Abstract
Non-LTR retrotransposons such as L1 elements are major components of the mammalian genome, but their mechanism of replication is incompletely understood. Like retroviruses and LTR-containing retrotransposons, non-LTR retrotransposons replicate by reverse transcription of an RNA intermediate. The details of cDNA priming and integration, however, differ between these two classes. In retroviruses, the nucleocapsid (NC) protein has been shown to assist reverse transcription by acting as a “nucleic acid chaperone,” promoting the formation of the most stable duplexes between nucleic acid molecules. A protein-coding region with an NC-like sequence is present in most non-LTR retrotransposons, but no such sequence is evident in mammalian L1 elements or other members of its class. Here we investigated the ORF1 protein from mouse L1 and found that it does in fact display nucleic acid chaperone activities in vitro. L1 ORF1p (i) promoted annealing of complementary DNA strands, (ii) facilitated strand exchange to form the most stable hybrids in competitive displacement assays, and (iii) facilitated melting of an imperfect duplex but stabilized perfect duplexes. These findings suggest a role for L1 ORF1p in mediating nucleic acid strand transfer steps during L1 reverse transcription.
LINE-1, or L1, is an abundant long interspersed nuclear element that has amplified to a high copy number in mammalian genomes by retrotransposition. In humans and mice for example, there are upwards of 100,000 copies of L1. In addition, it is likely that the even more abundant short interspersed nuclear elements and processed pseudogenes were created by the action of L1 machinery. Together these sequences account for approximately 30% of the mammalian genome. Transposition of these elements continues, occasionally leading to disease by insertional mutagenesis. Thus, L1 has been, and continues to be, a major dynamic force in modeling the mammalian genome (references 9 and 16 and references therein; 18).
L1 belongs to a larger group of mobile elements known as the non-long terminal repeat (LTR) retrotransposons (25). Members of this class are thought to use a unique mechanism for transposition called target site-primed reverse transcription (TPRT) (24). In TPRT, an element-encoded endonuclease cleaves the target site in genomic DNA to generate a 3′ OH. This hydroxyl acts as the primer for reverse transcription using element RNA as a template. The result is simultaneous reverse transcription and joining of the first-strand cDNA with the genome. The mechanism used to complete cDNA synthesis and integrate both ends into the chromosome has not been elucidated but may require additional proteins that are not encoded by the element.
TPRT-based replication differs from that of the other major class of retroelements, the retroviruses and related LTR retrotransposons. The latter elements use RNA primers to initiate reverse transcription instead of a genomic DNA 3′ end. Minus strand cDNA synthesis uses a cellular tRNA as the primer, while plus strand synthesis uses a fragment of element RNA. Reverse transcription further involves two template switches, ultimately yielding a linear, double-stranded cDNA molecule with blunt ends. The virus-encoded integrase protein then connects this viral cDNA to host DNA, forming the integrated provirus (for a review, see reference 4).
Transposition-competent versions of L1 recently have been isolated and studied. L1 elements are 6 to 7 kb long and encode two proteins that are necessary for retrotransposition, ORF1p and ORF2p (30). The endonuclease and the reverse transcriptase activities that are presumably required for TPRT reside within ORF2p (10, 29). The other protein, ORF1p, is essential for retrotransposition but its role is not well understood.
ORF1p copurifies with L1 RNA as a ribonucleoprotein complex (RNP) when extracts from mouse F9 embryonal carcinoma cells are fractionated through sucrose gradients (26). In addition, mouse ORF1p purified from Escherichia coli binds nonspecifically to RNA and single-stranded DNA, with apparent positive cooperativity (19). There is also evidence for a higher affinity binding to specific sequences within L1 RNA (15). Taken together, these properties are consistent with a role for ORF1p in the packaging of L1 RNA during retrotransposition.
These functions of ORF1p potentially parallel those of the retroviral gag proteins (reviewed in reference 4). Unlike L1 ORF1p, retroviral gag proteins are initially synthesized as a polyprotein. Following assembly of the gag precursor into particles, cleavage by the viral protease yields separate matrix, capsid, and nucleocapsid (NC) proteins. The NC protein in particular displays activities potentially related to those of ORF1p in L1 elements. The NC protein binds nucleic acids and contributes to multiple steps in viral nucleic acid metabolism, including (i) facilitating RNA dimerization, (ii) packaging of viral RNA during assembly, (iii) binding of the tRNA primer to the viral genomic RNA, (iv) facilitating two strand transfers during reverse transcription, and (v) potentially promoting cDNA integration (2, 3, 13, 21, 22, 34, 36, 38).
In vitro studies have begun to reveal how the NC protein facilitates these diverse reactions. Purified NC protein can greatly accelerate the annealing of complementary DNA sequences in dilute solution. The NC protein can also promote the formation of the most stable base-paired structure in a competitive annealing reaction, so-called nucleic acid chaperone activity (12, 22, 34, 38). These activities probably account for much of the function of NC during viral replication.
Since the initial recognition that L1 encoded a second protein in addition to reverse transcriptase, it has been tempting to speculate that ORF1p could be the functional homologue of retroviral gag (23). At odds with this view are the lack of sequence homology between L1 ORF1p and retroviral NC (14) and the striking differences between retroviral replication and TPRT. The nonhomologous ORF1 protein from the Drosophila non-LTR retrotransposon I was proposed to have an NC-like role after the protein was found to form large complexes with nucleic acids and to accelerate the annealing of complementary DNA fragments (8). ORF1p from the I element contains the CCHC motifs that are typical of retroviral NC proteins. In fact, of the 11 major clades of non-LTR retrotransposons, these motifs are found in six of the seven with an ORF1, with mammalian L1 being the sole exception (25). Since the CCHC domains are essential for proper function of the I element ORF1p as well as retroviral NC, their absence in mammalian L1 ORF1p is surprising. This raises the questions of whether NC-like function is required for L1 transposition and whether the divergent L1 ORF1p provides nucleic acid chaperone activities.
To examine the nucleic acid chaperone activities of mouse ORF1p, we studied the effects of purified recombinant protein on a series of DNA oligonucleotide substrates. L1 ORF1p was found to accelerate the annealing of complementary DNA oligonucleotides, as well as to increase the rate of formation of the most stable duplex when tested under strand-exchange conditions. These activities imply a much greater functional similarity between L1 ORF1p and retroviral NC proteins than has heretofore been recognized, in spite of their different sequences and the differences between retroviral reverse transcription-integration and TPRT. Finally, we propose a role for these chaperone activities of L1 ORF1p in TPRT.
