Abstract
Lesion mimic mutants resembling the hypersensitive response without pathogen attack are an ideal material to understand programmed cell death, the defence response, and the cross‐talk between defence response and development in plants. In this study, mic, a lesion mimic mutant from cultivar Yunyin treated with ethyl methanesulphonate (EMS), was screened. By map‐based cloning, a short‐chain alcohol dehydrogenase/reductase with an atypical active site HxxxK was isolated and designated as SDR7‐6. It functions as a homomultimer in rice and is localized at the endoplasmic reticulum. The lesion mimic phenotype of the mutant is light‐dependent. The mutant displayed an increased resistance response to bacterial blight, but reduced resistance to rice blast disease. The mutant and knockout lines showed increased reactive oxygen species, jasmonic acid content, antioxidant enzyme activity, and expression of pathogenicity‐related genes, while chlorophyll content was significantly reduced. The knockout lines showed significant reduction in grain size, seed setting rate, 1000‐grain weight, grain weight per plant, panicle length, and plant height. SDR7‐6 is a new lesion mimic gene that encodes a short‐chain alcohol dehydrogenase with atypical catalytic site. Disruption of SDR7‐6 led to cell death and had adverse effects on multiple agricultural characters. SDR7‐6 may act at the interface of the two defence pathways of bacterial blight and rice blast disease in rice.
Keywords: lesion mimic mutant, rice, SDR7‐6, short‐chain alcohol dehydrogenase/reductase
SDR7‐6, a short‐chain alcohol dehydrogenase/reductase family protein, negatively regulates cell death and the defence response to rice bacterial blight, but positively regulates the defence response to Magnaporthe oryzae.
1. INTRODUCTION
Lesion mimic mutants show a necrotic lesion phenotype in the absence of pathogen attack. The appearance of a lesion mimic phenotype is often, but not always, affected by environmental factors including light, temperature, day length, and humidity (Qiao et al., 2010; Zhu et al., 2016). Given the resemblance to the hypersensitive response, lesion mimics provide a tool to unravel the mechanisms of programmed cell death (PCD), defence responses (Beaugelin et al., 2019; Bruggeman et al., 2015; Ke et al., 2019; Lorrain et al., 2003), and the interaction between plant immunity and growth (Zhu et al., 2020). In rice, there are at least 56 lesion mimic genes documented in the Gramene database (Yang et al., 2019). The genes encode different functional proteins that participate in various physiological pathways, hinting at a complicated regulatory network in rice. Some of the genes function as defence‐signalling genes and exhibit nonspecific broad‐spectrum blast resistance (Li et al., 2020).
The short‐chain dehydrogenases/reductases (SDRs) are one of the largest superfamilies of enzymes known to date (Persson & Kallberg, 2013). They use NAD(H) or NADP(H) as a cofactor and have broad substrate spectra involved in primary and secondary metabolism (Moummou et al., 2012; Persson & Kallberg, 2013). The majority of SDRs are oligomeric, mostly homodimers or homotetramers (Kavanagh et al., 2008). Persson et al. (2009) used hidden‐Markov models (HMMs) to classify SDRs into seven groups (classical, extended, atypical, intermediate, divergent, complex, and unknown). Based on this result, Moummou combined HMMs and phylogenetic analysis methods to further categorize higher plant SDRs into 49 families. The SDR110C (C stands for “classical”) family has significant diversified function in vascular plants participating in terpenoids, alkaloids, phenolics metabolism, hormone biosynthesis, and flower development. Rice contains 227 SDRs; among them 110 belong to classical SDRs and 24 fall into the SDR110C family (Moummou et al., 2012). Five of them are induced by chitin and four are involved in the biosynthesis of diterpenoids (Kitaoka et al., 2016). Momilactone A synthase (OsMAS) oxidizes 3β‐hydroxy‐syn‐pimaradien‐19,6β‐olide to phytoalexin momilactone A (Atawong et al., 2002; Shimura et al., 2007), while OsSDR110C‐MS2 has been shown to have an efficient ability to catalyse this final step in vitro (Kitaoka et al., 2016; Miyamoto et al., 2016). OsSDR110C‐MI3 and OsSDR110C‐MS3 are involved in the biosynthesis of phytoalexin oryzalexins A–C (Kitaoka et al., 2016; Miyamoto et al., 2016). RMP8 is a glucose/ribitol dehydrogenase family protein, which is a microspore‐preferred protein (Nguyen et al., 2016). Another SDR110C is OsABA2, an abscisic acid (ABA) biosynthetic enzyme, responsible for the conversion of xanthoxin to abscisyl aldehyde (Endo et al., 2014). OsABA2 is also a negative regulator of cell death and the defence response (Liao et al., 2018).
In this study, a novel lesion mimic mutant, mic, was identified from an ethyl methanesulphonate (EMS)‐mutagenized japonica rice cultivar Yunyin population. The mutant exhibited light‐dependent necrosis at the seedling stage. By map‐based cloning, the causal gene was isolated and named as SDR7‐6 (the sixth SDR110C family protein of chromosome 7). SDR7‐6 can form a homomultimer and is a rice SDR110C with unknown function. It negatively regulates cell death and resistance to bacterial blight, but positively regulates resistance to the rice blast pathogen.
2. RESULTS
2.1. Phenotypic characterization of mic mutant
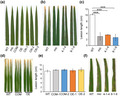
The mic mutant was first identified from an EMS‐mutagenized population of japonica rice cultivar Yunyin. Reddish‐brown lesions were initially observed on the leaf blade at the six‐leaf stage under natural light conditions. As it grew, the lesion number and intensity gradually increased. Then the lesions spread to the whole plant, including leaves, stems, sheaths, glumes, and spikelet branches (Figure 1a,b). Under light‐deficient conditions, the lesion mimic phenotype did not appear or was delayed in appearance. Therefore, we reasoned that the lesion mimic phenotype of this mutant may be light‐dependent like other lesion mimic mutants. To confirm this idea, we performed a shade treatment assay. We found that leaf blades wrapped with aluminium foil or the stem wrapped by its sheath showed no cell death phenotype (Figure 1c), whereas leaf areas exposed to light and stems extended out of the sheath exhibited numerous lesions. These results showed that the formation of lesions was light‐dependent in this mutant.
FIGURE 1.
The mic mutant showed a significant necrotic phenotype. (a) Phenotypes of the whole plant. Image of wild type (WT) and mutant mic at heading stage. Necrosis can be seen on the leaf, stem, sheath, and glumes of the mic mutant. Scale bar = 10 cm. (b) Necrosis on leaf (left), panicle (middle), and sheath (right). Red arrows indicate the necrosis on stem, glumes, and spikelet branches. (c) The impact of light on the necrosis phenotype. The area between the red dotted lines indicates the wrapped portions of leaf and stem
2.2. The mic mutant showed enhanced basal defence
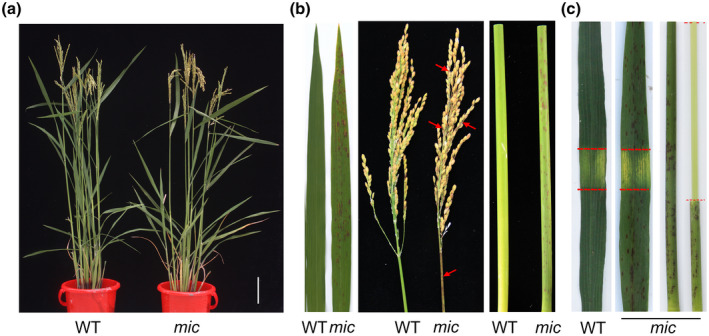
Because many of the reported lesion‐mimicking mutants exhibit spontaneous strong defence responses (Choi et al., 2019; Wang et al., 2017), we evaluated the defence response of the mic mutant to a compatible bacterial blight strain GD8988. At 15 days after inoculation, the infected part of mic plants was much shorter than that of wild‐type (WT) plants (Figure 2a). We further investigated the expression of pathogenesis‐related genes. The expression of PR1a and PR1b was elevated in the mutant; these two genes have been reported as the markers of rice responses in stress and disease (Agrawal et al., 2000). It is worth mentioning that PR1a is also commonly used as a marker gene for the salicylic acid (SA) signalling pathway (Yin et al., 2018). In line with the elevated expression of PR1a, the expression of another SA signalling pathway marker gene, PR5, was also increased in the mutant compared to WT (Choi et al., 2019). Meanwhile, PR4, a marker gene in the jasmonic acid (JA) signalling pathway, also showed significantly higher expression in the mic mutant than in the WT (Yin et al., 2018). These results indicate that the activation of SA and JA signalling pathways may contribute to the enhanced defence response of the mic mutant. PBZ1 and PR10 not only function as defence‐related genes but are also involved in the development of lesions (Kim et al., 2008; Takahashi et al., 1999). We measured the expression of PBZ1 and PR10 in the WT and mic mutant. The results showed that the expressions of PBZ1 and PR10 were dramatically up‐regulated in the leaves of the mic mutant compared to the WT (Figure 2b). Because the relative expression level of marker genes in the SA and JA pathways were elevated in the mutant, we measured total SA and JA contents using LC‐MS/MS. The results showed that the accumulated content of JA was significantly higher in the mic mutant compared to the WT, while the content of SA was comparable between the mutant and the WT (Figure 2c).
