Abstract
The process of assembling immunoglobulin and T-cell receptor genes from variable (V), diversity (D), and joining (J) gene segments, called V(D)J recombination, involves the introduction of DNA breaks at recombination signals. DNA cleavage is catalyzed by RAG-1 and RAG-2 in two chemical steps: first-strand nicking, followed by hairpin formation via direct transesterification. In vitro, these reactions minimally proceed in discrete protein-DNA complexes containing dimeric RAG-1 and one or two RAG-2 monomers bound to a single recombination signal sequence. Recently, a DDE triad of carboxylate residues essential for catalysis was identified in RAG-1. This catalytic triad resembles the DDE motif often associated with transposase and retroviral integrase active sites. To investigate which RAG-1 subunit contributes the residues of the DDE triad to the recombinase active site, cleavage of intact or prenicked DNA substrates was analyzed in situ in complexes containing RAG-2 and a RAG-1 heterodimer that carried an active-site mutation targeted to the same or opposite RAG-1 subunit mutated to be incompetent for DNA binding. The results show that the DDE triad is contributed to a single recombinase active site, which catalyzes the nicking and transesterification steps of V(D)J recombination by a single RAG-1 subunit opposite the one bound to the nonamer of the recombination signal undergoing cleavage (cleavage in trans). The implications of a trans cleavage mode observed in these complexes on the organization of the V(D)J synaptic complex are discussed.
Immunoglobulin and T-cell receptor genes are assembled from arrays of component variable (V), diversity (D), and joining (J) gene segments by a series of site-specific DNA rearrangements. This process, called V(D)J recombination, is directed by recombination signal sequences (RSSs) flanking the antigen receptor gene segments (24). The RSS is composed of conserved heptamer and nonamer elements, separated by a spacer whose sequence is nominally conserved but whose length is either 12 or 23 bp (12-RSS and 23-RSS, respectively); recombination normally occurs between gene segments whose RSSs carry dissimilar length spacers (the 12/23 rule). V(D)J recombination is initiated by the products of two recombination-activating genes (RAG-1 and -2) (31, 41) that together catalyze DNA double-strand breaks at pairs of RSSs between the heptamers and the adjacent coding gene segments. This process generates two distinct DNA ends: blunt 5′ phosphorylated signal ends and coding ends terminating in DNA hairpins (37, 38, 42). These products are a direct consequence of a two-step reaction that first involves the introduction of a nick at the 5′ end of the heptamer. In the second step, a 3′ OH at the terminus of the coding segment (exposed by the nicking step) is covalently linked to the phosphate group on the opposing DNA strand via direct transesterification (27, 50). The resulting signal ends are ligated heptamer to heptamer, and coding ends are joined to yield precise signal joints and imprecise coding joints (24). While the chemistry of the cleavage reaction renders suitable substrates for signal joint formation, hairpinned coding ends must be resolved before the ends can be joined. The RAG proteins may play a role in coding end processing, as they have been shown to catalyze hairpin opening and nicking of 3′ flap structures in vitro (5, 39, 44).
Several lines of evidence suggest that the V(D)J recombinase is most closely related to a family of enzymes that includes both bacterial transposases and retroviral integrases (32, 36). Features that the V(D)J recombinase shares with members of this family include (i) the catalysis of polynucleotidyl transfer reactions via direct transesterification (50); (ii) the reversibility of the cleavage reaction (28); (iii) the generation of DNA hairpin intermediates in recombination, as observed for the transposition of Tn10 (19) and Tn5 (6); (iv) the catalysis of transpositional recombination in vitro (1, 17); (v) the presence of a triad of conserved carboxylate residues that comprise the enzyme active site (13, 20, 22); and (vi) the existence of 3′ flap endonuclease activity, as observed in the Tn10 transposase (39). Further insight into the biochemical and structural similarities between the V(D)J recombinase and other members of the transposase-retroviral integrase family requires a greater understanding of how the DNA binding and catalytic activities of the RAG proteins are distributed among the individual protein components in RAG-RSS complexes.
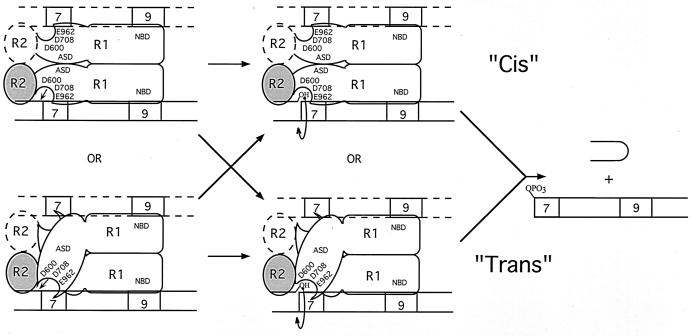
The biochemical properties of RAG-1 and RAG-2 both in the absence of DNA and bound to single or paired RSS substrates have been studied extensively (reviewed in reference 12). Working models of RAG interactions with a single RSS substrate have previously been developed, based on the characterization of single RSS complexes containing RAG-1 in the absence or presence of RAG-2 (46). In single RSS complexes containing both RAG proteins, RAG-1 is bound as a dimer, associating with one or two monomers of RAG-2 (Fig. 1). Nonamer interactions are mediated by the nonamer binding domain (NBD; residues 389 to 446) of RAG-1 (9, 45, 47). RAG-1 heterodimers bearing NBD mutations in a single RAG-1 subunit but not in both subunits assemble stable protein-DNA complexes containing both RAG-1 and RAG-2, suggesting that a single RAG-1 subunit is competent to support DNA binding and that a single copy of the RSS substrate is present in the heteromeric complex (46). However, the DNA stoichiometry in RAG complexes assembled in the presence of a single-length RSS has not been rigorously established. Therefore, whether complexes containing both RAG-1 and RAG-2 contain a single copy of the RSS substrate (Fig. 1, solid lines) or an identical pair of substrates (Fig. 1, dashed lines) is unclear. Compared to RAG-1–nonamer interactions, the specificity of RAG-1-toward-heptamer sequences is relatively low in the absence of RAG-2 (35), but when RAG-2 is present in the protein-DNA complex, stable RAG-1–heptamer contacts are readily observed (3, 47). However, whether these heptamer contacts are mediated by the same or opposite subunit of the RAG-1 dimer that is bound to the adjoining nonamer is unknown.
FIG. 1.
