Abstract
Staphylococcus aureus (S. aureus) is a causative agent of many hospital- and community-acquired infections with the tendency to develop resistance to all known antibiotics. Therefore, the development of novel antistaphylococcal agents is of urgent need. Sortase A is considered a promising molecular target for the development of antistaphylococcal agents. The main aim of this study was to identify novel sortase A inhibitors. In order to find novel antistaphylococcal agents, we performed phenotypic screening of a library containing 15512 compounds against S. aureus ATCC43300. The molecular docking of hits was performed using the DOCK program and 10 compounds were selected for in vitro enzymatic activity inhibition assay. Two inhibitors were identified, N,N-diethyl-N′-(5-nitro-2-(quinazolin-2-yl)phenyl)propane-1,3-diamine (1) and acridin-9-yl-(1H-benzoimidazol-5-yl)-amine (2), which decrease sortase A activity with IC50 values of 160.3 µM and 207.01 µM, respectively. It was found that compounds 1 and 2 possess antibacterial activity toward 29 tested multidrug resistant S. aureus strains with MIC values ranging from 78.12 to 312.5 mg/L. These compounds can be used for further structural optimization and biological research.
Keywords: Staphylococcus aureus, sortase A, molecular docking, inhibitor, antibiotic resistance
1. Introduction
Staphylococcus aureus (S. aureus) is a causative agent of the majority of skin infections, hospital-acquired infections and severe diseases such as bacteremia, sepsis, meningitis, osteomyelitis, endocarditis, and pneumonia [1].
S. aureus belongs to the group of high-priority dangerous “ESKAPE” pathogens, which includes the multidrug resistant isolates of Enterococcus, Staphylococcus, Klebsiella, Acinetobacter, Pseudomonas and Enterobacter that are currently considered as the greatest challenge in medicine. Many hospital-acquired S. aureus isolates have become resistant to most available antibiotics. Staphylococcal resistance to penicillin is mediated by penicillinase which hydrolyses the β-lactam ring of antibiotic. In clinical practice for treatment of staphylococcal infections, methicillin, chemically modified penicillin which cannot be hydrolyzed by penicillinase, has been widely used. At the present time, methicillin-resistant S. aureus (MRSA), which is resistant to all of the β-lactam antibiotics due to modification of penicillin-binding protein, has become one of the most dangerous pathogens. The antibiotic of choice in a case of MRSA-associated infection is vancomycin. However, vancomycin-resistant MRSA strains have emerged recently [2]. It should be noted that no other antibiotic to date has shown any superiority to vancomycin in the treatment of MRSA infections with the possible exception of linezolid in hospital-acquired pneumonia [3]. Therefore, the development of multidrug resistance in S. aureus has caused an urgent need for the search of novel effective targets and corresponding inhibitors to develop principally new antibiotics effective against strains which are resistant to already known antibiotics of choice.
Sortase A is considered as a promising molecular target for the development of antistaphylococcal agents. Sortase A is a membrane-bound transpeptidase which catalyzes the transfer and immobilization of essential virulence factors to the surface of microorganisms. Inhibitors of sortase A affect virulence and biofilm formation, therefore decreasing selective pressure which can cause the development of antibiotic resistance [4]. Sortase A is not presented in eukaryotic organisms, hence the inhibitors of this enzyme may possibly have less toxicity for human organisms. Since this enzyme is membrane-bound, the inhibitors do not have to penetrate into the cell.
To date, small-molecular inhibitors of sortase A have been reported among the derivatives of diarylacrylonitrile [5,6], aryl(β-amino)ethyl ketone [7], rhodanine, pyridazinone, pyrazolethione [8], morpholinobenzoate, aryl 3-acryloamides [9,10], dihydro-β-carboline [11], benzo[d]isothiazol-3(2H)-one-adamantane amine [12], 3,6-disubstituted triazolothiazole [13], 2-phenylbenzofuran-3-carboxamide [14], 2-phenylbenzo[d]oxazole-7-carboxamide [15], 2-(2-phenylhydrazinylidene)alkanoic acid [16], indolethiazolidine [17], pyrrolomycin [18], 2-phenylthiazole [19], 2,5-disubstituted thiadiazole [20], thiadiazolinedione [21], disulfanylbenzamide [22], and 1,2,4-oxadiazole topsentin analogs [23]. Furthermore, several inhibitors for sortase A have been identified among natural products such as β-sitosterol-3-O-glucopyranoside [24], berberine chloride [25], bis(indole)-alkaloid [26], isoaaptamine [27], flavonoids (kurarinol [28], myricetin [29], quercitrin [30], morin [31], eriodictyol [32], acacetin [33], 7-hydroxy-6-methoxy-flavanone and formononetin [34], dryocrassin ABBA [35]), curcumin [36], maltol-3-O-(4′-O-cis-p-cumaroyl-6′-O-(3-hydroxy-3-methylglutaroyl)-β-glucopyranoside [37], skyrin [38], aspermytin A [39], natural naphthoquinones [40], isovitexin [41], coumarines [42], taxifolin [43], erianin [44], quinone [45], chalcone [46], bis(indole) alkaloids [47], orientin [48], gallotannins [49] and peptides from the marine-derived fungi Aspergillus allahabadii [50]. Several review works describe important small organic compounds that act as potent sortase A inhibitors [51,52,53,54]. In the present article, we report two novel inhibitors of S. aureus sortase A belonging to novel chemical classes, the derivatives of acridin-9-yl-(1H-benzoimidazol-5-yl)-amine and 2-phenyl-quinazoline, which possess antimicrobial activity toward multidrug resistant S. aureus strains.