FIGURE 2.
The mic mutant showed enhanced basal defence. (a) The resistance of the mic mutant to rice bacterial blight. The flag leaves of wild type (WT) and mutant mic were inoculated with bacterial blight strain GD8988 and photographed at 15 days after inoculation. Scale bars = 1 cm. (b) The relative expression levels of PR genes. The reverse third leaves of WT and mic with necrosis were pooled as one biological replicate (n = 5). The experiments were conducted with four technical replicates and three biological replicates. The experiment was repeated twice with the similar results. (c) The total content of salicylic acid (SA) and jasmonic acid (JA). The values represent the mean ± SD of three biological replicates from one independent experiment (n = 5). (d) Trypan blue staining of leaves from WT and mutant mic. (e) 3,3′‐diaminobenzidine (DAB) staining. mic‐DAB, the leaf stained with DAB; mic‐EtOH, the leaf decoloured by ethanol. (f) H2O2 concentration in the mic mutant. FW, fresh weight. (g–k) Antioxidant enzyme activity and malondialdehyde (MDA) concentration determination. Student's t test was used to generate the p values. *p < 0.05, **p < 0.01 and ***p < 0.001 indicate the significant difference between WT and mic mutant. Data represent mean ± SD
In addition to playing significant roles in the plant resistance response, reactive oxygen species (ROS) are also critical in local lesion formation (Qiu et al., 2019; Zhu et al., 2020). We used histochemical marker trypan blue and 3,3′‐diaminobenzidine (DAB) to monitor the cellular ROS level in leaves of WT and lesion mimic mutant. The results showed that the mutant had more intense blue staining spots and DAB stained spots (Figure 2d,e). Consistent with this result, the H2O2 content was significantly elevated in the mutant compared to the WT (Figure 2f). We further investigated the activities of peroxide‐scavenging enzymes and the content of malondialdehyde (MDA) (Figure 2g–k). The activities of superoxide dismutase (SOD), ascorbate peroxidase (APX), and peroxidase (POD) were significantly increased in the mutant compared to the WT, indicating that the excessive accumulation of ROS in the lesion mimic mutant triggered the peroxide‐scavenging enzyme activity.
2.3. Genetic analysis and map‐based cloning of SDR7‐6
For genetic analysis of this lesion mimic mutant, the mutant was crossed with its WT parent Yunyin. All of the the F1 generation plants showed the WT phenotype and in the 271 F2 plants, 60 plants showed the lesion mimic phenotype, which matched the 3:1 segregation ratio (χ2 = 1.03 < 3.84). This result indicated that the lesion mimic phenotype was controlled by a single recessive gene. For map‐based cloning, the mutant was hybridized with cv. Lemont (Oryza sativa subsp. japonica) to construct the segregating population. Three hundred and forty‐nine plants with the lesion mimic phenotype were selected as a recessive population. The mutant locus was first mapped on the long arm of chromosome 7 between markers InDel 142 and InDel 144 by using 88 recessive plants. Then the locus was further narrowed down to a 26.8 kb region flanked by markers 7‐66 and 7‐84, which were designed using polymorphism between the parents, with one recombinant at each side. There are five predicted open reading frames within this region (Figure 3). Sequence analysis of the promoter and coding region of these genes revealed a single base mutation (G‐A) was present in the 821th nucleotide of Os07g0663900 (LOC_Os07g46860), resulting in a glycine (G)274 substituted by a glutamic acid (E)274. This gene was designated as SDR7‐6.
FIGURE 3.
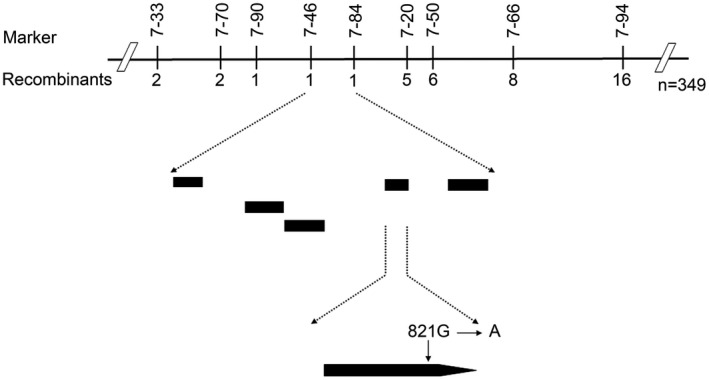
Map‐based cloning of SDR7‐6. SDR7‐6 was mapped between markers 7‐66 and 7‐84 on the long arm of chromosome 7 by using 349 recessive plants. The number below the marker means the number of F2 recombinants detected by the marker. Black squares indicate candidate genes between markers 7‐66 and 7‐84. Arrows indicate the target gene and the G to A substitution of sdr7‐6
2.4. SDR7‐6 negative regulates cell death and participates in the defence response
To confirm the mutation of SDR7‐6 is causal to the lesion mimic phenotype in the mutant, the complementary vector of pSDR7‐6::SDR7‐6, including 2164 bp upstream of the start codon, the coding region, and 517 bp downstream of the termination codon, and p35S::SDR7‐6 were constructed and transformed into mutant callus, and CRISPR/Cas9 technology was used to construct a SDR7‐6 knockout vector and transformed into the callus of the WT. The SDR7‐6 gene was highly expressed in complementation and overexpression lines, while the expression level of this gene in mic and CRISPR/Cas9 lines was comparable to the WT (Figure S1). The phenotypes of the complementation and overexpression lines showed that expression of the WT version of the SDR7‐6 gene in the mutant background fully rescued the lesion symptom of the mutant. When the WT gene was knocked out in the CRISPR/Cas9 T0 lines, the plants showed the lesion mimic phenotype (Figure 4a,b). These results confirmed that the lesion mimic phenotype is caused by the mutation in SDR7‐6. After inoculation with the compatible Xanthomonas oryzae pv. oryzae (bacterial blight) strain GD8988, the knockout lines also exhibited an elevated resistant phenotype like that of the mutant (Figure 4c,d). Meanwhile, the overexpression lines driven by the native promoter or the 35S promoter showed enhanced susceptibility to the pathogen compared to the WT (Figure 4e,f). This result further indicated that SDR7‐6 is involved in the defence response to bacterial blight.
FIGURE 4.
