Abstract
TSG-6 is a soluble protein secreted in the extracellular matrix by various cell types in response to inflammatory stimuli. TSG-6 interacts with extracellular matrix molecules, particularly hyaluronan (HA), and promotes cutaneous wound closure in mice. Between epidermal cells, the discrete extracellular matrix contains HA and a tiny amount of TSG-6. However, challenges imposed to keratinocytes in reconstructed human epidermis revealed strong induction of TSG-6 expression, after exposure to T helper type 2 cytokines to recapitulate the atopic dermatitis phenotype or after fungal infection that causes secretion of cytokines and antimicrobial peptides. After both types of challenge, enhanced release of TSG-6 happens simultaneously with increased HA production. TSG-6 deficiency in N/TERT keratinocytes was created by inactivating TNFAIP6 using CRISPR/Cas9. Some TSG-6–/– keratinocytes analyzed through scratch assays tend to migrate more slowly but produce reconstructed human epidermis that exhibits normal morphology and differentiation. Few significant alterations were noticed by transcriptomic analysis. Nevertheless, reduced HA content in TSG-6–/– reconstructed human epidermis was observed, along with enhanced HA release into the culture medium, and this phenotype was even more pronounced after the challenging conditions. Reintroduction of cells producing TSG-6 in reconstructed human epidermis reduced HA leakage. Our results show a role for TSG-6 in sequestering HA between epidermal cells in response to inflammation.
Abbreviations: AD, atopic dermatitis; crRNA, CRISPR RNA; ECM, extracellular matrix; GEO, Gene Expression Omnibus; HA, hyaluronan; HAS, hyaluronan synthase; KC, keratinocyte; KLK, kallikrein; RHE, reconstructed human epidermis; RNA-seq, RNA sequencing; Th, T helper type
Introduction
In the skin, although occupying most of the space in the dermis, the extracellular matrix (ECM) also plays crucial roles in the epidermis, where it is composed of basal lamina and 15–20 nm intercellular spaces around cell junctions. The main ECM components in the epidermis include various polysaccharides, particularly hyaluronan (HA), which is swollen by water. This environment creates a favorable milieu for the diffusion of nutrients, GFs, and cytokines (Sandjeu and Haftek, 2009; Toole, 2001). Langerhans cells and lymphocytes migrate through the epidermal ECM, bringing innate or adaptive immunity.
HA is a linear glycosaminoglycan composed of repeated disaccharides, namely D-glucuronic acid, linked to N-acetyl-D-glucosamine, forming polymers up to 25 μm in length and 105–107 Da in mass (Sakai et al., 2000; Toole, 2004). Half of the total body HA resides in the skin (Fraser et al., 1997; Meyer and Stern, 1994), mostly in the dermis but also in the living layers of the epidermis (Pasonen-Seppänen et al., 2003). The functions of epidermal HA are not fully understood. Roles in proliferation and differentiation of epidermal keratinocytes (KCs) are unlikely, but altered regulation of HA synthases (HASs) and increased HA production were described in inflammatory conditions such as atopic dermatitis (AD) (Malaisse et al., 2014).
Across the body, HA interacts with several partners, such as TSG-6 protein, secreted by fibroblasts on TNF-α stimulation (Jadin et al., 2014; Lee et al., 1990). TSG-6 is encoded by the TNFAIP6 gene located on human chromosome 2. The protein of 277 amino acids harbors a cleavable signal peptide, an HA-binding (LINK) domain typical of hyaladherins, and a CUB domain, typically part of ECM and plasma membrane proteins. Besides fibroblasts, several cell types secrete TSG-6 protein in response to proinflammatory stimuli (Baranova et al., 2011). Initially associated with inflammation, TSG-6 is also detected in conditions implying ECM remodeling (Fülöp et al., 2003) and in organs exerting a barrier function (Day and Milner, 2019; Tan et al., 2011). In healthy skin, TSG-6 is detected in the dermis and in living epidermal layers (Tan et al., 2011). Its expression is upregulated during skin injury (Shakya et al., 2020).
Soluble TSG-6 protein dimerizes or interacts with ITI, and thereby becomes able to bind HA’s N-acetyl glucosamine unit and favor cross-linking of HA chains in ECM (Baranova et al., 2011; Milner and Day, 2003; Milner et al., 2007). Interaction between TSG-6 and HA increases the affinity of HA for the CD44 receptor involved in KCs adhesion, proliferation, differentiation, and migration (Baranova et al., 2013; Bourguignon, 2014; Bourguignon et al., 2004; Kavasi et al., 2017; Lesley et al., 2004). CD44 also contributes to maintaining a pericellular matrix around KCs (Muto et al., 2019; Pasonen-Seppänen et al., 2012). Similar to CD44, TSG-6 holds HA in the pericellular matrix (Selbi et al., 2006), thereby controlling differentiation of, for example, fibroblasts (Simpson et al., 2009).
In this study, we investigated the human epidermis produced and challenged in vitro (Hennies and Poumay, 2021) to characterize the TSG-6 expression induced in such conditions. We then used genetic approaches to create TSG-6–/– in vitro reconstructed epidermis and analyze the potential role for TSG-6. Data show a crucial impact of TSG-6 deficiency on epidermal HA.
Results
TSG-6 is expressed and released at low levels in the culture medium of normal human epidermis
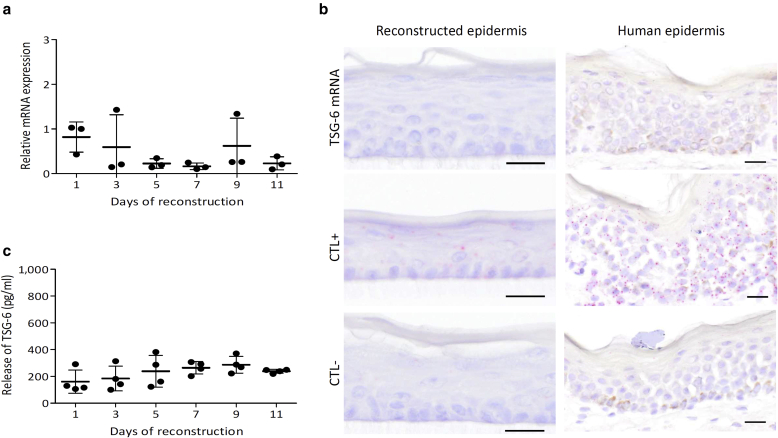
Reconstructed human epidermis (RHE) produced from primary KCs (Frankart et al., 2012) was analyzed for TSG-6-encoding mRNA expression and protein release in a culture medium during the whole reconstruction process (Figure 1). RT-qPCR showed that TSG-6-encoding mRNA expression remained very low (Cq > 32). No significant induction of TSG-6-encoding mRNA was observed (Figure 1a). Accordingly, in situ hybridization was unable to detect TSG-6-encoding mRNA in normal RHE and human epidermis (Figure 1b). Secreted TSG-6 protein levels were monitored, and small amounts were detected in the culture medium during the reconstruction process (Figure 1c).
Figure 1.
Weak TSG-6-encoding mRNA expression and protein release in a culture medium during the reconstruction of the normal epidermis. (a) TSG-6-encoding mRNA expression of RHE at 1, 3, 5, 7, 9, and 11 days of growth assessed by RT-qPCR. Reference gene is RPLP0 (mean ± SD; n = 3; one-way ANOVA). (b) TSG-6, PPIB (CTL+), and DapB (CTL–) mRNA detection using in situ hybridization and hemalun counterstaining of day 11 RHE and normal human epidermis. Bar = 20 μm. (c) TSG-6 protein concentration assayed using ELISA (mean ± SD; n = 4; one-way ANOVA). CTL, control; RHE, reconstructed human epidermis.
TSG-6 expression and release strongly increase on exposure of RHE to challenging conditions
To monitor TSG-6 regulation in inflammatory settings, two very different challenging conditions were imposed on RHE to analyze the tissue response.
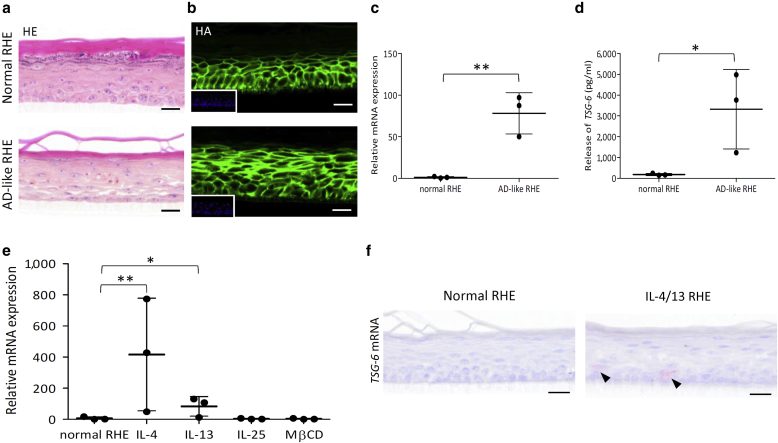
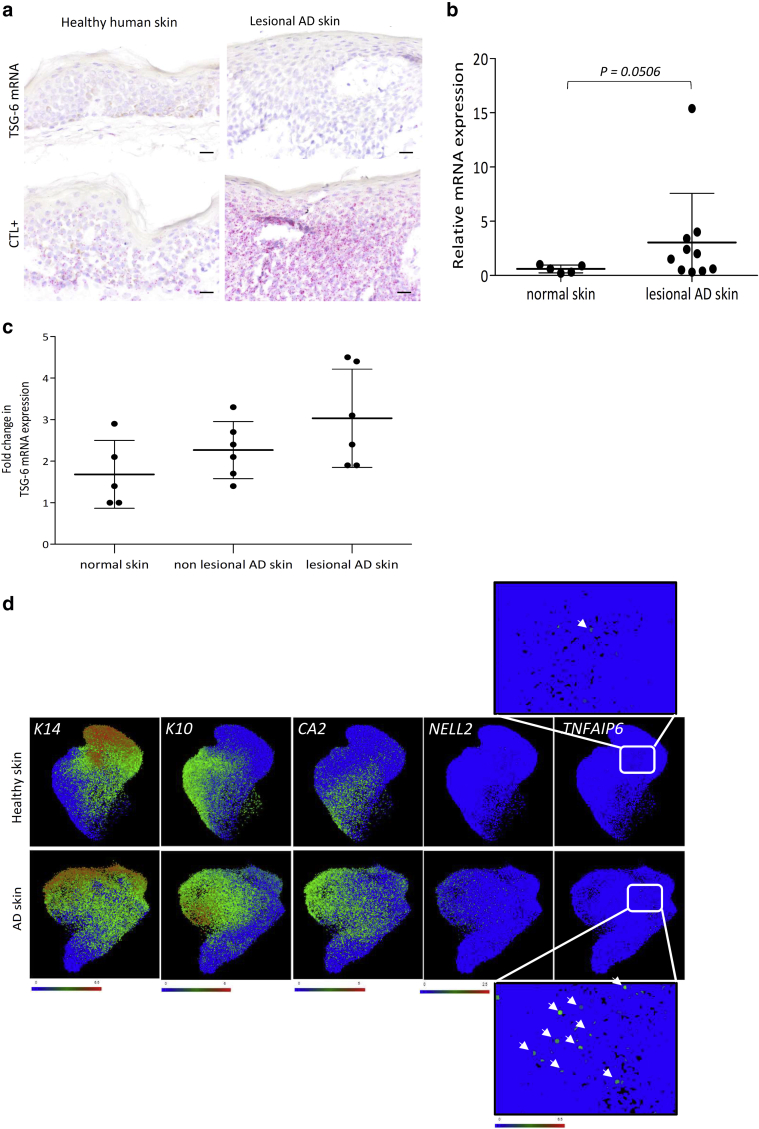
A first challenge, mimicking AD by deprivation of plasma membrane cholesterol followed by incubation with T helper type (Th) 2 ILs and exhibiting barrier alterations (De Vuyst et al., 2018) were selected on the basis of previous transcriptomic analysis (Hubaux et al., 2018; Park et al., 2021). In response to IL-4, IL-13, and IL-25 incubation for 48 hours, TNFAIP6 appeared as the sixth most upregulated gene, with a 27-fold increase in expression and a P-value of 1.37 × 10–8. Results are accessible at Gene Expression Omnibus (GEO) Series (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149638; GSE149639). Unlike control tissues, AD-like RHE exhibited spongiosis and hypogranulosis (Figure 2a) and brighter detection of HA around KCs (Figure 2b). Analysis of TSG-6-encoding mRNA expression revealed an approximately 90-fold induction in AD-like RHE compared with that of the controls (Figure 2c). The amount of TSG-6 protein released in the culture medium corroborated increased epidermal expression (Figure 2d). TSG-6-encoding mRNA expression in RHE incubated for 48 hours with either IL-4, IL-13, or IL-25 or with methyl-β-cyclodextrin identified IL-4 as the stronger inducer of TSG-6 expression, whereas IL-25 and methyl-β-cyclodextrin had no effect (Figure 2e). Thus, IL-4 and IL-13 were chosen to stimulate TSG-6 expression. In accordance with RT-qPCR data, TSG-6-encoding mRNA were detected using in situ hybridization in living layers of RHE challenged with IL-4 and IL-13 (Figure 2f). Conversely, addition of TNF-α in the culture medium of RHE did not induce TSG-6 expression (unpublished observations). In skin biopsies from lesional areas of patients with AD and from healthy controls, in situ hybridization was not sensitive enough to detect TSG-6-encoding mRNA (Figure 3a). However, RT-qPCR analysis of biopsies revealed increased TSG-6-encoding mRNA levels in lesional AD compared with that in the normal skin with a P-value as low as 0.0506 (Figure 3b). This increase in TSG-6 expression in the skin of patients suffering from AD is also detected in large-scale analyses. A search of GEO Profiles databases (Edgar et al., 2002) highlighted an increase in TSG-6 expression (GEO accession GSE5667 [Plager et al., 2010]) (Figure 3c). Likewise, transcriptomic analysis of a single-cell RNA sequencing (RNA-seq) dataset allowed a precise study of mRNA expression in undifferentiated and differentiated KCs of healthy and lesional AD skins (Reynolds et al., 2021; https://developmentcellatlas.ncl.ac.uk/datasets/ hca_skin_portal). As expected, expression of CA2 and NELL2, two typical hallmarks of AD lesions, was detected in more AD KCs than in normal KCs. In parallel, TSG-6-encoding mRNA was present in some undifferentiated AD KCs, whereas the signal was barely detectable in healthy cells (Figure 3d).
