + | 0.7 | 0.1 | |||
| HRS/SRp40 pGL0.1 | − | − | 1.0 | 0.2 | ||
| − | + | 0.5 | 0.1 | |||
| + | − | 2.0 | 0.4 | |||
| + | + | 0.3 | 0.0 | |||
| SMAD7 | − | − | 1.0 | 0.1 | ||
| − | + | 1.1 | 0.2 | |||
| + | − | 1.6 | 0.4 | |||
| + | + | 1.5 | 0.4 | |||
HeLa cells were transfected with the indicated reporter constructs (50 ng) in 24-well plates. For cotransfection experiments, 50 ng of pCMV-STAT3, 50 ng of pCMV-c-fos, 100 ng of DN-STAT3, or 50 ng of pcDNA-HNF-1α (aa 1 to 628, full length) was (+) or was not (−) used. The transfected cells were treated with IL-6 (100 ng/ml) for 4 h. Luciferase activity was expressed as fold induction relative to the basal activity of the reporter construct in the absence of IL-6 treatment and in the absence of any indicated expression plasmids. Nine independent determinants were made for each construct by performing triplicates in three separate experiments.
To determine if the STAT3-cytokine-mediated pathway was required to augment the expression of G6Pase and α-fibrinogen promoters, DN-STAT3 was overexpressed in HeLa cells. As shown (Table 3), the ability of the exogenous HNF-1α to activate the G6Pase and α-fibrinogen promoters was blocked by DN-STAT3. A similar repressive effect of DN-STAT3 was detected in promoters previously known to be transcriptionally upregulated by IL-6, such as the α-fibrinogen (32), AP-1, and HRS/SRp40 pGL0.1 (13) promoters. To further illustrate that DN-STAT3 is not a general transcription repressor, both PRL-1 pP1-Sma, which is transcriptionally upregulated by Egr-1 and is independent of IL-6 regulation (44, 57), and SMAD7, which is transcriptionally enhanced by transforming growth factor β, activin, and gamma interferon (41, 62), were included in the analyses. As shown (Table 3), the expression of PRL-1 pP1-Sma and SMAD7 promoters was unaffected by DN-STAT3.
DISCUSSION
HNF-1 binding sites are found in more than 100 different liver-specific genes (61). It has been shown that a highly efficient, liver-specific promoter can be obtained with only a TATA box and an HNF-1 site (34, 48, 59), as direct physical interaction between HNF-1 and TFIIB has been demonstrated and been implicated to play an important role during the formation of the preinitiation complexes (25). Liver-specific genes with known HNF-1 binding sites include hepatic metabolic genes like those encoding G6Pase and PEPCK and hepatic acute-phase response genes like those coding fibrinogen, α1-antitrypsin, and C-reactive protein (3, 5, 6, 7, 12, 47, 58, 61). α1-Antitrypsin, fibrinogen, and C-reactive protein are transcriptionally upregulated by IL-6 (29, 40, 63, 65) during the acute-phase inflammatory response to help restore homeostasis and restrict proteolytic and/or fibrogenic activity and tissue damage (40). Previously it was felt that this upregulation was mediated largely via STAT3 DNA binding elements, but our studies suggest the possible involvement of STAT3–HNF-1 protein-protein interactions as well.
In HepG2 cells, we showed that STAT3 and AP-1 (c-Fos/c-Jun) factors were needed to promote the maximal expression of the IGFBP-1 promoter via the HNF-1 site in the presence of IL-6. In nonhepatic cells, both an intact HNF-1 binding site and HNF-1 protein were required for IL-6/STAT3/AP-1-mediated transcriptional upregulation, as overexpression of STAT3 and/or c-Fos/c-Jun in the absence of HNF-1 had minimal effect in transactivating the IGFBP-1 promoter. We also identified the IL-6/STAT3 pathway as the main mediator pathway for IL-6-dependent activation of the IGFBP-1 promoter. Overexpression of DN-STAT3 blocked the IL-6 transcriptional enhancement found in HepG2 cells. The significance of the IL-6/STAT3 pathway in IGFBP-1 transcriptional upregulation by IL-6 was again demonstrated in HeLa cells, where overexpression of DN-STAT3 impeded the transactivation observed after overexpressing HNF-1α/AP-1 and HNF-1α/c-Fos. This was a general effect on hepatic gene transcription, as IL-6/STAT3/c-Fos/HNF-1α could also synergistically transactivate promoters like the G6Pase and α-fibrinogen promoters.