2. Results
In order to find novel antistaphylococcal agents, phenotypic screening of a library containing 15,512 compounds, provided by commercial supplier OTAVA Ltd., was performed by the Community for Open Antimicrobial Drug Discovery (CO-ADD), against methicillin-resistant S aureus strain ATCC43300. As a result, 250 compounds inhibiting growth of MRSA ATCC43300 at least by 30% at the concentration of 32 mg/L were found. The list of active compounds in SMILES format is available in the Supplementary Materials. To identify which compounds can potentially inhibit S. aureus sortase A, we performed molecular docking of 250 compounds into the active site of this enzyme using the DOCK program. According to the results of molecular docking calculations and visual inspection of the best-scored complexes, we selected 10 compounds for in vitro testing. The chemical structure of compounds and their antibacterial activity toward Staphylococcus aureus are presented in Table 1.
Table 1.
Structures and antibacterial activity (percentage growth inhibition) against S. aureus MRSA ATCC43300 for compounds 1–10 at 32 mg/L.
| No. | Structure | Concentration (μM) | Antibacterial Activity on S. aureus MRSA ATCC43300 (%) |
|---|---|---|---|
| 1 |
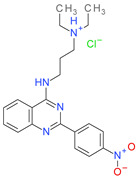
|
77 | 58.75 |
| 2 |
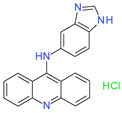
|
92 | 99.57 |
| 3 |
|
118 | 65.25 |
| 4 |
|
98 | 98.51 |
| 5 |
|
103 | 51.2 |
| 6 |
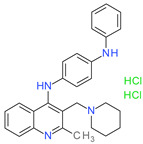
|
65 | 38.98 |
| 7 |
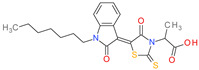
|
74 | 43.25 |
| 8 |
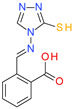
|
129 | 89.6 |
| 9 |
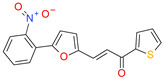
|
98 | 86.65 |
| 10 |

|
75 | 75.5 |
The tested compounds inhibited the growth of the methicillin-resistant S. aureus strain ATCC43300 with inhibition percentages ranging from 38.98 up to 99.57%. Compounds 2 and 4 almost completely inhibited the bacterial growth after exposure at 32 mg/L and could be used to develop new antibacterial substances.
2.1. Sortase A Activity Assay
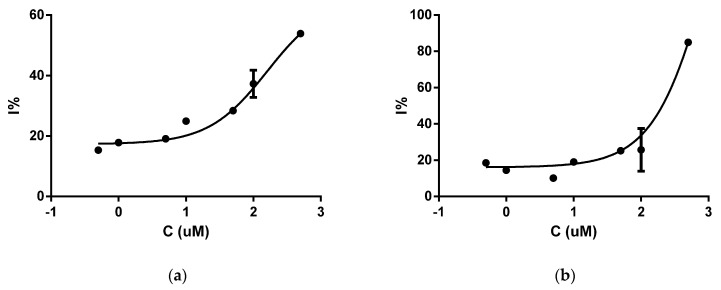
Among these ten investigated compounds, using in vitro sortase A activity assay we found two inhibitors of S. aureus sortase A, N,N-diethyl-N′-(5-nitro-2-(quinazolin-2-yl)phenyl)propane-1,3-diamine (1) and acridin-9-yl-(1H-benzoimidazol-5-yl)-amine (2), which decreased enzyme activity with IC50 values of 160.3 µM and 207.01 µM, respectively (Figure 1). The other eight compounds produced only a small inhibition. Therefore, these compounds possess other molecular mechanisms of action. Possibly, compound 4, with high antibacterial activity, should be investigated for inhibitory activity toward S. aureus DNA gyrase, since according to recent literature data, several indole derivatives demonstrate antistaphylococcal activity targeting this enzyme [55].
Figure 1.
The plotted sortase A inhibition (I%) against compounds logarithm of concentration (μM): (a) compound 1; (b) compound 2.
2.2. Molecular Docking
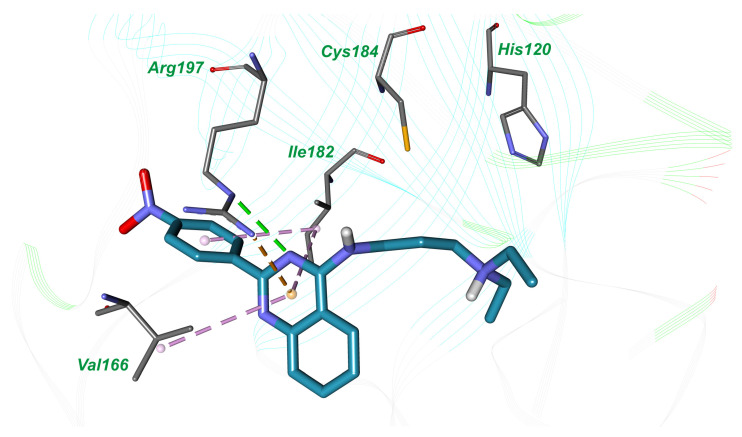
According to molecular docking results, the compound N,N-diethyl-N′-(5-nitro-2-(quinazolin-2-yl)phenyl)propane-1,3-diamine (1) is involved in hydrophobic interactions with amino acid residues Val166 and Ile182 in the active site of sortase A. Furthermore, the nitrogen atom of the quinazolin ring forms a hydrogen bond with conserved amino acid residue Arg197, which is important for catalysis and belongs to catalytic triad (His120, Cys184, and Arg197). Quinazolin heterocycle is also implicated in π-cation interaction with Arg197 (Figure 2).
Figure 2.
The binding mode of compound 1 in the active site of S. aureus sortase A; hydrogen bond is shown by the green dotted line, hydrophobic interactions are indicated by the magenta dotted lines and π-cation interaction is presented by orange dotted line.