Figure 2.
Increase of TSG-6-encoding mRNA and protein in challenged RHE (AD-like RHE model). RHEs were treated with MβCD and IL-4, IL-13, and IL-25 for 48 hours. (a) HE staining and (b) HA detection. Bar = 20 μm. (c) TSG-6-encoding mRNA expression of AD-like RHE assessed by RT-qPCR (mean ± SD; n = 3; Student t-test; ∗∗P < 0.01). (d) TSG-6 protein concentration of challenged RHE assayed using ELISA (mean ± SD; n = 3; Student t-test; ∗P < 0.05). (e) TSG-6-encoding mRNA expression of RHE treated for 48 hours by ILs or MβCD separately assessed by RT-qPCR (mean ± SD; n = 3; one-way ANOVA; ∗P < 0.05; ∗∗P < 0.01 vs. normal RHE). (f) TSG-6 and PPIB (CTL+) mRNA detection using in situ hybridization and hemalun counterstaining of RHE challenged or not challenged with IL-4 and IL-13. AD, atopic dermatitis; HA, hyaluronan; HE, hemalun/eosin; MβCD, methyl-β-cyclodextrin; RHE, reconstructed human epidermis.
Figure 3.
Increase of TSG-6-encoding mRNA in lesional AD skin. (a) TSG-6 and PPIB (CTL+) mRNA detection using in situ hybridization and hemalun counterstaining of healthy and lesional AD skin. Bar = 20 μm. (b) TSG-6-encoding mRNA expression in normal and lesional AD skin assessed by RT-qPCR. The reference gene is RPLP0 (mean ± SD; n = 5 for normal skin and n = 10 for AD skin; unpaired t-test with Welch’s correction). (c) TSG-6-encoding mRNA expression in normal, nonlesional AD, and lesional AD skins analyzed from GEO Profiles database (accession GSE5667; GPL96; 206026_s_at) (mean ± SD; n = 5 or 6; one-way ANOVA). (d) UMAP visualization of undifferentiated and differentiated KC in healthy and lesional AD skins found in single-cell RNA-seq dataset (https://developmentcellatlas.ncl.ac.uk/datasets/ hca_skin_portal) (n = 5). Normalized gene expressions of K14, K10, CA2, NELL2, and TNFAIP6 are represented in green for a low expression and in red for a strong expression. AD, atopic dermatitis; CTL, control; GEO, Gene Expression Omnibus; K, keratin; KC, keratinocyte; RNA-seq, RNA sequencing; UMAP, Uniform Manifold Approximation and Projection.
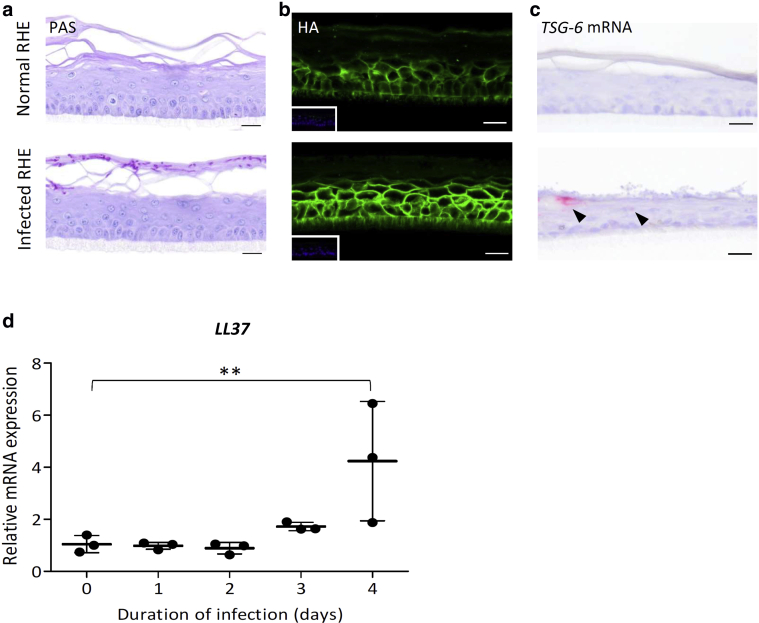
A second challenge imposed on RHE consisted in superficial infection by Trichophyton rubrum, which induces cytokine release in concomitance with the significant induction of expression and release of TSG-6 protein (Faway et al., 2019, 2017). After 4 days of infection, RHE becomes invaded by fungal elements only in the stratum corneum (Figure 4a). At the same time, the signal of HA seems higher around the KCs of infected RHE than around the control (Figure 4b). In parallel, TSG-6-encoding mRNA can be detected through in situ hybridization solely in infected RHE (Figure 4c) when its mRNA expression is significantly increased (induction of 46-fold) according to RT-qPCR analysis. Alongside, we found an interesting significant increase in the mRNA encoding LL37, an antimicrobial peptide that blocks the proinflammatory HA‒toll-like receptor 4‒CD44 axis (Morioka et al., 2008) (Figure 4d).
Figure 4.
Elevated TSG-6-encoding mRNA expression and protein release in RHE infected by Trichophyton rubrum arthroconidia. RHEs were infected or not infected by T. rubrum arthroconidia and maintained in culture for 4 additional days. (a) PAS staining with α-amylase pretreatment and hemalun counterstaining and (b) HA detection. Bar = 20 μm. (c) TSG-6-encoding mRNA detection using in situ hybridization and hemalun counterstaining. Bar = 20 μm. (d) LL37 mRNA expression assessed by RT-qPCR. The reference gene is RPLP0 (mean ± SD; n = 3; one-way ANOVA; ∗∗P < 0.01 vs. day 0). HA, hyaluronan; PAS, periodic-acid schiff; RHE, reconstructed human epidermis.
Generation, validation, and characterization of TSG-6–/– N/TERT clones
To investigate the potential roles of epidermal TSG-6, we used CRISPR/Cas9 to disrupt the TNFAIP6 gene in N/TERT KCs. RNA-seq analysis of epidermis reconstructed from N/TERT cells and stimulated with IL-4 and IL-13 showed a transcriptional profile similar to that of AD-like models made of primary KCs (Table 1). RNA-seq data are accessible in ArrayExpress (http://www.ebi.ac.uk/arrayexpresse; E-MTAB-10095).
Table 1.
Transcriptomic Analysis of Epidermis Reconstructed from N/TERT Cells and Stimulated or Not with IL-4 and IL-13 through RNA Sequencing
| Gene | Name | log2FC | Padj |
|---|---|---|---|
| CISH | Cytokine inducible SH2 containing protein | 3.07684025 | 0.000E+00 |
| ANO1 | Anoctamin 1 | 2.80591740 | 1.042E–219 |
| SLC16A14 | Solute carrier family 16 member 14 | 4.47550706 | 8.627E–192 |
| CA2 | Carbonic anhydrase 2 | 2.18654582 | 1.411E–188 |
| CTSC | Cathepsin C | 1.54462245 | 5.680E–123 |
| GLDC | Glycine decarboxylase | 6.48425833 | 7.039E–122 |
| SLC26A2 | Solute carrier family 26 member 2 | 1.42119675 | 7.896E–113 |
| AC100801.2 | 2.67582484 | 4.386E–86 | |
| C1QTNF1 | C1q and TNF related 1 | 2.76558625 | 2.474E–85 |
| TNFAIP6 | TNF alpha-induced protein 6 | 4.61410057 | 1.402E–80 |
| BCL2L15 | BCL2 like 15 | 4.38693821 | 1.861E–75 |
| DIO2 | Iodothyronine deiodinase 2 | –1.38620973 | 8.042E–75 |
| ST6GAL1 | ST6 beta-galactoside alpha-2,6-sialyltransferase 1 | 1.94904986 | 5.983E–73 |
| EGR3 | Early growth response 3 | –1.43824080 | 1.248E–70 |
| KRT16 | Keratin 16 | 1.76615476 | 1.724E–67 |
| HRH1 | Histamine receptor H1 | 2.04522544 | 1.800E–60 |
| POSTN | Periostin | 2.96569063 | 9.221E–60 |
| ETS2 | ETS proto-oncogene 2, transcription factor | –0.97990996 | 3.121E–59 |
| EGLN3 | Egl-9 family hypoxia inducible factor 3 | 1.90222718 | 7.334E–58 |
| APOL6 | Apolipoprotein L6 | 1.11187734 | 1.488E–57 |
| LBH | LBH regulator Of WNT signaling pathway | 1.63185691 | 3.760E–57 |
| MXRA5 | Matrix remodeling associated 5 | –1.75646450 | 7.512E–56 |
| RGMB | Repulsive guidance molecule BMP coreceptor B | 1.27097907 | 8.026E–56 |
| NEK6 | NIMA related kinase 6 | 1.46907088 | 4.158E–55 |
| C1orf68 | Chromosome 1 open reading frame 68 | –2.17535843 | 5.709E–54 |
| NELL2 | Neural EGFL like 2 | 2.77669873 | 1.809E–52 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | –2.48531923 | 2.546E–51 |
| GCNT3 | Glucosaminyl (N-acetyl) transferase 3, mucin type | 2.13974316 | 1.036E–50 |
| MAP3K14 | Mitogen-activated protein kinase 14 | 1.62240356 | 1.088E–50 |
| DUOX2 | Dual oxidase 2 | 2.12196856 | 3.027E–50 |
| PADI3 | Peptidyl arginine deiminase 3 | 1.83977015 | 5.216E–50 |
| CD59 | CD59 molecule (CD59 blood group) | 0.75327573 | 1.087E–49 |
| KRT4 | Keratin 4 | 1.99977131 | 1.734E–49 |
| BMP4 | Bone morphogenetic protein 4 | 2.53670445 | 1.151E–47 |
| BTBD3 | BTB domain containing 3 | 0.78079374 | 1.162E–47 |
| AREG | Amphiregulin | –1.47407342 | 7.747E–47 |
| PDE6A | Phosphodiesterase 6A | 2.22277344 | 1.398E–46 |
| DENND4A | DENN domain containing 4A | 0.83293501 | 1.443E–44 |
| CCN1 | Cellular communication network factor 1 | –1.81412009 | 2.467E–44 |
| SORL1 | Sortilin related receptor 1 | 0.96862192 | 5.845E–44 |
| BCL10 | BCL10 immune signaling adaptor | –1.01814850 | 9.519E–44 |
| RASGRP1 | RAS guanyl releasing protein 1 | 1.15867682 | 1.093E–43 |
| CXCL2 | C-X-C motif chemokine ligand 2 (IL-2) | –3.20712371 | 7.322E–43 |
| HBEGF | Heparin-binding EGF like growth factor | –1.92196976 | 1.046E–42 |
| FLT1 | Fms related receptor tyrosine kinase 1 | 2.52370748 | 1.357E–42 |
| LGALSL | Galectin like | –0.93501839 | 3.556E–42 |
| CASP14 | Caspase 14 | –1.20133651 | 6.805E–41 |
| CTSH | Cathepsin H | –1.48572279 | 8.798E–40 |
| TXN | Thioredoxin | 1.09419641 | 2.633E–39 |
Abbreviations: Padj, adjusted P; FC, fold change; RHE, reconstructed human epidermis.
List of 50 more regulated genes in IL-4‒ and IL-13‒challenged RHE compared with nonstimulated RHE based on Padj-value.
