Abstract
The novel coronavirus disease has spread rapidly and caused sustained pressure on economic and medical resources to many countries. Vaccines and effective drugs are needed to fight against the epidemic. Traditional Chinese Medicine (TCM) plays an important and effective role in the treatment of COVID-19. Therefore, the active components of TCM are potential structural basis for the discovery of antiviral drugs. Through screening by molecular docking, Oleanolic acid, Tryptanthrin, Chrysophanol and Rhein were found to have better spike protein and ACE2 inhibitory activity, which could block the invasion and recognition of SARS-CoV-2 at the same time, should be investigated as antiviral candidates.
Keywords: SARS-CoV-2, ACE2, Spike glycoprotein, Dual antagonists
1. Introduction
The novel coronavirus disease (COVID-19) which was caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Vogl et al., 2021) has brought out a worldwide epidemic (Hanaei and Rezaei, 2020) since its outbreak in 2019. SARS-CoV-2 is an enveloped positive-sense RNA single-stranded β-CoV (Facchetti et al., 2020), similar to severe acute respiratory syndrome of coronavirus (SARS-CoV) or the middle east respiratory syndrome-related coronavirus (MERS-CoV) in 2003 and 2012 (Mohamed et al., 2021). COVID-19 has caused persistent economic and medical pressure to many countries. Vaccine and effective drugs are the key tools to fight against COVID-19, especially in face of the continuous emergence of virus mutants.
The genomic RNA of SARS-CoV-2 is 29.9 kb, in which there are 11 open reading frames (ORF). ORF1a and ORF1ab encode the replicase polyproteins which are processed into approximately 15 non-structural proteins (Hussain et al., 2005). Other ORF is responsible for the production of four structural proteins (spike protein, nucleocapsid protein, membrane protein, and envelope protein) (Khailany et al., 2020). The spike protein (S) is a trimeric transmembrane glycoprotein, each monomer consists subunits S1 and S2.
The renin-angiotensin-aldosterone system (RAAS) is the major mechanisms of hypertension (Navar, 2010). Angiotensin converting enzyme (ACE) secreted by the lungs and the epithelial cells is the main regulator of RAAS, cleave the short peptide angiotensin I to angiotensin II which leads to arterioles constriction and blood pressure increase (Yee et al., 2010). Angiotensin-converting enzyme 2 (ACE2) distributed at the cellular surface of tissues, is a transmembrane protein with the extracellular N-terminal and intracellular C-terminal domains (Serfozo et al., 2020). The structure of ACE2 consists transmembrane domain, Zn2+ binding domain and a signal peptide (Donoghue et al., 2000). ACE2 is a potential regulator of RAAS (Gheblawi et al., 2020), which could convert Ang II to Angiotensin (1–7) (Vickers et al., 2002). ACE2 plays a protective role in reducing hypertension and activating anti-inflammatory pathways after tissue injury (Santos et al., 2018). ACE2 is the main receptor of SARS-CoV-2. Besides ACE2, DPP-4 may also be utilized by spike glycoproteins as entry receptor (Vankadari and Wilce, 2020). The first step for the invasion of virus is binding to ACE2 (Singh et al., 2021), followed by the division of S1 / S2 subunit (Ren et al., 2006) which priming the integration of the virus and host cell membranes (Çakır et al., 2021). The receptor-binding domain (RBD) of SARS-CoV-2 located in S1 subunit, includes a core domain known as the receptor-binding motif (RBM). RBM binds to the viral binding motif of ACE2 (Li, 2015), and mediates the recognition of virus to receptor ACE2 (Wan et al., 2020). SARS-CoV-2 recognizes ACE2 with RBD of S1, and downregulates the expression of ACE2 (Suh et al., 2021).
