Abstract
The demand for facial masks remains high. However, little is known about discarded masks as a potential refuge for contaminants and to facilitate enrichment and spread of antibiotic resistance genes (ARG) in the environment. We address this issue by conducting an in-situ time-series experiment to investigate the dynamic changes of ARGs, bacteria and protozoa associated with discarded masks. Masks were incubated in an estuary for 30 days. The relative abundance of ARGs in masks increased after day 7 but levelled off after 14 days. The absolute abundance of ARGs at 30 days was 1.29 × 1012 and 1.07 × 1012 copies for carbon and surgical masks, respectively. According to normalized stochasticity ratio analysis, the assembly of bacterial and protistan communities was determined by stochastic (NST = 62%) and deterministic (NST = 40%) processes respectively. A network analysis highlighted potential interactions between bacteria and protozoa, which was further confirmed by culture-dependent assays, that showed masks shelter and enrich microbial communities. An antibiotic susceptibility test suggested that antibiotic resistant pathogens co-exist within protozoa. This study provides an insight into the spread of ARGs through discarded masks and highlights the importance of managing discarded masks with the potential ecological risk of mask contamination.
Keywords: ARGs, Marine, Plastisphere, Microplastics, Microbiome
Graphical Abstract
1. Introduction
Coronavirus disease (COVID-19) was declared a worldwide pandemic in early 2020 (Huang et al., 2020) and poses a serious threat to human health. As airborne transmission is the primary route of virus spread (Chan et al., 2020; Li et al., 2020), personal health protection is an imperative to mitigate transmission, resulting in a burgeoning demand for personal protective equipment (PPE) especially prior to the rollout of effective vaccine programs (Dharmaraj et al., 2021, Haque et al., 2021). Face masks in particular are considered essential equipment as they can effectively filter aerosols carrying virus particles (Dietz et al., 2020; Patricio Silva et al., 2021). Masking policies are mandatory in most countries to curb the spread of COVID-19 (Feng et al., 2020). As a result, a total of 129 billion face masks will likely be used globally each month (Kalina and Tilley, 2020). Most surgical masks currently available are made from non-biodegradable polymers such as polypropylene, polyethylene and polyacrylonitrile, which are considered as plastic contaminants in the environment (Chen et al., 2021, Fadare and Okoffo, 2020, Kutralam-Muniasamy et al., 2020). Hence, discarded face masks have potential to increase the recognized environmental threat posed by plastic pollution (Prata et al., 2020). Prior to the pandemic, plastic pollution of both terrestrial and marine environments was already a global issue (Eriksen et al., 2014, Horton et al., 2017). Although national authorities have taken measures to dispose medical waste (Anbumani and Kakkar, 2018, Prata et al., 2020), it is estimated that around 3.4 billion masks are discarded daily with potential environmental consequences (Abbasi et al., 2020, Benson et al., 2021). Furthermore, masks may be carriers of chemical and biological contaminants, providing a long-term stable ecological niche, a so-called “plastisphere”, for microorganisms in marine ecosystems (Zettler et al., 2013).
The contamination of aquatic ecosystems with antibiotic resistance genes (ARGs) and antibiotic resistant bacteria (ARB) is of increasing concern (Su et al., 2018). ARGs can be acquired in the environment through horizontal gene transfer (HGT) between microorganisms, causing antibiotic-sensitive bacteria to evolve resistance, impeding antibiotic function (Baquero et al., 2008, Shi et al., 2013). Wastewater treatment plants, terrestrial runoff and aquaculture are the main sources of ARG enrichment and resistome contamination of marine ecosystems (Griffin et al., 2001, Kotlarska et al., 2015, Zheng et al., 2021). As plastics in the ocean can inevitably become a refuge for ARGs (Su et al., 2021), discarded masks may also serve as an “antibiotic resistant reef” providing an interface for the enrichment of ARGs. Furthermore, it may accelerate gene exchange between aquatic microorganisms (Arias-Andres et al., 2018). While the relationship between marine plastics and ARGs has been well documented (Imran et al., 2019, Yang et al., 2019), it remains to be determined whether discarded masks, will exacerbate the risk of enrichment and spread of resistance genes. In addition, the dynamics over time of microorganisms such as bacteria and protozoa, and ARGs colonizing the masks remain unclear, and may impact ecological risk assessments of ARGs.
An in-situ time-series experiment was established at Xinglin Bay, an estuary to Taiwan strait, where masks were exposed to brackish water for 30 days and sampled every 7 days. The estuary receives daily contamination from municipal and sewage outlets. To investigate the dynamic profiles of both ARGs and mobile genetic elements (MGEs) associated with discarded masks, high-throughput quantitative PCR was performed with 296 primer sets which targeted 283 ARGs, 12 MGEs and 1 16S rRNA gene. The aims of the current study are to (1) understand and evaluate the pattern of changes in ARGs, bacteria and protozoa, especially the persistence of ARGs on masks; (2) calculate the absolute abundance of ARGs on masks; (3) establish the link among ARGs, bacteria and protozoa, respectively.
2. Material and methods
2.1. Sample collection and experimental design
Two types of masks, activated carbon (C) and surgical (S) masks, were studied at a site located in Xinglin Bay, Xiamen city, Fujian, China (24° 34′ N, 118° 05′ E) and incubated for 30 days. Each mask was tied to a cotton rope (10 m in length) fixed to the shore and placed on the water surface of Xinglin Bay. Four replicates of both mask types were collected at 7, 14, 21 and 30 days after establishment of the experiment. Thus, a total of 8 masks were sampled at each time point. Two pieces (5 cm × 1 cm) were excised from each mask using sterile scissors and used for DNA extraction and protozoa collection. Four 500 ml water samples were simultaneously collected from each sample site also for DNA and protozoa extraction. In the current study, we used new rather than used masks because it is difficult to ensure that human-derived microorganisms are uniform across different users, and to ensure that the health of the authors was not compromised during the study.
2.2. DNA extraction from water and masks samples
200 ml water samples were initially filtered through a 0.22 μM cellulose membrane to capture the aquatic microbial community. Membranes and collected mask samples were cut into pieces by sterile scissors. A Fast DNA Spin Kit for Soil (MP Biomedicals) was used for DNA extraction following the manufacturer’s instruction. DNA was quantified and quality checked using a NanoDrop ND 1000 (Thermo Scientific, Waltham, MA) Extracted DNA was stored at − 20 °C prior to sample processing.
2.3. ARGs analysis
High-throughput quantitative PCR (HT-qPCR) was used to analyze antibiotic resistome profiles following the methods and primer sets outlined previously (Chen et al., 2017). The primer sets selected represent a panel of known ARGs and MGEs (Zhu et al., 2013), that allow rapid semi-quantitative assessment of the antibiotic resistome in samples. To ensure quality control, each primer set had three technical replicates and the PCR cycle threshold (CT) was set to 31, defined as the upper limit of detection for successful amplification, and was only considered successful if all three technical replicates successfully amplified. Relative and absolute abundance of ARGs and MGEs were calculated as previously described (Su et al., 2015).
2.4. Scanning electron microscopy (SEM)
Scanning electron microscopy (SEM) (HITACHI S-4800), was performed for photography of masks samples at day 7 and day 30. The protocol of SEM analysis was as previously described (Yang et al., 2020).
