Abstract
The dbl oncogene product (onco-Dbl) is the prototype member of a family of guanine nucleotide exchange factors (GEFs) for Rho GTPases. The Dbl homology (DH) domain of onco-Dbl is responsible for the GEF catalytic activity, and the DH domain, together with the immediately adjacent pleckstrin homology (PH) domain, constitutes the minimum module bearing transforming function. In the present study, we demonstrate that the onco-Dbl protein exists in oligomeric form in vitro and in cells. The oligomerization is mostly homophilic in nature and is mediated by the DH domain. Mutagenesis studies mapped the region involved in oligomerization to the conserved region 2 of the DH domain, which is located at the opposite side of the Rho GTPase interacting surface. Residue His556 of this region, in particular, is important for this activity, since the H556A mutant retained the GEF catalytic capability and the binding activity toward Cdc42 and RhoA in vitro but was deficient in oligomer formation. Consequently, the Rho GTPase activating potential of the H556A mutant was significantly reduced in cells. The focus-forming and anchorage-independent growth activities of onco-Dbl were completely abolished by the His556-to-Ala mutation, whereas the abilities to stimulate cell growth, activate Jun N-terminal kinase, and cause actin cytoskeletal changes were retained by the mutant. The ability of onco-Dbl to oligomerize allowed multiple Rho GTPases to be recruited to the same signaling complex, and such an ability is defective in the H556A mutant. Taken together, these results suggest that oligomerization of onco-Dbl through the DH domain is essential for cellular transformation by providing the means to generate a signaling complex that further augments and/or coordinates its Rho GTPase activating potential.
The dbl oncogene product (onco-Dbl) was originally isolated from a diffuse B-cell lymphoma (16). Over the past decade, a large group of proteins has joined the Dbl family by virtue of their structural similarity with onco-Dbl in an approximately 300-amino-acid region consisting of a Dbl homology (DH) domain and a pleckstrin homology (PH) domain. Many members of this family, including Vav, Ect2, Tim, Ost, Dbs, Lbc, Lfc, Lsc, and Net, possess a transformation or invasion capability like onco-Dbl has. Other members include proteins identified as gene products of sequences that are rearranged in human diseases (Bcr or FGD1) or as proteins with other catalytic functions, such as the Sos or RasGRF Ras guanine nucleotide exchange factors (GEFs) (for reviews, see references 8 and 58).
Onco-Dbl and the related yeast protein Cdc24 were among the first to be realized to function as Rho GTPase GEFs, i.e., to stimulate the replacement of bound GDP by GTP on specific members of the Rho family small GTPases (25, 62). Subsequent studies of individual Dbl-like molecules have found that Lbc, Lfc, and Lsc oncoproteins act as specific GEFs for Rho and cause cellular transformation through the Rho signaling pathway (19, 57, 64), the ost oncogene product shows GEF activity for Cdc42 and Rho and is capable of binding to the GTP-bound form of Rac1 (27), and the hematopoietic cell type-specific vav oncogene product functions as a Rac-specific GEF (12) and is involved in multiple pathways mediating T- or B-cell activation (6). The receptor tyrosine phosphatase LAR-associated molecule Trio contains two GEF modules that are specific to Rac and Rho (14), and its Caenorhaloditis elegans homolog Unc-73 (53) and Drosophila melanogaster homolog (41) have both been found to be Rac-specific activators that link axon guidance receptors to the growth cone cytoskeleton. The T-cell tumor invasive gene product Tiam-1, which appears to directly influence the invasive capacity of T-lymphoma cells (21), is known to activate Rac1 and Cdc42 in vitro and to stimulate the Rac-mediated pathways in cells (38), whereas the FGD1 protein, a mutation of which in the DH domain cosegregates with human faciogenital dysplasia (46), has been demonstrated to be a Cdc42-specific GEF in vitro and in vivo (44, 66). These and a large body of other studies (8, 56, 58) have helped establish that the biological functions of Dbl family members are intimately dependent upon their ability to interact and activate Rho GTPases, and the cellular effects of onco-Dbl and Dbl-like proteins, including actin cytoskeletal reorganization, cell growth stimulation, and transformation, are likely the consequences of coordinated activation of their immediate downstream substrates, the Rho family GTPases.
Current biochemical data have pointed to the conserved structural motif of the Dbl family, the DH domain, as the primary interactive site with Rho GTPases. The DH domain does not share significant sequence homology with other subtypes of small GTPase GEFs, such as the Cdc25 domain and the Sec7 domain, which are specific to Ras and ARF, respectively (5, 20), indicating that the DH-Rho protein interaction employs a distinct mechanism (9). Deletions or mutations within the DH domain have been reported to result in the loss of cellular function by the GEFs (26, 47, 50), suggesting that an intact DH domain, and likely its Rho GTPase interactive ability, is essential for the cellular effects of Dbl family members. The recently available three-dimensional structures of the DH domain (2, 35, 51) and systematic mutagenesis studies (67) have provided a preliminary model for how the DH-Rho GTPase interaction might occur: the DH domain is folded into a flattened, elongated α-helix bundle in which two of the three conserved regions, conserved region 1 (CR1) and conserved region 3 (CR3), are exposed near the center of one surface. CR1 and CR3, together with a part of α6 and the DH-PH junction site, constitute the Rho GTPase interacting pocket. These surface contact sites of the DH domain are responsible for Rho GTPase recognition and GEF catalysis, both of which appear to be essential for the transforming function of Dbl-like oncoproteins (67).
Many members of the Dbl family seem to exist in an inactive state prior to full activation. The incoming upstream signals, such as the heterotrimeric G protein Gα or Gβγ subunits, protein tyrosine or serine/threonine kinases, and phosphoinositol lipids, may contribute by varying degrees to the GEF activation processes (18, 23, 24, 29, 36, 42, 54). Currently available literature suggests that the inactive state may be maintained by one of three possible regulatory modes involving intra- or intermolecular interactions. The first is through the interdomain interaction between DH and PH motifs within the same GEF molecule. Examples of such an interaction include those of Vav and Sosl, in which cases binding to PIP3 by the PH domain seems to alleviate an inhibitory effect on the DH domain (13). The second mode of regulation is through the intramolecular interaction of a regulatory domain with the PH or DH domain of the GEF protein. Such interactions are expected to impose a constraint on the normal DH and/or PH domain function by masking the access site from the Rho GTPase substrate and/or by altering the PH domain's intracellular targeting. Examples of such regulation include proto-Vav, Vav3, Sos, and proto-Dbl (1, 3, 10, 40; our unpublished results). The third possible mode involves oligomerization through an intermolecular interaction between DH domains. This mode of regulation has been suggested only recently for RasGRF1 and RasGRF2 (4), two closely related Ras activators that also contain the DH-PH functional module. Little is known about how this oligomer formation would contribute to the regulation of their biological functions and whether oligomerization could occur for other Dbl family members.
In the present report, we describe the finding that the onco-Dbl protein forms oligomers in vitro and in mammalian cells. The oligomerization is mediated by the DH domain and is mostly homophilic in nature. Structural mapping by site-specific mutagenesis helps identify a central part of CR2 located at the opposite side of the Rho GTPase interacting surface as a critical site involved in oligomer formation. Detailed functional analysis revealed that while the GEF activity toward Rho GTPases and the oligomerization activity of the DH domain are two separable events, both of these biochemical activities are indispensable for the transforming function of onco-Dbl. Our results suggest that homo-oligomerization of onco-Dbl through the DH domain provides the means to generate a signaling complex that augments its Rho GTPase activating potential and therefore is essential for cellular transformation.
MATERIALS AND METHODS
Construction of mutant Dbl cDNA.
Constructs of pZipneo–onco-Dbl, pZipneoGST–DH-PH, and pKH3–DH-PH were described previously (67). The Flag-tagged Dbl constructs were generated by subcloning the DH-PH sequences (residues 498 to 825) into the pCMV2B vector. The expression plasmids expressing the DH domain and the PH domain of onco-Dbl were produced by PCR cloning of the cDNA sequences encoding residues 498 to 690 and 691 to 825 of onco-Dbl, respectively, into the pKH3 vector. The DH domain point mutants were generated by oligonucleotide-directed mutagenesis of onco-Dbl cDNA in a pBluescript vector by a PCR-based second extension amplification technique using the Pfu polymerase (Stratagene), with primers that contained the desired mutations (31). The DH-PH chimeras of the DH-PH module were produced by PCR using the Pfu polymerase, which generates blunt-ended DNA fragments in PCR reactions. The products amplified from cDNAs encoding two separate DH and PH fragments with primers sandwiching the respective domains were then coinserted into the BamHI sites of the pBluescript vector (31). The resulting chimeric or point mutant constructs were sequence proofed by automated sequencing before further subcloning into the mammalian expression vector pKH3. The BamHI fragments encoding the DH-PH module of onco-Dbl mutants were also subcloned into the BglII and BamHI sites of the pVL1392 vector, together with the cDNAs encoding the glutathione S-transferase (GST) or His6 sequences for insect cell expression (63), or into the BamHI site of the mammalian pZipneoGST vector for transfection into NIH 3T3 cells (67). The hemagglutinin (HA)-tagged TrioN, TrioC, Lbc, and Ost expression vectors were generated by PCR cloning of the respective coding cDNAs into the pKH3 vector.
Expression of recombinant proteins.
