Abstract
Sweet potato (Ipomoea batatas) is one of the most important crops in the world, and its production rate is mainly decreased by the sweet potato virus disease (SPVD) caused by the co‐infection of sweet potato chlorotic stunt virus (SPCSV) and sweet potato feathery mottle virus. However, methods for improving SPVD resistance have not been established. Thus, this study aimed to enhance SPVD resistance by targeting one of its important pathogenesis‐related factors (i.e., SPCSV‐RNase3) by using the CRISPR‐Cas13 technique. First, the RNA targeting activity of four CRISPR‐Cas13 variants were compared using a transient expression system in Nicotiana benthamiana. LwaCas13a and RfxCas13d had more efficient RNA and RNA virus targeting activity than PspCas13b and LshCas13a. Driven by the pCmYLCV promoter for the expression of gRNAs, RfxCas13d exhibited higher RNA targeting activity than that driven by the pAtU6 promoter. Furthermore, the targeting of SPCSV‐RNase3 using the LwaCas13a system inhibited its RNA silencing suppressor activity and recovered the RNA silencing activity in N. benthamiana leaf cells. Compared with the wild type, transgenic N. benthamiana plants carrying an RNase3‐targeted LwaCas13a system exhibited enhanced resistance against turnip mosaic virus TuMV‐GFP and cucumber mosaic virus CMV‐RNase3 co‐infection. Moreover, transgenic sweet potato plants carrying an RNase3‐targeted RfxCas13d system exhibited substantially improved SPVD resistance. This method may contribute to the development of SPVD immune germplasm and the enhancement of sweet potato production in SPVD‐prevalent regions.
Keywords: CRISPR‐Cas13, genetic improvement of SPVD resistance, LwaCas13a, SPCSV‐RNase3, RfxCas13d
Targeting of SPCSV‐RNase3 using CRISPR‐Cas13 (RfxCas13d) improves resistance against the sweet potato virus disease (SPVD) in transgenic sweet potato plants.

1. INTRODUCTION
Sweet potato (Ipomoea batatas) is rich in vitamins, minerals, and dietary fibre, and is one of the most important food crops, ranking seventh in terms of worldwide staple food production (Bovell‐Benjamin, 2007, 2010). According to the Food and Agriculture Organization of the United Nations (FAO) report, the global production of sweet potatoes was 92 million tonnes in 2018, half of which was produced by China (53 million tonnes) (FAOSTAT, 2018; http://www.fao.org/faostat/en/). In sub‐Saharan African countries, such as Nigeria, Tanzania, Ethiopia, and Uganda, orange‐fleshed sweet potato varieties that contain abundant β‐carotene are important food sources of provitamin A that can be used to combat vitamin A deficiency in local residents and thus are cultivated in large land areas (Bednarek et al., 2021; Jan et al., 2017). However, the average sweet potato production in these countries is low, at approximately a third of that in China (Bednarek et al., 2021; Jan et al., 2017).
As a vegetatively propagated root crop, sweet potato yield is largely reduced due to the continuous accumulation of viral diseases over generations (Loebenstein et al., 2015). The most devastating viral disease in sweet potatoes is known as sweet potato virus disease (SPVD), caused by the synergistic co‐infection by sweet potato chlorotic stunt virus (SPCSV; genus Crinivirus, family Closteroviridae) and sweet potato feathery mottle virus (SPFMV; genus Potyvirus, family Potyviridae) (Bednarek et al., 2021; Gibson et al., 2004; Karyeija et al., 2000; Loebenstein et al., 2015; Wang et al., 2019). SPVD can cause yield losses of 80%–90% in infected sweet potato plants (Bednarek et al., 2021; Loebenstein et al., 2015). However, the infection of neither SPCSV nor SPFMV alone produces disease‐related symptoms and yield loss (Bednarek et al., 2021; Loebenstein et al., 2015). Once SPCSV and SPFMV co‐infection occurs, the SPFMV titres increase by several hundred‐ to thousand‐fold in plants, leading to severe symptoms, including vein clearing, stunting, leaf malformation, and dramatic yield loss (Kreuze et al., 2005). The dsRNA‐specific class 1 RNase III endoribonuclease (RNase3) encoded by the SPCSV genome is an important pathogenesis‐related factor in SPVD (Cuellar et al., 2009). SPCSV‐RNase3 is a unique viral suppressor eliminating the basal antiviral defence based on posttranscriptional RNA interference (RNAi) in an endoribonuclease activity‐dependent manner (Cuellar et al., 2009; Weinheimer et al., 2014, 2015, 2016). In this process, 21–22‐nucelotide (nt) viral siRNA duplexes produced by Dicer‐like dsRNA‐specific endoribonucleases in plants are cleaved by SPCSV‐RNase3 and form short 14‐nt vsiRNA that cannot induce the host RNAi defence response (Cuellar et al., 2009). Therefore, previous attempts to improve SPVD resistance in sweet potatoes through RNAi‐based methods are ineffective (Kreuze et al., 2008; Sivparsad & Gubba, 2014). To date, no efficient method has been developed for the genetic improvement of SPVD resistance in sweet potatoes.
Clustered regularly interspaced short palindromic repeat (CRISPR)‐associated protein (CRISPR‐Cas) is an adaptive immunity system against phage invaders in prokaryotes (Jinek et al., 2012). According to the genomic architecture of the CRISPR array and the signature interference effector, these systems can be classified into two classes and several types or subtypes (Chen et al., 2019; Komor et al., 2017; Zhu et al., 2020). Class I requires multiple subunits for nucleic acid interference, and class II only requires one efficient single CRISPR‐Cas effector (Chen et al., 2019; Komor et al., 2017; Zhu et al., 2020). CRISPR systems target double‐stranded DNA, single‐stranded DNA, and single‐stranded RNA (Ali et al., 2018; Cong et al., 2013; Harrington et al., 2018). As RNA‐guided endonuclease systems, CRISPR–Cas9, Cas12a, and Cas12b have become widely used sequence‐specific nucleases in plant genome engineering (Cong et al., 2013; Yan et al., 2019; Zetsche et al., 2015). CRISPR‐Cas9 has also been harnessed as a tool to confer resistance against DNA viruses in plants (Ali et al., 2015; Ji et al., 2018). CRISPR‐Cas13 belongs to class‐II type‐VI CRISPR‐Cas systems and is an RNA‐guided RNA‐targeting immunity system against RNA and/or DNA bacteriophages in prokaryotes (Ali et al., 2018). Since 2016, several variants of Cas13 proteins belonging to different Cas13 subtypes (A–D) have been identified (Abudayyeh et al., 2016, 2017; Cox et al., 2017; Konermann et al., 2018). Different CRISPR‐Cas13 variants (LshCas13a, LwaCas13a, PspCas13b, BzCas13b, and RfxCas13d) have also been harnessed for targeting RNA viruses in various plants, including turnip mosaic virus (TuMV) and tobacco mosaic virus in Nicotiana benthamiana (Aman et al., 2018; Cao et al., 2021; Mahas et al., 2019), southern rice black‐streaked dwarf virus and rice stripe mosaic virus in rice (Hang et al., 2019), and potato virus Y in potatoes (Zhan et al., 2019). These successful examples indicate that CRISPR‐Cas13 may be a better strategy than RNAi to improve SPVD resistance in sweet potatoes.
In this study, the RNA targeting activities of four different Cas13 variants were compared using N. benthamiana as model plants to verify the application of CRISPR‐Cas13 for engineering SPVD resistance in sweet potatoes. LwaCas13a and RfxCas13d were identified as two efficient Cas13 orthologs for RNA virus targeting. Furthermore, targeting SPCSV‐RNase3 using LwaCas13a recovers the capability of RNAi in the host cells and improves the tolerance of N. benthamiana plants against SPCSV‐RNase3‐mediated viral synergism. Lastly, the generated RNase3‐targeted RfxCas13d transgenic sweet potato plants exhibited substantially enhanced SPVD resistance. This study may present a new strategy for the genetic improvement of sweet potatoes against SPVD.