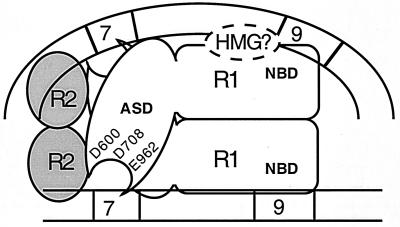
Modes of cleavage in protein-DNA complexes containing RAG-1 and RAG-2 bound to a single RSS. These models are adapted in part from those published previously (46). RAG-1 (R1) binds a 12-RSS as a dimer and interacts with one or two subunits of RAG-2 (R2); a single subunit of a RAG-1 dimer mediates nonamer binding via the NBD. If both RAG-1 subunits contain wild-type NBDs, nonamer binding could be mediated by either subunit. These complexes minimally contain one RSS substrate (solid lines), although the presence of a second, identical RSS substrate cannot be excluded (dashed lines). RAG-2 promotes heptamer occupancy by RAG-1. RAG-1 bears the recombinase active-site domain (ASD) containing three carboxylate residues essential for catalysis (D600, D708, and E962). In principle, the active site that catalyzes the nicking (left) and transesterification (right) steps of V(D)J recombination on a single RSS substrate may be donated by the same (in cis) or opposite (in trans) RAG-1 subunit that binds the nonamer of the RSS being cleaved. A single active site may or may not catalyze both reactions (arrows).
Because V(D)J recombination shares many biochemical similarities with bacterial transposition and retroviral integration, the active site of the V(D)J recombinase was speculated to contain a constellation of carboxylate residues similar to the DDE motif often observed in enzymes that catalyze the latter two forms of DNA rearrangement. Recently, a DDE triad of amino acids essential for the cleavage steps of V(D)J recombination has been identified in RAG-1 (Asp-600, Asp-708, and Glu-962); no essential carboxylate residues were found in RAG-2 (13, 20, 22). As in other transposase and retroviral integrase proteins (for review, see reference 14 and references therein), the DDE motif in RAG-1 appears to play a role in catalysis by directly coordinating divalent metal ions required as cofactors in the cleavage reaction (13, 20, 22). The localization of critical active-site residues to RAG-1 but not to RAG-2 is consistent with the major role that RAG-1 plays in mediating recognition near the DNA cleavage site and suggests that the region of RAG-1 which interacts with the heptamer-coding junction is proximal to the active site of the V(D)J recombinase. Therefore, defining the active-site organization in V(D)J initiation complexes provides a means to determine which RAG-1 subunit contacts the RSS near the cleavage site.
In principle, the DDE triad of amino acid residues may comprise part of a single active site on a single RAG-1 subunit (Fig. 1). Another formal possibility (not shown) is that some of the essential carboxylate residues are contributed by one RAG-1 subunit, while the remaining residues are donated by the other RAG-1 subunit, forming a composite active site. For simplicity, only the former possibility is considered in Fig. 1. In these models, the essential active-site residues may be contributed by the same RAG-1 subunit as the one bound to the nonamer of the RSS undergoing cleavage (in cis) (Fig. 1, top) or the opposite Rag-1 subunit (in trans) (Fig. 1, bottom). Theoretically, a single active site may or may not catalyze both the nicking and transesterification steps of V(D)J recombination (Fig. 1, left and right, respectively).
To distinguish between these possibilities, I have exploited a difference in DNA binding activity between wild-type RAG-1 and a form of RAG-1 bearing a mutant NBD to construct RAG-1 chimeras that contain an active-site mutation, a mutant NBD, or both. These chimeras were coexpressed in pairwise fashion, yielding RAG-1 heterodimers containing an active-site mutation targeted to the same or opposite RAG-1 subunit bearing a mutant NBD. Precleavage RAG-RSS complexes were assembled on intact or prenicked RSS substrates and separated by an electrophoretic mobility shift assay (EMSA). Catalysis was initiated in situ by adding an appropriate divalent metal cation, and the resulting cleavage products were recovered and analyzed. The results suggest that a single RAG-1 subunit donates the entire DDE motif to a single recombinase active site. In addition, both the nicking and transesterification steps of V(D)J recombination are catalyzed by a single active site contributed by the RAG-1 subunit opposite the one bound to the nonamer of the RSS undergoing cleavage (cleavage in trans). These findings provide insight into how the RAG-1 subunits may be organized in synaptic complexes and further highlight the biochemical similarities between the V(D)J recombinase and other members of the transposase-retroviral integrase family.
MATERIALS AND METHODS
Plasmid constructs and mutagenesis.
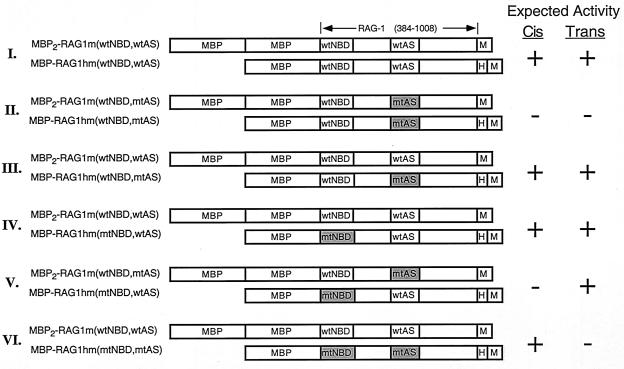
Expression constructs encoding core fragments of RAG-1 and RAG-2 fused at the amino terminus to one or two copies of the maltose binding protein (MBP) and possessing a carboxyl-terminal myc epitope and polyhistidine tag have been described (46, 47). Versions of these vectors containing the myc epitope but lacking the polyhistidine tag were also constructed using conventional techniques. Single alanine substitutions were introduced at Asp-600, Asp-708, or Glu-962 of RAG-1 by recombination PCR and verified by DNA sequencing (18). Primer sequences are available upon request. Expression vectors encoding single or double MBP–RAG-1 chimeras containing an alanine substitution in the NBD (residues 384 to 393), the active site of RAG-1 (D600A, D708A, or E962A), or both were constructed by subcloning from appropriate constructs described previously (46, 47). The encoded fusion proteins are designated in Fig. 2.
FIG. 2.
Heterodimer combinations expected from cotransfections of single and double MBP–RAG-1 expression constructs. Diagrams of the single and double MBP–RAG-1 combinations used to distinguish cis versus trans cleavage are depicted and designated at left. MBP, myc (M), and polyhistidine (H) sequences are indicated; core RAG-1 residues are numbered at the top. The positions of the NBD (wild type or mutant; wtNBD or mtNBD, respectively) and active site (wild type or mutant; wtAS or mtAS, respectively) are indicated. The mtNBD carries alanine substitutions at residues 384 to 393 (47); the mtAS carries either the D600A, D708A, or E962A mutation. Fusion proteins were coexpressed in the indicated combinations (I to VI) and purified as described in Materials and Methods. Homodimers of the single but not double MBP–RAG-1 fusion protein are copurified with the heterodimers depicted, as single but not double MBP–RAG-1 contains a polyhistidine tag. The predicted activity of the RAG-1 heterodimers in the presence of RAG-2 when substrate cleavage is catalyzed in cis or in trans is indicated at the right.
Protein purification.
