Abstract
AmpC beta-lactamases are cephalosporinases that confer resistance to a wide variety of β-lactam drugs and that may thereby create serious therapeutic problems. Although reported with increasing frequency, the true rate of occurrence of AmpC beta-lactamases in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis remains unknown. We tested a total of 1,286 consecutive, nonrepeat isolates of these three species and found that, overall, 45 (3.5%) yielded a cefoxitin zone diameter less than 18 mm (screen positive) and that 16 (1.2%) demonstrated AmpC bands by isoelectric focusing. Based on the species, of 683 E. coli, 371 K. pneumoniae, and 232 P. mirabilis isolates tested, 13 (1.9%), 28 (7.6%), and 4 (1.7%), respectively, demonstrated decreased zone diameters and 11 (1.6%), 4 (1.1%), and 1 (0.4%), respectively, demonstrated AmpC bands. Cefoxitin resistance was transferred for all but 8 (E. coli) of the 16 AmpC producers. We also describe a three-dimensional extract test, which was used to detect phenotypically isolates that harbor AmpC beta-lactamase. Of the 45 cefoxitin-resistant isolates, the three-dimensional extract test accurately identified all 16 AmpC producers and 28 of 29 (97%) isolates as non-AmpC producers. Interestingly, most (86%) isolates in the latter group were K. pneumoniae isolates. These data confirm that, at our institution, E. coli, K. pneumoniae, and P. mirabilis harbor plasmid-mediated AmpC enzymes.
Group 1 AmpC beta-lactamases are cephalosporinases that are poorly inhibited by clavulanic acid (9). They are clinically significant because they may confer resistance to a wide variety of β-lactam drugs, including α-methoxy-β-lactams, such as cefoxitin, narrow-, expanded-, and broad-spectrum cephalosporins, β-lactam–beta-lactamase inhibitor combinations, and aztreonam. Genes for AmpC beta-lactamases are commonly found on the chromosomes of several members of the family Enterobacteriaceae, including Enterobacter, Shigella, Providencia, Citrobacter freundii, Morganella morganii, Serratia marcescens, and Escherichia coli (17). Chromosomal expression is typically inducible except in E. coli and Shigella spp., in which it is usually constitutive and minimal (17, 27). Occasional isolates of E. coli (1 to 2%) (17) may produce large amounts of AmpC enzyme (27) and have a phenotype resembling that of a derepressed AmpC mutant Enterobacter sp. DNA sequencing data for five hyperproducing E. coli isolates showed that the ampC gene was preceded by a strong promoter, which resulted in increased transcription (25). Although chromosomal genes for group 2b beta-lactamases are common in Klebsiella pneumoniae (17), genes for AmpC beta-lactamases are notably absent. The first example of a chromosomally encoded AmpC-type beta-lactamase in Proteus mirabilis was reported only recently (8).
Genes for AmpC beta-lactamases have also recently been found on plasmids that transfer noninducible cephalosporin resistance to K. pneumoniae (5, 7, 13–15, 29, 31, 39; A. Bauernfeind, R. Jungwirth, I. Schneider, H. Sahly, and U. Ullmann, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-2, p. 69, 1998; A. Bauernfeind, S. Schweighart, K. Dornbusch, and H. Giamarellou, Program and Abstr. 30th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 190, p. 118, 1990; S. Boyer-Mariotte, L. Raskine, B. Hanau, A. Philippon, M. J. Sanson-LE Pors, and G. Arlet, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-7, p. 70, 1998), E. coli (3, 19, 30; C. Hoyen, L. B. Rice, and R. A. Bonomo, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-161, p. 115, 1998) and P. mirabilis (6). These enzymes are believed to have originated from the chromosomes of Enterobacter, Citrobacter, and Pseudomonas spp. (9, 35). In a recent survey, K. pneumoniae and E. coli isolates from patients from 8 of 20 intensive care units in the United States harbored transmissible AmpC-type beta-lactamases (G. A. Jacoby, P. Han, M. Alvarez, and F. Tenover, Abstr. 35th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C40, p. 46, 1995). Documentation of these enzymes in seven or more countries in a relatively short time period (since 1989) may portend future problems (4, 21, 20).
Although reported with increasing frequency in case isolates (5, 13–15, 19, 29, 30, 39), the true rate of occurrence of plasmid-mediated AmpC beta-lactamases in K. pneumoniae, E. coli, and P. mirabilis remains unknown. Many laboratories have difficulty detecting these enzymes in clinical isolates. In a recent study, 28 (74%) of 38 laboratories in Connecticut reported at least one nonsusceptible result with an extended-spectrum cephalosporin or aztreonam for an AmpC-producing strain of E. coli that was known to be resistant to these agents (34). These data suggest that the standard systems used in the study failed to detect resistance and that additional testing was not performed. Current National Committee for Clinical Laboratory Standards (NCCLS) guidelines for performing in vitro susceptibility testing (22–24) do not indicate either the phenotypic screening or confirmatory tests that should be used for isolates that harbor AmpC beta-lactamases. For this reason, a study was designed to determine the occurrence of plasmid-mediated AmpC beta-lactamases in K. pneumoniae, E. coli, and P. mirabilis at a veterans medical center. The study also included E. coli isolates that produced high levels of AmpC enzyme due to chromosome-mediated factors. In addition, we report on a phenotypic method for the detection of isolates that harbor these enzymes.