SARS-CoV-2 and SARS-CoV use the same entry receptor ACE2 (Hoffmann et al., 2020), however, the binding affinity of SARS-CoV-2 for ACE2 receptor appears to be 10 times stronger (Malhotra et al., 2020). Compared with SARS-CoV, SARS-CoV-2 is more infectious. In the SARS-CoV-2 infection, viral entry into the host cells depends on the presence of ACE2 and transmembrane protease serine type 2 (TMPRSS2). Cells with a relatively high expression of ACE2 should be considered as potential SARS-CoV-2 infection sites (Wu et al., 2020). Higher ACE2 expression is correlated with higher SARS-CoV-2 viral infectivity (Ou et al., 2020). Since ACE2 serves as the main entry point for SARS-CoV-2 infection, more severe symptoms may be related to higher viral load in affected cells due to easier viral entry (Yang et al., 2020). So targeting ACE2 expression can be used as a treatment for COVID-19 (Swaminathan et al., 2021). Inhibition the entry of SARS-CoV-2 into the host cell via the ACE 2 receptors maybe an effective strategy to antagonize COVID-19 (Lu et al., 2020). ACE inhibitors have been used in the treatment of hypertension. ACE2 and ACE share the similar protein sequences (Mehrabadi et al., 2021), however, the ACE inhibitors could not block ACE-2 (Tipnis et al., 2000). ACE inhibitors have been reported to increase ACE2 expression in RAAS (Oz et al., 2021). Therefore, it is necessary to find specific antagonists against ACE2 to resist SARS-CoV-2 infection. Several key residues of ACE2 may affected the interaction with spike protein of SARS-CoV-2. The N720D variant of ACE2 affected the stability and flexibility by increasing the level of motion in the loop region, resulting in a more favorable site for TMPRSS2 (Mohammad et al., 2020). ACE2 residues E564, R559, N556 were found in the interaction with spike protein (Rui et al., 2020). The S1 subunit of CoV could be further divided into an N-terminal domain (NTD) and a C-terminal domain (CTD), both NTD and CTD can be used as RBD to recognize the receptor of host cell. As to SARS-CoV-2, the viral CTD was applied to binding with ACE2 (Wang et al., 2020). The RBM has a high degree of structural plasticity to accommodate amino acid changes without disrupting ACE2 binding (Thomson et al., 2021). There are few variations at the RBM that drastically alter the binding affinity of RBD to the ACE2 in SARS-CoV2 (Chakraborty, 2021).
Traditional Chinese medicine (TCM) has been used to reduce symptoms and slow down the course of COVID-19 (Ren et al., 2021). The effectiveness and importance of TCM during the treatment of SARS-CoV-2 infection has already been demonstrated (Wang and Yang, 2021). A lot of plants belonging to TCM with natural antiviral effect, such as Forsythia suspense, Isatis root, Schisandra chinensis, Licorice and Lithospermum, etc. Quercetin distributed in a variety of TCM and vegetables is a flavonoid compound with anti-viral activity against a variety of viruses (Wenjiao et al., 2016; Huang et al., 2020). Glycyrrhizic acid is a compound isolated from the TCM licorice with significant antiviral and bactericidal effects (Farag and Wessjohann, 2012). Andrographolide is a component of TCM with wide biological activities including immunity regulation, antivirus, antibacterial (Kishore et al., 2016). Schizandrin, Ursolic acid and Baicalin (Ishfaq et al., 2019) also have different antioxidant, antiallergic and antiviral activity (H-y et al., 2015). In addition, Coumarin (Chen et al., 2018) and Radix Isatidis (Xiao et al., 2014) are also active ingredients with antiviral effect.
SARS-CoV2-ACE2 binding-directed approaches mainly consist of targeting receptor binding domain, and the domain is relatively conservative, which can be used as drug design targets. A putative target on the host transmembrane to interfere with the virus RBD is a potential site. Blocking or manipulating the SARS-CoV2-ACE2 binding interface may offer the best method to fight against the virus (Sharifkashani et al., 2020). A spike glycoprotein / ACE2 dual antagonist could bind to RBD, and compete with the binding of SARS-CoV-2 to ACE2 to prevent the virus entry by two mechanisms. In this study, components of TCM with antiviral activities, that can effectively inhibit spike protein as well as protect the viral binding site of ACE2 receptor were used to screen for the dual antagonists against COVID-19 by molecular docking.