2. RESULTS
2.1. Comparison of RNA targeting activity among different CRISPR‐Cas13 orthologs
To examine the efficiency of different CRISPR‐Cas13 systems, exogenous RNA targeting activity was compared among LshCas13a, LwaCas13a, PspCas13b, and RfxCas13d by using N. benthamiana as the model plant. In our previous work, we found that the SPLCV replicon‐based CRISPR‐Cas13 vector system exhibited higher RNA targeting activity than that of the regular vector in the transient experiments (Yu, Wang, et al., 2020). To maximize the RNA targeting activity of different Cas13 orthologs, we decided to use the SPLCV replicon‐based vector system (AC2m/AC4m) (Yu, Wang, et al., 2020). The construction of CRISPR‐Cas13 targeting vectors followed the principle of our previous work (Yu, Wang, et al., 2020). The nuclear localization signal (NLS) derived from simian virus 40 large T antigen was fused to the open reading frames of four Cas13 proteins. These Cas13 orthologs were then cloned into a SPLCV vector, and their expression was driven by the U4 promoter (Figure 1). Two gRNAs against the mGFP5 transcript were designed under the expression of the AtU6 promoter for each Cas13 protein, and the targeting vectors were recombined (Figure 1). The same vectors containing a nontargeting (NT) spacer were used as the control. mGFP5 interference experiments were performed as previously reported (Yu, Wang, et al., 2020). Although the RNA targeting activity of LwaCas13a has been reported in our previous work (Yu, Wang, et al., 2020), we further conducted a repeated experiment of LwaCas13a at the same time with other Cas13 systems to make the data of LwaCas13a comparable with three other Cas13 orthologs. After 4 days of infiltration, a broad range in the interference levels was observed among the different Cas13 variants (Figure 1). LwaCas13a (70% knockdown) exhibited the highest level of interference against mGFP5 compared with the other Cas13 orthologs (Figure 1). This RNA targeting activity of LwaCas13a was comparable with previous results using the same experimental system (Yu, Wang, et al., 2020).
FIGURE 1.

Comparison of RNA targeting activity of four CRISPR‐Cas13 orthologs using the transient mGFP5 knockdown assays. (a–d) Schematic of the targeting vectors of four Cas13 protein variants and corresponding crRNA structures. The different Cas13 variants are fused with NLS and 3× FLAG or 3× HA tags under the expression of the U4 promoter. gRNA expression was driven by either the AtU6 promoter or the CmYLCV promoter. DR, specific DRs of different Cas13 systems. The right panel represents the designed spacers complementary to the target RNA (mGFP5) sequences. (e–h) GFP monitoring and reverse transcription quantitative PCR analysis of mGFP5 knockdown to evaluate the Cas13‐mediated RNA interference activities in agroinfiltrated Nicotiana benthamiana leaves. Images and leaf samples were collected 4 days after infiltration. NT, vectors with NT spacer. For each Cas13 variant, the knockdown efficiency of each targeting vector is shown relative to the NT vector. Values are shown as mean ± SEM (n = 13–15)
The RNA targeting activity of CRISPR‐Cas13 in plants is believed to correlate with the expression level of gRNAs (Aman et al., 2018; Mahas et al., 2019). This work attempted to enhance RNA targeting activity by replacing the AtU6 promoter with a CmYLCV promoter, which exhibits high efficiency for expressing gRNA in CRISPR‐Cas9 system (Čermák et al., 2017). Our data show that the mGFP5 interference activities of LshCas13a, LwaCas13a, and PspCas13b were slightly enhanced when the AtU6 promoter was replaced with the CmYLCV promoter (Figure 1). The efficiency of mGFP5 knockdown in RfxCas13d reached 85%, which is much higher than that of the AtU6 promoter (approximately 50% knockdown) (Figure 1). Given its lowest RNA interference activity (Figure 1), LshCas13a was not used in the further experiments.
2.2. LwaCas13a and RfxCas13d exhibit efficient targeting activity against RNA viruses
To test the efficiency of different Cas13 systems for targeting a single RNA virus, TuMV targeting experiments were conducted (Aman et al., 2018; Mahas et al., 2019). Ten single gRNAs complementary to the TuMV‐GFP genome (Figure S4a; NIb or GFP) were designed and driven by the AtU6 promoter (LwaCas13a/PspCas13b) or CmYLCV promoter (RfxCas13d). These targeting vectors were then recombined (Figures 2a and S1a). Each of the targeting vectors and TuMV‐GFP infectious clones was codelivered into the leaves of wild‐type N. benthamiana plants via agroinfiltration to assay for the interference activity against the RNA virus of different Cas13 variants. An NT spacer with no complementarity to the TuMV‐GFP genome was used as the control. Our data show that PspCas13b substantially inhibited TuMV‐GFP accumulation in the agroinfiltrated leaves (ILs) and systemic leaves (SLs) with average interference efficiencies of 50% and 52%, respectively (Figure S1b,c). LwaCas13a and RfxCas13d almost completely abolished TuMV accumulation in ILs and SLs (Figure 2b). The average interference efficiencies of viral accumulation (NIb, GFP, and CP) in ILs and SLs reached 90% for LwaCas13a and 89% for RfxCas13d (Figure 2c,d), considerably higher than that of PspCas13b. Given its lower RNA virus interference activity (Figure S1), PspCas13b was not used in the subsequent experiments.
FIGURE 2.

RNA virus interference activity of LwaCas13a and RfxCas13d. (a) Schematic of the targeting vectors of LwaCas13a and RfxCas13d. gRNA expression was driven by the AtU6 promoter for LwaCas13a and CmYLCV promoter for RfxCas13d. DR, specific DRs of LwaCas13a or RfxCas13d. The lower panel represents the designed spacers complementary to the target sequences of TuMV‐GFP genome (NIb and GFP). (b) Cas13‐mediated interference against TuMV‐GFP virus. Nicotiana benthamiana plants were infiltrated with TuMV‐GFP infectious clone alone or combined with CRISPR‐Cas13 targeting vectors. At 6 days postinoculation (dpi), plants were imaged under fluorescent excitation lamp for GFP signal detection. EV, empty vector; NT, vectors with NT spacer; SL, systemic leaves; IL, infiltrated leaves. (c, d) Reverse transcription quantitative PCR analysis of TuMV‐GFP knockdown in the ILs and SLs, respectively. For each CRISPR‐Cas13 targeting vector (including NT), knockdown efficiency is shown relative to the EV. Values are shown as mean ± SEM (n = 6). For all histograms, the significant differences reach the level of p < 0.0001 between the targeting vectors and NT vectors
The designation of multiple targets against the genome of RNA viruses in a CRISPR‐Cas13 system is important for the avoidance of off‐targets (Mehta et al., 2019). To test whether LwaCas13a and RfxCas13d could target multiple RNA viruses simultaneously, another RNA virus was used in further experiments. A fluorescent marker (DsRed) was fused to the N terminal of CMV‐2b to create a CMV‐DsRed infectious clone. After co‐infection with TuMV‐GFP and CMV‐DsRed, the accumulation of the two RNA viruses can be monitored under different fluorescent excitation lamps and filters (Figure S2). We designed pre‐gRNAs that contain four 28‐ (LwaCas13a) or 30‐NT (RfxCas13d) spacers complementary to the TuMV‐NIb, TuMV‐GFP, CMV‐1a, and CMV‐2a and four 36‐ (LwaCas13a) or 30‐NT (RfxCas13d) DRs (LwaCas13a‐gRNA and RfxCas13d‐gRNA constructs) (Figure 3a). RfxCas13d CRISPR arrays harbouring NT sequences were used as the control (RfxCas13d‐NT construct). These constructs were co‐infiltrated with TuMV‐GFP and CMV‐DsRed infectious clones into N. benthamiana plants. After 6 days of infection, RfxCas13d‐NT‐inoculated plants exhibited green and red fluorescence in ILs and SLs, suggesting the successful infection and replication of TuMV and CMV (Figure 3b). However, TuMV and CMV accumulation in ILs and SLs were significantly reduced in LwaCas13a‐gRNA‐ and RfxCas13d‐gRNA‐infiltrated plants (Figure 3b). The average suppression efficiency of TuMV was 95% for LwaCas13a and 91% for RfxCas13d, and that of CMV was 68% for LwaCas13a and 66% for RfxCas13d (Figure 3c). These data suggest that LwaCas13a and RfxCas13d can process their own pre‐gRNAs to form multiple functional gRNAs for the multiplexed targeting of the same virus or for the simultaneous targeting of multiple RNA viruses in plants.