MATERIALS AND METHODS
Oligonucleotides.
Oligonucleotides were purchased from Genset (La Jolla, Calif.) or Integrated DNA Technologies (Coralville, Iowa). The sequences are given in Table 1. Oligonucleotides used in annealing, strand-exchange, and melting assays were purified by electrophoresis through denaturing polyacrylamide gels before use. The oligonucleotides tested included one pair with a region of the human immunodeficiency virus type 1 LTR and several others with sequences that were more biologically relevant to L1 retrotransposition (Table 1). One of the two complementary oligonucleotides was 5′ end labeled using [gamma-32P]ATP (Amersham) and T4 polynucleotide kinase (New England Biolabs). Unincorporated isotope was removed by gel filtration (QuickSpin TE; Boehringer Mannheim).
TABLE 1.
Oligonucleotides used for chaperone assays
| Namea | Sequence (5′ to 3′) |
|---|---|
| 29 | AAAAAGTACACAGTCTAACATCAACTCGC |
| c29 | GCGAGTTGATGTTAGACTGTGTACTTTTT |
| 38 | CAACAATTACTTTTCCTTAATATCTCTTAACATCAATG |
| c38 | CATTGATGTTAAGAGATATTAAGGAAAAGTAATTGTTG |
| 25 | AGTACACAGTCTAACATCAACTCGC |
| c25 | GCGAGTTGATGTTAGACTGTGTACT |
| mm29c | GCGAGTTGACGTCAGACCGTGCACTTTTTb |
| 48 | GCCGTCCAGTCTTCAGTTTAAAAAGTACACAGTCTAACATCAACTCGC |
| c48 | GCGAGTTGATGTTAGACTGTGTACTTTTTAAACTGAAGACTGGACGGC |
| 32 | ACTGCTAGAGATTTTCCACACGGATCCTAGGC |
| c32 | GCCTAGGATCCGTGTGGAAAATCTCTAGCAGT |
Oligonucleotides are named for their length, with “c” indicating the inverse complement. c25 has the same sequence as c29, except that it is 4 nt shorter on the 3′ end. The 38-nt region lies just downstream of the presumed translation initiation codon in ORF2. The homologous region in human LINE-1 was described as a sequence-specific RNA binding site for L1Hs ORF1p, although DNA interactions were not observed (15). All other oligonucleotides, except 32 and c32, were designed to mimic a DNA target site for a new L1 insertion (17).
The nucleotides in mm29c that form mismatches when hybridized to 29 are underlined.
Purification of ORF1p from E. coli.
The ORF1 region from pJS16 35) was amplified by PCR using the primers ORF1 start (5′-GGGGAATTCATGGCGAAAGGCAAACG) and ORF1 end (5′-GGGGAATTCGCTGTCTTCTTTTTGGTTTGTTGA). The product was digested with EcoRI and then cloned into pGEX-6P-1 (Amersham Pharmacia Biotech). Protein was produced in BL21 grown at 21°C for 16 h after induction with IPTG (isopropyl-β-d-thiogalactopyranoside). All subsequent steps were performed at 4°C. The cells were recovered, and a crude lysate was prepared as previously described (7). Nucleic acid was removed by precipitation with 0.5% polyethyleneimine (39). The protein-containing supernatant was adjusted to final concentrations of 5 mM dithiothreitol (DTT) and 8% glycerol and then bound to glutathione-Sepharose (Amersham Pharmacia Biotech). The column was washed three times with 10 ml of 50 mM Tris-HCl (pH 8.0)–150 mM NaCl–5 mM DTT–50 mM EDTA–8% glycerol and then two times with 5 ml of cleavage buffer (50 mM Tris-HCl [pH 7.0], 150 mM NaCl, 1 mM EDTA, 1 mM DTT). The glutathione-Sepharose was resuspended in 1 ml of cleavage buffer containing 80 U of PreScission protease (Amersham Pharmacia Biotech) and incubated overnight at 4°C. The column was drained and then rinsed 10 times with 1 ml of cleavage buffer. Fractions containing ORF1p (based upon sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE] analysis) were pooled, dialyzed against buffer A (20 mM HEPES [pH 7.6], 150 mM NaCl, 2 mM DTT, and 0.1 mM EDTA), and then applied to a CM Sepharose (Amersham Pharmacia Biotech) column. After washing with five column volumes of buffer A, proteins were eluted with a 150 mM to 1 M NaCl gradient. Fractions containing ORF1p were identified by SDS-PAGE, pooled, and then dialyzed into 20 mM HEPES (pH 7.6)–100 mM NaCl–2 mM DTT–0.1 mM EDTA–5% glycerol.
Purification of L1 ORF1p from SF9 cells.
Both A-type and TF-type ORF1p (p41 and p43, respectively, in a report by Kolosha and Martin [20]) were expressed as His-tagged fusion proteins. The fusion adds an N-terminal tag of 36 amino acid residues, which contains a hexahistidine block for purification by metal-chelating affinity chromatography. A cytoplasmic extract from SF9 cells infected with recombinant baculovirus was incubated in lysis buffer containing 50 mM sodium phosphate (pH 7.5), 4 mM 2-mercaptoethanol, 0.1% NP-40, and 1 mM phenylmethylsulfonyl fluoride (PMSF) for 30 min on ice followed by centrifugation at 20,000 × g for 30 min at 4°C. ORF1p in the supernatant was incubated with Ni-agarose (Qiagen) in binding buffer (50 mM sodium phosphate [pH 8.0], 500 mM NaCl, 25 mM imidazole, 0.2% Tween 20, 0.1% NP-40, 10 mM 2-mercaptoethanol, 1 mM PMSF) and 10 μg each of chymstatin, leupeptin, antipain, and pepstatin for 2 days at 4°C with gentle mixing. The Ni-agarose slurry with the bound protein was transferred to a column and then washed twice with 50 mM sodium phosphate (pH 8.0)–500 mM NaCl–0.2% Tween 20–0.1% NP-40–40 mM imidazole–10 mM 2-mercaptoethanol. The proteins were eluted in 50 mM sodium phosphate (pH 8.0)–300 mM NaCl–250 mM imidazole–1 mM PMSF. Fractions containing ORF1p were identified by Western blotting, pooled, and then dialyzed against ORF1 storage buffer (20 mM HEPES [pH 7.6], 100 mM NaCl, 2 mM DTT, 0.1 mM EDTA, and 5% glycerol).