MATERIALS AND METHODS
Tests for AmpC-producing isolates of K. pneumoniae, E. coli, and P. mirabilis.
A total of 1,286 consecutive, nonrepeat E. coli (n = 683), K. pneumoniae (n = 371), and P. mirabilis (n = 232) isolates were recovered at the McGuire Veterans Affairs Medical Center (VAMC) during a 14-month period (November 1995 to January 1997). Isolates were identified with the Vitek and API 20E systems (bioMerieux Vitek, Hazelwood, Mo.) and were tested for susceptibility by the standard disk diffusion method (23). A 30-μg cefoxitin disk (Becton Dickinson Microbiology Systems, Cockeysville, Md.) was placed on inoculated Mueller-Hinton agar (Remel, Lenexa, Kans.). By following the NCCLS criteria for nonsusceptible organisms (24), isolates with zone diameters less than 18 mm were selected for MIC and beta-lactamase testing.
The MICs of ampicillin, cefoxitin, cefotaxime, ceftazidime, aztreonam, cefepime, and imipenem were determined by the standard broth microdilution method (22). The MICs of ceftriaxone and cefpodoxime with and without 2 and 4 μg of clavulanic acid per ml (fixed concentrations) (37), respectively, as well as the MICs of cefoxitin and ceftriaxone in combination with the penicillanic acid sulfone Ro 48-1220 (Hoffmann-La Roche Ltd., Basel, Switzerland), were also determined. Ro 48-1220 is a novel beta-lactamase inhibitor that protects expanded-spectrum cephalosporins against strains that produce group 1 and group 2be enzymes (40). E. coli ATCC 25922 was used as a control strain.
Beta-lactamases.
Isolates were tested for AmpC activity by a three-dimensional extract method, which was an adaptation of procedures described previously for the detection of extended-spectrum beta-lactamases (ESBLs) (36, 41). Briefly, 50 μl of a 0.5 McFarland bacterial suspension prepared from an overnight blood agar plate was inoculated into 12 ml of tryptic soy broth and the culture was grown for 4 h at 35°C. The cells were concentrated by centrifugation, and crude enzyme preparations were made by freezing-thawing the cell pellets five times. The surface of a Mueller-Hinton agar plate (Remel) was inoculated with one each of two E. coli strains (ATCC 25922 and ATCC 11775) as described for the standard disk diffusion method (23); a 30-μg cefoxitin disk was placed on the inoculated agar. With a sterile scalpel blade, a slit beginning 5 mm from the edge of the disk was cut in the agar in an outward radial direction. By using a pipet, 25 to 30 μl of enzyme preparation was dispensed into the slit, beginning near the disk and moving outward. Slit overfill was avoided. The inoculated media were incubated overnight at 35°C. Enhanced growth of the surface organism at the point where the slit intersected the zone of inhibition was considered a positive three-dimensional test result and was interpreted as evidence for the presence of AmpC beta-lactamase (see Fig. 1). To test the extracts of P. mirabilis, MacConkey agar was also used to suppress the (swarming) growth of unlysed cells, which occasionally interfered with interpretation of results. E. coli strains which contained plasmid derivatives of the FOX-1, LAT-2, and MIR-1 AmpC beta-lactamases (Bush group 1) were tested as positive controls.
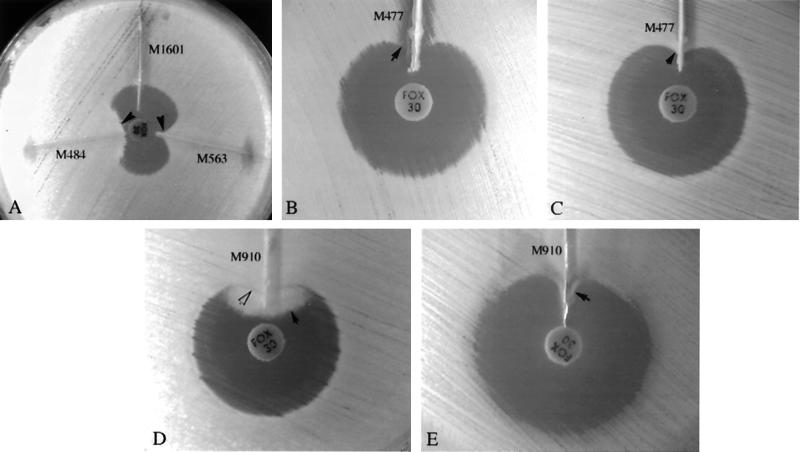
FIG. 1.
Three-dimensional extract test patterns for five isolates. (A) Enhanced growth of the surface organism, E. coli ATCC 25922, is seen near agar slits (arrows) that contain extracts of E. coli (M563) and K. pneumoniae (M484) test isolates, both of which are AmpC producers. The remaining slit contained an extract of a non-AmpC-producing E. coli isolate (M1601). The extract of AmpC-producing E. coli isolate M477 inhibited the growth of one surface organism, E. coli ATCC 25922 (B) (arrow), but did not interfere with the growth of the second surface organism, E. coli ATCC 11775 (C) (arrow). (D) Swarming growth (dark arrow) of unlysed cells in an extract of AmpC-producing P. mirabilis isolate M910 interfered with detection of growth of surface organism (white arrow) when Mueller-Hinton agar was used. (E) On MacConkey agar, growth of P. mirabilis was inhibited, and enhanced growth of the surface organism was easily seen (arrow).