FIGURE 3.

Multiplexed targeting of TuMV‐GFP and CMV‐DsRed viruses through LwaCas13a and RfxCas13d. (a) Schematic of the targeting vectors of LwaCas13a/RfxCas13d and corresponding pre‐gRNA structures. Pre‐gRNA expression was driven by the CmYLCV promoter. DR, specific DRs of LwaCas13a or RfxCas13d. The lower panel represents the designed spacers complementary to the target sequences of TuMV‐GFP (NIb and GFP) and CMV‐DsRed (1a and 2a) genomes. (b) LwaCas13a/RfxCas13d‐mediated simultaneous interference against TuMV‐GFP and CMV‐DsRed viruses. Nicotiana benthamiana plants were infiltrated with TuMV‐GFP and CMV‐DsRed alone or combined with CRISPR‐Cas13 targeting vectors. At 6 days postinoculation (dpi), plants were imaged under appropriate fluorescent excitation lamp for GFP and RFP signal detection. NT, vectors with NT spacer; SL, systemic leaves; IL, infiltrated leaves. (c) Reverse transcription quantitative PCR analysis of TuMV and CMV knockdown in the ILs and SLs. For each CRISPR‐Cas13 targeting vectors, knockdown efficiency is shown relative to the RfxCas13d‐NT vector. Values are shown as mean ± SEM (n = 6). For all histograms, the significant differences reach the level of p < 0.0001 between the targeting vectors and NT vectors
2.3. Targeting SPSCV‐RNase3 via LwaCas13a inhibits its RNA silencing suppressor activity and improves plant resistance against RNase3‐mediated viral synergism
To test whether targeting SPCSV‐RNase3 by CRISPR‐Cas13 could recover the RNA silencing activity in host cells, we conducted the following experiments: SPCSV‐RNase3 was isolated from SPVD‐infected sweet potato plants in Xuzhou, Jiangsu Province, China. Multiple sequence alignment of this RNase3 and its orthologs released in NCBI revealed that it belongs to WA strain and has six amino acid mutations outside the RNase3 signature motif (Figure S3). RNA silencing experiments proved that this isolated SPCSV‐RNase3 exhibited strong RNA silencing suppressor activity (Figure S4b). Two single gRNAs complementary to this RNase3 were also designed under the expression of CmYLCV promoter for LwaCas13a (Figure 4a). An NT spacer was used as the control. GFP silencing experiments were performed using 16C N. benthamiana plants. Six days after infiltration, a high GFP fluorescence accumulation was found in LwaCas13a‐NT, GFP, and RNase3 co‐infiltrated regions (left side), suggesting the strong RSS activity of RNase3 (Figure 4b). Compared with the left side of leaves infiltrated with LwaCas13a‐NT constructs, the right side infiltrated with RNase3‐targeting constructs exhibited a visible decline in GFP fluorescence (Figure 4b). Reverse transcription quantitative PCR (RT‐qPCR) data show that the interference efficiency of LwaCas13a‐gRNA constructs on RNase3 expression reached 80%, and the GFP transcripts were lowered by approximately 60% (Figure 4c). This finding suggests the recovery of RNA silencing activity in RNase3‐targeting N. benthamiana leaf cells.
FIGURE 4.

Targeting of SPCSV‐RNase3 using LwaCas13a recovers RNA silencing activity in Nicotiana benthamiana leaf cells. (a) Schematic of the targeting vectors of LwaCas13a. gRNA expression was driven by the CmYLCV promoter. DR, specific DRs of LwaCas13a. The lower panel represents the designed spacers complementary to the target sequences of SPCSV‐RNase3. NT LwaCas13a vector was used as control. (b) Two sectors (left and right) of a leaf of N. benthamiana 16C constitutively expressing GFP were agroinfiltrated to co‐express GFP, RNase3, and (left) NT vector or (right) targeting vectors. GFP silencing was observed by the disappearance of GFP fluorescence, and GFP fluorescence above the background level indicated the suppression of GFP silencing. The leaf was photographed under fluorescent excitation lamp at 6 days postinoculation (dpi). Similar results were obtained in six independent experiments. (c) Reverse transcription quantitative PCR analysis of SPCSV‐RNase3 and GFP expression levels. For each LwaCas13a targeting vectors, the expression levels of SPCSV‐RNase3 and GFP are shown relative to the LwaCas13a‐NT vector. Values are shown as mean ± SEM (n = 6)
An RNase3 heterologous expression system was constructed in N. benthamiana plants to test whether RNase3 targeting could enhance the resistance against RNase3‐mediated viral synergism in whole plants. RNase3 was fused to a truncated CMV‐2b (77 amino acids) to determine whether 2b‐RNase3‐fused protein could enhance the RSS activity in plants. As shown in Figure S4b, a delay of GFP decay was observed in GFP and 2b (77 amino acids) co‐infiltrated 16C N. benthamiana leaves, suggesting that basic RSS activity exists in this truncated 2b protein. However, 2b‐RNase3‐fused protein significantly enhanced the decay of GFP fluorescence compared with truncated 2b protein or 2b‐RNase3m, which was fused with a mutated and nonfunctional RNase3 (Figure S4b). This result suggests that 2b‐RNase3‐fused protein markedly enhances RSS activity compared with truncated 2b protein. RNase3 was then fused to CMV‐2b to form a CMV‐RNase3 infectious clone, and TuMV‐GFP (potyvirus) was selected as synergistic virus for this system (Figure S4a). The mutated RNase3 was also fused to CMV‐2b to form a CMV‐RNase3m infectious clone and used as the control (Figure S4a). These infectious clones were co‐delivered into the leaves of N. benthamiana plants via agroinfiltration. Six days after infiltration with TuMV‐GFP, GFP fluorescence was observed in the ILs and SLs, suggesting the successful infection and replication of TuMV. The GFP fluorescence in ILs and SLs did not show any difference between TuMV‐GFP‐ and TuMV‐GFP/CMV‐RNase3m‐infiltrated plants. However, a significantly enhanced accumulation of GFP fluorescence was observed in the ILs and SLs of TuMV‐GFP/CMV‐RNase3 co‐infected plants compared with that of TuMV‐GFP‐ or TuMV‐GFP/CMV‐RNase3m co‐infected plants (Figure S4c). The TuMV‐GFP/CMV‐RNase3 co‐infected N. benthamiana plants exhibited severe wilting compared with the other two groups 9 days after infection (Figure S4d). Moreover, RT‐qPCR data show that the viral titres of TuMV were significantly enhanced in N. benthamiana plants co‐infected with TuMV‐GFP/CMV‐RNase3 compared with those in the other two groups (Figure S4e). These results clearly show that CMV‐RNase3 markedly enhances the accumulation and virulence of TuMV in N. benthamiana plants.