The concentration of each protein was determined by Bradford assay (Bio-Rad). All proteins were stored in small aliquots at −80°C until use.
Other proteins.
Lambda repressor was a gift from S. Munroe and M. Ptashne. The protein was purified as described previously (32). E. coli single-strand binding (SSB) protein was purchased from New England Biolabs, and bovine serum albumin (BSA) was purchased from Pierce.
Annealing assay.
Ten-microliter annealing reaction mixtures contained 1× annealing buffer (20 mM HEPES [pH 7.6], 1 mM EDTA, 1 mM MgCl2, 1 mM DTT, 0.1% Triton X-100) and the indicated amount of each oligonucleotide and protein. Unless otherwise indicated, timing was initiated by addition of the radiolabeled oligonucleotide to a tube containing all of the other reaction components. Reactions were incubated for two minutes at 37°C and then stopped by the addition of 5 μl of stop mix (0.4 mg of tRNA per ml, 0.2% SDS, 15% Ficoll [type 400], 0.2% bromophenol blue, and 0.2% xylene cyanol blue). Double- and single-stranded oligonucleotides were resolved by electrophoresis through 15% native polyacrylamide gels in 1× TBE (100 mM Tris-HCl [pH 8.3], 100 mM boric acid, 2 mM EDTA) at 4°C. Gels were dried and then examined by phosphorimage analysis (Imagequant; Molecular Dynamics).
Strand-exchange assay.
A preannealed duplex was made by mixing 200 mM 32P-labeled oligonucleotide with its complement in water. The mixture was heated for 5 min at 95°C, NaCl was added to a concentration of 50 mM and the mixture was cooled slowly (∼ 2 h) to room temperature and then stored frozen until use. Preannealed duplex oligonucleotide was diluted in water and then mixed with a 50× molar excess of complementary single-stranded oligonucleotide in 1× annealing buffer containing 50 mM NaCl on ice. Timing was initiated upon the addition of protein. After incubation at 37°C, the reaction was stopped by the addition of 0.6 to 1 volume of ice-cold stop mix. Gels and analysis were performed as described for the annealing reactions.
Melting assay.
Preannealed duplex was mixed with protein in 45 μl of 1× annealing buffer containing 50 mM NaCl on ice. A 5-μl aliquot was removed and mixed with 5 μl of ice-cold stop mix after a 5-min incubation. The tube was transferred to an Eppendorf Thermomixer R and incubated for 5 min at the indicated temperature, and then another aliquot was removed. The temperature was increased by 5°C, and the sampling process was repeated at 5°C intervals through the temperatures indicated. Gels and analysis were performed as described for the annealing reactions.
RESULTS
Experimental plan.
We have investigated whether L1 ORF1, has an intrinsic nucleic acid chaperone activity. Because the CCHC motif is seen in ORF1 sequences from other non-LTR retrotransposons and because one of these, the I factor, has an ORF1p that accelerates annealing, we asked whether the L1 ORF1 protein displays nucleic acid chaperone activities in vitro.
L1 ORF1 proteins were purified after overexpression in either E. coli or baculovirus. A map of L1 is shown in Fig. 1, illustrating the ORF1 protein used. This TF-type mouse L1 ORF1 protein was purified from the soluble fraction after overexpression of a glutathione S-transferase fusion protein in E. coli. Some of the assays were also performed with a His-tagged A- or TF-type ORF1p protein that was purified following expression in baculovirus-infected insect cells. To assess nucleic acid chaperone activities, purified ORF1 protein was assayed for the ability to (i) accelerate annealing of complementary DNA strands in dilute solution, (ii) promote formation of the most stable DNA duplex in annealing exchange reactions, and (iii) melt DNA duplexes.
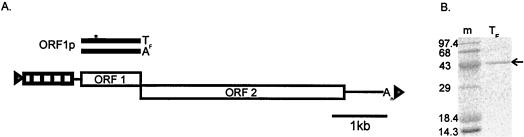
FIG. 1.
Map of L1 and reagents used in this study. (A) A full-length, active L1 element from the mouse genome is made up of approximately 6.7 kb. It contains a repeating promoter motif (tandem boxes, each about 200 nt long), a short 5′ noncoding region (line), the ORF1 and ORF2 coding sequences (rectangles), and a 3′ noncoding region (line) ending with a poly(A) tail (An). Each element is bounded by short target site duplications (triangles). A length polymorphism in ORF1 shows the addition of 14 amino acids to the TF-type compared to the A-type ORF1 protein. (B) TF-type ORF1p (1.0 μg) isolated from E. coli was fractionated by SDS-PAGE and stained with Coomassie blue. m, molecular size marker.
Accelerated annealing by ORF1p.
To assay the acceleration of annealing, pairs of complementary oligonucleotides were mixed in dilute solution, and duplex formation was monitored (Fig. 2A). One oligonucleotide of each pair was labeled on the 5′ end with 32P. Four pairs of oligonucleotides were studied, each containing a different DNA sequence and ranging in length from 29 to 48 base pairs (Table 1). These included a region from L1 that lies within ORF2 sequences (Fig. 2B, D, and E), an L1 insertion target half-site (Fig. 2C and F to H), an L1 insertion target full-site (data not shown), and an irrelevant sequence (data not shown). Products of annealing reactions were separated on native polyacrylamide gels and visualized by phosphorimage analysis. The mobilities of the single-stranded and duplex forms were determined by coelectrophoresis of end-labeled oligonucleotide alone or of end-labeled oligonucleotide that had been annealed to its complement by boiling followed by slow cooling, respectively.
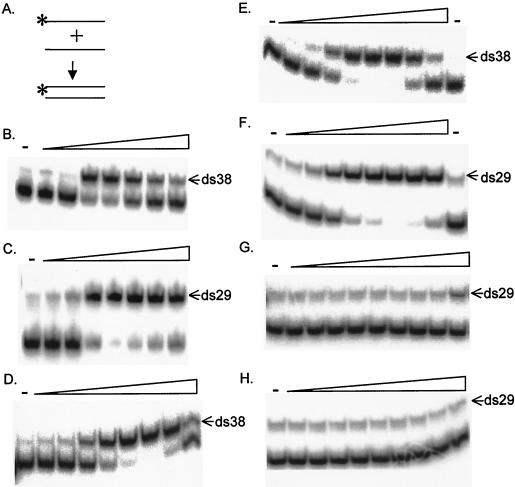
FIG. 2.