2.5. PCR assays and amplicon sequencing
The V4-V5 hypervariable region was selected for sequencing the bacterial community using primer set: 515F/907R (Turner et al., 1999). The V4 region of the protist 18S rRNA gene was amplified using primer set TAReukFWD1F (CCAGCASCYGCGGTAATTCC) and TAReukREV3R (ACTTTCGTTCTTGATYRA) (Stoeck et al., 2010). Both PCR protocols were performed by Majorbio (Shanghai, China), as previously described (Zhou et al., 2021). For 18S rRNA, each PCR reaction comprised: a 20 μl total volume of 4 μl 5 × reaction buffer, 2 μl dNTPs (2.5 mM), 0.8 μl primer (5 uM), FastPfu Polymerase 0.4 μl, 2 μl BSA, 10 ng DNA template, and the remainder ddH2O. PCR conditions for 18S rRNA were: an initial denaturation step at 95 °C for 3 min followed by 37 cycles at 95 °C for 30 s, annealing at 53 °C for 30 s, extension at 72 °C for 45 s and a final elongation step at 72 °C for 10 min. Bacterial DNA was amplified using universal primer set 27F/1493R (Baker et al., 2003). PCR reactions were described previously (Zhou et al., 2018).
Sequence analysis of both microbial and protist communities used Quantitative Insights Into Microbial Ecology 2 (QIIME 2) (Bolyen et al., 2019). Chimeric sequences were identified and discarded using “USEARCH” (Edgar, 2013). Amplicon sequence variants (ASVs) were generated using a DADA2 pipeline. Silva v138 was used to annotate bacterial taxonomy, whereas eukaryotic ASVs were taxonomically assigned by blasting against the Protist Ribosomal Reference (PR2) database version 4.12.0 (https://github.com/pr2database/pr2database) using blast-2.9.0 (http://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/2.9.0/). Valid protist sequences were retained after removing mitochondrion, chloroplast sequences, and those of unknown taxa. All sequencing data are accessible at National Center for Biotechnology Information (NCBI). Under accession numbers PRJNA739507 and PRJNA739512 for bacteria and protozoa, respectively.
2.6. Cell sorting and antibiotic sensitivity test
300 ml coastal water samples were filtered through 70 µm cell strainers to avoid clogging up the flow cytometry. Filtrate was passed through a 3 µm filter to enrich protists and were subsequently washed with 5 ml 0.01 M sterile Page's amoeba saline (PAS).
Five cm2 of the mask samples were repeatedly washed with 5 ml 0.01 M sterile PAS to collect any microorganisms that had colonized the masks. The resultant slurry was also filtered with 70 µm cell strainers to remove large particles. Processed samples (mask and water) were incubated for 10 min with Lysotracker Green DND-26 (75 nmol l-1; Invitrogen, Carlsbad, CA, USA), a pH-sensitive green fluorescing probe that stains food vacuoles in protists (Rose et al., 2004).
Target cells were identified and sorted using flow cytometry (Beckman-Coulter, Brea, CA, USA) equipped with a 488 nm laser for excitation. Before sorting each sample, the cytometer was cleaned thoroughly with bleach. A 1% NaCl solution (0.2 mm filtered and ultraviolet-treated) was used as sheath fluid. Beads (2.5 µm) were used to establish a minimum size for protists on the green fluorescence (FL1) cytogram versus forward scatter (FSC) according to Rose et al. (2004). Heterotrophic protists were identified by the presence of Lysotracker fluorescence and absence of chlorophyll fluorescence (Sintes and del Giorgio, 2010). Target cells were collected, followed by incubation for 1 h at room temperature with 400 µg ml-1 gentamicin. Gentamicin was used to kill bacteria external to the protist cells without harming the bacteria within protists (Rønn et al., 2017). After removing the remaining gentamicin, 0.4% Triton X-100 was added to lyse the protist cells, followed by plating 100 μl of relevant dilutions on LB agar plates in triplicate and incubated for 16 h at 37 °C. A total of 30 bacteria isolates from protists were purified and identified.
Antibiotic susceptibility of bacterial isolates followed a disk diffusion method (CLSI, 2019). The bacterial solution was initially adjusted to OD600 ∼ 0.08–0.135 (0.5 McFarland standard), then inoculated onto a Muller–Hinton agar (MHA) plate with antibiotic disks (erythromycin, meropenem and tetracycline) and incubated for a 16–18 h. Susceptibility was evaluated by measuring the diameter of the inhibition zone.
2.7. Statistical analyses
Standard errors, sum and means of ARGs, bacterial and protistan data were calculated using Microsoft EXCEL 2016. Principal Coordinate Analysis (PCoA), Procrustes analysis and Mantel tests were calculated based on Bray-Curtis dissimilarity distances and visualized by R (ver. 3.6.3) packages “vegan”, and “ggplot2” (Dixon, 2003, Wickham et al., 2020). SPSS 21 performed the significance test (ANOVA), with P < 0.05 considered significant. Bar charts were visualized by Originlab 2018. Venn diagrams were generated using Venny (2.1.0) (https://bioinfogp.cnb.csic.es/tools/venny/index.html). The neutral community model (NCM) (Burns et al., 2016) was conducted in R and follows the concepts previously reported (Sloan et al., 2006). The normalized stochasticity ratio (NST) was conducted to quantify the relative importance of stochastic and deterministic processes in the assembly of bacterial and protistan communities, with 50% as the threshold for determining the dominance of deterministic or stochastic processes (Ning et al., 2019). Network analysis was calculated and adjusted using R packages “Hmisc” and “fdrtool”, respectively (Strimmer, 2008, Yadav and Roychoudhury, 2018) and drawn using Gephi (0.9.2). Structural equation models were calculated and visualized by AMOS graphic (IBM, USA) to analysis the contribution of time, number of mask layer, water resistome, MGEs in masks, bacterial and protistan α-diversity to the resistome profiles of masks. For the model, we assumed that: (1) time, water resistome, number of mask layers and mask MGEs directly affect the resistome profile of masks; (2) bacterial and protistan α-diversity indirectly affects the mask resistome by directly affect mask MGEs; and (3) the bacterial α-diversity had a negative effect on the mask resistome. The model could only be considered as valid if all indicators were satisfied as follows: chi-square (P > 0.05), high goodness of fit index (GFI > 0.9), and the root mean square error of approximation (RMSEA < 0.05) (Schermelleh-Engel et al., 2003). R scripts used for the data analysis have been deposited on Github (https://github.com/looopooong/ARGs_Mask_2021.git).