Isolates with decreased cefoxitin zone diameters were tested for the presence of beta-lactamases by isoelectric focusing (IEF) of cell extracts as described previously (10). The cells were grown in 50 ml of tryptic soy broth (Becton Dickinson Microbiology Systems) for 4 h and were washed in 0.1 M phosphate buffer (pH 7). The centrifuged cells were resuspended in 300 μl of phosphate buffer and were frozen at −70°C. The cells were sonicated with a Branson Cell Disruptor 200 (Branson Ultrasonics Corp., Danbury, Conn.) for 10 s and were then cooled with ice for 10 s; this cycle was repeated four times. Cellular debris was removed by centrifugation. The quantities of proteins in the preparations were not determined. Enzyme activity on the focused gels was detected with molten agar containing nitrocefin (50 μg/ml). Filter paper strips moistened with one of two different inhibitors at 1 mM were briefly applied to the focused gel surface prior to the addition of the molten agar (33). AmpC beta-lactamases are inhibited by cloxacillin, and preparations with IEF patterns that demonstrated the loss of a nitrocefin band after application of a cloxacillin-moistened strip were interpreted to contain AmpC enzyme and show AmpC activity by IEF.
All isolates that demonstrated AmpC activity by IEF were tested for the ability to transfer resistance to recipient strains. Each donor strain was tested by filter mating with two or more of the following recipient E. coli strains: CGSC 1867, C600, and 26R793. The selective medium contained 400 μg of sodium azide per ml and 4 μg of aztreonam per ml, 512 μg of nalidixic acid per ml and 25 μg of cefoxitin per ml, or 512 μg of rifampin per ml and either 4 μg of aztreonam per ml or 50 or 75 μg of cefoxitin per ml.
RESULTS
Occurrence of AmpC-producing organisms.
Of the 1,286 isolates that were tested, 45 (3.5%) yielded cefoxitin zone diameters less than 18 mm (screen positive), and 16 of these (1.2%) demonstrated AmpC bands by IEF. Based on the species, of 683 E. coli, 371 K. pneumoniae, and 232 P. mirabilis isolates tested, 13 (1.9%), 28 (7.6%), and 4 (1.7%), respectively, demonstrated decreased zone diameters and 11 (1.6%), 4 (1.1%), and 1 (0.4%), respectively, demonstrated AmpC bands.
All 16 AmpC-producing isolates yielded a positive three-dimensional test result with at least one of the two surface organisms. For most isolates, the growth patterns of both surface organisms were similar and relatively easy to interpret (Fig. 1A). The extract of one E. coli isolate, however, inhibited the growth of one surface organism uniformly along the entire length of the slit (Fig. 1B) and thereby interfered with interpretation of the test result. In contrast, no inhibition was observed along the slit with the second surface organism (Fig. 1C). MacConkey agar markedly suppressed growth from unlysed Proteus cells (Fig. 1D), and its use allowed easier interpretation of test results (Fig. 1E). Positive test results were seen with extracts of the three control strains. An extract of only 1 of the 29 non-AmpC-producing isolates that yielded decreased cefoxitin zone diameters was associated with a positive three-dimensional test result. The extract source was a K. pneumoniae isolate that harbored two ESBLs (of the SHV type). The positive three-dimensional test result was partially reversed when a disk containing clavulanic acid (Augmentin; Becton Dickinson Microbiology Systems) was added to the extract (140 μl) prior to injection into the slit.
Table 1 lists the beta-lactamase isoelectric points and the susceptibilities of the 16 AmpC-producing isolates. Although the MIC patterns for several E. coli and K. pneumoniae isolates were similar, all isolates were unique by typing by pulsed-field gel electrophoresis (data not shown). In addition to the cloxacillin-inhibited group 1 AmpC enzymes, some isolates harbored other beta-lactamases that were inhibited by clavulanic acid but not by cloxacillin. However, the addition of clavulanic acid to cefpodoxime or ceftriaxone resulted in no change in MIC greater than twofold relative to the MIC of the β-lactam alone. In contrast, for 11 of the 16 AmpC producers, the addition of 4 μg of Ro 48-1220 per ml to cefoxitin decreased the MIC at least fourfold relative to the MIC of the drug alone (Table 1). Fourfold or greater differences in MICs were achieved for the remaining five isolates, one E. coli isolate (M752) and four K. pneumoniae isolates, in the presence of higher concentrations of the inhibitor Ro 48-1220 (8 and 32 μg of Ro 48-1220 per ml, respectively) (data not shown). Fourfold or greater differences in MICs were also obtained with ceftriaxone and 4 μg of the inhibitor Ro 48-1220 per ml for the same 11 isolates compared with those of cefoxitin (data not shown).
TABLE 1.