HA-tagged wild-type Cdc42, Rac1, and RhoA were produced by subcloning the BamHI-EcoRI fragment of cDNAs encoding the full-length GTPases into the pKH3 vector and were expressed in Cos-7 cells. Expression and purification of small GST fusion GTP binding proteins (GST-Cdc42, GST-RhoA, GST-N17Cdc42, and GST-N19RhoA) from pGEX vector-transformed Escherichia coli were carried out as described previously (26). Production and purification of the Sf9 insect cell-expressed GST-Dbl and DH mutants or His6-Dbl were performed similarly to a previously described method (63). The concentration and integrity of purified proteins were estimated by Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using bovine serum albumin as a standard.
Cell culture and transfection.
Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum (NIH 3T3 and Swiss 3T3 cells) or 10% fetal bovine serum (Cos-7 cells). Transfections were carried out using the Lipofectamine method (Gibco Life Sciences, Inc.) with 0.5 μg of plasmid DNA in 60-mm-diameter dishes. To generate stable cell lines, NIH 3T3 cells were transfected with pZipneoGST constructs and were selected in DMEM supplemented with 5% calf serum and 350 μg of G418 per ml. The drug-resistant colonies were cloned and subcultured in the same medium after 18 days. To measure Jun N-terminal kinase (JNK) activation, a transient expression reporter gene assay (PathDetect; Stratagene, San Diego, Calif.) was employed. NIH 3T3 cells were transfected with the pKH3 construct of wild-type or mutant Dbl, together with a c-Jun fusion trans-activator plasmid and the pFR-Luc reporter plasmid, according to the supplier's instructions (Stratagene). Analysis of luciferase expression in these cells was performed at 48 h posttransfection with enhanced chemiluminescent reagents and a Monolight 2010 luminometer (Analytical Luminescence, San Diego, Calif.). Retroviral expression of wild-type and mutant Dbl in Swiss 3T3 cells was carried out following the published protocols using the pMX-IRES-GFP vector and ecotropic Phoenix viral packaging cells (67).
In vitro GDP-GTP exchange assay.
The time courses for [3H]GDP-GTP exchange of Rho family GTPases in the presence or absence of GST or GST-Dbl were determined as previously described using the nitrocellulose filtration method (63). The GEF reaction buffer contained [3H]GDP-loaded Rho proteins with 20 mM Tris-HCl (pH 7.6), 100 mM NaCl, 10 mM MgCl2, 0.5 mM GTP, and 1 mM dithiothreitol supplemented with GST, GST-Dbl, or Dbl mutants. To extrapolate the kinetics parameters of the Dbl mutant-catalyzed exchange, the initial GDP dissociation rates (V0) were determined at increasing concentrations of Cdc42-GDP in the GEF reaction buffer. The resulting hyperbolic curves were analyzed by best fitting the data into a modified Michaelis-Menten equation as described before (61).
Complex formation assay.
Cos-7 cells were transfected with various Dbl constructs as described previously (67). At 48 h posttransfection, complex formation between HA-Dbl or the DH mutants and GST-fused dominant-negative Cdc42 (Cdc42T17N) or RhoA (RhoAT19N) or between HA-Dbl or the DH mutants and the GST-Dbl protein were carried out similarly to a previously described method by incubation of the Dbl-expressing cell lysates with the immobilized GST fusion proteins (26). The coprecipitation complexes were visualized by chemiluminescence reagents (Amersham Pharmacia) after SDS-PAGE and Western blotting with the indicated antibodies.
In vivo Rho GTPase activation assay.
The glutathione-agarose-immobilized GST-PAK1, which contains the p21-binding domain of human PAK1 (residues 51 to 135), and GST-PKN, which contains the site required for RhoA-GTP recognition of protein kinase N (residues 1 to 128), were expressed and purified in E. coli by using the pGEX-KG vector, as previously described (32). The active, GTP-bound form of HA-Cdc42 or HA-RhoA in fresh Cos-7 cell lysates coexpressing the small GTPase and various Dbl constructs was captured by incubation with the GST-fused effector domains and detected by anti-HA immunoblotting (67).
Cell growth and transformation assay.
To measure cell growth rates, the stably transfected cells were plated at a density of 5,000/30-mm-diameter culture dish and grown in DMEM with 2% calf serum. Cell numbers were quantified at 2-day intervals. Measurement of the capability of cells to grow in soft agar was carried out as previously described (33). Briefly, 2 × 104 cells were suspended in DMEM supplemented with 10% calf serum and 0.3% agarose and plated on top of solidified DMEM with 10% calf serum and 0.5% agarose. Cells were fed weekly by the addition of 1 ml of DMEM supplemented with 10% calf serum and 0.3% agarose. Two-and-a-half weeks after plating, colonies larger than 50 μm were scored under a microscope. To assay transforming activity, NIH 3T3 cells were transfected with the pZipneoGST-Dbl constructs by the Lipofectamine method following the instructions from Gibco Life Sciences, Inc. The transfected NIH 3T3 cells were fed every 2 days with fresh DMEM supplemented with 10% calf serum. At 12 to 14 days posttransfection, the cell culture dishes were either visualized directly under a microscope for focus formation or were stained with a 2% solution of Giemsa for focus scoring (67).
Fluorescence microscopy.
Log-phase growing fibroblasts were seeded at a density of 3 × 104 per 12-mm-diameter coverslip (Fisher Scientific) overnight before fixation in phosphate-buffered saline containing 4% paraformaldehyde for 10 min at room temperature. The cells were permeabilized in Tris-buffered saline (pH 7.4) containing 0.2% Triton X-100 for 5 min and were stained for F-actin using rhodamine-phalloidin (Molecular Probes). Coverslips were mounted onto slides in 50% glycerol–Tris-buffered saline. Stained cells were analyzed by using a conventional fluorescence microscope (Olympus BX60) (67).
RESULTS
Onco-Dbl forms oligomers in vitro and in vivo.
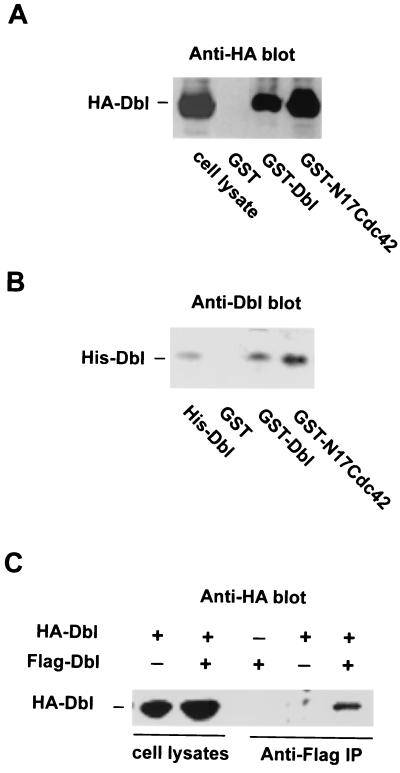
The Dbl family members RasGRF1 and RasGRF2 were reported to oligomerize through their DH domain in yeast two-hybrid assays and in mammalian cells (4). We wished to examine whether the oligomer formation property of RasGRFs could be extended to the onco-Dbl protein. Initially, we used glutathione-agarose-immobilized GST-Dbl (residues 498 to 825, making up the DH-PH module, which retains the wild-type onco-Dbl GEF activity and cell transformation capability) (26) as a probe to detect possible complex formation with the HA-tagged onco-Dbl protein expressed in Cos-7 cells. Anti-HA Western blot analysis of the GST-Dbl coprecipitates from the Cos-7 cell lysates revealed that HA-Dbl readily complexed with GST-Dbl without detectable association with GST (Fig. 1A), which is similar to what happens when using the immobilized GST-N17Cdc42, a dominant-negative mutant of Cdc42 bearing a Thr17-to-Asn mutation that is able to bind onco-Dbl tightly (Fig. 1A) (26). To determine if the complex formation between Dbl molecules is mediated by a direct contact, we subsequently used the purified components, GST-Dbl and His6-Dbl, in a glutathione-agarose pull-down assay. As shown in Fig. 1B, His6-Dbl specifically coprecipitated with GST-Dbl (Fig. 1A), indicating that oligomerization between onco-Dbl is mediated by a direct physical association. To demonstrate that oligomerization of Dbl occurs in cells, we transiently cotransfected the onco-Dbl cDNAs tagged with an HA epitope or a Flag epitope into Cos-7 cells. Immunoprecipitation with antibodies against the Flag epitope showed a specific coprecipitation of the HA-Dbl protein (Fig. 1C), indicating that complex formation among different onco-Dbl populations could occur in mammalian cells. Taken together, these results indicate that onco-Dbl may exist in oligomer form in vitro and in vivo, and the oligomerization is mediated by a direct physical association between onco-Dbl molecules.
FIG. 1.
Oligomer formation of onco-Dbl. (A) GST-Dbl forms a stable complex with HA-Dbl expressed in Cos-7 cells. HA-Dbl was transiently expressed in Cos-7 cells for 48 h before cells were lysed and analyzed by GST-glutathione affinity precipitation with GST, GST-Dbl, or GST-N17Cdc42. After three washes with ice-cold cell lysis buffer, the coprecipitates were visualized by anti-HA Western blotting. (B) Oligomerization of onco-Dbl is mediated through direct interaction between Dbl molecules. Purified His6-Dbl was incubated with glutathione-agarose-immobilized GST, GST-Dbl, or GST-N17Cdc42 for 30 min before separation by centrifugation. The coprecipitates were detected by Western blotting with anti-Dbl antibody. (C) The oligomerization of onco-Dbl occurs in cells. The Flag-tagged onco-Dbl protein was transiently expressed in Cos-7 cells with or without HA-Dbl, and the cell lysates were subjected to anti-Flag immunoprecipitation followed by anti-HA Western blotting.