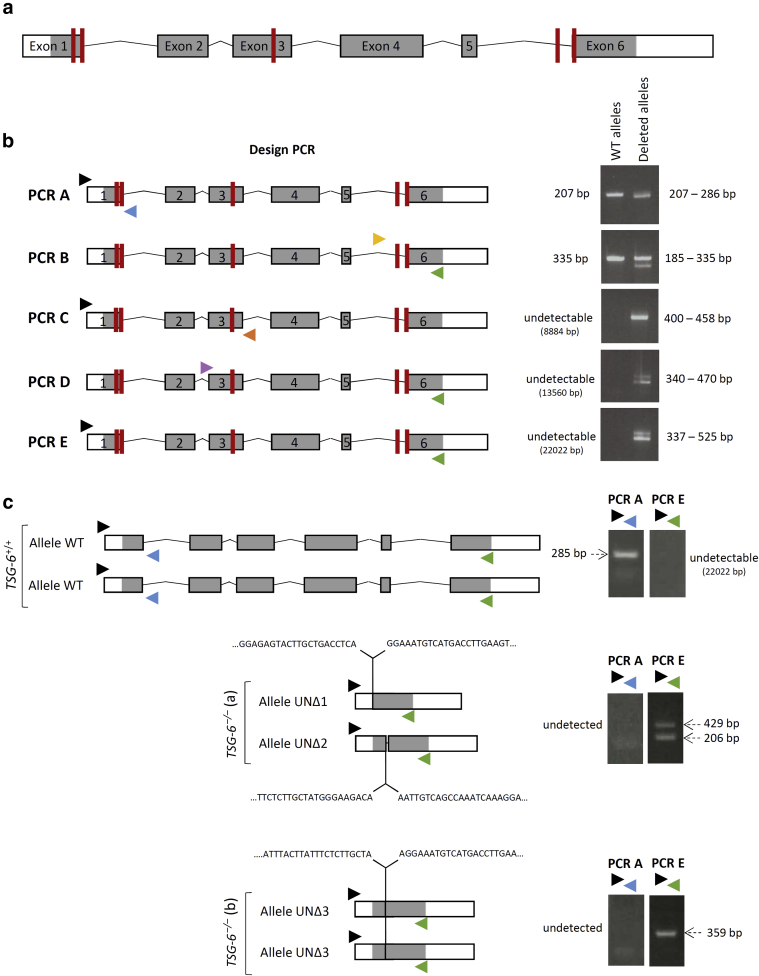
To generate deletions in the TNFAIP6 gene, five targets were chosen in the first, third, and sixth exons (Figure 5a). DNA extracted from the pool of N/TERT KCs electroporated with Cas9/ribonucleoprotein was analyzed by PCR to characterize deletions. Figure 5b illustrates the PCR data that revealed deletions at various sites. Among 100 clones sequenced and presenting many different mutations of several base pairs (not shown), two were selected for large deletions in both alleles of the TNFAIP6 gene leading to probable null alleles. Genomic positions were defined by numbering nucleotide A of the methionine initiation codon as 1. The first clone TNFAIP6UNΔ1/UNΔ2 named N/TERT TSG-6–/– (a) exhibits a UNΔ1 deletion in allele 1 that encompasses nucleotide ‒120 (i.e., 44 nucleotides upstream of exon 1) to +21,696 and a UNΔ2 deletion in allele 2 encompassing nucleotide +44 (in exon 1) to +21,635 (62 bases upstream from exon 6). The second clone TNFAIP6UNΔ3/UNΔ3 named N/TERT TSG-6–/– (b) exhibits identical deletions in both alleles encompassing nucleotide +33 (in exon 1) to +21,695 (two nucleotides upstream from exon 6) (Figure 5c). Both clones encode putative peptides lacking LINK and CUB domains.
Figure 5.
Deletions in TNFAIP6 gene using CRISPR/Cas9 genome editing in N/TERT KC. (a) Genomic organization of TNFAIP6 gene using the localization of five sites targeted by crRNA (red bars). (b) PCR screening of deletions in DNA from a pool of N/TERT cells electroporated with a mixture of five crRNA (red bars) and Cas9. Primers are indicated by arrowheads. Obtained amplicon sizes of WT and deletant alleles are shown on the right. (c) Genomic deletions in TSG-6–/– clone (a) and clone (b) using primer combinations PCR A and PCR E illustrated in panel b. All PCR products have been sequenced. Only the sequences of the junctions are shown. bp, base pair; crRNA, CRISPR RNA; KC, keratinocyte; WT, wild-type
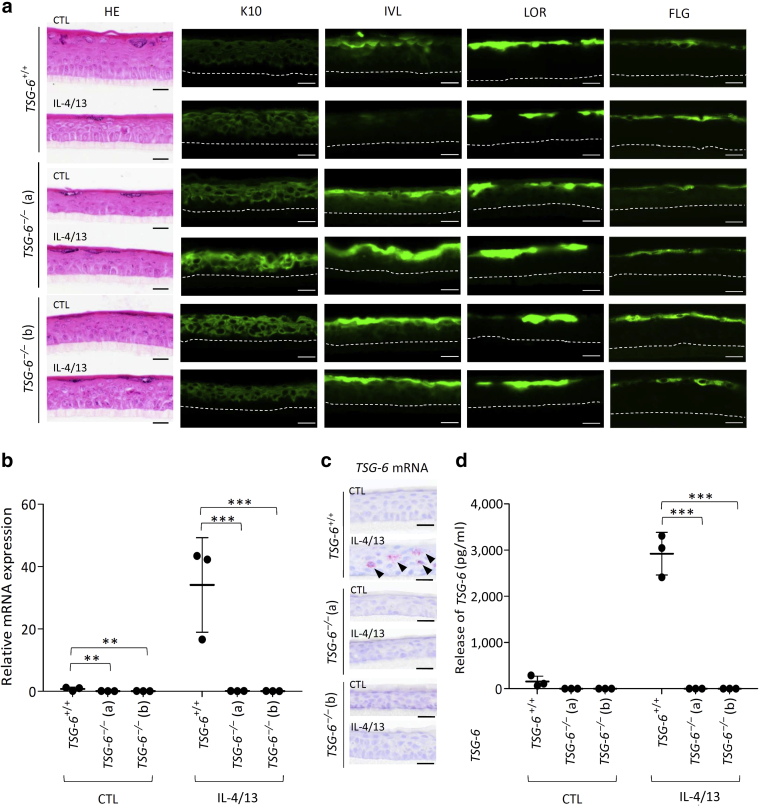
RHEs were produced using unedited TSG-6+/+ or deletants TSG-6–/– (a) and TSG-6–/– (b) N/TERT cells and were treated or not treated for 48 hours with IL-4 and IL-13. Both mutant TSG-6–/– (a) and TSG-6–/– (b) clones produced RHE with a functional barrier (not shown) and exhibited identical morphological features compared with unedited N/TERT cells, including localization of keratin 10, involucrin, loricrin, and FLG (Figure 6a). As expected, TSG-6-encoding mRNA was only overexpressed in the RHE produced with TSG-6+/+ N/TERT cells challenged with IL-4 and IL-13. Conversely, TSG-6-encoding mRNA was undetectable in the RHE produced with TSG-6–/– (a) or TSG-6–/– (b) N/TERT cells (Figure 6b). Using in situ hybridization, TSG-6-encoding mRNA were solely detected in living layers of TSG-6+/+ N/TERT RHE challenged with IL-4 and IL-13 and were absent in knockout RHE (Figure 6c). A strong increase in TSG-6 protein level was found in the medium of challenged TSG-6+/+ N/TERT but not in mutant RHE (Figure 6d). Transcriptomes of TSG-6–/– (a) and TSG-6–/– (b) RHE challenged with IL-4 and IL-13 were compared with those of TSG-6+/+ RHE (http://www.ebi.ac.uk/arrayexpresse; E-MTAB-10095), confirming suppressed TNFAIP6 expression. Deletion of TSG-6 resulted in weak upregulation and downregulation in a few genes (Table 2). Among them, suppressed expression of kallikreins (KLKs) 5 and 8, confirmed by RT-qPCR analysis in clone TSG-6–/– (b) (Figure 7), could indicate consequences on epidermal homeostasis and migration processes. Weak alterations in the gene expression observed after knocking out TSG-6 concurred with the normal morphology of TSG-6–/– RHE.
Figure 6.
Validation and characterization of the two clones exhibiting large deletions in the TNFAIP6 gene.TSG-6+/+ and TSG-6–/– (a and b) RHE stimulated or not stimulated for 48 hours with IL-4 and IL-13. (a) HE staining and immunofluorescence (in green) of K10, IVL, LOR, and FLG. Bar = 20 μm. (b) TSG-6-encoding mRNA expression of TSG-6+/+ and TSG-6–/– RHE assessed by RT-qPCR. The reference gene is RPLP0 (mean ± SD; n = 3; two-way ANOVA; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. TSG-6+/+). (c) TSG-6-encoding mRNA detection using in situ hybridization and hemalun counterstaining of TSG-6+/+ and TSG-6–/– RHE. Bar = 20 μm. (d) TSG-6 protein concentration of TSG-6+/+ and TSG-6–/– RHE assayed using ELISA (mean ± SD; n = 3; two-way ANOVA; ∗∗∗P < 0.001 vs. TSG-6+/+). CTL, control; HE, hemalun/eosin; IVL, involucrin; K, keratin; LOR, loricrin; RHE, reconstructed human epidermis.
Table 2.
Transcriptomic Analysis of TSG-6–/– Epidermis through RNA Sequencing
| TSG-6–/– (a) |
TSG-6–/– (b) |
||||
|---|---|---|---|---|---|
| ID | Gene | log2FC | Padj | log2FC | Padj |
| ENSG00000123610 | TNFAIP6 | –7.41503 | 2.96E–32 | –7.32736 | 3E–30 |
| ENSG00000234444 | ZNF736 | –4.44636 | 2.81E–22 | –4.69333 | 3.14E–18 |
| ENSG00000170430 | MGMT | –4.33712 | 8.06E–16 | –4.18849 | 3.18E–12 |
| ENSG00000105750 | ZNF85 | –3.87594 | 3.01E–37 | –2.40597 | 9.47E–27 |
| ENSG00000144115 | THNSL2 | –3.86052 | 2.91E–15 | –3.78559 | 1.02E–13 |
| ENSG00000187210 | GCNT1 | –3.0438 | 6E–50 | –2.61444 | 6.09E–38 |
| ENSG00000011465 | DCN | –2.82165 | 1.1E–84 | –1.5491 | 3.33E–29 |
| ENSG00000188257 | PLA2G2A | –2.7461 | 1.93E–24 | –2.00917 | 1.94E–14 |
| ENSG00000177990 | DPY19L2 | –2.37162 | 3.21E–10 | –2.42415 | 6.58E–09 |
| ENSG00000153802 | TMPRSS11D | –2.35278 | 9.53E–15 | –1.24544 | 0.00015 |
| ENSG00000129455 | KLK8 | –2.27628 | 3.22E–18 | –1.70997 | 1.05E–10 |
| ENSG00000122691 | TWIST1 | –2.26396 | 2.75E–06 | –2.35514 | 8.97E–08 |
| ENSG00000103546 | SLC6A2 | –2.23051 | 1.73E–11 | –1.01528 | 0.002253 |
| ENSG00000102837 | OLFM4 | –2.10858 | 1.07E–09 | –2.29476 | 3.6E–09 |
| ENSG00000188868 | ZNF563 | –2.01096 | 1.75E–17 | –2.394 | 7.67E–21 |
| ENSG00000244242 | IFITM10 | –1.93999 | 6.41E–12 | –1.9349 | 1.62E–10 |
| ENSG00000153292 | ADGRF1 | –1.93659 | 6.88E–08 | –1.08517 | 0.008469 |
| ENSG00000206195 | DUXAP8 | –1.88083 | 3.76E–05 | –2.0841 | 6.63E–07 |
| ENSG00000213096 | ZNF254 | –1.8603 | 1.41E–38 | –1.78 | 1.78E–33 |
| ENSG00000260027 | HOXB7 | –1.72059 | 2.43E–05 | –2.21449 | 1.82E–05 |
| ENSG00000081803 | CADPS2 | –1.64989 | 2.05E–05 | –1.67567 | 1.61E–05 |
| ENSG00000237440 | AC008554.1 | –1.61936 | 0.000136 | –2.20067 | 5.76E–08 |
| ENSG00000105357 | MYH14 | –1.48954 | 1.63E–16 | –1.0035 | 3.51E–07 |
| ENSG00000101017 | CD40 | –1.47799 | 5.85E–06 | –2.08413 | 1.64E–08 |
| ENSG00000172478 | MAB21L4 | –1.44783 | 9.05E–36 | –1.82477 | 1.8E–52 |
| ENSG00000197647 | ZNF433 | –1.36208 | 5.15E–08 | –2.48496 | 3.31E–17 |
| ENSG00000167754 | KLK5 | –1.34785 | 2.03E–16 | –1.92299 | 1.2E–30 |
| ENSG00000198182 | ZNF607 | –1.344 | 3.92E–17 | –1.09462 | 7.89E–11 |
| ENSG00000250644 | AC068580.4 | –1.33136 | 8.21E–07 | –1.14364 | 0.000121 |
| ENSG00000160284 | SPATC1L | –1.27579 | 0.001851 | –1.65145 | 0.000263 |
| ENSG00000196263 | ZNF471 | –1.20779 | 9.05E–05 | –1.20674 | 0.000435 |
| ENSG00000077063 | CTTNBP2 | –1.16597 | 7.2E–06 | –2.10572 | 5.59E–12 |
| ENSG00000235584 | AC008268.1 | –1.15826 | 2.67E–07 | –1.34538 | 1.76E–08 |
| ENSG00000091656 | ZFHX4 | –1.14386 | 0.00455 | –1.27665 | 0.005149 |
| ENSG00000196172 | ZNF681 | –1.13207 | 3.24E–16 | –1.08222 | 1.35E–13 |
| ENSG00000150687 | PRSS23 | –1.04503 | 8.83E–14 | –1.57255 | 8.84E–28 |
| ENSG00000139445 | FOXN4 | –1.03112 | 0.000567 | –1.48352 | 3.45E–06 |
| ENSG00000219665 | ZNF433-AS1 | –1.0184 | 0.003964 | –1.37053 | 0.000331 |
| ENSG00000130038 | CRACR2A | 4.155504 | 4.42E–37 | 1.339632 | 3.03E–05 |
| ENSG00000007038 | PRSS21 | 2.825615 | 4.82E–27 | 1.229979 | 3.46E–05 |
| ENSG00000248370 | LINC02434 | 2.61115 | 6.57E–43 | 1.637452 | 8.7E–16 |
| ENSG00000150722 | PPP1R1C | 2.394706 | 1.72E–10 | 1.089967 | 0.003993 |
| ENSG00000182389 | CACNB4 | 2.179791 | 1.85E–23 | 1.175044 | 1.83E–06 |
| ENSG00000131771 | PPP1R1B | 2.1777 | 1.76E–11 | 1.057933 | 0.004363 |
| ENSG00000141753 | IGFBP4 | 2.083609 | 1.45E–42 | 1.219661 | 3.22E–13 |
| ENSG00000108176 | DNAJC12 | 2.057347 | 2.89E–15 | 1.800988 | 4.46E–11 |
| ENSG00000234929 | AC018685.2 | 1.77764 | 1.76E–10 | 1.325183 | 1.51E–05 |
| ENSG00000166897 | ELFN2 | 1.660626 | 3.69E–06 | 1.268515 | 0.000785 |
| ENSG00000153064 | BANK1 | 1.481794 | 5.99E–08 | 1.306159 | 1.21E–05 |
| ENSG00000100968 | NFATC4 | 1.471711 | 2.94E–11 | 1.093399 | 9.51E–06 |
| ENSG00000251003 | ZFPM2-AS1 | 1.354186 | 3.59E–20 | 1.06091 | 2.67E–11 |
| ENSG00000131634 | TMEM204 | 1.305841 | 1.52E–10 | 2.388995 | 4.19E–33 |
| ENSG00000164684 | ZNF704 | 1.215393 | 4.55E–09 | 1.085985 | 1.47E–06 |
| ENSG00000141622 | RNF165 | 1.162247 | 7.5E–05 | 1.046022 | 0.001585 |
| ENSG00000179914 | ITLN1 | 1.144014 | 0.000195 | 1.104532 | 0.001204 |
| ENSG00000279108 | AC008537.3 | 1.114701 | 0.001084 | 1.41162 | 0.000283 |
| ENSG00000225968 | ELFN1 | 1.04966 | 0.002312 | 1.356617 | 0.000548 |
| ENSG00000172005 | MAL | 1.00604 | 0.000887 | 1.881428 | 3.6E–10 |
Abbreviations: Padj, adjusted P; FC, fold change; RHE, reconstructed human epidermis.