Isoelectric points and susceptibilities of AmpC-producing isolates
| Isolate | pIa | MICb (μg/μl)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | FOX | FOX-Ro(4) | CPD | CPD-Cl(4) | CRO | CRO-Cl(2) | CTX | CAZ | AZT | CEF | IMI | ||
| E. coli | |||||||||||||
| M477 | 9.1 | 256 | 32 | 8 | 128 | 128 | 16 | 16 | 8 | 128 | 16 | 4 | 0.13 |
| M491 | 9.1 | 512 | 64 | 16 | 128 | 128 | 4 | 4 | 8 | 32 | 16 | 0.13 | 0.13 |
| M483 | 9.1 | 512 | 32 | 8 | 512 | 256 | 16 | 16 | 16 | 64 | 16 | 0.5 | 0.13 |
| M540 | 9.0, 5.4 | 4,096 | 128 | 32 | 128 | 128 | 2 | 1 | 4 | 4 | 16 | 0.13 | 0.06 |
| M545B | 9.0 | 512 | 64 | 16 | 128 | 64 | 1 | 1 | 4 | 4 | 16 | 0.06 | 0.06 |
| M563c | 9.1 | 265 | 32 | 2 | 128 | 64 | 1 | 0.5 | 2 | 4 | 8 | 0.03 | 0.13 |
| M656 | 9.1 | 64 | 8 | 2 | 8 | 4 | 0.13 | 0.13 | 0.5 | 0.5 | 1 | 0.06 | 0.13 |
| M683 | 9.0 | 64 | 8 | 2 | 8 | 8 | 0.13 | 0.06 | 0.5 | 0.5 | 1 | 0.02 | 0.06 |
| M752 | 7.2, 5.6 | >4,096 | 64 | 32 | 128 | 128 | 2 | 2 | 8 | 32 | 1 | 0.06 | 0.13 |
| M804 | 9.0 | 256 | 32 | 4 | 128 | 64 | 1 | 0.5 | 2 | 2 | 2 | 0.03 | 0.13 |
| M809 | 9.0 | 512 | 128 | 4 | 512 | 512 | 32 | 32 | 16 | 32 | 16 | 0.13 | 0.13 |
| K. pneumoniae | |||||||||||||
| M484 | 7.2, 5.6 | >4,096 | 1,024 | 1,024 | 2,048 | 2,048 | 64 | 32 | 32 | 256 | 8 | 4 | 1.0 |
| M621 | 7.2, 5.6, 7.8 | >4,096 | 64 | 64 | 128 | 128 | 8 | 4 | 16 | 64 | 16 | 0.25 | 0.13 |
| M625B | 7.2, 5.6, 7.8 | >4,096 | 128 | 64 | 256 | 128 | 8 | 4 | 8 | 64 | 32 | 0.5 | 0.13 |
| M846 | 7.2, 5.6 | >4,096 | 64 | 64 | 256 | 256 | 8 | 8 | 16 | 64 | 4 | 0.5 | 0.25 |
| P. mirabilisM910 | 9.0 | 128 | 16 | 2 | 256 | 256 | 2 | 2 | 4 | 1 | 0.25 | 0.13 | 4.0 |
The first isoelectric points correspond to those for the cloxacillin-inhibited band (AmpC); additional pIs correspond to those for clavulanic acid-inhibited bands.
AMP, ampicillin; FOX, cefoxitin; FOX-Ro(4), cefoxitin plus 4 μg of Ro 48-1220 per ml; CPD, cefpodoxime; CPD-Cl(4), cefpodoxime plus 4 μg of clavulanic acid per ml; CRO, ceftriaxone; CRO-Cl(2), ceftriaxone plus 2 μg clavulanic acid per ml; CTX, cefotaxime; CAZ, ceftazidime; AZT, aztreonam; CEF, cefepime; IMI, imipenem.
Resistance was transferred for the underlined isolates.
Cefoxitin resistance was transferred for three E. coli, four K. pneumoniae, and one P. mirabilis isolates (Table 1). The ampicillin and cefoxitin MICs for each transconjugant were at least 256- and 16-fold, respectively, greater than the corresponding MICs for the recipient strain (data not shown). The pI of the AmpC band for each transconjugant was the same as the pI of the AmpC band for the corresponding donor.
Because a relatively large number (86%) of the K. pneumoniae isolates that showed decreased susceptibility to cefoxitin (zone diameter, ≤17 mm; MIC, ≥16 μg/ml) demonstrated no cloxacillin-inhibited band by IEF, the MICs of several drugs were determined for these organisms. Table 2 shows the MICs at which 50% of isolates are inhibited (MIC50s), MIC90s, and MIC100s of these drugs.
TABLE 2.
MICs for non-AmpC-producing K. pneumoniae isolates with decreased susceptibility to cefoxitin (n = 24)
| MIC | MICa (μg/ml)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | FOX | CPD | CPD-Cl(4) | CRO | CRO-Cl(2) | CTX | CAZ | AZT | CEF | IMI | |
| 50% | 256 | 32 | 1 | 0.25 | 0.25 | 0.13 | 0.5 | 1 | 0.25 | 0.25 | 0.13 |
| 90% | >4,096 | 64 | 16 | 1 | 16 | 0.5 | 16 | 128 | 128 | 2 | 0.25 |
| 100% | >4,096 | 512 | 512 | 1 | 128 | 1 | 512 | 2,048 | 2,048 | 32 | 0.5 |
See footnote b of Table 1 for definitions of abbreviations.