2. Methods and materials
The interactions of amino acid residues near the binding site of SARS-CoV-2 RBD determines the affinity with receptor. The residues L455, F486, Q493, S494, N501 and Y505 of RBD played a key role in the recognition and affinity of CoV to ACE-2 (Yan et al., 2020). Amino acid residues of RBD and the amino acid motif of ACE2 responsible for the interactions were used for molecular screening design. Effective ACE2 receptor antagonists would block the binding of SARS-CoV-2 through competition.
The model of SARS-CoV-2 S-ACE2 complex (PDB ID: 7DF4, http://www1.rcsb.org) was used as the model for molecular docking. 7DF4 was a complex of spike protein trimer and the human ACE2 receptor protein, the structure was solved by electron microscopy with 3.80 Å resolution (Xu et al., 2020). Spike protein and ACE2 were separated from the complex and used as independent receptors for molecular docking screening, respectively. The study was confined to molecular docking without validation by molecular dynamics (MD) simulations.
2.1. RBD of Spike glycoprotein structure analysis
The RBD of SARS-CoV-2 includes amino acid residues 318–510 (Gurung et al., 2021). According to the protein-protein interface of complex 7DF4, amino acid residues VAL-503, GLY-502, GLN-506, THR-500, PRO-499, PHE-497, LEU-455, GLY-446, ALA-475, SER-477, PHE-486, GLY-476, PHE-490, GLN-474, GLY-447, TYR-505, GLN-493, GLY-49, LEU455, PHE486, SER494, ASN501 and TYR505 were in the active sites of docking for screening antagonists against spike.
2.2. ACE2 modeling and structure analysis
At present, there are no specific ACE2 antagonists, and existing ACE antagonists cannot bind to ACE2. Based on the interface of complex 7DF4, the key amino acid residues of ACE2 antagonists were composed of amino acid residues THR-20, ALA-386, LYS-353, GLY-352, PRO-389, PRO-499, ALA-387, GLN-325, GLN-388, LYS-353, SER-19, LYS-31, GLN-24, GLY-354, GLY-326, ASP-355, SER-19, GLY-354, THR-371, GLU-375 and ALA-348.
2.3. Molecular docking
The molecular docking experiment was carried out by AutoDock Vina software. Before docking experiments, water was removed from spike protein and ACE2 receptor protein, then hydrogens were added to the macromolecular receptor, followed by charge calculation and atoms type assign. M2M was used to optimize the structure of ligand molecules to obtain the lowest energy conformation. After docking, the conformations with the lowest binding energies were chosen antagonist candidates. The dual antagonists were screened in 28 natural plant active ingredients.