3. Results
3.1. Profiles of antibiotic resistance genes on discarded masks
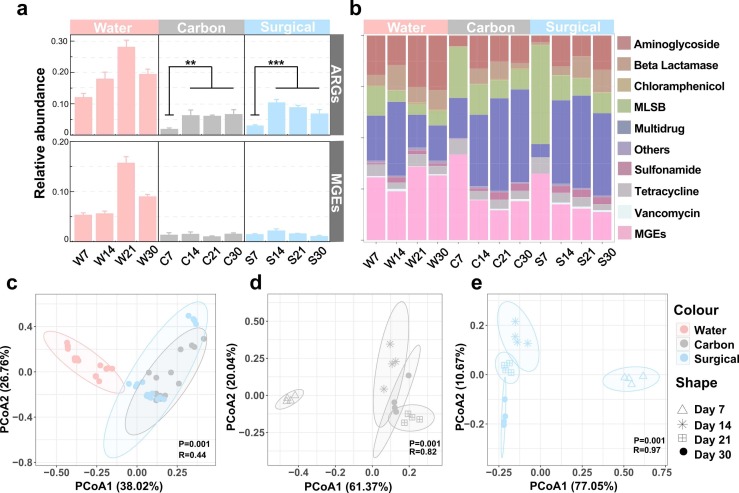
A total of 216 ARGs and 11 MGEs were detected from water and masks samples. ARGs detected from water, activated carbon and surgical mask samples were from 9 classes (Aminoglycoside, Beta Lactam, Chloramphenicol, Macrolide lincosamide streptogramin B resistance (MLSB), Multidrug, Sulfonamide, Tetracycline, Vancomycin and Others). The relative abundance of both ARGs and MGEs in water samples was highest at day 21 ( Fig. 1a) (P < 0.01). At each sampling, the relative abundance of ARGs and MGEs was significantly higher in water samples than either mask type (P < 0.05). In activated carbon masks, the relative abundance of ARGs significantly increased on day 14 and remained constant throughout (P < 0.01). ARGs from surgical masks showed the same trend as activated carbon masks (Fig. 1, P < 0.001). The relative abundance of MGEs in both types of masks did not significantly change with time. The absolute abundance of ARGs and MGEs in both mask types followed the same trend as relative abundance with absolute abundance of ARGs lowest at day 7 and stabilized after day 14. No significant changes in the absolute abundance of MGEs were found (P > 0.05, Fig. S1). The percentage of MLSB resistance genes declined (P < 0.05) whereas Multidrug resistance genes increased significantly (P < 0.05) with time for both mask types (Fig. 1b). The composition of ARGs from water samples was distinct for both mask types (P = 0.001, R = 0.44) (Fig. 1c). The composition of ARGs changed over time with samples from activated carbon masks on day 7 different to those on days 14, 21 and 30 (P = 0.001, R = 0.82) (Fig. 1d). Surgical mask samples on day 7 separated from other samples along the PCoA1 axis, (P = 0.001, R = 0.97) (Fig. 1e).
Fig. 1.
ARG profiles throughout the experimental period. (a) Relative abundance of ARGs and MGEs in water, carbon and surgical mask samples. (b) Percentage of each class of ARGs and MGEs in all samples. (c), (d) and (e) Principal coordinate analysis (PCoA) of ARGs in all samples based on a Bray-Curtis dissimilarity matrix. Colors indicates samples, while triangle, cross, square and circle represent sample time points.
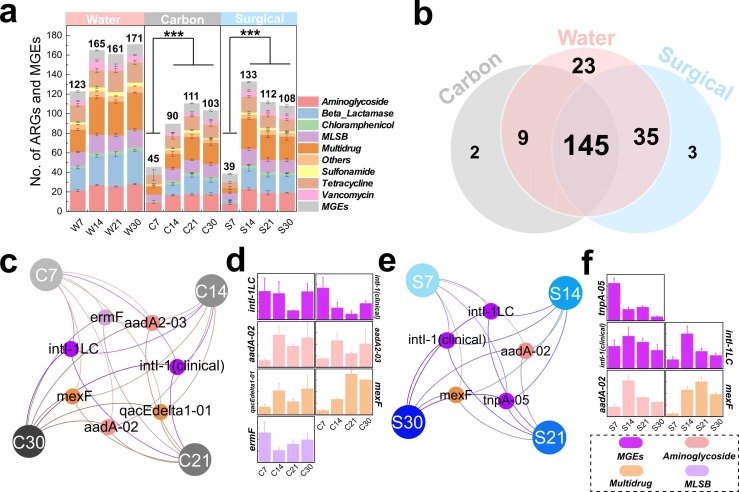
The average number of ARGs and MGEs ranged from 123 to 171 (water samples), 45–111 (activated carbon mask samples) and 39–133 (surgical mask samples) ( Fig. 2a). The number of ARGs and MGEs from both mask types increased significantly (P < 0.001) compared to day 7. Activated carbon and surgical masks shared 154 and 180 ARGs with water samples, while only two and three ARGs, respectively were unique to each mask type (Fig. 2b). Five ARGs and two MGEs persisted after 30 days of incubation in activated carbon masks (Fig. 2c). Of those, the relative abundance of intl-1LC decreased significantly (P = 0.034) on day 21 but returned to the previous level on day 30 (P = 0.036). Both aadA-02 (Aminoglycoside) and mexF (Multidrug) genes increased significantly after 14 days (Fig. 2d). For surgical mask samples, three MGEs and two ARGs persisted throughout (Fig. 2e). The relative abundance of intl-1 (clinical) (MGEs) gene remained consistent throughout the experimental period. However, tnpA-05 decreased significantly after 7 days (Fig. 2f) whereas Multidrug resistance gene mexF increased (P < 0.001) after 14 days (Fig. 2f).
Fig. 2.
(a) Detected number of ARGs and MGEs throughout the experimental period. (b) Shared and unique number of ARGs from different samples. (c) and (d) persistent ARGs and MGEs from carbon mask samples at each sampling time point, and the relative abundance of each persistent ARGs with time. (e) and (f) persistent ARGs and MGEs from surgical mask samples at each sampling time point, and the relative abundance of each persistent ARGs with time.
3.2. The microbial community on masks
Activated carbon and surgical masks consisted of four and three layers respectively. Samples for SEM observation were collected after 3- and 30-days of incubation. Microbial colonizers penetrated the masks and constructed a biofilm. On each layer of the activated carbon mask, less microbial biomass was observed after 3 days incubation compared to 30 days (Fig. S2a–h). The trend for the surgical mask samples was similar to carbon mask samples (Fig. S2i–n).
A total of 2,157,909 and 3,063,566 high quality bacterial and protist sequences were generated, ranging from 31,961 to 88,282 and 38,775–88,738 sequences per sample. The Shannon index at family level for bacteria detected from both carbon and surgical masks was significantly higher than in water samples (P < 0.001) (Fig. S3a). The relative abundance of Comamonadaceae, Rhodocyclaceae, Ectothiorhodospiraceae and Ilumatobacteraceae in water samples was significantly higher than in both mask types (P < 0.05). In contrast, Cyclobacteriaceae and Xanthomonadaceae were higher in both mask types than in water samples (P < 0.05) (Fig. S3b). For protists, no significant differences were found in alpha diversity across all samples (Fig. S3c). The 20 dominant protist families accounted for over 75% of protists detected. Suctoria_X, Stentoridae and Zoothamniidae were dominant in mask samples, and had a significantly higher abundance than in water samples (P < 0.05).
A Principal Coordinate Analysis (PCoA) based on Bray-Curtis dissimilarity values, separated mask and water bacterial communities (Fig. S4a), while masks samples were similar (Fig. S4d). Bacterial communities from both mask types changed over time. The composition of bacteria at day 7 differed from other sampling times along PCoA1 (activated carbon, Fig. S4b) and at days 21 and 30 (surgical, Fig. S4c). The protistan communities showed the same trends as bacterial communities for both mask types, with PCoA1 explaining 41.8% (carbon) and 53.7% (surgical) of the total variation.