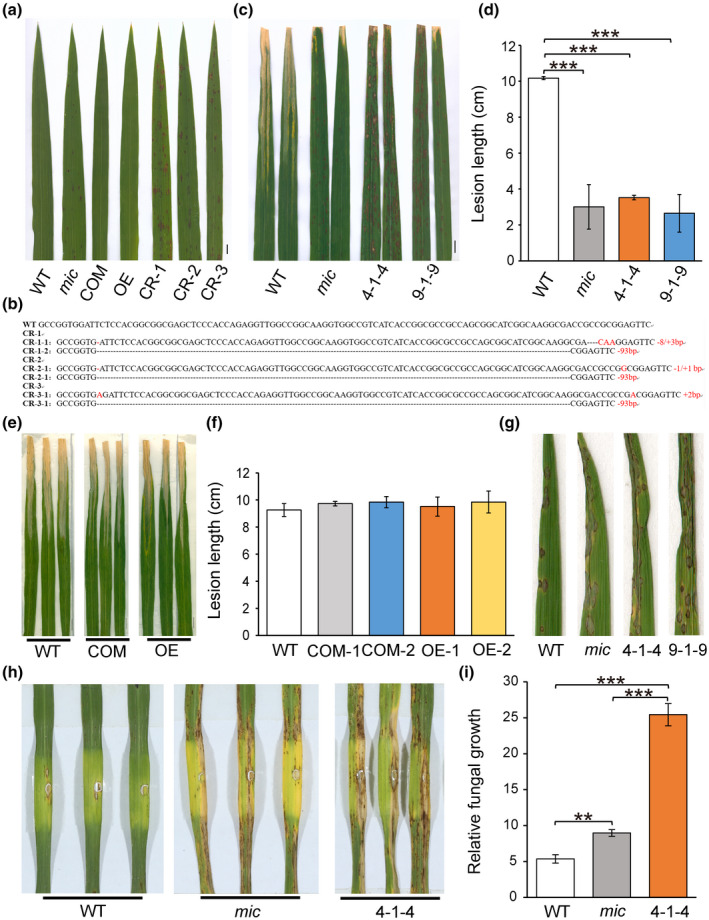
SDR7‐6 negatively regulates cell death and participates in the defence response. (a) The phenotype of complementary lines, overexpression lines, and knockout lines of SDR7‐6. COM, complementary line transformed with pSDR7‐6::SDR7‐6; OE, overexpression line transformed with p35S::SDR7‐6; CR, knockout lines using the CRISPR/Cas9 system. (b) The deleted/inserted sequences of three T0 knockout lines. WT, wild‐type sequence; CR‐1, CR‐2, CR‐3, three T0 knockout lines; CR‐1‐1, CR‐1‐2, the heterozygous sequence of CR‐1; CR‐2‐1, CR‐2‐2, the heterozygous sequence of CR‐2; CR‐3‐1, CR‐3‐2, the heterozygous sequence of CR‐3. The dotted line means deleted base. Characters in red indicate inserted bases. The number on the right is the total number of deleted/inserted bases. (c) Disease symptoms of WT, mic, and two homozygous knockout lines inoculated with Xanthomonas oryzae pv. oryzae GD8988. The leaves were photographed at 15 days after inoculation. 4‐1‐4, homozygous line of CR‐1‐1; 9‐1‐9, homozygous line of CR‐2‐1. Scale bar = 1 cm. (d) Lesion length of WT, mic, and two knockout homozygous lines. The values represent the mean ± SD of three biological replicates from one independent experiment (n = 5, ***p < 0.001, Student's t test). The experiments were repeated three times with similar results. (e) Disease symptoms of WT, complementation line, and overexpression line. Scale bar = 1 cm. (f) Lesion length of WT, two complementation lines, and two overexpression lines. The values represent the mean ± SD of three biological replicates from one independent experiment (n = 5). Student's t test was used to generate the p value. (g) Disease symptoms of WT, mic, and two homozygous knockout lines inoculated with Magnaporthe oryzae 43‐ZB‐15‐68 using the spraying method. Images were photographed at 6 days after inoculation. Two independent experiments were conducted with similar results. (h) Disease symptoms of punch inoculation with M. orzyae 43‐ZB‐15‐68. Images were taken at 9 days after inoculation. (i) Fungal biomass of punch inoculation. Data represent mean ± SD with three biological replicates. **p < 0.01, ***p < 0.001, Student's t test
Many lesion mimic mutants show an enhanced immunity response to both rice bacterial blight and rice blast disease (Qiao et al., 2010). To investigate whether the mutation of this gene results in enhanced immunity to rice blast disease, we sprayed the WT, the mutant, and two homozygous knockout lines with compatible Magnaporthe oryzae 43‐ZB‐15‐68. To our surprise, the mutant line and knockout lines showed more susceptibility to 43‐ZB‐15‐68 than the WT (Figure 4g). To confirm whether the appearance of the lesion mimic necrosis contributes to the immunity response, we used the punch inoculation method to inoculate after the lesion mimic appeared (Park et al., 2012). The punch inoculation results showed that the lesion was larger in the mutant and knockout lines compared to the WT (Figure 4h). We further calculated the relative fungal biomass (Park et al., 2012). The results showed that the relative fungal biomass was significantly greater in the mutant and knockout lines than in the WT (Figure 4i). These results show that the mutant and knockout lines demonstrate a reduced resistance to rice blast pathogen, suggesting that SDR7‐6 plays different roles in plant immunity to bacterial blight and rice blast diseases.
2.5. The mutation of SDR7‐6 has an adverse effect on agronomic traits in rice
Because many lesion mutants have unfavourable side effects on plant growth, we investigated the agronomic traits of the WT and two knockout lines (9‐1‐9, 4‐1‐4) to evaluate whether the mutation of SDR7‐6 had an impact on the yield. Under field conditions, the grain length of the two knockout lines did not show significant differences from the WT. However, the grain width and thickness of two knockout lines were significantly lower than in the WT (Figure 5a–c). The setting rate of the two knockout lines was 81% of that in the WT (Figure 5d). The 1000‐grain weight, grain weight per plant, panicle length, and plant height were also reduced as compared to the WT (Figure 5e–h). These results show that mutation of SDR7‐6 has adverse effects on agronomic traits including plant height, grain yield, and grain width and thickness.
FIGURE 5.
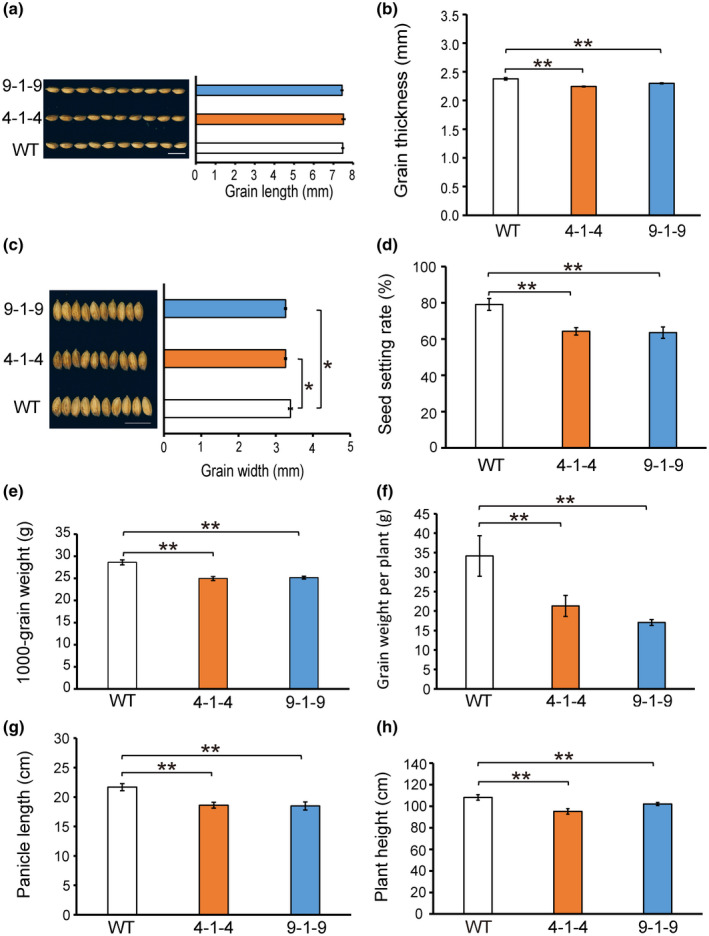
Agronomic traits of wild type (WT) and two knockout lines. (a–c) Phenotypic data for the grain length, grain thickness, and grain width of the WT and two knockout lines (n = 30). The data were measured with three individual plants. (d–h) Phenotypic data of seed setting rate, 1000‐grain weight, grain weight per plant, panicle length, and plant height (n = 5). Student's t test was used to generate the p values. *p < 0.05, **p < 0.01 indicate significant differences. Data represent mean ± SD. Scale bars = 1 cm
Under field conditions, we found that the density of the necrotic lesions in the mic mutant were less than in the CRISPR/Cas9 lines (Figure S2a). As in many lesion mimic mutants, the spots can lead to a reduction in photosynthetic pigments (Ma et al., 2019; Zhu et al., 2016). We then determined the content of chlorophyll in WT, mic and two CRISPR/Cas9 lines with the lesion mimic phenotype. The chlorophyll content was decreased in mic and CRISPR/Cas9 lines compared to the WT. In addition, the chlorophyll content was less in the CRISPR/Cas9 lines than in the mic mutant (Figure S2b). The same phenotype also occurred at the seedling stage. Twenty‐five days after sowing, the bottom leaves of CRISPR/Cas9 lines were withered or had small lesions while the mutant had no cell death symptom (Figure S2c,d). This situation might be because this gene in the mutant merely has a G to A substitution with a single amino acid change while in the CRISPR/Cas9 strain, 4‐1‐4 is a −8 bp/+3 bp mutation and 9‐1‐9 is a −1 bp/+1 bp mutation (Figure 4b), both of which result in frame‐shift mutation. Therefore, we speculated that the mutated sdr7‐6 might still be partially functional.