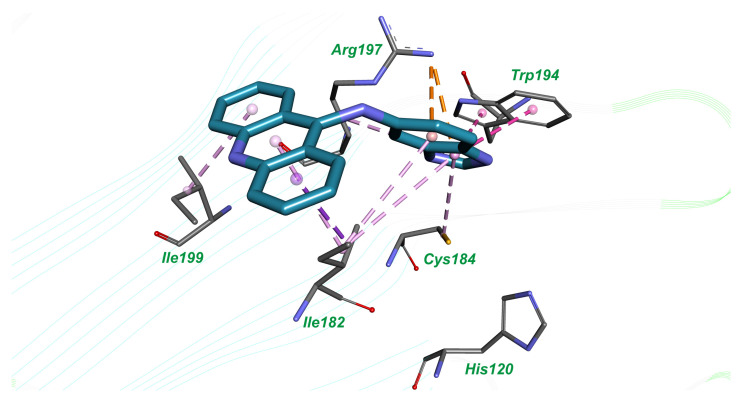
The inhibitor acridin-9-yl-(1H-benzoimidazol-5-yl)-amine is involved in hydrophobic interactions with amino acid residues Ile199, Ile182, Cys184, Trp194 and benzoimidazol heterocycle forms π-cation interactions with Arg197 (Figure 3).
Figure 3.
The binding mode of compound 2 in the active site of S. aureus sortase A; hydrogen bond is shown by the green dotted line, hydrophobic interactions are indicated by the magenta dotted lines and π-cation interactions are presented by the orange dotted lines.
2.3. Antibacterial Assay
The compounds 1 and 2 were extensively screened for antibacterial activity toward multidrug resistant S. aureus strains isolated in Ukrainian hospitals. The minimum inhibitory concentration (MIC) values for compounds 1 and 2 toward S. aureus isolates are presented in the Table 2.
Table 2.
The antimicrobial activity of compounds 1 and 2 toward multidrug resistant S. aureus strains.
| S. aureus Strains | MIC (mg/L) | |
|---|---|---|
| Compound 1 | Compound 2 | |
| S. aureus 29213 | 78.12 | 156.2 |
| MR 433 | 78.12 | 156.2 |
| S. aureus 854 | 156.2 | 312.5 |
| S. aureus 887 | 156.2 | 156.2 |
| S. aureus 890 | 156.2 | 312.5 |
| S. aureus 892 | 156.2 | 312.5 |
| S. aureus 938 | 156.2 | 156.2 |
| S. aureus 964 | 312.5 | 156.2 |
| S. aureus 997 | 312.5 | 156.2 |
| S. aureus 1012 | 78.12 | 156.2 |
| S. aureus 1013 | 312.5 | 156.2 |
| S. aureus 1021 | 156.2 | 156.2 |
| S. aureus 584 | 156.2 | 156.2 |
| S. aureus 585 | 156.2 | 156.2 |
| S. aureus 586 | 156.2 | 156.2 |
| S. aureus 522 | 156.2 | 156.2 |
| S. aureus 523 | 156.2 | 156.2 |
| S. aureus 524 | 156.2 | 156.2 |
| S. aureus 501 | 78.12 | 156.2 |
| S. aureus 502 | 78.12 | 156.2 |
| S. aureus 503 | 156.2 | 156.2 |
| S. aureus 504 | 78.12 | 156.2 |
| S. aureus 505 | 156.2 | 156.2 |
| S. aureus 506 | 78.12 | 156.2 |
| S. aureus 507 | 156.2 | 156.2 |
| S. aureus 508 | 156.2 | 156.2 |
| S. aureus 509 | 156.2 | 156.2 |
| S. aureus 510 | 78.12 | 156.2 |
| S. aureus 511 | 78.12 | 156.2 |
The MIC values ranged between 78.12 and 312.5 mg/L for compound 1 and in the range of 156.2 and 312.5 mg/L for compound 2. Compound 1 was more active than compound 2 on 12 of the 29 bacterial strains, while compound 2 had a higher antimicrobial effect on 3 strains. This observation is correlated with the higher inhibitory effect of compound 1 on the bacterial sortase.
The bacterial strains were investigated for sensitivity to antibiotics by the disco diffusion method using Mueller–Hinton Agar. The visual analysis of antibiotic sensitivity was performed according to EUCAST (European Committee on Antimicrobial Susceptibility Testing) recommendations [56] and the results are presented in Table 3.
Table 3.
The sensitivity of S. aureus strains to antibiotics.
| Strain | Penicillin | Oxacillin | Erythromycin | Clindamycin | Gentamicin | Chloramphenicol | Ciprofloxacin | Cefazolin | Azithromycin | Vancomycin | Linezolid | Teicoplanin | Moxifloxacin |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus 854 | R | IR | IR | S | IR | S | S | IR | IR | R | S | IR | S |
| S. aureus 887 | R | IR | S | S | IR | S | S | IR | IR | IR | S | IR | S |
| S. aureus 890 | R | R | R | R | R | R | IR | R | R | R | S | IR | IR |
| S. aureus 892 | R | IR | S | S | IR | S | IR | S | IR | IR | S | IR | S |
| S. aureus 938 | R | IR | S | S | IR | S | S | IR | IR | IR | S | IR | S |
| S. aureus 964 | R | R | R | R | IR | IR | IR | S | R | IR | S | IR | IR |
| S. aureus 997 | R | S | R | R | IR | IR | IR | S | IR | R | R | R | IR |
| S. aureus 1012 | S | S | S | S | IR | S | S | IR | IR | R | S | IR | S |
| S. aureus 1013 | R | S | IR | IR | IR | R | S | S | IR | IR | S | IR | S |
| S. aureus 1021 | R | S | S | S | IR | IR | IR | IR | IR | IR | IR | IR | S |
| S. aureus 584 | R | IR | R | S | IR | S | S | IR | IR | IR | S | S | S |
| S. aureus 585 | R | R | IR | S | R | S | R | IR | R | IR | S | IR | IR |
| S. aureus 586 | R | R | IR | S | R | R | R | R | IR | IR | S | IR | IR |
| S. aureus 522 | R | R | IR | IR | R | R | R | IR | IR | IR | S | IR | IR |
| S. aureus 523 | R | R | IR | IR | R | R | R | IR | IR | IR | S | IR | IR |
| S. aureus 524 | R | IR | R | R | IR | S | IR | IR | R | S | S | S | S |
| S. aureus 501 | R | R | R | R | IR | S | IR | S | R | S | S | IR | S |
| S. aureus 502 | R | R | R | R | IR | S | IR | S | IR | IR | S | IR | S |
| S. aureus 503 | R | R | R | IR | R | S | IR | S | R | R | S | IR | S |
| S. aureus 504 | R | R | IR | IR | IR | S | S | S | R | IR | S | IR | S |
| S. aureus 505 | R | R | R | R | IR | S | IR | S | R | IR | S | IR | S |
| S. aureus 506 | R | R | R | IR | IR | S | IR | S | R | IR | S | S | S |
| S. aureus 507 | R | R | R | R | IR | S | IR | S | R | S | S | S | S |
| S. aureus 508 | R | R | R | IR | IR | S | IR | S | R | IR | S | IR | S |
| S. aureus 509 | R | R | R | S | R | S | S | S | R | IR | S | IR | S |
| S. aureus 510 | R | R | R | IR | IR | S | S | IR | R | IR | S | IR | S |
| S. aureus 511 | R | IR | R | S | R | S | IR | IR | R | IR | S | IR | S |
R—resistance; IR—intermediate resistance; S—susceptibility.