In mouse livers, HNF-1α/STAT3 complexes were more readily detected than HNF-1α/c-Fos complexes. Possible explanations for the relative weakness of the HNF-1α–c-Fos interaction in vivo could be explained by the fact that there is a relatively small amount of c-Fos protein at this time point after IL-6 injection (8). To further discern the interaction, we carried out coimmunoprecipitation experiments in both HepG2 and HeLa cells using Myc-tagged HNF-1α and Flag-tagged STAT3 constructs. In HepG2 and HeLa cells, unlike liver cells, we observed a stronger association between HNF-1α and c-Fos than HNF-1α/STAT3 after overexpressing STAT3, c-Fos, and HNF-1α. An enhanced interaction between HNF-1α(1–295) and STAT3 was seen after IL-6 stimulation. The IL-6-dependent association between HNF-1α and STAT3 or HNF-1α and c-Fos might have been difficult to detect using whole-cell extracts as in our assays. A major aspect of IL-6-mediated regulation involves the cytoplasmic-nuclear transport of STAT3 after IL-6 stimulation, which would allow de novo nuclear STAT3 to associate with preexisting nuclear proteins like HNF-1α, c-Fos, and others. The observation that some complexes (i.e., STAT3/HNF-1) showed an IL-6-dependent association suggests the possibility of enhanced interactions in the presence of phosphorylated or dimerized STAT3, posttranslational modifications in STAT3 which occur in response to IL-6.
It is known that transcriptional activation of mammalian genes is orchestrated by a complex array of transcription factors and may involve transcriptional coactivator molecules such as p300 and CREB binding protein (CBP) (20, 21, 22). CBP/p300 can interact with a variety of transcription factors and components of the basal transcription machinery such as c-Fos, c-Jun, STAT3, CREB, and Src-1 (20, 21). Even though we observed a twofold increase in IGFBP-1 promoter activity after overexpressing p300 coupled with IL-6 treatment, and an additional twofold induction after overexpressing both STAT3 and p300 in the presence of IL-6 in HepG2 cells (data not shown), the degree of enhancement was not as significant as the IL-6/STAT3/AP-1 effect. In HeLa cells, even in the presence of overexpressed HNF-1α, overexpression of p300 did not further enhance IGFBP-1 promoter activity (data not shown). Soutoglou et al. (51) recently showed that HNF-1α can physically interact with CBP, p300/CBP-associated factor (P/CAF), Src-1, and RAC3, and that these coactivators increase HNF-1-dependent transcription in a synergistic manner. Although we could not detect similar synergistic CBP/P/CAF/HNF-1α enhancement in HeLa cells (data not shown), we cannot rule out a role for coactivators in this response. Nonetheless, we have shown a clear amplification of hepatic promoter activities, G6Pase, IGFBP-1, and α-fibrinogen, by growth-induced factors STAT3 and AP-1 and tissue-specific factor HNF-1α.
Studies have shown that simultaneous expression of multiple transcription factors provides numerous opportunities for the complex regulation seen during liver regeneration and other regulated physiologic processes. However, here we have described a novel mechanism whereby a tissue-specific transcription factor and DNA binding element present in the promoter of many liver-specific genes may interact with induced transcription factors in response to specific external signals. In this report, we have proposed a mechanism by which HNF-1α coordinates the interaction of STAT3/IL-6 and c-Fos, leading to synergistic transcriptional upregulation of promoters like the IGFBP-1, G6Pase, and α-fibrinogen promoters. Since coordinated expression and regulation of hepatic growth and differentiation transcription factors enable the liver to maintain metabolic homeostasis during times of growth and repair, it is conceivable that our finding may represent a general transcriptional adaptive mechanism that the liver and other organs potentially employ postinjury or after stress.
ACKNOWLEDGMENTS
We thank Gerald R. Crabtree for the pBJ5-HNF-1α and pBJ5-HNF-1β constructs, Curt M. Horvath for DN-STAT3, and Yan Chen for the SMAD7 promoter (−408/+112).
DNA sequencing was performed by the University of Pennsylvania sequencing facility, supported in part by the University of Pennsylvania Center for the Molecular Study of Digestive Diseases (P30 DK50306). This work was supported in part by Digestive and Liver Center grant P30 DK50306 (technical support) and grants DK49210 and DK49629 (R.T.).