ORF1p accelerates annealing of complementary DNA oligonucleotides. (A) Schematic of the annealing assay. The asterisk indicates the 32P end label, here and throughout the figures. (B to H) Phosphorimages of oligonucleotides separated by electrophoresis. Increasing amounts of protein are indicated by triangles in panels B to H, and a dash above a lane indicates that no protein was added. The double-stranded oligonucleotide is indicated by the arrow; the other band is the single-stranded, end-labeled oligonucleotide, based upon the migration of known standards through the same or similar gels. For panels B and C, a 1.8 nM concentration of labeled oligonucleotide and a 2 nM concentration of its reverse complement were incubated with threefold serial dilutions (0.34 to 250 nM) of E. coli TF-type ORF1p. For panels D to H, 150 pg of each oligonucleotide was incubated with threefold serial dilutions of baculovirus-produced TF-type ORF1p (0.3 to 639 nM) (D), A-type ORF1p (0.5 to 1,100 nM) (E and F), lambda repressor (0.13 to 820 nM) (G), or BSA (0.45 to 3,000 nM) (H).
Little annealing is seen in the absence of protein, but significant annealing is observed when ORF1p purified from E. coli (Fig. 2B and C) or baculovirus (Fig. 2D to F) is added to the reaction. The ORFp proteins from both the TF-type and A-type mouse L1 subfamilies share this activity (Fig. 2D and E). The activity is not specific for any particular oligonucleotide, as the proteins act on all of the oligonucleotide pairs tested (Fig. 2B to F; Table 1). The annealing activity was present in four independent preparations of ORF1p; two of these preparations were TF-type ORF1p produced in E. coli, and the other two were an A- and a TF-type ORF1p produced in baculovirus. Because of the different source and purification methods used to make the baculovirus- versus E. coli-expressed proteins, the observed annealing activity is highly likely to be intrinsic to L1 ORF1p and not a property of any minor contaminants in the protein preparations. Not all proteins facilitate annealing under these conditions. We also tested two other nucleic acid binding proteins, lambda repressor and E. coli SSB. Lambda repressor, which binds a specific sequence in DNA with high affinity, fails to accelerate annealing until very high protein concentrations are reached (Fig. 2G); 50% annealing is observed at a 2.5 μM repressor concentration. No double-stranded form of the labeled oligonucleotide is detected using E. coli SSB in this assay at concentrations ranging between 0.18 nM and 10.8 μM. However, at a 44 nM SSB concentration, 50% of the labeled oligonucleotide shifts to the well, where all of it is found at all higher concentrations (data not shown). BSA has no effect on the oligonucleotides in these reaction conditions at any concentration tested (Fig. 2H). We conclude that accelerating annealing is not a property of DNA binding proteins generally.
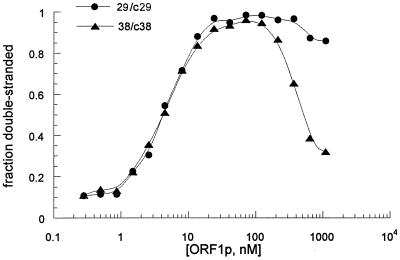
As the concentration of ORF1p is increased, the reaction shows a sudden transition, with annealing becoming apparent only after the protein concentration is approximately equimolar to the oligonucleotide concentration. At higher protein concentrations, a greater fraction of the labeled DNA remains single stranded. This is more pronounced with the 38-nucleotide (nt) pair than it is with the 29-nt pair of complementary oligonucleotides (Fig. 3). The differences in behavior between these two oligonucleotide pairs is not due simply to their different lengths, because in annealing assays performed with a 48-mer duplex, 45% of the product was double stranded at the highest protein concentration, compared to 25% with the 38-mer duplex and 89% with the 29-mer duplex (data not shown). Thus, the difference in annealing efficiency at high ORF1p concentrations appears to be due to the nucleotide sequence of the different oligonucleotide pairs.
FIG. 3.
Comparison of ORF1p annealing activity on two oligonucleotide pairs. Baculovirus-produced ORF1p (0.28 to 1,100 nM) was incubated with 150 pg of each oligonucleotide, either [32P]29-c29 or [32P]38-c38.
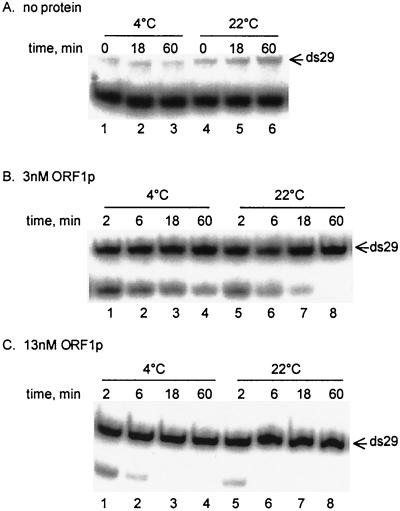
The time and temperature dependence of the annealing reaction were also tested. ORF1p facilitates annealing on ice and at 22°C, as well as at 37°C (Fig. 4, compare panels B and C to panel A), although the reaction proceeds more slowly at the lower temperatures. With low concentrations of protein, complete conversion to the double-stranded form occurs at 22°C in 60 min but not at 4°C (Fig. 4B, lanes 4 and 8) or with shorter incubation times (Fig. 4B, lanes 1 to 3 and 5 to 7; also compare the same incubation times in the absence of protein in panel A, lanes 1 to 3 and 4 to 6). With a higher concentration of ORF1p, annealing is complete in 18 min at 4°C and 6 min at 22°C (Fig. 4C, lanes 3 and 6).
FIG. 4.
Time and temperature dependence of the annealing activity of ORF1p. Aliquots from each reaction were removed at the indicated times after the addition of the radiolabeled oligonucleotide. Incubations were carried out at either 4 or 22°C. Reactions contained 1.8 nM [32P]29, 2 nM c29, and either no protein (A) or 3 or 13 nM ORF1p made from E. coli (B and C, respectively).