3. Results and discussion
3.1. spike protein
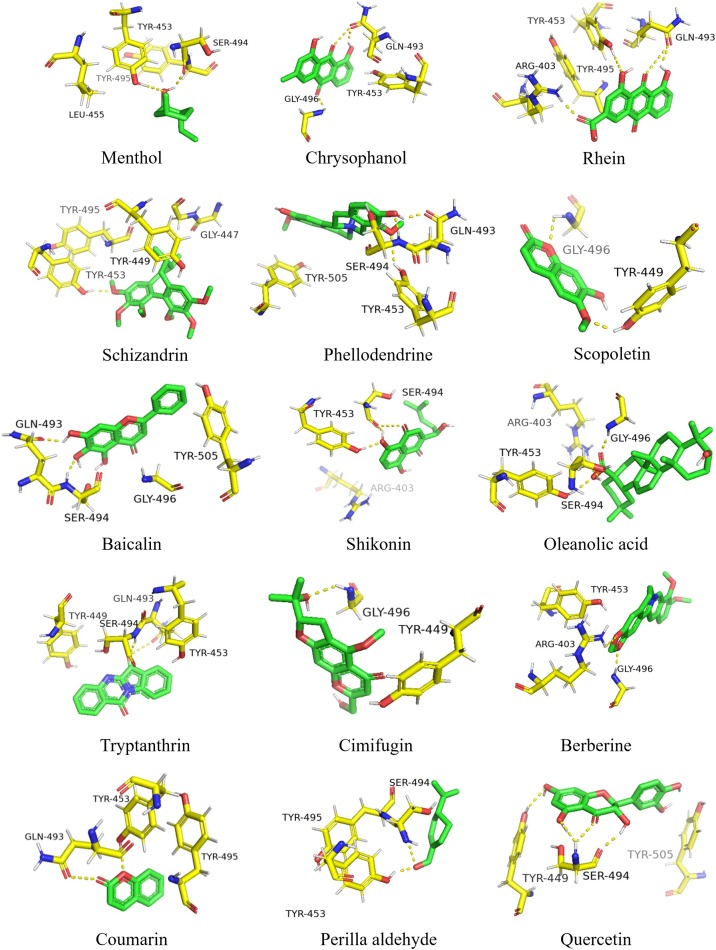
Menthol formed 2 hydrogen bonds with the RBD by SER-494 and TYR-495, formed π-π interaction with TYR-453 and hydrophobic interaction with LEU-455. Chrysophanol binds to the almost same site with Menthol. It formed 3 hydrogen bonds with GLN-493 and GLY-496, formed π-π interaction with TYR-453. Rhein interacted with RBD through 3 hydrogen bonds and π-π interaction. The 3 hydrogen bonds were formed between Rhein and GLN-493, TYR-495 and ARG-403. Schizandrin formed one hydrogen bond with TYR-453, as well as 3 hydrophobic interaction with TYR-495, GLY-447 and TYR-449. Phellodendrine interacted with GLN-493, SER-494 and TYR-453 through 3 hydrogen bonds and TYR-505 by π-π interaction. Scopoletin formed hydrogen bonds with GLY-496 and TYR-449. Baicalin formed hydrogen bonds with GLN-493 and SER-494, hydrophobic force with GLY-496, π-π interaction with TYR-505. Shikonin interacted with TYR-453 and SER-494 through hydrogen bonds, formed hydrophobic interaction with ARG-403. Oleanolic acid formed hydrogen bond interaction with TYR-453 and GLY-496, hydrophobic forces with ARG-403 and SER-494. Tryptanthrin made hydrogen interaction with GLN-493 and SER-494, π-π interaction with TYR453 and TYR-449. Cimifugin and Berberine all formed hydrogen bonds with GLY-496. Coumarin interacted with GLN-493 and TYR-453 through hydrogen bonds. Perilla aldehyde formed hydrogen forces with TYR-453 and SER-494. Quercetin interacted with SER-494 and TYR-449 by hydrogen bonds, formed π-π interaction with TYR-505.
The key amino acid residues of RBD involved in the formation of hydrogen bond are TYR-453, SER-494, GLN-493, GLY-496, TYR-495, TYR-449 and ARG-403. Besides hydrogen forces, π-π interaction plays an significant role in the interaction. The RBD binding sites of 15 compounds were shown in Fig. 1 .
Fig. 1.
Binding region of RBD.
3.2. ACE2
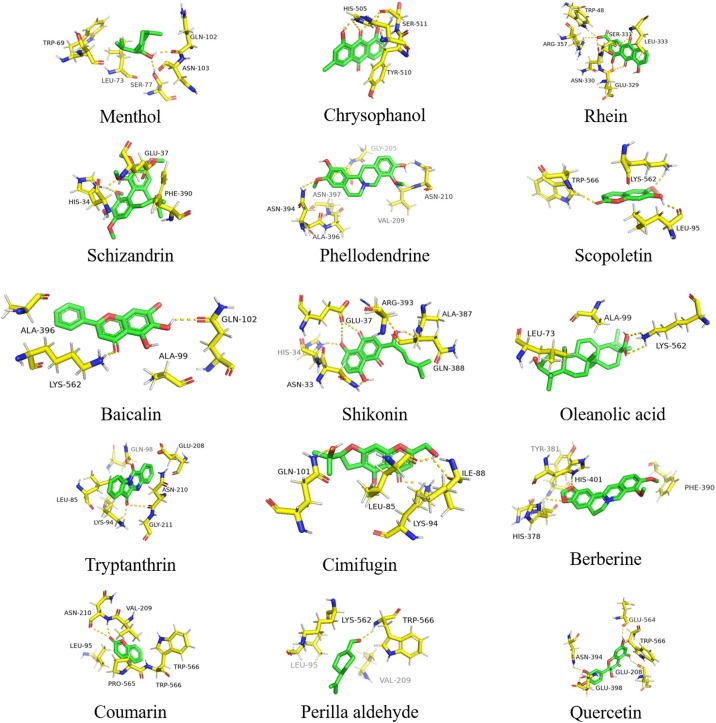