3. Discussion
Using molecular docking techniques, we identified two novel inhibitors of S. aureus sortase A, N,N-diethyl-N′-(5-nitro-2-(quinazolin-2-yl)phenyl)propane-1,3-diamine (1), and acridin-9-yl-(1H-benzoimidazol-5-yl)-amine (2), which possess moderate enzyme inhibitory activity with IC50 values of 160.3 µM and 207.01 µM, respectively. When comparing the binding mode of highly potent benzisothiazolinone-based inhibitor in the crystal structure of S. aureus sortase A (PDB ID: 2MLM) [12], which was used for molecular docking, and docked complexes of compounds 1 (Figure 1) and 2 (Figure 2), it can be concluded that the simultaneous formation of a hydrogen bond with conservative Arg197 and tight hydrophobic interactions with Trp194, which were established in the co-crystal structure, can be important for inhibitory potency. It should be noted that in our study, compounds 1 and 2 form only one from these two intermolecular interactions—compound 1 builds hydrogen bond with Arg197 and compound 2 forms hydrophobic interactions with Trp194. Possibly, further chemical optimization of compounds 1 and 2 should be performed in order to reach both these types of ligand–receptor interactions.
Antibacterial studies revealed that compounds 1 and 2 inhibit growth of a number of tested multidrug resistant S. aureus strains with MIC values in the range from 78.12 to 312.5 mg/L (Table 2). The antibacterial activity of sortase A inhibitors is low in comparison to known antistaphylococcal antibiotics such as vancomycin and linezolid, which have MIC values about 1 mg/L. Taking into account high levels of resistance to standard antibiotics, the development of antibiotics with novel mechanisms of action is of urgent need.
As it can be seen from the Table 2, compound 1 is more profitable than compound 2 and reveals the antimicrobial activity toward S. aureus strains which have different profiles of antibiotic sensitivity with MIC values in the range from 78.12 to 312.5 mg/L, while compound 2 demonstrates antimicrobial activity with MIC values in the range from 156.2 to 312.5 mg/L (Table 3).
Compound 1 possess the highest antibacterial activity with MIC value of 78.12 mg/L toward multidrug resistant S. aureus strains 1012, 501, 502, 504, 506, 510, 511. All these strains have susceptibility to chloramphenicol, moxifloxacin and linezolid. Vice versa, it was revealed that the compound 1 has the lowest antibacterial activity toward S. aureus isolates 964, 997 and 1013 which all have resistance to chloramphenicol; two of them (964, 997) have resistance to moxifloxacin and one of them (997) has resistance to linezolid. Compound 2 has antibacterial activity toward tested S. aureus strains, mostly with a MIC value of 156.2 mg/L, except three isolates, 854, 890 and 892, which all are susceptible to linezolid. Therefore, compounds 1 and 2 possess different effectiveness toward S. aureus strains with various antibiotic resistance profiles and can be useful for further optimization and development of novel lead compounds with antibacterial activity toward multidrug resistant S. aureus strains.
4. Materials and Methods
4.1. Sortase A Activity Assay
The inhibitory activity of compounds was determined by quantifying the fluorescence intensity upon 5-FAM/QXL® substrate cleavage using the SensoLyte® 520 Sortase A Activity Assay Kit (Anaspec, San Jose, CA, USA). The compounds were dissolved in dimethyl sulfoxide (DMSO) and diluted with distilled water until the concentration of DMSO was 1% and the solution’s intrinsic fluorescence was checked. Each compound was tested at 7 concentration levels in the range of 100–0.1 µM. According to kit protocol, the assay was performed in a 96-well plate, each well containing 10 µL test compound solution, 40 µL enzyme solution and 50 µL substrate solution. We used as controls the enzyme, a 1% DMSO solution, the substrate solution, and 4-hydroxymercuribenzoic acid (HMB) as the positive control. The enzymatic assay was performed for 60 min at room temperature and analyzed fluorometrically (SpectraMAX Gemini XS, San Jose, CA, USA) at Ex/Em = 490 nm/520 nm. All reported values are the means of duplicate experiments.
4.2. Molecular Docking
The molecular docking was carried out with DOCK program [57,58,59,60]. As a receptor we used crystal structure of S. aureus sortase A with PDB ID: 2MLM [12]. The geometry of ligands was calculated using YFF force field [61]. The hydrogen atoms were added with Open Babel v 2.4.0 [62]. Partial atomic charges of the ligands were added with Kirchhoff method [63].
Docking parameters were set as described earlier [64] with several modifications. In our experiments, as the active site atoms we selected the atoms of amino acid residues within 10 Å from the reference ligand—benzo[d]isothiazol-3-one. The spheres in the active site for semi-flexible ligand docking were set with DOCK sphgen software. Grid maps were calculated using Grid program, with grid spacing 0.3 Å. Proteins were represented by the all atom model. We used ‘multiple anchors’ parameter for virtual screening, the minimum of heavy atoms in the anchor was set to 6, and the maximum number of orientations was 1000.