A significant correlation between bacterial and protistan communities (ASV level) for both mask types was found (Procrustes analysis and Mantel test, M2 = 0.2731, Permutations = 9999, P = 0.0001 and r = 0.81) (Fig. S5a). A network analysis evaluated the co-occurrence of bacteria and protozoa. Positive and negative correlations were found in both mask types, with more co-occurrences associated with surgical (nodes: 108, edges: 695) than activated carbon masks (nodes: 87, edges: 152) (Spearman’s ρ > 0.8, P < 0.001) (Fig. S5b, c).
Using flow cytometry, a total of 30 bacterial endosymbionts in protists were isolated from water, carbon and surgical mask samples. Bacteria identified included Escherichia, Aeromonas and Klebsiella. Different bacterial strains from each sample type were selected for antibiotic susceptibility testing. Three strains, Aeromonas caviae (carbon and surgical masks, #4 and #22) and Aeromonas sp. strain PI-20 (water samples, #18), were considered resistant to tetracyclines (Fig. S6).
3.3. Drivers of microbial community assembly associated with masks
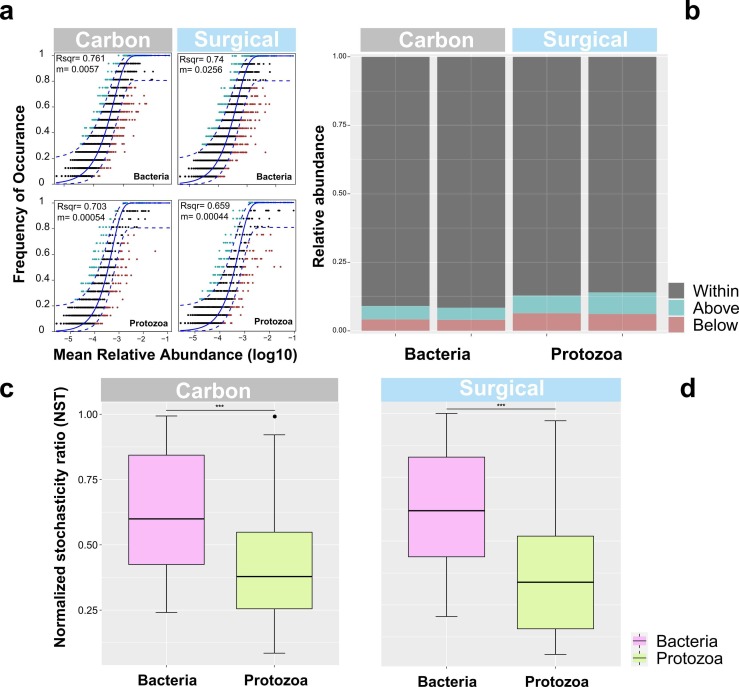
Both bacterial and protistan communities of mask samples were fitted to a neutral community model (NCM). Bacterial communities had a better goodness fit than protistan communities (0.761 VS 0.703 and 0.740 VS 0.659 in carbon and surgical masks, respectively). The migration rate (m) of bacterial communities was greater than protistan (0.0057 VS 0.00054 and 0.0256 VS 0.00044 in carbon and surgical masks, respectively), indicating that the bacteria were less affected by dispersal processes ( Fig. 3a). The majority of microbial ASVs (~ 90% of bacterial taxa and ~ 86% of protistan taxa) in both masks were predicted by NCM (Fig. 3b). In addition, a normalized stochasticity ratio (NST) was performed to quantify the role of deterministic and stochastic processes between bacterial and protistan communities (Fig. 3c and d). The NST value was above a 50% threshold for bacterial communities in carbon masks with an average of 61.8%, indicating that the stochastic processes dominated the assembly of bacterial communities. While deterministic process dominated in assembling protistan communities with an average of 42.3%. A similar result for bacteria and protists was also observed in surgical mask samples. In both types of masks, the NST values of the bacterial communities were significantly higher than those of the protozoa (P < 0.001).
Fig. 3.
Fit of neutral community model (NCM) of bacterial and protistan communities for mask samples (a). The solid black and dashed blue lines indicate the best fit to the model and the 95% confidence intervals, respectively. The R square and m indicate the goodness of model prediction and estimated migration rate, respectively. The bar charts (b) depict the relative abundance of the over-represented (blue dots), best fit (black dots) and under-represented (red dots) ASV around the model prediction. The comparison of normalized stochasticity ratio (NST) between bacterial and protistan communities (*** P < 0.001) (c) and (d).
3.4. Contribution of microbial communities to ARGs
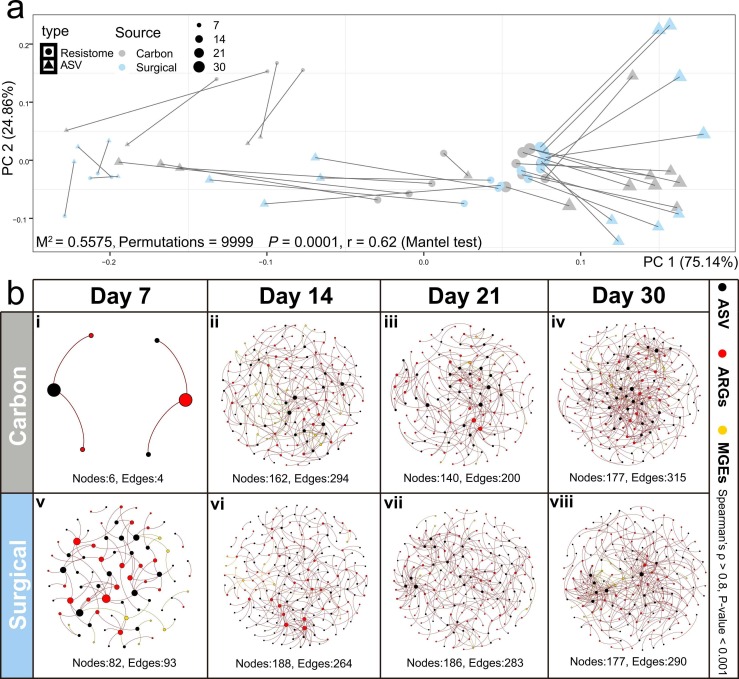
The antibiotic resistome was significantly associated with bacterial ASVs for both mask types (Procrustes analysis and Mantel test, M2 = 0.5575, Permutations = 9999, P = 0.0001 and r = 0.62) ( Fig. 4a). After FDR adjustment for P values (Spearman’s ρ > 0.8, P < 0.001), network analysis highlighted the dynamic co-occurrence patterns among ARGs, MGEs and bacterial ASVs in both mask types during the experimental period (Fig. 4b). Both mask types had fewest nodes and edges at day 7 (P < 0.05) (Fig. 4b i and v), with more complex relationships from day 14 onwards.
Fig. 4.
Procrustes analysis and Mantel test between antibiotic resistome and bacterial ASV (M2 = 0.5575, Permutations = 9999, P = 0.0001 and r = 0.62) (a). Triangle and dots represent ASV and antibiotic resistome, respectively. Size of the dot indicates the different sampling time point. Co-occurrence patterns among bacterial ASV, ARGs and MGEs with time (Spearman’s ρ > 0.8, P < 0.001). Black, red and yellow dots represent bacterial ASV, ARGs, and MGEs respectively (b).