List of commonly downregulated and upregulated genes in both IL-4‒ and IL-13‒challenged TSG-6–/– RHE compared with challenged TSG-6+/+ RHE (FC ≥ 2; P < 0.01).
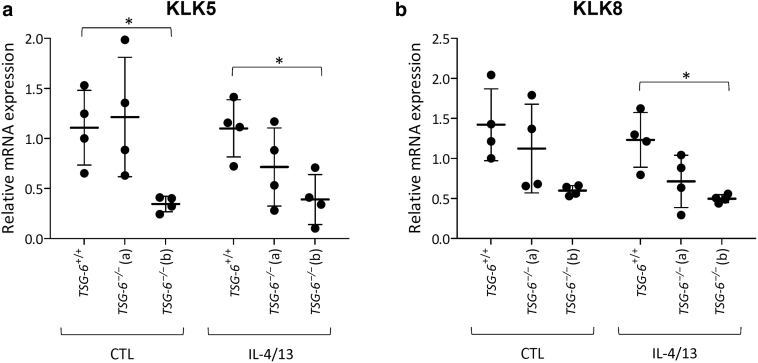
Figure 7.
mRNA levels of KLK genes are decreased in TSG-6–/– RHE.TSG-6+/+ and TSG-6–/– (a and b) RHEs were stimulated or not stimulated for 48 hours with IL-4 and IL-13. (a) KLK5 and (b) KLK8 mRNA expression assessed by RT-qPCR. The reference gene is RPLP0 (mean ± SD; n = 4; two-way ANOVA; ∗P < 0.05 vs. TSG-6+/+). CTL, control; KLK, kallikrein; RHE, reconstructed human epidermis.
Deficiency in TSG-6 protein slows down KC migration
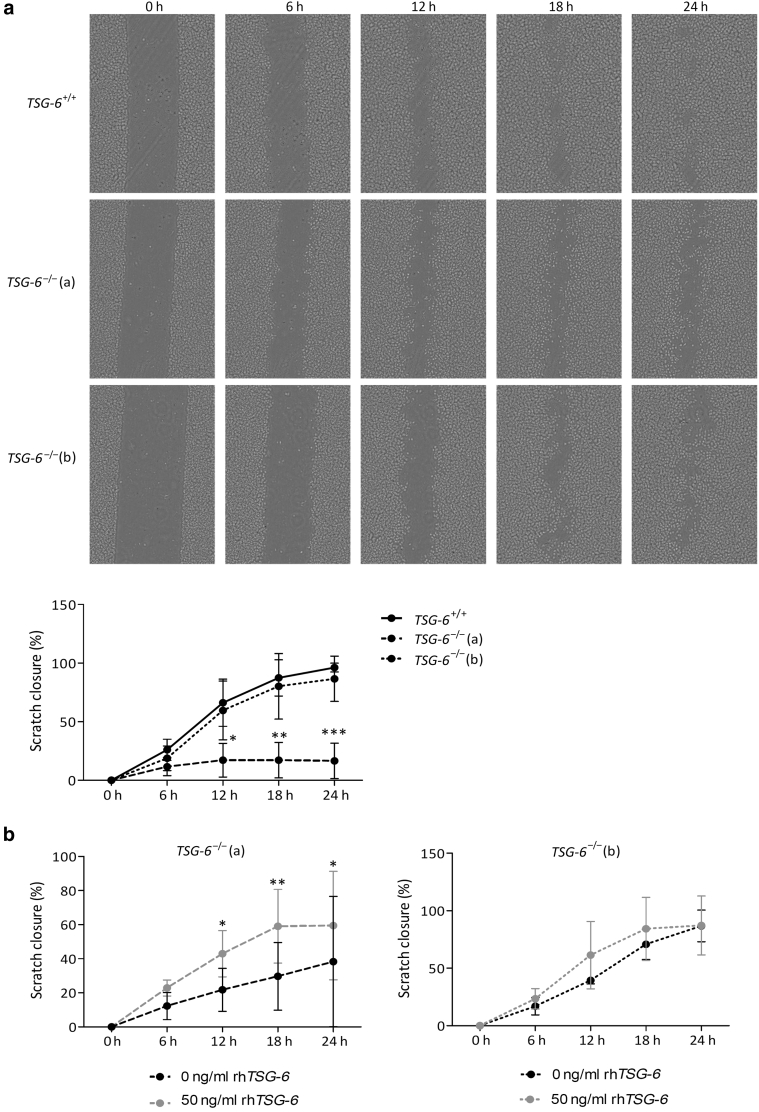
Because some genes implicated in migration (KLK5 and 8) were suppressed in TSG-6–/– RHE and because Tsg-6–/– mice show delayed wound closure (Shakya et al., 2020), the migrative ability of knocked-out TSG-6 KCs was monitored in a scratch assay. TSG-6–/– (a) N/TERT KCs migrated more slowly than TSG-6+/+ KCs, being unable to close the scratch within 24 hours. Migration of TSG-6–/– (b) N/TERT KCs was also slower than that of nonedited N/TERT cells (Figure 8a). The migration defect was partially restored in the presence of recombinant TSG-6 (Figure 8b).
Figure 8.
Lack of TSG-6 slows down KCs migration. Monolayers cultures of TSG-6+/+ or TSG-6–/– (a and b) N/TERT KCs at confluence stage treated with IL-4 and IL-13 for 24 h. KCs were growth arrested by mitomycin C before performing a scratch. KCs migration was monitored for 24 h in the presence of IL-4 and IL-13. (a) Scratch closure was analyzed every 6 h by Cytonote and was quantified by ImageJ software (mean ± SD; n = 5; two-way ANOVA; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. TSG-6+/+). (b) Recombinant human TSG-6 protein (50 ng/ml) was added in the culture medium just after scratching monolayers cultures of TSG-6–/– (a) and (b) cells (mean ± SD; n = 3 for TSG-6–/– (a) and n = 4 for TSG-6–/– (b); two-way ANOVA; ∗P < 0.05; ∗∗P < 0.01 vs. 0 ng/ml rhTSG-6). Scratch closure was monitored by Cytonote and quantified by ImageJ software. h, hour; KC, keratinocyte; rhTSG-6, recombinant human TSG-6.
Deficiency in TSG-6 protein is responsible for enhanced leakage of HA from the epidermis
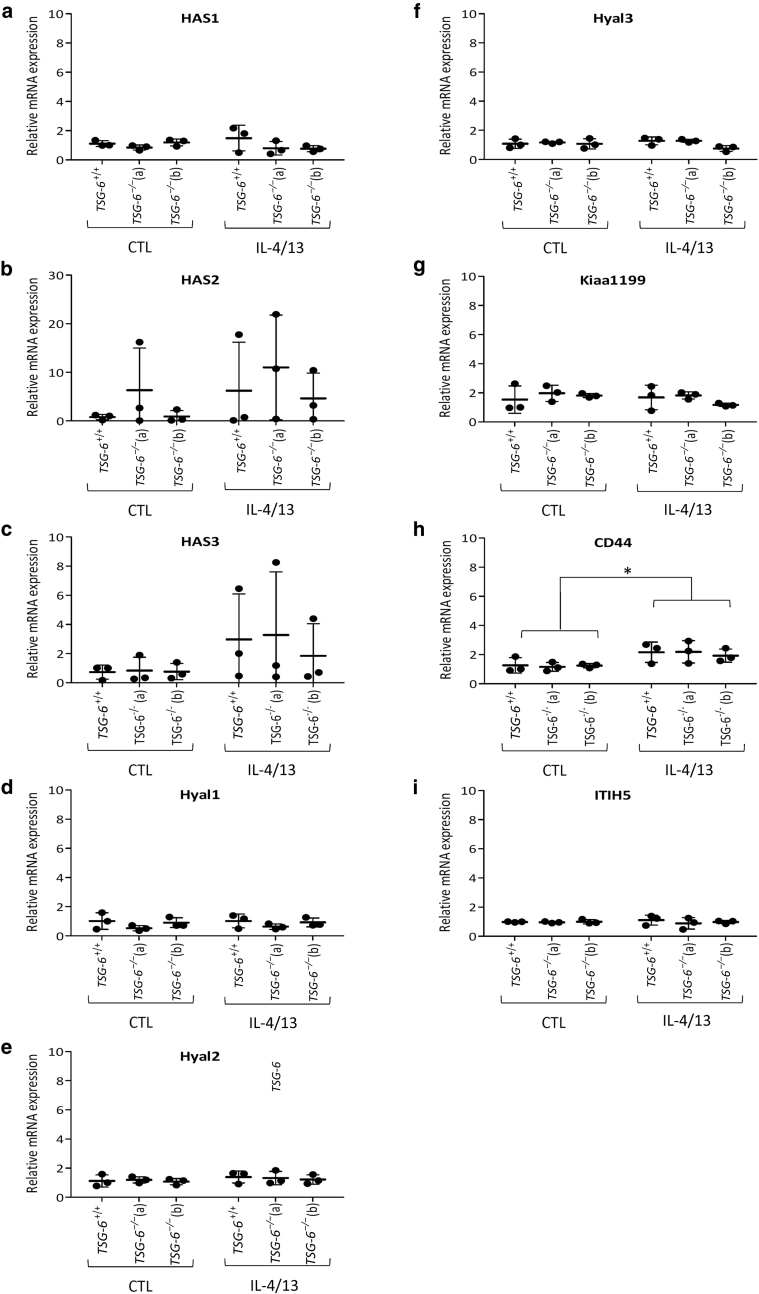
The effect of TSG-6 deficiency on the synthesis and degradation of HA as well as on HA partners (CD44 and ITIH5) was investigated at the mRNA level. No alteration in knock-out tissues was observed (Figure 9). CD44 and HAS3 appeared overexpressed in both TSG-6+/+ and TSG-6–/– tissues stimulated with IL-4 and IL-13, in accordance with previous results (De Vuyst et al., 2018; do Nascimento Pedrosa et al., 2017; Malaisse et al., 2014).
Figure 9.
mRNA levels of genes related to HA are unchanged in TSG-6–/– RHE.TSG-6+/+ and TSG-6–/– (a and b) RHEs were stimulated or not stimulated for 48 hours with IL-4 and IL-13. (a) HAS1, (b) HAS2, (c) HAS3, (d) Hyal1, (e) Hyal2, (f) Hyal3, (g) Kiaa1199, (h) CD44, and (i) ITIH5 mRNA expression assessed by RT-qPCR. The reference gene is RPLP0 (mean ± SD; n = 3; two-way ANOVA; ∗P < 0.05). CTL, control; HA, hyaluronan; RHE, reconstructed human epidermis.
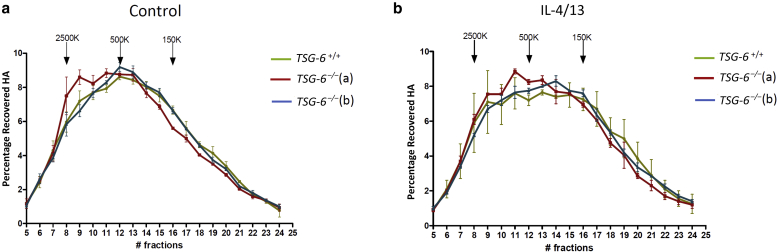
Because HAS3 is enhanced in RHE exposed to IL-4 and IL-13 and produces proinflammatory low-molecular-weight HA fragments (Weigel et al., 1997), HA released in the culture medium was analyzed using size exclusion chromatography (Figure 10). In control conditions, both TSG-6+/+ and TSG-6–/– molecular-weight distribution of HA revealed similar profiles with a peak around 500 kDa (Figure 10a). When RHEs were stimulated with IL-4 and IL-13, the size of HA slightly shifted to lower molecular weights in both nonedited and knock-out tissues (Figure 10b).
Figure 10.