Visual inspection of the complexes of compounds with sortase A was performed using Discovery Studio Visualizer 4.0 [65].
4.3. Antibacterial Assay
All bacteria were cultured in Cation-adjusted Mueller Hinton broth (CAMHB) at 37 °C overnight. A sample of each culture was then diluted 40-fold in fresh broth and incubated at 37 °C for 1.5–3 h. The resultant mid-log phase cultures were diluted (CFU/mL measured by OD600), then added to each well of the compound-containing plates, giving a cell density of 5 × 105 CFU/mL and a total volume of 50 μL. All the plates were covered and incubated at 37 °C for 18 h without shaking.
Growth inhibition of all bacteria was determined measuring absorbance at 600 nm (OD600), using a Tecan M1000 Pro monochromator plate reader. The percentage of growth inhibition was calculated for each well, using the negative control (media only) and positive control (bacteria without inhibitors) on the same plate as references. The growth rates for bacteria had a variation of ±10%, which is within the reported normal distribution of bacterial growth.
The MIC was determined as the lowest concentration at which the growth was fully inhibited, defined by an inhibition ≥80%. In addition, the maximal percentage of growth inhibition is reported as DMax, indicating any compounds with partial activity.
Acknowledgments
Antimicrobial screening toward methicillin-resistant Staphylococcus aureus strain ATCC43300 was performed by CO-ADD (The Community for Open Antimicrobial Drug Discovery), funded by the Wellcome Trust (UK) and The University of Queensland (Australia).
Supplementary Materials
The following are available online, list of active compounds in SMILES format.
Author Contributions
Conceptualization, V.B.; Methodology, G.V., H.V., G.N., G.M.N., A.U., D.M., O.M., V.B., I.K., O.I., S.T. and S.Y.; Supervision, S.Y.; Writing—original draft, G.V., H.V., G.N., G.M.N., A.U., D.M., O.M., V.B., I.K., O.I., S.T. and S.Y.; Writing—review and editing, I.K. and S.Y. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by the authors. Antimicrobial screening toward methicillin-resistant Staphylococcus aureus strain ATCC43300 was performed by CO-ADD (The Community for Open Antimicrobial Drug Discovery), funded by the Wellcome Trust (UK) and The University of Queensland (Australia).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds 1–10 are available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jenkins A., An Diep B., Mai T.T., Vo N.H., Warrener P., Suzich J., Kendall Stover C., Sellman B.R. Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. mBio. 2015;6:e02272-14. doi: 10.1128/mBio.02272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes N.E., Tong S.Y.C., Davis J.S., Hal S.J.V. Treatment of methicillin-resistant staphylococcus aureus: Vancomycin and beyond. Semin. Respir. Crit. Care Med. 2015;36:17–30. doi: 10.1055/s-0034-1397040. [DOI] [PubMed] [Google Scholar]
- 3.Cong Y., Yang S., Rao X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. J. Adv. Res. 2020;21:169–176. doi: 10.1016/j.jare.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nitulescu G., Margina D., Zanfirescu A., Olaru O.T., Nitulescu G.M. Targeting bacterial sortases in search of anti-virulence therapies with low risk of resistance development. Pharmaceuticals. 2021;14:415. doi: 10.3390/ph14050415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh K.B., Oh M.N., Kim J.G., Shin D.S., Shin J. Inhibition of sortase-mediated Staphylococcus aureus adhesion to fibronectin via fibronectin-binding protein by sortase inhibitors. Appl. Microbiol. Biotechnol. 2006;70:102–106. doi: 10.1007/s00253-005-0040-8. [DOI] [PubMed] [Google Scholar]
- 6.Oh K.B., Kim S.H., Lee J., Cho W.J., Lee T., Kim S. Discovery of Diarylacrylonitriles as a Novel Series of Small Molecule Sortase A Inhibitors. J. Med. Chem. 2004;47:2418–2421. doi: 10.1021/jm0498708. [DOI] [PubMed] [Google Scholar]
- 7.Maresso A.W., Wu R., Kern J.W., Zhang R., Janik D., Missiakas D.M., Duban M.E., Joachimiak A., Schneewind O. Activation of inhibitors by sortase triggers irreversible modification of the active site. J. Biol. Chem. 2007;282:23129–23139. doi: 10.1074/jbc.M701857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suree N., Yi S.W., Thieu W., Marohn M., Damoiseaux R., Chan A., Jung M.E., Clubb R.T. Discovery and structure–activity relationship analysis of Staphylococcus aureus sortase A inhibitors. Bioorg. Med. Chem. 2009;17:7174–7185. doi: 10.1016/j.bmc.2009.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chenna B.C., King J.R., Shinkre B.A., Glover A.L., Lucius A.L., Velu S.E. Synthesis and structure activity relationship studies of novel Staphylococcus aureus Sortase A inhibitors. Eur. J. Med. Chem. 2010;45:3752–3761. doi: 10.1016/j.ejmech.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chenna B.C., Shinkre B.A., King J.R., Lucius A.L., Narayana S.V.L., Velu S.E. Identification of novel inhibitors of bacterial surface enzyme Staphylococcus aureus Sortase A. Bioorg. Med. Chem. Lett. 2008;18:380–385. doi: 10.1016/j.bmcl.