REFERENCES
- 1.Babajko S, Groyer A. Interplay of liver-enriched trans-acting factors, DBP and HNF1 in the transactivation of human IGFBP-1 promoter. Biochem Biophys Res Commun. 1993;196:480–486. doi: 10.1006/bbrc.1993.2275. [DOI] [PubMed] [Google Scholar]
- 2.Babajko S, Tronche F, Groyer A. Liver-specific expression of human insulin-like growth factor binding protein-1: functional role of transcription factor HNF-1 in vivo. Proc Natl Acad Sci USA. 1993;90:272–276. doi: 10.1073/pnas.90.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumhueter S, Courtois G, Crabtree G R. A variant nuclear protein in dedifferentiated hepatoma cells binds to the same functional sequences in the β fibrinogen gene promoter as HNF-1. EMBO J. 1988;7:2485–2493. doi: 10.1002/j.1460-2075.1988.tb03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 1996;10:267–282. [PubMed] [Google Scholar]
- 5.Costa R H, Grayson D R, Xanthopoulos K G, Darnell J E., Jr A liver specific DNA binding protein recognizes multiple nucleotide sites in regulatory regions of transthyretin, α1-antitrypsin, albumin, and simian virus 40 genes. Proc Natl Acad Sci USA. 1988;85:3840–3844. doi: 10.1073/pnas.85.11.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courtois G, Baumhueter S, Crabtree G R. Purified hepatocyte nuclear factor 1 interacts with a family of hepatocyte specific promoters. Proc Natl Acad Sci USA. 1988;85:7937–7941. doi: 10.1073/pnas.85.21.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courtois G, Morgan J G, Campbell L A, Fourel G, Crabtree G R. Interaction of a liver specific nuclear factor with the fibrinogen and α-1 antitrypsin promoters. Science. 1987;238:688–692. doi: 10.1126/science.3499668. [DOI] [PubMed] [Google Scholar]
- 8.Cressman D E, Greenbaum L E, DeAngelis R A, Ciliberto G, Furth E E, Poli V, Taub R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;22:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 9.Crissey M S, Leu J I, DeAngelis R A, Greenbaum L E, Scearce L M, Kovalovich K, Taub R. Liver specific and proliferation-induced deoxyribonuclease I hypersensitive sites in the mouse insulin-like growth factor binding protein-1 gene. Hepatology. 1999;30:1187–1197. doi: 10.1002/hep.510300520. [DOI] [PubMed] [Google Scholar]
- 10.Curran T. The fos oncogene. In: Reddy E P, Skalka A M, Curran T, editors. The oncogene handbook. Amsterdam, The Netherlands: Elsevier; 1988. pp. 307–325. [Google Scholar]
- 11.Dalmon J, Laurent M, Courtois G. The human beta fibrinogen promoter contains a hepatocyte nuclear factor 1-dependent interleukin-6-responsive element. Mol Cell Biol. 1993;13:1183–1193. doi: 10.1128/mcb.13.2.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Simone V, Ciliberto G, Hardon E, Paonessa G, Palla F, Lundberg L, Cortese R. Cis- and trans-acting elements responsible for the cell-specific expression of the human α1-antitrypsin gene. EMBO J. 1987;6:2759–2766. doi: 10.1002/j.1460-2075.1987.tb02570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du K, Leu J I, Peng Y, Taub R. Transcriptional up-regulation of the delayed early gene HRS/SRp40 during liver regeneration. J Biol Chem. 1998;273:35208–35215. doi: 10.1074/jbc.273.52.35208. [DOI] [PubMed] [Google Scholar]
- 14.Haber B A, Chin S, Chuang E, Buikhuisen W, Naji A, Taub R. High levels of glucose-6-phosphatase gene and protein expression in proliferating liver and diabetes. J Clin Investrs. 1995;95:832–841. doi: 10.1172/JCI117733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardon E, Frain M, Paonessa G, Corteso R. Two distinct factors interact with the promoter regions of several liver specific genes. EMBO J. 1988;7:1711–1719. doi: 10.1002/j.1460-2075.1988.tb03000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins G M, Anderson R M. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:656–665. [Google Scholar]