DISCUSSION
E. coli, K. pneumoniae, and P. mirabilis are the species in the family Enterobacteriaceae that are most commonly isolated in the clinical laboratory (16, 26). However, few studies have assessed the occurrence of AmpC beta-lactamases among these species. Gazouli et al. (11) tested 2,133 E. coli isolates from 10 Greek hospitals and found that 63 (3%) had cefoxitin zone diameters of ≤14 mm. Eight isolates lacked an outer membrane protein, and 55 (2.6%) contained AmpC beta-lactamases on the basis of the results of hydrolysis and inhibition studies and hybridization tests with AmpC-specific probes. Cefoxitin resistance was transferred for only a “few” isolates. These results mimic our results for E. coli, wherein 1.9 and 1.6% of the isolates demonstrated decreased susceptibility to cefoxitin and AmpC bands, respectively. Similar data have not been reported for K. pneumoniae, but our results indicate that this species harbors AmpC enzymes less frequently (1.1%) than E. coli does. The higher incidence of AmpC beta-lactamases in E. coli may reflect two modes of production: hyperproduction of chromosome-mediated AmpC and plasmid-mediated AmpC beta-lactamases. On a comparative note, these enzymes were present in E. coli and K. pneumoniae isolates at our institution less frequently than ESBLs (4 and 19%, respectively), as reported previously (10). Reports of plasmid-mediated AmpC in P. mirabilis are rare (6, 42), while the first isolation of a chromosomally encoded AmpC in this species was reported only in 1998 (8).
The current NCCLS documents do not indicate the screening and confirmatory tests that should be used for the detection of AmpC beta-lactamases in K. pneumoniae and E. coli (24). We used the standard disk diffusion breakpoint for cefoxitin (zone diameter, <18 mm) to screen isolates and the three-dimensional extract test as a confirmatory test. Our results indicate that the disk diffusion test has poor specificity, especially with K. pneumoniae isolates. Of the 45 isolates with decreased cefoxitin susceptibility, 29 (64%) were non-AmpC producers, and 24 (83%) of these were K. pneumoniae. Had we used a cefoxitin zone diameter of ≤14 mm as the criterion for the screening of isolates (11), the number of non-AmpC producers would have decreased from 29 to 10 (all K. pneumoniae), but we also would have failed to detect 2 (E. coli) of the 16 (13%) AmpC producers. Cefoxitin resistance in non-AmpC producers may be due to a lack of permeation of porin (28) (see below). The results of this study underscore the need for reliable laboratory tests that confirm the presence of AmpC beta-lactamases in clinical isolates.
All 16 of the AmpC-producing and 1 of the 29 non-AmpC-producing isolates were positive by the three-dimensional extract test. An extract of one of the AmpC producers inhibited the growth of one surface organism (Fig. 1B). Because this test is a confirmatory test and the additional cost is minimal, the use of two indicator organisms is recommended. The reason for the false-positive result with the non-AmpC producer was unclear and is the focus of ongoing studies. A limitation of methods used to detect the AmpC enzyme is that an increasing number of clinical isolates have multiple beta-lactamases, which in turn can make inhibition patterns complex and difficult to interpret (37, 38). The isolate with the discrepant extract test result harbored two SHV-type ESBLs. However, group 2be beta-lactamases usually are not active against cephamycins. Interestingly, two other non-AmpC-producing K. pneumoniae isolates also harbored two SHV-type ESBLs each but were negative by the extract test.
Several features of the in vitro susceptibility testing results (Table 1) were worthy of note. The results of screening of two AmpC-producing E. coli isolates (M656 and M683, Table 1) by the disk diffusion method were borderline, with initial cefoxitin zone diameter readings of 17 mm and readings of 18 mm on repeat testing (data not shown). By using the current breakpoint of 18 mm (24) as the cutoff criterion in the screening test, these isolates nearly missed detection. These results also suggest that the true frequency of AmpC producers may be somewhat higher than the 1.2% stated above. The cefoxitin MICs for these isolates were within the susceptible range, as were the MICs of ceftriaxone, cefotaxime, ceftazidime, aztreonam, cefepime, and imipenem (Table 1). Interestingly, Bauernfeind et al. (2) recently isolated a clinically significant strain of K. pneumoniae that harbored a novel type of AmpC beta-lactamase and that also demonstrated a low level of activity against cephamycins (cefoxitin MIC, 4 μg/ml). These data suggest that although screening methods which use cefoxitin in standardized methods to detect AmpC-harboring isolates are useful, they are not perfect.
The MICs of β-lactams for the other AmpC-producing isolates tested in this study were variable but were generally lower than those reported elsewhere for K. pneumoniae and E. coli isolates that harbor plasmid-mediated AmpC enzymes (Table 1) (7, 11, 17). These results may be due to the screening method, which was designed to include isolates with borderline susceptibility to cephamycins. As seen in early reports, several AmpC producers were resistant to many expanded-spectrum β-lactams including cephamycins but were susceptible to “fourth-generation” cephalosporins (e.g., cefepime) and carbapenems. Given that AmpC producers are typically resistant to cephamycins and susceptible to fourth-generation cephalosporins and that ESBL producers are frequently susceptible to cephamycins and variably resistant to fourth-generation cephalosporins (17), it is of therapeutic interest for a clinical laboratory to distinguish between these beta-lactamases. These issues become more significant with the ever increasing number of reports of AmpC- or ESBL-producing organisms for which the MICs of expanded-spectrum β-lactams are low and which are associated with clinical disease (2, 32).