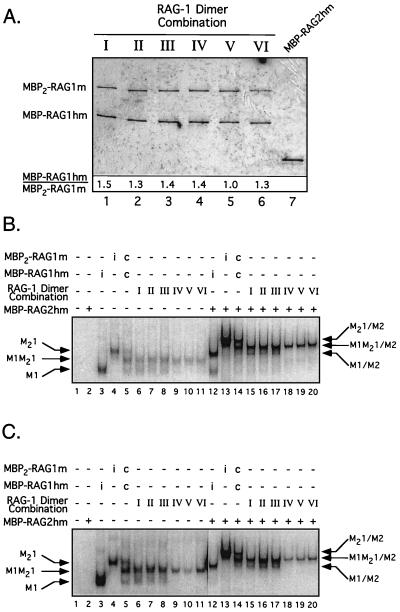
Single or double MBP-RAG fusion proteins were expressed individually or coexpressed (where noted) in 293 cells and purified by amylose affinity chromatography as previously described (25, 47). For preparations of heterodimeric single and double MBP–RAG-1 fusion proteins, eluates from the amylose resin were diluted 1:2 in buffer B (20 mM Tris [pH 7.9], 0.5 M NaCl, 2 mM β-mercaptoethanol, 20 mM imidazole), applied to a Ni2+ chelating resin (ProBond; Invitrogen), washed with 10 volumes of buffer B, eluted in buffer B containing 250 mM imidazole, and dialyzed against buffer C (25 mM Tris [pH 8.0], 150 mM KCl, 2 mM dithiothreitol, 10% glycerol) as described (27). The protein preparations were judged to be >95% pure by silver staining (Fig. 3). RAG-1 was immunoblotted with affinity-purified rabbit anti-RAG-1 polyclonal antibody 307 (raised against residues 507 to 524) (26), detected by chemiluminescence using peroxidase-conjugated anti-rabbit immunoglobulin G and the ECL Plus reagent (Amersham Pharmacia Biotech), and quantified with a Molecular Imager using the CH screen for chemiluminescent imaging (Bio-Rad).
FIG. 3.
Purification and DNA binding activity of the heterodimer preparations. (A) Purified single and double MBP–RAG-1 fusion proteins coexpressed in the combinations indicated in Fig. 2 (I to VI, lanes 1 to 6) as well as MBP-RAG-2hm (lane 7) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by silver staining. The positions of the single and double MBP–RAG-1 fusion proteins are indicated at left. The ratio of the single to double MBP–RAG-1 fusion in each preparation (indicated below the gel) was determined from immunoblots using anti-RAG-1 antibody 307 (26). (B and C) EMSA of the D708A series using an intact (B) or prenicked (C) 32P-end-labeled 12-RSS substrate. The DNA substrate was incubated without (−) protein (lane 1), with (+) RAG-2 alone (lane 2), or with single or double MBP–RAG-1 fusion proteins expressed individually (i) (lanes 3 and 4 and 12 and 13) or coexpressed (c) (lanes 5 and 14) and each heterodimer preparation (Fig. 2, I to VI) in the absence (lanes 3 to 11) or presence (lanes 12 to 20) of RAG-2 under binding conditions as indicated at the top. Positions of protein-DNA complexes containing only single MBP–RAG-1 subunits, double MBP–RAG-1 subunits, or both in the absence (M1, M21, or M1M21, respectively) or presence (M1/M2, M21/M2, or M1M21/M2, respectively) of RAG-2 are designated at left or right, respectively. Note that in panel C, lanes 1 to 11 are derived from the same gel as lanes 12 to 20 but are taken from a longer exposure to avoid overexposure of lanes 12 to 20. Longer exposure of lanes 18 to 20 in panels B and C does not show significant formation of the M1/M2 complex (data not shown).
Oligonucleotide binding and cleavage assays.
The standard substrate used in binding and cleavage assays was a 50-bp duplex containing a single 12-RSS, formed by annealing the radiolabeled oligonucleotide DAR39 to unlabeled DAR40 (27) and purified as described previously (25). The prenicked substrate was made by annealing the radiolabeled oligonucleotide DAR42 and DG10, which contained a nonradioactive 5′ phosphate introduced during its chemical synthesis, to unlabeled DAR40 (27). Intact and prenicked 23-RSS substrates were similarly assembled from oligonucleotides DG61, DG62, and DG4 (27).
Binding reactions (10 μl) containing purified heterodimeric single and double MBP–RAG-1 fusion proteins (∼20 ng), RAG-2 (∼20 ng, where indicated), and either an intact or prenicked RSS substrate (0.02 pmol) were assembled in the presence of Ca2+ and analyzed by EMSA as previously described (47), except that samples were incubated at 25°C. For in situ cleavage experiments, binding reactions were scaled up fivefold. For samples containing 23-RSS substrates, HMG-1 (gift of Y.-M. Yen, B. Wong, and R. Johnson) was added to a final concentration of 1 μg/ml (48). RAG-RSS complexes were separated by an EMSA (the duration of electrophoresis was extended to enhance separation of RSS complexes containing both RAG-1 and RAG-2), and the gel was subsequently submerged in cleavage buffer (25 mM morpholinepropanesulfonic acid [MOPS]-KOH [pH 7.0], 60 mM potassium glutamate) containing either MgCl2 (intact substrates) or MnCl2 (prenicked substrates) to a final concentration of 5 mM. After incubation for 1 h at 37°C to initiate cleavage, the DNA was electrophoretically transferred to DEAE-cellulose paper (DE81; Whatman), visualized by autoradiography, and recovered from discrete RAG-RSS complexes as described (47). The cleavage products were fractionated by denaturing gel electrophoresis, visualized by autoradiography, and quantified with a phosphorimager using the Molecular Analyst software (Bio-Rad).
RESULTS
Targeting active-site mutations in a V(D)J initiation complex.
Determining which subunit of the RAG-1 dimer contributes the essential residues of the DDE motif to the recombinase active site in single RSS complexes requires targeting an active-site mutation to the same or opposite RAG-1 subunit that binds the nonamer of the RSS being followed for cleavage. Since mutations within the DDE motif do not significantly impair RSS binding (13, 20, 22), RAG-1 heterodimers bearing an active-site mutant on one subunit could theoretically bind the RSS substrate via the NBD on either RAG-1 subunit. Hence, either configuration could potentially be responsible for the generation of any cleavage products observed, precluding the possibility of distinguishing between cis and trans cleavage. In principle, this difficulty could be overcome by pairing an active-site mutation in one RAG-1 subunit with an NBD mutation in the same or opposite RAG-1 subunit that impairs RSS binding by disrupting RAG-1–nonamer interactions (9, 45, 47). In these heterodimers, the position of the active-site mutation is necessarily enforced in cis or in trans to the RAG-1 subunit bound to the nonamer (via the single intact NBD) of the RSS targeted for cleavage. Analyzing the cleavage activity in the resulting heteromeric protein-DNA complexes should resolve whether the residues of the DDE motif are contributed to the recombinase active site by the same (in cis) or opposite (in trans) RAG-1 subunit bound to the nonamer of the RSS undergoing cleavage.