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 11.Lee H.S., Yoon K.M., Han Y.R., Lee K.J., Chung S.C., Kim T.I., Lee S.H., Shin J., Oh K.B. 5-Hydroxyindole-type alkaloids, as Candida albicans isocitrate lyase inhibitors, from the tropical sponge Hyrtios sp. Bioorg. Med. Chem. Lett. 2009;19:1051–1053. doi: 10.1016/j.bmcl.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Zhulenkovs D., Rudevica Z., Jaudzems K., Turks M., Leonchiks A. Discovery and structure–activity relationship studies of irreversible benzisothiazolinone-based inhibitors against Staphylococcus aureus sortase A transpeptidase. Bioorg. Med. Chem. 2014;22:5988–6003. doi: 10.1016/j.bmc.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J., Liu H., Zhu K., Gong S., Dramsi S., Wang Y.-T., Li J., Chen F., Zhang R., Zhou L., et al. Antiinfective therapy with a small molecule inhibitor of Staphylococcus aureus sortase. Proc. Natl. Acad. Sci. USA. 2014;111:13517–13522. doi: 10.1073/pnas.1408601111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He W., Zhang Y., Bao J., Deng X., Batara J., Casey S., Guo Q., Jiang F., Fu L. Synthesis, biological evaluation and molecular docking analysis of 2-phenyl-benzofuran-3-carboxamide derivatives as potential inhibitors of Staphylococcus aureus Sortase A. Bioorg. Med. Chem. 2017;25:1341–1351. doi: 10.1016/j.bmc.2016.12.030. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Bao J., Deng X.X., He W., Fan J.J., Jiang F.Q., Fu L. Synthesis, biological evaluation and molecular docking of 2-phenyl-benzo[d]oxazole-7-carboxamide derivatives as potential Staphylococcus aureus Sortase A inhibitors. Bioorg. Med. Chem. Lett. 2016;26:4081–4085. doi: 10.1016/j.bmcl.2016.06.074. [DOI] [PubMed] [Google Scholar]
- 16.Maggio B., Raffa D., Raimondi M.V., Cascioferro S., Plescia F., Schillaci D., Cusimano M.G., Leonchiks A., Zhulenkovs D., Basile L., et al. Discovery of a new class of sortase a transpeptidase inhibitors to tackle gram-positive pathogens: 2-(2-phenylhydrazinylidene)alkanoic acids and related derivatives. Molecules. 2016;21:241. doi: 10.3390/molecules21020241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raj K.K., Ganesh Kumar V., Leela Madhuri C., Pardhasaradhi M., Durga Lakshmi R., Ravi M., Sri Ramudu B., Venkata Rao S.V., Ramachandran D. Designing of potential inhibitors against Staphylococcus aureus sortase A: Combined analogue and structure based approach with in vitro validation. J. Mol. Graph. Model. 2015;60:89–97. doi: 10.1016/j.jmgm.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Raimondi M.V., Listro R., Cusimano M.G., La Franca M., Faddetta T., Gallo G., Schillaci D., Collina S., Leonchiks A., Barone G. Pyrrolomycins as antimicrobial agents. Microwave-assisted organic synthesis and insights into their antimicrobial mechanism of action. Bioorg. Med. Chem. 2019;27:721–728. doi: 10.1016/j.bmc.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Oniga S., Araniciu C., Palage M., Popa M., Chifiriuc M.-C., Marc G., Pirnau A., Stoica C., Lagoudis I., Dragoumis T., et al. New 2-Phenylthiazoles as Potential Sortase A Inhibitors: Synthesis, Biological Evaluation and Molecular Docking. Molecules. 2017;22:1827. doi: 10.3390/molecules22111827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wehrli P.M., Uzelac I., Olsson T., Jacso T., Tietze D., Gottfries J. Discovery and development of substituted thiadiazoles as inhibitors of Staphylococcus aureus Sortase A. Bioorg. Med. Chem. 2019;27:115043. doi: 10.1016/j.bmc.2019.115043. [DOI] [PubMed] [Google Scholar]
- 21.Yang T., Zhang T., Guan X.N., Dong Z., Lan L., Yang S., Yang C.G. Tideglusib and Its Analogues As Inhibitors of Staphylococcus aureus SrtA. J. Med. Chem. 2020;63:8442–8457. doi: 10.1021/acs.jmedchem.0c00803. [DOI] [PubMed] [Google Scholar]
- 22.Barthels F., Marincola G., Marciniak T., Konhäuser M., Hammerschmidt S., Bierlmeier J., Distler U., Wich P.R., Tenzer S., Schwarzer D., et al. Asymmetric Disulfanylbenzamides as Irreversible and Selective Inhibitors of Staphylococcus aureus Sortase A. ChemMedChem. 2020;15:839–850. doi: 10.1002/cmdc.201900687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parrino B., Carbone D., Cascioferro S., Pecoraro C., Giovannetti E., Deng D., Di Sarno V., Musella S., Auriemma G., Cusimano M.G., et al. 1,2,4-Oxadiazole topsentin analogs as staphylococcal biofilm inhibitors targeting the bacterial transpeptidase sortase A. Eur. J. Med. Chem. 2021;209:112892. doi: 10.1016/j.ejmech.2020.112892. [DOI] [PubMed] [Google Scholar]
- 24.Kim S.H., Shin D.S., Oh M.N., Chung S.C., Lee J.S., Chang I.M., Oh K.B. Inhibition of Sortase, a Bacterial Surface Protein Anchoring Transpeptidase, by β-Sitosterol-3- O -glucopyranoside from Fritillaria verticillata. Biosci. Biotechnol. Biochem. 2003;67:2477–2479. doi: 10.1271/bbb.67.2477. [DOI] [PubMed] [Google Scholar]
- 25.Kim S.H., Shin D.S., Oh M.N., Chung S.C., Lee J.S., Oh K.B. Inhibition of the Bacterial Surface Protein Anchoring Transpeptidase Sortase by Isoquinoline Alkaloids. Biosci. Biotechnol. Biochem. 2004;68:421–424. doi: 10.1271/bbb.68.421. [DOI] [PubMed] [Google Scholar]