Menthol formed hydrogen bonds with GLN-102 and SER-77, formed hydrophobic interaction with LEU-73, TRP-69 and ASN-103. Chrysophanol binds to ACE2 by hydrogen bonds with SER-511 and HIS-505, formed π-π interaction with TRY-510. Rhein formed hydrogen bonds with GLU-329, LEU-333, TRP-48 and ARG-357, formed hydrophobic interaction with ASN-330 and SER-331. Schizandrin formed one hydrogen bond with HIS-34, interacted with GLU-37 and PHE-390 by hydrophobic force. Scopoletin formed hydrogen bonds with TRP-566, LEU-95 and LYS-562. Baicalin formed hydrogen bonds with GLN-102 and LYS-562, hydrophobic force with ALA-396 and ALA-99. Shikonin interacted with GLU-37, HIS-34, ARG-393, ASN-33 and ALA-387 through hydrogen bonds, formed hydrophobic interaction with GSN-388. Oleanolic acid formed hydrogen bond interaction with LYS-562, hydrophobic forces with LEU-73 and ALA-99. Tryptanthrin made hydrogen interaction with LYS-94 and GLY-211, hydrophobic interaction with GLN-98, GLU-208, ASN-210 and LEU-85. Cimifugin formed hydrogen bonds with GLN-101, ILE-88, LEU-85 and LYS-94. Berberine interacted with HIS-378 and HIS-401, interacted with PHE-390 and PHE-40 by π-π interaction. Coumarin formed hydrogen bond with ASN-210. Perilla aldehyde formed hydrogen forces with TRP-566. Quercetin formed hydrogen force with GLU-564, GLU-208, ASN-394 and GLU-398, formed π-π interaction with TRP-566.
According to the analysis of amino acids at the binding sites, histidine and lysine are more involved in the formation of hydrogen bonds. The major amino acid residues involved in the formation of hydrogen bonds are LYS-562, LYS-94, HIS-34, GLN-102 and TRP-566. The ACE2 receptor binding sites of 15 compounds were shown in Fig. 2 .
Fig. 2.
Binding region of ACE2.
3.3. Binding energy
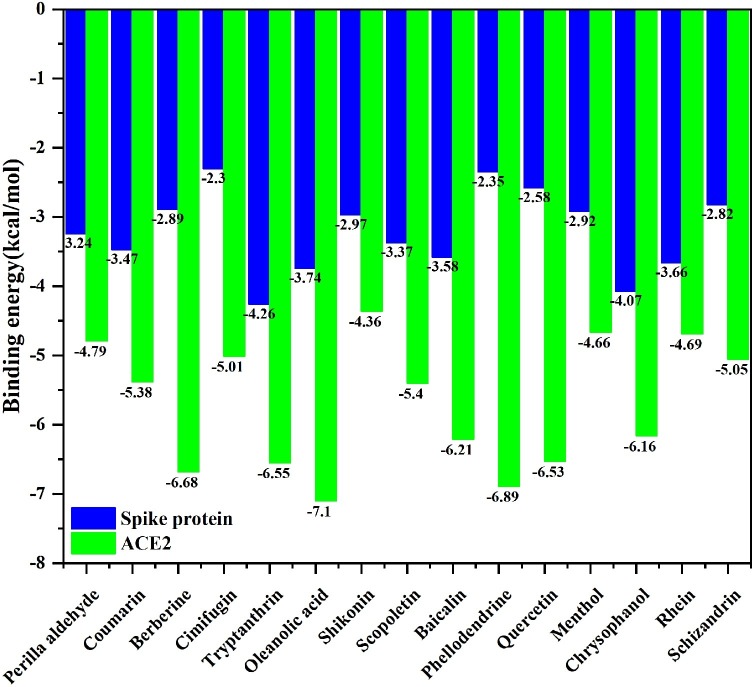
As to the ACE2 receptor, the lowest binding energy was got by Oleanolic acid with -7.1 kcal/mol, followed by Phellodendrine, Berberine, Tryptanthrin and Quercetin. For spike protein, the binding energy of Tryptanthrin -4.37 kcal/mol was the lowest, followed by Chrysophanol, Rhein, Scopoletin and Oleanolic acid. The number of hydrogen bonds is related to the binding energy. The more the hydrogen bonds, the lower the binding energy. Hydrogen bond has a strong stabilizing effect on the binding of antagonists to receptors. From the perspective of binding energy, Oleanolic acid, Tryptanthrin, Chrysophanol and Rhein are potential dual antagonists against SARS-CoV-2 By simultaneously blocking the virus spike protein and virus receptor ACE2, respectively.