Toward this end, mammalian expression vectors encoding single or double MBP fusions of RAG-1 that carry an alanine substitution within the NBD (residues 384 to 393), the active site (D600A, D708A, or E962A), or both were constructed (either with [single MBP–Rag-1] or without [double MBP–Rag-1] a carboxyl-terminal polyhistidine tag) (Fig. 2). These constructs were contransfected in pairwise combinations into 293 cells (Fig. 2, pairs I to VI) and purified first by amylose affinity chromatography, followed by affinity chromatography over a Ni2+ chelating resin. This two-step process resulted in selective recovery of RAG-1 dimers in which one or both subunits bear a polyhistidine tag. The heterodimer composition expected from the individual transfections and their predicted activity in the presence of RAG-2 when cleavage occurs in cis or in trans are shown in Fig. 2. Note that homodimers of the single but not double MBP–RAG-1 fusion protein would be copurified with the heterodimers, as the latter fusion protein lacks a polyhistidine tag. Therefore, assuming RAG-1 dimerization is random and not influenced by the appended fusion partner(s), the single MBP–RAG-1 fusion protein is expected to be in twofold molar excess over the double MBP–RAG-1 fusion protein after the second affinity purification step. Analysis of the protein preparations by silver staining and immunoblotting shows that the ratio of the single to double MBP–RAG-1 fusion proteins is close but lower than the predicted value, as seen previously (Fig. 3A) (46, 47). The greater solubility of the double MBP–RAG-1 fusion proteins relative to their single MBP counterparts (46; my personal observation), as well as variations in the solubility of the mutant proteins, may explain why the representation of the double MBP–RAG-1 fusion protein in the preparation is generally larger than expected. Heterodimers containing mutant NBDs in both subunits were not analyzed, because RAG-1 homodimers bearing NBD mutations in both subunits fail to form stable RSS complexes containing both RAG proteins (see references 46 and 47 and Fig. 3).
The purified proteins were examined by an EMSA for their ability to bind an intact or prenicked 32P-labeled 12-RSS substrate (Fig. 3B or C, respectively). A representative example is shown for the panel of heterodimers prepared to analyze the contribution of Asp-708. In both cases, consistent with previous results (47), no binding was seen in reactions lacking protein or containing RAG-2 alone (Fig. 3B or C, lanes 1 and 2). In the absence of RAG-2, a species of retarded mobility (M1M21) was observed in all RAG-1 samples purified from transfections I to VI (Fig. 3B and C, lanes 6 to 11). This species corresponds to a heterodimer containing single and double MBP–RAG-1 subunits, as it comigrates with the intermediate of three species formed when MBP-RAG1hm (wtNBD, wtAS) and MBP2-RAG1m (wtNBD, wtAS) are coexpressed and purified only by amylose affinity chromatography (Fig. 3B and C, lane 5). A faster-migrating species (M1) is easily observed in samples purified from transfections I to III (Fig. 3B and C, lanes 6 to 8) but is less abundant in samples derived from transfections IV to VI (Fig. 3B and C, lanes 9 to 11). This species corresponds to a 12-RSS complex containing a homodimer of the single MBP–RAG-1 fusion protein, as it comigrates with a species formed in the presence of the MBP-RAG1hm (wtNBD, wtAS) alone (Fig. 3B and C, lane 3). The decreased abundance of the M1 species in lanes 9 to 11 reflects the presence of NBD mutations on both RAG-1 subunits, shown previously to impair DNA binding (46, 47). Importantly, a species comigrating with the homodimer of MBP2-RAG1m (wtNBD, wtAS) bound to the 12-RSS was not observed in any of the heterodimer preparations (Fig. 3B and C, lane 4), demonstrating inefficient subunit exchange under these conditions.
As expected from previous results, both the RAG-1 homo- and heterodimers obtained from transfections I to III assemble RSS complexes containing RAG-2 of the expected mobility (M1/M2 and M1M21/M2, respectively) (Fig. 3B and C, compare lanes 12 to 14 to lanes 15 to 17), as both species contain at least one RAG-1 subunit with an intact NBD. In contrast, no M1/M2 complex is observed in samples from transfections IV to VI (Fig. 3B and C, lanes 18 to 20), because the copurified homodimer bears mutant NBDs on both subunits and hence is unable to form stable protein-DNA complexes containing RAG-2 (46, 47). Similar results were obtained for the other two panels of heterodimers (data not shown).
The DDE triad acts in trans in the nicking step of V(D)J recombination.
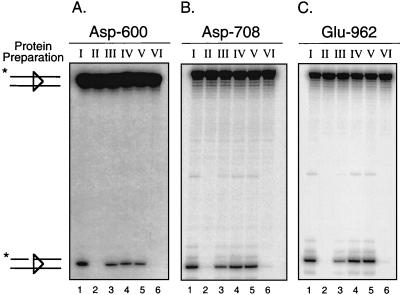
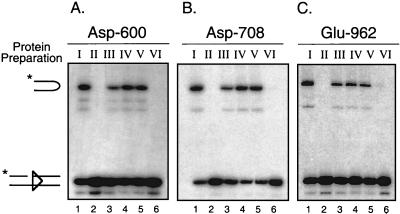
To determine how the residues of the DDE motif are contributed to the active site that catalyzes the nicking step of V(D)J recombination, heterodimer samples prepared from transfections I to VI were incubated with an intact 32P-end-labeled 12-RSS substrate in the presence of Ca2+ and RAG-2 on a preparative scale. Discrete protein-DNA complexes were separated by an EMSA as described for Fig. 3, and the gel was subsequently soaked in cleavage buffer containing Mg2+ for 1 h at 37°C to permit catalysis of nicking in situ. Cleavage products attributed to the heteromeric M1M21/M2 complex were analyzed for panels of heterodimers prepared for each of the three active-site mutants D600A, D708A, and E962A (Fig. 4A to C, respectively, and Table 1). As expected, heterodimers containing a wild-type NBD and active site in both subunits were able to catalyze nicking in the presence of RAG-2 (Fig. 4A to C, lane 1), whereas heterodimers containing the D600A, D708A, or E962A mutation in both subunits were essentially inactive (Fig. 4A to C, lane 2). Heterodimers bearing either an active site or NBD mutation (but not both) in only one subunit are active in complexes containing RAG-2 (Fig. 4A to C, lanes 3 and 4).
FIG. 4.
The DDE triad acts in trans in the catalysis of nicking. (A to C) In situ cleavage analysis for D600A, D708A, and E962A mutant heterodimer preparations, respectively. Preparative binding reactions containing an intact 32P-end-labeled 12-RSS substrate, RAG-2, and each RAG-1 heterodimer sample (Fig. 2, I to VI) were assembled, and protein-DNA complexes were fractionated by EMSA. Cleavage products generated in situ in the presence of Mg2+ were recovered from the M1M21/M2 EMSA complex derived from each heterodimer preparation (I to VI) and were fractionated by denaturing gel electrophoresis. The positions of full-length and nicked species are indicated at left. Quantitative analysis of substrate cleavage is found in Table 1.