To test whether targeting RNase3 by CRISPR‐Cas13 could improve plant resistance against RNase3‐mediated viral synergism, we generated RNase3‐targeted transgenic N. benthamiana plants. A construct was constructed containing an LwaCas13a expression cassette under the expression of the 35S promoter, a pre‐gRNA expression cassette containing three RNase3‐targeting spacers under the expression of AtU6 promoter, and a kanamycin resistance gene (Figure 5a). This construct was transformed into N. benthamiana plants through leaf disc transformation, and seven positive lines were obtained (Figure S5). Three T0 lines (RNase3‐targeting lines 1–3: RTL1‐3) with normal growth and robust LwaCas13a protein expression (anti‐FLAG tag) were selected for the collection of T1 seeds (Figure 5b). The T1 transgenic seedlings of RTL1–3 were screened on kanamycin‐containing medium prior to TuMV‐GFP/CMV‐RNase3 infection experiments. Compared with the wild type (WT), RTL1–3 seedlings exhibited a significantly decreased accumulation of GFP fluorescence in the ILs and SLs 6 and 9 days after co‐infection with TuMV‐GFP and CMV‐RNase3 (Figure 5c). RTL1–3 seedlings did not exhibit wilting symptoms that occurred in WT plants after 9 days of infection (Figure 5c). RT‐qPCR data showed that the RNA level of RNase3, TuMV‐CP, and TuMV‐GFP was remarkably reduced in the ILs and SLs after 6 and 9 days of infection (Figure 5d–f). Western blot data also showed that the protein accumulation of RNase3, TuMV‐CP, and TuMV‐GFP correspondingly decreased in the SLs of RTL1–3 compared with that in the WT (Figure 5g). These results reveal that RNase3 targeting using LwaCas13a enhances the resistance of N. benthamiana plants against RNase3‐mediated viral synergism. Thus, this CRISPR‐Cas13‐based strategy shows great potential to improve SPVD resistance in sweet potatoes.
FIGURE 5.
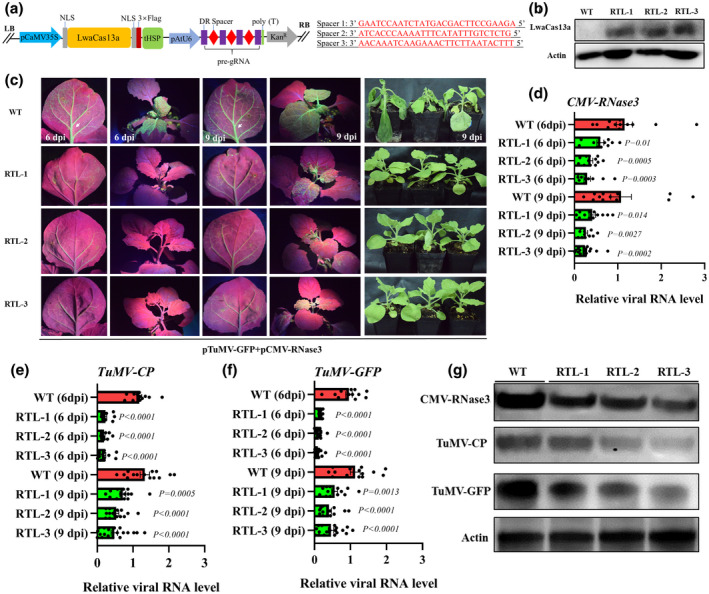
Targeting of SPCSV‐RNase3 via LwaCas13a enhances resistance against the RNase3‐mediated viral synergism in transgenic Nicotiana benthamiana plants. (a) Schematic of the RNase3‐targeted LwaCas13a vector. LwaCas13a expression was driven by the CaMV 35S promoter, and that of pre‐gRNAs was driven by the AtU6 promoter. DR, specific DRs of LwaCas13a. The right panel represents the designed spacers complementary to the target sequences of SPCSV‐RNase3. (b) Western blot detection of the LwaCas13a protein (anti‐FLAG tag) in wild‐type (WT) and three RNase3‐targeted transgenic lines (RTL1–3). (c) RTLs antagonize the CMV–RNase3‐stimulated TuMV accumulation. N. benthamiana seedlings (WT and RTL1–3) were co‐infiltrated with TuMV‐GFP and CMV‐RNase3 infectious clones. At 6 and 9 days postinoculation (dpi), the GFP signals in the infiltrated leaves (ILs) and systemic leaves (SLs) were imaged. Images showing the growth phenotypes of WT and RTLs at 9 dpi. (d–f) Reverse transcription quantitative PCR analysis of RNase3, TuMV‐GFP, and TuMV‐CP transcripts in the SLs. For each RTL, the expression levels of viral RNA are shown relative to the WT. Columns are shown as mean ± SEM (n = 9–12). (g) Representative western blot results showing RNase3 (anti‐RNase3), TuMV‐GFP (anti‐GFP), and TuMV‐CP (anti‐TuMV‐CP) accumulation in the SL of WT and RTLs plants at 9 dpi
2.4. Targeting SPCSV‐RNase3 via RfxCas13d enhances SPVD resistance in transgenic sweet potato
To test whether SPCSV‐RNase3‐targeted CRISPR‐Cas13 system could enhance SPVD resistance in sweet potato, we further generated transgenic sweet potato plants. A construct was designed containing an RfxCas13d expression cassette under the expression of 2 × 35S promoter, a pre‐gRNA expression cassette containing three RNase3‐targeting spacers under the expression of the AtU6 promoter, and a hygromycin resistance gene under the expression of enhanced 35S promoter (RfxCas13d‐RNase3; Figure 6a). A similar construct without the pre‐gRNA expression cassette was used as the control vector (RfxCas13d‐NT; Figure 6a). An LwaCas13a‐RNase3 vector was also constructed in accordance with the same principle as the RfxCas13d‐RNase3 vector. The construction of these vectors for sweet potato was prior to the experiments of the CmYLCV promoter in N. benthamiana plants. Thus, the AtU6 promoter was used for sweet potato experiments.
FIGURE 6.

Targeting of SPCSV‐RNase3 using RfxCas13d improves the resistance against the SPVD in transgenic sweet potato plants. (a) Schematic of the RNase3‐targeted RfxCas13d vector (RfxCas13d‐RNase3). RfxCas13d expression was driven by the 35S promoter, and that of pre‐gRNAs was driven by the AtU6 promoter. A similar vector without the crRNA was used as the control (RfxCas13d‐NT). DR, specific DRs of RfxCas13d. The lower panel represents the designed spacers complementary to the target sequences of SPCSV‐RNase3. (b) Growth conditions of stem tips in the grafting experiments. The virus‐free stem tips from RfxCas13d‐NT and RfxCas13d‐RNase3 transgenic sweet potato plantlets were grafted onto the same rootstock infected with SPVD. (c) Reverse transcription quantitative PCR (RT‐qPCR) analysis of SPCSV‐RNase3 and SPFMV‐CP transcripts in tender leaves of scions after 14 and 21 days of grafting. The expression levels of viral RNA in RfxCas13d‐RNase3 transgenic scions are shown relative to the RfxCas13d‐NT transgenic scions. Columns are shown as mean ± SEM (n = 6). (d) SPVD symptoms of young leaves from wild‐type (WT), RfxCas13d‐NT, and RfxCas13d‐RNase3 transgenic sweet potato plants after 6 months of vegetative propagation under natural infection conditions. (e) RT‐qPCR analysis of SPCSV‐RNase3 and SPFMV‐CP accumulation in the young leaves from the three genotypes. The expression levels of viral RNA in transgenic plants are shown relative to the WT plants. Columns are shown as mean ± SEM (n = 6). (f) Representative western blot results showing RNase3 (anti‐RNase3) and SPFMV‐CP (anti‐SPFMV‐CP) accumulation in the young leaves of the three genotypes
These constructs were transformed into a cultivated sweet potato (Xushu 29) using the embryonic callus‐based transformation method (Kim et al., 2012; Yu, Xuan, et al., 2020). Four and three transgenic sweet potato lines transformed with the RfxCas13d‐NT vector and the RfxCas13d‐RNase3 vector were obtained, respectively. However, we did not obtain regenerated sweet potato plantlets transformed with the LwaCas13a‐RNase3 vector (data not shown). Robust RfxCas13d protein expression (anti‐FLAG tag) was detected in two transgenic sweet potato lines (RfxCas13d‐NT L1 and RfxCas13d‐RNase3 L1), and these lines were selected for further experiments (Figure S6).