Similar molecular-weight distribution of HA released in the culture medium of normal and challenged TSG-6–/– RHE. The culture medium of TSG-6+/+ and TSG-6–/– (a and b) RHE (a) control or (b) treated with IL-4 and IL-13 for 48 hours was collected and chromatographed on a Sephacryl S-1000 column. HA fragments of 2,500, 500, and 150 kDa were used as standards. Graphs compared HA size distribution in each fraction (mean ± SD; n = 2). #, number of; HA, hyaluronan; K, thousand; RHE, reconstructed human epidermis.
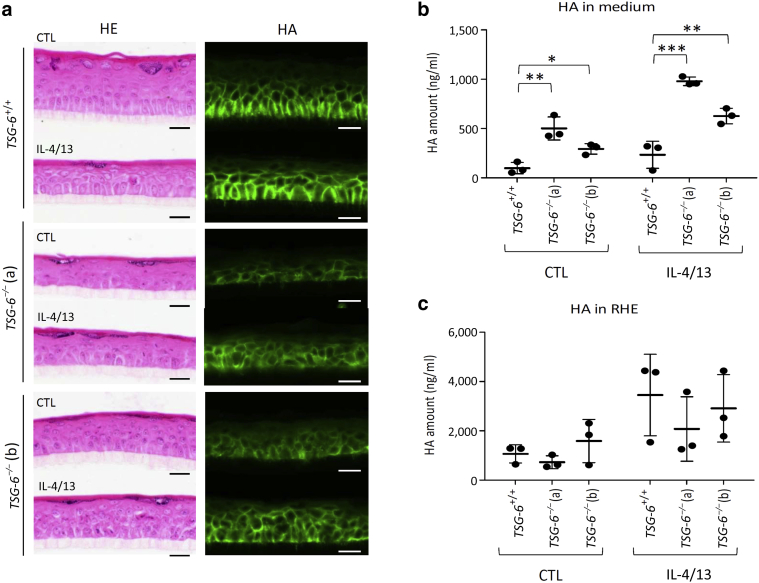
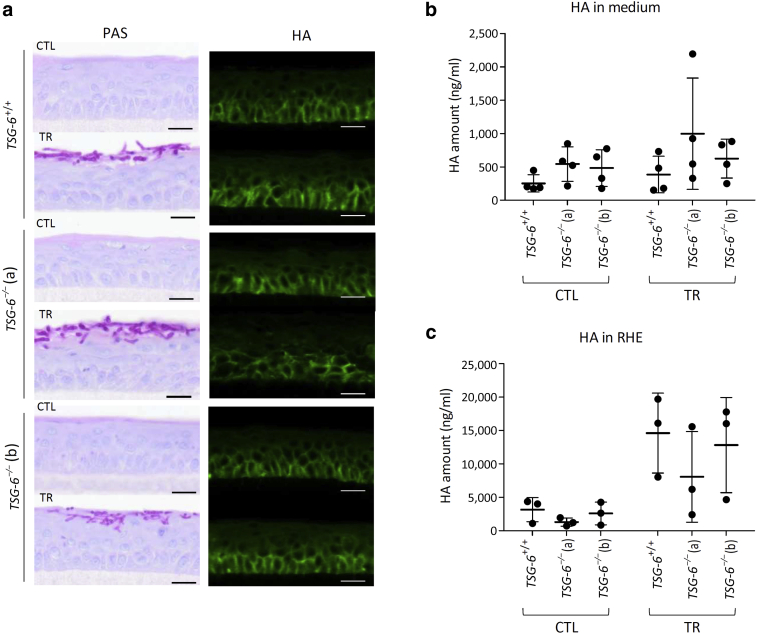
Morphologically, HA distribution inside TSG-6+/+ and TSG-6–/– tissues appeared similar. However, the intensity of the HA fluorescent signal was weaker in TSG-6–/– than in TSG-6+/+ RHE (Figure 11a). This concurred with a larger HA amount released into the culture medium from TSG-6–/– than that from TSG-6+/+ RHE, especially after IL-4 and IL-13 treatment (Figure 11b). Simultaneously, the amount of HA inside TSG-6–/– RHE was reduced, although it was nonsignificant (Figure 11c). The distribution of HA was similar in TSG-6+/+ and TSG-6–/– RHE infected with dermatophytes, and HA leakage was increased from infected TSG-6–/– RHE, concomitantly with a reduced content inside tissues. However, the differences were nonsignificant (Figure 12).
Figure 11.
The change of HA localization in TSG-6–/– RHE is enhanced in Th2-challenged conditions.TSG-6+/+ and TSG-6–/– (a and b) RHEs were stimulated or not stimulated for 48 hours with IL-4 and IL-13. (a) HE staining and HA detection (green). Bar = 20 μm. (b) HA released in culture medium measured by ELISA-like assay (mean ± SD; n = 3; two-way ANOVA; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. TSG-6+/+). (c) HA remaining inside RHE (mean ± SD; n = 3; two-way ANOVA). CTL, control; HA, hyaluronan; HE, hemalun/eosin; RHE, reconstructed human epidermis; Th, T helper type.
Figure 12.
The change of HA localization in TSG-6–/– RHE seems to be enhanced by TR infection.TSG-6+/+ and TSG-6–/– (a and b) RHE infected or not infected with TR and recovered 4 days after infection. (a) PAS staining with α-amylase pretreatment and hemalun counterstaining and HA detection. Bar = 20 μm. (b) HA released in culture medium measured by ELISA-like assay (mean ± SD; n = 4; two-way ANOVA). (c) HA remaining inside RHE (mean ± SD; n = 3; two-way ANOVA). CTL, control; HA, hyaluronan; PAS, periodic-acid schiff; RHE, reconstructed human epidermis; TR, Trichophyton rubrum.
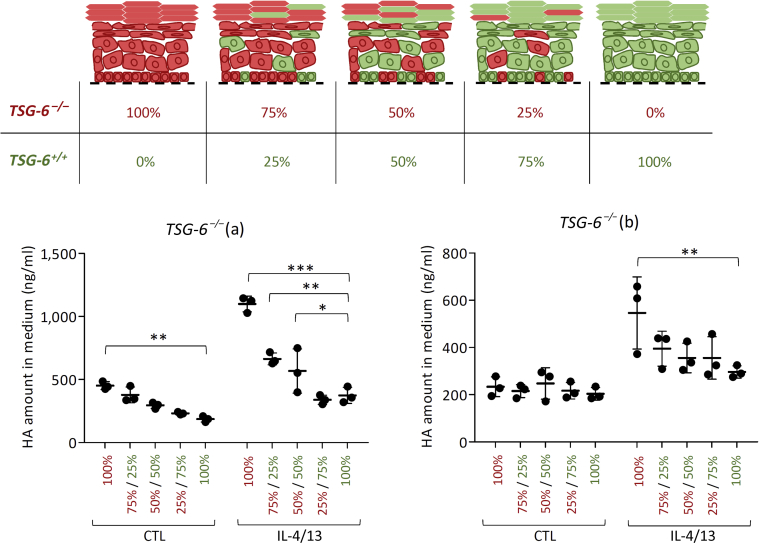
To reintroduce TSG-6 in knocked-out tissues, knocked-out cells were mixed with nonedited N/TERT cells in varying proportions while reconstructing tissues (Figure 13). Interestingly, HA leakage from TSG-6–/– tissues was rescued depending on the proportion of TSG-6‒producing cells (Figure 13).
Figure 13.
Introduction of TSG-6+/+ cells in TSG-6–/– RHE can rescue HA leakage. Schematic representation of TSG-6–/– rescue test: epidermis were reconstructed from different ratios of TSG-6+/+ (green) and TSG-6–/– (red) KCs. HA released in the culture medium of epidermis reconstructed from mixtures of KCs TSG-6+/+ and TSG-6–/– (a) or TSG-6–/– (b) was measured by ELISA-like assay (mean ± SD; n = 3; two-way ANOVA; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 vs. TSG-6+/+ 100%). CTL, control; HA, hyaluronan; KC, kerationcyte; RHE, reconstructed human epidermis.
For yet unidentified reason, clone TSG-6–/– (a) constantly releases a higher amount of HA than clone TSG-6–/– (b) as shown in Figures 11b and 14a despite similar expression levels of HASs and hyaluronidases (Figure 9).
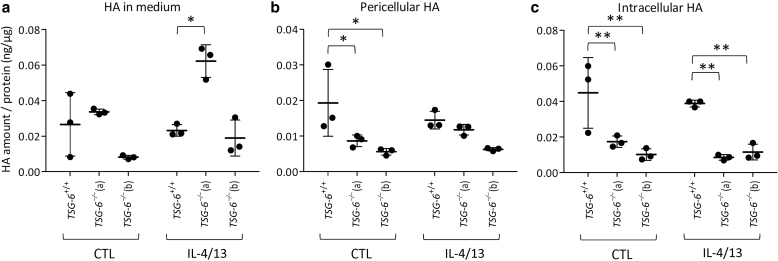
Figure 14.
Change of HA localization in TSG-6–/– KCs cultured in monolayers. Autocrine monolayers cultures of TSG-6+/+ or TSG-6–/– (a and b) N/TERT KCs at confluence stage treated or not treated with IL-4 and IL-13 for 48 hours. HA amount (a) released in the culture medium present (b) in pericellular spaces and (c) in intercellular compartment measured by ELISA-like assay and relative to the total amount of protein (mean ± SD; n = 3; two-way ANOVA; ∗P < 0.05; ∗∗P < 0.01 vs. TSG-6+/+). CTL, control; HA, hyaluronan; KC, keratinocyte.
In addition, pericellular HA was quantified. The RHE model does not allow to distinguish pericellular from intracellular compartment. Therefore, HA distribution was studied using monolayer cultures stimulated or not with IL-4 and IL-13 for 48 hours at the confluence. Whereas HA released in the culture medium of TSG-6–/– monolayers (Figure 14a) was increased in response to treatment, pericellular (Figure 14b) or intracellular (Figure 14c) HA was significantly reduced.
Discussion
In normal conditions, TSG-6 is weakly expressed in mouse and human skin (Shakya et al., 2020; Tan et al., 2011). During in vitro reconstruction of the human epidermis, TSG-6 expression remains very weak as well.
In infected RHE, TSG-6 expression is induced in response to recognition of fungal elements by toll-like receptors, leading to production of antimicrobial peptides and cytokines such as TNF-α (Faway et al., 2019; Lee et al., 1990). TSG-6 expression is also induced in RHE exposed to Th2 cytokines to mimic typical features of AD epidermis (do Nascimento Pedrosa et al., 2017), and HA production simultaneously increases as a result of HAS3 overexpression (De Vuyst et al., 2018; Malaisse et al., 2014). In vivo, TSG-6 expression is increased in lesional AD skin and in contact dermatitis (GEO accession GSE57225 [Quaranta et al., 2014] and GEO accession GSE711996 [Tan et al., 2017]) and is upregulated in Th1/Th17 diseases such as psoriasis (GEO accession GSE166388) and wound healing (GEO accession GSE50425). Altogether, these observations show the involvement of TSG-6 in many inflammatory conditions.
To understand the role of epidermal TSG-6, deletions in the TNFAIP6 gene were carried out by CRISPR/Cas9 in N/TERT KCs. This immortalized human cell line was chosen because it allows for proper reconstruction of epidermis able to respond to Th2 cytokines (Dickson et al., 2000; Moran et al., 2021; Rheinwald et al., 2002; Smits et al., 2017). Recently, genome editing was performed in N/TERT cells using Cas9 plasmids for studies of Harlequin ichthyosis (Enjalbert et al., 2020) to target the genes linked to glycan metabolism (Dabelsteen et al., 2020) and to knockout claudin 1 and CD40 (Moran et al., 2021). In this study, ribonucleoprotein/Cas9 complexes were preferred to plasmid to reach higher efficiency of gene editing and benefit from a lower occurrence of off targets (Farboud et al., 2018). Indeed, all analyzed N/TERT cell clones harbored mutations. Among them, two clones exhibiting major deletions that conduct to null alleles were chosen for further studies. No off-target effect was found by screening RNA-seq results.
TSG-6–/– KCs were used for RHE production. These tissues exhibit normal morphology in accordance with the unaltered epidermal phenotype of Tsg-6-knockout mice (Fülöp et al., 2003). In control and challenging conditions, deficiency of TSG-6 has no severe impact on gene expression levels as revealed by transcriptomic analysis. We noticed that the expression of decorin, a proteoglycan playing a role in cutaneous inflammatory reactions (Bocian et al., 2013), is downregulated. Of interest, KLK5 and KLK8 involved in skin homeostasis, KCs migration, and wound remodeling (Kishibe et al., 2012; Nauroy and Nyström, 2020) are also downregulated.
In the scratch assay, whereas TSG-6–/– N/TERT clone (b) shows a very discreet slowdown, the second clone (a) migrates more slowly than control cells, reminiscent of the delayed wound closure in Tsg-6–/– mouse skin (Shakya et al., 2020). The cell migration rate is partially restored by reintroduction of soluble recombinant TSG-6 in the medium, as reported with epithelial corneal cells (Lee et al., 2015).
Because the amount of HA is decreased in the pericellular matrix of TSG-6–/– KCs, we hypothesize that the defect in migration could result from poor pericellular HA organization. A disorganized ECM was described around preovulatory follicles in Tsg-6‒knockout mice (Fülöp et al., 2003). The HA interaction with its CD44 membrane receptor is facilitated by TSG-6 (Baranova et al., 2013), and this interaction promotes HA internalization together with proliferation and migration (Bourguignon, 2014; Csoka et al., 2001; Pasonen-Seppänen et al., 2012). TSG-6 deficiency may therefore influence the HA‒CD44 signaling axis and pathways located downstream of other receptors such as EGFR (Perez et al., 2013).