Oligomerization of onco-Dbl is homophilic in nature and is mediated by the DH domain.
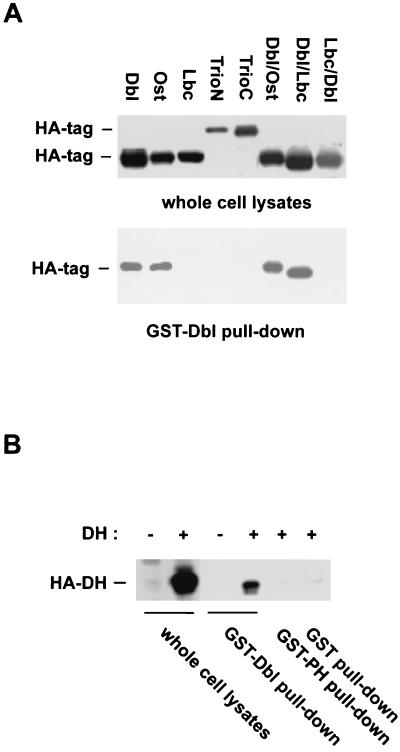
To examine if onco-Dbl is capable of forming a stable complex with other Dbl family members, we overexpressed the HA-tagged cDNA constructs encoding Ost, Lbc, TrioN, and TrioC in Cos-7 cells and sought for a coprecipitation pattern of these proteins with the glutathione-agarose-immobilized GST-Dbl (Fig. 2A). Of the panel of four GEFs tested, Ost coprecipitated with GST-Dbl, like onco-Dbl did, whereas Lbc, TrioN, and TrioC failed to form a stable complex with GST-Dbl. Thus, oligomerization among the Dbl family proteins appears to be selective. Because a RasGRF1 mutant generated at the DH domain was previously found to be defective in oligomer formation, suggesting a role for the DH domain in RasGRF1 oligomerization (4), we next examined the possible involvement of the DH domain of Dbl in the oligomerization process. The DH domain of Dbl alone expressed in Cos-7 cells readily formed a stable complex with GST-Dbl consisting of the DH-PH module, like onco-Dbl did, but it failed to complex with the GST-PH domain of Dbl (Fig. 2B), implicating that it is the DH domain that contributes primarily to the oligomerization activity. A further test of the complex formation patterns of a few DH-PH chimeras, DHDbl-PHOst, DHDbl-PHLbc, and DHLbc-PHDbl, revealed that while both DHDbl-PHOst and DHDbl-PHLbc, which contain the DH domain of onco-Dbl, remained capable of complex formation with GST-Dbl, the DHLbc-PHDbl protein, which contains only the PH portion of onco-Dbl, was inactive in binding to GST-Dbl (Fig. 2A). These results establish that the oligomerization activity of onco-Dbl is mediated by the DH domain. Furthermore, the oligomer formation is likely homophilic in nature, since the DH domain of Ost, which is capable of complex formation with Dbl, shares ∼66% sequence identity with that of Dbl, compared to the DH domains of Lbc, TrioN, and TrioC, which are 28, 46, and 44% identical to that of Dbl, respectively, and are inactive in binding to onco-Dbl.
FIG. 2.
Oligomerization of onco-Dbl is homophilic in nature and is mediated by the DH domain. (A) Complex formation of GST-Dbl with the HA-tagged Dbl family members and DH-PH chimeras. HA-tagged Dbl, Ost, Lbc, TrioN, TrioC, and the HA-tagged DH domain-PH domain chimeras made between Dbl and Ost (Dbl/Ost), between Dbl and Lbc (Dbl/Lbc), and between Lbc and Dbl (Lbc/Dbl) were expressed in Cos-7 cells. The cell lysates were incubated with glutathione-agarose-immobilized GST-Dbl for 30 min. The GST-Dbl coprecipitates, as well as the input cell lysates, were subjected to anti-HA Western blotting analysis. (B) The DH domain is responsible for complex formation with GST-Dbl. Cell lysates expressing the HA-DH domain of Dbl were subjected to a GST-Dbl, GST-PH, or GST pull-down assay. The input lysates and the glutathione-agarose coprecipitates were visualized by an anti-HA Western blot.
CR2 of the DH domain is involved in oligomerization.
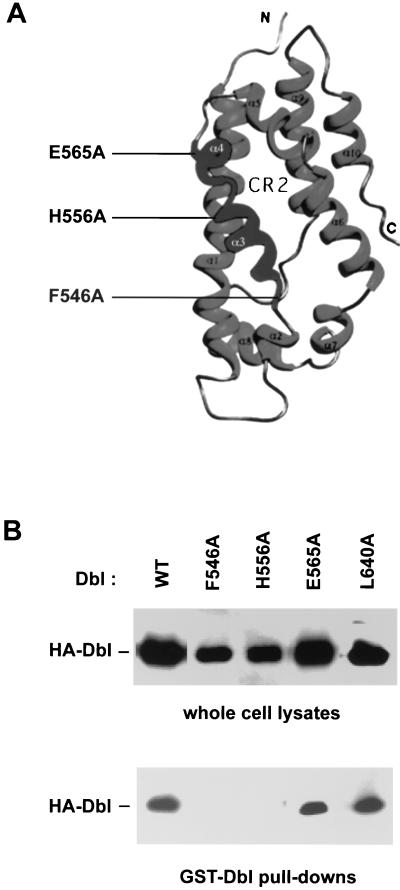
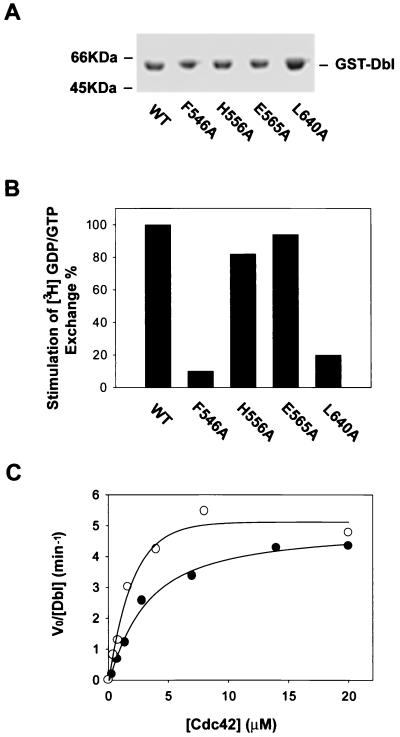
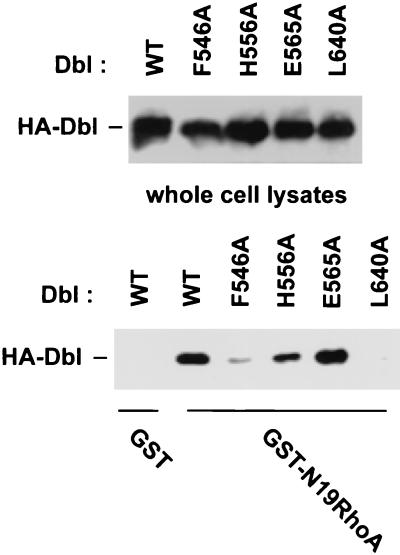
Since purified onco-Dbl protein displays constitutively active GEF activity toward RhoA and Cdc42, even at tens of a micromolar concentrations (data not shown), we reasoned that oligomer formation by onco-Dbl would not interfere with its Rho GTPase interacting ability. Recent structural and alanine substitution mutagenesis studies have identified the Rho GTPase interactive surface of the DH domain along the shallow groove formed by the N terminus of α1 (CR1) and the middle section of α9 (CR3), extending to a part of α6 and the DH-PH junction site (2, 35, 67). We therefore examined the opposite side of the flattened α-helix bundle of the DH domain in a search for possible sites involved in oligomer formation (Fig. 3A). CR2 of the DH domain is located at the surface of this side, and its function among Dbl family members has not yet been assessed. Mutation of two relatively conserved residues in CR2, F546 and H556, to alanine residues led to a complete loss of the oligomerization activity by onco-Dbl, while the E565A mutant made at the C-terminal end of CR2 had no effect on this activity (Fig. 3B). To rule out the possibility that the effects of CR2 mutations were due to disrupted structural folding of the DH domain, the three mutants (F546A, H556A, and E565A), as well as the wild type and the CR3 L640A mutant, were expressed in Sf9 insect cells and purified to homogeneity (Fig. 4A). By using RhoA as the substrate, the GEF catalytic activities of the H556A and E565A mutants were observed to be similar to that of the wild type, while the GEF activity of the F546A mutant was severely impaired, like that of the L640A mutant (Fig. 4B), which was previously shown to affect Rho binding and catalysis (67). A more stringent kinetic analysis of the H556A mutant revealed that by using Cdc42 as a substrate, its GEF reaction Km is 2.84 μM and its kcat is 4.56 min−1, which is similar to the GEF reaction efficiency of wild-type onco-Dbl (Km, 1.68 μM; kcat, 5.07 min−1) (Fig. 4C). When the ability of the mutants to interact with Rho GTPases was further examined by the GST–dominant-negative Rho pull-down assay, it became clear that both the H556A and the E565A mutants behave similarly to wild-type onco-Dbl in interacting with the dominant-negative form of the Rho GTPase, while the F546A mutant has lost most of the Rho GTPase binding activity (Fig. 5). Therefore, of the three CR2 mutants, the F546A mutant was unable to maintain proper DH folding, which resulted in the loss of Rho GTPase binding and oligomer-forming activities; the E565A mutant retained both of the activities of the wild-type DH domain; and the H556A mutant selectively retained the Rho protein interactive function while losing the oligomerization activity. We conclude that the H556 site in CR2 is involved in oligomerization of onco-Dbl.