In the grafting experiments, the virus‐free stem tips of RfxCas13d‐NT and RfxCas13d‐RNase3 transgenic sweet potato plants were grafted onto the SPVD‐infected rootstocks for 21 days (Figure 6b). RT‐qPCR data showed that the RNA levels of SPCSV‐RNase3 and SPFMV‐CP were significantly lower in RfxCas13d‐RNase3 transgenic scions than in RfxCas13d‐NT scions, which were grafted onto the same root stock (Figure 6c). The interference efficiency of RfxCas13d‐RNase3 transgenic scions on the accumulation of SPCSV and SPFMV reached 95% and 80%, respectively, 21 days after grafting (Figure 6c). This result shows that SPCSV‐RNase3‐targeting by RfxCas13d inhibits SPCSV and SPFMV accumulation in sweet potato plants.
A 6‐month natural infection experiment was performed to test whether RfxCas13d‐RNase3 transgenic sweet potato plants exhibit enhanced SPVD resistance under long‐term growth conditions. Uniform virus‐free WT, RfxCas13d‐NT, and RfxCas13d‐RNase3 transgenic Xushu 29 plants (Figure S8) were placed alongside the SPVD‐infected sweet potato plants in a greenhouse with whiteflies and aphids, which are insect vectors of SPCSV and SPFMV, respectively, for 6 months. The WT and RfxCas13d‐NT transgenic Xushu 29 plants exhibited severe SPVD symptoms, including leaf chlorosis, malformation, and mosaic (Figure 6d). The RfxCas13d‐RNase3 transgenic Xushu 29 plants did not show any SPVD symptom compared with the other genotypes (Figure 6d). RT‐qPCR data show that SPCSV and SPFMV titres were significantly reduced in the young leaves of RfxCas13d‐RNase3 transgenic Xushu 29 compared with those in the WT and RfxCas13d‐NT transgenic lines (Figure 6e). The accumulation of viral proteins (SPCSV‐RNase3 and SPFMV‐CP) was largely inhibited in the young leaves of RfxCas13d‐RNase3 transgenic Xushu 29 compared with that in other two genotypes (Figure 6f). These results prove that SPCSV‐RNase3‐targeting via RfxCas13d enhances SPVD resistance in transgenic sweet potato plants.
3. DISCUSSION
In the present study, the RNA targeting activities of various CRISPR‐Cas13 systems were compared, and these systems were used for SPCSV‐RNase3 interference to improve the resistance of sweet potato against SPVD. The interference efficiency of different Cas13 variants in plants is correlated to the variable crRNA scaffolds, Cas13 protein sizes, catalytic activities, and subcellular localization (Aman et al., 2018; Mahas et al., 2019). Mahas and colleagues conducted systematic research and revealed that CasRX (RfxCas13d) has the highest interference activities against RNA virus among the tested Cas13 variants (Mahas et al., 2019). In addition, RfxCas13d has been reported as the most efficient knockdown platform to inhibit the expression of endogenous transcripts in animal embryos and Drosophila melanogaster cell lines (Huynh et al., 2020; Kushawah et al., 2020). Inconsistent with a previous report, RfxCas13d did not show efficient RNA targeting activity when the AtU6 promoter was used to drive the expression of gRNAs in the present study (Figure 1d) (Mahas et al., 2019). The CmYLCV promoter contributes to the efficient expression of gRNAs in the CRISPR‐Cas9 system (Čermák et al., 2017). Our results showed that using the CmYLCV promoter to drive the expression of gRNAs remarkably enhanced the targeting activity of RfxCas13d compared with the use of the AtU6 promoter (Figure 1d). However, RNA targeting activity was only slightly enhanced for the other Cas13 variants (Figure 1), indicating that the restriction of RNA targeting activity of CRISPR‐Cas13 in our system is due to the inefficient expression of gRNAs for RfxCas13d and the inefficient catalytic activity for other Cas13 orthologs. This result suggests that the designation of appropriate expression cassettes for expressing gRNAs is important in improving the interference activity of special CRISPR‐Cas13 systems (Čermák et al., 2017). Given that RfxCas13d and LwaCas13a reached a high knockdown of viral RNA accumulation when the CmYLCV promoter was used to drive the expression of gRNAs (>80%), we suggest that RfxCas13d and LwaCas13a are two excellent candidates for RNA virus interference applications in plants. These variants can be used to target two RNA viruses or multiple targets within one RNA virus genome simultaneously (Figures 3 and 4), thus suggesting their potential use in crops for the interference of mixed infections of RNA viruses under natural conditions (Mahas et al., 2019). Compared with RNAi, which often suffers from low efficiencies or off‐target effects, and the CRISPR‐Cas9‐based CRISPRi, which requires a protospacer adjacent motif (Rosa et al., 2018; Zhang et al., 2018), CRISPR‐Cas13 may represent a flexible and compelling alternative to existing methods for antagonizing RNA viruses or interference of endogenous transcripts in plants.
RNA silencing serves as a primary antiviral defence mechanism via the production of vsiRNAs that interfere with the replicative ability of viruses (Li & Wang, 2019). SPCSV‐RNase3 encoded by the SPCSV genome can provoke viral synergism in sweet potato plants by processing 21‐ to 22‐nt vsiRNAs into 14‐nt vsiRNAs and thus destroying the host RNA silencing machinery (Cuellar et al., 2009). Transgenic sweet potato plants carrying SPFMV segments exhibited resistance to SPFMV in the greenhouse while losing resistance following infection with SPCSV in the field (Kreuze et al., 2008). In addition, transgenic sweet potato plants carrying segments derived from various viruses exhibited resistance to corresponding viruses in the absence of SPCSV (Sivparsad & Gubba, 2014). In the current study, SPCSV‐RNase3 targeting via LwaCas13a successfully recovered the RNA silencing activity in host cells (Figure 4). Compared with that in the WT N. benthamiana plants, the accumulation of untargeted TuMV was largely reduced in RNase3‐targeting plants when co‐infected with CMV‐RNase3 and TuMV‐GFP (Figure 5). This phenomenon may contribute to the recovered RNA silencing activity in RNase3‐targeting plants, which in turn may be beneficial to the improved resistance of RNase3‐targeting plants against the co‐infection of CMV‐RNase3 and TuMV‐GFP (Figure 5). Grafting and natural infection experiments showed that RfxCas13d provided robust interference against SPCSV‐RNase3 and theoretically recovered the RNA silencing capability of the transgenic sweet potato plants, thus significantly limiting SPFMV accumulation in the leaves (Figure 6). Although the AtU6 promoter cannot confer the RNA targeting activity of RfxCas13d to the maximum extent, molecular analysis corroborated our phenotypic data and indicated that the resistance of transgenic sweet potato against the SPVD was significantly enhanced (Figure 6). Cas13, especially variants from the Cas13d family, display minimal off‐target tendencies (Huynh et al., 2020; Konermann et al., 2018; Kushawah et al., 2020; Mahas et al., 2019) and thus might help quell concerns regarding the immune escape of plant viruses in nature (Mehta et al., 2019). This CRISPR‐Cas13‐based SPCSV‐RNase3 interference system can be improved further by optimizing the promoter for gRNA expression or by designing multiple targets that are against SPFMV and SPCSV genomes simultaneously. In the future, SPVD‐immune sweet potato germplasm could be created using the CRISPR‐Cas13 technique. This germplasm would benefit sweet potato farming in sub‐Saharan African countries, such as Nigeria, Tanzania, Ethiopia, and Uganda, where SPVD can cause yield losses of 80%–90% in infected sweet potato plants (Bednarek et al., 2021; Loebenstein, 2015).
In conclusion, LwaCas13a and RfxCas13d enable efficient RNA virus interference in plants, and the selection of promoters for gRNA expression is important to optimize the RNA targeting activity of RfxCas13d. Targeting SPSCV‐RNase3 via CRISPR‐Cas13 significantly recovered the RNA silencing activity of host cells and enhanced SPVD resistance in transgenic sweet potatoes. CRISPR‐Cas13 has great potential for the optimization and development of creating SPVD‐immune sweet potato germplasm in the future.