Nucleic acid chaperone activity.
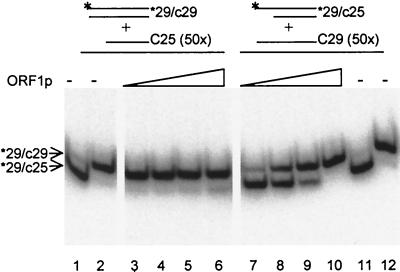
The demonstration of the ability of L1 ORF1p to accelerate annealing of complementary oligonucleotides raises the question of whether the protein has additional activities that are characteristic of nucleic acid chaperone proteins. To test whether ORF1p facilitates the rate at which nucleic acids anneal to form the thermodynamically most stable form, performed duplexes were challenged with a 50-fold molar excess of a complementary single-stranded oligonucleotide. Two conditions were tested: (i) the 29-c29 perfect duplex, challenged with the shorter c25, and (ii) the 29-c25 duplex, challenged with c29. In both cases, the starting 29-mer strand was labeled on the 5′ end with 32P. Reaction products were separated on native acrylamide gels, and DNAs were visualized by phosphorimage analysis. In this experiment, the 29-nt perfect duplex migrated more slowly than the 29-c25 duplex. No single-stranded oligonucleotide was detected.
In the absence of ORF1p, the [32P]29-c29 duplex remained stable when incubated for 15 min at 37°C without protein (Fig. 5, lane 2), even in the presence of a 50-fold excess of the 25-mer strand. However, exchange to form the favored 29-c25 duplex was observed when the mixture was heated to 95°C for 5 min and then incubated at 45°C for 15 min (Fig. 5, lane 1). Addition of ORF1 protein to preannealed [32P]29-c29 in the presence of excess single-stranded c25 resulted in little exchange of the label to form the 29-c25 duplex (Fig. 5, lanes 3 to 6). This result was expected, because the labeled oligonucleotide was already in its thermodynamically most favored form in the starting duplex.
FIG. 5.
Strand-exchange activity of ORF1p. Preannealed duplex DNA ([32P]29-c29 or [32P]29-c25) was challenged with a 50× molar excess of the complementary single strand as indicated. Controls contained all components except protein. Lanes 2 and 11, untreated duplex oligonucleotides; lanes 1 and 12, three oligonucleotides in the same proportion as the lanes with protein but heated for 5 min at 95°C and then for 15 min at 45°C. All other samples were incubated for 15 min at 37°C. Recombinant ORF1p from E. coli was added to a final concentration of 17 nM (lanes 3 and 7), 52 nM (lanes 4 and 8), 155 nM (lanes 5 and 9), or 466 nM (lanes 6 and 10). The concentrations of duplex oligonucleotides were 1.6 nM and of single-stranded oligonucleotides were 80 nM.
In contrast, if a preformed [32P]29-c25 duplex is challenged by the addition of excess c29, strand exchange to form the more stable [32P]29-c29 duplex occurs in the presence of ORF1p. Increased amounts of the [32P]29-c29 duplex were formed as the concentration of ORF1p in the reaction increased (Fig. 5, lanes 7 to 10), whereas no exchange of the [32P]29-c25 duplex was observed in the absence of protein (Fig. 5, lane 11). Thus, ORF1p promotes the exchange of strands to form the most stable hybrid.
As was observed for the annealing reactions, the extent of the exchange reaction increased with increasing time and temperature. Longer incubations are required at lower temperatures to achieve the same degree of exchange. Longer incubation times also increased the extent of exchange. For example, essentially all of a 1 nM [32P]29-c25 duplex was exchanged to form the [32P]29-c29 duplex when 50 nM c29 was incubated with a 50 nM protein concentrations for 1 h at 37°C (data not shown).
Comparison of mismatched and partially single-stranded substrates.
The mechanism of the nucleic acid chaperone activity was next probed. Because the [32P]29-c25 duplex leaves 4 nt of single-stranded DNA on one end that can base pair with c29, it is possible that the exchange reaction requires this single-strand overhang to nucleate base pairing. Once the three oligonucleotides form a joint structure, branch migration of the 29-mer strand may displace the 25-mer strand while simultaneously forming the fully complementary 29-mer strand.
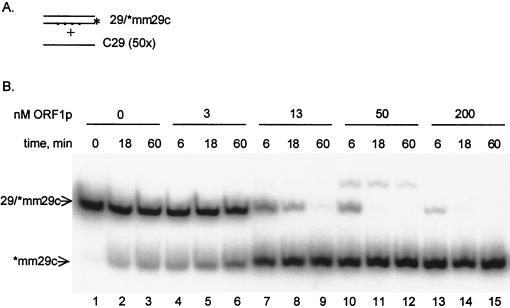
To investigate this model, we assayed whether the free single-stranded region in the 29-25 hybrid is essential for strand exchange by observing the chaperone activity on a 29-mer–29-mer duplex containing four internal mismatches (29-[32P]mm29c). Assays of stable hybrid formation were carried out as described above by adding the perfectly matched complementary 29-mer strand. In the presence of ORF1p the perfect complement, c29, readily displaced the 32P-labeled mm29c from this duplex (Fig. 6). As was observed above for the annealing reactions, the appearance of single-stranded [32P]mm29c increased with increasing times of incubation (e.g., compare lanes 1 to 3 with lanes 7 to 9 in Fig. 6) and increasing concentrations of ORF1p (e.g., compare lanes 2, 5, 8, 11, and 14, the 18-min incubation times with increasing amounts of protein, in Fig. 6). In rare instances an additional band was observed (note the minor band in Fig. 6, lane 11, which migrated more slowly than the duplex). This band is an alternative structure of the 32P-labeled mm29c. After the kinase reaction, approximately 60% of this oligonucleotide was in the slow-migrating form. A 5-min incubation at 95°C converted it completely to the fast-migrating form. The slow-migrating structure of mm29c appeared after its displacement from the duplex only when the concentration of ORF1p in the reaction was roughly equimolar to that of the oligonucleotides. The displacement of [32P]mm29c from its duplex form with 29 by 29c indicates that the exchange reaction does not require nucleation of base pairing at a single-stranded complementary overhang.
FIG. 6.