- 17.Hu C-H, Harris J E, Davie E W, Chung D W. Characterization of the 5′-flanking region of the gene for the α chain of human fibrinogen. J Biol Chem. 1995;270:28342–28349. doi: 10.1074/jbc.270.47.28342. [DOI] [PubMed] [Google Scholar]
- 18.Huber P, Dalmon J, Courtois G, Laurent M, Assouline Z, Marguerie G. Characterization of the 5′-flanking region for the human fibrinogen beta gene. Nucleic Acids Res. 1987;15:1615–1625. doi: 10.1093/nar/15.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber P, Laurent M, Dalmon J. Human beta-fibrinogen gene expression. Upstream sequences involved in its tissue specific expression and its dexamethasone and interleukin 6 stimulation. J Biol Chem. 1990;265:5695–5701. [PubMed] [Google Scholar]
- 20.Janknecht R, Hunter T. A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 21.Janknecht R, Hunter T. Transcriptional control: versatile molecular glue. Curr Biol. 1996;6:951–954. doi: 10.1016/s0960-9822(02)00636-x. [DOI] [PubMed] [Google Scholar]
- 22.Janknecht R, Hunter T. Nuclear fusion of signaling pathways. Science. 1999;284:443–444. doi: 10.1126/science.284.5413.443. [DOI] [PubMed] [Google Scholar]
- 23.Kaptein A, Paillard V, Saunders M. Dominant negative STAT3 mutant inhibits interleukin-6 induced Jak-STAT signal transduction. J Biol Chem. 1996;271:5961–5964. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- 24.Kovalovich K, DeAngelis R A, Li W, Furth E, Ciliberto G, Taub R. Increased toxin-induced liver injury and fibrosis in interleukin-6 deficient mice. Hepatology. 2000;31:149–159. doi: 10.1002/hep.510310123. [DOI] [PubMed] [Google Scholar]
- 25.Ktistaki E, Talianidis, I. I. Modulation of hepatic gene expression by hepatic nuclear factor 1. Science. 1997;277:109–112. doi: 10.1126/science.277.5322.109. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Greenbaum L, Haber B A, Nagle D, Lee V, Miles V, Mohn K L, Bucan M, Taub R. Structure and localization of the IGFBP-1 gene and its expression during liver regeneration. Hepatology. 1994;19:656–665. doi: 10.1002/hep.1840190317. [DOI] [PubMed] [Google Scholar]
- 27.Lee PDK, Giudice L C, Conover C A, Powell D R. Insulin-like growth factor binding protein-1: recent findings and new directions. Proc Soc Exp Bio Med. 1997;216:319–357. doi: 10.3181/00379727-216-44182. [DOI] [PubMed] [Google Scholar]
- 28.Lee, Hintz Y R, James P, Lee P, Shively J, Powell D R. Insulin-like growth factor (IGF) binding protein complementary deoxyribonucleic acid from human HEP G2 hepatoma cells: fredicted sequence suggests an IGF binding domain different from those of the IGF-I and IGF-II receptors. Mol Endocrinol. 1988;2:404–411. doi: 10.1210/mend-2-5-404. [DOI] [PubMed] [Google Scholar]
- 29.Li S P, Goldman N D. Regulation of human C-reactive protein gene expression by two synergistic IL-6 responsive elements. Biochemistry. 1996;35:9060–9068. doi: 10.1021/bi953033d. [DOI] [PubMed] [Google Scholar]
- 30.Lin B, Morris D W, Chou J Y. Hepatocyte nuclear factor 1alpha is an accessory factor required for activation of glucose-6-phosphatase gene transcription by glucocorticoids. DNA Cell Biol. 1998;17:967–974. doi: 10.1089/dna.1998.17.967. [DOI] [PubMed] [Google Scholar]
- 31.Lin B, Morris D W, Chou J Y. The role of HNF1α, HNF3γ, and cyclic AMP in glucose-6-phosphatase gene activation. Biochemistry. 1998;36:14096–14106. doi: 10.1021/bi9703249. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Fuller G M. Detection of a novel transcription factor for the A alpha-fibrinogen gene in response to interleukin-6. J Biol Chem. 1995;270:7580–7586. doi: 10.1074/jbc.270.13.7580. [DOI] [PubMed] [Google Scholar]