The molecular dynamics simulations of antagonists have not been carried out yet, the study did not consider the possible flexibility and the exposition (Behloul et al., 2021) of the interface residues of RBD predictions. The binding energy of potential dual antagonists were shown in Fig. 3 .
Fig. 3.
Binding energy of 15 antagonists.
4. Conclusion
TCM and natural plants play an important role in the fight against COVID-19. Natural products are rich sources of antiviral compounds. The target models of spike protein RBD and ACE2 receptor binding site were established on the complete structure of SARS-CoV-2 and ACE2 complex to improve the accuracy of molecular docking. 28 compounds selected from the natural products with antiviral activity were screened for effective antagonists. Oleanolic acid, Tryptanthrin, Chrysophanol and Rhein are potential dual antagonists, which could block the invasion and recognition of SARS-CoV-2 and become antiviral candidates. The molecular dynamics simulation of complexes should be further developed. At present, there are no exclusive inhibitors for ACE2, and ACE inhibitors cannot inhibit ACE2. Therefore, screening ACE2 receptor inhibitors is of great significance for finding antagonists to all virus with ACE2 as receptor.
Data availability
Data will be made available on request.
Author statement
Ran Yu: Conceptualization, Methodology, Writing - Original Draft
Peng Li: Writing - Review & Editing
Declaration of Competing Interest
None.
Acknowledgements
Authors wish to thank general projects of scientific research plan of Beijing Municipal Commission of Education for financial support under the project KM202010858002.
References
- Behloul N., Baha S., Guo Y., Yang Z., Shi R., Meng J. In silico identification of strong binders of the SARS-CoV-2 receptor-binding domain. Eur. J. Pharmacol. 2021;890 doi: 10.1016/j.ejphar.2020.173701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çakır B., Okuyan B., Şener G., Tunali-Akbay T. Investigation of beta-lactoglobulin derived bioactive peptides against SARS-CoV-2 (COVID-19): in silico analysis. Eur. J. Pharmacol. 2021;891 doi: 10.1016/j.ejphar.2020.173781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S. Evolutionary and structural analysis elucidates mutations on SARS-CoV2 spike protein with altered human ACE2 binding affinity. Biochem. Biophys. Res. Commun. 2021;538:97–103. doi: 10.1016/j.bbrc.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-C., Liu L., Shen Y.-F., Hu Y., Ling F., Wang G.-X., et al. A new coumarin derivative plays a role in rhabdoviral clearance by interfering glycoprotein function during the early stage of viral infection. Cell. Signal. 2018;51:199–210. doi: 10.1016/j.cellsig.2018.08.007. [DOI] [PubMed] [Google Scholar]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., et al. A novel angiotensin-converting enzyme–Related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Facchetti F., Bugatti M., Drera E., Tripodo C., Sartori E., Cancila V., et al. SARS-CoV2 vertical transmission with adverse effects on the newborn revealed through integrated immunohistochemical, electron microscopy and molecular analyses of Placenta. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag M.A., Wessjohann L.A. Volatiles profiling in medicinal licorice roots using steam distillation and solid‐phase microextraction (SPME) coupled to chemometrics. J. Food Sci. 2012;77:C1179–C1184. doi: 10.1111/j.1750-3841.2012.02927.x. [DOI] [PubMed] [Google Scholar]
- Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.-C., Turner A.J., et al. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung A.B., Ali M.A., Lee J., Farah M.A., Al-Anazi K.M. Identification of potential SARS-CoV-2 entry inhibitors by targeting the interface region between the spike RBD and human ACE2. J. Infect. Public Health. 2021;14:227–237. doi: 10.1016/j.jiph.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaei S., Rezaei N. COVID-19: Developing from an Outbreak to A Pandemic. Arch. Med. Res. 2020;51 doi: 10.1016/j.arcmed.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.052. 271-80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Li Y., Leung E.L.-H., Liu X., Liu K., Wang Q., et al. A review of therapeutic agents and Chinese herbal medicines against SARS-COV-2 (COVID-19) Pharmacol. Res. 2020;158 doi: 10.1016/j.phrs.2020.104929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain Snawar, Ji’an Pan, Yu Chen, et al. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:5288–5295. doi: 10.1128/JVI.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- H-y Zhu, Han L., X-l Shi, B-l Wang, Huang H., Wang X., et al. Baicalin inhibits autophagy induced by influenza A virus H3N2. Antiviral Res. 2015;113:62–70. doi: 10.1016/j.antiviral.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Ishfaq M., Chen C., Bao J., Zhang W., Li J. Baicalin ameliorates oxidative stress and apoptosis by restoring mitochondrial dynamics in the spleen of chickens via the opposite modulation of NF-κB and Nrf2/HO-1 signaling pathway during Mycoplasma gallisepticum infection. Poult. Sci. 2019 doi: 10.3382/ps/pez406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19 doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore V., Yarla N.S., Bishayee A., Putta S., Dhananjaya B.L. Multi-targeting andrographolide and its natural analogs as potential therapeutic agents. Curr. Top. Med. Chem. 2016;16 doi: 10.2174/1568026616666160927150452. [DOI] [PubMed] [Google Scholar]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Chatterjee S., Xiao K., Riedel I., Wang Y., Foo R., et al. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J. Mol. Cell. Cardiol. 2020;148:46–49. doi: 10.1016/j.yjmcc.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A., Hepokoski M., McCowen K.C., Y-J Shyy J. ACE2, Metformin, and COVID-19. IScience. 2020;23 doi: 10.1016/j.isci.2020.101425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabadi M.E., Hemmati R., Tashakor A., Homaei A., Yousefzadeh M., Hemmati K., et al. Induced dysregulation of ACE2 by SARS-CoV-2 plays a key role in COVID-19 severity. Biomed. Pharmacother. 2021:111363. doi: 10.1016/j.biopha.2021.111363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed K., Yazdanpanah N., Saghazadeh A., Rezaei N. Computational drug discovery and repurposing for the treatment of COVID-19: a systematic review. Bioorg. Chem. 2021;106 doi: 10.1016/j.bioorg.2020.104490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad A., Marafie S.K., Alshawaf E., Abu-Farha M., Abubaker J., Al-Mulla F. Structural analysis of ACE2 variant N720D demonstrates a higher binding affinity to TMPRSS2. Life Sci. 2020;259 doi: 10.1016/j.lfs.2020.118219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navar L.G. Counterpoint: activation of the intrarenal renin-angiotensin system is the dominant contributor to systemic hypertension. J. Appl. Physiol. 2010;109:1998–2000. doi: 10.1152/japplphysiol.00182.2010a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oz M., Lorke D.E., Kabbani N. A comprehensive guide to the pharmacologic regulation of angiotensin converting enzyme 2 (ACE2), the SARS-CoV-2 entry receptor. Pharmacol. Ther. 2021;221 doi: 10.1016/j.pharmthera.2020.107750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Glende J., Al-Falah M., de Vries V., Schwegmann-Wessels C., Qu X., et al. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J. Gen. Virol. 2006;87:1691–1695. doi: 10.1099/vir.0.81749-0. [DOI] [PubMed] [Google Scholar]
- Ren W., Liang P., Ma Y., Sun Q., Pu Q., Dong L., et al. Research progress of traditional chinese medicine against COVID-19. Biomed. Pharmacother. 2021:111310. doi: 10.1016/j.biopha.2021.111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L., Haonan L., Wanyi C. Silico analysis of interaction between full-length SARS-CoV2 S protein with human Ace2 receptor: modelling, docking, MD simulation. Biophys. Chem. 2020;267 doi: 10.1016/j.bpc.2020.106472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R.A.S., Sampaio W.O., Alzamora A.C., Motta-Santos D., Alenina N., Bader M., et al. The ACE2/Angiotensin-(1–7)/MAS Axis of the renin-angiotensin system: focus on angiotensin-(1–7) Physiol. Rev. 2018;98:505–553. doi: 10.1152/physrev.00023.