In TSG-6–/– RHE, a change in the localization of HA is observed. A large amount of HA diffuses into the subepidermal compartment, suggesting a role for TSG-6 in maintaining HA inside the tissue. HA leakage from the epidermis was already reported in skin equivalent models and was attributed to an improper organization of the basement membrane (Iriyama et al., 2021; Tammi et al., 2000). Nonetheless, Rosdy et al. (1993) had illustrated typical basal lamina elements in RHE. Because TSG-6 is not a constituent of the basement membrane, enhanced release of HA from RHE made of TSG-6–/– cells cannot be attributed to a lack of integrity or to an altered composition of the basement membrane. We propose that the lack of HA retention in ECM is a more likely explanation.
Future studies are required to complement the data obtained with our single-cell type in vitro model. Understanding the consequences of massive HA release due to the absence of TSG-6 on underlying dermal fibroblasts and immune cells is of interest. For instance, recruitment and migration of immune cells to an inflammatory site depend on the CD44-mediated organization of HA into cables (Jokela et al., 2008; Kobayashi et al., 2020).
Our serum-free culture conditions do not allow to assess the enzymatic activity of TSG-6. Adding one source of ITI is required to investigate HA covalent cross-linking through a heavy chain. More precisely, the involvement of heavy chain 5 in ECM stabilization was shown in AD and psoriasis and should therefore be further analyzed (Huth et al., 2020).
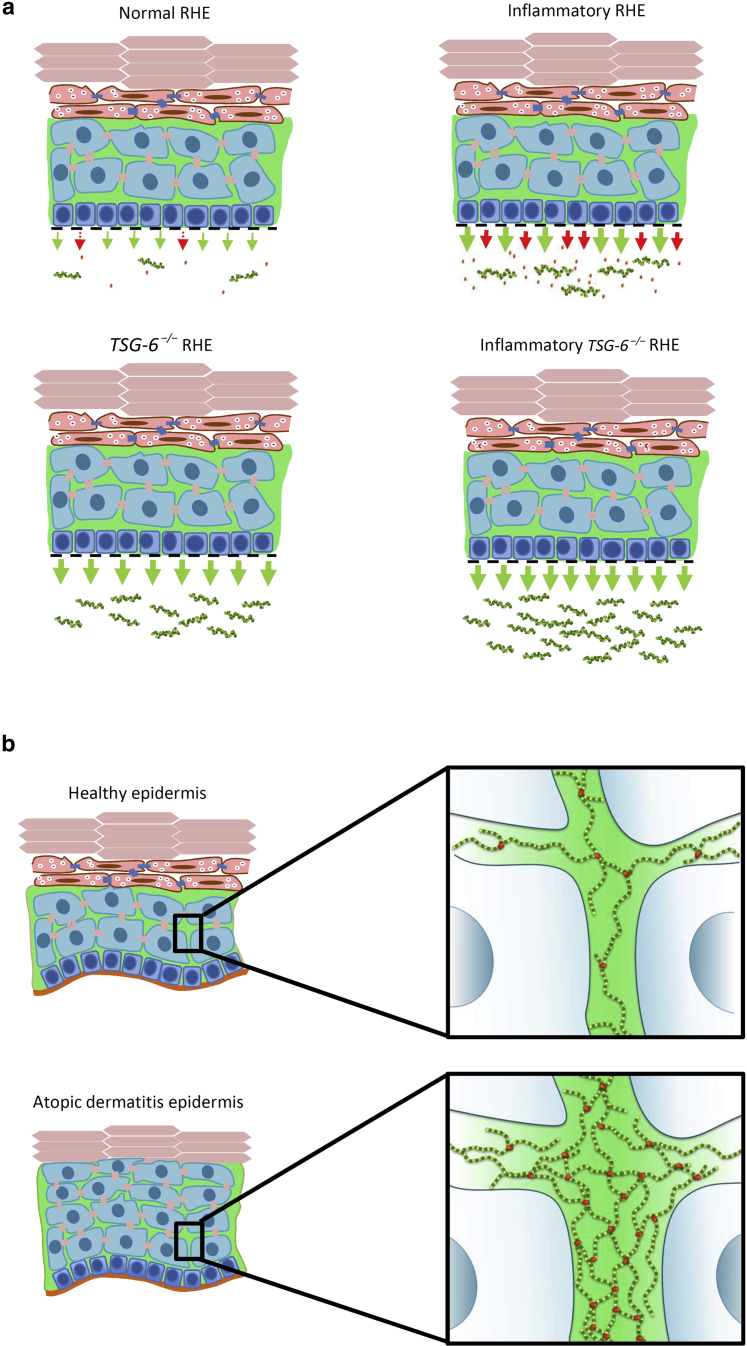
In conclusion, our data suggest a role for TSG-6 in retaining HA in inflamed epidermis by interaction and cross-linking in the ECM, as shown in other tissues (Day and Milner, 2019). In AD lesions, Th2-stimulated production of TSG-6 could participate in HA accumulation between KCs (Malaisse et al., 2014) (Figure 15). More broadly, when interacting with ECM components, TSG-6 potentially modulates the presentation of GFs and cytokines to neighboring receptors. In the epidermis, it could influence the signaling pathways related to HA (Kavasi et al., 2017) and play roles in inflammatory skin conditions.
Figure 15.
TSG-6 retains HA in the ECM of the epidermis. (a) Schematic representations of in vitro observations in TSG-6+/+ and TSG-6–/– RHE challenged or not challenged. In control conditions, weak amounts of HA (green) and TSG-6 (red) are released in the culture medium. In inflammatory conditions, increased TSG-6 secretion goes along with enhanced HA release. In the absence of TSG-6, HA release is significantly increased, especially after the challenge. (b) Hypothetical role of TSG-6 in HA distribution and organization in healthy and lesional atopic dermatitis epidermis. In the healthy epidermis, the weak amount of secreted TSG-6 is involved in ECM organization by HA cross-linking. In atopic dermatitis lesions, enlarged intercellular spaces go along with enhanced HA production. Secreted TSG-6 links HA cables to form a dense matrix. ECM, extracellular matrix; HA, hyaluronan; RHE, reconstructed human epidermis.
Materials and Methods
KCs origins, cultures, and reconstruction of the epidermis
Skin biopsies (University Hospital in Liège, Belgium) from patients with AD were collected on lesional zones as described (Mathay et al., 2011). Healthy skin biopsies and normal human KCs were isolated from abdominoplasties (Clinique St Luc, Namur, Belgium) as described (Minner et al., 2010). All samples were obtained after written informed consent, in accordance with Medical Ethical Committees of the University Hospital in Liège, Clinique St-Luc in Namur, and Saint-Louis Hospital in Paris (France) and according to the Declaration of Helsinki Principles. Immortalized human KCs (N/TERT-1) were obtained from J.G. Rheinwald’s laboratory (Dickson et al., 2000). Cells authenticity was assessed at ATCC (Manassas, VA) (http://www.atcc.org) by a comparison of an original frozen vial of cells (Bertrand-Vallery et al., 2010) with current cultured N/TERT cells. Normal and immortalized KCs were cultured as autocrine monolayer cultures (Minner et al., 2010) or were grown on a polycarbonate filter and exposed to the air‒liquid interface for 11 days to produce a reconstructed epidermis (De Vuyst et al., 2014; Frankart et al., 2012). Whereas autocrine monolayers were cultured in EpiLife culture medium supplemented by amino acids (Sigma-Aldrich, St. Louis, MO) and hydrocortisone (0.18 μg/ml; Sigma-Aldrich), RHEs were grown in EpiLife medium containing Human Keratinocytes Growth Supplement (Cascade Biologics, Mansfield, United Kingdom) and calcium ion (1.5 mM), KC GF (10 ng/ml) (R&D Systems, Abingdon, United Kingdom), and vitamin C (50 mg/ml). Both cultures were maintained at 37 °C and 5% carbon dioxide.
Challenges applied to RHE to mimic AD or dermatophytosis
A described procedure was used to mimic AD alterations in RHE (De Vuyst et al., 2018). Briefly, on the 11th day of reconstruction and after cholesterol depletion with methyl-β-cyclodextrin (7.5 mM; Sigma-Aldrich, Diegem, Belgium) for 2 hours, RHEs were incubated for 48 hours with IL-4 (50 ng/ml), IL-13 (50 ng/ml), and IL-25 (20 ng/ml) (PeproTech, London, United Kingdom). RHEs were eventually challenged with IL-4 and IL-13 only for 48 hours to produce AD-like alterations in a simpler way (do Nascimento Pedrosa et al., 2017).
Another challenge was applied using infection with T. rubrum arthroconidia (Faway et al., 2017). Briefly, topical application of dermatophyte arthroconidia in suspension in PBS over RHE was performed on the 11th day of reconstruction. After 4 hours of exposure to these dermatophytes, RHEs were washed with PBS and then re-exposed to air for 4 additional days of culturing. This protocol was adapted for infection of RHE using N/TERT KCs. Owing to a weaker barrier, an inoculum of 850 arthroconidia per cm2 was used instead of 1,700 per cm2.
Scratch assay
KCs migration was studied using a scratch assay in culture (Mound et al., 2017). Confluent monolayers were treated 1 hour with 10 μg/ml mitomycin C (#sc 3514B; Santa Cruz Biotechnology, Dallas, TX) before scraping using a p200 micropipette tip and then washed and incubated with fresh medium for 24 hours. Rescue was performed by adding 50 ng/ml recombinant human TSG-6 (#2104-TS; R&D Systems) in the culture medium during the 24 hours of scratch closure. The scratch closure was monitored by time-lapse imaging using an Iprasense Cytonote device (Iprasense, Montpellier, France) and analyzed by ImageJ software (National Institutes of Health, Bethesda, MD).
Histological staining, immunofluorescence, and HA labeling
For morphological analysis, RHE and skin biopsies were fixed in a 4% formaldehyde, 70% ethanol, and 5% acetic acid solution (Lin et al., 1997) and then dehydrated and embedded in paraffin to prepare 7 μm tissue sections using a microtome. Tissue sections were stained with hemalun-eosin(-saffron). Infected RHEs were stained by periodic-acid schiff after incubation of sections with α-amylase (Faway et al., 2017).
For immunolabeling, tissue sections were incubated 1 hour at room temperature with primary anti-keratin 10 (1:100 dilution, Dako, Glostrup, Denmark), anti-involucrin (1:200 dilution, Sigma-Aldrich), and anti-loricrin (1:100 dilution, Abcam. Cambridge, United Kingdom) or overnight at 4 °C with primary anti-FLG (1:75 dilution, Thermo Fisher Scientific, Waltham, MA) antibodies and then incubated 1 hour at room temperature with secondary antibodies conjugated with Alexa fluorochrome A488: anti-mouse diluted 1:200 for keratin 10, involucrin, and FLG and anti-rabbit diluted at 1:500 for loricrin (Life Technologies, Carlsbad, CA).
HA was localized on tissue sections using an overnight incubation at 4 °C with a biotinylated HA-binding protein at 1:100 dilution (Calbiochem-Merck, Darmstadt, Germany), followed by 1-hour incubation at room temperature with fluorescein-labeled streptavidin (Vector Laboratories, Burlingame, CA).
Sections were mounted with coverslips using Mowiol (Sigma-Aldrich) and analyzed under an Olympus BX63 microscope.
RNA extraction and RT-qPCR
Total RNA was extracted using a High Pure RNA isolation kit (Roche, Basel, Switzerland) for monolayers and an RNeasy kit (Qiagen, Hilden, Germany) for RHE. First-strand cDNA was synthesized using SuperScript III (Invitrogen, Carlsbad, CA) and amplified by FastStart Universal SYBR Green Master (Roche) in the Light Cycler (Light Cycler 96 Instrument; Roche). Relative mRNA expression level was normalized to RPLP0 reference gene (Minner and Poumay, 2009) and calculated using the ΔΔCq method (Livak and Schmittgen, 2001). Specifics primers (Eurogentec, Liège, Belgium) were used at 300 nM and are listed in Table 3.
Table 3.
The Sequence of Primer Pairs Used for RT-qPCR
| Gene Symbol | Forward Primer | Reverse Primer |
|---|---|---|
| RPLP0 | ATCAACGGGTACAAACGAGTC | CAGATGGATCAGCCAAGAAGG |
| TSG-6 | GCTAGAGGCAGCCAGAAAAA | ATCCAACTCTGCCCTTAGCC |
| HAS1 | CACCTCACCAACCGCATGCT | GTCTGCTGGCTCAGCCACCG |
| HAS2 | CCTCATCATCCAAAGCCTGT | GATGCAAAGGGCAACTGTTT |
| HAS3 | GCGATTCGGTGGACTACAT | GGATCCTCCTCCAGGACTC |
| Hyal1 | TCCAGATCTTCTATGACACGACAAA | TCCTTGATGGCCTGACATGA |
| Hyal2 | GCACTCCCAGTCTACGTCTTCA | GCACTCTCGCCAATGGTAGAG |
| Hyal3 | GATCTGGGAGGTTCCTGTCC | AGAGCTGGAGAGGCTCAGGT |
| Kiaa1199 | TACTGGGACGAGGACTCAGG | TCACAGCCTTTCATGGAGCA |
| CD44 | CATTGCAGTCAACAGTCGAAGAA | ATTGCCACTGTTGATCACTAGCTT |
| ITIH5 | ACTGTCGCTGGAGAACTGTG | CGGGGTCCTGATTTCATCGT |
| LL37 | CCAGGACGACACAGCAGTCA | CTTCACCAGCCCGTCCTTC |
RNA-seq
Total RNA was extracted using the ReliaPrep RNA Tissue Miniprep System (Promega, Madison, WI) before checking integrity through an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). RNA-seq and standard analysis were performed by the GIGA-Genomics platform (University of Liège, Belgium). RNA-seq data are available in the ArrayExpress database at EMBL’s European Bioinformatics Institute (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-10095.