2.6. SDR7‐6 encodes an atypical short‐chain alcohol dehydrogenase/reductase protein and is polymerized as a homomultimer
The open reading frame of SDR7‐6 is predicted to encode a short‐chain dehydrogenase/reductase family protein of 288 amino acids in length according to the rice annotation project database. According to Persson and Kallberg's (2013) nomenclature methods (http://www.sdr‐enzymes.org/), SDR7‐6 encodes a short‐chain alcohol dehydrogenase belonging to the SDR110C “classical” subfamily. The protein contains a conserved TGxxxGxG coenzyme binding site and an Asp residue around position 60, preferring NAD(H) as cofactor over NADP(H) (Kitaoka et al., 2016; Tanaka et al., 2001). However, YxxxK, a typical conserved active site in this family, is replaced by HxxxK in SDR7‐6 (Figure S3). Comparing the SDR7‐6 sequence with the other 22 SDR110C in rice (Moummou et al., 2012), we found that this atypical active site only exists in SDR7‐6 (Figure S4). We then constructed a phylogenetic tree of all the rice SDR110C subfamily, in which SDR7‐6 resided in the same clade of OsSDR110C‐MI3 and OsSDR110C‐MI4 (Figure S5). OsSDR110C‐MI3 has catalytic activity for the synthesis of labdane‐related diterpenoid phytoalexins, while the function of OsSDR110C‐MI4 has not been determined (Kitaoka et al., 2016; Miyamoto et al., 2016). Comparison of the amino acid sequence of SDR7‐6 against the National Center for Biotechnology Information (NCBI) predicted it to be a momilactone A synthase‐like protein. In addition, SDR7‐6 was in the same clade as OsSDR110C‐MI3, which could also convert 3β‐hydroxy‐syn‐pimaradien‐19,6β‐olide to momilactone A. We conducted an enzyme activity determination assay using OsMAS as a positive control. The results showed that OsMAS could convert 3β‐hydroxy‐syn‐pimaradien‐19,6β‐olide to momilactone A, while SDR7‐6 and sdr7‐6 could not catalyse this reaction (Figure S6).
Because most SDRs form homomultimer structures, we used yeast two‐hybrid (Y2H) and co‐immunoprecipitation (Co‐IP) assays to test whether SDR7‐6 or sdr7‐6 could interact with itself. The full lengths of SDR7‐6 and sdr7‐6 were used to construct bait and prey proteins. Interactions were examined using SD medium lacking Ade, His, Leu, and Trp. The results revealed that SDR7‐6 or sdr7‐6 could form a homocomplex (Figure 6a). To validate the interactions, we fused SDR7‐6 and sdr7‐6 with green fluorescent protein (GFP) and FLAG tags. The Co‐IP result was consistent with the Y2H assays. In addition, we noticed that the sdr7‐6 had slower migrating bands than SDR7‐6 in SDS‐PAGE (Figure 6b). To further examine the homomultimer formation, we separated the proteins by native PAGE. Full‐length SDR7‐6 or sdr7‐6 was inserted into pCAMIA2300‐3FLAG vector digested with SalⅠ and then transformed into rice protoplasts. The results showed that SDR7‐6 or sdr7‐6 formed homomultimers. Meanwhile, sdr7‐6 had slower electrophoretic mobility than SDR7‐6, which was in line with the Co‐IP result (Figure 6c). These results indicate that SDR7‐6 can form a homomultimer. The mutation of (G)274 to (E)274 in sdr7‐6 did not affect the ability to form a homomultimer but influenced its electrophoretic mobility.
FIGURE 6.
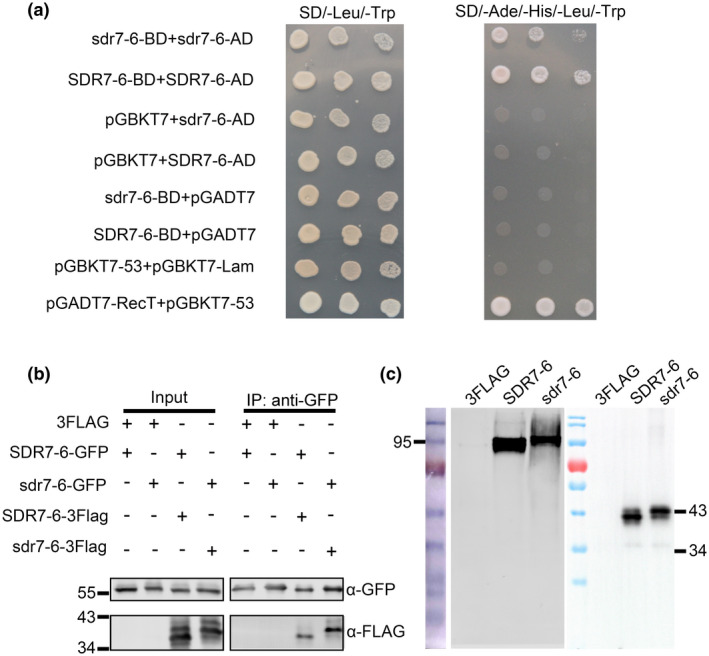
SDR7‐6 and sdr7‐6 can form a homomultimer. (a) Interaction between SDR7‐6 and sdr7‐6 in yeast. The full lengths of SDR7‐6 and sdr7‐6 were inserted into pGADT7 and pGBKT7 to construct SDR7‐6‐AD, SDR7‐6‐BD, sdr7‐6‐AD, and sdr7‐6‐BD. (b) Interaction between SDR7‐6 and sdr7‐6 in a Co‐IP assay. SDR7‐6‐3FLAG mixed with SDR7‐6‐GFP and sdr7‐6‐3FLAG mixed with sdr7‐6‐GFP were transformed into rice protoplasts. Total proteins were extracted and samples were immunoprecipitated with anti‐GFP antibody. Input proteins and immunoprecipitated proteins were blotted with corresponding anti‐FLAG or anti‐GFP antibodies. The numbers on the left represent molecular weights. (c) Verification of the homomultimer between SDR7‐6 and sdr7‐6 by native PAGE. The left gel is native PAGE; the right gel is SDS‐PAGE. 3FLAG, pCAMBIA2300‐3FLAG; SDR7‐6, SDR7‐6‐3FLAG; sdr7‐6, sdr7‐6‐3FLAG. The numbers represent protein molecular weights (kDa)
2.7. SDR7‐6 was localized at the endoplasmic reticulum
To investigate the subcellular localization of SDR7‐6 and whether the mutation of sdr7‐6 would affect its localization, we generated GFP fusion proteins p35S::SDR7‐6‐GFP and p35S::sdr7‐6‐GFP. We transiently expressed them with endoplasmic reticulum (ER) marker mCherry‐HDEL in rice protoplasts. The fluorescent signals were observed under confocal microscopy. The results showed that both SDR7‐6‐GFP and sdr7‐6‐GFP were colocalized with the ER marker (Figure 7). The mutation of the glycine (G)274 to the glutamic acid (E)274 did not affect the subcellular localization of sdr7‐6.
FIGURE 7.