FIG. 3.
Disruption of onco-Dbl oligomer formation by CR2 mutants of the DH domain. (A) Ribbon depiction of the positions of CR2 mutations in the three-dimensional structure of the DH domain. The Rho GTPase interacting CR1 and CR3 are located at the opposite side of CR2. (B) Complex formation of GST-Dbl with the CR2 mutants of onco-Dbl. HA-tagged mutants were transiently expressed in Cos-7 cells. The GST-Dbl coprecipitates from the cell lysates, as well as the input cell lysates, were visualized by an anti-HA Western blot. The L640A mutation is located at the center of CR3. WT, wild type.
FIG. 4.
Effect of CR2 mutations on the GEF activity of onco-Dbl. The cDNAs encoding the wild-type DH-PH module or the module bearing mutations in the DH domain were cloned into an insect cell transfer vector with an N-terminal GST fusion tag for functional expression in Sf9 insect cells. (A) Coomassie blue-stained SDS-PAGE gel of the insect cell-expressed, glutathione-agarose affinity-purified GST fusion mutants. WT, wild type. (B) Relative GEF activities of the recombinant DH domain mutants on RhoA. Approximately 0.2 μg of purified GST-Dbl or Dbl mutant was incubated with 1 μg of [3H]GDP-loaded RhoA in the GEF reaction buffer for 5 min before termination of the reaction by nitrocellulose filtration. The percent retention of RhoA-bound [3H]GDP catalyzed by the mutants was normalized to that catalyzed by wild-type Dbl. (C) Derivation of the kinetic parameters of wild-type Dbl and the H556A mutant using Cdc42 as substrate. The V0s were determined in the presence of 20 nM Dbl or H556A mutant at 1-min intervals with various concentrations of Cdc42-GDP. The resulting V0 and substrate concentration data were best fitted into a modified Michaelis-Menton equation, with corrections being made for basal GDP dissociation from Cdc42.
FIG. 5.
Interaction of the DH mutants with dominant-negative RhoA. Wild-type Dbl and various DH mutants were expressed as HA-tagged proteins in Cos-7 cells by transient transfection. Glutathione-agarose-immobilized GST or GST-N19RhoA (5 μg/sample) was incubated with the respective Cos-7 cell lysates for 1 h followed by centrifugation and three washes. The expression of the respective HA-DH mutants in the cell lysates and their coprecipitation patterns with GST or GST-N19RhoA were detected by anti-HA Western blotting. WT, wild type.
Rho GTPase activating potential of the oligomerization-deficient DH mutants in cells.
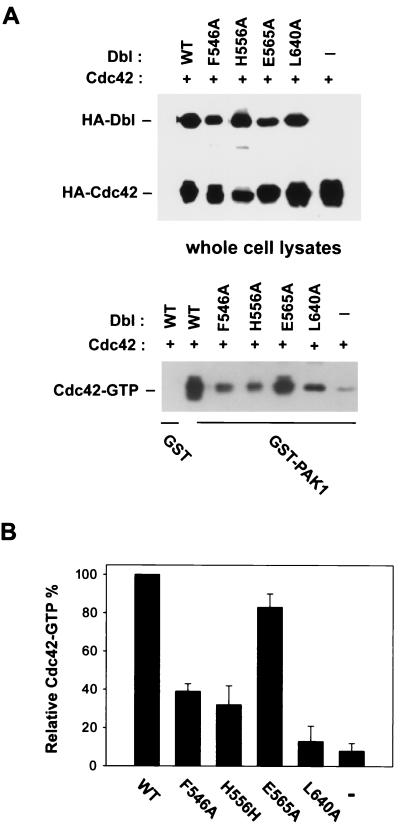
Given that the in vitro GEF activity of the H556A mutant is mostly intact, but with impaired oligomerization activity, we next set out to determine whether its Rho GTPase activating potential in cells is affected. Transient cotransfection of the HA-tagged Dbl constructs with HA-tagged, wild-type Cdc42 or RhoA in Cos-7 cells allowed a direct comparison of the expression levels of DH mutants and Cdc42 or RhoA with that of the wild-type onco-Dbl situation (Fig. 6A). The relative amounts of activated GTPases in the cell lysates were measured by a GST-PAK1 or GST-PKN pull-down assay which specifically recognizes and stabilizes the GTP-bound form of Cdc42 or RhoA (67). As shown in Fig. 6B, the F546A and H556A mutants demonstrated 2 two- to fourfold lower Cdc42 activating potentials than the wild-type onco-Dbl, while the E565A mutant was almost as active as the wild type, and the CR3 L640A mutant was inactive in Cdc42 activation. Similar observations were also made when RhoA was examined as an in vivo substrate (data not shown). These results suggest that the cellular Rho GTPase activating potential of the Dbl mutants does not necessarily correlate with the in vitro GEF activity and that oligomerization of onco-Dbl is important for optimal GEF activity in cells.
FIG. 6.
Cdc42 activation potential of the DH mutants in cells. (A) The HA-tagged wild-type Dbl or the DH mutants were cotransfected with HA-tagged Cdc42 in Cos-7 cells. At 48 h posttransfection, cell lysates were subjected to GST-PAK1 affinity precipitation. The coprecipitated Cdc42-GTP was detected by anti-HA Western blotting. A sample that was 10% of the amount of whole cell lysates used for GST-PAK1 incubations was also subjected to anti-HA blotting in parallel. WT, wild type. (B) Quantification of the Cdc42-GTP pull-down assays by densitometry measurement. The amount of Cdc42-GTP coprecipitate for the wild-type Dbl cotransfected cells was treated as 100%. The data represent results from four independent experiments.
Biological activity of the oligomerization-deficient DH mutants of onco-Dbl.
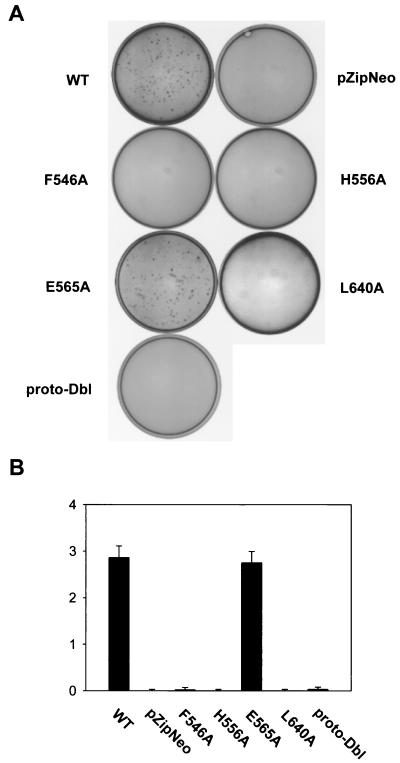
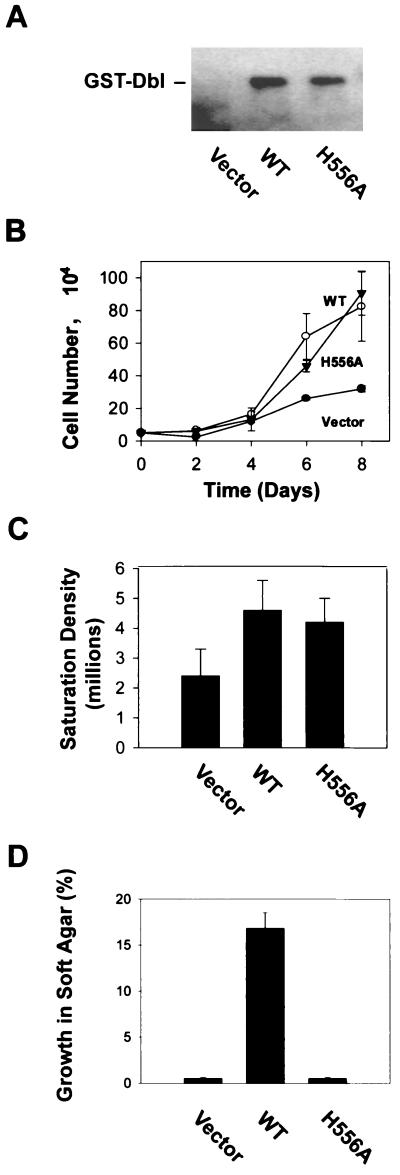
To determine the effect of the oligomerization-deficient DH mutations on the cellular transformation activity of onco-Dbl, NIH 3T3 cells were transfected with wild-type Dbl, one of the three CR2 mutants, or the CR3 L640A mutant which is defective in Rho interaction in the DH-PH module, as well as with proto-Dbl. At 14 days posttransfection, induced foci became visible under a microscope (Fig. 7A). Consistent with a previous report, proto-Dbl displayed an ∼60- to 80-fold weaker transforming activity than onco-Dbl (Fig. 7B). Of the CR2 mutants, the F546A and H556A mutants were transformation defective, like the L640A mutant, while the E565A mutant was as potent in focus induction as wild-type onco-Dbl (Fig. 7B). The transforming activities of the mutants mirrored the cellular Rho GTPase activating potentials shown in Fig. 6. The fact that the H556A mutant appeared to be even less transforming than proto-Dbl suggests that they might adopt different mechanisms of regulation. To further investigate the cellular functioning of the H556A mutant, NIH 3T3 cells stably expressing GST-H556A, as well as the cells expressing GST-Dbl, were generated by transfection with the pZipneoGST constructs followed by G418 drug selection. The expression of the GST fusion proteins in the cell clones was detected by anti-GST Western blotting (Fig. 8A). The H556A mutant-expressing cells grew as fast as the wild-type Dbl transfectants in low-serum conditions (Fig. 8B), and the cells reached a onefold higher saturation density over the mock-transfected cells, similar to the wild-type Dbl-expressing cells (Fig. 8C). However, consistent with the transformation results, the H556A-expressing cells were incapable of anchorage-independent growth in soft agar (Fig. 8D). The results for the H556A mutant indicate that maintaining the Rho GTPase interacting activity and a basal Rho activating potential by monomeric onco-Dbl may lead to the cell growth stimulatory effect, but the effect is insufficient for cell transformation. A fully activated state of Dbl comprised of oligomer rather than monomer appears to be essential for eliciting the transforming function, likely by enhancing the Rho protein activating potential of monomeric onco-Dbl molecules in cells.