4. EXPERIMENTAL PROCEDURES
4.1. Vector construction
SPCSV‐RNase3 (XZ) was cloned from SPVD‐infected sweet potato and then sequenced (File S2) and aligned. The DNA sequences of plant codon‐optimized LshCas13a (Abudayyeh et al., 2017), LwaCas13a (Abudayyeh et al., 2017), PspCas13b (Cox et al., 2017), and RfxCas13d (Konermann et al., 2018) with fusions of the NLS encoding sequence at the C or N terminal were artificially synthesized (File S1) and subsequently cloned into the pSPLCV (AC2m/AC4m) vector under the expression of the NbU4 promoter by using a ClonExpress MultiS One‐step Cloning Kit (Vazyme Biotech Co., Ltd) to construct the SPCSV‐RNase3, mGFP5, and RNA virus‐targeted CRISPR‐Cas13 vectors for transient assays. The ortholog's corresponding direct repeat (DR) with the targeting or NT sequences was designed and artificially synthesized (General Biol Co., Ltd). crRNAs were ligated into the pSPLCV (AC2m/AC4m) expression vectors at reserved KpnI/BamHI restriction sites by T4 ligase. All the Cas13 variant sequences and crRNA sequences were confirmed in pSPLCV (AC2m/AC4m) vectors with Sanger sequencing. In some experiments, the AtU6 promoter of crRNAs was replaced by the CmYLCV promoter. The DNA sequences of plant codon‐optimized LwaCas13a were directly cloned into the linearized pRI201‐AN binary vector that was digested by restriction endonucleases NdeI/SalI to construct the RNase3‐targeted LwaCas13a vector for the stable transformation of N. benthamiana. The crRNA including an AtU6 promoter and an RNase3‐targeted pre‐gRNA were artificially synthesized (General Biol Co., Ltd) and then cloned into the linearized pRI201‐AN binary vector at the KpnI/EcoRI restriction sites.
The DNA sequences of plant codon‐optimized RfxCas13d were cloned from the above constructed pSPLCV (AC2m/AC4m) vector and then cloned into the linearized pCambia1380 binary vector under the expression of the 2 × 35S promoter at the StuI/XbaI restriction sites to construct the RNase3‐targeted RfxCas13d vector used for the stable transformation of sweet potatoes. The crRNA including an AtU6 promoter and an RNase3‐targeted pre‐gRNA were artificially synthesized (General Biol Co., Ltd) and directly cloned into the linearized pCambia1380 binary vector at the KpnI/BamHI restriction sites.
The genome sequences of TuMV‐GFP were synthesized as three DNA fragments (approximately 3500 bp for each fragment) to construct the TuMV‐GFP infectious clone (GenBank EF028235.1) (Aman et al., 2018; Suehiro et al., 2004). The multiple DNA fragments of TuMV‐GFP genome sequences were ligated into the pCambia0390 binary vector under the expression of the 2 × 35S promoter (at HindIII/EcoRI restriction sites) by using a ClonExpress MultiS One Step Cloning Kit (Vazyme Biotech Co., Ltd). The DNA sequences of DsRed were amplified from the PBI121‐DsRed vector to construct the CMV‐DsRed infectious clone. The PCR product was purified and fused to truncated CMV‐2b (Wang et al., 2016), which was digested by the restriction endonucleases KpnI/XbaI. The DNA fragment of SPCSV‐RNase3 was fused to the truncated CMV‐2b at the KpnI/XbaI restriction sites to generate the CMV‐RNase3 infectious clone. For RNA targeting and GFP silencing experiments, other transient expression vectors (including mGFP5, GFP, RNase3, RNase3m, 2b‐RNase3, and 2b‐RNase3m) were constructed on the basis of the pRI201‐AN binary vector.
4.2. Generation of transgenic lines
Leaf discs of N. benthamiana were infected with Agrobacterium tumefaciens GV3101 carrying RNase3‐targeted LwaCas13a vector for 3 days and then transferred to Murashige & Skoog (MS) medium containing cefotaxime (500 mg/ml) and kanamycin (50 mg/ml) to generate transgenic N. benthamiana plants. The regenerated kanamycin‐resistant shoots of 2–3 cm height were cut off and placed on rooting medium for rooting. Transgenic‐positive lines were confirmed by genomic DNA PCR and western blot analysis of the leaf samples collected from the regenerated plantlets. Three positive lines (T0, RTL1 to RTL3) were selected. The seeds harvested from the three lines were screened on kanamycin‐containing MS medium. Surviving seedlings were used for experiments.
RNase3‐targeted RfxCas13d or NT construct was introduced into A. tumefaciens EHA105, grown overnight on a shaker at 200 rpm in YPD liquid medium containing 200 μM acetosyringone and 12.5 mg/L hygromycin at 28°C and used to transform sweet potato embryogenic callus (Xushu 29) using the A. tumefaciens‐mediated transformation method to generate transgenic sweet potato plants (Kim et al., 2012; Yu, Xuan, et al., 2020; Zang et al., 2009). Transgenic calli were screened on hygromycin‐containing medium. Transgenic‐positive lines were confirmed by genomic DNA PCR and western blot analysis of the leaf samples collected from the regenerated plants. Two transgenic lines (one for NT vector and one for RNase3‐targeted RfxCas13d vector) were selected, and the virus‐free plantlets were propagated via shoot meristem culture. Grafting and natural infection experiments were initiated when a specific amount of virus‐free plantlets was obtained.
4.3. Agroinfiltration of N. benthamiana leaves
Three‐week‐old wild‐type 16C or LwaCas13a‐transgenic N. benthamiana plants grown under long‐day conditions (16 h light, 8 h dark at 25°C) were used for experiments. The above CRISPR‐Cas13 vectors, viral infectious clones (TuMV‐GFP, CMV‐DsRed, and CMV‐RNase3), and other transient expression vectors were transformed into A. tumefaciens GV3101. Single colonies grown overnight in a selective medium were centrifuged and suspended in an infiltration medium (10 mM MES pH 5.7, 10 mM MgCl2, 200 μM acetosyringone) and incubated at ambient temperature for 4 h. For infiltration into N. benthamiana leaves, the cultures were mixed with the same volume in accordance with the experimental design at a final OD600 of 0.5 for all the constructs. Healthy and fully developed leaves were agroinfiltrated using a 1‐ml needleless syringe (Horsch et al., 1985).
4.4. Sweet potato grafting experiments
Sweet potato plants exhibiting typical SPVD symptoms were collected from the field and screened with RT‐PCR (RNA virus) or PCR (DNA virus). One plant co‐infected with SPFMV and SPCSV but apparently free from other tested viruses was selected and propagated vegetatively under a fly net for 2 months (Figure S7). The seedlings propagated from this SPVD‐infected plant were used as rootstocks in grafting experiments or as viral source in natural infection experiments. One virus‐free stem tip from the NT transgenic plants and another from the RNase3‐targeted RfxCas13d transgenic plants were grafted onto the same rootstock. The growth conditions of scions were recorded, and the fully expanded young leaves of scions were collected for RT‐qPCR analysis 14 and 21 days after grafting.
4.5. Natural infection experiments
The SPVD‐infected sweet potato seedlings were cultured along with the virus‐free WT sweet potato plants, NT transgenic sweet potato plants, and RNase3‐targeted RfxCas13d transgenic sweet potato plants in a greenhouse that contained insect vectors (whiteflies for SPCSV and aphids for SPFMV) for 6 months. The young shoots were cut off every 2 months for propagation. After 6 months, the phenotypes of the young shoots collected from the three genotypes were photographed. The tender leaves were collected for RT‐qPCR and western blot analysis.