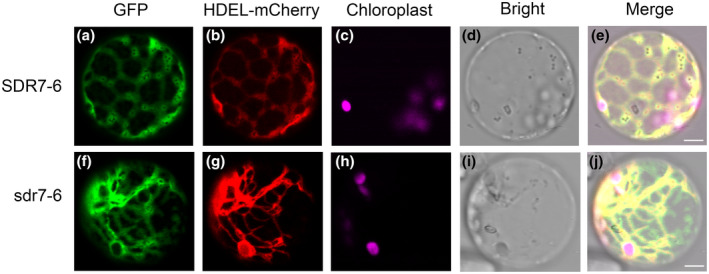
SDR7‐6 and sdr7‐6 localized in the endoplasmic reticulum (ER). SDR7‐6‐GFP or sdr7‐6‐GFP was cotransformed with mCherry‐HDEL in rice protoplasts. mCherry‐HDEL is the ER marker. Merge is the combination image of GFP, mCherry, and chloroplast fluorescence. Scale bar = 8 µm
3. DISCUSSION
3.1. SDR7‐6 is an SDR110C with undefined function
In this study, we isolated a new lesion mimic gene SDR7‐6, which encodes a short‐chain alcohol dehydrogenase/reductase family protein. In classical SDRs, catalytic activity relies on the highly conserved catalytic tetrad N‐S‐YxxxK. The Tyr and Lys residues in this tetrad are crucial for coenzyme binding. In addition, the tyrosine residue is the central acid‐base catalyst that donates or accepts a proton to or from the substrate (Buysschaert et al., 2013). Although SDR7‐6 belongs to the SDR110C family, it has an atypical active site HxxxK. In recent years, some atypical active sites have been identified in SDRs that lose their enzyme activity but play a role in RNA metabolism and transcriptional regulation (Link et al., 2012). For example, MxxxK in NmrA, PglF and CapE (Miyafusa et al., 2013; Riegert et al., 2017; Stammers et al., 2001), LxxxK in HCF244 (Link et al., 2012), TxxxR in Q9HYA2 (Huether et al., 2010), VxxxK in ROQH1 (Amstutz et al., 2020), and CSP41 and SDRvv, which lack the catalytic tetrad (Baker et al., 1998; Buysschaert et al., 2013). In PglF, MxxxK does not act as a catalytic base like YxxxK; it is Thr 395 and Asp 396 that function together to remove the proton (Riegert et al., 2017). NmrA, devoid of conserved catalytic tyrosine, shows a nonenzymatic function in transcriptional regulation (Stammers et al., 2001). HCF244 is essential for Arabidopsis psbA mRNA translational initiation and stabilization (Link et al., 2012). In this study, although SDR7‐6 falls into the SDR110C family, it has an atypical active site HxxxK different from the other 22 SDR110C family members in rice. Phylogenetic analysis of all rice SDR110C family members showed that SDR7‐6 resides in the same clade as OsSDR110C‐MI3 and OsSDR110C‐MI4. Although OsSDR110C‐MI3 has weaker activity to convert 3β‐hydroxy‐syn‐pimaradien‐19,6β‐olide to momilactone A than OsMAS1 and OsMAS2, it is reported to be an oxidase of 3β‐hydroxy‐syn‐pimaradien‐19,6β‐olide and oryzalexin D (Kitaoka et al., 2016; Miyamoto et al., 2016). We found that neither SDR7‐6 nor sdr7‐6 were able to catalyse the conversion of 3β‐hydroxy‐syn‐pimaradien‐19,6β‐olide to momilatone A. Therefore, SDR7‐6 may have different functions to OsSDR110C‐MI3. The catalytic activity and mechanism, substrates, and the upstream regulator of SDR7‐6 remain to be elucidated. In our study, we also observed that a single base substitution in sdr7‐6 could slow down the protein's electrophoretic mobility, but did not affect homomultimer formation and subcellular localization. When GST‐tagged SDR7‐6 and sdr7‐6 were expressed in Escherichia coli BL21, the electrophoretic mobility of these two proteins were almost the same (unpublished results), so we hypothesize that SDR7‐6 might be posttranslationally modified in rice. The mutated amino acid might affect this posttranslational modification in SDR7‐6. Because the density of necrosis of CRISPR/Cas9 lines were higher than that in the mic mutant, we deduced that sdr7‐6 mutant protein may still have partial function. The reason for the slower electrophoretic mobility of sdr7‐6 remains to be elucidated.
3.2. SDR7‐6 negatively regulates cell death and participates in the defence response
Complementation assays revealed that the expression of SDR7‐6 in the mutant could rescue the lesion mimic phenotype and knockout of SDR7‐6 in the WT could cause the necrosis phenotype. The knockout lines also showed increased resistance to bacterial blight. These results indicated that impairment of SDR7‐6 could lead to cell death and a spontaneous strong defence response to rice bacterial blight. ROS are important for initiation of local lesion formation, which can be erased by treatment with exogenous antioxidants (Qiu et al., 2019). In addition, ROS also act as a signal to activate defence and acclimation mechanisms to abiotic stresses. In this study, by staining and measuring the content of H2O2, ROS was confirmed to accumulate more in the mic mutant than in the WT. Because the antioxidant system strictly regulates the ROS content, we measured the antioxidant enzyme activities. SOD, POD, and APX activities were significantly elevated in the mutant. Although H2O2 accumulates in many lesion mimic mutants, the activity of antioxidant enzymes varies in different lesion mimic mutants. For example, in spl36, the activities of POD and SOD are reduced (Rao et al., 2021). In spl40, the activities of CAT, SOD, and POD increase (Sathe et al., 2019). This implies that the ROS scavenging system may be regulated differently in different lesion mimic mutants. Whether the spontaneous necrosis formation and activation of defence response in mic mutant were due to excessive ROS accumulation remains to be elucidated.
The up‐regulated expression of PR5, PR10, and PBZ1 is a common feature of the lesion mimic mutants (Chen et al., 2013). PBZ1 and PR10 not only function as defence‐related genes, but their expression is also correlated with development of lesions (Kim et al., 2008; Takahashi et al., 1999). In our study, we observed the expressions of PR5, PR10, and PBZ1 were elevated in the mutant with a lesion mimic phenotype. The expressions of PR1a, PR1b, and PR4 were also elevated in the mutant. Because PR1a and PR5 are marker genes in the SA signalling pathway and PR4 in the JA signalling pathway, we deduced that the SA and JA pathways may be involved in the spontaneous activation of defence response in the mic mutant. In many lesion mimic mutants, the SA and JA pathways have been proved to participate in the formation of the lesion and defence response (Beaugelin et al., 2019; Miao et al., 2019). To our surprise, by measuring the total contents of SA and JA, we found that JA accumulated significantly, while SA did not, in the mutant. In lesion mimic mutants, the necrosis formation can be SA‐dependent or SA‐independent (Miao et al., 2019). In ssn and GbTRP1 knockout lines, although the relative expression of marker pathogenicity‐related (PR) genes for the SA response is up‐regulated, the content of SA is not elevated (Miao et al., 2019; Sun et al., 2014). The lesion mimic formation and up‐regulated expression of the PR genes in the mic mutant may be SA‐independent or the SA signalling may be induced but not the level of SA due to the higher basal level of SA in rice (Yin et al., 2018).
Studying lesion mimic mutants sheds light on their relationship to the defence response. Some mutants increase resistance to multiple pathogens, while others are resistant to certain diseases or pathogen races (Qiao et al., 2010). SPL11 interacts with SPIN6 to transduce the defence signal to OsRac1 (Liu et al., 2015). Disruption of SPL11 displays nonrace‐specific resistance to both rice blast disease and rice bacterial blight (Zeng et al., 2004), while cdr1, cdr2, and Cdr3 are only resistant to a single blast isolate (Takahashi et al., 1999). Both OsSPL7 overexpression and knockout plants might enhance resistance to rice blast disease and rice bacterial blight through OsWRKY45 overexpression (Hoang et al., 2019). MoSM1‐OE plants confer broad‐spectrum resistance against rice blast and rice bacterial blight disease, but not increased resistance to sheath blight disease (Hong et al., 2017). In this study, the mic mutant showed increased resistance to rice bacterial blight but a reduced defence response to rice blast. This indicates that SDR7‐6 might function at the interface of the two defence pathways of bacterial blight and rice blast disease in rice. How SDR7‐6 regulates the defence response remains to be studied.
4. EXPERIMENTAL PROCEDURES
4.1. Plant materials and growth conditions
Yunyin rice is a japonica cultivar and is a landrace of Yunnan province. The mic mutant was isolated from the WT treated with EMS and the necrosis phenotype was stably inherited. Lemont is a japonica cultivar in the USA. All plants were grown under natural paddy fields in Fuzhou city in the Fujian province of China and in Sanya city in the Hainan province of China.