FIG. 7.
Effect of DH mutations on the transforming activity of Dbl. cDNAs encoding the wild-type DH-PH domain module of Dbl (residues 498 to 825), the DH mutation-bearing DH-PH modules, and proto-Dbl were subcloned into the pZipneoGST vector and assayed for focus-forming activity in NIH 3T3 cells. Foci were quantified at 14 days posttransfection by Giemsa staining. WT, wild type. (A) Tissue culture dishes transfected with 0.1 μg of pZipneoGST-Dbl cDNA were visualized directly by a video camera. (B) Normalized focus-forming activities (103 foci/μg of DNA) of the DH mutants made in CR2 compared to those of wild-type Dbl, proto-Dbl, and the CR3 L640A mutant.
FIG. 8.
Growth properties of the H556A mutant-expressing NIH 3T3 cells. (A) Mock-transfected NIH 3T3 cells (pZipneoGST vector) and the cell clones stably expressing wild-type (WT) GST-Dbl or GST-H556A were analyzed by anti-GST Western blotting. (B) The cell growth rate of the H559A mutant-expressing cells was compared with that of wild-type Dbl-expressing or mock-transfected cells. Cell growth was initiated at a density of 5,000/35-mm-diameter culture dish at day 0 in DMEM supplemented with 2% calf serum. The number of cells in the dishes was counted in 2-day intervals. (C) Cells were plated at a density of 50,000/100-mm-diameter dish at day 0. The saturation densities of the cells were determined after the cell growth was stopped, at day 9. (D) The ability of the transfectants to grow on soft agar was measured in DMEM supplemented with 10% calf serum and 0.3% agarose on top of solidified DMEM with 0.5% agarose. Colonies were scored at 3 weeks postplating under a microscope.
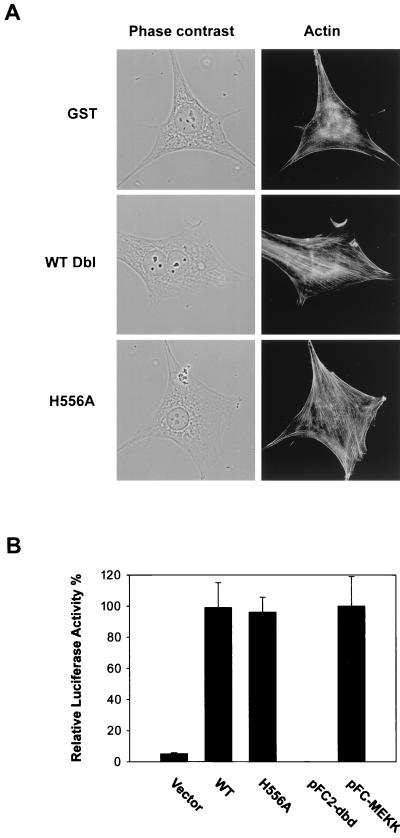
Onco-Dbl is known to induce both actin cytoskeletal changes and to stimulate signaling pathways to the nucleus (34, 43, 44, 56). The H556A mutant behaved similarly to wild-type Dbl in the first aspect, since both the H556A-expressing NIH 3T3 cells and the onco-Dbl-expressing NIH 3T3 cells led to significantly enhanced actin stress fibers compared to the GST-expressing cells after serum starvation, as revealed by rhodamine-labeled phalloidin staining (Fig. 9A). A distinction between these cells, however, is that a higher proportion of onco-Dbl transfectants displayed a multinucleus phenotype, which was lacking in the H556A-expressing cells. When assayed in Swiss 3T3 cells by retroviral induction, the H556A mutant was found to be as potent in stimulating membrane ruffling as the wild type (data not shown). To examine the possible effect on signal transduction to the nucleus, we compared the ability of the H556A mutant to activate JNK, a known target for onco-Dbl (11, 39), with that of the wild type by a luciferase-coupled c-Jun reporter assay. As shown in Fig. 9B, the H556A mutant appeared to be as potent an activator of JNK as wild-type Dbl, increasing its activity over 20-fold. It is therefore likely that although the reduced Rho protein activating potential of the monomeric form of Dbl (H556A) might have resulted in the loss of the transforming function and the lack of a stimulatory effect on cytokinesis, it remained capable of transducing a subset of signals to alter cell actin structures and to activate the JNK pathway.
FIG. 9.
Effect of the oligomer-deficient H556A mutant on actin cytoskeletal structure and JNK activation. (A) The morphology and actin structures of H556A mutant-or wild-type (WT) Dbl-expressing NIH 3T3 cells, as well as of the mock-transfected cells, were visualized under a phase-contrast or fluorescence microscope after actin staining with rhodamine-conjugated phalloidin. (B) Various pKH3 constructs (0.4 μg) or controls (pFC-MEKK and pFC2-dbd plasmids) (0.1 μg) were transiently cotransfected into NIH 3T3 cells together with the pFR-Luc reporter plasmid (1 μg) and pFA2-cJun plasmid (0.1 μg). At 48 h posttransfection, the cells were washed and harvested for the measurement of luciferase activities.
Onco-Dbl oligomer is capable of recruiting multiple Rho GTPases into the same signaling complex.
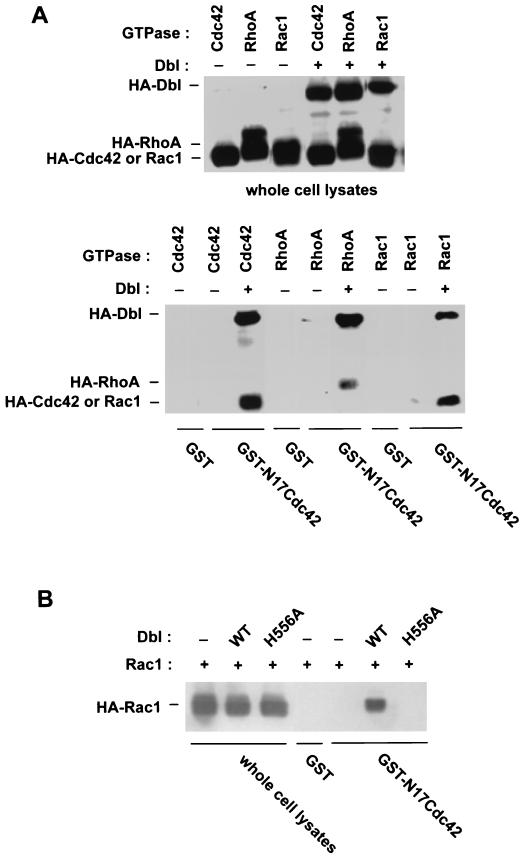
Since oligomerization of onco-Dbl does not affect the in vitro GEF activity of Rho GTPases and since the site mediating oligomerization is opposite from the Rho protein interactive site of the DH domain, we reasoned that the oligomer complex of onco-Dbl should be able to bind to multiple Rho proteins simultaneously. To test this hypothesis, we used immobilized GST-N17Cdc42 as a probe to complex with HA-Dbl and HA-Rho GTPase coexpressed in Cos-7 cells. The dominant-negative form of Cdc42 bound tightly to HA-Dbl, as expected, but did not form a detectable complex with HA-Cdc42, HA-Rac1, or HA-RhoA when they were expressed alone in the cells (Fig. 10A). When both HA-Dbl and an HA-tagged Rho GTPase were coexpressed in the cells, GST-N17Cdc42 was able to pull down the Rho GTPases together with Dbl, as revealed by Western blotting analysis (Fig. 10A), suggesting the formation of a complex consisting of GST-N17Cdc42, a Dbl oligomer, and the respective HA-Rho GTPase. When the H556A mutant was coexpressed with a Rho GTPase, Rac1, however, the HA-tagged Rho protein failed to coprecipitate with GST-N17Cdc42, whereas it readily formed a complex with GST-N17Cdc42 in the presence of wild-type onco-Dbl (Fig. 10B). This can be attributed to the deficiency in oligomerization activity by the H556A mutant, because the H556A mutant remained capable of forming a stable complex with dominant-negative Rho GTPase (Fig. 5) and with wild-type Rho GTPase (data not shown). These results suggest that homo-oligomerization of onco-Dbl through the DH domain provides the means to recruit multiple Rho family GTPases into the same signaling complex. Such a complex may serve to coordinate the activation of multiple Rho GTPases and/or to further augment the Rho GTPase activating potentials.