ELISA assay
TSG-6 protein levels were quantified in a culture medium using a homemade ELISA assay. Plates were coated overnight with an anti‒TSG-6 capture antibody (#A38.1.20; Santa Cruz Biotechnology) and were blocked for 1 hour with 0.5% BSA in PBS. After 2 hours of incubation in the presence of samples of culture medium or in the presence of recombinant human TSG-6 protein (#2104-TS; R&D Systems) samples at different concentrations chosen to calibrate the assay, TSG-6 was secondarily detected using an anti‒TSG-6 biotinylated antibody (#BAF2104; R&D Systems), horseradish peroxidase-streptavidin, and chromogenic substrate (R&D Systems). The concentration of TSG-6 protein is proportional to absorbance (λ: 450–570 nm) measured by spectrophotometer (Molecular Device, Berkshire, United Kingdom).
Procedures for HA analysis
The amount of HA present in the culture medium, in pericellular, and in intracellular compartments was analyzed using Hyaluronan DuoSet kit (R&D Systems) as described by Malaisse et al. (2014).
To measure the size of HA fragments, the total amount of HA recovered in the RHE culture medium was assessed using the Hyaluronan DuoSet kit. Samples were then prepared to load an identical amount of HA (10 ng of HA per sample) on a Sephacryl S-1000 column (Amersham Biosciences, Uppsala, Sweden). Samples were eluted at 70 μl/min with 0.15 M sodium acetate and 0.1% CHAPS (Sigma-Aldrich, Steinheim, Germany) to collect 30 fractions from each sample. Standard HA fragments were chromatographed in identical conditions.
In situ hybridization
RNAscope kit was used (Advanced Cell Diagnostics, Silicon Valley, CA) in accordance with the manufacturer’s instructions (RNAscope 2.5 HD assay Red). Tissue sections were targeted by specific probes Homo sapiens-TNFAIP6 (reference 444421). Homo sapiens-PPIB (Peptidyl-prolyl cis-trans isomerase B) probe (reference 313901) was used as the positive control, and DapB (Bacillus subtilis dihydrodipicolinate reductase) probe (reference 310043) as the negative control. Fast Red staining in sections counterstained by hemalun (blue) reveals sequence hybridization as described by Wang et al. (2012).
Genome editing by CRISPR/Cas9
N/TERT KCs genome was edited to generate TNFAIP6-knockout cells. The procedure is a modified version of what is described in the user guide provided with the Alt-R CRISPR-Cas9 system (Integrated DNA Technologies, Coralville, IA).
Practically, CRISPR RNA (crRNA) trans-activating crRNA duplexes were formed by mixing crRNA (200 μM) and trans-activating crRNA (200 μM, #1072532; Integrated DNA Technologies), hybridizing them by heating up to 95 °C for 5 minutes, and then cooling them down to room temperature. Five crRNAs were designed using http://crispor.tefor.net/, https://zlab.bio/guide-design-resources and eu.idtdna.com website and are listed as follows: #Exon1 crRNA Up 5′-TTGCTATGGGAAGACACTCA AGG-3′, #Exon1 crRNA Down 5′-AAGGTTCTTACCAAGCCATA TGG-3′, #Exon3 crRNA Up 5′-ATTATGGAATCCGTCTCAAT AGG-3′, #Exon6 crRNA Up 5′-GAGAATCGACCTCTGAATTC TGG-3′, and #Exon6 crRNA Down 5′-GGTCATGACATTTCCTGTTG GGG-3' (Figure 5a). Duplexes were then mixed to Cas9 enzyme (#1081058; Integrated DNA Technologies) to form ribonucleoprotein complexes, which were electroporated (Amaxa Nucleofector I device, #AAD-1001; Lonza, Bâle, Switzerland) in proliferative N/TERT KCs according to the manufacturer’s instructions (Amaxa Human Keratinocyte Nucleofector Kit of Lonza, #VAPD-1002). After 48 hours of recovery, edited cells were seeded in 96-well plates at low density to obtain clonal populations (limiting dilution).
The mutations in the TNFAIP6 gene were amplified by PCR. PCR products corresponding to homologous alleles of TSG-6 were isolated by TA cloning and sequenced by Sanger sequencing (Genewiz company, Takeley, United Kingdom). PCR primers (Eurogentec) used at 10 μM were exon 1 forward 5′-ACAGCGGAAAATGCCATTGG-3′, exon 1 reverse 5′-TCTCAACAAGAGGCTCATGAGT-3′, exon 3 forward 5′-TTGGAAGTGGCTATCTTTGCT-3′, exon 3 reverse 5′-GGAACACAACTCAATGTCGGG-3′, exon 6 forward 5′-CATGCGGCAATGAAACCACA-3′, and exon 6 reverse 5′-AAGTGGCTAAATCTTCCAGCT-3' (Figure 5b).
Statistical analyses
All data were analyzed using SigmaPlot software, version 11.0 (Systat Software, Chicago, IL). Statistical differences were evaluated by Student t-test, one-way ANOVA, and two-way ANOVA repeated measure tests, and error bars represented SD. All graphs were performed using GraphPad Prism 5 software (GraphPad Software, San Diego, CA).
Data availability statement
Dataset related to this article can be found at https://data.mendeley.com/datasets/4fbgg2syzk/1, an open-source online data repository hosted at Mendeley Data.
ORCIDs
Céline Evrard: http://orcid.org/0000-0002-7450-0759
Emilie Faway: http://orcid.org/0000-0003-3406-1103
Evelyne De Vuyst: http://orcid.org/0000-0001-7152-1370
Olivier Svensek: http://orcid.org/0000-0001-9254-0060
Valérie De Glas: http://orcid.org/0000-0001-7366-2601
David Bergerat: http://orcid.org/0000-0003-0013-6621
Michel Salmon: http://orcid.org/0000-0002-1728-8448
Olivier De Backer: http://orcid.org/0000-0002-1490-4508
Bruno Flamion: http://orcid.org/0000-0003-4458-7627
Hélène Le-Buanec: http://orcid.org/0000-0002-2139-0357
Catherine Lambert de Rouvroit: http://orcid.org/0000-0002-0273-8995
Yves Poumay: http://orcid.org/0000-0001-5200-3367
Author Contributions
Conceptualization: CE, CLDR, BF, YP; Data Curation: CE, CLDR, YP; Formal Analysis: CE; Funding Acquisition: CLDR, YP; Investigation: CE, VDG, EF, MS, DB; Methodology: CE, EF, EDV, OS, ODB, DB, HLB, CLDR, YP; Project Administration: CE, CLDR, BF, YP; Supervision: CE, CLDR, YP; Validation: CE, CLDR, YP; Visualization: CE, CLDR, YP; Writing - Original Draft Preparation: CE, CLDR, YP; Writing - Review and Editing: CE, CLDR, YP
Acknowledgments
We acknowledge B. Bienfait and J.S. Blairvacq for providing skin samples and A. Nikkels for providing atopic dermatitis biopsies. We gratefully thank K. De Swert, V. Bielarz, and D. Desnoeck for their technical support; M. Caruso for the bioinformatics analyses; J. Malaisse for schemas and advice; and Marc Thiry for his skills in transmission electron microscopy analysis. EF was financially supported by the Région Wallonne (MYCEPI, convention n°1910074), and OS is a research fellow funded by FRIA (Fund for Research training in Industry and Agriculture), Belgium.
Conflict of Interest
The authors state no conflict of interest.
accepted manuscript published online XXX; corrected proof published online XXX
Footnotes
Cite this article as: JID Innovations 2021;X:100054
References
- Baranova N.S., Foulcer S.J., Briggs D.C., Tilakaratna V., Enghild J.J., Milner C.M., et al. Inter-α-inhibitor impairs TSG-6-induced hyaluronan cross-linking. J Biol Chem. 2013;288:29642–29653. doi: 10.1074/jbc.M113.477422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova N.S., Nilebäck E., Haller F.M., Briggs D.C., Svedhem S., Day A.J., et al. The inflammation-associated protein TSG-6 cross-links hyaluronan via hyaluronan-induced TSG-6 oligomers. J Biol Chem. 2011;286:25675–25686. doi: 10.1074/jbc.M111.247395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand-Vallery V., Boilan E., Ninane N., Demazy C., Friguet B., Toussaint O., et al. Repeated exposures to UVB induce differentiation rather than senescence of human keratinocytes lacking p16(INK-4A) Biogerontology. 2010;11:167–181. doi: 10.1007/s10522-009-9238-y. [DOI] [PubMed] [Google Scholar]
- Bocian C., Urbanowitz A.K., Owens R.T., Iozzo R.V., Götte M., Seidler D.G. Decorin potentiates interferon-γ activity in a model of allergic inflammation. J Biol Chem. 2013;288:12699–12711. doi: 10.1074/jbc.M112.419366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon L.Y. Matrix hyaluronan-activated CD44 signaling promotes keratinocyte activities and improves abnormal epidermal functions. Am J Pathol. 2014;184:1912–1919. doi: 10.1016/j.ajpath.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon L.Y., Singleton P.A., Diedrich F. Hyaluronan-CD44 interaction with Rac1-dependent protein kinase N-gamma promotes phospholipase Cgamma1 activation, Ca(2+) signaling, and cortactin-cytoskeleton function leading to keratinocyte adhesion and differentiation. J Biol Chem. 2004;279:29654–29669. doi: 10.1074/jbc.M403608200. [DOI] [PubMed] [Google Scholar]
- Csoka A.B., Frost G.I., Stern R. The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 2001;20:499–508. doi: 10.1016/s0945-053x(01)00172-x. [DOI] [PubMed] [Google Scholar]
- Dabelsteen S., Pallesen E.M.H., Marinova I.N., Nielsen M.I., Adamopoulou M., Rømer T.B., et al. Essential functions of glycans in human epithelia dissected by a CRISPR-Cas9-Engineered human organotypic skin model. Dev Cell. 2020;54:669–684.e7. doi: 10.1016/j.devcel.2020.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A.J., Milner C.M. TSG-6: a multifunctional protein with anti-inflammatory and tissue-protective properties. Matrix Biol. 2019;78–79:60–83. doi: 10.1016/j.matbio.2018.01.011. [DOI] [PubMed] [Google Scholar]
- De Vuyst E., Charlier C., Giltaire S., De Glas V., de Rouvroit C.L., Poumay Y. Reconstruction of normal and pathological human epidermis on polycarbonate filter. Methods Mol Biol. 2014;1195:191–201. doi: 10.1007/7651_2013_40. [DOI] [PubMed] [Google Scholar]
- De Vuyst É., Giltaire S., Lambert de Rouvroit C., Malaisse J., Mound A., Bourtembourg M., et al. Methyl-β-cyclodextrin concurs with interleukin (IL)-4, IL-13 and IL-25 to induce alterations reminiscent of atopic dermatitis in reconstructed human epidermis. Exp Dermatol. 2018;27:435–437. doi: 10.1111/exd.13113. [DOI] [PubMed] [Google Scholar]
- Dickson M.A., Hahn W.C., Ino Y., Ronfard V., Wu J.Y., Weinberg R.A., et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Nascimento Pedrosa T., De Vuyst E., Mound A., Lambert de Rouvroit C., Maria-Engler S.S., Poumay Y. Methyl-β-cyclodextrin treatment combined to incubation with interleukin-4 reproduces major features of atopic dermatitis in a 3D-culture model. Arch Dermatol Res. 2017;309:63–69. doi: 10.1007/s00403-016-1699-7. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert F., Dewan P., Caley M.P., Jones E.M., Morse M.A., Kelsell D.P., et al. 3D model of harlequin ichthyosis reveals inflammatory therapeutic targets. J Clin Invest. 2020;130:4798–4810. doi: 10.1172/JCI132987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B., Jarvis E., Roth T.L., Shin J., Corn J.E., Marson A., et al. Enhanced genome editing with Cas9 ribonucleoprotein in diverse cells and organisms. J Vis Exp. 2018;135:57350. doi: 10.3791/57350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faway E., Cambier L., De Vuyst E., Evrard C., Thiry M., Lambert de Rouvroit C., et al. Responses of reconstructed human epidermis to Trichophyton rubrum infection and impairment of infection by the inhibitor PD169316. J Invest Dermatol. 2019;139:2080–2089.e6. doi: 10.1016/j.jid.2019.03.1147. [DOI] [PubMed] [Google Scholar]
- Faway É., Cambier L., Mignon B., Poumay Y., Lambert de Rouvroit C. Modeling dermatophytosis in reconstructed human epidermis: a new tool to study infection mechanisms and to test antifungal agents. Med Mycol. 2017;55:485–494. doi: 10.1093/mmy/myw111. [DOI] [PubMed] [Google Scholar]
- Frankart A., Malaisse J., De Vuyst E., Minner F., de Rouvroit C.L., Poumay Y. Epidermal morphogenesis during progressive in vitro 3D reconstruction at the air-liquid interface. Exp Dermatol. 2012;21:871–875. doi: 10.1111/exd.12020. [DOI] [PubMed] [Google Scholar]
- Fraser J.R., Laurent T.C., Laurent U.B. Hyaluronan: its nature, distribution, functions and turnover. J Intern Med. 1997;242:27–33. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- Fülöp C., Szántó S., Mukhopadhyay D., Bárdos T., Kamath R.V., Rugg M.S., et al. Impaired cumulus mucification and female sterility in tumor necrosis factor-induced protein-6 deficient mice. Development. 2003;130:2253–2261. doi: 10.1242/dev.00422. [DOI] [PubMed] [Google Scholar]
- Hennies H.C., Poumay Y. Skin disease models in vitro and inflammatory mechanisms: predictability for drug development. Handb Exp Pharmacol. 2021;265:187–218. doi: 10.1007/164_2020_428. [DOI] [PubMed] [Google Scholar]