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfozo P., Wysocki J., Gulua G., Schulze A., Ye M., Liu P., et al. Ang II (Angiotensin II) conversion to angiotensin-(1-7) in the circulation is POP (Prolyloligopeptidase)-Dependent and ACE2 (Angiotensin-Converting enzyme 2)-Independent. Hypertension. 2020;75:173–182. doi: 10.1161/HYPERTENSIONAHA.119.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifkashani S., Bafrani M.A., Khaboushan A.S., Pirzadeh M., Kheirandish A., Yavarpour_Bali H., et al. Angiotensin-converting enzyme 2 (ACE2) receptor and SARS-CoV-2: Potential therapeutic targeting. Eur. J. Pharmacol. 2020;884 doi: 10.1016/j.ejphar.2020.173455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Choudhari R., Nema V., Khan A.A. ACE2 and TMPRSS2 polymorphisms in various diseases with special reference to its impact on COVID-19 disease. Microb. Pathog. 2021;150 doi: 10.1016/j.micpath.2020.104621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J.-S., Kim H.-S., Kim T.-J. Development of a SARS-CoV-2-derived receptor-binding domain-based ACE2 biosensor. Sens. Actuators B Chem. 2021:129663. doi: 10.1016/j.snb.2021.129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S., Dehghan M., Raj J.M., Thomas T., Yusuf S. Associations of cereal grains intake with cardiovascular disease and mortality across 21 countries in Prospective Urban and Rural Epidemiology study: prospective cohort study. BMJ (online). 2021:m4948. doi: 10.1136/bmj.m4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021 doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme: cloning and functional expression as a captopril-insensitive carboxypeptidase*. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- Vankadari N., Wilce J.A. Emerging COVID-19 coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., et al. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase *. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- Vogl T., Leviatan S., Segal E. SARS-CoV-2 antibody testing for estimating COVID-19 prevalence in the population. Cell Reports Medicine. 2021;2 doi: 10.1016/j.xcrm.2021.100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS Coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang L. Chinese herbal medicine: fighting SARS-CoV-2 infection on all fronts. J. Ethnopharmacol. 2021;270 doi: 10.1016/j.jep.2021.113869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.045. 894-904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenjiao Wu, Richan Li, Xianglian Li, et al. Quercetin as an antiviral agent inhibits influenza a virus (IAV) entry. Viruses. 2016 doi: 10.3390/v8010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-Y., Lin Y.-S., Yang Y.-H., Shu L.-H., Cheng Y.-C., Liu H.T. GB-2 inhibits ACE2 and TMPRSS2 expression: in vivo and in vitro studies. Biomed. Pharmacother. 2020;132 doi: 10.1016/j.biopha.2020.110816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao P., Huang H., Chen J., Li X. In vitro antioxidant and anti-inflammatory activities of Radix isatidis extract and bioaccessibility of six bioactive compounds after simulated gastro-intestinal digestion. J. Ethnopharmacol. 2014;157:55–61. doi: 10.1016/j.jep.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Xu C., Wang Y., Liu C., Zhang C., Han W., Hong X., et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. bioRxiv. 2020;2020 doi: 10.1126/sciadv.abe5575. 06.30.177097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367 doi: 10.1126/science.abb2762. eabb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Li Y., Xiao S.-Y. Differential expression of ACE2 in the respiratory tracts and its relationship to COVID-19 pathogenesis. EBioMedicine. 2020;60 doi: 10.1016/j.ebiom.2020.103004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee A.H., Burns J.D., Wijdicks E.F.M. Cerebral salt wasting: pathophysiology, diagnosis, and treatment. Neurosurg. Clin. N. Am. 2010;21:339–352. doi: 10.1016/j.nec.2009.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.