Tzouvelekis et al. (40) reported that Ro 48-1220, a potent AmpC enzyme inhibitor, at a concentration of 4 μg/ml protected ceftriaxone and ceftazidime against organisms that produced group 1 or 2be beta-lactamases. In our study, this inhibitor at the same concentration protected cefoxitin against most AmpC producers. However, up to eight times greater inhibitor concentration was required to ensure protection against five isolates that harbored at least one other beta-lactamase in addition to AmpC (Table 1). Ro 48-1220 inhibits both extended-spectrum and AmpC beta-lactamases, and this may account for the increased inhibitor concentrations needed to ensure protection against these isolates.
In our study, 64% of all isolates with decreased susceptibility to cefoxitin failed to harbor an AmpC beta-lactamase. Because most of these isolates (83%) were K. pneumoniae, MICs were determined (Table 2) and were compared to the MICs for AmpC-producing K. pneumoniae. Some overlap in MIC endpoints was seen between isolates that produced AmpC enzymes and isolates that did not produce these enzymes (Table 1), thereby making it difficult to distinguish both groups on the basis of phenotypic results. The MIC100 data demonstrate that for some non-AmpC producers cefoxitin MICs are greater than those for the majority of the AmpC producers. These data corroborate the results of the cefoxitin disk test, which was used to initially screen isolates for AmpC production; this test was nonspecific (see above). Cephamycin resistance in non-AmpC-producing K. pneumoniae strains is often due to porin-deficient mutants (1, 28). Hernandez-Alles et al. (12) demonstrated that interruption of a porin gene by insertion sequences is a common type of mutation that causes the loss of porin expression and increased cefoxitin resistance in K. pneumoniae. In our study, large differences (≥3 twofold dilutions) in MICs between cefpodoxime or ceftriaxone with and without clavulanic acid were seen for 7 of the 24 Klebsiella non-AmpC producers, and all differences were attributed to the presence of ESBLs (data not shown).
In summary, we have demonstrated that at our institution the overall rate of occurrence of relatively high levels of AmpC beta-lactamase production in nonrepeat E. coli, K. pneumoniae, and P. mirabilis isolates was 1.2%. Cefoxitin resistance was transferred for half of the 16 AmpC producers. This is significant in light of recent reports which suggest that these new plasmid-mediated enzymes may create serious therapeutic problems in the future (18, 31). For a relatively large number of cefoxitin-resistant K. pneumoniae isolates (86%), cephamycin resistance was not associated with the AmpC enzyme. The three-dimensional extract test was a reliable method of detection of isolates that harbor the AmpC enzyme.
ACKNOWLEDGMENTS
We thank Patricia A. Bradford for providing E. coli DH5α(pCLL3414), which expressed the ACT-1 β-lactamase and which was used as a standard in IEF testing (7). We also thank Michael W. Climo for the testing of isolates by pulsed-field gel electrophoresis.
REFERENCES
- 1.Ardanuy C, Linares J, Dominguez M A, Hernandez-Alles S, Benedi V J, Martinez-Martinez L. Outer membrane profiles of clonally related Klebsiella pneumoniae isolates from clinical samples and activities of cephalosporins and carbapenems. Antimicrob Agents Chemother. 1998;42:1636–1640. doi: 10.1128/aac.42.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind A, Schneider I, Jungwirth R, Sahly H, Ullmann U. A novel type of AmpC β-lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob Agents Chemother. 1999;43:1924–1931. doi: 10.1128/aac.43.8.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Wagner S, Jungwirth R, Schneider I, Meyer D. A novel class C β-lactamase (FOX-2) in E. coli conferring resistance to cephamycins. Antimicrob Agents Chemother. 1997;41:2041–2046. doi: 10.1128/aac.41.9.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauernfeind A, Chong Y, Lee K. Plasmid-encoded AmpC β-lactamases: how far have we gone 10 years after their discovery? Yonsei Med J. 1998;39:520–525. doi: 10.3349/ymj.1998.39.6.520. [DOI] [PubMed] [Google Scholar]
- 5.Bauernfeind A, Chong Y, Schweighart S. Extended broad spectrum β-lactamase in Klebsiella pneumoniae including resistance to cephamycins. Infection. 1989;17:316–321. doi: 10.1007/BF01650718. [DOI] [PubMed] [Google Scholar]