- Hubaux R., Bastin C., Salmon M. On the relevance of an in vitro reconstructed human epidermis model for drug screening in atopic dermatitis. Exp Dermatol. 2018;27:1403–1407. doi: 10.1111/exd.13810. [DOI] [PubMed] [Google Scholar]
- Huth S., Huth L., Marquardt Y., Fietkau K., Dahl E., Esser P.R., et al. Inter-alpha-trypsin inhibitor heavy chain 5 (ITIH5) is a natural stabilizer of hyaluronan that modulates biological processes in the skin. Skin Pharmacol Physiol. 2020;33:198–206. doi: 10.1159/000509371. [DOI] [PubMed] [Google Scholar]
- Iriyama S., Nishikawa S., Hosoi J., Amano S. Basement membrane helps maintain epidermal hyaluronan content. Am J Pathol. 2021;191:1010–1019. doi: 10.1016/j.ajpath.2021.03.007. [DOI] [PubMed] [Google Scholar]
- Jadin L., Huang L., Wei G., Zhao Q., Gelb A.B., Frost G.I., et al. Characterization of a novel recombinant hyaluronan binding protein for tissue hyaluronan detection. J Histochem Cytochem. 2014;62:672–683. doi: 10.1369/0022155414540176. [DOI] [PubMed] [Google Scholar]
- Jokela T.A., Lindgren A., Rilla K., Maytin E., Hascall V.C., Tammi R.H., et al. Induction of hyaluronan cables and monocyte adherence in epidermal keratinocytes. Connect Tissue Res. 2008;49:115–119. doi: 10.1080/03008200802148439. [DOI] [PubMed] [Google Scholar]
- Kavasi R.M., Berdiaki A., Spyridaki I., Corsini E., Tsatsakis A., Tzanakakis G., et al. HA metabolism in skin homeostasis and inflammatory disease. Food Chem Toxicol. 2017;101:128–138. doi: 10.1016/j.fct.2017.01.012. [DOI] [PubMed] [Google Scholar]
- Kishibe M., Bando Y., Tanaka T., Ishida-Yamamoto A., Iizuka H., Yoshida S. Kallikrein-related peptidase 8-dependent skin wound healing is associated with upregulation of kallikrein-related peptidase 6 and PAR2. J Invest Dermatol. 2012;132:1717–1724. doi: 10.1038/jid.2012.18. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Chanmee T., Itano N. Hyaluronan: metabolism and function. Biomolecules. 2020;10:1525. doi: 10.3390/biom10111525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.J., Kim D.H., Ryu J.S., Ko A.Y., Ko J.H., Kim M.K., et al. Topical TSG-6 administration protects the ocular surface in two mouse models of inflammation-related dry eye. Invest Ophthalmol Vis Sci. 2015;56:5175–5181. doi: 10.1167/iovs.14-16307. [DOI] [PubMed] [Google Scholar]
- Lee T.H., Lee G.W., Ziff E.B., Vilcek J. Isolation and characterization of eight tumor necrosis factor-induced gene sequences from human fibroblasts. Mol Cell Biol. 1990;10:1982–1988. doi: 10.1128/mcb.10.5.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley J., Gál I., Mahoney D.J., Cordell M.R., Rugg M.S., Hyman R., et al. TSG-6 modulates the interaction between hyaluronan and cell surface CD44. J Biol Chem. 2004;279:25745–25754. doi: 10.1074/jbc.M313319200. [DOI] [PubMed] [Google Scholar]
- Lin W., Shuster S., Maibach H.I., Stern R. Patterns of hyaluronan staining are modified by fixation techniques. J Histochem Cytochem. 1997;45:1157–1163. doi: 10.1177/002215549704500813. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Malaisse J., Bourguignon V., De Vuyst E., Lambert de Rouvroit C., Nikkels A.F., Flamion B., et al. Hyaluronan metabolism in human keratinocytes and atopic dermatitis skin is driven by a balance of hyaluronan synthases 1 and 3. J Invest Dermatol. 2014;134:2174–2182. doi: 10.1038/jid.2014.147. [DOI] [PubMed] [Google Scholar]
- Mathay C., Pierre M., Pittelkow M.R., Depiereux E., Nikkels A.F., Colige A., et al. Transcriptional profiling after lipid raft disruption in keratinocytes identifies critical mediators of atopic dermatitis pathways. J Invest Dermatol. 2011;131:46–58. doi: 10.1038/jid.2010.272. [DOI] [PubMed] [Google Scholar]
- Meyer L.J., Stern R. Age-dependent changes of hyaluronan in human skin. J Invest Dermatol. 1994;102:385–389. doi: 10.1111/1523-1747.ep12371800. [DOI] [PubMed] [Google Scholar]
- Milner C.M., Day A.J. TSG-6: a multifunctional protein associated with inflammation. J Cell Sci. 2003;116:1863–1873. doi: 10.1242/jcs.00407. [DOI] [PubMed] [Google Scholar]
- Milner C.M., Tongsoongnoen W., Rugg M.S., Day A.J. The molecular basis of inter-alpha-inhibitor heavy chain transfer on to hyaluronan. Biochem Soc Trans. 2007;35:672–676. doi: 10.1042/BST0350672. [DOI] [PubMed] [Google Scholar]
- Minner F., Herphelin F., Poumay Y. Study of epidermal differentiation in human keratinocytes cultured in autocrine conditions. Methods Mol Biol. 2010;585:71–82. doi: 10.1007/978-1-60761-380-0_6. [DOI] [PubMed] [Google Scholar]
- Minner F., Poumay Y. Candidate housekeeping genes require evaluation before their selection for studies of human epidermal keratinocytes. J Invest Dermatol. 2009;129:770–773. doi: 10.1038/jid.2008.247. [DOI] [PubMed] [Google Scholar]
- Moran M.C., Pandya R.P., Leffler K.A., Yoshida T., Beck L.A., Brewer M.G. Characterization of human keratinocyte cell lines for barrier studies. JID Innov. 2021;1:100018. doi: 10.1016/j.xjidi.2021.100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka Y., Yamasaki K., Leung D., Gallo R.L. Cathelicidin antimicrobial peptides inhibit hyaluronan-induced cytokine release and modulate chronic allergic dermatitis. J Immunol. 2008;181:3915–3922. doi: 10.4049/jimmunol.181.6.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mound A., Lozanova V., Warnon C., Hermant M., Robic J., Guere C., et al. Non-senescent keratinocytes organize in plasma membrane submicrometric lipid domains enriched in sphingomyelin and involved in re-epithelialization. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:958–971. doi: 10.1016/j.bbalip.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Muto J., Sayama K., Gallo R.L., Kimata K. Emerging evidence for the essential role of hyaluronan in cutaneous biology. J Dermatol Sci. 2019;94:190–195. doi: 10.1016/j.jdermsci.2019.01.009. [DOI] [PubMed] [Google Scholar]
- Nauroy P., Nyström A. Kallikreins: essential epidermal messengers for regulation of the skin microenvironment during homeostasis, repair and disease. Matrix Biol Plus. 2020;6–7:100019. doi: 10.1016/j.mbplus.2019.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A.Y., Bourtembourg M., Chrétien A., Hubaux R., Lancelot C., Salmon M., et al. Modulation of gene expression in a sterile atopic dermatitis model and inhibition of Staphylococcus aureus adhesion by fucoidan. Dermatopathology (Basel) 2021;8:69–83. doi: 10.3390/dermatopathology8020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasonen-Seppänen S., Hyttinen J.M., Rilla K., Jokela T., Noble P.W., Tammi M., et al. Role of CD44 in the organization of keratinocyte pericellular hyaluronan. Histochem Cell Biol. 2012;137:107–120. doi: 10.1007/s00418-011-0883-2. [DOI] [PubMed] [Google Scholar]
- Pasonen-Seppänen S., Karvinen S., Törrönen K., Hyttinen J.M., Jokela T., Lammi M.J., et al. EGF upregulates, whereas TGF-beta downregulates, the hyaluronan synthases Has2 and Has3 in organotypic keratinocyte cultures: correlations with epidermal proliferation and differentiation. J Invest Dermatol. 2003;120:1038–1044. doi: 10.1046/j.1523-1747.2003.12249.x. [DOI] [PubMed] [Google Scholar]
- Perez A., Neskey D.M., Wen J., Pereira L., Reategui E.P., Goodwin W.J., et al. CD44 interacts with EGFR and promotes head and neck squamous cell carcinoma initiation and progression. Oral Oncol. 2013;49:306–313. doi: 10.1016/j.oraloncology.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plager D.A., Kahl J.C., Asmann Y.W., Nilson A.E., Pallanch J.F., Friedman O., et al. Gene transcription changes in asthmatic chronic rhinosinusitis with nasal polyps and comparison to those in atopic dermatitis. PLoS One. 2010;5:e11450. doi: 10.1371/journal.pone.0011450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaranta M., Knapp B., Garzorz N., Mattii M., Pullabhatla V., Pennino D., et al. Intraindividual genome expression analysis reveals a specific molecular signature of psoriasis and eczema. Sci Transl Med. 2014;6:244ra90. doi: 10.1126/scitranslmed.3008946. [DOI] [PubMed] [Google Scholar]
- Reynolds G., Vegh P., Fletcher J., Poyner E.F.M., Stephenson E., Goh I., et al. Developmental cell programs are co-opted in inflammatory skin disease. Science. 2021;371 doi: 10.1126/science.aba6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald J.G., Hahn W.C., Ramsey M.R., Wu J.Y., Guo Z., Tsao H., et al. A two-stage, p16(INK4A)- and p53-dependent keratinocyte senescence mechanism that limits replicative potential independent of telomere status. Mol Cell Biol. 2002;22:5157–5172. doi: 10.1128/MCB.22.14.5157-5172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosdy M., Pisani A., Ortonne J.P. Production of basement membrane components by a reconstructed epidermis cultured in the absence of serum and dermal factors. Br J Dermatol. 1993;129:227–234. doi: 10.1111/j.1365-2133.1993.tb11839.x. [DOI] [PubMed] [Google Scholar]
- Sakai S., Yasuda R., Sayo T., Ishikawa O., Inoue S. Hyaluronan exists in the normal stratum corneum. J Invest Dermatol. 2000;114:1184–1187. doi: 10.1046/j.1523-1747.2000.00992.x. [DOI] [PubMed] [Google Scholar]
- Sandjeu Y., Haftek M. Desmosealin and other components of the epidermal extracellular matrix. J Physiol Pharmacol. 2009;60(Suppl. 4):23–30. [PubMed] [Google Scholar]
- Selbi W., Day A.J., Rugg M.S., Fülöp C., de la Motte C.A., Bowen T., et al. Overexpression of hyaluronan synthase 2 alters hyaluronan distribution and function in proximal tubular epithelial cells. J Am Soc Nephrol. 2006;17:1553–1567. doi: 10.1681/ASN.2005080879. [DOI] [PubMed] [Google Scholar]
- Shakya S., Mack J.A., Alipour M., Maytin E.V. Cutaneous wounds in mice lacking TSG-6 exhibit delayed closure and an abnormal inflammatory response. J Invest Dermatol. 2020;140:2505–2514. doi: 10.1016/j.jid.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R.M., Meran S., Thomas D., Stephens P., Bowen T., Steadman R., et al. Age-related changes in pericellular hyaluronan organization leads to impaired dermal fibroblast to myofibroblast differentiation. Am J Pathol. 2009;175:1915–1928. doi: 10.2353/ajpath.2009.090045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits J.P.H., Niehues H., Rikken G., van Vlijmen-Willems I.M.J.J., van de Zande G.W.H.J.F., Zeeuwen P.L.J.M., et al. Immortalized N/TERT keratinocytes as an alternative cell source in 3D human epidermal models. Sci Rep. 2017;7:11838. doi: 10.1038/s41598-017-12041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammi R.H., Tammi M.I., Hascall V.C., Hogg M., Pasonen S., MacCallum D.K. A preformed basal lamina alters the metabolism and distribution of hyaluronan in epidermal keratinocyte "organotypic" cultures grown on collagen matrices. Histochem Cell Biol. 2000;113:265–277. doi: 10.1007/s004180000128. [DOI] [PubMed] [Google Scholar]
- Tan K.T., McGrouther D.A., Day A.J., Milner C.M., Bayat A. Characterization of hyaluronan and TSG-6 in skin scarring: differential distribution in keloid scars, normal scars and unscarred skin. J Eur Acad Dermatol Venereol. 2011;25:317–327. doi: 10.1111/j.1468-3083.2010.03792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q., Thomassen M., Burton M., Mose K.F., Andersen K.E., Hjelmborg J., et al. Generalized correlation coefficient for non-parametric analysis of microarray time-course data. J Integr Bioinform. 2017;14:20170011. doi: 10.1515/jib-2017-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toole B.P. Hyaluronan in morphogenesis. Semin Cell Dev Biol. 2001;12:79–87. doi: 10.1006/scdb.2000.0244. [DOI] [PubMed] [Google Scholar]
- Toole B.P. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- Wang F., Flanagan J., Su N., Wang L.C., Bui S., Nielson A., et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel P.H., Hascall V.C., Tammi M. Hyaluronan synthases. J Biol Chem. 1997;272:13997–14000. doi: 10.1074/jbc.272.22.13997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Dataset related to this article can be found at https://data.mendeley.com/datasets/4fbgg2syzk/1, an open-source online data repository hosted at Mendeley Data.