TABLE 1.
Quantitative analysis of in situ cleavage experiments
| Mutant heterodimer preparation | RSS substrate | Metal | % Conversiona
|
trans preference (V/VI ratio) | |||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | ||||
| D600A | Intact 12 (Fig. 4A) | Mg | 8.7 | 0.3 | 5.6 | 6.6 | 6.6 | 0.3 | 19 |
| Intact 12 (expt 2) | Mg | 17 | 0.2 | 7.8 | 21 | 22 | 0.4 | 54 | |
| Prenicked 12 (Fig. 5A) | Mn | 39 | 0.6 | 18 | 32 | 28 | 0.7 | 38 | |
| Prenicked 12 (expt 2) | Mn | 38 | 0.5 | 16 | 42 | 40 | 0.9 | 47 | |
| Prenicked 12 | Mg | 1.1 | 0.06 | 0.36 | 0.78 | 0.58 | 0.03 | 18 | |
| Intact 23b | Mg | 2.8 | 0.1 | 1.0 | 1.5 | 1.5 | 0.2 | 7 | |
| Prenicked 23b | Mn | 17 | 0.1 | 5.0 | 1.5 | 7.0 | 0.1 | 54 | |
| D708A | Intact 12 (Fig. 4B) | Mg | 11 | 0.2 | 4.0 | 8.7 | 9.0 | 0.2 | 43 |
| Intact 12 (expt 2) | Mg | 23 | 0.3 | 20 | 14 | 19 | 0.4 | 47 | |
| Prenicked 12 (Fig. 5B) | Mn | 59 | 1.0 | 31 | 55 | 52 | 1.1 | 45 | |
| Prenicked 12 (expt 2) | Mn | 44 | 0.4 | 13 | 41 | 42 | 0.6 | 73 | |
| Prenicked 12 | Mg | 0.73 | 0.00 | 0.31 | 0.71 | 0.65 | 0.03 | 21 | |
| Intact 23b | Mg | 12 | 0.2 | 6.4 | 6.6 | 6.1 | 0.2 | 27 | |
| Prenicked 23b | Mn | 35 | 0.7 | 11 | 24 | 23 | 0.2 | 104 | |
| E962A | Intact 12 (Fig. 4C) | Mg | 16 | 0.1 | 4.0 | 14 | 15 | 0.2 | 78 |
| Intact 12 (expt 2) | Mg | 15 | 0.1 | 3.7 | 13 | 12 | 0.2 | 57 | |
| Intact 12 (expt 3)c | Mg | 16 | 5.7 | 9.4 | 17 | 13 | 4.2 | 3 | |
| Prenicked 12 (Fig. 5C) | Mn | 29 | 0.7 | 13 | 15 | 15 | 1.1 | 13 | |
| Prenicked 12 (expt 2) | Mn | 59 | 0.3 | 11 | 57 | 53 | 0.7 | 77 | |
| Intact 23b | Mg | 2.7 | 0.2 | 0.9 | 1.4 | 1.2 | 0.1 | 9 | |
| Prenicked 23b | Mn | 20 | 0.9 | 8.1 | 7.7 | 7.0 | 0.2 | 35 | |
Measured as percentage of predominant nicked (intact) or hairpin (prenicked) species present relative to total DNA in samples recovered from complexes containing RAG-1 heterodimers (I to VI) (Fig. 2).
RAG-RSS complexes assembled in the presence of HMG-1 (see Materials and Methods).
Nicked products found in samples II and VI likely arose from cross-contamination since they are found at similar levels in both samples and are not reproducibly observed in two other experiments. However, the possibility that they are attributed to Glu-962 independent activity, observed by others at elevated metal ion concentrations (20), cannot be formally excluded. In both cases, the actual trans preference may be much greater than threefold, either because the amount of cleaved product recovered overrepresents the actual cleavage activity in these samples (the former case) or because the cleavage product observed represents a basal activity present in every sample that can be subtracted from all the samples (the latter case).
Importantly, RAG-1 heterodimers containing a mutant NBD on one subunit and a mutant active site on the other supported nicking at levels similar to those for heterodimers containing a single mutant NBD (Fig. 4A to C, compare lanes 4 and 5). In contrast, RAG-1 heterodimers containing both a mutant NBD and a mutant active site on a single subunit did not catalyze significant nicking in all three mutant series (Fig. 4A to C, lane 6). These results indicate that the V(D)J recombinase uses a single active site on a single RAG-1 subunit to catalyze hydrolysis of an intact 12-RSS substrate. In addition, the subunit opposite the one bound to the nonamer of the RSS undergoing cleavage donates the DDE triad to the active site (in trans). Finally, these data suggest that only a single copy of the RSS substrate is present in complexes containing RAG-2 and a RAG-1 heterodimer with a mutant NBD on a single subunit. If two RSS substrates were present in the complex (one bound to each RAG-1 subunit, despite the NBD mutation on one of the subunits), significant cleavage of the RSS substrate would be detected in both the cis and trans configurations (e.g., preparations V and VI) in all three mutant panels, which clearly is not observed. Indeed, the difference in the cleavage activity between the two configurations likely reflects the difference in the relative affinities between the wild-type and mutant RAG-1 subunits for the RSS.
Hairpin formation is catalyzed in trans by the DDE triad.
To determine whether hairpin product formation is catalyzed in trans, experiments similar to those described for Fig. 4 were performed using a prenicked 12-RSS substrate, except that Mg2+ was replaced by Mn2+ in the cleavage buffer to support hairpin formation (Fig. 5A to C). The results obtained from these experiments were qualitatively similar to those shown for Fig. 4 with respect to the cleavage activity of the individual heterodimer preparations. Importantly, heterodimers containing a mutant NBD on one RAG-1 subunit and a mutant active site on the opposite subunit but not on the same subunit supported hairpin formation at levels similar to those for heterodimers containing a single mutant NBD (Fig. 5A to C, compare lane 4 to lanes 5 and 6).
FIG. 5.
Hairpin formation is catalyzed in trans by the DDE motif. In situ cleavage of a 32P-end-labeled prenicked 12-RSS substrate was performed for each active-site mutant series (A to C) (D600A, D708A, and E962A, respectively) as described for Fig. 4, except that Mn2+ rather than Mg2+ was used as the divalent metal ion cofactor in the cleavage reaction. The positions of nicked and hairpin species are indicated at left. The data are otherwise displayed as shown for Fig. 4. See Table 1 for quantitative analysis of substrate cleavage.