4.2. Shading treatment
To test whether the lesion mimic phenotype was light‐dependent, a shading assay was performed. Before necrosis was initiated, the mutant leaves were wrapped with 3‐cm wide aluminium foil. The phenotypes in the wrapped areas of the leaf and stem were observed when lesion spots appeared in the unwrapped parts.
4.3. Pathogen inoculation
Xanthomonas oryzae pv. oryzae GD8988 was used in this study. For the leaf blight test, GD8988 was cultivated on YEB medium (sucrose 5 g, tryptone 10 g, yeast extract 5 g, MgSO4.7H2O, agar 8 g per litre) at 28℃ for 3 days, then scraped with sterile water to OD600 = 0.5. The fully expanded leaves were inoculated by the leaf‐clipping method and investigated 15 days after inoculation. For the rice blast disease resistance assay, M. oryzae 43‐ZB‐15‐68 was used. Spraying, punch inoculation, and relative fungal biomass calculation were conducted according to Park et al. (2012). Three punched leaves were pooled as one biological replicate to calculate the relative fungal growth (Park et al., 2012).
4.4. Gene expression analysis
The reverse third leaves of five plants of the WT and the mutant with the spotted phenotype were pooled as one biological repeat and ground in the liquid nitrogen. Total RNA was extracted using TransZol Up reagent (Transgen). One microgram of RNA was used for first‐strand cDNA synthesis using the ReverTra Ace qPCR RT Master Mix with gDNA Remover kit (FSQ‐301; Toyobo). Reverse transcription‐quantitative PCR (RT‐qPCR) was performed with four technical replicates and three biological replicates using FastStart Universal SYBR Green Master (ROX) reagent (Roche) on QuanStudio 6 Flex (Thermo). Relative expressions were calculated using the 2−Δ C t method. The primer sequences are listed in Table S1.
4.5. Histochemical assay, H2O2, and antioxidant enzyme activity measurement
Fresh leaves of WT and the mutant with necrosis were selected in the staining experiments. The trypan blue and DAB staining was performed as described by Qiao et al. (2010). The content of H2O2, MDA, and antioxidant enzyme activities were measured with three biological replicates following the corresponding assay kit instructions (Solarbio).
4.6. Population construction and map‐based cloning
To construct the genetic analysis population, the mutant was crossed with the WT. The ratio of lesion mimic phenotype plants was calculated in the F2 population and analysed using the χ2 test. To construct the mapping population, the mutant was crossed with cv. Lemont (O. sativa subsp. japonica), 10 WT individuals and 10 lesion mimic phenotype individuals were pooled, then bulked‐segregant analysis was carried out. Plants with the lesion mimic phenotype were applied to recessive class analysis (Zhang et al., 1994). Two hundred and thirteen InDels, designed by the differential between 9311 and Nipponbare, almost evenly distributed on the 12 chromosomes, were selected to screen the differential between the parents. The polymorphism primers were screened for further linkage analysis. To fine‐map the causal gene, the InDels were designed based on the polymorphism between the japonica and indica sequence (http://www.gramene.org) and the polymorphism markers between the parents were designed by using Primer3 (Koressaar & Remm, 2007; Untergasser et al., 2012). The primers in Figure 3 are listed in Table S1.
4.7. Complementation test
To confirm the putative causal gene, pSDR7‐6::SDR7‐6 and p35S::SDR7‐6 vectors were constructed. The full length of SDR7‐6, including 2164 bp upstream of the start codon, the coding region, and 517 bp downstream of the termination codon, was amplified and inserted into pCAMBIA1300 digested with EcoRⅠ and BamHⅠ to construct pSDR7‐6::SDR7‐6. The coding region of SDR7‐6 was amplified and inserted into pCAMBIA1302 digested with NcoI and BstEII to construct p35S::SDR7‐6. Then the vectors were transformed into mutant calli by the Agrobacterium‐mediated method. The primers for constructs of pSDR7‐6::SDR7‐6 and p35S::SDR7‐6 are listed in Table S1.
4.8. Knockout vector construction
SDR7‐6 knockout lines were obtained using the CRISPR/Cas9 system (Xing et al., 2014). Two 20‐bp sequences (5′‐GTTCCTGATGAACTC CGCGG‐3′ and 5′‐GTGGATTCTCCACGGCGGCG‐3′) were selected by searching the target sequence on CRISPR‐PLANT (https://www.genome.arizona.edu/crispr/CRISPRsearch.html). After self‐crossing, two homozygous genotype lines (4‐1‐4 and 9‐1‐9) with different deletions were selected for further study. The primers SDR7‐6‐CR for construction of the CRISPR/Cas9 lines are listed in Table S1.
4.9. Pigment measurement
To measure the content of chlorophyll, the reverse third leaf of the WT and the mutant with lesion mimic phenotype were detached. The leaves were cut into pieces then soaked in 10 ml of ice‐cold 80% acetone and extracted in the dark at 4℃ for 36 h to decolour the leaf. Chlorophyll a was measured at 662 nm and chlorophyll b was measured at 646 nm by the spectrophotometer (MULTISKAN GO; Thermo Scientific) (Arnon, 1949). The measure was conducted in three technical replicates and three biological replicates.
4.10. Agronomic traits measurement
The WT and two knockout lines (4‐1‐4 and 9‐1‐9) were grown in a paddy field at Sanya city, Hainan province. Seed setting rate, 1000‐grain weight, weight per plant, panicle length, and plant height were measured for five individual plants. Grain length, grain width, and grain thickness were calculated for 30 grains of three individual plants.
4.11. Subcellular localization of SDR7‐6 and sdr7‐6
To determine the subcellular localization of SDR7‐6 and sdr7‐6, the coding regions of SDR7‐6 and sdr7‐6 without a stop codon were amplified and inserted into pBIN: GFP plasmid digested with SalⅠ. The constructed vectors were cotransformed with an ER marker (mCherry‐HDEL, Nelson et al., 2007) into the rice protoplast (Bart et al., 2006). After incubation at 28℃ for 16 h, fluorescence was detected under confocal laser scanning microscopy (DMi8; Leica).
4.12. Y2H assay
The coding region of SDR7‐6 and sdr7‐6 was cloned into pGBKT7 and pGADT7 vectors digested with EcoRⅠ separately to construct bait plasmids and prey plasmids. The Y2H assay was performed according to the Yeast Protocols Handbook (Clontech Laboratories, Inc.).
4.13. Co‐immunoprecipitation assay
For homomultimer test, the coding region of SDR7‐6 and sdr7‐6 was amplified and inserted into pCAMBIA2300‐3FLAG vector digested with SalI to construct SDR7‐6‐3FLAG and sdr7‐6‐3FLAG vectors. SDR7‐6‐3FLAG was cotransformed with p35S::SDR7‐6‐GFP into rice protoplasts. The sdr7‐6‐3FLAG was cotransformed with p35S::sdr7‐6‐GFP into rice protoplasts. The total proteins were extracted with extraction buffer (50 mM Tris‐HCl pH7.5, 150 mM NaCl, 0.1% NP‐40, 5 mM MgCl2, 2 mM CaCl2, 10% vol/vol glycerol) and separated by 10% SDS‐PAGE. Anti‐GFP antibodies were obtained from Abmart (M20004m); anti‐FLAG antibodies were obtained from Proteintech (20543‐1‐AP). Primers are listed in Table S1.
4.14. Phylogenetic analysis of rice SDR110C
According to Moummou et al. (2012), except for LOC_Os07g46900, which cannot be achieved, the amino acid sequences of the other 23 rice SDR110C were retrieved from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/index.shtml) and aligned by ClustalX v. 1.83. The neighbour‐joining tree of rice SDR110C was constructed using MEGA 4 with a bootstrap value of 1000 replicates.
4.15. Enzyme asssay
The full lengths of OsMAS, SDR7‐6, and sdr7‐6 were subcloned into pGEX‐6P‐1 to construct recombinant proteins tagged with glutathione S‐transferase (GST). The vectors were transformed into E. coli BL21. As a control, pGEX‐6P‐1 was transformed into E. coli BL21. The recombinant proteins and GST control were purified using glutathione sepharose 4B according to the manufacturer's instructions (GE Healthcare) and confirmed by western blot. The enzyme assay was conducted according to Shimura et al. (2007) using 3‐hydroxy‐9H‐pimara‐7,15‐dien‐19,6‐olide as a substrate.