4.6. Observation of fluorescent proteins
For the expression analysis of different Cas13 proteins, the epidermis of agroinfiltrated N. benthamiana leaves was peeled off. GFP fluorescence was visualized under an Olympus BX63 epifluorescence microscope. For the observation of transiently expressed mGFP5, TuMV‐expressed GFP, and CMV‐expressed DsRed in the ILs, the agroinfiltrated leaves were cut off after 4, 6, and 9 days postinfiltration (dpi) in different experiments. The corresponding fluorescence was excited by different handheld fluorescent excitation lamps (blue or green light) and photographed with different filters in the dark (Aman et al., 2018; Mahas et al., 2019). For the observation of TuMV‐expressed GFP or CMV‐expressed DsRed in the SLs, the whole plants were photographed at 6 or 9 dpi (Aman et al., 2018; Mahas et al., 2019).
4.7. RNA extraction and RT‐qPCR analysis
Total RNA was extracted from the ILs and SLs of N. benthamiana plants or the young leaves of WT and transgenic sweet potato plants by using FastPure Plant Total RNA Isolation Kit (Polysaccharides & Polyphenolics –rich) (TIANGEN Biochemical Technology) following the manufacturer's instructions. Reverse transcription was performed using oligo(dT)23VN primers and HiScript II 1st Strand cDNA Synthesis Kit (Vazyme). RNA was quantified with RT‐qPCR using the SYBR Premix Ex TaqTM II Kit (TaKaRa). All RT‐qPCRs were performed in 20‐μl reactions with three technical replicates in 96‐well format and read out using a qTOWER2 Real‐time PCR System (Analytik Jena). Primers are listed in Table S1. Relative expression levels were calculated using the 2−∆∆ C t method. The housekeeping reference genes used in this study were NbActin (AY179605.1) for N. benthamiana and IbUBQ (cloned in our laboratory) for sweet potatoes.
4.8. Western blot analysis
Total proteins were extracted from 100 mg of leaf samples by using the Plant Total Protein Extraction Kit (Sigma), and protein concentrations were quantified by bicinchoninic acid (BCA) spectrophotometry. Proteins were separated by 6%–12% SDS‐PAGE. Immunoblot analysis was conducted using mouse α‐GFP antibody (1:2000) (Sangon Biotech [Shanghai] Co., Ltd) for the detection of TuMV‐expressed GFP protein; rat 3× FLAG antibody (Sangon Biotech [Shanghai] Co., Ltd) for the detection of LwaCas13a and RfxCas13d, and rat‐TuMV‐CP antibody, rat‐SPCSV‐RNase3 antibody, and rat SPFMV‐CP antibody (polyclonal antibodies prepared by ProbeGene Life Science Co., Ltd) for the detection of TuMV‐CP, SPCSV‐RNase3, and SPFMV‐CP, respectively. Horseradish peroxidase (HRP)‐labelled goat anti‐rabbit IgG (H + L) (1:1000) (Beyotime Biotechnology) was used as the secondary antibody. Antigens were detected by chemiluminescence using an ECL‐detecting reagent (GeneScript Biotechnology) and a chemiluminescence instrument (FluorChem M; GE). For each experiment, three independent biological replicates were conducted.
4.9. Statistical analysis
Data were subjected to analysis of variance (ANOVA). The p value is marked in the appropriate position in figures.
CONFLICT OF INTERESTS
The authors have no conflict of interests.
Supporting information
ACKNOWLEDGEMENTS
This work was supported by China Agriculture Research System of MOF and MARA (CARS‐10‐B03), the National Natural Science Foundation of China (31871684), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (18KJA180004), the Key R & D Program of Xuzhou (KC19127), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). We appreciate Professor Tao Zhou from China Agriculture University for kindly providing the CMV infectious clone.
Yu, Y. , Pan, Z. , Wang, X. , Bian, X. , Wang, W. , Liang, Q. , et al. (2022) Targeting of SPCSV‐RNase3 via CRISPR‐Cas13 confers resistance against sweet potato virus disease. Molecular Plant Pathology, 23, 104–117. 10.1111/mpp.13146
Yicheng Yu, Zhiyuan Pan and Xiao Wang contributed equally to the article.
Contributor Information
Zongyun Li, Email: zongyunli@jsnu.edu.cn.
Jian Sun, Email: sunjian@jsnu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author on reasonable request.
REFERENCES
- Abudayyeh, O.O. , Gootenberg, J.S. , Essletzbichler, P. , Han, S. , Joung, J. , Belanto, J.J. et al. (2017) RNA targeting with CRISPR‐Cas13. Nature, 550, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh, O.O. , Gootenberg, J.S. , Konermann, S. , Joung, J. , Slaymaker, I.M. , Cox, D.B.T. et al. (2016) C2c2 is a single‐component programmable RNA‐guided RNA‐targeting CRISPR effector. Science, 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, Z. , Abulfaraj, A. , Idris, A. , Ali, S. , Tashkandi, M. , Mahfouz, M.M. et al. (2015) CRISPR/Cas9‐mediated viral interference in plants. Genome Biology, 16, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, Z. , Mahas, A. & Mahfouz, M. (2018) CRISPR/Cas13 as a tool for RNA interference. Trends in Plant Science, 23, 374–378. [DOI] [PubMed] [Google Scholar]
- Aman, R. , Ali, Z. , Butt, H. , Mahas, A. , Aljedaani, F. , Khan, M.Z. et al. (2018) RNA virus interference via CRISPR/Cas13a system in plants. Genome Biology, 19, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek, R. , David, M. , Fuentes, S. , Kreuze, J. & Fei, Z. (2021) Transcriptome analysis provides insights into the responses of sweet potato to sweet potato virus disease (SPVD). Virus Research, 295, 198293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovell‐Benjamin, A.C. (2007) Sweet potato: a review of its past, present, and future role in human nutrition. Advances in Food and Nutrition Research, 52, 1–59. [DOI] [PubMed] [Google Scholar]
- Bovell‐Benjamin, A.C. (2010) Sweet potato utilization in human health, industry and animal feed systems. In Ray, R.C. & Tomlins, K. (Eds.), Sweet potato: post harvest aspects in food, feed and industry. New York: Nova Science Publishers Inc, pp. 193–224. [Google Scholar]
- Cao, Y. , Zhou, H. , Zhou, X. & Li, F. (2021) Conferring resistance to plant RNA viruses with the CRISPR/CasRx system. Virologica Sinica, 36, 814–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák, T. , Curtin, S.J. , Gil‐Humanes, J. , Čegan, R. , Kono, T.J.Y. , Konečná, E. et al. (2017) A multipurpose toolkit to enable advanced genome engineering in plants. The Plant Cell, 29, 1196–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. , Wang, Y. , Zhang, R. , Zhang, H. & Gao, C. (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annual Review of Plant Biology, 70, 667–697. [DOI] [PubMed] [Google Scholar]
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D.B.T. , Gootenberg, J.S. , Abudayyeh, O.O. , Franklin, B. , Kellner, M.J. , Joung, J. et al. (2017) RNA editing with CRISPR‐Cas13. Science, 358, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar, W.J. , Kreuze, J.F. , Rajamäki, M.L. , Cruzado, K.R. , Untiveros, M. & Valkonen, J.P.T. (2009) Elimination of antiviral defense by viral RNase III. Proceedings of the National Academy of Sciences USA, 106, 10354–10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT (2018). Crops and livestock products. Available at: http://www.fao.org/faostat/en/ [Accessed 29th September 2021].