To test whether the composition of the divalent metal ion influenced the preference for the trans configuration, the same experiment was performed with the D600A and D708A panels, substituting Mg2+ for Mn2+. Because Mg2+ does not support catalysis of the hairpin reaction as efficiently as Mn2+ on single RSS substrates (49), the degree of substrate conversion in the presence of Mg2+ was quite low compared to that for reactions performed in the presence of Mn2+ (≤1% versus 15 to 50%, respectively) (Table 1). Despite the low levels of conversion, a ∼20-fold preference for the trans configuration was observed in both the D600A and D708A panels (Table 1), suggesting that in these complexes, the active-site organization is not selected by the divalent metal ion cofactor assisting the cleavage reaction. However, since the protein-DNA complexes being examined were assembled in the presence of Ca2+, which supports binding but not cleavage of the RSS by the RAG proteins (16), the possibility that Ca2+ directs the organization of active sites in a manner unlike that of Mg2+ or Mn2+ cannot be formally excluded. Although the trans preference was qualitatively reproducible for both the intact and prenicked 12-RSS substrates, some quantitative variation in the levels of substrate cleavage (both within a given sample and between samples) was observed in independent experiments (Table 1).
These results, together with cleavage data obtained using intact RSS substrates, provide evidence that a RAG-1 dimer contains two active sites, one on each subunit; a single active site is responsible for catalyzing both the nicking and hairpin formation steps of V(D)J recombination. Moreover, the DDE triad comprising an essential part of the active site is contributed in trans by the RAG-1 subunit opposite the subunit bound to the nonamer of the RSS being cleaved.
A 23-RSS substrate is cleaved in trans.
To examine whether the trans cleavage mode observed in 12-RSS complexes extends to complexes containing a 23-RSS substrate, experiments similar to those described in Fig. 4 and 5 were performed on RAG complexes assembled on intact or prenicked 23-RSS substrates (Table 1). Besides the RAG proteins, the binding reactions also included the high-mobility group protein 1 (HMG-1), which has been shown to stimulate RAG bending and binding of 23-RSS substrates (2, 48). The 23-RSS complexes containing both RAG proteins displayed a pattern of electrophoretic mobility similar to that for those formed using 12-RSS substrates (data not shown). The cleavage activity of the individual heterodimer preparations on these substrates was qualitatively similar to that obtained using 12-RSS substrates, as shown for Fig. 4 and 5. Importantly, for both the intact and prenicked 23-RSS substrates, a strong preference for the trans configuration was observed (Table 1, last column), suggesting that the basis for how the active site is organized within the V(D)J initiation complex does not depend on the length of the RSS spacer arm.
DISCUSSION
The DDE triad acts in trans in both cleavage steps of V(D)J recombination.
Previous studies identified a DDE triad of conserved carboxylate residues in RAG-1 essential for the catalytic activity of the V(D)J recombinase (13, 20, 22). Mutations in any one of the residues in the DDE motif impair both the nicking and hairpin reactions, suggesting that a single active site catalyzes both of these steps in V(D)J recombination. How the residues comprising the DDE motif are organized within a V(D)J initiation complex was not determined. In this study, single alanine mutations were introduced in each of the essential DDE triad of acidic amino acid residues of RAG-1 to probe the active-site organization in discrete protein-DNA complexes assembled on an intact or prenicked single RSS substrates containing dimeric RAG-1 and one or two monomers of RAG-2. The results presented here extend previous studies by demonstrating that (i) a RAG-1 dimer contains two active sites, one on each subunit; (ii) the entire DDE triad is contributed to a single active site by a single RAG-1 subunit, rather than being distributed between the subunits to form a composite active site; and (iii) a single active site on the RAG-1 subunit opposite the one bound to the nonamer of the RSS undergoing cleavage catalyzes both the hydrolysis (nicking) and transesterification (hairpin) steps of V(D)J recombination in the presence of RAG-2 (cleavage in trans).
Implications for regulation of V(D)J recombination.
The data presented here help refine models of RAG-RSS interactions with single RSS substrates developed previously, based on a combination of DNA footprinting, photocross-linking, and protein stoichiometry analysis (46, 47), by providing evidence that recognition of the heptamer-coding junction by RAG-1 and its subsequent cleavage in these complexes occur in trans. Modification interference footprinting and photocross-linking studies suggest that RAG-2-dependent RAG-1 interactions with the heptamer region appear to extend from the 5′ end of the spacer arm to the heptamer-coding junction (11, 46, 47). Whether RAG-1 interacts in trans with this entire region or just the heptamer-coding junction cannot be determined from data presented here but might be ascertained by using photocross-linking methods.
Whether synaptic complexes containing both a 12- and 23-RSS, expected to be the physiologically relevant complexes supporting V(D)J recombination in vivo, also follow a trans mode of cleavage remains unknown but is of considerable interest. However, determining the active-site organization in synaptic complexes poses some difficulty for two reasons. First, although the synaptic complex is speculated to contain at least one RAG-1 dimer (and perhaps a pair of dimers), the stoichiometry of RAG-1 (and RAG-2) in these complexes has not yet been formally demonstrated, precluding analysis of the active-site configuration at the present time. Second, and perhaps more important, RAG-1 exists in dimeric form both in solution and bound to DNA (4, 35, 46). Therefore, since both RAG-1 subunits must have intact DNA binding domains to form bona fide synaptic complexes, targeting an active-site mutation to a particular RAG-1 subunit in order to follow cleavage at a specific RSS is problematic because the RSS being followed could be bound by either subunit of the RAG-1 dimer.
Nevertheless, several observations drawn from studies of single RSS complexes containing both RAG-1 and RAG-2 provide a basis to speculate on the composition and organization of the RAG proteins in the synaptic complex (Fig. 6). In modeling the arrangement of RAG-1 in the synaptic complex, three observations were considered. (i) RAG-1 is able to contact both the heptamer and nonamer elements (3, 9, 11, 35, 45–47). (ii) RAG-1 retains its dimeric configuration on single RSS substrates regardless of whether RAG-2 is present (4, 46). (iii) A single RAG-1 dimer contains two active sites, one contributed by each subunit (this study). Thus, the most economical model of the synaptic complex is one that contains a single RAG-1 dimer; each RAG-1 subunit is bound to a separate RSS nonamer via the NBD. Interactions with the RSS heptamers would be mediated by RAG-1 but depend on the presence of RAG-2. By symmetry considerations and mobility shift assays on single RSS complexes (4, 46), RAG-2 would exist in this speculative model of the synaptic complex as a pair of monomers. The region of RAG-1 interacting with the heptamer-coding junction would contain the DDE triad that facilitates catalysis of both cleavage steps of V(D)J recombination.
FIG. 6.