- 6.Bobrowski M M, Mathew M, Barth P T, Datta N, Grinter N J, Jacob A E, Kontomichalou P, Dale J W, Smith J T. Plasmid-determined β-lactamase indistinguishable from the chromosomal β-lactamase of Escherichia coli. J Bacteriol. 1976;123:149–157. doi: 10.1128/jb.125.1.149-157.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford P A, Urban C, Mariano N, Projan S J, Rahal J J, Bush K. Imipenem resistance in Klebsiella pneumoniae is associated with the combinations of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob Agents Chemother. 1997;41:563–569. doi: 10.1128/aac.41.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bret L, Chanal-Claris C, Sirot D, Chaibi E B, Labia R, Sirot J. Chromosomally encoded AmpC-type β-lactamase in a clinical isolate of Proteus mirabilis. Antimicrob Agents Chemother. 1998;42:1110–1114. doi: 10.1128/aac.42.5.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coudron P E, Moland E S, Sanders C C. Occurrence and detection of extended-spectrum β-lactamases in members of the family Enterobacteriaceae at a veterans medical center: seek and you may find. J Clin Microbiol. 1997;35:2593–2597. doi: 10.1128/jcm.35.10.2593-2597.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gazouli M, Tzouvelekis L S, Vatopoulos A C, Tzelepi E. Transferable class C β-lactamases in Escherichia coli strains isolated in Greek hospitals and characterization of two enzyme variants (LAT-3 and LAT-4) closely related to Citrobacter freundii AmpC β-lactamase. J Antimicrob Chemother. 1998;42:419–425. doi: 10.1093/jac/42.4.419. [DOI] [PubMed] [Google Scholar]
- 12.Hernandex-Alles S, Benedi V J, Martinez-Martinez L, Pascual A, Aguilar A, Tomas J M, Alberti S. Development of resistance during antimicrobial therapy caused by insertion sequence interruption of porin genes. Antimicrob Agents Chemother. 1999;43:937–939. doi: 10.1128/aac.43.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horii T, Arakawa Y, Ohta M, Ichiyama L, Wacharotayankun R, Kato N. Plasmid-mediated AmpC-type β-lactamase isolated from Klebsiella pneumoniae confers resistance to broad-spectrum β-lactams, including moxalactam. Antimicrob Agents Chemother. 1993;37:984–990. doi: 10.1128/aac.37.5.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenks P J, Hu Y M, Danel F, Mehtar S, Livermore D M. Plasmid-mediated production of class 1 (AmpC) β-lactamase by two Klebsiella pneumoniae isolates in the UK. J Antimicrob Chemother. 1995;35:235–236. doi: 10.1093/jac/35.1.235. [DOI] [PubMed] [Google Scholar]
- 15.Leiza M G, Perez-Diaz J C, Ayala J, Casellas J M, Martinez-Beltran J, Bush K, Baquero F. Gene sequences and biochemical characterization of FOX-1 from Klebsiella pneumoniae, a new AmpC-type plasmid-mediated β-lactamase with two molecular variants. Antimicrob Agents Chemother. 1994;38:2150–2157. doi: 10.1128/aac.38.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P Y F, Gur D, Hall L M C, Livermore D M. Survey of the prevalence of β-lactamases amongst 1000 gram-negative bacilli isolated consecutively at the Royal London Hospital. J Antimicrob Chemother. 1992;30:429–447. doi: 10.1093/jac/30.4.429. [DOI] [PubMed] [Google Scholar]
- 17.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Martinez L, Pascual A, Hernandez-Alles S, Alvarez-Diaz D, Suarez A I, Tran J, Benedi V J, Jacoby G A. Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob Agents Chemother. 1999;43:1669–1673. doi: 10.1128/aac.43.7.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto Y, Ikeda F, Kamimura T, Yokota Y, Mine Y. Novel plasmid-mediated β-lactamase from Escherichia coli that inactivated oxyiminocephalosporins. Antimicrob Agents Chemother. 1988;32:1243–1246. doi: 10.1128/aac.32.8.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morosini M, Negri M, Shoichet B, Baquero M, Baquero F, Blazquez J. An extended-spectrum AmpC-type β-lactamase obtained by in vitro antibiotic selection. FEMS Microbiol Lett. 1998;165:85–90. doi: 10.1111/j.1574-6968.1998.tb13131.x. [DOI] [PubMed] [Google Scholar]
- 21.M'Zali F H, Heritage J, Gascoyne-Binzi D M, Denton M, Todd N J, Hawkey P M. Transcontinental importation into the UK of Escherichia coli expressing a plasmid-mediated AmpC-type beta-lactamase exposed during an outbreak of SHV-5 extended-spectrum beta-lactamase in a Leeds hospital. J Antimicrob Chemother. 1997;40:823–831. doi: 10.1093/jac/40.6.823. [DOI] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. 1997. Approved standard M7-A4 (M100-S7). National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. 6th ed. 1997. Approved standard M2-A6 (M100-S7). National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing; ninth informational supplement (M100-S9). Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 25.Nelson E C, Elisha B G. Molecular basis of AmpC hyperproduction in clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1999;43:957–959. doi: 10.1128/aac.43.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicolas-Chanoine M H, Chardon H, Avril J L, Cattoen Y, Croix J C, Dabernat H, Etienne J, Fosse T, Ghnassia J C, Lecaillon E, Marmonier A, Roussel-Delvallez M, Soussy J C, Trevoux A, Sirot J. Susceptibility of Enterobacteriaceae to beta-lactams and fluoroquinolones: a French multicentre study. Clin Microbiol Infect. 1997;3(Suppl. 2):74–75. doi: 10.1046/j.1469-0691.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- 27.Normark S, Grunstrom T, Bergstrom S. Susceptibility to penicillins and cephalosporins in β-lactamase producing strains of E. coli and relative amount of β-lactamase produced from these strains. Scand J Infect Dis. 1980;25:23–29. [PubMed] [Google Scholar]