FIG. 10.
The oligomeric complex of onco-Dbl is capable of recruiting multiple Rho GTPases. (A) HA-tagged Cdc42, RhoA, or Rac1 was expressed in Cos-7 cells alone or together with HA-Dbl. The cell lysates were incubated with glutathione-agarose-immobilized GST or GST-N17Cdc42 for 30 min. The input cell lysates and the GST fusion coprecipitates were analyzed in parallel by anti-HA Western blotting. (B) HA-Rac1 was expressed alone, together with wild-type (WT) onco-Dbl, or together with the H556A mutant of onco-Dbl in Cos-7 cells. The cell lysates and the GST or GST-N17Cdc42 coprecipitates from the cell lysates were probed with anti-HA antibody in a Western blot.
DISCUSSION
The Dbl family GEFs for Rho GTPases include over 40 cell growth regulatory molecules (56). Their cellular functions appear to intimately depend on their ability to interact and activate specific Rho GTPases in various physiological situations, including the processes of cytokinesis, cell movement, cell proliferation, and apoptosis. Given our current knowledge of the involvement of Rho GTPases in multiple cell growth pathways (22, 56), it is not surprising that many of the Dbl family proteins were initially identified as oncogene products by virtue of their ability to transform fibroblast cells. Although most Dbl family members contain diverse multifunctional motifs, they all share the structural array of a central DH domain in tandem at the carboxyl terminus with a PH domain. Previous studies have established that while the DH domain in these proteins is primarily responsible for the Rho GTPase binding and the GEF activities, the PH domain is involved in intracellular targeting and/or modulation of the DH domain function, and together with the DH domain, constitutes the minimum structural module required for the transforming function of the GEFs (13, 26, 45, 57, 65). Aside from the PH domain-mediated regulation of DH function, an additional mode of regulation of the DH domain could be provided by the structural elements residing outside the DH-PH module of the GEF molecules through intramolecular interactions. Examples of such a mode of regulation include the proto-Vav, proto-Dbl, and Ost proteins, which utilize their unique N-terminal sequences to suppress the DH and/or PH function (1, 3, 27; our unpublished results), and the proto-Lbc and GEF-H1 molecules, in which the C-terminal sequences appear to supply the constraining elements (49, 52). A recent characterization of the regulatory mechanism for RasGRF1 and RasGRF2, two closely related Dbl family members whose primary role is to activate Ras GTPase through their Cdc25 (RasGEF) catalytic domains, has suggested that they can form oligomers via their respective DH domains (4), raising the possibility that yet another mode of regulation, intermolecular oligomerization, may be involved in the regulation of certain Dbl-related proteins. For the present study, we have examined the intermolecular interaction between onco-Dbl proteins in detail. Our results indicate that homo-oligomerization of onco-Dbl through the DH domain is essential for its cellular transforming function. We propose that onco-Dbl utilizes the oligomerization mechanism to form a large signaling complex in augmenting and/or coordinating its Rho GTPase activating potential.
By using a glutathione-agarose pull-down assay, we have shown that the GST-Dbl protein can directly form a stable complex with HA-tagged onco-Dbl. Moreover, the results from mammalian cells confirmed that the complex formation between onco-Dbl molecules could occur for two distinct populations of Dbl, suggesting that the oligomerization phenomenon of onco-Dbl is physiologically relevant. An initial estimation of the oligomerization binding affinity put the dissociation constant within 100 nM (our unpublished results). Further examination of the complex formation pattern between onco-Dbl and other Dbl-related GEFs revealed that onco-Dbl could also form a stable complex with Ost, which shares ∼67% sequence identity in the DH domain with Dbl but not with Lbc, TrioN, or TrioC, which are significantly more divergent from Dbl, implying that the interaction is mostly homophilic in nature. The facts that the DH domain of Dbl is sufficient to oligomerize with onco-Dbl and that the DH-PH chimeras made between Dbl and other Dbl family members can form a complex with onco-Dbl only when the intact DH domain of Dbl is present further indicate that the DH domain constitutes the necessary and sufficient structural unit responsible for the homophilic oligomerization activity. This is similar to the reported cases of RasGRF1 and RasGRF2, which can form oligomeric complexes among themselves mediated by their respective DH domains but fail to interact with the more divergent onco-Dbl protein (4). Whether the oligomer complex between onco-Dbl molecules contains dimers or multimers remains to be seen, but our structural mapping results of the DH domain favor a dimer configuration, as discussed below.
The tertiary structure of the DH domain is depicted as a flattened, elongated α-helix bundle in which two of the three conserved regions, CR1 and CR3, are exposed near the center of one surface (2, 35, 51). Previous sequence analysis and alanine substitution studies have provided clues that the surface defined by CR1, CR3, a part of α6, and the DH-PH junction site is involved in the formation of a Rho GTPase interactive pocket (35, 67). In addition, purified onco-Dbl proteins are constitutively active as Rho GEFs, even at high concentrations when most are expected to form oligomers, suggesting that oligomer formation would not compromise the Rho GTPase interacting capability. This rationale led us to test the hypothesis that a site of the DH domain located opposite from the Rho GTPase interactive site is involved in the DH-DH contact. To identify the site on the DH domain contributing to oligomer formation, we have focused on CR2, which consists of the α3 and α4 helices and is opposite from the Rho protein binding surface. Of the three CR2 mutants examined, the F545A mutant suffered loss of the Rho GEF activity, the Rho binding activity, and the oligomerization activity, suggesting that this mutation most likely adopted a misfolded conformation. The E545A mutant, on the other hand, retained the wild-type activities of catalyzing guanine nucleotide exchange, binding to Rho proteins, and oligomerizing with onco-Dbl, and it was transformation competent like wild-type Dbl, indicating that the E565 residue does not contribute to any of the tested functions. The H556A mutant, however, behaved similarly to wild-type onco-Dbl in the in vitro GEF reactions and in binding to the Rho GTPases but appeared to be oligomerization deficient. Although we could not completely rule out the possibility that the H556A mutation has an effect on an as yet unknown function of the DH domain, our results strongly support the notion that residue H556 of CR2 constitutes a critical site involved in oligomer formation. On a similar note, residue L263 in the DH domain of RasGRF1, the mutation of which to Gln resulted in deficiencies in both oligomerization and transformation (4), is unlikely to be a DH-DH interaction site because it is located in the central CR1 region, which is expected to be involved in Rho GTPase recognition. The L263Q mutation of RasGRF1, therefore, may have caused a disruption of the normal DH structure, leading to the loss of function. Recently, the crystal structure of Tiam1 in complex with Rac1 was solved (60). In this structure, the DH domain of Tiam1 forms a dimer with the DH domain of an adjacent Tiam1 molecule, and an extended region of the DH domain opposite from the Rac1 interactive site and including CR2 is responsible for the intermolecular contact. The back-to-back dimer configuration in the complex does not affect Rac1 interaction at the G protein binding pocket of the DH domain. These observations, combined with the current mutagenesis results, favor a model in which the DH domain of onco-Dbl presents two independent biochemical functions on two distinct tertiary surfaces, one being the GEF catalytic activity toward Rho proteins and the other involving a direct contact with adjacent Dbl molecules to form functional oligomers.
To investigate the functional relevance of oligomerization by onco-Dbl, we have further analyzed the cellular activities of the H556A mutant, which acts like the wild-type Dbl in vitro, except it lacks the oligomer formation activity. It turned out that although the H556A mutant is fully active as a GEF for Rho GTPase and remains capable of binding Rho GTPases in vitro, the mutant is significantly impaired in the Rho GTPase activating potential in cells. This decreased Rho GTPase activating potential may be attributed to an altered subcellular distribution pattern or, more likely, to lower levels of GEF activity in cells. The oligomerization-deficient mutant lacked any detectable transforming activity in NIH 3T3 cells, similar to the misfolded CR2 F545A mutant and to the CR3 L640A mutant, which was capable of oligomerization but was unable to bind to Rho GTPase. These results, combined with recent mutagenesis studies that have demonstrated the requirement of maintaining a threshold of GEF catalytic activity in onco-Dbl transformation (67), indicate that oligomerization may contribute to the maintenance of the threshold of GEF activity in vivo, which is essential for transformation.
Aside from the lack of transforming activity, the H556A mutant behaved like wild-type onco-Dbl in stimulating cell growth, in enabling cells to reach higher saturation density, in inducing actin stress fiber formation and membrane ruffling, and in stimulating INK activity. These observations indicate that the mutant remains partially active in vivo, but the remaining functions are not sufficient for transformation. It is possible that to acquire oncogenicity, onco-Dbl needs to achieve a higher threshold of activation potential for Rho GTPases in order to stimulate additional pathways required for transformation. These may include the recently characterized MEK and NF-κB pathways (59).