Speculative model of the synaptic complex that supports V(D)J recombination. The synaptic complex, adapted from reference 46, is proposed to contain dimeric RAG-1 (R1) and two monomers of RAG-2 (R2). The NBD of each RAG-1 subunit interacts with a separate nonamer element in the 12- or 23-RSS (depicted as straight or curved lines, respectively). The active-site domain (ASD) proximal to a given heptamer-coding junction is donated by the RAG-1 subunit bound to the nonamer of the opposing RSS (in trans). A single active site catalyzes both the nicking and transesterification steps of V(D)J recombination. The RSSs are oriented heptamer to heptamer, based on evidence that the minimum intersignal distance supporting coupled cleavage and recombination is shortest when the signals have this configuration (10, 43). This model is not meant to imply a particular location where the domains bearing the active site or nonamer binding activity in RAG-1 cross over on the RSS, nor is it intended to convey a specific organization, localization, or function of the RAG-2 monomers. The position of the HMG protein(s), which facilitates RAG-RSS synaptic complex assembly (15) and coupled cleavage (48) in vitro, is shown bridging the NBD of RAG-1 and the 23-RSS, thereby enhancing RAG bending and binding of the 23-RSS (2).
The trans cleavage mode adopted by the RAG proteins in single RSS complexes might reasonably be expected to extend to the synaptic complex for three reasons (Fig. 6). First, trans cleavage is observed for both cleavage steps in the presence of Mg2+, which supports 12/23-regulated cleavage of RSS pairs in vitro and is considered the relevant divalent metal ion cofactor in vivo (10, 51). Second, both 12- and 23-RSS substrates are individually cleaved in trans. Third, this arrangement enables one RAG-1 subunit to interact with both RSSs in a synaptic complex. One advantage that this organization may provide is greater control over the possibility of single-site cleavage, which is more easily visualized if the RAG-1 subunit bound to a given RSS is also the one that cleaves it. A second advantage is that the RAG-1 subunit bound to a given RSS controls each cleavage step performed on the partner RSS, thereby providing a more direct means of ensuring that only RSSs with spacers of different lengths are cleaved by the V(D)J recombinase.
Of course, I certainly acknowledge the possibility that the synaptic complex may adopt a cis mode of cleavage. In this scenario, the decision to initiate cleavage at a given RSS is made by the RAG-1 subunit bound to it. Thus, protein-protein interactions between the RAG-1 subunits would be the only means to convey the information necessary to enforce the 12/23 rule. If the RAG-1 subunits are found to be arranged in the synaptic complex in cis, the data presented here suggest the possibility that major remodeling of the protein-DNA complexes formed at isolated RSSs accompanies the process involved in bringing a pair of RSSs together in a synaptic complex.
How the RAG-1 subunits are able to discriminate spacer length within the context of a synaptic complex is unclear. Moreover, how RAG-2 and HMG-1 and HMG-2, which facilitate synaptic complex assembly (15) and activity (48), are involved in this process remains to be elucidated. Because the HMG proteins promote RAG binding to the 23-RSS (48) and interact with the NBD of RAG-1 (2), it is tempting to speculate that ternary interactions among RAG-1, the 23-RSS, and the HMG protein(s) may provide a key sensing mechanism that coordinates synaptic complex formation in a 12/23-regulated fashion (Fig. 6).
DNA cleavage in trans: a common theme underlying bacterial transposition and V(D)J recombination?
DNA transactions performed by the V(D)J recombinase share many biochemical similaries to reactions catalyzed by a family of enzymes that includes bacterial transposases and retroviral integrases. In these enzymes, a scissile phosphodiester is first hydrolyzed by an activated water molecule. As a result, a 3′ hydroxyl group is exposed, which subsequently attacks the scissile phosphodiester on the target DNA, becoming covalently linked via a direct transesterification reaction. The V(D)J recombinase resembles this class of enzymes with respect to the mechanism of DNA strand scission (28, 50), the composition of the residues essential for catalysis (13, 20, 22), its associated, non-sequence-dependent activities (e.g., transposition [1, 17] and 3′ flap endonuclease activity [39]), and predicted active-site folding topology (13).
The transposases from bacteriophage Mu, Tn5, Tn10, and Tn7 have been characterized with respect to their active-site organization. Catalysis of the donor cleavage and strand transfer reactions by the MuA transposase occurs within a protein-DNA complex containing a tetramer of MuA (23). A single active site on one MuA monomer catalyzes both reactions on a single Mu end; both reactions are performed by the MuA monomer bound to the partner Mu end (cleavage in trans) (29, 52). In Tn10 transposition, a single transposase active site sequentially catalyzes the cleavage of the transferred and nontransferred strands of the donor DNA (via hairpin formation on the transposon end), as well as the subsequent strand transfer reaction; all chemical reactions at one transposon end are performed by a single transposase monomer (7, 19). Whether the Tn10 transposase monomer that catalyzes the cleavage reactions at a given transposase end is the one bound to that end (in cis) or the one bound to the partner end (in trans) is not yet known. However, recent biochemical and structural studies of a similar transposase from Tn5, which also mediates transposition via a DNA hairpin intermediate (6), suggest that strand cleavage and strand transfer reactions are catalyzed by this transposase in trans (8, 30). Unlike the previous three examples, the DNA cleavage and joining activities underlying Tn7 transposition are distributed between two proteins, TnsA and TnsB; each contains a discrete active site and performs distinct DNA processing reactions at the transposon ends (40). The V(D)J recombinase resembles the Tn5, Tn10, and Mu transposase in the use of a single active site containing a DDE triad to catalyze sequential DNA cleavage reactions (7, 8, 52). Moreover, like both the Mu transposase and the Tn5 transposase, the V(D)J recombinase cleaves DNA in trans.
Both Tn5 and Tn10 transpositions share additional features with V(D)J recombination, including (i) the generation of DNA hairpin intermediates (6, 19); (ii) the presence of a single enzyme binding site at each DNA end undergoing cleavage (21, 34); (iii) an unusually large spacing between the second Asp and third Glu residue of the DDE motif comprising the enzyme active site (134 residues in Tn5 [8], 131 residues in Tn10 transposase [7], and 254 residues in RAG-1 [20, 22]), compared to the more typical 35- to 55-residue spacing observed in many transposases and retroviral integrases, including MuA and TnsB (33); and (iv) the use of a single active site to catalyze the sequential nicking and hairpin formation steps of the DNA cleavage reaction (7, 19).
Despite many similarities between RAG-1 and the Tn5 and Tn10 transposases, two important distinctions between V(D)J recombination and these two transposition systems are worth noting. First, unlike Tn5 and Tn10 transposition, which generates hairpin intermediates formed on the transposon end (6, 19), V(D)J recombination produces hairpin intermediates at the coding end (equivalent to the donor DNA). Second, whereas only one protein, the transposase, is able to perform all cleavage reactions involved in transposition, RSS recognition and cleavage in V(D)J recombination require two proteins, RAG-1 and RAG-2. Although RAG-2 does not appear to be directly involved in catalysis, RAG-2 may play a key role in directing which DNA strand gets nicked at the heptamer-coding junction and/or enforcing the cleavage mode of the V(D)J recombinase. This may be achieved directly by tethering the RAG-1 active site to the cleavage site through RAG-2 interactions with the heptamer-coding junction or more indirectly by serving as a structural cofactor whose primary role is stabilizing the association of RAG-1 with the RSS near the cleavage site. While these models are not mutually exclusive, recent photocross-linking studies demonstrating a physical proximity between RAG-2 and the heptamer-coding junction in RSS complexes containing both RAG proteins are more consistent with the former possibility (11, 46). Further efforts to distinguish between these possibilities will provide insight into how the RAG proteins direct the cleavage steps of V(D)J recombination to the appropriate DNA strand in trans.