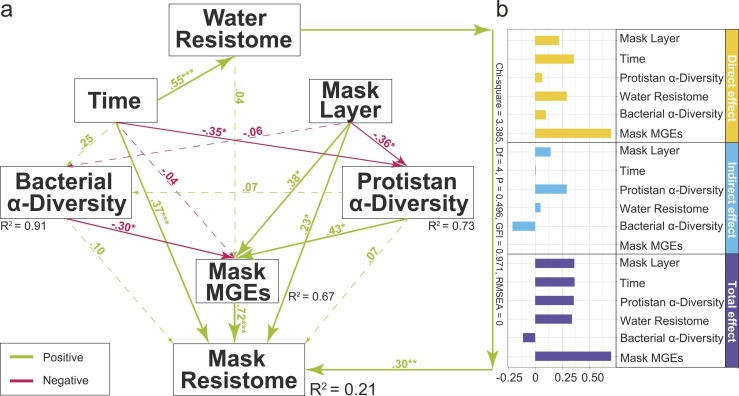
Structural equation modelling showed the direct and indirect effects of sampling time, water resistome, number of mask layers, bacterial α-diversity, protist α-diversity and mask MGEs on composition of the mask resistome ( Fig. 5). The above factors explained 21% of the mask resistome. Individually, time (λ = 0.37, P < 0.001), mask layer (λ = 0.23, P < 0.05), mask MGEs (λ = 0.72, P < 0.001) and water resistome (λ = 0.30, P < 0.01) directly and positively impacted the mask resistome.
Fig. 5.
(a) and (b) Structural equation modelling indicating the direct and indirect impact of time, water resistome, mask MGEs, mask layer bacterial and protist α-diversity on the composition of antibiotic resistome in mask samples (Chi-square = 3.385, P = 0.496, df = 4, GFI = 0.971 and RMSEA = 0). Red and Green arrows indicate the positive and negative correlation between factors. Path coefficients are noted for each arrow. Solid and dotted lines demonstrate the significant effects on the different targets (* P ≤ 0.05,** ≤ 0.01,*** P ≤ 0.001). The variance explained by the different factors on the target is indicated by R2.
4. Discussion
4.1. Discarded masks are an artificial microbial refuge in marine ecosystem
Due to the multi-layer design, masks can prevent harmful microorganisms from being inhaled (Zorko et al., 2020). However, the penetrable surface of the masks is also susceptible to microbial colonization in aquatic environments and can function as a so-called “microbial reef”. According to SEM, each layer of both mask types was colonized by microorganisms during the first 3 days, while after 30 days the microbial biomass increased significantly (Fig. S2). The bacterial Shannon index of both mask types were significantly higher than water samples, which further suggested that discarded masks in marine systems are likely microbial shelters and provide a stable environment for microbial survival and propagation.
Both deterministic and stochastic processes were important for forming the microbial communities in both mask types. We applied a neutral community model to understand the assembly process of microbial communities in discarded masks. Although the distribution of most of the bacterial and protistan ASVs could be predicted, bacteria had a better goodness fit than protozoa, indicating that bacterial communities may be formed mainly by passive dispersal or ecological drift (Adair et al., 2018). The remaining unpredicted taxa suggested that masks may act as environmental filters affecting the microbial community. A stronger influence of stochastic processes on bacterial assembly was suggested by NCM, which was further supported by NST analysis. This may be due to the differences in body size (bacteria vs protozoa), resulting in protozoa being more vulnerable to environmental filtration (Liu et al., 2015). Thus, organism size may play a role in ecological determinism (De Bie et al., 2012).
Both bacterial and protistan mask communities were significantly different from water samples (Fig. S4), suggesting that masks provide a distinct niche to harbor microorganisms from the surrounding aquatic environment (De Tender et al., 2017). The composition of mask bacterial and protistan communities changed with time, suggesting that selective pressure occurs during microbial colonization. For instance, mask layers may be an obstacle that hinders propagation of protozoa. Unlike some bacteria (e.g., Xanthomonadaceae and Comamonadaceae) that can bind tightly to the mask surface through the fimbriae or pili (Yang et al., 2020), protozoa have a wider range of activity and require constant feeding (Behnke et al., 2010). The protistan communities may also be affected by the availability, abundance, and quality of bacteria (Hirakata et al., 2020; Sherr and Sherr, 2002). Both positive and negative complex relationships were identified by network analysis (Fig. S5), suggesting that potential microbial food webs occur in these novel niches (Sherr and Sherr, 2002).
4.2. The persistence of ARGs in discarded mask
The current study provided a comprehensive pattern of ARGs in discarded carbon and surgical masks exposed to the marine environment.
A total of 156 and 183 resistant genes were detected in carbon and surgical masks respectively, suggesting that masks enrich the antibiotic resistome. In particular, the relative abundance, absolute abundance, and detected number of ARGs increased at day 7 and stabilized after day 14, highlighting that masks can accumulate diverse antibiotic resistomes derived from marine systems (Yang et al., 2019), but after reaching saturation they stabilize. Aquatic environments are unequivocally in this study the main source of resistome in masks (Fig. 2b), however, the distinct pattern of resistome profiles between water and masks samples suggested that ARGs and MGEs were selectively enriched in mask samples. Although the mask resistome profile changed from day 7 to day 30, the patterns tended to be stable over time (Fig. 1c, d and e). These results may be attributed to the colonization of microorganisms and their dynamic profiles.
Resistome members aadA-02, mexF, intl-1(clinical) and intl-1LC represent Aminoglycoside, multidrug, MLSB and MGEs and were persistent in both carbon and surgical masks throughout the experimental period. In particular, the persistence of MGEs in masks may result in mediating ARGs dissemination through HGT (Gaze et al., 2011). In addition, the multidrug resistance gene, mexF, has been reported to code for a cytoplasmic membrane efflux pump (Kohler et al., 1997). Both relative and absolute abundance of mexF were significantly increased in both mask samples, suggesting a possible increased number of multidrug resistant bacteria with time.
4.3. Biotic and abiotic factors influencing ARGs in mask
The composition of ARGs and bacterial communities shared the same trend with time, suggesting that ARGs profiles may be closely associated with changes in bacterial communities. The ARG profile remained relatively stable once bacterial colonization reached saturation. A network analysis supported the correlation between ARGs and bacteria which was relatively stable after day 14. A likely explanation is the detachment of the biofilm that limits microbial accumulation (VanLoosdrecht et al., 1997) with a proportion of cells escaping from biofilms due to fluid frictional forces (Petrova and Sauer, 2016). As we suspended the mask in water during the experiment, it may have been impacted by the water flow, causing some of the biofilm that was not tightly attached to the surface of the masks to detach.
A key finding of this study was the isolation of bacterial endosymbionts from protozoa in both water and mask samples. These were subjected to antibiotic sensitivity tests and a total of three strains were found to be resistant to tetracycline. The three resistant strains were identified as belonging to the genus Aeromonas (Seshadri et al., 2006), known pathogens of fish (Wang et al., 1996). Structural equation model suggested that protozoa directly and indirectly effected MGEs and ARGs, respectively (Fig. 5). This links ARGs, bacteria and protozoa, suggesting that protozoa may provide shelter for the persistence of antibiotic resistant bacteria or ARG, or even carry out HGT. For example, previous studies have reported that endosymbionts utilize their hosts as a survival strategy to mitigate impacts from harsh environments (Adekambi et al., 2006, El-Etr et al., 2009).