The ability of Dbl molecules to oligomerize without interference with the Rho GTPase binding activity and GEF catalysis suggests that onco-Dbl may induce the formation of a large signaling complex consisting of multiple Rho GTPases. Indeed, we were able to detect the binding of two distinct populations of Rho GTPases to the same Dbl oligomer complex, which could not be achieved with the oligomerization-deficient H556A mutant of Dbl. The fact that the H556A mutant displayed a significantly reduced Rho protein activating potential in cells while retaining the wild-type GEF catalytic capability in vitro further suggests that oligomerization by onco-Dbl may contribute to the coordination and/or augmentation of Rho GTPase activation in vivo. The Dbl oligomer-Rho GTPase complex may have a synergistic advantage for one particular type of Rho protein activation, e.g., activation of multiple Cdc42s at the same time and place, which would be advantageous for growth on soft agar (48). In addition, the complex could activate two distinct downstream pathways simultaneously, e.g., the Cdc42-PAK pathway and the Rho-ROK pathway, both of which are important for cell growth (56), which would be favorable for focus induction. Such a coordinated or augmented activation of Rho GTPases appears to be essential for onco-Dbl transformation but is not required for cell growth stimulation or induction of actin cytoskeletal reorganization, as evidenced by the behaviors of the H556A mutant. This interpretation is consistent with the apparent involvement of multiple Rho family members, i.e., Cdc42, Rac1, and RhoA, in mediating the onco-Dbl transforming activity (34, 43) and with the previous observation that onco-Dbl is a much more potent transforming agent than any of the activated forms of Rho proteins alone (28). Thus, oligomerization of the DH domain introduces an additional layer of regulation to the onco-Dbl regulatory mechanism, and such a mode of intermolecular interaction may be utilized by other Dbl family GEFs (e.g., RasGRFs) in further fine tuning of their downstream signal intensities.
One important issue remaining to be addressed is whether oligomerization of proto-Dbl can occur in vivo. Given that the N-terminal constraining motif of proto-Dbl may mask the access site of DH and PH domains (our unpublished results), it is possible that only the open form of the DH-PH module, and not the autoinhibitory full-length molecule, is capable of oligomerization. The lack of oligomer formation by proto-Dbl, compared with the oligomerization capability and the full biological activity displayed by the DH-PH module, would suggest that induction of oligomerization is an important step in GEF activation. Alternatively, like for RasGRF1 and RasGRF2 (4), oligomerization by proto-Dbl may be constitutive. In such a scenario, the upstream signals that mediate proto-Dbl activation would be required only for the alleviation of constraints imposed by the N-terminal regulatory sequences independent from the oligomerization process.
It will be of particular interest to see whether oligomerization is a generalized mechanism for GEF regulation, since it seems to provide an efficient way to amplify the signal flows upstream of the small GTP binding proteins coordinately. Besides onco-Dbl and the RasGRFs, a few additional GEFs for Ras-like small GTPases, including the yeast Ras GEF, Cdc25p (7), and the ARF-specific activators, BIG1 and BIG2 (55), have been reported recently to form oligomers. Given the structural divergence of these molecules from Dbl, their mechanisms of oligomerization are likely to be different. Whether the oligomer formation is a required element in their cellular functions similar to the herein-described case of onco-Dbl remains to be determined. It is an attractive hypothesis that these and certain other GEFs for the Ras superfamily GTPases may behave like onco-Dbl in forming oligomers in order to provide a control for a quantitative threshold of the signaling pathways in the small GTPase cascades, the variation of which may lead to different cellular effects (15). An additional functional consequence of oligomerization among GEFs, not unlike that of many small GTPase effectors, such as Raf (17, 37) and PAK1 (30), would be to create an interconnected network of proteins to modulate the final signal outcome of the small G protein pathways.
ACKNOWLEDGMENTS
We thank Michel Streuli for the Trio cDNA clone.
This work was supported by National Institutes of Health grant GM 53943 to Y.Z.
REFERENCES
- 1.Abe K, Whitehead I P, O'Bryan J P, Der C J. Involvement of NH2-terminal sequences in the negative regulation of Vav signaling and transforming activity. J Biol Chem. 1999;274:30410–30418. doi: 10.1074/jbc.274.43.30410. [DOI] [PubMed] [Google Scholar]
- 2.Aghazadeh B, Zhu K, Kubiseski T J, Liu G A, Pawson T, Zheng Y, Rosen M K. Structure and mutagenesis of the Dbl homology domain. Nat Struct Biol. 1998;12:1098–1107. doi: 10.1038/4209. [DOI] [PubMed] [Google Scholar]
- 3.Aghazadeh B, Lowry W E, Huang X-Y, Rosen M K. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell. 2000;102:625–633. doi: 10.1016/s0092-8674(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 4.Anborgh P H, Qian X, Papageorge A G, Vass W C, DeClue J E, Lowy D R. Ras-specific exchange factor GRF: oligomerization through its Dbl homology domain and calcium-dependent activation of Raf. Mol Cell Biol. 1999;19:4611–4622. doi: 10.1128/mcb.19.7.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boriack-Sjodin P A, Margarit S M, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 6.Bustelo X R. Regulatory and signaling properties of the Vav family. Mol Cell Biol. 2000;20:1461–1477. doi: 10.1128/mcb.20.5.1461-1477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camus C, Geminat M, Garreau H, Baudet-Nessler S, Jacqet M. Dimerization of Cdc25p, the guanine nucleotide exchange factor for Ras from Saccharomyces cerevisiae, and its interaction with Sdc25p. Eur J Biochem. 1997;247:703–708. doi: 10.1111/j.1432-1033.1997.00703.x. [DOI] [PubMed] [Google Scholar]
- 8.Cerione R A, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 9.Cherfils J, Chardin P. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- 10.Corbalan-Garcia S, Margarit S M, Galron D, Yang S-S, Bar-Sagi D. Regulation of Sos activity by intramolecular interactions. Mol Cell Biol. 1998;18:880–886. doi: 10.1128/mcb.18.2.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coso O A, Chiatiello M, Yu J-C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 12.Crespo P, Schuebel K E, Ostrom A A, Gutkind J S, Bustelo X R. Phosphotyrosine dependent activation of Rac1 GDP/GTP exchange by the vav proto-oncogene product. Nature. 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 13.Das B, Shu X, Day G J, Han J, Krishna M, Falck J R, Broek D. Control of intramolecular interactions between the pleckstrin homology and Dbl homology domains of Vav and Sosl regulates Rac binding. J Biol Chem. 2000;275:15074–15081. doi: 10.1074/jbc.M907269199. [DOI] [PubMed] [Google Scholar]
- 14.Debant A, Serra-Pages C, Seipel K, O'Brien S, Tang M, Park S H, Streuli M. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc Natl Acad Sci USA. 1996;93:5466–5471. doi: 10.1073/pnas.93.11.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Downward J. Control of Ras activation. Cancer Surv. 1996;27:87–100. [PubMed] [Google Scholar]
- 16.Eva A, Aaronson S A. Isolation of a new human oncogene from a diffuse B-cell lymphoma. Nature. 1985;316:273–275. doi: 10.1038/316273a0. [DOI] [PubMed] [Google Scholar]
- 17.Farrar M A, Alberolaila J, Perlmutter R M. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature. 1996;383:178–181. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]
- 18.Fleming I N, Elliott C M, Buchanan F G, Downes C P, Exton J H. Ca2+/calmodulin-dependent protein kinase II regulates Tiam1 by reversible protein phosphorylation. J Biol Chem. 1999;274:12753–12758. doi: 10.1074/jbc.274.18.12753. [DOI] [PubMed] [Google Scholar]
- 19.Glaven J A, Whitehead I P, Normanbhoy T, Kay R, Cerione R A. Lfc and Lsc oncoproteins represent two new guanine nucleotide exchange factors for the Rho GTP-binding protein. J Biol Chem. 1996;271:27374–27381. doi: 10.1074/jbc.271.44.27374. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg J. Structural basis for activation of ARF GTPase: mechanism of guanine nucleotide exchange and GTP-myristoyl switch. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 21.Habets G G M, Scholtes E H M, Zuydgeest D, van der Kammen R, Stam J C, Berns A, Collard J G. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 22.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller R, Murali K, Falck J R, White M A, Broek D. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- 24.Hart M J, Jiang X, Kozasa T, Roscoe W, Singer W D, Gilman A G, Sterweis P C, Bollag G. Direct stimulation of the guanine nucleotide exchange activity of p115RhoGEF by Gα13. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 25.Hart M J, Eva A, Evans T, Aaronson S A, Cerione R A. Catalysis of guanine nucleotide exchange on the Cdc42Hs protein by the dbl oncogene product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 26.Hart M J, Eva A, Zangrilli D, Aaronson S A, Evans T, Cerione R A, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- 27.Horii Y, Beeler J F, Sakaguchi K, Tachibana M, Miki T. A novel oncogene, ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signaling pathways. EMBO J. 1994;13:4776–4786. doi: 10.1002/j.1460-2075.1994.tb06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khosravi-Far R, Solski P A, Kinch M S, Burridge K, Der C J. Activation of Rac and Rho, and mitogen activated protein kinases, are required for Ras transformation. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiyono M, Kaziro Y, Satoh T. Induction of Rac-guanine nucleotide exchange activity of RasGRF1/Cdc25Mm following phosphorylation by the nonreceptor tyrosine kinase Src. J Biol Chem. 2000;275:5441–5446. doi: 10.1074/jbc.275.8.5441. [DOI] [PubMed] [Google Scholar]