Strand-exchange activity of ORF1p. (A) Preannealed duplex DNA (29-[32P]-mm29c [1 nM]) was challenged with 50 nM c29 in the presence or absence of ORF1p made in E. coli. (B) The times of incubation and concentrations of protein are indicated. For this reaction, the two duplexes, 29-c29 and 29-mm29c, are not resolved by size, and only mm29c is labeled with 32P. Thus, the exchange reaction leads to the disappearance of the 29-[32P]mm29c duplex and the appearance of single-stranded [32P]mm29c. The two major bands resolved in this gel are the 29-[32P]mm29c duplex and the denatured form of [32P]mm29c. The minor band, migrating slower than the duplex, is a structured form of the single-stranded [32P]mm29c (see Results).
DNA melting by ORF1p.
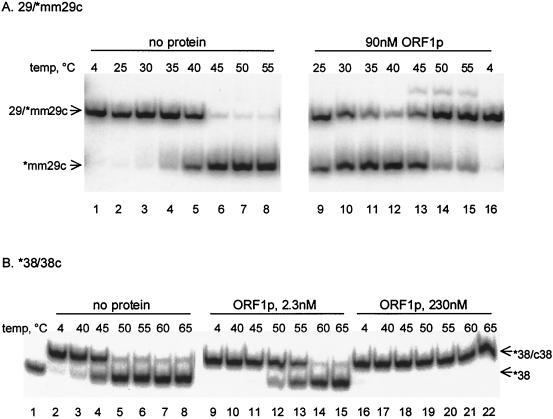
To further probe the mechanism of the nucleic acid chaperone activity, ORF1p was tested for its effects on DNA melting. The mismatched duplex, 29-[32P]mm29c, shows the typical transition from a duplex to the single-stranded form over a narrow temperature range in the absence of protein (Fig. 7). The addition of ORF1p lowers the temperature at which half of the radiolabeled oligonucleotide (mm29c) is found in duplex form (Tm) from 41°C to 23 to 28°C (Fig. 7A, compare lanes 4 and 11). ORF1p greatly extends the range of temperatures in which both single- and double-stranded forms are present (Fig. 7A, compare lanes 2 to 6 to lanes 9 to 13). ORF1p also stabilizes the double-stranded form at higher temperatures (Fig. 7A, compare lanes 7 and 8 to lanes 14 and 15). These effects on the Tm of the 29-mm29c duplex are seen at protein concentrations ranging between 10 and 250 nM, although only the 90-nM results are shown.
FIG. 7.
Effect of ORF1p on melting temperature of DNA duplexes. (A) Tm of an imperfect duplex. Lanes contain aliquots removed after a 5-min incubation at the indicated temperatures from a mixture with 1 nM 29-[32P]mm29c and either no protein (lanes 1 to 8) or 90 nM ORF1p purified from E. coli (lanes 9 to 16). (B) Tm of a perfect duplex. Lanes contain aliquots removed after a 5-min incubation at the indicated temperatures from a mixture with 1.3 nM [32P]38-c38 and either no protein (lanes 2 to 8) or 2.3 nM (lanes 9 to 15) or 230 nM (lanes 16 to 22) ORF1p purified from E. coli. Lane 1 contains the single-stranded [32P]38 oligonucleotide as a mobility standard.
In contrast, when ORF1p is added to perfect duplexes, they are stabilized by the presence of protein. This was observed for the 25-c25, 29-c29, and 38-c38 duplexes; results obtained using [32P]38-c38 are shown in Fig. 7B. Even at a low concentration of protein, 2.3 nM, the Tm of this duplex shifts from 46 to 55°C (Fig. 7B, compare lanes 3 to 6 with lanes 10 to 13). At higher ORF1p concentrations, the duplex did not melt over the temperatures tested (Fig. 7B, lanes 16 to 22). Thus, the three perfect duplexes are stabilized by the protein, whereas the mismatched duplexes are destabilized. It appears that small regions of internal mismatches are sensed and melted by ORF1p.
DISCUSSION
These studies indicate that the ORF1p protein from mouse L1 is able to facilitate rearrangements of nucleic acids into more stable conformations. Previous studies have named this nucleic acid chaperone activity (34), though we note that the analogy between nucleic acid and protein chaperones is only partial. Unlike protein chaperones, nucleic acid chaperones are not ATPases and do not turn over catalytically; rather, they remain associated with the annealed nucleic acid products. Nucleic acid chaperone activity has been reported in retrovirus (38) and retroelement (6) NC proteins and in the I element non-LTR retrotransposon ORF1p protein (8). The NC and I element ORF1p proteins contain the CCHC zinc knuckle sequence, raising the question of whether other retroelements, such as mammalian L1, that lack the CCHC sequence also require nucleic acid chaperone activity. We report here that L1 ORF1p does indeed show such activity. These findings strongly support the idea that nucleic acid chaperone activity is a general requirement for retroelement replication (6). Based on these findings, we speculate that other retroelements lacking CCHC-containing proteins, such as the foamy retroviruses and hepatitis B viruses, also encode or recruit nucleic acid chaperone proteins to promote reverse transcription.
How does ORF1p accelerate the annealing of complementary DNA molecules? ORF1p is known to bind to single-stranded DNA molecules and form multimers (19), so it may bring complementary strands together via protein-protein interactions. Binding of the basic ORF1 protein will also neutralize the negative charges of the acidic DNA phosphate backbone, facilitating strand association by charge shielding. However, the DNA in the complex must be able to sample different potential pairing partners, so the DNAs must be able to change positions relative to one another. Thus, the addition of ORF1p allows the system to achieve its equilibrium position more rapidly. Many basic proteins are known to accelerate annealing, including histone H1 (5), yeast Rad52 (31), hnRNPA1 protein (33), I element ORF1 (8), and retroviral NC (22, 38). Not all proteins, however, accelerate annealing, including other DNA binding proteins. For example, we tested lambda repressor and found that annealing was not facilitated until the protein was in large (∼100-fold) excess over nucleic acid and that neither E. coli SSB nor BSA accelerated annealing. In a separate series of experiments, we found that the nucleic acid interaction domain of ORF1p lies in its carboxy-terminal third. This domain also accelerates annealing, indicating that the chaperone activity is contained within the nucleic acid binding domain (28). Similar properties were reported recently for the C-terminal portion of the TYA1 protein (6).