Incubation time, the number of mask layers and the water resistome positively, and significantly directly affected the mask resistome, suggesting that ARGs enrichment in masks in marine systems is multicausal. As the COVID-19 pandemic continues, the demand for facial masks will remain high (Verbeek et al., 2020). Thus, contamination from discarded masks may be long-term and thus monitoring of ARGs contamination associated with masks is necessary to inform future risk assessment.
5. Conclusions
A total of 216 ARGs and 11 mobile genetic elements were detected from two common types of facial masks used to mitigate Covid-19 infection, suggesting that discarded masks may provide a potential refuge for the enrichment of antibiotic resistome in marine systems. Resistome abundance reached saturation after 14 days incubation. Masks harbored microbial communities, including antibiotic resistant pathogens. As discarded masks persist in the environment an ecological risk assessment is required to better understand their long-term impact on marine ecosystems.
CRediT authorship contribution statement
Shu-Yi-Dan Zhou: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. Chenshuo Lin: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. Kai Yang: Conceptualization, Methodology, Writing – review & editing. Le-Yang Yang: Investigation, Writing – review & editing. Xiao-Ru Yang: Funding acquisition, Writing – review & editing. Fu-Yi Huang: Conceptualization, Funding acquisition, Resources, Project administration, Writing – review & editing. Roy Neilson: Funding acquisition, Writing – review & editing. Jian-Qiang Su: Resources, Methodology, Writing – review & editing. Yong-Guan Zhu: Conceptualization, Funding acquisition, Resources, Project administration, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (41807460, 41977210, 42021005 and U1805244). The James Hutton Institute receives financial support from Scottish Government Rural and Environment Science and Analytical Services (RESAS).
Editor: Dr. G. Jianhua
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jhazmat.2021.127774.
Appendix A. Supplementary material
Supplementary material.
.
References
- Abbasi S.A., Khalil A.B., Arslan M. Extensive use of face masks during COVID-19 pandemic: (micro-)plastic pollution and potential health concerns in the Arabian Peninsula. Saudi J. Biol. Sci. 2020;27:3181–3186. doi: 10.1016/j.sjbs.2020.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair K.L., Wilson M., Bost A., Douglas A.E. Microbial community assembly in wild populations of the fruit fly Drosophila melanogaster. Isme J. 2018;12:959–972. doi: 10.1038/s41396-017-0020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adekambi T., Ben Salah S., Khlif M., Raoult D., Drancourt M. Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl. Environ. Microbiol. 2006;72:5974–5981. doi: 10.1128/AEM.03075-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbumani S., Kakkar P. Ecotoxicological effects of microplastics on biota: a review. Environ. Sci. Pollut. Res. 2018;25:14373–14396. doi: 10.1007/s11356-018-1999-x. [DOI] [PubMed] [Google Scholar]
- Arias-Andres M., Klumper U., Rojas-Jimenez K., Grossart H.P. Microplastic pollution increases gene exchange in aquatic ecosystems. Environ. Pollut. 2018;237:253–261. doi: 10.1016/j.envpol.2018.02.058. [DOI] [PubMed] [Google Scholar]
- Baker G.C., Smith J.J., Cowan D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Baquero F., Martinez J.L., Canton R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008;19:260–265. doi: 10.1016/j.copbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Behnke A., Barger K.J., Bunge J., Stoeck T. Spatio-temporal variations in protistan communities along an O-2/H2S gradient in the anoxic Framvaren Fjord (Norway) FEMS Microbiol. Ecol. 2010;72:89–102. doi: 10.1111/j.1574-6941.2010.00836.x. [DOI] [PubMed] [Google Scholar]
- Benson N.U., Bassey D.E., Palanisami T. COVID pollution: impact of COVID-19 pandemic on global plastic waste footprint. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., Bai Y., Bisanz J.E., Bittinger K., Brejnrod A., Brislawn C.J., Brown C.T., Callahan B.J., Caraballo-Rodriguez A.M., Chase J., Cope E.K., Da Silva R., Diener C., Dorrestein P.C., Douglas G.M., Durall D.M., Duvallet C., Edwardson C.F., Ernst M., Estaki M., Fouquier J., Gauglitz J.M., Gibbons S.M., Gibson D.L., Gonzalez A., Gorlick K., Guo J.R., Hillmann B., Holmes S., Holste H., Huttenhower C., Huttley G.A., Janssen S., Jarmusch A.K., Jiang L.J., Kaehler B.D., Bin Kang K., Keefe C.R., Keim P., Kelley S.T., Knights D., Koester I., Kosciolek T., Kreps J., Langille M.G.I., Lee J., Ley R., Liu Y.X., Loftfield E., Lozupone C., Maher M., Marotz C., Martin B.D., McDonald D., McIver L.J., Melnik A.V., Metcalf J.L., Morgan S.C., Morton J.T., Naimey A.T., Navas-Molina J.A., Nothias L.F., Orchanian S.B., Pearson T., Peoples S.L., Petras D., Preuss M.L., Pruesse E., Rasmussen L.B., Rivers A., Robeson M.S., Rosenthal P., Segata N., Shaffer M., Shiffer A., Sinha R., Song S.J., Spear J.R., Swafford A.D., Thompson L.R., Torres P.J., Trinh P., Tripathi A., Turnbaugh P.J., Ul-Hasan S., vander Hooft J.J.J., Vargas F., Vazquez-Baeza Y., Vogtmann E., von Hippel M., Walters W., Wan Y.H., Wang M.X., Warren J., Weber K.C., Williamson C.H.D., Willis A.D., Xu Z.Z., Zaneveld J.R., Zhang Y.L., Zhu Q.Y., Knight R., Caporaso J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns A.R., Stephens W.Z., Stagaman K., Wong S., Rawls J.F., Guillemin K., Bohannan B.J.M. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. Isme J. 2016;10:655–664. doi: 10.1038/ismej.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Yuan S.F., Kok K.H., To K.K.W., Chu H., Yang J., Xing F.F., Liu J.L., Yip C.C.Y., Poon R.W.S., Tsoi H.W., Lo S.K.F., Chan K.H., Poon V.K.M., Chan W.M., Ip J.D., Cai J.P., Cheng V.C.C., Chen H.L., Hui C.K.M., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.L., Li H., Zhou X.Y., Zhao Y., Su J.Q., Zhang X., Huang F.Y. An underappreciated hotspot of antibiotic resistance: the groundwater near the municipal solid waste landfill. Sci. Total Environ. 2017;609:966–973. doi: 10.1016/j.scitotenv.2017.07.164. [DOI] [PubMed] [Google Scholar]
- Chen X., Chen X., Liu Q., Zhao Q., Xiong X., Wu C. Used disposable face masks are significant sources of microplastics to environment. Environ. Pollut. 2021;285 doi: 10.1016/j.envpol.2021.117485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie T., De Meester L., Brendonck L., Martens K., Goddeeris B., Ercken D., Hampel H., Denys L., Vanhecke L., Van der Gucht K., Van Wichelen J., Vyverman W., Declerck S.A.J. Body size and dispersal mode as key traits determining metacommunity structure of aquatic organisms. Ecol. Lett. 2012;15:740–747. doi: 10.1111/j.1461-0248.2012.01794.x. [DOI] [PubMed] [Google Scholar]