- 33.Maione D, Di Carlo E, Li W, Musiani P, Modesti A, Peters M, Rose-John S, Della Rocca C, Tripodi M, Lazzaro D, Taub R, Savino R, Ciliberto G. Coexpression of IL-6 and soluble IL-6 receptor causes nodular regenerative hyperplasia and adenomas of the liver. EMBO J. 1998;17:5588–5597. doi: 10.1093/emboj/17.19.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maire P, Wuarin J, Schibler U. The role of cis-acting promoter elements in tissue-specific albumin gene expression. Science. 1989;244:343–346. doi: 10.1126/science.2711183. [DOI] [PubMed] [Google Scholar]
- 35.Michalopoulos G K, DeFrances M C. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 36.Mizuguchi J, Hu C-H, Cao Z, Loeb K R, Chung D W, Davie E W. Characterization of the 5′ flanking region of the gene for the γ chain of human fibrinogen. J Biol Chem. 1995;270:28350–28356. doi: 10.1074/jbc.270.47.28350. [DOI] [PubMed] [Google Scholar]
- 37.Mohn K L, Laz T M, Melby A E, Taub R. Immediate early gene expression differs between regenerating liver, insulin-stimulated H-35 cells, and mitogen-stimulated 3T3 cells: liver specific induction patterns of gene 33, PEPCK, and the jun, fos, and egr families. J Biol Chem. 1990;265:21914–21921. [PubMed] [Google Scholar]
- 38.Mohn K L, Melby A E, Tewari D S, Laz T M, Taub R. The gene encoding rat insulin like growth factor binding protein 1 is rapidly and highly induced in regenerating liver. Mol Cell Biol. 1991;11:1393–1401. doi: 10.1128/mcb.11.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan J I, Curran T. Stimulus-transcription coupling in the nervous system: involvement of the inducible protooncogenes fos and jun. Annu Rev Neurosci. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- 40.Moshage H. Cytokines and the hepatic acute phase response. J Pathol. 1997;181:257–266. doi: 10.1002/(SICI)1096-9896(199703)181:3<257::AID-PATH756>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 41.Nagarajan R P, Zhang J, Li W, Chen Y. Regulation of Smad7 promoter by direct association with Smad3 and Smad4. J Biol Chem. 1999;274:33412–33418. doi: 10.1074/jbc.274.47.33412. [DOI] [PubMed] [Google Scholar]
- 42.Nordlie R C, Foster J D, Lange A J. Regulation of glucose production by the liver. Annu Rev Nutr. 1999;19:379–406. doi: 10.1146/annurev.nutr.19.1.379. [DOI] [PubMed] [Google Scholar]
- 43.Peng Y, Genin A, Spinner N B, Diamond R H, Taub R. The gene encoding human nuclear protein tyrosine phosphatase, PRL-1. J Biol Chem. 1998;273:17286–17295. doi: 10.1074/jbc.273.27.17286. [DOI] [PubMed] [Google Scholar]
- 44.Peng Y, Du K, Ramirez S, Diamond R H, Taub R. Mitogenic up-regulation of the PRL-1 protein-tyrosine phosphatase gene by Egr-1. Egr-1 activation is an early event in liver regeneration. J Biol Chem. 1999;274:4513–4520. doi: 10.1074/jbc.274.8.4513. [DOI] [PubMed] [Google Scholar]
- 45.Poli V, Balena R, Fattori E, Markatos A, Yamamoto M, Tanaka H, Ciliberto G, Rodan G A, Costantini F. Interleukin-6 deficient mice are protected from bone loss caused by estrogen depletion. EMBO J. 1994;13:1189–1196. doi: 10.1002/j.1460-2075.1994.tb06368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Powell D R, Suwanichkul A. HNF1 activates transcription of the human gene for insulin-like growth factor binding protein-1. DNA Cell Biol. 1993;12:283–289. doi: 10.1089/dna.1993.12.283. [DOI] [PubMed] [Google Scholar]
- 47.Rollini P, Fournier R E K. The HNF-4/HNF-1α transactivation cascade regulates gene activity and chromatin structure of the human serine protease inhibitor gene cluster at 14q32.1. Proc Natl Acad Sci USA. 1999;96:10308–10313. doi: 10.1073/pnas.96.18.10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryffel G U, Kugler W, Wagner U, Kaling M. Liver-specific gene transcription in vitro: the promoter element HP1 and a TATA box are necessary and sufficient to generate a liver-specific promoter. Nucleic Acids Res. 1989;17:939–953. doi: 10.1093/nar/17.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samstein B, Hoimes M L, Fan J, Frost R A, Gelato M C, Lang C H. IL-6 stimulation of insulin-like growth factor binding protein (IGFBP)-1 production. Biochem Biophys Res Commun. 1996;228:611–615. doi: 10.1006/bbrc.1996.1705. [DOI] [PubMed] [Google Scholar]