The oligonucleotides remain in the single-stranded form at high concentrations of ORF1p. This was also observed for other proteins that accelerate annealing, such as NC. One explanation is that the protein aggregates, effectively lowering the concentration of free ORF1p. This seems unlikely, since aggregation was not evident experimentally and would affect all oligonucleotide pairs equally. A more likely alternative explanation is that ORF1p coats the single-stranded DNA at higher concentrations, thereby retaining it in its single-stranded form. According to this idea, stabilization of single-stranded DNA will only take place in the presence of enough ORF1p to coat a substantial fraction of the DNA strand. The differences observed among oligonucleotide pairs in our annealing assays may be explained by different affinities of ORF1p for the various oligonucleotides. If this is the case, stabilization of single-stranded DNA at high concentrations would represent a change in the position of the equilibrium of the system due to the tight binding of ORF1p to single strands.
How does ORF1p lower the melting temperature of the mismatched duplex but raise the melting temperature of a perfect duplex? It seems likely that the internal mismatches of the mismatched substrate are recognized as local single-stranded regions. This would make them a substrate for relatively high-affinity binding of ORF1p, which has been reported previously to bind at least 100-fold more tightly to single-stranded than to double-stranded DNA (19). Since the mismatches were spaced 3 to 5 base pairs apart in the substrate tested, this may have created a favorable setting for cooperative binding along the length of the mismatch-containing region, thereby facilitating melting. The stabilization of double-stranded DNA, in contrast, may be due to charge shielding by the ORF1 protein. ORF1p may prevent DNA denaturation by reducing electrostatic repulsion between the two DNA strands, as is seen with increasing salt concentrations. Why ORF1p stabilizes the mismatch-containing duplex at high temperatures is unclear. Possibly the protein denatures at high temperature, yielding a form capable of charge shielding but not selective binding to single DNA strands.
Results of the DNA annealing and melting studies suggest models for the DNA chaperone activity of ORF1p. In the case of the mismatched duplex, exchange may be mediated by active unpairing, thereby permitting association of the perfect complement. It is unclear whether complete melting is required before the mm29c oligonucleotide can be replaced by the perfect complement. If the mismatched duplex is completely melted, charge shielding and ORF1p protein-protein interactions could simply promote annealing. Alternatively, if the duplex is only partially melted, strand exchange would have to proceed through partial base pairing followed by displacement. This may also be the mechanism used to replace the 29-c25 duplex with the complementary 29-mer strand following association with the free single-stranded region of the duplex. In the case of the 29-c25 duplex, there are no mismatches to nucleate active unpairing, yet the 25-mer strand in this duplex is readily replaced by the 29-mer perfect complement.
The finding that the mouse L1 ORF1 protein contains nucleic acid chaperone activity strongly emphasizes the importance of this function for the replication of retroelements. The primary sequence of L1 ORF1p is unique in that it lacks the multiple NC-type CCHC zinc fingers typical of the other non-LTR retrotransposon ORF1s (25). The replacement of 5′ end sequences is a probable consequence of the non-LTR replication mechanism. Non-LTR retrotransposons often lose their 5′ sequences during transposition, leading to the formation of an inactive, truncated element (16). It may be that the non-LTR retrotransposons have acquired their variable ORF1 sequences by truncation followed by capture of a cellular protein-coding sequence that substituted for the lost activity. That is, the phylogenetic comparison provides a natural domain swap experiment documenting the need for an NC-like activity.
What is the role of the nucleic acid chaperone activity in non-LTR retrotransposon replication? None of the steps of retroviral replication that require NC chaperone activity are shared with TPRT. In TPRT, there is no need to dimerize the RNA, anneal a tRNA primer, facilitate strong-stop strand transfers during reverse transcription, or promote cDNA integration. However, a strand transfer reaction is required for TPRT once the target site DNA is cleaved. T-rich sequences at the cleaved target site anneal with the poly(A) tail of the L1 transcript. The 3′ end of the cleaved target DNA then acts as a primer for first-strand cDNA synthesis. We propose that the ORF1p strand transfer activity may be important for this primer transfer reaction. This pathway involves the exchange of a DNA-DNA duplex in the target site for an RNA-DNA duplex, resulting in the formation of a more stable hybrid.
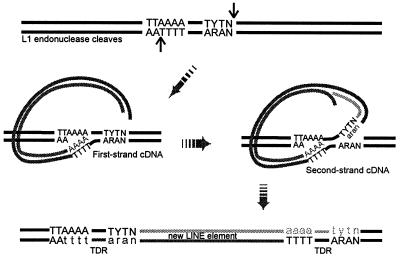
The nucleic acid chaperone activity may also be useful later in the reverse transcription pathway. The available in vitro TPRT systems do not carry out the late stages of the reaction, leaving the steps involved uncertain (24). However, a similar strand transfer reaction may allow the other strand of the cleaved target DNA to base pair with first-strand L1 cDNA, thereby acting as the primer for second-strand cDNA synthesis (Fig. 8). The nucleic acid chaperone activity may be important in this second annealing event as well.
FIG. 8.
Possible role for the strand-exchange activity of ORF1p during L1 retrotransposition. This model is adapted based on sequences of the L1 target site duplication deduced by Jurka (17) and from the model proposed for the TPRT reaction of the R1Bm element (11), which encodes an endonuclease related to the L1 endonuclease (10). Two strand-exchange reactions are required for the completion of cDNA synthesis and integration of a newly retrotransposed copy of L1. Both involve melting the duplex DNA of the genomic target site: the first exchanges the bottom strand of the DNA-DNA duplex to anneal with the L1 RNA in order to prime first-strand cDNA synthesis. The second exchanges the top strand of the target site duplex onto the first-strand cDNA, where it anneals to prime second-strand cDNA synthesis. Fill-in of the staggered, cleaved ends of chromosomal DNA yields a direct repeat of the original sequence (target site direct repeat [TDR]). Newly synthesized DNA is represented by lowercase letters, and the original nucleotides are capitalized. LINE, long interspersed nuclear element.