- De Tender C., Devriese L.I., Haegeman A., Maes S., Vangeyte J., Cattrijsse A., Dawyndt P., Ruttink T. Temporal dynamics of bacterial and fungal colonization on plastic debris in the North Sea. Environ. Sci. Technol. 2017;51:7350–7360. doi: 10.1021/acs.est.7b00697. [DOI] [PubMed] [Google Scholar]
- Dharmaraj S., Ashokkumar V., Hariharan S., Manibharathi A., Show P.L., Chong C.T., Ngamcharussrivichai C. The COVID-19 pandemic face mask waste: a blooming threat to the marine environment. Chemosphere. 2021;272 doi: 10.1016/j.chemosphere.2021.129601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz L., Horve P.F., Coil D.A., Fretz M., Eisen J.A., Van Den Wymelenberg K. 2019 Novel coronavirus (COVID-19) pandemic: built environment considerations to reduce transmission. Msystems. 2020;5:15. doi: 10.1128/mSystems.00375-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. [Google Scholar]
- Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- El-Etr S.H., Margolis J.J., Monack D., Robison R.A., Cohen M., Moore E., Rasley A. Francisella tularensis Type A strains cause the rapid encystment of acanthamoeba castellanii and survive in amoebal cysts for three weeks postinfection. Appl. Environ. Microbiol. 2009;75:7488–7500. doi: 10.1128/AEM.01829-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksen M., Lebreton L.C.M., Carson H.S., Thiel M., Moore C.J., Borerro J.C., Galgani F., Ryan P.G., Reisser J. Plastic pollution in the world’s oceans: more than 5 trillion plastic pieces weighing over 250,000 Tons afloat at sea. PLoS One. 2014;9:15. doi: 10.1371/journal.pone.0111913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadare O.O., Okoffo E.D. Covid-19 face masks: a potential source of microplastic fibers in the environment. Sci. Total Environ. 2020;737:4. doi: 10.1016/j.scitotenv.2020.140279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Shen C., Xia N., Song W., Fan M.Z., Cowling B.J. Rational use of face masks in the COVID-19 pandemic. Lancet Respir. Med. 2020;8:434–436. doi: 10.1016/S2213-2600(20)30134-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaze W.H., Zhang L.H., Abdouslam N.A., Hawkey P.M., Calvo-Bado L., Royle J., Brown H., Davis S., Kay P., Boxall A.B.A., Wellington E.M.H. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. Isme J. 2011;5:1253–1261. doi: 10.1038/ismej.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D.W., Lipp E.K., McLaughlin M.R., Rose J.B. Marine recreation and public health microbiology: quest for the ideal indicator. Bioscience. 2001;51:817–825. [Google Scholar]
- Haque M.S., Uddin S., Sayem S.M., Mohib K.M. Coronavirus disease 2019 (COVID-19) induced waste scenario: a short overview. J. Environ. Chem. Eng. 2021;9:14. doi: 10.1016/j.jece.2020.104660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakata Y., Hatamoto M., Oshiki M., Watari T., Araki N., Yamaguchi T. Food selectivity of anaerobic protists and direct evidence for methane production using carbon from prey bacteria by endosymbiotic methanogen. Isme J. 2020;14:1873–1885. doi: 10.1038/s41396-020-0660-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton A.A., Walton A., Spurgeon D.J., Lahive E., Svendsen C. Microplastics in freshwater and terrestrial environments: evaluating the current understanding to identify the knowledge gaps and future research priorities. Sci. Total Environ. 2017;586:127–141. doi: 10.1016/j.scitotenv.2017.01.190. [DOI] [PubMed] [Google Scholar]
- Huang C.L., Wang Y.M., Li X.W., Ren L.L., Zhao J.P., Hu Y., Zhang L., Fan G.H., Xu J.Y., Gu X.Y., Cheng Z.S., Yu T., Xia J.A., Wei Y., Wu W.J., Xie X.L., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J.G., Wang G.F., Jiang R.M., Gao Z.C., Jin Q., Wang J.W., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imran M., Das K.R., Naik M.M. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Chemosphere. 2019;215:846–857. doi: 10.1016/j.chemosphere.2018.10.114. [DOI] [PubMed] [Google Scholar]
- Kalina M., Tilley E. “This is our next problem”: cleaning up from the COVID-19 response. Waste Manag. 2020;108:202–205. doi: 10.1016/j.wasman.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T., MicheaHamzehpour M., Henze U., Gotoh N., Curty L.K., Pechere J.C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- Kotlarska E., Luczkiewicz A., Pisowacka M., Burzynski A. Antibiotic resistance and prevalence of class 1 and 2 integrons in Escherichia coli isolated from two wastewater treatment plants, and their receiving waters (Gulf of Gdansk, Baltic Sea, Poland) Environ. Sci. Pollut. Res. 2015;22:2018–2030. doi: 10.1007/s11356-014-3474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutralam-Muniasamy G., Perez-Guevara F., Elizalde-Martinez I., Shruti V.C. Review of current trends, advances and analytical challenges for microplastics contamination in Latin America. Environ. Pollut. 2020;267:15. doi: 10.1016/j.envpol.2020.115463. [DOI] [PubMed] [Google Scholar]
- Li Q., Guan X.H., Wu P., Wang X.Y., Zhou L., Tong Y.Q., Ren R.Q., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X.S., Xiang N.J., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W.X., Chen C.D., Jin L.M., Yang R., Wang Q., Zhou S.H., Wang R., Liu H., Luo Y.B., Liu Y., Shao G., Li H., Tao Z.F., Yang Y., Deng Z.Q., Liu B.X., Ma Z.T., Zhang Y.P., Shi G.Q., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z.J. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. New Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li Y., Ruan Z., Fu G., Chen X., Sadiq R., Deng Y. A new method to construct co-author networks. Phys. A: Stat. Mech.Appl. 2015;419:29–39. [Google Scholar]
- Ning D., Deng Y., Tiedje J.M., Zhou J. A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. USA. 2019;116:16892–16898. doi: 10.1073/pnas.1904623116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricio Silva A.L., Prata J.C., Walker T.R., Duarte A.C., Ouyang W., Barcelo D., Rocha-Santos T. Increased plastic pollution due to COVID-19 pandemic: challenges and recommendations. Chem. Eng. J. (Lausanne, Switz.: 1996) 2021;405 doi: 10.1016/j.cej.2020.126683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova O.E., Sauer K. Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr. Opin. Microbiol. 2016;30:67–78. doi: 10.1016/j.mib.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prata J.C., Silva A.L.P., Walker T.R., Duarte A.C., Rocha-Santos T. COVID-19 pandemic repercussions on the use and management of plastics. Environ. Sci. Technol. 2020;54:7760–7765. doi: 10.1021/acs.est.0c02178. [DOI] [PubMed] [Google Scholar]