Two microlitres of each sample of the solution and momilactone A authentic sample were subjected to high performance liquid chromatography (HPLC) using a Xevo TQ‐S mass spectrometer (Waters). Liquid chromatographic separation of the analytes was achieved with an ACQUITY UPLC HSS T3 column (1.8 µm, 2.1 × 100 mm; Waters Co.) with a binary gradient of 5 mM ammonium acetate including 0.1% aqueous formic (solvent A) and 100% MeCN (solvent B) with a flow rate of 0.4 ml/min at 40°C as follows: 80% vol/vol A (0 min), 30% vol/vol A (2 min), 5% vol/vol A (3 min), 5% vol/vol A (5.8 min), 80% vol/vol A (6 min), 80% vol/vol A (8 min). Samples were analysed by ESI‐MS/MS in the MRM mode with 8 min retention time. The electrospray capillary was set at 3.59 kV and the source temperature was 600℃. The products were determined with m/z combinations (precursor/product ions) of 315/271 for momilactone A.
4.16. Detection of phytohormones
The reversed third leaves of five different plants with necrotic phenotype were pooled as one biological replicate. Phytohormones contents were detected by MetWare (http://www.metware.cn/) based on the AB Sciex QTRAP 6500 LC‐MS/MS platform.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
ACKNOWLEDGEMENTS
The work was supported by the Special Foundation of Non‐Profit Research Institutes of Fujian Province (no. 2018R1021‐10) and the Youth Technology Innovation Team of the Fujian Academy of Agricultural Sciences (no. STIT2017‐3‐3). The bacterial blight strain was kindly provided by Xiaoyun Zhu from the Guangdong Academy of Agricultural Sciences, China. Plasmids for CRISPR/Cas9 were kindly provided by Qijun Chen from China Agricultural University, China. 3‐hydroxy‐9H‐pimara‐7,15‐dien‐19,6‐olide and momilactone A were kindly provided by Morifumi Hasegawa from Ibaraki University, Japan.
Zheng, Y. , Zhu, Y. , Mao, X. , Jiang, M. , Wei, Y. , Lian, L. et al. (2022) SDR7‐6, a short‐chain alcohol dehydrogenase/reductase family protein, regulates light‐dependent cell death and defence responses in rice. Molecular Plant Pathology, 23, 78–91. 10.1111/mpp.13144
Contributor Information
Guodong Lu, Email: lgd@fafu.edu.cn.
Jianfu Zhang, Email: jianfzhang@163.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Agrawal, G.K. , Rakwal, R. & Jwa, N.S. (2000) Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signals. Biochemical and Biophysical Research Communications, 278, 290–298. [DOI] [PubMed] [Google Scholar]
- Amstutz, C.L. , Fristedt, R. , Schultink, A. , Merchant, S. , Niyogi, K.K. , Malnoë, A. et al. (2020) An atypical short‐chain dehydrogenase‐reductase functions in the relaxation of photoprotective qH in Arabidopsis . Nature Plants, 6, 154–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon, D.I. (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris . Plant Physiology, 24, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atawong, A. , Hasegawa, M. & Kodama, O. (2002) Biosynthesis of rice phytoalexin: enzymatic conversion of 3β‐hydroxy‐9β‐pimara‐7,15‐dien‐19,6β‐olide to momilactone A. Bioscience Biotechnology and Biochemistry, 66, 566–570. [DOI] [PubMed] [Google Scholar]
- Baker, M.E. , Grundy, W.N. & Elkan, C.P. (1998) Spinach CSP41, an mRNA‐binding protein and ribonuclease, is homologous to nucleotide‐sugar epimerases and hydroxysteroid dehydrogenases. Biochemical and Biochemical and Biophysical Research Communications, 248, 250–254. [DOI] [PubMed] [Google Scholar]
- Bart, R. , Chern, M. , Park, C.J. , Bartley, L. & Ronald, P.C. (2006) A novel system for gene silencing using siRNAs in rice leaf and stem‐derived protoplasts. Plant Methods, 2, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaugelin, I. , Chevalier, A. , D'Alessandro, S. , Ksas, B. , Novák, O. , Strnad, M. et al. (2019) OXI1 and DAD regulate light‐induced cell death antagonistically through jasmonate and salicylate levels. Plant Physiology, 180, 1691–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman, Q. , Raynaud, C. , Benhamed, M. & Delarue, M. (2015) To die or not to die? Lessons from lesion mimic mutants. Frontiers in Plant Science, 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buysschaert, G. , Verstraete, K. , Savvides, S.N. & Vergauwen, B. (2013) Structural and biochemical characterization of an atypical short‐chain dehydrogenase/reductase reveals an unusual cofactor preference. The FEBS Journal, 280, 1358–1370. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Fu, S. , Zhang, P. , Gu, Z. , Liu, J. , Qian, Q. et al. (2013) Proteomic analysis of a disease‐resistance enhanced lesion mimic mutant spotted leaf 5 in rice. Rice, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Strickler, S.R. & Richards, E.J. (2019) Loss of CRWN nuclear proteins induces cell death and salicylic acid defense signaling. Plant Physiology, 179, 1315–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong, A. , Calderon‐Urrea, A. & Dellaporta, S.L. (1993) Sex determination gene TASSELSEED2 of maize encodes a short‐chain alcohol dehydrogenase required for stage‐specific floral organ abortion. Cell, 74, 757–768. [DOI] [PubMed] [Google Scholar]
- Endo, A. , Nelson, K.M. , Thoms, K. , Abrams, S.R. , Nambara, E. , Sato, Y. et al. (2014) Functional characterization of xanthoxin dehydrogenase in rice. Journal of Plant Physiology, 171, 1231–1240. [DOI] [PubMed] [Google Scholar]
- Hoang, T.V. , Vo, K.T.X. , Rahman, M.M. , Choi, S.H. & Jeon, J.S. (2019) Heat stress transcription factor OsSPL7 plays a critical role in reactive oxygen species balance and stress responses in rice. Plant Science, 289, 110273. [DOI] [PubMed] [Google Scholar]
- Hong, Y. , Yang, Y. , Zhang, H. , Huang, L. , Li, D. & Song, F. (2017) Overexpression of MoSM1, encoding for an immunity‐inducing protein from Magnaporthe oryzae, in rice confers broad‐spectrum resistance against fungal and bacterial diseases. Scientific Reports, 7, 41037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huether, R. , Mao, Q.L. , Duax, W.L. & Umland, T.C. (2010) The short‐chain oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 contains an atypical catalytic center. Protein Science, 19, 1097–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg, Y. , Oppermann, U. , Jörnvall, H. & Persson, B. (2002) Short‐chain dehydrogenase/reductase (SDR) relationshhips: a large family with eight clusters common to human, animal, and plant genomes. Protein Science, 11, 636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh, K.L. , Jörnvall, H. , Persson, B. & Oppermann, U. (2008) Medium‐ and short‐chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cellular and Molecular Life Sciences, 65, 3895–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, S. , Liu, S. , Luan, X. , Xie, X.M. , Hsieh, T.F. & Zhang, X.Q. (2019) Mutation in a putative glycosyltransferase‐like gene causes programmed cell death and early leaf senescence in rice. Rice, 12, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.T. , Kim, S.G. , Kang, Y.H. , Wang, Y. , Kim, J.Y. , Rakwal, R. et al. (2008) Proteomics analysis of rice lesion mimic mutant (spl1) reveals tightly localized probenazole‐induced protein (PBZ1) in cells undergoing programmed cell death. Journal of Proteome Research, 7, 1750–1760. [DOI] [PubMed] [Google Scholar]
- Kitaoka, N. , Wu, Y.S. , Zi, J.C. & Peters, R.J. (2016) Investigating inducible short‐chain alcohol dehydrogenases reductases clarifies rice oryzalexin biosynthesis. The Plant Journal, 88, 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar, T. & Remm, M. (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics, 23, 1289–1291. [DOI] [PubMed] [Google Scholar]