By analogy with retrovirus NC, we speculate that the ORF1p protein of L1 may be involved in several additional steps in L1 retrotransposition. The RNA binding activity of ORF1p likely allows it to coat and protect the RNA intermediate (26) as it waits in the cytoplasm for access to the DNA target for TPRT. Although ORF1p has only been detected in the cytoplasm to date (1, 27, 37), it could still be involved in later steps of retrotransposition, including TPRT. It may be that access to the DNA for insertion occurs in mitosis during nuclear envelope breakdown or that only a tiny fraction of the total L1 RNP (and ORF1p) ever enters the nucleus. During the later stages of L1 retrotransposition, the ORF1p nucleic acid chaperone activity is predicted to facilitate the strand transfer required to anneal the DNA primer from the target site to the L1 RNA to prime reverse transcription. ORF1p may help during reverse transcription as well, perhaps by permitting polymerization through secondary structures in the RNA. The unpairing of mismatches may also be biologically relevant, since it may promote repair of mismatched sites during cDNA synthesis. Finally, ORF1p may repeat these functions during priming and reverse transcription of the second strand. Understanding these nucleic acid chaperone activities of ORF1p and its inclusion in in vitro assays will contribute to our ability to detail the steps involved in the TPRT of L1 and other non-LTR retrotransposons.
ACKNOWLEDGMENTS
We thank J. Weisz for pJS28, V. Kolosha for purified A- and TF-type mouse ORF1 protein from baculovirus, and P. Hagerman, L. Orgel, and S. Carteau for helpful discussion.
This work utilized the DNA Sequencing and Tissue Culture Cores of the UCHSC Cancer Center (CA46934) and was supported by NSF grant MCB-9806033 and NIH grant GM40367 to S.L.M. and NIH grants GM56553 and AI34786 to F.D.B., the James B. Pendleton Charitable Trust, and the Berger Foundation. F.D.B. is a Scholar of the Leukemia and Lymphoma Society of America.
REFERENCES
- 1.Branciforte D, Martin S L. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carteau S, Batson S C, Poljak L, Mouscadet J-F, Rocquigny H, Darlix J-L, Roques B P, Kas E, Auclair C. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J Virol. 1997;71:6225–6229. doi: 10.1128/jvi.71.8.6225-6229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carteau S, Gorelick R J, Bushman F D. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 5.Cox M M, Lehman I R. Renaturation of DNA: a novel reaction of histones. Nucleic Acids Res. 1981;9:389–400. doi: 10.1093/nar/9.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cristofari G, Ficheux D, Darlix J L. The gag-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem. 2000;275:19210–19217. doi: 10.1074/jbc.M001371200. [DOI] [PubMed] [Google Scholar]
- 7.Cull M, McHenry C S. Preparation of extracts from procaryotes. Methods Enzymol. 1990;182:147–153. doi: 10.1016/0076-6879(90)82014-s. [DOI] [PubMed] [Google Scholar]
- 8.Dawson A, Hartswood E, Paterson T, Finnegan D J. A LINE-like transposable element in Drosophila, the I factor, encodes a protein with properties similar to those of retroviral nucleocapsids. EMBO J. 1997;16:4448–4455. doi: 10.1093/emboj/16.14.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 10.Feng Q, Moran J V, Kazazian H H, Jr, Boeke J D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 11.Feng Q, Schumann G, Boeke J D. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc Natl Acad Sci USA. 1998;95:2083–2088. doi: 10.1073/pnas.95.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y X, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorelick R J, Gagliardi T D, Bosche W J, Wiltrout T A, Coren L V, Chabot D J, Lifson J D, Henderson L E, Arthur L O. Strict conservation of the retroviral nucleocapsid (NC) protein zinc-finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant NC zinc-coordinating sequences. Virology. 1999;256:92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- 14.Hohjoh H, Singer M F. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 15.Hohjoh H, Singer M F. Sequence specific single-strand RNA-binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchison C A, III, Hardies S C, Loeb D D, Shehee W R, Edgell M H. LINEs and related retroposons: long interspersed repeated sequences in the eucaryotic genome. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. p. 593-617. [Google Scholar]
- 17.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazazian H H, Jr, Moran J V. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 19.Kolosha V O, Martin S L. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci USA. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolosha V O, Martin S L. Polymorphic sequences encoding the first open reading frame protein from LINE-1 ribonucleoprotein particles. J Mol Biol. 1995;270:2868–2873. doi: 10.1074/jbc.270.6.2868. [DOI] [PubMed] [Google Scholar]
- 21.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix J-L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix J-L. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loeb D D, Padgett R W, Hardies S C, Shehee W R, Comer M B, Edgell M H, Hutchison C A., III The sequence of a large L1Md element reveals a tandemly repeated 5′ end and several features found in retrotransposons. Mol Cell Biol. 1986;6:168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luan D D, Korman M H, Jakubezak J L, Eickbush T H. Reverse transcription of R2Bm RNA is primed by a nick at the chomosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 25.Malik H S, Burke W D, Eickbush T H. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 26.Martin S L. Mol. Cell. Biol. 11:4804–4807. 1991. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin S L, Branciforte D. Synchronous expression of LINE-1 RNA and protein in mouse embryonal carcinoma cells. Mol Cell Biol. 1993;13:5383–5392. doi: 10.1128/mcb.13.9.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin S L, Li J, Weisz J A. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J Mol Biol. 2000;304:11–20. doi: 10.1006/jmbi.2000.4182. [DOI] [PubMed] [Google Scholar]
- 29.Mathias S L, Scott A F, Kazazian H H, Jr, Boeke J D, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 30.Moran J V, Holmes S E, Naas T P, DeBerardinis R J, Boeke J D, Kazazian H H., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 31.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pabo C O, Sauer R T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 33.Pontius B W, Berg P. Renaturation of complementary DNA strands mediated by purified mammalian heterogeneous nuclear ribonucleoprotein A1 protein: implications for a mechanism for rapid molecular assembly. Proc Natl Acad Sci USA. 1990;87:8403–8407. doi: 10.1073/pnas.87.21.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rein A, Henderson L E, Levin J G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 35.Saxton J A, Martin S L. Recombination between subtypes creates a mosaic lineage of LINE-1 that is expressed and actively retrotransposing in the mouse genome. J Mol Biol. 1998;280:611–622. doi: 10.1006/jmbi.1998.1899. [DOI] [PubMed] [Google Scholar]
- 36.Tanchou V, Decimo D, Pechoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix J-L. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trelogan S A, Martin S L. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci USA. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type I nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You J C, McHenry C S. HIV nucleocapsid protein. J Biol Chem. 1993;268:16519–16527. [PubMed] [Google Scholar]