- Gibson, R.W. , Aritua, V. , Byamukama, E. , Mpembe, I. & Kayongo, J. (2004) Control strategies for sweet potato virus disease in Africa. Virus Research, 100, 115–122. [DOI] [PubMed] [Google Scholar]
- Hang, T. , Zhao, Y. , Ye, J. , Cao, X. , Xu, C. & Chen, B. (2019) Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnology Journal, 17, 1185–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, L.B. , Burstein, D. , Chen, J.S. , Paez‐Espino, D. , Ma, E. , Witte, I.P. et al. (2018) Programmed DNA destruction by miniature CRISPR‐Cas14 enzymes. Science, 362, 839–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch, R. , Fry, J. , Hoffmann, N. , Eichholtz, D. , Rogers, S. et al. (1985) A simple and general method for transferring genes into plants. Science, 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Huynh, N. , Depner, N. , Larson, R. & King‐Jones, K. (2020) A versatile toolkit for CRISPR‐Cas13‐based RNA manipulation in Drosophila . Genome Biology, 21, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, W.L. , Robert, O.M. , Andrade, M. , Carey, E. & Ball, A. (2017) Tackling vitamin A deficiency with biofortified sweet potato in sub‐Saharan Africa. Global Food Security, 14, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, X. , Si, X. , Zhang, Y. , Zhang, H. , Zhang, F. , Gao, C. et al. (2018) Conferring DNA virus resistance with high specificity in plants using virus‐inducible genome‐editing system. Genome Biology, 19, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. & Charpentier, E. (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karyeija, R.F. , Kreuze, J.F. , Gibson, R.W. & Valkonen, J.P.T. (2000) Synergistic interactions of a potyvirus and a phloem‐limited crinivirus in sweet potato plants. Virology, 269, 26–36. [DOI] [PubMed] [Google Scholar]
- Kim, S.H. , Ahn, Y.O. , Ahn, M.J. , Lee, H.S. & Kwak, S.S. (2012) Down‐regulation of β‐carotene hydroxylase increases β‐carotene and total carotenoids enhancing salt stress tolerance in transgenic cultured cells of sweet potato. Phytochemistry, 74, 69–78. [DOI] [PubMed] [Google Scholar]
- Komor, A.C. , Badran, A.H. & Liu, D. (2017) CRISPR‐based technologies for the manipulation of eukaryotic genomes. Cell, 169, 559. [DOI] [PubMed] [Google Scholar]
- Konermann, S. , Lotfy, P. , Brideau, N.J. , Oki, J. , Shokhirev, M.N. & Hsu, P.D. (2018) Transcriptome engineering with RNA‐targeting type VI‐D CRISPR effectors. Cell, 173, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuze, J.F. , Klein, I.S. , Lazaro, M.U. , Chuquiyuri, W.J. , Morgan, G.L. , Mejía, P.G. et al. (2008) RNA silencing‐mediated resistance to a crinivirus (Closteroviridae) in cultivated sweet potato (Ipomoea batatas L.) and development of sweet potato virus disease following co‐infection with a potyvirus. Molecular Plant Pathology, 9, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuze, J.F. , Savenkov, E.I. , Cuellar, W. , Li, X. & Valkonen, J.P.T. (2005) Viral class 1 RNase III involved in suppression of RNA silencing. Journal of Virology, 79, 7227–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushawah, G. , Hernandez‐Huertas, L. , Abugattas‐Nuñez del Prado, J. , Martinez‐Morales, J.R. , DeVore, M.L. , Hassan, H. et al. (2020) CRISPR‐Cas13d induces efficient mRNA knockdown in animal embryos. Developmental Cell, 54, 805–817. [DOI] [PubMed] [Google Scholar]
- Li, F.F. & Wang, A.M. (2019) RNA‐targeted antiviral immunity: more than just RNA silencing. Trends in Microbiology, 27, 792–805. [DOI] [PubMed] [Google Scholar]
- Loebenstein, G. (2015) Control of sweet potato virus diseases. Advance in Virus Research, 91, 33–45. [DOI] [PubMed] [Google Scholar]
- Mahas, A. , Aman, R. & Mahfouz, M. (2019) CRISPR‐Cas13d mediates robust RNA virus interference in plants. Genome Biology, 20, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, D. , Stürchler, A. , Anjanappa, R.B. , Zaidi, S.‐E.‐A. , Hirsch‐Hoffmann, M. , Gruissem, W. et al. (2019) Linking CRISPR‐Cas9 interference in cassava to the evolution of editing‐resistant geminiviruses. Genome Biology, 20, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa, C. , Kuo, Y.W. , Wuriyanghan, H. & Falk, B.W. (2018) RNA interference mechanisms and applications in plant pathology. Annual Review in Phytopathology, 56, 581–610. [DOI] [PubMed] [Google Scholar]
- Sivparsad, B.J. & Gubba, A. (2014) Development of transgenic sweet potato with multiple virus resistance in South Africa (SA). Transgenic Research, 23, 377–388. [DOI] [PubMed] [Google Scholar]
- Suehiro, N. , Natsuaki, T. , Watanabe, T. & Okuda, S. (2004) An important determinant of the ability of Turnip mosaic virus to infect Brassica spp. and/or Raphanus sativus is in its P3 protein. Journal of General Virology, 85, 2087–2098. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Poque, S. & Valkonen, J.P.T. (2019) Phenotyping viral infection in sweet potato using a high‐throughput chlorophyll fluorescence and thermal imaging platform. Plant Methods, 15, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Yang, X. , Wang, N. , Liu, X. , Nelson, R.S. , Li, W. et al. (2016) An efficient virus‐induced gene silencing vector for maize functional genomics research. Plant Journal, 86, 102–115. [DOI] [PubMed] [Google Scholar]
- Weinheimer, I. , Boonrod, K. , Moser, M. , Wassenegger, M. , Krczal, G. , Butcher, S.J. et al. (2014) Binding and processing of small dsRNA molecules by the class 1 RNase III protein encoded by sweet potato chlorotic stunt virus. Journal of General Virology, 95, 486–495. [DOI] [PubMed] [Google Scholar]
- Weinheimer, I. , Haikonen, T. , Ala‐Poikela, M. , Moser, M. , Streng, J. , Rajamäki, M.‐L. et al. (2016) Viral RNase3 co‐localizes and interacts with the antiviral defense protein SGS3 in plant cells. PLoS One, 11, e0159080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinheimer, I. , Jiu, Y. , Rajamäki, M.L. , Matilainen, O. , Kallijärvi, J. , Cuellar, W.J. et al. (2015) Suppression of RNAi by dsRNA‐degrading RNase III enzymes of viruses in animals and plants. PLoS Pathogens, 11, e1004711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, W.X. , Hunnewell, P. , Alfonse, L.E. , Carte, J.M. , Keston‐Smith, E. , Sothiselvam, S. et al. (2019) Functionally diverse type V CRISPR‐Cas systems. Science, 363, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Wang, X. , Sun, H. , Liang, Q. , Wang, W. , Zhang, C. et al. (2020) Improving CRISPR‐Cas‐mediated RNA targeting and gene editing using SPLCV replicon‐based expression vectors in Nicotiana benthamiana . Plant Biotechnology Journal, 18, 1993–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Xuan, Y. , Bian, X. , Zhang, L. , Pan, Z. , Kou, M. et al. (2020) Overexpression of phosphatidylserine synthase IbPSS1 affords cellular Na+ homeostasis and salt tolerance by activating plasma membrane Na+/H+ antiport activity in sweet potato roots. Horticulture Research, 7, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang, N. , Zhai, H. , Gao, S. , Chen, W. , He, S.Z. & Liu, Q.C. (2009) Efficient production of transgenic plants using the bar gene for herbicide resistance in sweetpotato. Scientia Horticulturae, 122, 649–653. [Google Scholar]
- Zetsche, B. , Gootenberg, J. , Abudayyeh, O. , Slaymaker, I. , Makarova, K. , Essletzbichler, P. et al. (2015) Cpf1 is a single RNA‐guided endonuclease of a class 2 CRISPR‐Cas system. Cell, 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, X. , Zhang, F. , Zhong, Z. , Chen, R. , Wang, Y. , Chang, L. et al. (2019) Generation of virus‐resistant potato plants by RNA genome targeting. Plant Biotechnology Journal, 17, 1814–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Zheng, Q. , Yi, X. , An, H. , Zhao, Y. , Ma, S. et al. (2018) Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnology Journal, 16, 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H. , Li, C. & Gao, C. (2020) Applications of CRISPR‐Cas in agriculture and plant biotechnology. Nature Review Molecular Cell Biology, 11, 661–677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.