- 50.Scearce L M, Cressman D E, Taub R. Early transcriptional changes in regenerating liver reflect triggering mechanisms. Death Differ. 1996;3:47–55. [Google Scholar]
- 51.Soutoglou E, Papafotiou G, Katrakili N, Talianidis I. Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins. J Biol Chem. 2000;275:12515–12520. doi: 10.1074/jbc.275.17.12515. [DOI] [PubMed] [Google Scholar]
- 52.Streeper R S, Svitek C A, Goldman J K, O'Brian R M. Differential role of hepatocyte nuclear factor-1 in the regulation of glucose-6-phosphatase catalytic subunit gene transcription by cAMP in liver- and kidney-derived cell lines. J Biol Chem. 2000;275:12108–12118. doi: 10.1074/jbc.275.16.12108. [DOI] [PubMed] [Google Scholar]
- 53.Suwanichkul A, Cubbage M L, Powell D R. The promoter region of the human gene for insulin-like growth factor binding protein-1. J Biol Chem. 1990;265:21185–21193. [PubMed] [Google Scholar]
- 54.Taub R. Liver regeneration 4: transcriptional control of liver regeneration. FASEB J. 1996;10:413–427. [PubMed] [Google Scholar]
- 55.Taub R. Liver regeneration in health and disease. Clin Lab Med. 1996;16:341–360. [PubMed] [Google Scholar]
- 56.Taub R. Interleukin-6 and liver growth regulation. In: Fleig X, editor. Proceedings of the International Falk Workshop: Normal and Malignant Liver Cell Growth. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 68–74. [Google Scholar]
- 57.Taub R, Greenbaum L E, Peng Y. Transcriptional regulatory signals define cytokine-dependent and independent pathways in liver regeneration. Semin Liver Dis. 1999;19:17–127. doi: 10.1055/s-2007-1007104. [DOI] [PubMed] [Google Scholar]
- 58.Toniatti C, Demartis A, Monaci P, Nicosia A, Ciliberto G. Synergistic trans-activation of the human C-reactive protein promoter by transcription factor HNF1 binding at two distinct sites. EMBO J. 1990;9:4467–4475. doi: 10.1002/j.1460-2075.1990.tb07897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tronche F, Rollier A, Bach I, Weiss M C, Yaniv M. The rat albumin promoter: cooperation with upstream elements is required when binding of APF/HNF1 to the proximal element is impaired by mutations or bacterial methylation. Mol Cell Biol. 1989;9:4759–4766. doi: 10.1128/mcb.9.11.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tronche F, Yaniv M. HNF1, a homeoprotein member of the hepatic transcription regulation network. Bioessays. 1992;14:579–587. doi: 10.1002/bies.950140902. [DOI] [PubMed] [Google Scholar]
- 61.Tronche F, Ringeisen F, Blumenfeld M, Yaniv M, Pontoglio M. Analysis of the distribution of binding sites for a tissue-specific transcription factor in the vertebrate genome. J Mol Biol. 1997;266:231–245. doi: 10.1006/jmbi.1996.0760. [DOI] [PubMed] [Google Scholar]
- 62.Ulloa L, Doody J, Massague J. Inhibition of transforming growth factor-β/SMAD signaling by the interferon-γ/STAT pathway. Nature. 1999;397:710–713. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]
- 63.Zhang D, Sun M, Samols D, Kushner I. STAT3 participates in transcriptional activation of the C-reactive protein gene by interleukin-6. J Biol Chem. 1996;271:9503–9509. doi: 10.1074/jbc.271.16.9503. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X, Wrzeszczynska M H, Horvath C M, Darnell J E., Jr Interaction regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol Cell Biol. 1999;19:7138–7146. doi: 10.1128/mcb.19.10.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z, Fuentes N L, Fuller G M. Characterization of the IL-6 responsive elements in the γ fibrinogen gene promoter. J Biol Chem. 1995;270:24287–24291. doi: 10.1074/jbc.270.41.24287. [DOI] [PubMed] [Google Scholar]