- 30.Lei M, Lu W, Meng W, Parrini M-C, Eck M J, Mayer B J, Harrison S C. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- 31.Li R, Zheng Y. Residues of the Rho family GTPases Rho and Cdc42Hs that specify sensitivity to Dbl-like guanine nucleotide exchange factors. J Biol Chem. 1997;272:4671–4681. doi: 10.1074/jbc.272.8.4671. [DOI] [PubMed] [Google Scholar]
- 32.Li R, Debreceni B, Jia B, Gao Y, Tigyi G, Zheng Y. Localization of the PAK1-, WASP-, and IQGAP1-specifying regions of the small GTPase Cdc42. J Biol Chem. 1999;274:29648–29654. doi: 10.1074/jbc.274.42.29648. [DOI] [PubMed] [Google Scholar]
- 33.Lin R, Bagrodia S, Cerione R A, Manor D. A novel Cdc42Hs mutant induces cellular transformation. Curr Biol. 1997;7:794–797. doi: 10.1016/s0960-9822(06)00338-1. [DOI] [PubMed] [Google Scholar]
- 34.Lin R, Cerione R A, Manor D. Specific contributions of the small GTPases Rho, Rac, and Cdc42 to Dbl transformation. J Biol Chem. 1999;274:23633–23641. doi: 10.1074/jbc.274.33.23633. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Wang H, Eberstadt M, Schnuchel A, Olejniczak E T, Meadows R P, Schkeryantz J M, Janowick D A, Harlan J E, Harris E A S, Staunton D E, Fesik S W. NMR structure and mutagenesis of the N-terminal Db1 homology domain of the nucleotide exchange factor Trio. Cell. 1998;95:269–277. doi: 10.1016/s0092-8674(00)81757-2. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Lago M, Lee H, Cruz C, Movilla N, Bustelo X R. Tyrosine phosphorylation mediates both activation and downmodulation of the biological activity of Vav. Mol Cell Biol. 2000;20:1678–1691. doi: 10.1128/mcb.20.5.1678-1691.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo Z, Tzivion G, Belshaw P J, Vavvas D, Marshall M, Avruch J. Oligomerization activates c-Raf-1 through a Ras-dependent mechanism. Nature. 1996;383:181–185. doi: 10.1038/383181a0. [DOI] [PubMed] [Google Scholar]
- 38.Michiels F, Habets G G M, Stam J C, van der Kammen R A, Collard J G. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- 39.Minden A, Lin A, Claret F-X, Abo A, Karin M. Selective activation of the JNK signaling cascade and Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 40.Movilla N, Bustelo X R. Biological and regulatory properties of Vav3, a new member of the vav family of oncoproteins. Mol Cell Biol. 1999;19:7870–7885. doi: 10.1128/mcb.19.11.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newsome T P, Schmidt S, Dietzl G, Keleman K, Asling B, Debant A, Dickson B J. Trio combines with Dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell. 2000;101:283–294. doi: 10.1016/s0092-8674(00)80838-7. [DOI] [PubMed] [Google Scholar]
- 42.Nishida K, Kaziro Y, Satoh T. Association of the proto-oncogene product Db1 with G protein βγ subunits. FEBS Lett. 1999;459:186–190. doi: 10.1016/s0014-5793(99)01244-2. [DOI] [PubMed] [Google Scholar]
- 43.Olivo C, Vanni C, Mancini P, Silengo L, Torris M R, Tarone G, Defilippi P, Eva A. Distinct involvement of Cdc42 and RhoA GTPases in actin organization and cell shape in untransformed and Dbl oncogene transformed NIH 3T3 cells. Oncogene. 2000;19:1428–1436. doi: 10.1038/sj.onc.1203440. [DOI] [PubMed] [Google Scholar]
- 44.Olson M F, Pasteris N G, Gorski J L, Hall A. Faciogenital dysplasia protein (FGD1) and Vav, two related proteins required for normal embryonic development, are upstream regulators of Rho GTPases. Curr Biol. 1996;6:1628–1633. doi: 10.1016/s0960-9822(02)70786-0. [DOI] [PubMed] [Google Scholar]
- 45.Olson M F, Sterpetti P, Nagata K, Toksoz D, Hall A. Distinct roles for the DH and PH domains in the Lbc oncogene. Oncogene. 1997;15:2827–2831. doi: 10.1038/sj.onc.1201594. [DOI] [PubMed] [Google Scholar]
- 46.Pasteris N G, Cadle A, Logie L J, Porteous M E M, Schwartz C E, Stevenson R E, Glover T W, Wilroy R S, Gorski J L. Isolation and characterization of the faciogenital dysplasia (Arskog-Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell. 1994;79:669–678. doi: 10.1016/0092-8674(94)90552-5. [DOI] [PubMed] [Google Scholar]
- 47.Qian X, Vass W C, Papageorge A G, Anborgh P H, Lowy D R. N terminus of Sos1 Ras exchange factor: critical roles for the Dbl and pleckstrin homology domains. Mol Cell Biol. 1998;18:771–778. doi: 10.1128/mcb.18.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu R-G, Abo A, McCormick F, Symons M. Cdc42 regulates anchorage-independent growth and is necessary for Ras transformation. Mol Cell Biol. 1997;17:3449–3458. doi: 10.1128/mcb.17.6.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren Y, Li R, Zheng Y, Busch H. Cloning and characterization of GEF-H1, a microtubule associated guanine-nucleotide exchange factor for Rac and Rho GTPases. J Biol Chem. 1998;273:34954–34962. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- 50.Ron D, Zannini M, Lewis M, Wickner R B, Hunt L T, Graziani G, Trinick S R, Aaronson S A, Eva A. A region of proto-Dbl essential for its transforming activity shows sequence similarity to a yeast cell-cycle gene, Cdc24, and the human break point cluster gene, bcr. New Biol. 1991;3:372–379. [PubMed] [Google Scholar]
- 51.Soisson S M, Nimnual A S, Uy M, Bar-Sagi D, Kuriyan J. Crystal structure of the Dbl and pleckstrin homology domains from the human son of sevenless protein. Cell. 1998;95:259–268. doi: 10.1016/s0092-8674(00)81756-0. [DOI] [PubMed] [Google Scholar]
- 52.Sterpetti P, Hack A A, Bashar M P, Park B, Cheng S-D, Knoll J H M, Urano T, Feig L A, Toksoz D. Activation of the Lbc Rho exchange factor proto-oncogene by truncation of an extended C terminus that regulates transformation and targeting. Mol Cell Biol. 1999;19:1334–1345. doi: 10.1128/mcb.19.2.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steven R, Kubiseski T J, Zheng H, Kulkarni S, Mancillas J, Ruiz Morales A, Hogue C W, Pawson T, Culotti J. Unc-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- 54.Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human Ect2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M. Purification and cloning of a brefeldin A-induced guanine nucleotide exchange protein for ADP-ribosylation factors. J Biol Chem. 1999;274:12308–12315. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- 56.Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 57.Whitehead I, Kirk H, Tognon C, Trigo-Gonzalez G, Kay R. Expression cloning of Lfc, a novel oncogene with structural similarities to guanine nucleotide exchange factors and to the regulatory region of protein kinase C. J Biol Chem. 1995;270:18388–18395. doi: 10.1074/jbc.270.31.18388. [DOI] [PubMed] [Google Scholar]
- 58.Whitehead I P, Campbell S, Rossman K L, Der C J. Dbl family proteins. Biochim Biophys Acta. 1997;1332:F1–F23. doi: 10.1016/s0304-419x(96)00040-6. [DOI] [PubMed] [Google Scholar]
- 59.Whitehead I P, Lambert Q T, Glaven J A, Abe K, Rossman K L, Mahon G M, Trzaskos J M, Kay R, Campbell S L, Der C J. Dependence of Dbl and Dbs transformation on MEK and NF-κB activation. Mol Cell Biol. 1999;19:7759–7770. doi: 10.1128/mcb.19.11.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Worthylake, D. K., and J. Sondek. The structural basis of the activation of Rac1 by Tiam1. Nature, in press.
- 61.Zhang B, Zhang Y, Wang Z, Zheng Y. The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP-hydrolysis reactions of Rho family GTPases. J Biol Chem. 2000;275:25299–25307. doi: 10.1074/jbc.M001027200. [DOI] [PubMed] [Google Scholar]
- 62.Zheng Y, Cerione R A, Bender A. Control of the yeast bud-site assembly GTPase Cdc42—catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994;269:2369–2372. [PubMed] [Google Scholar]
- 63.Zheng Y, Hart M, Cerione R A. Guanine nucleotide exchange catalyzed by dbl oncogene product. Methods Enzymol. 1995;256:77–84. doi: 10.1016/0076-6879(95)56011-4. [DOI] [PubMed] [Google Scholar]
- 64.Zheng Y, Olson M, Hall A, Cerione R A, Toksoz D. Direct involvement of the small GTP-binding protein Rho in lbc oncogene function. J Biol Chem. 1995;270:9031–9034. doi: 10.1074/jbc.270.16.9031. [DOI] [PubMed] [Google Scholar]
- 65.Zheng Y, Zangrilli D, Cerione R A, Eva A. The pleckstrin homology domain mediates transformation by oncogenic Dbl through specific intracellular targeting. J Biol Chem. 1996;271:19017–19020. doi: 10.1074/jbc.271.32.19017. [DOI] [PubMed] [Google Scholar]
- 66.Zheng Y, Fischer D J, Santos M F, Tigyi G, Pasteris N G, Gorski J L, Xu Y. The faciogenital dysplasia gene product FGD1 functions as a Cdc42Hs-specific guanine-nucleotide exchange factor. J Biol Chem. 1996;271:33169–33172. doi: 10.1074/jbc.271.52.33169. [DOI] [PubMed] [Google Scholar]
- 67.Zhu K, Debreceni B, Li R, Zheng Y. Identification of Rho GTPase-dependent sites in the DH domain of oncogenic Dbl that are required for transformation. J Biol Chem. 2000;275:25993–26001. doi: 10.1074/jbc.M003780200. [DOI] [PubMed] [Google Scholar]