- Rønn R., Hao X., Lüthje F., German N., Li X., Huang F., Kisaka J., Huffman D., Alwathnani H., Zhu Y.-G., Rensing C. Bacterial survival in dictyostelium. Bio-Protoc. 2017:7. doi: 10.21769/BioProtoc.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J.M., Caron D.A., Sieracki M.E., Poulton N. Counting heterotrophic nanoplanktonic protists in cultures and aquatic communities by flow cytometry. Aquat. Microb. Ecol. 2004;34:263–277. [Google Scholar]
- Schermelleh-Engel K., Moosbrugger H., Müller H. Evaluating the fit of structural equation models: tests of significance and descriptive goodness-of-fit measures. MPR-Online. 2003;8:23–74. [Google Scholar]
- Seshadri R., Joseph S.W., Chopra A.K., Sha J., Shaw J., Graf J., Haft D., Wu M., Ren Q.H., Rosovitz M.J., Madupu R., Tallon L., Kim M., Jin S.H., Vuong H., Stine O.C., Ali A., Horneman A.J., Heidelberg J.F. Genome sequence of Aeromonas hydrophila ATCC 7966(T): jack of all trades. J. Bacteriol. 2006;188:8272–8282. doi: 10.1128/JB.00621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr E.B., Sherr B.F. Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2002;81:293–308. doi: 10.1023/a:1020591307260. [DOI] [PubMed] [Google Scholar]
- Shi P., Jia S.Y., Zhang X.X., Zhang T., Cheng S.P., Li A.M. Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water. Water Res. 2013;47:111–120. doi: 10.1016/j.watres.2012.09.046. [DOI] [PubMed] [Google Scholar]
- Sintes E., del Giorgio P.A. Community heterogeneity and single-cell digestive activity of estuarine heterotrophic nanoflagellates assessed using lysotracker and flow cytometry. Environ. Microbiol. 2010;12:1913–1925. doi: 10.1111/j.1462-2920.2010.02196.x. [DOI] [PubMed] [Google Scholar]
- Sloan W.T., Lunn M., Woodcock S., Head I.M., Nee S., Curtis T.P. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- Stoeck T., Bass D., Nebel M., Christen R., Richards T.A. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 2010;19(Suppl. 1):S21–S31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- Su J.Q., An X.L., Li B., Chen Q.L., Gillings M.R., Chen H., Zhang T., Zhu Y.G. Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China (vol 5, 84, 2017) Microbiome. 2018;(6):1. doi: 10.1186/s40168-017-0298-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J.-Q., Wei B., Ou-Yang W.-Y., Huang F.-Y., Zhao Y., Xu H.-J., Zhu Y.-G. Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ. Sci. Technol. 2015;49:7356–7363. doi: 10.1021/acs.est.5b01012. [DOI] [PubMed] [Google Scholar]
- Su Y.L., Zhang Z.J., Zhu J.D., Shi J.H., Wei H.W., Xie B., Shi H.H. Microplastics act as vectors for antibiotic resistance genes in landfill leachate: the enhanced roles of the long-term aging process. Environ. Pollut. 2021;270:12. doi: 10.1016/j.envpol.2020.116278. [DOI] [PubMed] [Google Scholar]
- Turner S., Pryer K.M., Miao V.P.W., Palmer J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small submit rRNA sequence analysis. J. Eukaryot. Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- VanLoosdrecht M.C.M., Picioreanu C., Heijnen J.J. A more unifying hypothesis for biofilm structures. FEMS Microbiol. Ecol. 1997;24:181–183. [Google Scholar]
- Verbeek J.H., Rajamaki B., Ijaz S., Sauni R., Toomey E., Blackwood B., Tikka C., Ruotsalainen J.H., Balci F.S.K. Personal protective equipment for preventing highly infectious diseases due to exposure to contaminated body fluids in healthcare staff. Cochrane Database Syst. Rev. 2020:148. doi: 10.1002/14651858.CD011621.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Tyler K.D., Munro C.K., Johnson W.M. Characterization of cytotoxic, hemolytic Aeromonas caviae clinical isolates and their identification by determining presence of a unique hemolysin gene. J. Clin. Microbiol. 1996;34:3203–3205. doi: 10.1128/jcm.34.12.3203-3205.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H., Chang, W., Henry, L., Pedersen, L., Thomas., Takahashi, K., Wilke, C., Woo, K., Yutani, H., Dunnington, D., 2020. ggplot2: Create elegant data visualisations using the grammar of graphics, 3.3.1 ed, pp. A system for 'declaratively' creating graphics, based on "The Grammar of Graphics". You provide the data, tell 'ggplot2' how to map variables to aesthetics, what graphical primitives to use, and it takes care of the details.
- Yadav M.L., Roychoudhury B. Handling missing values: a study of popular imputation packages in R. Knowl. Based Syst. 2018;160:104–118. [Google Scholar]
- Yang K., Chen Q.-L., Chen M.-L., Li H.-Z., Liao H., Pu Q., Zhu Y.-G., Cui L. Temporal dynamics of antibiotic resistome in the plastisphere during microbial colonization. Environ. Sci. Technol. 2020;54:11322–11332. doi: 10.1021/acs.est.0c04292. [DOI] [PubMed] [Google Scholar]
- Yang Y.Y., Liu G.H., Song W.J., Ye C., Lin H., Li Z., Liu W.Z. Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes. Environ. Int. 2019;123:79–86. doi: 10.1016/j.envint.2018.11.061. [DOI] [PubMed] [Google Scholar]
- Zettler E.R., Mincer T.J., Amaral-Zettler L.A. Life in the “plastisphere”: microbial communities on plastic marine debris. Environ. Sci. Technol. 2013;47:7137–7146. doi: 10.1021/es401288x. [DOI] [PubMed] [Google Scholar]
- Zheng D., Yin G., Liu M., Chen C., Jiang Y., Hou L., Zheng Y. A systematic review of antibiotics and antibiotic resistance genes in estuarine and coastal environments. Sci. Total Environ. 2021;777 doi: 10.1016/j.scitotenv.2021.146009. [DOI] [PubMed] [Google Scholar]
- Zhou G.W., Yang X.R., Su J.Q., Zheng B.X., Zhu Y.G. Bacillus ferrooxidans sp nov., an iron(II)-oxidizing bacterium isolated from paddy soil. J. Microbiol. 2018;56:472–477. doi: 10.1007/s12275-018-7543-3. [DOI] [PubMed] [Google Scholar]
- Zhou S.-Y.-D., Zhang Q., Neilson R., Giles M., Li H., Yang X.-R., Su J.-Q., Zhu Y.-G. Vertical distribution of antibiotic resistance genes in an urban green facade. Environ. Int. 2021;152 doi: 10.1016/j.envint.2021.106502. [DOI] [PubMed] [Google Scholar]
- Zhu Y.-G., Johnson T.A., Su J.-Q., Qiao M., Guo G.-X., Stedtfeld R.D., Hashsham S.A., Tiedje J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA. 2013;110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorko D.J., Gertsman S., O’Hearn K., Timmerman N., Ambu-Ali N., Dinh T., Sampson M., Sikora L., McNally J.D., Choong K. Decontamination interventions for the reuse of surgical mask personal protective equipment: a systematic review. J. Hosp. Infect. 2020;106:283–294. doi: 10.1016/j.jhin.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.