- 26.Oh K.B., Mar W., Kim S., Kim J.Y., Oh M.N., Kim J.G., Shin D., Sim C.J., Shin J. Bis(indole) alkaloids as sortase A inhibitors from the sponge Spongosorites sp. Bioorg. Med. Chem. Lett. 2005;15:4927–4931. doi: 10.1016/j.bmcl.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 27.Jang K.H., Chung S.C., Shin J., Lee S.H., Kim T.I., Lee H.S., Oh K.B. Aaptamines as sortase A inhibitors from the tropical sponge Aaptosaaptos. Bioorganic Med. Chem. Lett. 2007;17:5366–5369. doi: 10.1016/j.bmcl.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Oh I., Yang W.Y., Chung S.C., Kim T.Y., Oh K.B., Shin J. In vitro sortase A inhibitory and antimicrobial activity of flavonoids isolated from the roots of Sophora flavescens. Arch. Pharm. Res. 2011;34:217–222. doi: 10.1007/s12272-011-0206-0. [DOI] [PubMed] [Google Scholar]
- 29.Nitulescu G., Nicorescu I., Olaru O., Ungurianu A., Mihai D., Zanfirescu A., Nitulescu G., Margina D. Molecular Docking and Screening Studies of New Natural Sortase A Inhibitors. Int. J. Mol. Sci. 2017;18:2217. doi: 10.3390/ijms18102217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B., Chen F., Bi C., Wang L., Zhong X., Cai H., Deng X., Niu X., Wang D. Quercitrin, an inhibitor of sortase A, interferes with the adhesion of Staphylococcal aureus. Molecules. 2015;20:6533–6543. doi: 10.3390/molecules20046533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang P., Hu P., Zhou S.Y., Li Q., Chen W.M. Morin inhibits sortase A and subsequent biofilm formation in Streptococcus mutans. Curr. Microbiol. 2014;68:47–52. doi: 10.1007/s00284-013-0439-x. [DOI] [PubMed] [Google Scholar]
- 32.Wang L., Li Q., Li J., Jing S., Jin Y., Yang L., Yu H., Wang D., Wang T., Wang L. Eriodictyol as a Potential Candidate Inhibitor of Sortase A Protects Mice From Methicillin-Resistant Staphylococcus aureus-Induced Pneumonia. Front. Microbiol. 2021;12:268. doi: 10.3389/fmicb.2021.635710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bi C., Dong X., Zhong X., Cai H., Wang D., Wang L. Acacetin protects mice from Staphylococcus aureus bloodstream infection by inhibiting the activity of sortase A. Molecules. 2016;21:1285. doi: 10.3390/molecules21101285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho H., Chung B., Kim C.K., Oh D.C., Oh K.B., Shin J. Spatholobus suberectus Dunn. constituents inhibit sortase A and Staphylococcus aureus cell clumping to fibrinogen. Arch. Pharm. Res. 2017;40:518–523. doi: 10.1007/s12272-016-0884-8. [DOI] [PubMed] [Google Scholar]
- 35.Zhang B., Wang X., Wang L., Chen S., Shi D., Wang H. Molecular Mechanism of the Flavonoid Natural Product Dryocrassin ABBA against Staphylococcus aureus Sortase A. Molecules. 2016;21:1428. doi: 10.3390/molecules21111428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park B.S., Kim J.G., Kim M.R., Lee S.E., Takeoka G.R., Oh K.B., Kim J.H. Curcuma longa L. constituents inhibit sortase A and Staphylococcus aureus cell adhesion to fibronectin. J. Agric. Food Chem. 2005;53:9005–9009. doi: 10.1021/jf051765z. [DOI] [PubMed] [Google Scholar]
- 37.Yang W.Y., Won T.H., Ahn C.H., Lee S.H., Yang H.C., Shin J., Oh K.B. Streptococcus mutans sortase A inhibitory metabolites from the flowers of Sophora japonica. Bioorg. Med. Chem. Lett. 2015;25:1394–1397. doi: 10.1016/j.bmcl.2015.02.051. [DOI] [PubMed] [Google Scholar]
- 38.Thappeta K.R.V., Zhao L.N., Nge C.E., Crasta S., Leong C.Y., Ng V., Kanagasundaram Y., Fan H., Ng S.B. In-silico identified new natural sortase a inhibitors disrupt S. aureus biofilm formation. Int. J. Mol. Sci. 2020;21:8601. doi: 10.3390/ijms21228601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park S.C., Chung B., Lee J., Cho E., Hwang J.-Y., Oh D.-C., Shin J., Oh K.-B. Sortase A-Inhibitory Metabolites from a Marine-Derived Fungus Aspergillus sp. Mar. Drugs. 2020;18:359. doi: 10.3390/md18070359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nitulescu G., Mihai D.P., Nicorescu I.M., Olaru O.T., Ungurianu A., Zanfirescu A., Nitulescu G.M., Margina D. Discovery of natural naphthoquinones as sortase A inhibitors and potential anti-infective solutions against Staphylococcus aureus. Drug Dev. Res. 2019;80:1136–1145. doi: 10.1002/ddr.21599. [DOI] [PubMed] [Google Scholar]
- 41.Mu D., Xiang H., Dong H., Wang D., Wang T. Isovitexin, a potential candidate inhibitor of sortase a of Staphylococcus aureus USA300. J. Microbiol. Biotechnol. 2018;28:1426–1432. doi: 10.4014/jmb.1802.02014. [DOI] [PubMed] [Google Scholar]
- 42.Park J.S., Chung B., Lee W.H., Lee J., Suh Y., Oh D.C., Oh K.B., Shin J. Sortase A-Inhibitory Coumarins from the Folk Medicinal Plant Poncirus trifoliata. J. Nat. Prod. 2020;83:3004–3011. doi: 10.1021/acs.jnatprod.0c00551. [DOI] [PubMed] [Google Scholar]