ACKNOWLEDGMENTS
This work was supported by the Health Future Foundation.
I thank Stephen Desiderio for reagents and support and Mark Schlissel for critically reviewing a version of the manuscript.
REFERENCES
- 1.Agrawal A, Eastman Q M, Schatz D G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 2.Aidinis V, Bonaldi T, Beltrame M, Santagata S, Bianchi M E, Spanopoulou E. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol Cell Biol. 1999;19:6532–6542. doi: 10.1128/mcb.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akamatsu Y, Oettinger M A. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailin T, Mo X, Sadofsky M J. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol Cell Biol. 1999;19:4664–4671. doi: 10.1128/mcb.19.7.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besmer E, Mansilla-Soto J, Cassard S, Sawchuk D J, Brown G, Sadofsky M, Lewis S M, Nussenzweig M C, Cortes P. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol Cell. 1998;2:817–828. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- 6.Bhasin A, Goryshin I Y, Reznikoff W S. Hairpin formation in Tn5 transposition. J Biol Chem. 1999;274:37021–37029. doi: 10.1074/jbc.274.52.37021. [DOI] [PubMed] [Google Scholar]
- 7.Bolland S, Kleckner N. The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell. 1996;84:223–233. doi: 10.1016/s0092-8674(00)80977-0. [DOI] [PubMed] [Google Scholar]
- 8.Davies D R, Goryshin I Y, Reznikoff W S, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 9.Difilippantonio M J, McMahan C J, Eastman Q M, Spanopoulou E, Schatz D G. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 10.Eastman Q M, Leu T M, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 11.Eastman Q M, Villey I J, Schatz D G. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fugmann S D, Lee A I, Shockett P E, Villey I J, Schatz D G. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 13.Fugmann S D, Villey I J, Ptaszek L M, Schatz D G. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 14.Haren L, Ton-Hoang B, Chandler M. Integrating DNA: transposases and retroviral integrases. Annu Rev Microbiol. 1999;53:245–281. doi: 10.1146/annurev.micro.53.1.245. [DOI] [PubMed] [Google Scholar]
- 15.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 16.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 17.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 18.Jones D H, Winistorfer S C. Recombinant circle PCR and recombination PCR for site-specific mutagenesis without PCR product purification. BioTechniques. 1992;12:528–534. [PubMed] [Google Scholar]
- 19.Kennedy A K, Guhathakurta A, Kleckner N, Haniford D B. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 20.Kim D R, Dai Y, Mundy C L, Yang W, Oettinger M A. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleckner N, Chalmers R M, Kwon D, Sakai J, Bolland S. Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. Curr Top Microbiol Immunol. 1996;204:49–82. doi: 10.1007/978-3-642-79795-8_3. [DOI] [PubMed] [Google Scholar]
- 22.Landree M A, Wibbenmeyer J A, Roth D B. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavoie B D, Chan B S, Allison R G, Chaconas G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Swanson P, Desiderio S. RAG-1- and RAG-2-dependent assembly of functional complexes with V(D)J recombination substrates in solution. Mol Cell Biol. 1997;17:6932–6939. doi: 10.1128/mcb.17.12.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin W C, Desiderio S. Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science. 1993;260:953–959. doi: 10.1126/science.8493533. [DOI] [PubMed] [Google Scholar]
- 27.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 28.Melek M, Gellert M, van Gent D C. Rejoining of DNA by the RAG1 and RAG2 proteins. Science. 1998;280:301–303. doi: 10.1126/science.280.5361.301. [DOI] [PubMed] [Google Scholar]
- 29.Namgoong S Y, Harshey R M. The same two monomers within a MuA tetramer provide the DDE domains for the strand cleavage and strand transfer steps of transposition. EMBO J. 1998;17:3775–3785. doi: 10.1093/emboj/17.13.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naumann T A, Reznikoff W S. Trans catalysis in Tn5 transposition. Proc Natl Acad Sci USA. 2000;97:8944–8949. doi: 10.1073/pnas.160107997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 32.Plasterk R. V(D)J recombination. Ragtime jumping. Nature. 1998;394:718–719. doi: 10.1038/29389. [DOI] [PubMed] [Google Scholar]
- 33.Polard P, Chandler M. Bacterial transposases and retroviral integrases. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 34.Reznikoff W S, Bhasin A, Davies D R, Goryshin I Y, Mahnke L A, Naumann T, Rayment I, Steiniger-White M, Twining S S. Tn5: a molecular window on transposition. Biochem Biophys Res Commun. 1999;266:729–734. doi: 10.1006/bbrc.1999.1891. [DOI] [PubMed] [Google Scholar]
- 35.Rodgers K K, Villey I J, Ptaszek L, Corbett E, Schatz D G, Coleman J E. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 1999;27:2938–2946. doi: 10.1093/nar/27.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth D B, Craig N L. VDJ recombination: a transposase goes to work. Cell. 1998;94:411–414. doi: 10.1016/s0092-8674(00)81580-9. [DOI] [PubMed] [Google Scholar]
- 37.Roth D B, Menetski J P, Nakajima P B, Bosma M J, Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 38.Roth D B, Zhu C, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santagata S, Besmer E, Villa A, Bozzi F, Allingham J S, Sobacchi C, Haniford D B, Vezzoni P, Nussenzweig M C, Pan Z Q, Cortes P. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol Cell. 1999;4:935–947. doi: 10.1016/s1097-2765(00)80223-3. [DOI] [PubMed] [Google Scholar]
- 40.Sarnovsky R J, May E W, Craig N L. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- 41.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 42.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 43.Sheehan K M, Lieber M R. V(D)J recombination: signal and coding joint resolution are uncoupled and depend on parallel synapsis of the sites. Mol Cell Biol. 1993;13:1363–1370. doi: 10.1128/mcb.13.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shockett P E, Schatz D G. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol Cell Biol. 1999;19:4159–4166. doi: 10.1128/mcb.19.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spanopoulou E, Zaitseva F, Wang F H, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 46.Swanson P C, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanson P C, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 48.van Gent D C, Hiom K, Paull T T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 50.van Gent D C, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 51.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 52.Williams T L, Jackson E L, Carritte A, Baker T A. Organization and dynamics of the Mu transpososome: recombination by communication between two active sites. Genes Dev. 1999;13:2725–2737. doi: 10.1101/gad.13.20.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]