- 28.Pangon B, Bizet C, Bure A, Pichon F, Philippon A, Regnier B, Gutmann L. In vivo selection of a cephamycin-resistant, porin-deficient mutant of Klebsiella pneumoniae producing a TEM-3 β-lactamase. J Infect Dis. 1989;159:1005–1006. doi: 10.1093/infdis/159.5.1005. [DOI] [PubMed] [Google Scholar]
- 29.Papanicolaou G A, Medeiros A A, Jacoby G A. Novel plasmid-mediated β-lactamase (MIR-1) conferring resistance to oxyimino- and alpha-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990;34:2200–2209. doi: 10.1128/aac.34.11.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payne D J, Woodford N, Amyes S G B. Characterization of the plasmid-mediated β-lactamase BIL-1. J Antimicrob Chemother. 1992;30:119–127. doi: 10.1093/jac/30.2.119. [DOI] [PubMed] [Google Scholar]
- 31.Pornull K J, Rodrigo G, Dornbusch K. Production of a plasmid-mediated AmpC-like β-lactamase by a Klebsiella pneumoniae septicemia isolate. J Antimicrob Chemother. 1994;34:943–954. doi: 10.1093/jac/34.6.943. [DOI] [PubMed] [Google Scholar]
- 32.Rasheed J K, Jay C, Metchock B, Berkowitz F, Weigel L, Crellin J, Steward C, Hill B, Medeiros A A, Tenover F C. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob Agents Chemother. 1997;41:647–653. doi: 10.1128/aac.41.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders C C, Sanders W E, Jr, Moland E S. Characterization of β-lactamases in situ on polyacrylamide gels. Antimicrob Agents Chemother. 1986;30:951–952. doi: 10.1128/aac.30.6.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenover F C, Mohammed M J, Gorton T S, Dembek Z F. Detection and reporting of organisms producing extended-spectrum β-lactamases: survey of laboratories in Connecticut. J Clin Microbiol. 1999;37:4065–4070. doi: 10.1128/jcm.37.12.4065-4070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomson K S, Prevan A M, Sanders C C. Novel plasmid-mediated β-lactamases in Enterobacteriaceae: emerging problems for new B-lactam antibiotics. In: Remington J S, Swartz M N, editors. Current clinical topics in infectious diseases. Vol. 16. Cambridge, Mass: Blackwell Science, Inc.; 1996. pp. 151–163. [PubMed] [Google Scholar]
- 36.Thomson K S, Sanders C C. Detection of extended-spectrum β-lactamases in members of the family Enterobacteriaceae: comparison of the double-disk and three-dimensional tests. Antimicrob Agents Chemother. 1992;36:1877–1882. doi: 10.1128/aac.36.9.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson K S, Sanders C C, Moland E S. Use of microdilution panels with and without β-lactamase inhibitors as a phenotypic test for β-lactamase production among Escherichia coli, Klebsiella spp., Enterobacter spp., Citrobacter freundii, and Serratia marcescens. Antimicrob Agents Chemother. 1999;43:1393–1400. doi: 10.1128/aac.43.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tzouvelekis L S, Vatopoulos A C, Katsanis G, Tzelepi E. Rare case of failure by an automated system to detect extended-spectrum β-lactamase in a cephalosporin-resistant Klebsiella pneumoniae isolate. J Clin Microbiol. 1999;37:2388. doi: 10.1128/jcm.37.7.2388-2388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tzouvelekis L S, Tzelepi E, Mentis A F, Tsakris A. Identification of novel plasmid-mediated β-lactamase with chromosomal cephalosporinase characteristics from Klebsiella pneumoniae. J Antimicrob Chemother. 1993;31:645–654. doi: 10.1093/jac/31.5.645. [DOI] [PubMed] [Google Scholar]
- 40.Tzouvelekis L S, Gazouli M, Prinarakis E E, Tzelepi E, Legakis N J. Comparative evaluation of the inhibitory activities of the novel penicillanic acid sulfone Ro 48-1220 against β-lactamases that belong to groups 1, 2b, and 2be. Antimicrob Agents Chemother. 1997;41:475–477. doi: 10.1128/aac.41.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vercauteren E, Descheemaeker P, Ieven M, Sanders C C, Goossens H. Comparison of screening methods for detection of extended-spectrum β-lactamases and their prevalence among blood isolates of Escherichia coli and Klebsiella spp. in a Belgian teaching hospital. J Clin Microbiol. 1997;35:2191–2197. doi: 10.1128/jcm.35.9.2191-2197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verdet C, Arlet G, Ben Redjeb S, Ben Hassen A, Lagrange P H, Philippon A. Characterisation of CMY-4, an AmpC-type plasmid-mediated beta-lactamase, in a Tunisian clinical isolate of Proteus mirabilis. FEMS Microbiol Lett. 1998;169:235–240. doi: 10.1111/j.1574-6968.1998.tb13323.x. [DOI] [PubMed] [Google Scholar]