- 43.Wang L., Wang G., Qu H., Wang K., Jing S., Guan S., Su L., Li Q., Wang D. Taxifolin, an Inhibitor of Sortase A, Interferes With the Adhesion of Methicillin-Resistant Staphylococcal aureus. Front. Microbiol. 2021;12:1876. doi: 10.3389/fmicb.2021.686864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouyang P., He X., Yuan Z.-W., Yin Z.-Q., Fu H., Lin J., He C., Liang X., Lv C., Shu G., et al. Erianin against Staphylococcus aureus Infection via Inhibiting Sortase A. Toxins. 2018;10:385. doi: 10.3390/toxins10100385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou X., Wang M., Wen Y., Ni T., Guan X., Lan L., Zhang N., Zhang A., Yang C.-G. Quinone skeleton as a new class of irreversible inhibitors against Staphylococcus aureus sortase A. Bioorg. Med. Chem. Lett. 2018;28:1864–1869. doi: 10.1016/j.bmcl.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Zhang B., Teng Z., Li X., Lu G., Deng X., Niu X., Wang J. Chalcone Attenuates Staphylococcus aureus Virulence by Targeting Sortase A and Alpha-Hemolysin. Front. Microbiol. 2017;8:1715. doi: 10.3389/fmicb.2017.01715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park J.S., Cho E., Hwang J.-Y., Park S.C., Chung B., Kwon O.-S., Sim C.J., Oh D.-C., Oh K.-B., Shin J. Bioactive Bis(indole) Alkaloids from a Spongosorites sp. Sponge. Mar. Drugs. 2020;19:3. doi: 10.3390/md19010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L., Jing S., Qu H., Wang K., Jin Y., Ding Y., Yang L., Yu H., Shi Y., Li Q., et al. Orientin mediates protection against MRSA-induced pneumonia by inhibiting Sortase A. Virulence. 2021;12:2149–2161. doi: 10.1080/21505594.2021.1962138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hricovíniová Z., Mascaretti Š., Hricovíniová J., Čížek A., Jampílek J. New Unnatural Gallotannins: A Way toward Green Antioxidants, Antimicrobials and Antibiofilm Agents. Antioxidants. 2021;10:1288. doi: 10.3390/antiox10081288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang J.-Y., Lee J.-H., Park S.C., Lee J., Oh D.-C., Oh K.-B., Shin J. New Peptides from The Marine-Derived Fungi Aspergillus allahabadii and Aspergillus ochraceopetaliformis. Mar. Drugs. 2019;17:488. doi: 10.3390/md17090488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cascioferro S., Raffa D., Maggio B., Raimondi M.V., Schillaci D., Daidone G. Sortase A Inhibitors: Recent Advances and Future Perspectives. J. Med. Chem. 2015;58:9108–9123. doi: 10.1021/acs.jmedchem.5b00779. [DOI] [PubMed] [Google Scholar]
- 52.Guo Y., Cai S., Gu G., Guo Z., Long Z. Recent progress in the development of sortase A inhibitors as novel anti-bacterial virulence agents. RSC Adv. 2015;5:49880–49889. doi: 10.1039/C5RA07568H. [DOI] [Google Scholar]
- 53.Ha M.W., Yi S.W., Paek S.-M. Design and Synthesis of Small Molecules as Potent Staphylococcus aureus Sortase A Inhibitors. Antibiotics. 2020;9:706. doi: 10.3390/antibiotics9100706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alharthi S., Alavi S.E., Moyle P.M., Ziora Z.M. Sortase A (SrtA) inhibitors as an alternative treatment for superbug infections. Drug Discov. Today. 2021;26:2164–2172. doi: 10.1016/j.drudis.2021.03.019. [DOI] [PubMed] [Google Scholar]
- 55.Liu H., Chu Z.-W., Xia D.-G., Cao H.-Q., Lv X.-H. Discovery of novel multi-substituted benzo-indole pyrazole schiff base derivatives with antibacterial activity targeting DNA gyrase. Bioorg. Chem. 2020;99:103807. doi: 10.1016/j.bioorg.2020.103807. [DOI] [PubMed] [Google Scholar]
- 56.EUCAST . EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance, Version 1.0, December 2013. EUCAST; Växjö, Sweden: 2013. [Google Scholar]
- 57.Ewing T.J.A., Makino S., Skillman A.G., Kuntz I.D. DOCK 4.0: Search strategies for automated molecular docking of flexible molecule databases. J. Comput.-Aided Mol. Des. 2001;15:411–428. doi: 10.1023/A:1011115820450. [DOI] [PubMed] [Google Scholar]
- 58.Ring C.S., Sun E., McKerrow J.H., Lee G.K., Rosenthal P.J., Kuntz I.D., Cohen F.E. Structure-based inhibitor design by using protein models for the development of antiparasitic agents. Proc. Natl. Acad. Sci. USA. 1993;90:3583–3587. doi: 10.1073/pnas.90.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bodian D.L., Yamasaki R.B., Buswell R.L., Stearns J.F., White J.M., Kuntz I.D. Inhibition of the Fusion-Inducing Conformational Change of Influenza Hemagglutinin by Benzoquinones and Hydroquinones. Biochemistry. 1993;32:2967–2978. doi: 10.1021/bi00063a007. [DOI] [PubMed] [Google Scholar]
- 60.Shoichet B.K., Stroud R.M., Santi D.V., Kuntz I.D., Perry K.M. Structure-based discovery of inhibitors of thymidylate synthase. Science. 1993;259:1445–1450. doi: 10.1126/science.8451640. [DOI] [PubMed] [Google Scholar]
- 61.Yakovenko O.Y., Oliferenko A.A., Golub A.G., Bdzhola V.G., Yarmoluk S.M. The new method of distribution integrals evaluations for high throughput virtual screening. Ukr. Bioorg. Acta. 2007;1:52–62. [Google Scholar]
- 62.O’Boyle N., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allouche A.-R. Gabedit-A graphical user interface for computational chemistry softwares. J. Comput. Chem. 2011;32:174–182. doi: 10.1002/jcc.21600. [DOI] [PubMed] [Google Scholar]
- 64.Bursulaya B.D., Totrov M., Abagyan R., Brooks C.L. Comparative study of several algorithms for flexible ligand docking. J. Comput.-Aided Mol. Des. 2003;17:755–763. doi: 10.1023/B:JCAM.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- 65.Accelrys Discovery Studio Visualizer 4.0. Accelrys; San Diego, CA, USA: 2012. [(accessed on 20 November 2021)]. Available online: http://www.accelrys.com. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.