- Li, W. , Deng, Y.W. , Ning, Y.S. , He, Z.H. & Wang, G.L. (2020) Exploiting broad‐spectrum disease resistance in crops: from molecular dissection to breeding. Annual Review of Plant Biology, 71, 575–603. [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Bai, Q. , Xu, P. , Wu, T. , Guo, D. , Peng, Y. et al. (2018) Mutation in rice abscisic acid2 results in cell death, enhanced disease‐resistance, altered seed dormancy and development. Frontiers in Plant Science, 9, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, S. , Engelmann, K. , Meierhoff, K. & Westhoff, P. (2012) The atypical short‐chain dehydrogenases HCF173 and HCF244 are jointly involved in translational initiation of the psbA mRNA of Arabidopsis . Plant Physiology, 160, 2202–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Park, C.H. , He, F. , Nagano, M. , Wang, M.O. , Bellizzi, M. et al. (2015) The RhoGAP SPIN6 associates with SPL11 and OsRac1 and negatively regulates programmed cell death and innate immunity in rice. PLoS Pathogens, 11, e1004629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain, S. , Vailleau, F. , Balague, C. & Roby, D. (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends in Plant Science, 8, 263–271. [DOI] [PubMed] [Google Scholar]
- Ma, J. , Wang, Y. , Ma, X. , Meng, L. , Jing, R. , Wang, F. et al. (2019) Disruption of gene SPL35, encoding a novel CUE domain‐containing protein, leads to cell death and enhanced disease response in rice. Plant Biotechnology Journal, 17, 1679–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, Y.H. , Xu, L. , He, X. , Zhang, L. , Shaban, M. , Zhang, X.L. et al. (2019) Suppression of tryptophan synthase activates cotton immunity by triggering cell death via promoting SA synthesis. The Plant Journal, 98, 329–345. [DOI] [PubMed] [Google Scholar]
- Miyafusa, T. , Caaveiro, J.M. , Tanaka, Y. , Tanner, M.E. & Tsumoto, K. (2013) Crystal structure of the capsular polysaccharide synthesizing protein CapE of Staphylococcus aureus . Bioscience Reports, 33, e00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, K. , Fujita, M. , Shenton, M.R. , Akashi, S. , Sugawara, C. , Sakai, A. et al. (2016) Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice. The Plant Journal, 87, 293–304. [DOI] [PubMed] [Google Scholar]
- Moummou, H. , Kallberg, Y. , Tonfack, L.B. , Persson, B. & Van Der Rest, B. (2012) The plant short‐chain dehydrogenase (SDR) superfamily: genome‐wide inventory and diversification patterns. BMC Plant Biology, 12, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B.K. , Cai, X. & Nebenführ, A. (2007) A multicolored set of in vivo organelle markers for co‐localization studies in Arabidopsis and other plants. The Plant Journal, 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nguyen, T.D. , Moon, S. , Nguyen, V.N.T. , Gho, Y. , Chandran, A.K.N. , Soh, M.S. et al. (2016) Genome‐wide identification and analysis of rice genes preferentially expressed in pollen at an early developmental stage. Plant Molecular Biology, 92, 71–88. [DOI] [PubMed] [Google Scholar]
- Park, C.H. , Chen, S. , Shirsekar, G. , Zhou, B. , Chang, H.K. , Songkumarn, P. et al. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. The Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, B. & Kallberg, Y. (2013) Classification and nomenclature of the superfamily of short‐chain dehydrogenases/reductases (SDRs). Chemico‐Biological Interactions, 202, 111–115. [DOI] [PubMed] [Google Scholar]
- Persson, B. , Kallberg, Y. , Bray, J.E. , Bruford, E. , Dellaporta, S.L. , Favia, A.D. et al. (2009) The SDR (short‐chain dehydrogenase/reductase and related enzymes) nomenclature initiative. Chemico‐Biological Interactions, 178, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Y. , Jiang, W. , Lee, J. , Park, B. , Choi, M.S. , Piao, R. et al. (2010) SPL28 encodes a clathrin‐associated adaptor protein complex 1, medium subunit μ1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytologist, 185, 258–274. [DOI] [PubMed] [Google Scholar]
- Qiu, Z. , Zhu, L. , He, L. , Chen, D. , Zeng, D. , Chen, G. et al. (2019) DNA damage and reactive oxygen species cause cell death in the rice local lesions 1 mutant under high light and high temperature. New Phytologist, 222, 349–365. [DOI] [PubMed] [Google Scholar]
- Rao, Y. , Jiao, R. , Wang, S. , Wu, X. , Ye, H. , Pan, C. et al. (2021) SPL36 encodes a receptor‐like protein kinase that regulates programmed cell death and defense responses in rice. Rice, 14, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegert, A.S. , Thoden, J.B. , Schoenhofen, I.C. , Watson, D.C. , Young, N.M. & Tipton, P.A. et al. (2017) Structural and biochemical investigation of PglF from Campylobacter jejuni reveals a new mechanism for a member of the short chain dehydrogenase/reductase superfamily. Biochemistry, 56, 6030–6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe, A.P. , Su, X. , Chen, Z. , Chen, T. , Wei, X. , Tang, S. et al. (2019) Identification and characterization of a spotted‐leaf mutant spl40 with enhanced bacterial blight resistance in rice. Rice, 12, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura, K. , Okada, A. , Okada, K. , Jikumaru, Y. , Ko, K.W. , Toyomasu, T. et al. (2007) Identification of a biosynthetic gene cluster in rice for momilactones. Journal of Biological Chemistry, 282, 34013–34018. [DOI] [PubMed] [Google Scholar]
- Stammers, D.K. , Ren, J. , Leslie, K. , Nichols, C.E. , Lamb, H.K. , Cocklin, S. et al. (2001) The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short‐chain dehydrogenase/reductases. The EMBO Journal, 20, 6619–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Zhu, L. , Xu, L. , Yuan, D. , Min, L. & Zhang, X. (2014) Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nature Communications, 5, 5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A. , Kawasaki, T. , Henmi, K. , Shii, K. , Kodama, O. , Satoh, H. et al. (1999) Lesion mimic mutants of rice with alterations in early signaling events of defense. The Plant Journal, 17, 535–545. [DOI] [PubMed] [Google Scholar]
- Tanaka, N. , Nonaka, T. , Nakamura, K.T. & Hara, A. (2001) SDR: structure, mechanism of action, and substrate recognition. Current Organic Chemistry, 5, 89–111. [Google Scholar]
- Untergasser, A. , Cutcutache, I. , Koressaar, T. , Ye, J. , Faircloth, B.C. , Remm, M. et al. (2012) Primer3‐new capabilities and interfaces. Nucleic Acids Research, 40, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Lei, C. , Wang, J. , Ma, J. , Tang, S. , Wang, C. et al. (2017) SPL33, encoding an eEF1A‐like protein, negatively regulates cell death and defense responses in rice. Journal of Experimental Botany, 68, 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, H.L. , Dong, L.I. , Wang, Z.P. , Zhang, H.Y. , Han, C.Y. , Liu, B. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biology, 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.B. , Meng, W.L. , Zhao, M.J. , Zhang, A.X. , Liu, W. , Xu, Z.S. et al. (2019) Proteomics analysis to identify proteins and pathways associated with the novel lesion mimic mutant E40 in rice using iTRAQ‐based strategy. International Journal of Molecular Sciences, 20, 1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, X. , Zou, B. , Hong, X. , Gao, M. , Yang, W. , Zhong, X. et al. (2018) Rice copine genes OsBON1 and OsBON3 function as suppressors of broad‐spectrum disease resistance. Plant Biotechnology Journal, 16, 1476–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L.R. , Qu, S. , Bordeos, A. , Yang, C. , Baraoidan, M. , Yan, H.Y. et al. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U‐Box/Armadillo repeat protein endowed with E3 Ubiquitin ligase activity. The Plant Cell, 16, 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Shen, B.Z. , Dai, X.K. , Mei, M.H. , Saghai Maroof, M.A. & Li, Z.B. (1994) Using bulked extremes and recessive class to map genes for photoperiod‐sensitive genic male sterility in rice. Proceedings of the National Academy of Sciences USA, 91, 8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Yin, J. , Liang, S. , Liang, R. , Zhou, X. , Chen, Z. et al. (2016) The multivesicular bodies (MVBs)‐localized AAA ATPase LRD6‐6 inhibits immunity and cell death likely through regulating MVBs‐mediated vesicular trafficking in rice. PLoS Genetics, 12, e1006311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Ze, M. , Mawsheng, C. , Chen, X. & Wang, J. (2020) Deciphering rice lesion mimic mutants to understand molecular network governing plant immunity and growth. Rice Science, 27, 278–288. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


